Abstract
The chicken reference genome contains 2 endogenous avian leukosis virus subgroup E (ALVE) insertions, but gaps and unresolved repetitive sequences in previous assemblies have hindered their precise characterization. Detailed analysis of the most recent reference genome (GRCg6a) now shows both ALVEs within contiguous chromosome assemblies for the first time. ALVE6 (ALVE-JFevA) and ALVE-JFevB are both located on chromosome 1, with ALVE6 close to the p-arm telomere. ALVE-JFevB is a structurally intact element containing the ALVE gag, pol, and env genes and is capable of forming replication competent viruses. In contrast, ALVE6 contains a 3,352 bp 5′ truncation and lacks the entire 5′ long terminal repeat and gag gene. Despite this, ALVE6 remains able to produce intact envelope protein, likely due to a mutation in the recognition site for a known inhibitory miRNA (miR-155). Whole genome resequencing data sets from layers, broilers, and 3 independent sources of wild-caught red junglefowl were surveyed for the presence of each of these reference genome ALVEs. ALVE-JFevB was found in no other chicken or red junglefowl genomes, whereas ALVE6 was identified in some layers, broilers, and native breeds but not within any other red junglefowl genome. Improved assembly contiguity has facilitated better characterization of the 2 ALVEs of the chicken reference genome. However, both the limited ALVE content and unique presence of ALVE-JFevB suggests that the reference individual is unrepresentative of ancestral Gallus gallus ALVE diversity.
Key words: ALVE, reference genome, ERV, ALVE6, ALVE-JFevB
Introduction
Endogenous retroviruses (ERV) constitute approximately 3% of the chicken (Gallus gallus) genome, a consequence of millions of years of retroviral integrations into the germline (Mason et al., 2016). The avian leukosis virus (ALV) is the only known chicken retrovirus with recurrent exogenous and endogenous activity, with the endogenous subgroup E (the ALVE, historically identified as ev genes) limited to the domestic chicken and its wild progenitor, the red junglefowl (RJF) (Frisby et al., 1979, Borysenko et al., 2008, Payne and Nair, 2012). Owing to their recent genome integration, ALVEs are typically present in low copy numbers, but many retain some structural integrity, facilitating persistent retroviral gene expression, and recombination with other ERV or exogenous retroviruses (Katzourakis et al., 2005, Payne and Nair, 2012). Recent in-depth studies have revealed the great diversity present across chicken populations, with more than 400 different ALVEs described to date (Benkel, 1998, Rutherford et al., 2016, Mason, 2018).
In commercial populations, ALVE-induced viremia elicits reductions in growth rate and total body weight in broilers (Fox and Smyth, 1985, Ka et al., 2009), and egg weight, specific gravity, and lifetime egg production in layers (Kuhnlein et al., 1989, Gavora et al., 1991). Expression of replication-competent proviruses (Crittenden et al., 1984, Gavora et al., 1995), or even gag glycoproteins alone (Astrin and Robinson, 1979, Robinson et al., 1981), can induce tolerance to novel ALV infections, resulting in delayed immune response and a higher incidence of lymphoid tumors. Furthermore, coinfection with Marek's disease virus, including attenuated vaccine viruses, has been shown to reactivate otherwise silenced ALVE in the genome and increase the incidence of spontaneous lymphoid tumors (Cao et al., 2015). However, ALVE effects are complex as expression of env glycoproteins prohibits some of these effects by receptor interference (Smith et al., 1990, Smith et al., 1991).
Despite extensive research into the effects of ALVEs, the 2 ALVEs present in the chicken reference genome remain incompletely described, most likely due to their locations within repetitive DNA, including one near the telomere of chromosome 1 (Benkel and Rutherford, 2014, Mason, 2018). The release of an updated, highly contiguous assembly (GRCg6a) provides a new opportunity to fully describe the ALVEs of the chicken reference genome. This study characterizes the location and structural integrity of both ALVE6 (ALVE-JFevA) and ALVE-JFevB in the GRCg6a assembly and determines their abundance in diverse chicken populations.
Materials and methods
ALVEs were detected in the new chicken genome assembly (GRCg6a; GenBank: GCA_000002315.5) by BLASTn (Altschul et al., 1990) using the ALVE1 reference sequence (GenBank: AY013303.1). Open reading frames (ORFs) were predicted with GLIMMER3 (Delcher et al., 2007), and the miR-155 AGCATTA recognition site (Hu et al., 2016) was annotated by the EMBOSS fuzznuc tool (Rice et al., 2000). Sequence surrounding each ALVE was annotated for other repetitive elements using RepBase CENSOR (Kohany et al., 2006) and identified repeat abundance was assessed by BLASTn.
