Abstract
Newcastle disease is an acute and highly contagious disease of poultry caused by Newcastle disease virus infection, which does great harm to the poultry industry all over the world. To diagnose the disease simply and quickly, 2 detection methods were established based on reverse transcription recombinase–aided amplification (RT-RAA) technology. One is reverse transcription recombinase–aided amplification-lateral flow dipstick (RT-RAA-LFD) that is to combine RT-RAA with lateral flow dipstick; the other is real-time fluorescence-based reverse transcription recombinase–aided amplification (RF-RT-RAA) that is the combination of RT-RAA and exo probe. In this study, the reaction conditions such as reaction temperature and reaction time of the 2 methods were optimized, and their specificity and sensitivity were tested. The results showed that the RT-RAA-LFD method could be used to complete reaction within 23 min, and its lowest detectable limit was 102 copies/μL, 10 times higher than that of the conventional PCR method (103 copies/μL); the RF-RT-RAA method could be used to complete reaction within 26 min, and its lowest detectable limit was 10 copies/μL, 100 times higher than that of conventional PCR method (103 copies/μL), and it was as sensitive as real-time fluorescence–based quantitative PCR (10 copies/μL). The 2 methods had no cross reaction to the nucleic acid of other avian pathogens and showed good specificity. A total of 86 clinical samples suspected of the Newcastle disease virus were tested by conventional PCR, real-time fluorescence–based quantitative PCR, RT-RAA-LFD, and RF-RT-RAA. Based on the commonly used conventional PCR method, the other 3 detection methods had a coincidence rate of higher than 93%. In summary, RT-RAA-LFD and RF-RT-RAA had high specificity, sensitivity, and efficiency, which were suitable for clinical and laboratory diagnosis, respectively, and provided technical support for the prevention and control of Newcastle disease.
Key words: Newcastle disease virus, reverse transcription recombinase–aided amplification-lateral flow dipstick, real-time fluorescence–based-reverse transcription recombinase-aided amplification, detection
Introduction
Newcastle disease (ND), also known as pseudo-fowl pest, is caused by the Newcastle disease virus (NDV). It can occur all the year round, especially in early spring and winter (Ganar et al., 2014). Newcastle disease spreads rapidly primarily through the digestive tract and respiratory tract and water, feed used or touched by infected chickens as well as their feces may carry the virus to infect other healthy chickens (Gao et al., 2019). The poultry, especially chickens, are vulnerable to the NDV, and the infection rate of ducks and geese is also increasing year by year. Young chickens are most susceptible to the NDV, and the diseased chickens show respiratory signs including elevated body temperature, dyspnea, coughing, greenish watery diarrhea, thin-shelled eggs, and reduced egg production. Even after recovery, it will have an impact on the growth and development of chickens and brings serious economic losses to the poultry industry (Liu et al., 2019).
The clinical symptoms of ND are similar to those of some other diseases including low pathogenic avian influenza, infectious laryngotracheitis, and infectious bronchitis, all showing the respiratory sign of dyspnea, so they may be confused with each other in the clinical diagnosis (Chumbe et al., 2017, Giovanni et al., 2017). A specific, sensitive, and convenient diagnostic method can provide strong support for the prevention and treatment of this disease. At present, a variety of NDV detection methods have been established at home and abroad, and serological methods include hemagglutination-inhibition test, agar gel diffusion test, neutralization test, immunofluorescence test, immunohistochemical technique, ELISA, and so on. However, at present, most chicken are vaccinated with NDV, leading to the existence of antibodies in healthy chickens, thus affecting the judgment results. The diagnostic methods of molecular biology include conventional PCR, real-time fluorescence–based quantitative PCR (RFQ-PCR), loop-mediated isothermal amplification, and so on (Lin et al., 1991, Zhang et al., 2018).
Reverse transcription recombinase-aided amplification (RT-RAA) is a new isothermal nucleic acid amplification technique in vitro (Kim and Easley, 2011). Reverse transcription recombinase–aided amplification is mainly used for the detection of RNA with simple operation, high sensitivity, and high specificity. It can be used to detect a variety of pathogens, and the current RAA technology has been successfully used for avian influenza virus (AIV) (Wang et al., 2019), infectious bronchitis virus (Wang et al., 2020), African swine fever virus (Lin et al., 2019), rabies virus (Tan et al., 2019), and so on.
The reverse transcription recombinase-aided amplification-lateral-flow dipstick (RT-RAA-LFD) detection method is to add an nfo probe with a length of 46 bp to 52 bp based on RAA reaction. The 5′end of this probe is labeled with 6-carboxyfluorescein (FAM), a position more than 30 bp from the 5′ end is labeled with tetrahydrofuran (THF) residue, and the 3′ end is blocked by phosphorylation, more than 15 bp from FAM residue. After the reaction, the double-labeled product is formed, and the rapid detection in the field is realized by the lateral flow dipstick (LFD) coated with biotin and FAM antibodies. Real-time fluorescence-based reverse transcription recombinase-aided amplification (RF-RT-RAA) adds a fluorescence-labeled probe based on RT-RAA, which contains a base analog (THF), and dT-fluorophores and the corresponding dT-quenchers are connected on both sides, and it is connected with a blocking group (such as C3 spacer) at the 3′end. In the natural state, the probe is single strand, and when fluorophores approach quenchers, there is fluorescence resonance energy transfer and the fluorescence signal emitted by fluorophores is absorbed by quenchers, so the fluorescence signal cannot be detected. Once it is identified that THF is cut off by the target sequence exo, FAM is in a free state. With the continuous generation of fluorescence signal in the amplification reaction, the enhancement of fluorescence signal is positively correlated with the accumulation of amplification products, and the amplification products can be monitored in real time (Piepenburg et al., 2006). The 2 methods have good specificity and high sensitivity and can be used for rapid detection in the field, and RF-RT-RAA is more sensitive and can be used both in laboratory and in the field. In this experiment, 2 detection methods were established based on the RT-RAA principle to improve reaction efficiency and reduce cost. The 2 methods are simple, specific, and sensitive, providing a new technical method for NDV detection.
