Abstract
The field of microfluidics allows for the precise spatial manipulation of small amounts of fluids. Within micro-structures, laminar flow of fluids can be exploited to control the diffusion of small molecules, creating desired microenvironments for cells. Cellular neuroscience has benefited greatly from devices designed to fluidically isolate cell bodies and axons. Microfluidic devices specialized for neuron compartmentalization are made of poly-dimethysiloxane (PDMS) which is gas permeable, compatible with fluorescence microscopy, and low cost. These devices are commonly used to study signals initiated exclusively on axons, somatodendritic compartments, or even single synapses. We have also found that microfluidic devices allow for rapid, reproducible interrogation of axon degeneration. Here we describe the methodology for assessing axonal degeneration in microfluidic devices. We describe several use cases, including enucleation (removal of cell bodies) and trophic deprivation to investigate axon degeneration in pathological and developmental scenarios, respectively.
Keywords: Microfluidic devices, PDMS, axon degeneration, injury, NGF deprivation, Wallerian degeneration
1. Introduction
Axons are the primary information conduits of the nervous system. Failure to maintain the integrity of axons is a feature of many inherited and acquired neurological disorders and can be attributed to roughly 16.8% of death, worldwide (2015) [1]. In response to pathological insult, such as injury (spinal cord injury, traumatic brain injury), exposure to toxic substances (Aβ, α-synuclein), or chemotherapies like vincristine, a process of axonal fragmentation, or Wallerian degeneration (WD) occurs, often resulting in permanent loss of neural function [2, 3]. WD of injured axons has been observed in various species and its molecular mechanisms are well conserved from mice to flies [4], but not in worms [5].
Perhaps counterintuitively, regressive/degenerative processes are essential for the formation of the vertebrate nervous system. During nervous system development, neurons, axon branches, and synapses are produced in excess. Components that receive sufficient trophic support stabilize, and those that don’t are eliminated via apoptosis, non-apoptotic degenerative mechanisms, or cytoskeletal rearrangement, respectively [6, 7]. Developmental degeneration involves the elimination of significant portions of primary axons and major collaterals. Importantly, this process is evolutionarily conserved and has been observed in cortical layer 5 projection neurons in mouse, elimination of missorted optic axons during retinotectal development in zebrafish, and mushroom body γ neuron pruning during drosophila metamorphosis [8–11]. Despite the differences between developmental and pathological axon degeneration, understanding the cellular basis of both processes holds promise for therapeutic application. Thus, a highly adaptable and reproducible system will expedite our understanding of axon degeneration.
Various in vitro and in vivo models allow the study of morphological and biochemical changes associated with pathological and developmental degeneration. In vivo models such as sciatic nerve transection, optic nerve crush, and lateral fluid percussion allow for simulation of degeneration after trauma [12, 13]. Drosophila mushroom body remodeling, terminal arbor pruning at the neuromuscular junction, and retinal ganglion projections during mouse retinotopic mapping provide experimental models for investigating developmental axon degeneration [14, 15]. On the other hand, In vitro models such as trophic factor deprivation, mechanical axotomy, and glutamate excitotoxicity allow rapid interrogation of the molecular mechanisms underlying degeneration. These classic in vitro and in vivo approaches are limited in their ability to apply insults locally (i.e., soma versus axon), which is likely a closer approximation of physiological and pathological degenerative triggers. Moreover, these classic approaches often do not allow for examining biochemical events associated with degeneration in specific subcellular locales. Compartmentalized culture approaches overcome both of these barriers to progress.
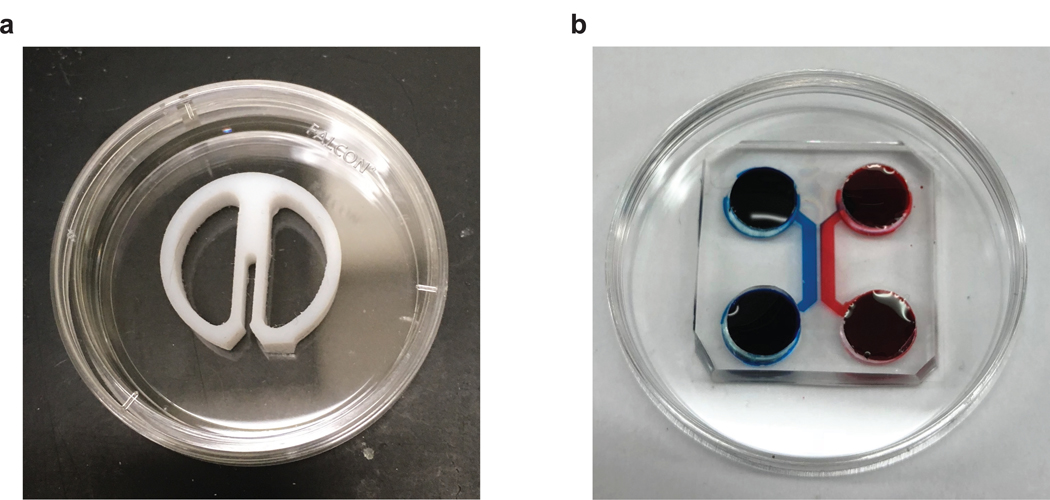
The compartmentalized “Campenot” chamber is the first device developed to allow local treatment of axons and soma. It consists of a reusable Teflon piece, that is affixed to a petri dish with a thin layer of silicone vacuum grease [16] (Fig.1a). These chambers have revolutionized the way the field considers long-distance signaling (i.e., from the retrograde signaling endosome) but suffered from a few non-trivial limitations including: 1. they are only suitable for neurons that project axons robust enough to cross the vacuum grease barrier. As such, most neurons of the central nervous system (CNS) cannot be grown in these devices. 2. Because these devices are plated on plastic and are opaque they are incompatible with high resolution fluorescence microscopy [17]. 3. Finally, preparing Campenot chambers is labor intensive, has a high failure rate (e.g., leaks), and is highly variable with respect to when axons cross to the barrier.
