Abstract
Cathelicidins represent a major group of host defense peptides (HDPs) that share a highly conserved cathelin-like domain. In birds, this gene family has been identified in many species. However, no information was available in the goose until now. In this study, we present the molecular characterization of 2 goose cathelicidin genes, namely goose CATH2 and goose CATH3, for the first time. The complete cDNA of goose CATH2 and goose CATH3 were 571 bp and 573 bp in length, respectively, and the deduced amino acid sequences exhibited high similarity with other avian cathelicidins. Furthermore, evolutionary analyses indicated that all known cathelicidins form 3 distinct clusters from reptiles, while the oldest cathelicidin member, which is known as CATHB1, is very likely absent in the goose genome. Meanwhile, highly expressed goose CATH2 and goose CATH3 were also observed in primary and secondary lymphoid tissues, same as the observations in other avian species. In addition, chemically synthesized mature peptides of the 2 cathelicidins exerted optimal antimicrobial abilities to a range of gram-negative and gram-positive bacteria. The discovery and characterization of goose cathelicidins complete the knowledge for goose HDPs and might contribute to understanding the evolution of avian cathelicidins as well as for the development of antibacterial agents.
Key words: cathelicidin, goose, gene expression, phylogenetic analysis, antimicrobial activity
Introduction
Antimicrobial peptides, better known as host defense peptides (HDPs), are a large family of cationic peptides with a highly diverse structure (Shafee et al., 2016, Haney et al., 2017, Yu et al., 2018a). Similar to defensins, cathelicidins serve as a typical kind of effector molecule in immunity. They not only exhibit broad-spectrum antimicrobial activity against various microorganisms primarily through nonspecific physical interactions and membrane disruption but also act as immunomodulators and possess other protective activities (Zanetti, 2004, Kościuczuk et al., 2012). With optimal antimicrobial, immunomodulatory, and many additional activities, cathelicidins are being developed as novel pharmaceutical agents to fight against the continued problem of antimicrobial resistance (Hancock and Sahl, 2006, Bechinger and Gorr, 2017).
A typical cathelicidin gene consists of 4 exons separated by 3 introns. The first exon corresponds to the 5′UTR, the short signal peptide, and part of the highly conserved cathelin domain. The second and third exons code the majority of the cathelin domain, which contains a 4-cysteine motif, and the last exon is mainly composed of the largely varied mature peptide (Zhu and Gao, 2009, Young-Speirs et al., 2018, Zhang et al., 2019b). In birds, the first complete repertoire of the cathelicidin gene family was reported in the chicken. Four chicken cathelicidin genes, designated as CATH1 (fowlicidin 1), CATH2 (fowlicidin 3), CATH3 (fowlicidin 3), and CATHB1, are clustered densely within a 7.5-kb distance on chromosome 2 and flanked by the KLHL18 and TBRG4 genes (Xiao et al., 2006, Goitsuka et al., 2007). After that, multiple avian cathelicidins have been reported in a wide range of avian species, including the ring-necked pheasant (Wang et al., 2011), quail (Feng et al., 2011), duck (Gao et al., 2015), and pigeon (Yu et al., 2015). Recent cross-species genomic analyses of 48 birds suggest that species-specific gene duplication and loss simultaneously occurred in the evolution of avian cathelicidins. That is, the CATH2, CATH3, and CATHB1 were fixed in the avian genome before species divergence. The duplication events of CATH1 have occurred in Galliformes, whereas some members of the cathelicidin gene family may have been lost in some specific clades (Cheng et al., 2015). More recently, the reference genome of the goose has been released (Lu et al., 2015). However, no information on goose cathelicidins could be retrieved from the current database. Therefore, it is very interesting to explore the presence of cathelicidin genes in the goose and characterize their basic information.
In the present study, we amplified and cloned full-length mRNA sequences of 2 cathelicidin genes from the goose for the first time. We further characterized their expression patterns and evolutionary relationships. Moreover, the deduced mature peptide of goose CATH2 and CATH3 were synthesized to study the antimicrobial activity against different bacterial strains. The results yielded by this work will increase our understanding of this gene family.
Materials and methods
Data Mining and Gene Identification
Two approaches were used to obtain potential cathelicidin sequences from the target genomes, as we previously described (Zhang et al., 2019a). First, the hidden Markov model (HMM) file of cathelicidin (accession number: PF00666.16) was used as a query to search against translated goose nonredundant sequences, expressed sequence tags, and protein sequences by using the HMMer (V3.1) program (Mistry et al., 2013) with default settings. All potential hits with both E-values (full sequence and best 1 domain) < 0.1 and the 7 known amino acid sequences of chicken and duck cathelicidin genes were then used to conduct an iterative TBLASTN (Altschul et al., 1990) search against the above databases and the goose genome (version 1.0) with the cutoff E value as 1.0.
Tissue Sampling and Preparation
Six healthy 7-day-old and 6 healthy 42-day-old male Sichuan white geese (Anser cygnoides) were obtained from a goose farm in Nanchong, Sichuan, China, and then euthanized with carbon dioxide and cervically dislocated. A range of tissues, including proventriculus, tongue, esophagus, gizzard, duodenum, jejunum, ileum, cecum, colon, heart, liver, kidney, lung, spleen, bursa of Fabricius, and thymus (only from 7-day-old geese) were collected and immediately frozen in liquid nitrogen until homogenization and RNA extraction. All procedures in this study were approved by the animal ethics committee of China West Normal University (no. 467234).