Sixteen whole-genome resequencing (WGS) data sets (totaling 142 chickens; summarized in Table 1), which were previously analyzed for their unassembled ALVE content (Mason, 2018), were used for this study. These samples included commercially used elite layer lines (White Leghorn, White Plymouth Rock, and Rhode Island Red breeds), Indonesian native breeds (Black Java, Black Sumatra, Kedu Hitam, and Sumatera), wild-caught RJF from Java, Sumatra, and Tibet, and an experimental research broiler line. WGS data were reanalyzed to specifically detect the presence of both ALVE6 (ALVE-JFevA) and ALVE-JFevB. Paired-end reads from each WGS data set were mapped to the GRCg6a assembly using BWA-mem v0.7.10 (Li, 2013), filtering out reads with a mapping quality less than 20. In all cases, average genome coverage across the assembled chromosome exceeded 10X. The presence of ALVE6 and ALVE-JFevB was detected by identifying reads with sequence homology to both the ALVE and the neighboring genome sequence. Such reads reflect contiguous sequence in the host genome and the presence of that specific ALVE insertion (Mason, 2018).
Table 1.
Summary of whole genome resequencing datasets surveyed for ALVE6 and ALVE-JFevB.
| Name | Library preparation | Reference/Accession |
|---|---|---|
| Hy-Line International elite layer lines | Kranis et al., 2013 | |
| 5 x White Leghorn | 5 x Pool of 10 | |
| 2 x White Plymouth Rock | 2 x Pool of 10 | |
| 1 x Rhode Island Red | 1 x Pool of 10 | |
| Indonesian natives | DDBJ: DRA003951 | |
| Black Java | Pool of 10 | |
| Black Sumatra | Pool of 10 | |
| Kedu Hitam | Pool of 10 | |
| Sumatera | Pool of 5 | |
| Red junglefowl from Java | Pool of 3 | |
| Red junglefowl from Sumatra | Pool of 2 | |
| INRA experimental broiler line | 16 individuals | ENA: PRJNA247952 |
| Red junglefowl from Tibet | 6 individuals | ENA: PRJNA241474 |
Abbreviation: ALVE, avian leukosis virus subgroup E.
Results and discussion
For the first time, the current chicken genome assembly (GRCg6a) contains both the endogenous ALV integrations present in the reference genome RJF (International Chicken Genome Sequencing Consortium, 2004). Previous assemblies had correctly assigned ALVE-JFevB to chromosome 1 (1p2.3), but ALVE6 (ALVE-JFevA) is located near the telomere of chromosome 1 (1p2.10), so remained unassembled and incompletely sequenced (Benkel and Rutherford, 2014). With the improvements in the current assembly, both ALVEs can now be more completely described.
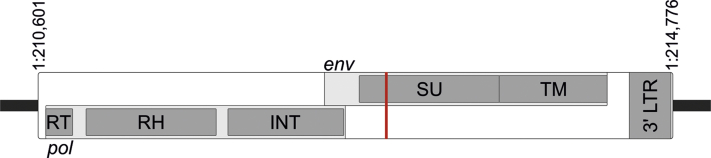
ALVE6 (1:210601-214776) is a 5′ truncated, 4,176 bp insertion in the forward orientation, with the previously identified target site duplication GGCGCT (Benkel, 1998) assembled at the 3′ end (Figure 1). The 5′ truncation has deleted 3,352 bp of the ALVE, without any associated flanking genomic sequence deletion, removing the 5′ long terminal repeat (LTR), gag domain and 67 bp of reverse transcriptase. The remaining sequence has 2 ORFs. The first (ALVE6:43-2013, first frame) encodes the reverse transcriptase thumb domain, RNaseH, and integrase, and the second (ALVE6:1877-3736, second frame) encodes an intact envelope. Chickens containing ALVE6 have long been known to express high titers of envelope glycoproteins (Robinson et al., 1981), perhaps due, in part, to a previously undescribed mutation in the recognition site of miR-155 ([A > G]GCATTA), a miRNA which typically regulates ALVE envelope expression by targeting transcripts for degradation (Hu et al., 2016).
Figure 1.
ALVE6 integration site showing 2 putative open reading frames. ALVE6 pol is truncated at the 5′ end but encodes the reverse transcriptase (RT) thumb domain, intact RNaseH (RH), and integrase (INT) domains. The envelope ORF is complete and features a mutated recognition site for the known inhibitory miRNA miR-155 (vertical bar). Abbreviations: ALVE, avian leukosis virus subgroup E; LTR, long terminal repeat; ORF, open reading frame; SU, surface; TM, transmembrane.