Figure 1: Campenot chamber and two chamber microfluidic device.
a. Campenot chamber. A 35 mm petri dish divided into three chambers by a Teflon divider.
b. Two chamber microfluidic device. A PDMS microfluidic device is attached to the 22×22 mm coverslip in a 35 mm petri dish.
Microfluidic compartmentalized neuronal devices have channels with micrometer dimensions allowing for the manipulation of small volumes and control of the net flow of diffusion [18] (Fig.1b). By incorporating microfluidic channels and precise control over factors bathing distinct neuronal regions, compartmentalized microfluidic platforms have been developed and used to study neuronal injury and axon degeneration [19–21]. These have several advantages over Campenot chambers: 1. Neurons from a wide range of sources including, dorsal root ganglia, cortex, and hippocampus can be grown in these devices. 2. The usage of transparent PDMS offers high efficiency, good biological compatibility, and ability to integrate with other research techniques like electrophysiology and high-resolution microscopy [22]. 3. The process of making microfluidic devices is fast, inexpensive and highly reproducible.
There are three steps in the fabrication of microfluidic devices for neuronal culture: 1) design and print out the mask; 2) fabrication of the master mold using photolithography; 3) production of microfluidics using poly-dimethysiloxane (PDMS) (Section 3) [20, 23, 24]. A rudimentary two chamber device has been used to great effect for studies ranging from axonal transport of endosomes to axon regeneration after injury [19, 25]. This design has been modified into platforms with multi-reserviors for multiple drug testing on axons [26–28], vertically-layered platforms for 3D neuron/glia co-culture [29], valve-based chambers for a compressive injury model [30, 31], and neuronal behavior analysis chips for small animals like C. elegans [32, 33].
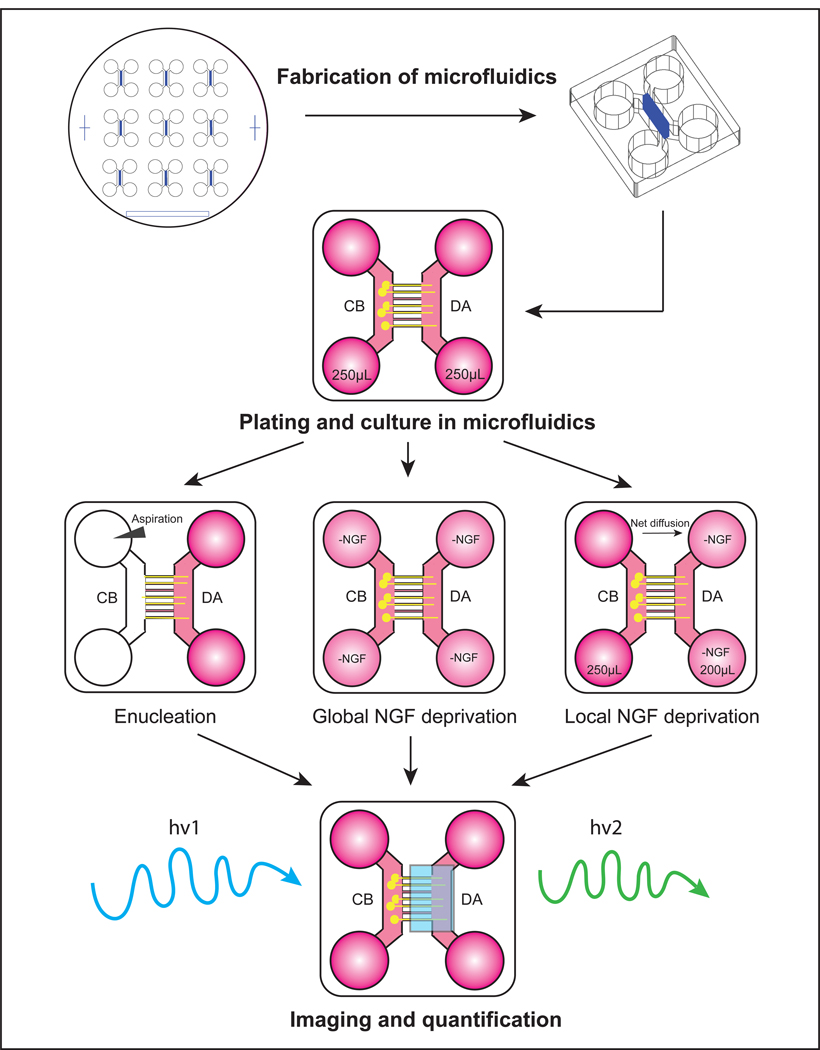
Based on a protocol to study axon regeneration [20], we have used a similar approach of aspirating cell bodies away from axons to study the molecular mechanisms of Wallerian degeneration [21]. Here we describe our protocol for examining axon degeneration after injury or trophic deprivation in microfluidic devices using neurons from mouse superior cervical ganglia (SCG) (Fig.2).