Materials and methods
Materials
Both the NDV and AIV were preserved in our laboratory (from clinical cases, identified by PCR), the experimental procedure was approved by the Institutional Animal Care and Ethics Committee of Hebei Agricultural University. The infectious laryngotracheitis virus (ILTV) (AV195) and infectious bronchitis virus (IBV) (AV1511) were purchased from the China Institute of Veterinary Drug Control. A viral genomic DNA/RNA extraction kit was purchased from Tiangen Biotech (Beijing) Co., Ltd.; an RT-RAA nucleic acid amplification kit (LFD) and RT-RAA nucleic acid amplification kit (fluorescence method) were purchased from Jiangsu Qitian Gene Biological Technology Co., Ltd.; TB Green Premix and T-Vector Pmd20 plasmids were purchased from Takara Biomedical Technology (Beijing) Co., Ltd.; the primers and probes were synthesized and labeled by Shanghai Sangon Biotech Co., Ltd.; a LightCycler 96 RFQ-PCR instrument was purchased from Roche, and and ABI 2720 PCR instrument was purchased from Applied Biosystems.
Methods
Extraction of Genomes
In accordance with the instructions of DNA/RNA extraction kit, the RNA of the NDV, AIV and IBV was extracted, and the DNA of the ILTV was extracted. The viral RNA was extracted from 86 suspected NDV clinical samples and reversely transcribed into cDNA for subsequent experiments.
Screening of Primers
Four pairs of primers were designed on the basis of GenBank accession number JQ015296.1 (Table 1). The specificity of the primers was detected by RT-RAA agarose electrophoresis.
Table 1.
Primers selected for NDV reaction by RT-RAA.
| Primers | Sequence (5′-3′) | Gene localization |
|---|---|---|
| NDV-1-F | TGATTCGTGGACAGACAGTAAGGAAGACTCGG | 3431-3466 |
| NDV-1-R | TGAAAGTAACTCGTGCTTGGGATTATCGCTG | 3542-3573 |
| NDV-2-F | TGGACGTGGTCCCGAAGAGCCCGTTAGTCA | 3930-3960 |
| NDV-2-R | CGAGCACATCACTGAGCCCGACAGATAAAT | 4090-4120 |
| NDV-3-F | CAGCAAGCCATCTTATCTATCAAAGTGTCAACATC | 1108-1143 |
| NDV-3-R | TGTCCCTACTGTGAGAACTCTGCCTTCGGC | 1227-1257 |
| NDV-4-F | ACGTCTTTATTACTCCTGAGCTTGTCATTGTGA | 9008-9041 |
| NDV-4-R | TGATTACCTAAGTCCTTTGCCAGAGCATCTACT | 9227-9260 |
Abbreviations: NDV, Newcastle disease virus; RT-RAA, reverse transcription recombinase–aided amplification.
The reaction system contains the following: VI buffer, 25 μL; purified water, 16.8 μL; upstream primer, (10 μmol) 2.1 μL; downstream primer, (10 μmol) 2.1 μL; template, 1.5 μL; magnesium acetate, 2.5 μL. After the reaction product was purified as per the DNA product purification kit at 39°C for 30 min, the RT-RAA reaction product was detected by 2% agarose gel electrophoresis, and the most suitable primers were selected for the establishment of RT-RAA-LFD and RF-RT-RAA diagnostic methods for the NDV.
Establishment of Standard
The PCR primer containing the target gene fragment was designed, NDV-F: 5′-ACGTCTTTATTACTCCTGAGCTTGT-3′ and NDV-R: 5′-TGATTACCTAAGTCCTTTGCCAG-3′. In 50 μL PCR reaction system, the purified target fragment was ligated to pMD20 plasmid after gel extraction of PCR products. After introducing the competent cells, the white colonies were selected by blue and white screening, and the plasmids were extracted after overnight culture and identification. The DNA copy number in the standard plasmid was calculated as per Moore's law.
The standard plasmid constructed was diluted to a concentration gradient of 108–100 copies/μL by 10-fold dilution method and stored for later use.
Establishment of RT-RAA-LFD Detection Method
In accordance with the optimal primers selected, the 5′ end of downstream primer was labeled with biotin. The probe was designed after comparing the sequences (5′-AAGTTCACATGCCTCACCCAGGAACTTGTA/idSp/TGATGTATGCGGATA-3′), and the probe was labeled with FAM at the 5′end.
The reaction system contains the following: fluorescent basic buffer, 25 μL; upstream primer, (10 μmol) 2.1 μL; downstream primer (10 μmol) 2.1 μL; probe, 0.6 μL; template, 2 μL; purified water, 15.7 μL; magnesium acetate, 2.5 μL. After reaction at 39°C for 30 min, 10 μL of RT-RAA amplification product was put in the test area, the dipstick was inserted into the EP tube containing 100 μL of buffer, and the results were observed after 3 min.