ALVE-JFevB (1:32724216-32731739) is an intact, 7,524 bp insertion in the forward orientation, with a GGCTTG target site duplication assembled at both ends (Figure 2). The ALVE-JFevB LTRs retain 100% identity and share 97.8% identity with the ALVE1 LTRs, with no variants affecting the TATA box or transcription factor binding sites, including the transcription start site. ALVE-JFevB contains intact ORF for gag-pol (ALVE-JFevB:479-5364), taking into account the ribosomal -1 frameshift just before the gag termination codon (Leblanc et al., 2013), and the envelope domain (ALVE-JFevB:5228-7084). Taken together, the intactness of ALVE-JFevB supports a recent integration and the ability to form replication competent viral particles. However, the presence of the intact miR-155 site within the envelope domain may inhibit complete expression of ALVE-JFevB.
Figure 2.
ALVE-JFevB integration site within a GGERV20 element. ALVE-JFevB is intact with gag, pol, and env ORF with detectable subunits. GGERV20 is in the negative orientation and contains putative gag and pol ORFs. pol is truncated at the 3′ end, but the 4 core catalytic components remain unaffected. Abbreviations: ALVE, avian leukosis virus subgroup E CA, capsid; INT, integrase; LTR, long terminal repeat; MA, matrix; NC, nucleocapsid; ORF, open reading frame; PR, protease; RH, RNaseH; RT, reverse transcriptase; SU, surface; TM, transmembrane.
The ALVE-JFevB integration site is complex because of its location within another transposable element: GGERV20, an ERV related to spumaviruses (Huda et al., 2008, Benkel and Rutherford, 2014). GGERV20 is a 5,827-bp element, which retains the ability to retrotranspose within the genome and is therefore polymorphic between chicken populations, with at least 65 full-length copies throughout the GRCg6a assembly. ALVE-JFevB has inserted within a reverse orientation GGERV20, 801 bp from the GGERV20 3′LTR. While this does disrupt the 3′ end of the GGERV20 pol ORF, the longer 5′ fragment contains all the core polymerase catalytic domains (Figure 2) and therefore may retain functional activity if expressed.
The Prevalence of ALVE6 and ALVE-JFevB Among Chickens
ALVE-JFevB was detected in no other analyzed dataset, including the wild-caught RJF from Java, Sumatra, and Tibet. Furthermore, the GGERV20 element in which ALVE-JFevB has inserted was also not found in any other data set, suggesting a sequential GGERV20 retrotransposition followed by ALVE-JFevB integration in the reference RJF lineage. Crucially, the LTR pairs of both ALVE-JFevB and the outer GGERV20 share 100% identity, supporting evolutionarily recent integrations.
ALVE6 was also not found in any other RJF genome in this study. However, ALVE6 was identified in the INRA broiler population, 4 Hy-Line elite layer lines (2 White Leghorn and 2 White Plymouth Rocks), and in the Black Java and Black Sumatra birds from Indonesia. Although this distribution is quite broad, it does not unambiguously support the presence of ALVE6 in the common G. gallus ancestor.
Recent work has revealed the large diversity of chicken ALVEs within and between populations. Noncommercial chicken populations harbor large numbers of low frequency and lineage-specific ALVEs, with individual bird genomes typically containing more than 6 ALVE loci (Rutherford et al., 2016, Mason, 2018). The presence of only 2 ALVEs in the RJF reference genome is another measure of how unrepresentative this ‘reference’ bird is of extant G. gallus genomic diversity (Ulfah et al., 2016). Consequently, great care needs to be taken when using the reference genome as a background for the study of ALVEs, such as the recent work by Sun et al., 2017, who identified postdomestication piRNA-mediated defense against these elements. Such comparative genomic approaches, particularly those attempting to unpick the complex chicken domestication process (Rubin et al., 2010), should only be undertaken once the complete ALVE complement of that bird or line has been ascertained by methods such as obsERVer (Mason, 2018). This is indicative of a wider issue of confidence in reference genomes, as ALVEs are just one marker, which highlights how unrepresentative of chicken diversity the reference genome can be.
The GRCg6a assembly contiguity improvements facilitate more comprehensive ALVE and broader structural variant identification from WGS projects. However, ALVEs may still be missed when present on the largely incomplete, or absent, assemblies of chromosomes 29, 34–38 and W, or in the poorly assembled centromeres and telomeres.