Figure 2:
Schematic presentation of studying axon degeneration in microfluidics
2. Materials
2.1. Fabrication of microfluidic molds and devices
Silicon wafers
Sylgard 184 silicone elastomer base and curing agent
¼” hollow punch
Razor blade or X-ACTO knife
Vinyl tape
Desiccator
SU-8 5 and SU-8 50 photoresist
Propylene glycol monomethyl ether acetate (PGMEA, photoresist developer)
Isopropanol (IPA)
Hot plates and/or ovens
Kapton tape
Photomask
2.2. Plating solutions/reagents
Coating solution: 50 μg/mL poly-D-lysine and 1 μg/mL laminin. Add 1 mL poly-D-lysine (10x stock) and 100 μL laminin (100x stock) to 8.9 mL 1x PBS.
22×22–1.5 glass coverslips
35 mm Petri dishes
Cell culture medium (varies depending on the cell type).
β-D-arabinofuranosie (Ara-C)
2.3. Immunostaining solutions
4% PFA: Add 160 mL water to flask and warm it up to 50–60°C. Add 16g paraformaldehyde to 160 mL water in chemical hood, stirring for 15 min. Add 200 μL of 10N NaOH solution. Add 20 mL 10x PBS. Keep the heat and stir for at least 5 hrs until the solution is clear. Test and adjust the pH to 7.2–7.4. Make up to final volume of 200 mL with water. Cool to room temperature and aliquot. Make 1:1 dilution with 1x PBS. Store at −20°C (see Note 1).
4% PPS (PFA-PHEM-Sucrose): 4% PFA with 120 mM sucrose, 2 mM MgCl2, 10 mM EGTA, 25 mM HEPES, and 60 mM PIPES. Weigh and add 1.82 g of PIPES, 0.60 g of HEPES, 0.38 g of EGTA, 0.019 g of MgCl2, and 4.11 g of sucrose to 100 mL 4% PFA. Test and adjust the pH to 7.4. Aliquot and store at −20°C.
Blocking solution: 5% normal goat serum, 0.05% Triton-X-100 in 1x PBS. Freshly prepared.
3. Methods
3.1. Fabrication of master mold
The steps to make master mold for microfluidics are described here.
Design and manufacture two high-resolution photomasks containing a desired microgroove and main chamber patterns with alignment marks, respectively. The autocad files for these masks can be found in supplemental material.
Clean the silicon wafer with IPA. Dry using pressurized insert gas.
Taking care to avoid air bubbles, add about 3 mL of SU-8 5 photoresist on a cleaned silicon wafer. Spin at 3,000 rpm for 1 min (3 μm thick).
Bake at 65°C for 1 min then at 95°C for 1 min.
Expose the wafer to a photomask containing a microgroove pattern and alignment marks for 30 s at 200W.
Bake at 65°C for 1 min then at 95°C for 1 min.
Develop with PGMEA for 3–5 min. Rinse with IPA (see Note 2). Dry using pressurized insert gas.
Bake at 65°C for 1 min then at 95°C for 10 min.
Cover the alignment marks with Kapton tape (PDMS strips or scotch tape are effective alternatives).
Taking care to avoid air bubbles, add about 3 mL of SU-8 50 photoresist on the silicon wafer. Spin at 1,000 rpm for 1 min (100 μm thick).
Remove the Kapton tape. Soft bake at 65°C for 10 min then at 95°C for 30 min.
Align the chamber mask to the grooves patterned on the wafer using the alignment marks. Expose for 2 min at 200W.
Bake at 65°C for 1 min then at 95°C for 10 min.