The criteria are as follows: It was positive when 2 red bands appeared (one in quality control area and the other in test area), indicating that nucleic acid fragments to be detected were contained in the sample; it was negative if only one red band appeared (in quality control area), indicating that it did not contain the test fragment.
Optimization of RT-RAA-LFD Reaction Conditions
When using RT-RAA amplification kit for amplification, the reaction temperature, primer concentration, and reaction time were optimized. The reaction temperature was set to 33°C, 35°C, 37°C, 39°C, and 41°C. The reaction time gradient was controlled to 13, 15, 17, 19, 21, 23, and 25 min. The primer concentration gradient was 10,000, 5,000, 2,500, 1,250, and 625 nmol/L.
The criteria for the best reaction conditions are as follows: Clear and obvious test line, shortest reaction time, and lowest primer or probe concentration.
Establishment of RF-RT-RAA Detection Method
A probe was designed and labeled on the sequence between the selected upstream and downstream primers. Probe sequence: 5′-TGGAAGGCAGGGACATGGTTAATATAATATC/i6FAMdT//idSp/C/iBHQ1dT/ACAGCAGCACATCTC-3′ (C3 Spacer).
The RF-RT-RAA reaction system contains the following: VI buffer, 25 μL; purified water, 15.7 μL; upstream primer, (10 μmol) 2.1 μL; downstream primer, (10 μmol) 2.1 μL; probe, 0.6 μL; template, 2 μL.
Optimization of RF-RT-RAA Reaction Conditions
In accordance with the previous reaction system, the temperature was set to 37°C, 38°C, 39°C, 40°C, and 41°C, and the reaction time was set to 31 min.
The criteria for the best reaction conditions are shortest reaction time and strongest fluorescence signal.
Specificity Detection of RT-RAA-LFD and RF-RT-RAA
As per the established reaction system, the common avian viruses (NDV, IBV, AIV, and ILTV) were selected as templates, with water for negative control.
In accordance with the established reaction system, the template was identical with RT-RAA-LFD. The liquid was transferred into the reaction tubes after mixing, 2.5 μL of magnesium acetate was added before reaction, and it was put into the RFQ-PCR instrument to react for 27 min. 6-Carboxyfluorescein signals were collected during the reaction.
Comparison of Sensitivity of Four Methods: RT-RAA-LFD, RF-RT-RAA, Conventional PCR, and RFQ-PCR
In accordance with the optimized reaction conditions and reaction system, the templates were 106–100 copies/μL and 104–100 copies/μL, respectively. With water as a blank control, the lowest detectable limit was observed.
Water as a negative control was set with 107–100 copies/μL plasmid as template. The 25-μL PCR reaction system contains the following: 2×Taq Mix, 12.5 μL; DNA template, 2 μL; upstream primer, (10 μmol) 0.5 μL; downstream primer, (10 μmol) 0.5 μL; replenished with 9.5 μL of ddH2O. The reaction system is predenatured at 94°C for 5 min; denatured at 94°C for 45 s; annealed at 52°C for 45 s; extended at 72°C for 60 s, a total of 30 cycles; and extended at 72°C for 5 min, stored at 4°C. The results were observed by 2% agarose electrophoresis.
Using 106–100 copies/μL plasmid as template, 25 μL RFQ-PCR system (the primer was the same as that of conventional PCR) was as follows: TB Green Premix DimerEraser (2X), 12 μL; upstream primer, (10 μmol) 0.75 μL; downstream primer, (10 μmol) 0.75 μL; template, 2 μL; the final volume was made up to 25 μL with water. The reaction system is predenatured at 95°C for 30 s; denatured at 95°C for 5 s; annealed at 55°C for 30 s; and extended at 72°C for 30 s, and the results are to be observed after a total of 40 cycles.
Detection of Clinical Samples
A total of 86 suspected ND samples were detected by RT-RAA-LFD, RF-RT-RAA, conventional PCR, and RFQ-PCR, and the coincidence rates of these methods were compared.