Conclusions
The current chicken genome assembly (GRCg6a) now shows both the genomic location and complete sequence of the 2 ALVEs known to exist with the reference RJF. ALVE-JFevB is structurally intact and is unique to the reference genome. ALVE6 (ALVE-JFevA), while truncated by nearly half its length, is found across diverse chicken breeds and is capable of producing envelope protein, potentially due to a mutation identified here in the miR-155 recognition site, a miRNA which typically marks envelope transcript for degradation. Examination of genome sequences from diverse chicken populations and wild-caught RJF did not reveal the universal presence of either of these 2 ALVEs, and showed that, on average, individual chicken genomes typically contain over 6 ALVE integrations. These 2 observations suggest that the reference genome is not representative of G. gallus ALVE abundance and diversity and is unlikely to represent ALVE content in ancestral RJF. Therefore, caution must be applied when using the current reference genome as a baseline for the predomesticated state.
Acknowledgments
This work was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) as part of an Impact Accelerator Award (BB/GCRF-IAA/25). The authors also acknowledge the efforts of Ashlee Lund to generate high-throughput detection assays for these ALVEs.
Conflict of Interest Statement: The authors have no conflicts of interest to declare.
References
- Altschul S.F., Gish W., Miller W., Myers E., Lipman D. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Astrin S.M., Robinson H. Gs, an Allele of chickens for endogenous avian leukosis viral Antigens Segregates with ev3, a Genetic Locus that contains structural genes for virus. J. Virol. 1979;31:420–425. doi: 10.1128/jvi.31.2.420-425.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkel B.F., Rutherford K. Endogenous avian leukosis viral loci in the Red Jungle Fowl genome assembly. Poult. Sci. 2014;93:2988–2990. doi: 10.3382/ps.2014-04309. [DOI] [PubMed] [Google Scholar]
- Benkel B.F. Locus-specific diagnostic tests for endogenous avian leukosis-type viral loci in chickens. Poult. Sci. 1998;77:1027–1035. doi: 10.1093/ps/77.7.1027. [DOI] [PubMed] [Google Scholar]
- Borysenko L., Stepanets V., Rynditch A.V. Molecular characterization of full-length MLV-related endogenous retrovirus ChiRV1 from the chicken, Gallus gallus. Virology. 2008;376:199–204. doi: 10.1016/j.virol.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Cao W., Mays J., Kulkarni G., Dunn J., Fulton R.M., Fadly A. Further observations on serotype 2 Marek’s disease virus-induced enhancement of spontaneous avian leukosis virus-like bursal lymphomas in ALVA6 transgenic chickens. Avian Pathol. 2015;44:23–27. doi: 10.1080/03079457.2014.989195. [DOI] [PubMed] [Google Scholar]
- Crittenden L.B., Smith E.J., Fadly A.M. Influence of endogenous viral (ev) gene expression and strain of exogenous avian leukosis virus (ALV) on mortality and ALV infection and shedding in chickens. Avian Dis. 1984;28:1037–1056. [PubMed] [Google Scholar]
- Delcher A.L., Bratke K.A., Powers E.C., Salzberg S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox W., Smyth J.R.J. The effects of recessive white and dominant white genotypes on early growth rate. Poult. Sci. 1985;64:429–433. doi: 10.3382/ps.0640429. [DOI] [PubMed] [Google Scholar]
- Frisby D.P., Weiss R.A., Roussel M., Stehelin D. The distribution of endogenous chicken retrovirus sequences in the DNA of galliform birds does not coincide with avian phylogenetic relationships. Cell. 1979;17:623–634. doi: 10.1016/0092-8674(79)90270-8. [DOI] [PubMed] [Google Scholar]
- Gavora J.S., Kuhnlein U., Crittenden L.B., Spencer J.L., Sabour M.P. Endogenous viral genes: Association with reduced egg production rate and egg size in white Leghorns. Poult. Sci. 1991;70:618–623. doi: 10.3382/ps.0700618. [DOI] [PubMed] [Google Scholar]
- Gavora J.S., Spencer J.L., Benkel B.F., Gagnon C., Emsley A., Kulekamp A. Endogenous viral genes influence infection with avian leukosis virus. Avian Pathol. 1995;24:653–664. doi: 10.1080/03079459508419105. [DOI] [PubMed] [Google Scholar]
- Hu X., Zhu W., Chen S., Liu Y., Sun Z., Geng T., Wang X., Gao B., Song C., Qin A., Cui H. Expression of the env gene from the avian endogenous retrovirus ALVE and regulation by miR-155. Arch. Virol. 2016;161:1623–1632. doi: 10.1007/s00705-016-2833-8. [DOI] [PubMed] [Google Scholar]