Cool the wafer to room temperature (about 1 min). Develop with PGMEA for 5–10 min as needed (see Note 2). Rinse three times with fresh developer solution. Rinse with IPA. Dry using pressurized insert gas.
Bake at 65°C for 1 min then at 95°C for 10 min. Store in clean case.
3.2. Fabrication of microfluidic devices
Fashion a dish out of aluminum foil 20% larger than the master mold (see Note 3). Place the master mold (silicon wafer) in foil dish and place it in a 150 mm petri dish (Fig.3a–c).
Measure 24 g of silicone elastomer solution with 2.67 g of curing agent in a weighing boat (see Note 4). Mix well by stirring.
Place the plate into the leveled vacuum chamber or desiccator to remove bubbles for 30–60 min (Fig.3d, see Note 5).
Cure the elastomer at 60°C overnight in the oven, which will solidify the PDMS (see Note 6).
-
Take out each PDMS mold and cut out the square block of nine chambers with a razor blade (Fig.3e, see Note 7). Punch holes directly over the circle reservoirs using sharpened stainless-steel punch (Fig.3f).
Cut each individual chamber such that its dimensions are approximately 1”x1”, which can fit on to a square coverslip (22mmx22mm). Cut the corners to help with handling (Fig.3g–h).
Use vinyl tape to remove dust sticking to the chamber on the groove-side.
Always keep groove-side up and store the microfluidics in closed plate at room temperature (Fig. 3i, see Note 8).
Figure 3: Fabrication of microfluidics.
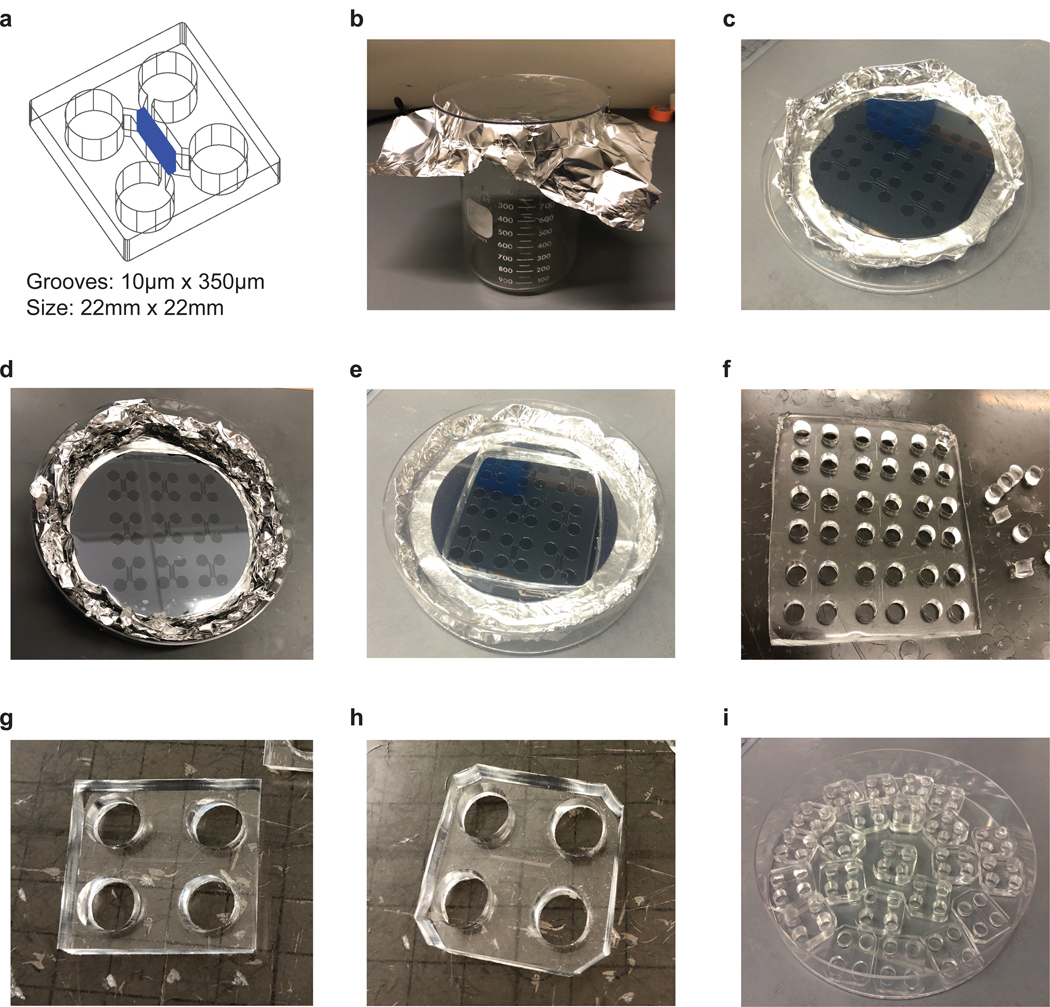
a. Design of two chamber microfluidics. The microgoove is 10 μm wide, 350 μm long. The size of microfluidic device is 22×22 mm, which fits the square glass coverslips.
b. Picture of foil cup.
c. Picture of the master mold.
d. Picture of PDMS mold after debubbling.
e. Picture of PDMS mold after cutting out the square block.
f. Picture of punching holes over the circle reserviors.
g. Picture of individual microfluidic device.
h. Picture of individual microfluidic device after cutting corners.
i. Picture of PDMS microfluidics.