Results
Results of Primer Screening
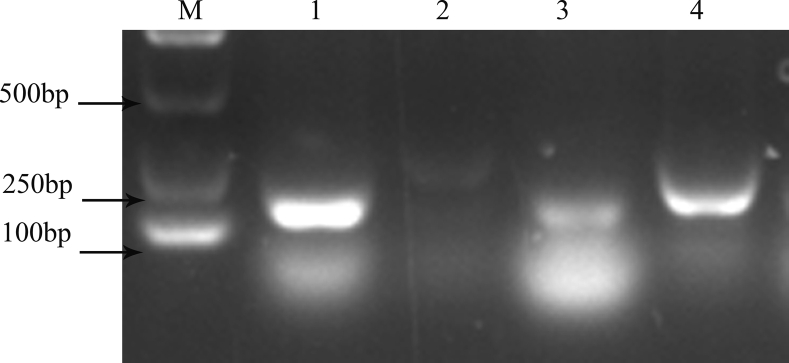
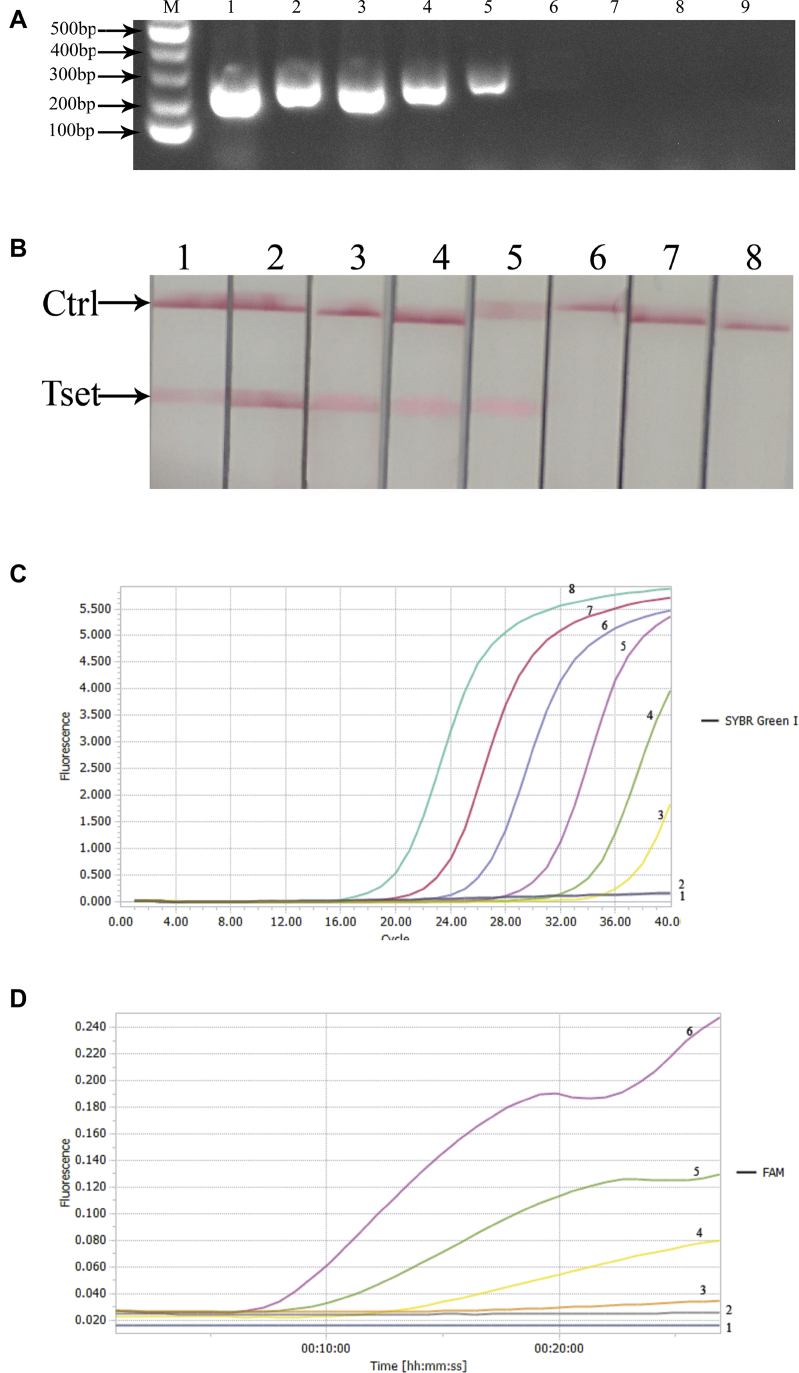
In accordance with the requirements of the RT-RAA nucleic acid amplification kit, 4 pairs of primers were designed. The products were amplified using the RT-RAA nucleic acid amplification kit and purified, and 2% agarose gel electrophoresis was used for screening. The results are shown in Figure 1. The results showed that a single target band could be amplified in the first and fourth primers, but the band amplified from primer NDV-4-F/NDV-4-R was clear and there was no obvious primer dimer. Therefore, NDV-4-F/NDV-4-R was chosen as the primer for the follow-up experiment.
Figure 1.
Screening of primers for NDV detection by RT-RAA. M: D2000 Marker; 1–4: amplification products of primer NDV 1–4. Abbreviations: NDV, Newcastle disease virus; RT-RAA, reverse transcription recombinase–aided amplification.