- Huda A., Polavarapu N., Jordan I.K., McDonald J.F. Endogenous retroviruses of the chicken genome. Biol. Direct. 2008;3 doi: 10.1186/1745-6150-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Ka S., Kerje S., Bornold L., Lijegren U., Siegel P.B., Andersson L., Hallböök F. Proviral integrations and expression of endogenous avian leucosis virus during long term selection for high and low body weight in two chicken lines. Retrovirology. 2009;6:68. doi: 10.1186/1742-4690-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A., Rambaut A., Pybus O.G. The evolutionary dynamics of endogenous retroviruses. Trends Microbiol. 2005;13:463–468. doi: 10.1016/j.tim.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Kohany O., Gentles A.J., Hankus L., Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranis A., Gheyas A.A., Boschiero C., Turner F., Yu L., Smith S., Talbot R., Pirani A., Brew F., Kaiser P., Hocking P.M. Development of a high density 600K SNP genotyping array for chicken. BMC Genomics. 2013;14:59. doi: 10.1186/1471-2164-14-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnlein U., Sabour M., Gavora J.S., Fairfull R.W., Bernon D.E. Influence of selection for egg production and Marek’s disease Resistance on the incidence of endogenous viral genes in white Leghorns. Poult. Sci. 1989;68:1161–1167. doi: 10.3382/ps.0681161. [DOI] [PubMed] [Google Scholar]
- Leblanc J., Weil J., Beemon K. Posttranscriptional regulation of retroviral gene expression: primary RNA transcripts play three roles as pre-mRNA, mRNA, and genomic RNA. Wiley Interdiscip. Rev. RNA. 2013;4:567–580. doi: 10.1002/wrna.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv preprint arXiv:1303.3997. [Google Scholar]
- Mason A.S., Fulton J.E., Hocking P.M., Burt D.W. A new look at the LTR retrotransposon content of the chicken genome. BMC Genomics. 2016;17:688. doi: 10.1186/s12864-016-3043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason A.S. The University of Edinburgh; Edinburgh, UK: 2018. The Abundance and Diversity of Endogenous Retroviruses in the Chicken Genome. Doctoral Dissertation. [Google Scholar]
- Payne L.N., Nair V. The long view: 40 years of avian leukosis research. Avian Pathol. 2012;41:11–19. doi: 10.1080/03079457.2011.646237. [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Robinson H.L., Astrin S.M., Senior A.M., Salazar F.H. Host Susceptibility to endogenous viruses: defective, glycoprotein-expressing proviruses interfere with infections. J. Virol. 1981;40:745–751. doi: 10.1128/jvi.40.3.745-751.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin C.-J., Zody M.C., Eriksson J., Meadows J.R.S., Sherwood E., Webster M.T., Jiang L., Ingman M., Sharpe T., Ka S., Hallböök F. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- Rutherford K., Meehan C.J., I Langille M.G., Tyack S.G., McKay J.C., McLean N.L., Benkel K., Beiko R.G., Benkel B.F. Discovery of an expended set of avian leukosis subroup E proviruses in chickens using Vermillion, a novel sequence capture and analysis pipeline. Poult. Sci. 2016;95:2250–2258. doi: 10.3382/ps/pew194. [DOI] [PubMed] [Google Scholar]
- Smith E.J., Fadly A.M., Crittenden L.B. Interactions between endogenous virus loci ev6 and ev21: 1. Immune response to exogenous avian leukosis virus infection. Poult. Sci. 1990;69:1244–1250. doi: 10.3382/ps.0691244. [DOI] [PubMed] [Google Scholar]
- Smith E.J., Fadly A.M., Levin I., Crittenden L.B. The influence of ev6 on the immune response to avian leukosis virus infection in Rapid-Feathering Progeny of Slow- and Rapid-Feathering Dams. Poult. Sci. 1991;70:1673–1678. doi: 10.3382/ps.0701673. [DOI] [PubMed] [Google Scholar]
- Sun Y.H., Xie L.H., Zhuo X., Chen Q., Ghoneim D., Zhang B., Jagne J., Yang C., Li X.Z. Domestic chickens activate a piRNA defense against avian leukosis virus. Elife. 2017;6:1–24. doi: 10.7554/eLife.24695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulfah M., Kawahara-Miki R., Farajalllah A., Muladona M., Dorshorst B., Martin A., Kono T. Genetic features of red and green junglefowls and relationship with Indonesian native chickens Sumatera and Kedu Hitam. BMC Genomics. 2016;17:320. doi: 10.1186/s12864-016-2652-z. [DOI] [PMC free article] [PubMed] [Google Scholar]