3.3. Neuron plating and culture in microfluidics
Carry out all procedures in sterile environment (tissue culture hood).
Coat the square glass coverslips (22×22–1.5) with coating solution at 37°C 5–10% CO2 incubator overnight (see Note 9). Put the coated coverslips in 35 mm petri dish. Wash them with sterile water 3 times. Air dry the coverslips.
Sterilize the microfluidic devices by dipping them in pure ethanol. Let the devices completely dry (1–2 hrs). Attach the microfluidic device to the coated coverslip (see Note 10).
Prepare P0-P2 mouse superior cervical ganglion (SCG) dissociated cell suspension. Other neuronal cells such as embryonic DRG neurons have also been successfully cultured in microfluidics. Add 3 μL of cell suspension (1000–5000 neurons) to cell body side channel of each microfluidic device. Tap the plate to make sure the fluid connects two cell body side wells.
Put the plates into 37°C 5–10% CO2 incubator for 5 min (see Note 11).
Add about 125 μL of medium to each cell body side well without flushing out the cells. Incubate at 37°C 5–10% CO2 for 30 min.
Push 200 μL of medium into distal axon side channel to make sure that the two distal axon side wells are connected. Add more medium to the distal axon side wells to achieve 125 μL of medium per well.
Replace the medium in the wells next day. Add 5 μM cytosine β-D-arabinofuranosie (Ara-C) into the medium to remove glia for 48–72 hrs at 37°C 5–10% CO2.
Change medium in the wells every 2 days for following experiments (see Note 12, 13).
3.4. Live imaging in microfluidic cultures
For live imaging, instead of plating and culturing the neurons on the glass coverslips in petri dish, attach the PDMS microfluidic device to glass bottom tissue culture dish. Carry out all procedures in tissue culture hood unless otherwise specified.
Wash the microfluidic culture with live imaging medium (e.g., DMEM/F-12, Phenol Red free) for 3× 5 min.
Replace Phenol Red free medium containing live imaging dye, (e.g., Fluo4-AM, 1 μM). Incubate cells at 37°C 5–10% CO2 for 30 min prior to imaging.
To remove excess dye, replace live imaging medium.
Setup desired microscope system and paradigms to perform live imaging.
3.5. Inducing axon degeneration
Always check the plates under microscope to ensure that neurons project their axons to the axonal chamber (3–7 DIV). Few examples of degeneration paradigms are described below. These procedures are also applicable to chemotherapeutics and axon regeneration studies. Carry out all procedures in tissue culture hood unless otherwise specified.
3.5.1. Axotomy
Remove medium in the cell body side wells.
Use glass Pasteur pipet connected to the vacuum to aspirate 3 mL of 1x PBS through the cell body chamber, leaving the axons intact in their respective chamber (see Note 14).
Check the enucleating condition under microscope. Aspirate additional 1–3 mL of 1x PBS through the cell body chamber if necessary.
Remove all the remaining medium in 4 wells. Add pre-warmed medium without NGF to 4 wells. Incubate at 37°C 5–10% CO2 for designated times.
3.5.2. Global NGF deprivation
Wash the microfluidic culture with pre-warmed medium without NGF for 3×5 min.
Replace the medium in all 4 wells with NGF deficient medium containing 1 μg/mL anti-NGF function neutralizing antibody. Incubate at 37°C 5–10% CO2 for designated time.
3.5.3. Local NGF deprivation
Wash the microfluidic culture with pre-warmed medium without NGF for 3×5 min.
Add 200 μL of NGF deficient medium containing 1 μg/mL anti-NGF antibody to distal axon chamber.
Add about 250 μL of medium with NGF to cell body chamber (see Note 15). Incubate at 37°C 5–10% CO2 for designated time. Check the level of medium in each well every 8 hrs. Re-establish the volume differential if necessary. Rapid loss of volume differential indicates an improper seal rendering the device inappropriate for differential treatment.
3.6. Immunostaining in microfluidic cultures
Carry out all procedures in plates with PDMS microfluidic devices attached to the coverslips. It’s not necessary to remove the microfluidic chamber for immunostaining.
-
1.
Remove all the medium from 4 wells of the microfluidics.
-
2.
To fix axons, add 4% PFA to all 4 wells. Fix at room temperature for 20 min. To fix delicate structures like growth cones or axon spheroids, add pre-warmed 4% PPS to wells and fix at 37°C for 20 min (see Note 16).