Optimization of RT-RAA-LFD Reaction Conditions
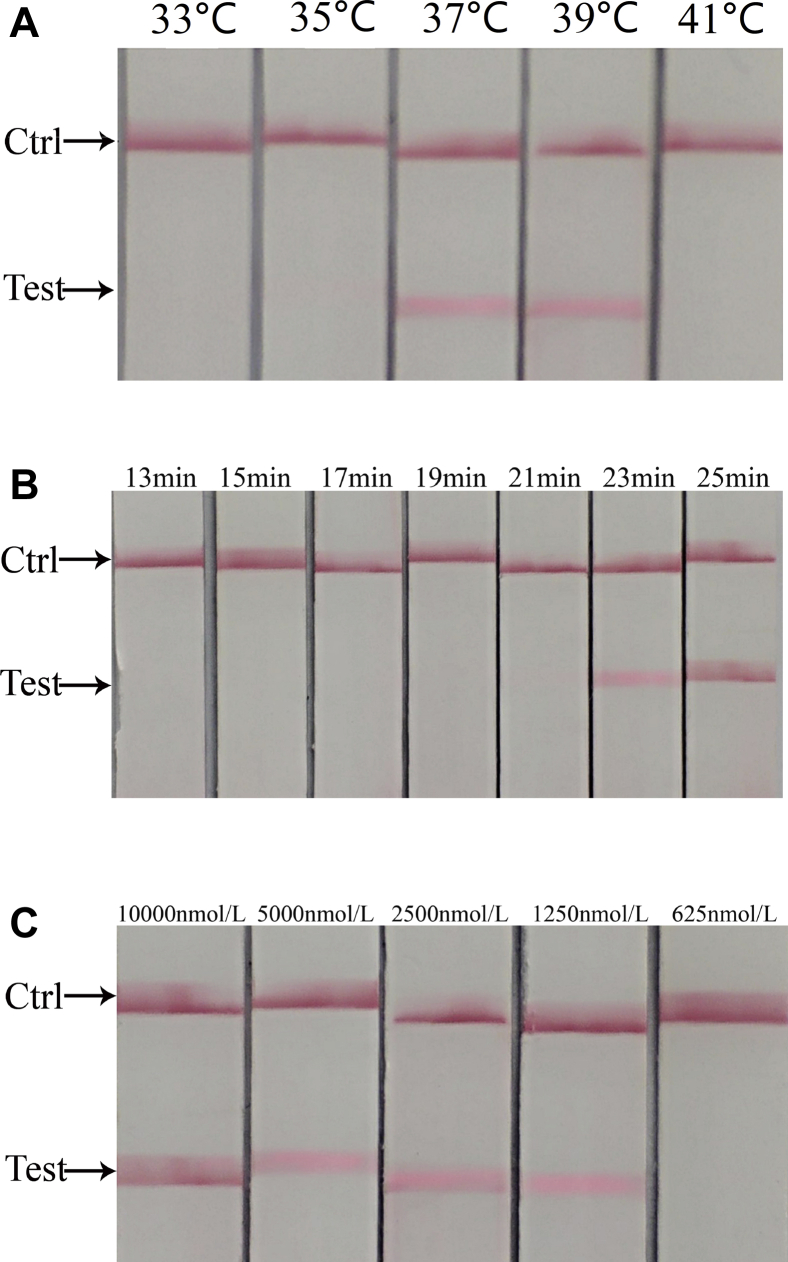
In this experiment, the reaction time, temperature, and primer concentration were optimized. The criteria for the best reaction conditions are as follows: Clear and obvious test line, lowest reaction temperature, shortest reaction time, and lowest primer or probe concentration. Under the conditions of 37°C, 23 min, and a primer concentration of 1,250 nmol/L, the obvious test line can be observed. Finally, the optimum reaction conditions were determined as follows: reaction temperature, 37°C; reaction time, 23 min; and primer concentration, 1,250 nmol/L. The results are shown in Figure 2.
Figure 2.
Optimization of reaction conditions for NDV detection by RT-RAA-LFD. (A) Screening of reaction temperature. The band appeared in the test area at 37°C and 39°C but not at 33°C, 35°C, and 41°C. As per the criteria for the best reaction conditions (obvious test line and lowest reaction temperature), 37°C was determined as the best reaction temperature. (B) Screening of reaction time. There was band in the test area at 23 and 25 min but not at 13, 15, 17, 19, and 21 min. As per the criteria for the best reaction conditions (obvious test line and shortest reaction time), 23 min was determined as the best reaction time (C) Screening of primer and probe concentration. 10,000, 5,000, 2,500, and 1,250 nmol/L test area showed bands, and no bands were found in 625 nmol/L test area. As per the criteria for the best reaction conditions (obvious test line and lowest primer or probe concentration), 1,250 nmol/L was determined as the best primer and probe concentration. Abbreviations: NDV, Newcastle disease virus; RT-RAA-LFD, reverse transcription recombinase–aided amplification-lateral flow dipstick.
Optimization of RF-RT-RAA Reaction Conditions
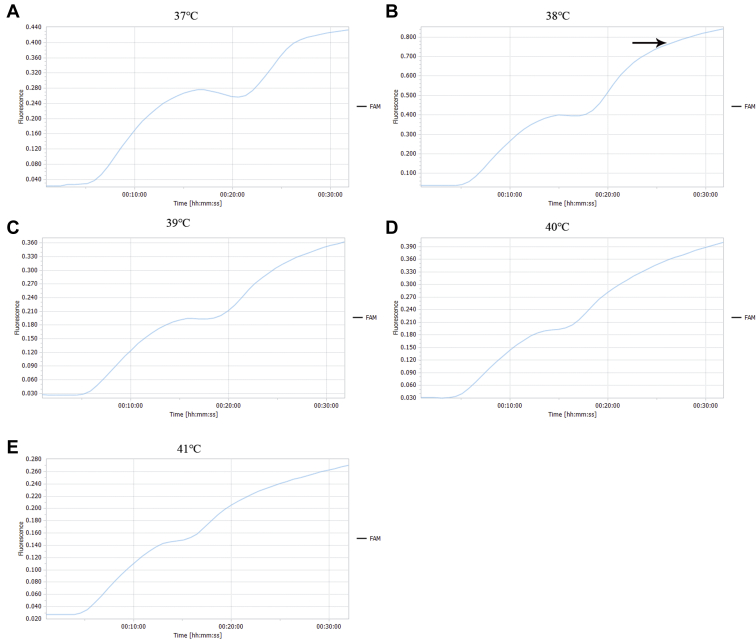
As per the peak time and fluorescence value of the amplification curve, the fluorescence value of the reaction was the highest at 38°C, and the reaction efficiency decreased after 26 min. Therefore, the optimum reaction temperature was 38°C and the reaction time was 26 min. The results are shown in Figure 3.
Figure 3.
Screening of reaction temperature and reaction time for NDV detection by RF-RT-RAA. Reaction time was set to 26 min, (A–E) 37°C to 41°C, respectively. As per the peak time and fluorescence value of the amplification curve, 38°C was determined as the optimum reaction temperature and 26 min was determined as the optimum reaction time. Abbreviations: NDV, Newcastle disease virus; RF-RT-RAA, real-time fluorescence-based reverse transcription recombinase–aided amplification.
Specificity Detection of RT-RAA-LFD
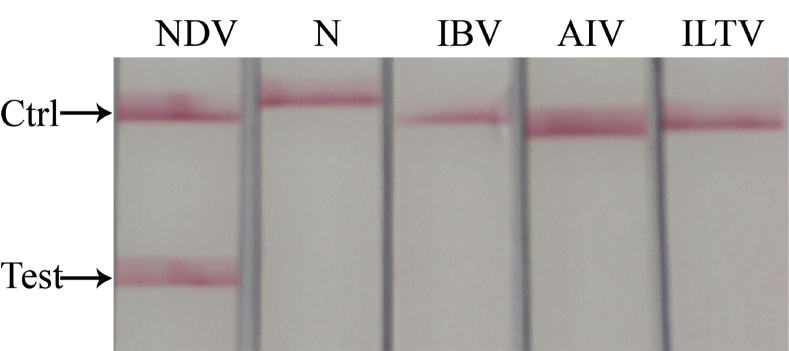
After LFD detection, only the RT-RAA product of the NDV showed both test line and quality control line on the dipstick. However, the products of other viruses and negative control could only show quality control line, and no other nonspecific amplification was found. The results are shown in Figure 4. It suggested that the specificity of RT-RAA-LFD was good, and this method was tenable.
Figure 4.
Specificity of RT-RAA-LFD in NDV detection. Bands appeared in the NDV test area; no bands were found in N (negative control), IBV, AIV, and ILTV test area. Abbreviations: AIV, avian influenza virus; IBV, infectious bronchitis virus; ILTV, infectious laryngotracheitis virus; NDV, Newcastle disease virus; RT-RAA-LFD, reverse transcription recombinase–aided amplification-lateral flow dipstick.
Specificity Detection of RF-RT-RAA
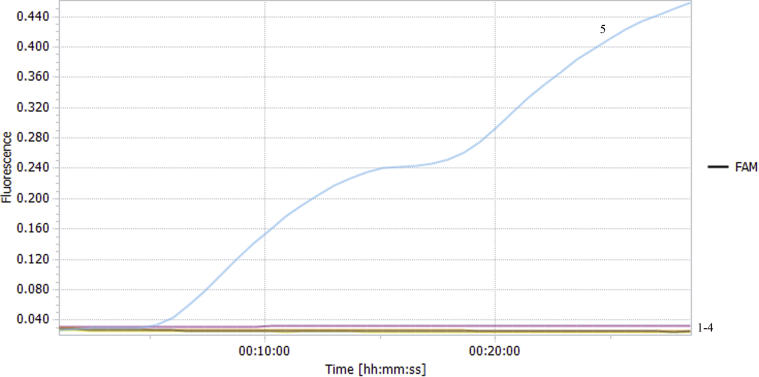
The templates were NDV, IBV, AIV, and ILTV, with water as a negative control. The liquid was transferred into the reaction tubes after mixing, and it was put into the RFQ-PCR instrument. 6-Carboxyfluorescein signals were collected during the reaction. The results showed that there was no fluorescence curve for other viral nucleic acids and negative control except for the NDV, which indicated that the RF-RT-RAA method could specifically detect the NDV and had no cross reaction with nucleic acids of other avian viruses, with strong specificity (Figure 5).
Figure 5.
Specificity of RF-RT-RAA in NDV detection. 1–4: Negative control, ILTV, AIV and IBV, respectively; 5: NDV. Bands appeared in the NDV test area; no fluorescence curve for other viral nucleic acids and negative control except for NDV. Abbreviations: AIV, avian influenza virus; IBV, infectious bronchitis virus; ILTV, infectious laryngotracheitis virus; NDV, Newcastle disease virus; RF-RT-RAA, real-time fluorescence-based reverse transcription recombinase–aided amplification.
Comparison of Sensitivity of Four Methods
In this experiment, the sensitivity of PCR, RFQ-PCR, RT-RAA-LFD, and RF-RT-RAA was compared. Among the 4 methods, the conventional PCR showed the minimum template concentration of 103 copies/μL for the occurrence of the obvious single band (Figure 6A). The RT-RAA-LFD showed the minimum template concentration of 102 copies/μL for the occurrence of the obvious test line was (Figure 6B), whereas the minimum template concentration for the occurrence of obvious amplification curve was 10 copies/μL in RFQ-PCR and RF-RT-RAA (Figure 6C, 6D). The results showed that RF-RT-RAA and RFQ-PCR had the highest sensitivity (10 copies/μL), followed by RT-RAA-LFD (102 copies/μL) and PCR (103 copies/μL).
Figure 6.