-
2.
Wash the plate with 1x PBS for 3×5 min.
-
3.
Replace 1x PBS in the wells with blocking buffer. Incubate at room temperature for 1 hr.
-
4.
Replace the blocking buffer in wells with desired primary antibody diluted in blocking buffer (e.g., Tuj1, 1:1000). Incubate at 4°C overnight.
-
5.
Wash the plate with 1x PBS for 3×5 min.
-
6.
Replace 1x PBS in the wells with fluorescent conjugated secondary antibody diluted in blocking buffer (i.e. Goat anti-mouse Alexa 488, 1:800). Incubate at room temperature in dark for 1 hour.
-
7.
Wash the plate with 1x PBS for 3×5 min in dark.
-
8.
Carefully remove the microfluidic device with coverslip attached from petri dish and place it directly onto an inverted microscope stage for square coverslip (see Note 17). Perform imaging.
3.7. Quantification of axon degeneration
Axon degeneration in microfluidic culture can be quantified from β-III tubulin immunostained fluorescence images either automatically or manually. Loss of positive staining axons can be quantified by ImageJ software using “threshold” function or Stereo Investigator software using spaceballs or petrimetrics stereological probe in unbiased manner [34]. However, in areas with dense axons, the individual axons may appear to merge after applying the threshold analysis. Parameters and settings vary depending on the staining and experimental groups. Algorithms like AxonQuant provide a fast, microfluidic optimized way to measure “axonal continuity” and morphology in automated manner independent of neuronal or axonal density [35]. Alternatively, manual quantification of axon degeneration has been used in many studies. Below is an example of the manual quantification, which is based on the analysis of morphological characteristics (i.e. beads/blebs) of degenerating axons [36].
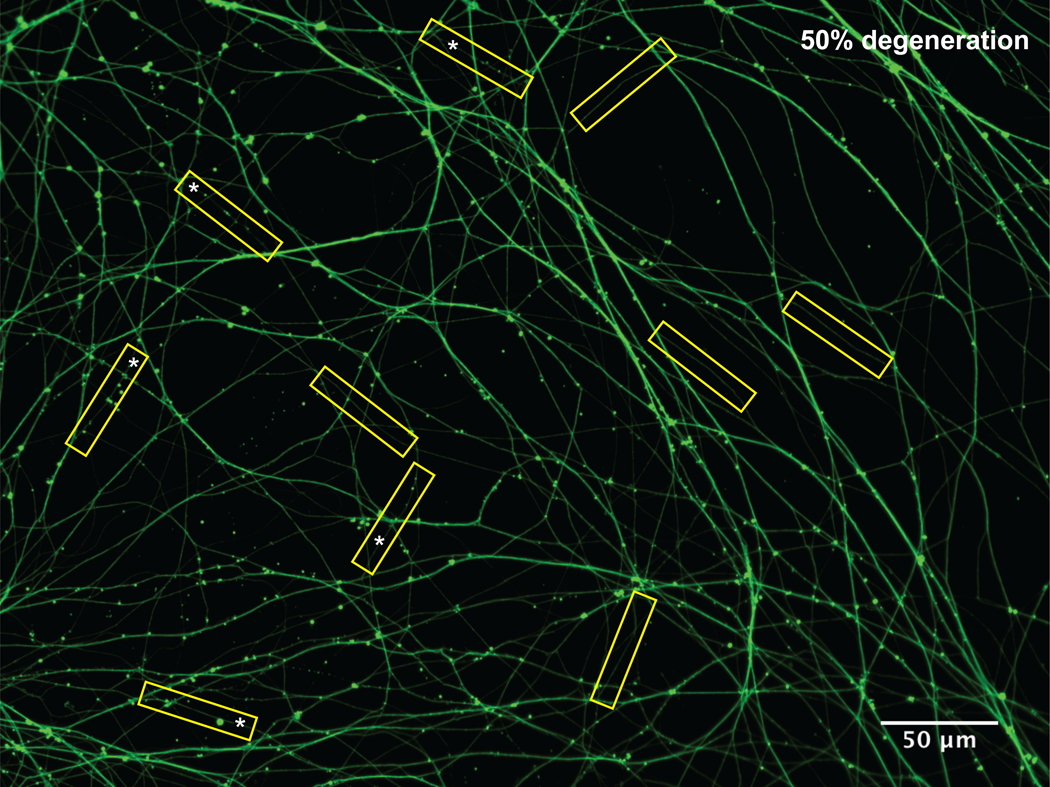
Investigator A takes 10 representative images of axons in microfluidics per experimental condition (or biological replicate). Blind the images.
Blinded Investigator B randomly assigns 10 50 μm-long rectangular boxes to single axons on each image (see Note 18).