Comparison of sensitivity of 4 methods in NDV detection. (A) Sensitivity test of PCR. 1–8: 107–100 copies/μL, respectively; 9: Negative control. The results showed the minimum template concentration for the occurrence of the obvious single band was 103 copies/μL, which was the LDL for PCR. (B) Sensitivity test of RT-RAA-LFD. 1–7: 106–100 copies/μL, respectively; 8: Negative control. The results showed the minimum template concentration for the occurrence of the obvious test line was 102 copies/μL, which was the LDL for RT-RAA-LFD. (C) Sensitivity test of RFQ-PCR. 1: Negative control; 2–8: 100–106 copies/μL. The results showed the minimum template concentration for the occurrence of obvious amplification curve was 10 copies/μL, which was the LDL for RFQ-PCR. (D) Sensitivity test of RF-RT-RAA. 1: Negative control; 2–6: 100–104 copies/μL, respectively. The results showed the minimum template concentration for the occurrence of obvious amplification curve was 10 copies/μL, which was the LDL for RF-RT-RAA. Abbreviations: LDL, lowest detectable limit; NDV, Newcastle disease virus; RT-RAA-LFD, reverse transcription recombinase–aided amplification-lateral flow dipstick; RF-RT-RAA, real-time fluorescence-based reverse transcription recombinase–aided amplification; RFQ-PCR, real-time fluorescence–based quantitative PCR.
Clinical Sample Detection
A total of 86 clinically suspected NDV samples were detected by PCR, RFQ-PCR, RT-RAA-LFD, and RF-RT-RAA. The results are shown in Table 2. With RT-PCR as the standard, the coincidence rates of RFQ-PCR, RT-RAA-LFD, and RF-RT-RAA were 94.19, 95.35, and 93.02%, respectively.
Table 2.
Clinical test results of 4 methods (n = 86).
| Methods | Positive cases | Negative cases | Positive coincidence rate (%) | Negative coincidence rate (%) | Total consistent rate (%) |
|---|---|---|---|---|---|
| PCR | 45 | 41 | - | - | - |
| RFQ-PCR | 50 | 36 | 100 | 87.80 | 94.19 |
| RT-RAA-LFD | 49 | 37 | 100 | 90.24 | 95.35 |
| RF-RT-RAA | 51 | 35 | 100 | 85.37 | 93.02 |
Abbreviations: RF-RT-RAA, real-time fluorescence-based reverse transcription recombinase–aided amplification; RFQ-PCR, real-time fluorescence–based quantitative PCR; RT-RAA-LFD, reverse transcription recombinase–aided amplification-lateral flow dipstick.
Discussion
At present, among the NDV detection methods, PCR and RFQ-PCR are the most conventional clinical detection methods, which have the characteristics of high specificity and low price, and RFQ-PCR can also help us to realize the absolute quantitative analysis of genes (Jang et al., 2011, Zhang et al., 2019), but both methods require expensive PCR instruments and take a long time and have higher technical requirements for operators. RAA is a new isothermal amplification technique in vitro, which avoids the disadvantages of rapid rise and fall of reaction temperature and reduces the complexity of instruments. In areas with poor conditions, the reaction can be completed by a simple thermostatic device, and the reagent used for RT-RAA is generally freeze-dried powder without cryopreservation, which ensures its stability in the process of transportation and preservation and reduces the cost of preservation, so RT-RAA is very suitable for on-site detection on farm.
In this experiment, RT-RAA was combined with dipstick and fluorescent probe. The RT-RAA-LFD detection method needed no traditional agarose electrophoresis and visualized the detection results. In this method, the reaction could be completed at 37°C for 23 min, and the reaction product only needed to be put onto the dipstick for 3 min to observe the results. The sensitivity of this method could reach 102 copies/μL, which was 10 times higher than that of PCR (103 copies/μL). The reaction did not need other special instruments, and detection could be quickly completed in the field at the temperature of human body under extremely harsh conditions.
At reaction temperature of 38°C and reaction time of 26 min, the sensitivity of RF-RT-RAA could reach 10 copies/μL, which was equivalent to that of RFQ-PCR and 100 times higher than that of PCR (103 copies/μL), but the reaction time was only 33% of that of RFQ-PCR. The whole reaction took a short time and the result was clear, so it could be used for qualitative detection. It is worth mentioning that RF-RT-RAA can come with a portable fluorescent thermostatic amplification instrument, which is only the size of a notebook and can be rechargeable, making the detection feasible in the field. And, it is cheaper than the RFQ-PCR (portable fluorescent thermostatic amplification instrument cost $2,000–$3,000, RFQ-PCR cost $50,000–$60,000) instrument, which can reduce the capital investment to buy the device. Real-time fluorescence-based reverse transcription recombinase–aided amplification can also realize detection by RFQ-PCR instrument, with no need for the portable fluorescent thermostatic amplification instrument. Moreover, the 2 detection methods of RT-RAA-LFD and RF-RT-RAA do not need to carry out RNA reverse transcription separately, and the experiment is completed by one-step method, which saves time and resources.
Reverse transcription recombinase–aided amplification-lateral flow dipstick and RF-RT-RAA have their own advantages and disadvantages. Although RF-RT-RAA has high sensitivity, it needs to come with instruments, which is relatively suitable for laboratory diagnosis. The sensitivity of RT-RAA-LFD is lower, but there is no need to buy other devices for clinical use, so it is more suitable for clinical diagnosis.
Conclusion
Reverse transcription recombinase–aided amplification-lateral flow dipstick was established to detect the NDV, with reaction time of 23 min and lowest detectable limit of 102 copies/μL, 10 times higher than that of RTQ-PCR (103 copies/μL). Real-time fluorescence-based reverse transcription recombinase–aided amplification was established to detect the NDV, only taking 26 min, with LDL of 10 copies/μL, equivalent to that of RFQ-PCR and 100 times higher than that of conventional PCR (103 copies/μL). The 2 detection methods had good specificity and had no cross-reaction with the nucleic acids of other avian diseases.
Acknowledgments
This work was supported by Open Fund of State Key Laboratory of Veterinary Etiological Biology (No. Y2017PT44) from Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, China; Shijiazhuang Key R&D Projects (No. 201500412A) from Bureau of Science & Technology, Shijiazhuang city, China; and Key R&D Plan of Science and Technology Department of Hebei Province (Nos. 202030504040006 and 202030504030005) from Science & Technology Department, Hebei Province, China.
Conflict of Interest: The authors did not provide any conflict of interest statement.
References
- Chumbe A., Izquierdo-Lara R., Calderon K., Fernandez-Diaz M., Vakharia V.N. Development of a novel Newcastle disease virus (NDV) neutralization test based on recombinant NDV expressing enhanced green fluorescent protein. Virol. J. 2017;14:232. doi: 10.1186/s12985-017-0900-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganar K., Das M., Sinha S., Kumar S. Newcastle disease virus: current status and our understanding. Virus Res. 2014;184:71–81. doi: 10.1016/j.virusres.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Zhao Y., Yu J., Wang X., Zheng D., Cai Y., Liu H., Wang Z. Comparison between class I NDV and class II NDV in aerosol transmission under experimental condition. Poult. Sci. 2019;98:5040–5044. doi: 10.3382/ps/pez233. [DOI] [PubMed] [Google Scholar]
- Giovanni F., Paola M., Maria T.C., Ilaria B., Giovanni T., Laura F., Massimo C., Antonio L., Mattia C., Ana M. Think globally, act locally: phylodynamic reconstruction of infectious bronchitis virus (IBV) QX genotype (GI-19 lineage) reveals different population dynamics and spreading patterns when evaluated on different epidemiological scales. PLoS One. 2017;12:e0184401. doi: 10.1371/journal.pone.0184401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J., Hong S.H., Kim I.H. Validation of a real-time RT-PCR method to quantify Newcastle disease virus (NDV) titer and comparison with other quantifiable methods. J. Microbiol. Biotechnol. 2011;21:100–108. doi: 10.4014/jmb.1006.06006. [DOI] [PubMed] [Google Scholar]
- Kim J., Easley C.J. Isothermal DNA amplification in bioanalysis: strategies and applications. Bioanalysis. 2011;3:227–239. doi: 10.4155/bio.10.172. [DOI] [PubMed] [Google Scholar]
- Lin Y.X., Cao C.F., Zeng S.L., Hua Q.J., Yang J.X., Huang C.H., Shi W.J., Hua Q.Y. Establishment and preliminary application of a fluorescent RPA method for rapid detection of African classical swine fever virus. Vet. Sci. China. 2019;49:1090–1095. [Google Scholar]
- Lin Z., Kato A., Kudou Y., Ueda S. A new typing method for the avian infectious bronchitis virus using polymerase chain reaction and restriction enzyme fragment length polymorphism. Arch. Virol. 1991;116:19. doi: 10.1007/BF01319228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Song Y., Yang Y., Bu Y., Cheng J., Zhang G., Xue J. Hemagglutinin-Neuraminidase and fusion genes are determinants of NDV thermostability. Vet. Microbiol. 2019;228:53–60. doi: 10.1016/j.vetmic.2018.11.013. [DOI] [PubMed] [Google Scholar]
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006;4:1115–1121. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L.Q., Zheng X.C., Wang C.P., Li W.S., Qin Z.F. Establishment of real-time fluorescence RT-RPA isothermal detection method for rabies virus. Chin. J. Vet. Med. 2019;55:44–47. [Google Scholar]
- Wang W., Wang C., Zhang P., Yao S., Liu J., Zhai X., Zhang T. Reverse transcription recombinase-aided amplification assay combined with a lateral flow dipstick for detection of avian infectious bronchitis virus. Poult. Sci. 2020;99:89–94. doi: 10.3382/ps/pez559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yang P.P., Zhang Y.H., Tian K.Y., Bian C.Z., Zhao J. Development of a reverse transcription recombinase polymerase amplification combined with lateral-flow dipstick assay for avian influenza H9N2 HA gene detection. Transbound. Emerg. Dis. 2019;66:546–551. doi: 10.1111/tbed.13063. [DOI] [PubMed] [Google Scholar]
- Zhang T., Chen J., Wang C., Shi W., Li D. The therapeutic effect of Yinhuangerchen mixture on Avian infectious laryngotracheitis. Poult. Sci. 2018;97:2690–2697. doi: 10.3382/ps/pey125. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Liu D., Hu J., Sun W., Liu K., Li J., Xu H., Liu J., He L., Jiang D., Gu M., Hu S., Wang X., Liu X., Liu X. Multiplex one-step real-time PCR assay for rapid simultaneous detection of velogenic and mesogenic Newcastle disease virus and H5-subtype avian influenza virus. Arch. Virol. 2019;164:1111–1119. doi: 10.1007/s00705-019-04180-6. [DOI] [PubMed] [Google Scholar]