The blinded Investigator B counts the number of beads/blebs (n) in each box to determine the degenerating axons. If there are 3 or more beads, blebs or breaks the boxed axon is considered a degenerating axon (Fig.2). The percentage of degeneration per picture is determined by the number of degenerating boxed axons (i.e., If there are 5 boxed axons out of 10 counted as degenerating axons, the percentage of degeneration is 50%).
Investigator A unblinds all the images and performs statistical analysis to determine mean, standard error and other parameters of axon degeneration in each experimental condition.
Supplementary Material
Figure 4: Quantification of axon degeneration.
P0 superior cervival ganglion neurons were cultured in microfluidics. At DIV 5, neurons were treated with global NGF deprivation for 19 hours, and then fixed and immunostained for Tuj1. 10 yellow 50 μm-long rectangular boxes were randomly assigned to signle axons in the image. Asterisks marked the degenerated axons. The percentage of degeneration in this field is 50%.
5. Acknowledgement
We thank Nadine Ly for technical assistance, Kanchana Gamage and Shayla Clark for helpful suggestions and comments on the chapter. We thank Prof. Brian Pierchala (University of Michigan) for Campenot chamber image.
Footnotes
Notes
To avoid repeated thawing, leave one aliquot of 4% FBS at 4°C for current use.
If the grooves or edges of chamber look cloudy after IPA rinse, it requires more time to develop. Change into fresh developer if necessary.
To make casting dishes, you may use the bottom of a 500 mL or 1 L beaker mold the aluminum foil.
The first time pouring, requires about two times more silicone elastomer and curing agent to achieve appropriate coverage and thickness. The proportion of silicone elastomer to curing agent should be 9:1.
This step takes approximately 1–2 hrs depending on the vacuum and how many plates are loaded. Take care that all microbubbles are removed curing in the oven.
Do not stack the plates. The oven should be leveled.
Too much pressure during cutting will crack the mold.
Protect the microfluidics from dust. Avoid touching the groove-side as they are very delicate. Check the devices before attaching to the coated coverslips to make sure the grooves and channels are connected.
Coverslips in coating solution should be incubated at least 2 hrs, and can be used up to 2 weeks.
Press hard on microfluidic devices to ensure proper attachment and prevent leakage, use light pressure for the middle of the chamber (grooves).
You may skip this step if there are too many plates to handle. However, it may affect the seeding of neurons in the microfluidics.
When changing medium, point the pipet tip away from the connecting channel of the microfluidics to avoid disturbing soma and axons.
To maintain the humidity of long-term in vitro cultures, store the 35mm plates in a 150mm plate with one water dish.
When enucleating neurons, point the pipet tip close to the connecting channel of the cell body side.
For local NGF deprivation on cell body side, switch the volume for the two compartments. To maintain fluidic isolation, reset a volume difference of 30–50μL between the two compartments if necessary.
To fix axonal spheroids, slowly add 4% PPS to microfluidic chambers along the edge of the wells. To maintain the surface tension, don’t remove all of the medium in each well.
To remove the petri dish, leaving coverslips attached to microfluidic device, squeeze the plastic petri dish and use forceps to take out the microfluidics.
The investigator should take care not to box bundles of axons, which may confound analysis.
6 References
- 1.Feigin VL, Abajobir AA, Abate KH, et al. (2017) Global, regional, and national burden of neurological disorders during 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Neurol 16:877–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vargas ME, Barres BA (2007) Why Is Wallerian Degeneration in the CNS So Slow? Annu Rev Neurosci 30:153–179. [DOI] [PubMed] [Google Scholar]
- 3.Waller A (1850) Experiments on the Section of the Glossopharyngeal and Hypoglossal Nerves of the Frog, and Observations of the Alterations Produced Thereby in the Structure of Their Primitive Fibres. Philos Trans R Soc London 140:423–429. [PMC free article] [PubMed] [Google Scholar]
- 4.Coleman MP, Freeman MR (2010) Wallerian Degeneration, Wld S, and Nmnat. Annu Rev Neurosci 33:245–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nichols ALA, Meelkop E, Linton C, et al. (2016) The Apoptotic Engulfment Machinery Regulates Axonal Degeneration in C. elegans Neurons. Cell Rep 14:1673–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cusack CL, Swahari V, Hampton Henley W, et al. (2013) Distinct pathways mediate axon degeneration during apoptosis and axon-specific pruning. Nat Commun 4:1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solomon F, Magendantz M (1981) Cytochalasin separates microtubule disassembly from loss of asymmetric morphology. J Cell Biol 89:157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo L, O’Leary DDM (2005) Axon Retraction and Degeneration in Development and Disease. Annu Rev Neurosci 28:127–156. [DOI] [PubMed] [Google Scholar]
- 9.Neukomm LJ, Freeman MR (2014) Diverse cellular and molecular modes of axon degeneration. Trends Cell Biol 24:515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geden MJ, Deshmukh M (2016) Axon degeneration: Context defines distinct pathways. Curr Opin Neurobiol 39:108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poulain FE, Bin Chien C(2013) Proteoglycan-Mediated Axon Degeneration Corrects Pretarget Topographic Sorting Errors. Neuron 78:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheriyan T, Ryan DJ, Weinreb JH, et al. (2014) Spinal cord injury models: A review. Spinal Cord 52:588–595. [DOI] [PubMed] [Google Scholar]
- 13.Xiong Y, Mahmood A, Chopp M (2013) Animal models of traumatic brain injury. Nat Rev Neurosci 14:128–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watts RJ, Schuldiner O, Perrino J, et al. (2004) Glia Engulf Degenerating Axons during Developmental Axon Pruning. Curr Biol 14:678–684. [DOI] [PubMed] [Google Scholar]
- 15.Low LK, Cheng HJ (2006) Axon pruning: An essential step underlying the developmental plasticity of neuronal connections. Philos Trans R Soc B Biol Sci 361:1531–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campenot RB (1977) Local control of neurite development by nerve growth factor. Proc Natl Acad Sci U S A 74:4516–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JW, Kim HJ, Byun JH, et al. (2009) Novel microfluidic platform for culturing neurons: Culturing and biochemical analysis of neuronal components. Biotechnol J 4:1573–1577. [DOI] [PubMed] [Google Scholar]
- 18.Whitesides GM (2006) The origins and the future of microfluidics. Nature 442:368–373. [DOI] [PubMed] [Google Scholar]
- 19.Taylor AM, Blurton-Jones M, Rhee SW, et al. (2005) A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods 2:599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park JW, Vahidi B, Taylor AM, et al. (2006) Microfluidic culture platform for neuroscience research. Nat Protoc 1:2128–36. [DOI] [PubMed] [Google Scholar]
- 21.Gamage KK, Cheng I, Park RE, et al. (2017) Death Receptor 6 Promotes Wallerian Degeneration in Peripheral Axons. Curr Biol 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Millet LJ, Gillette MU (2012) New perspectives on neuronal development via microfluidic environments. Trends Neurosci 35:752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beebe DJ, Mensing GA, Walker GM (2002) Physics and Applications of Microfluidics in Biology. Annu Rev Biomed Eng 4:261–286. [DOI] [PubMed] [Google Scholar]
- 24.Gross PG, Kartalov EP, Scherer A, Weiner LP (2007) Applications of microfluidics for neuronal studies. J Neurol Sci 252:135–143. [DOI] [PubMed] [Google Scholar]
- 25.Barford K, Keeler A, McMahon L, et al. (2018) Transcytosis of TrkA leads to diversification of dendritic signaling endosomes. Sci Rep 8:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J, Koito H, Li J, Han A (2012) Multi-compartment neuron-glia co-culture platform for localized CNS axon-glia interaction study. Lab Chip 12:3296–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jocher G, Mannschatz SH, Offterdinger M, Schweigreiter R (2018) Microfluidics of Small-Population Neurons Allows for a Precise Quantification of the Peripheral Axonal Growth State. Front Cell Neurosci 12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kilinc D, Peyrin JM, Soubeyre V, et al. (2011) Wallerian-like degeneration of central neurons after synchronized and geometrically registered mass axotomy in a three-compartmental microfluidic chip. Neurotox Res 19:149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi M, Majumdar D, Gao Y, et al. (2013) Glia co-culture with neurons in microfluidic platforms promotes the formation and stabilization of synaptic contacts. Lab a Chip - Miniaturisation Chem Biol 13:3008–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park JW, Kim HJ, Kang MW, Jeon NL (2013) Advances in microfluidics-based experimental methods for neuroscience research. Lab Chip 13:509–521. [DOI] [PubMed] [Google Scholar]
- 31.Shrirao AB, Kung FH, Omelchenko A, et al. (2018) Microfluidic platforms for the study of neuronal injury in vitro. Biotechnol Bioeng 115:815–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chronis N, Zimmer M, Bargmann CI (2007) Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat Methods 4:727–731. [DOI] [PubMed] [Google Scholar]
- 33.Ben-Yakar A, Chronis N, Lu H (2009) Microfluidics for the analysis of behavior, nerve regeneration, and neural cell biology in C. elegans. Curr Opin Neurobiol 19:561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kneynsberg A, Collier TJ, Manfredsson FP, Kanaan NM (2016) Quantitative and semi-quantitative measurements of axonal degeneration in tissue and primary neuron cultures. J Neurosci Methods 266:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Yang M, Huang Z, et al. (2014) AxonQuant: A microfluidic chamber culture-coupled algorithm that allows high-throughput quantification of axonal damage. NeuroSignals 22:14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhai Q, Wang J, Kim A, et al. (2003) Involvement of the ubiquitin-proteasome system in the early stages of Wallerian degeneration. Neuron 39:217–225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.