RNA Extraction and Real-Time PCR
According to the manufacturer's instructions, total RNA was extracted from each tissue sample using a TRIzol Plus RNA Purification Kit (Invitrogen, Carlsbad, CA). RNA integrity and purity were assessed by Nanodrop after being electrophoresed in formaldehyde gel (Jin et al., 2018, Yan et al., 2019). The genomic DNA contamination of RNA samples was removed by using a gDNA Eraser (Takara Bio Inc., Dalian, China), and the PrimeScript RT reagent Kit (Takara Bio Inc.) was subsequently used for reverse transcription reaction and performed according to the manufacturer's instructions. The cDNA was then diluted 10 times with RNase-free water before real-time PCR analysis. Real-time PCR was performed on each sample by using CFX96 real-time PCR Detection System (Bio-Rad Laboratories, Richmond, VA), and the amplification was conducted in a total volume of 20 μL, containing 10 μL of SYBR Premix Ex Taq II (Takara Bio Inc.), 7 μL of RNase-free water, 1 μL of the diluted cDNA, and 0.5 μL of each primer (Table 1). The optimum real-time PCR program was 95°C for 1 min, followed by 40 cycles of 95°C for 15 s and 60°C for 25 s. Relative quantification of mRNA transcripts was accomplished using the 2−ΔΔCt method (Livak and Schmittgen, 2001), with glyceraldehyde-3-phosphate dehydrogenase being used as the endogenous control gene. Simultaneously, 3 samples were used as calibrators, and the expression of each gene was reported as fold increase relative to the average value of the calibrators.
Table 1.
Primer sequences used for gene clone and real-time PCR analysis.
| Primer name | Primer sequences (5′to3′) | Product size (bp) | Purpose |
|---|---|---|---|
| GAPDH-F | CTGCCCAGAACATTATCCCAGCAT | 137 | Real-time PCR |
| GAPDH-R | GCAGGTCAGGTCCACGACAGA | ||
| CATHL2-F | CCGCCGACGACTGCGACTT | 78 | Real-time PCR |
| CATHL2-R | GGGTGCCCTGCTGGAACGT | ||
| CATHL3-F | GCAGGCCGTGGACACCTACA | 104 | Real-time PCR |
| CATHL3-R | GCAGGGAGCTGAGCTGGACAT | ||
| mCATHL2-F | CCYGAGSTGCASAAYGCCTTC | 298 | Coding region amplification |
| mCATHL2-R | GYCGGAYCTTBCYCAGGAAGC | ||
| rCATHL2-F1 | GGTGCAGAACGCCTTCAGGCTGCTC | 335 | 3′-RACE |
| rCATHL2-F2 | GGGCGCTCAACTTCACCATAATGGAGACA | ||
| rCATHL2-R1 | GGCCCCGTTCTCCTTGAAGTCGCAGT | 280 | 5′-RACE |
| rCATHL2-R2 | GCCCGGGGCACAGTCTGTCTCCATTA | ||
| mCATHL3-F | GSCCYGAGGTGCASAAYGCCTTC | 261 | Coding region amplification |
| mCATHL3-R | GAGTCCACGCAGGTGACATCGA | ||
| rCATHL3-R1 | CCGTCCTCCTTGAAGTCGCAGGTGTC | 212 | 5′-RACE |
| rCATHL3-R2 | GGTTCCGCAGGGAGCTGAGCTGGAC | ||
| rCATHL3-F1 | CCTGCGGAACCTCAACTTCACCATCA | 309 | 3′-RACE |
| rCATHL3-F2 | CCGACACCTGCGACTTCAAGGAGGAC |
5′and 3′ Rapid Amplification of the cDNA Ends
Based on the predicted partial sequences of goose cathelicidins and their counterparts in the chicken, 2 pairs of degenerate primers were designed to amplify the middle coding region of goose CATH2 and CATH3 using cDNA from the spleen, respectively. Subsequently, a cDNA amplification kit (Clontech) was used to amplify the 5′ and 3′ regions of the goose cathelicidin genes. The gene-specific primers were summarized in Table 1. All PCR products were separated by electrophoresis on 3% agarose gel and recovered using the E.Z.N.A. Gel Extraction Kit (Omega Bio-Tek, Norcross, GA). The purified fragments were cloned into the pMD-19T vector (Takara Bio, Inc.), and 6 clones of each gene were selected for sequencing as previously described (Yu et al., 2018b, Xiao et al., 2019).
Phylogenetic and Evolutionary Analyses
Amino acid sequences were aligned using the MUSCLE program and visualized by using Boxshade (https://embnet.vital-it.ch/software/BOX_form.html). A neighbor-joining tree was constructed from the p-distance of amino acid sequences from representative avian and reptile cathelicidins by using Mega 7.0 (Kumar, et al., 2016). The reliability of each branch was tested by bootstrap method with 1 000 replications.
Peptide Synthesis and Antimicrobial Assay
The deduced mature peptides of goose CATH2 (LVQRGRFGRFLGRIRRFRPRVSIRVQAHGTIRFG) and CATH3 (RVKRFWPLVPVVINTVAAGINLYKAIRRK) were synthesized by Shanghai Apeptide Co., Ltd. (Shanghai, China) and analyzed by high-performance liquid chromatography and electrospray ionization mass spectrometry to confirm the purity was higher than 95%. Three gram-negative bacteria (Escherichia coli CICC 10389, Salmonella typhimurium ATCC 14028, and Shigella flexneri ATCC 12022) and gram-positive bacteria (Staphylococcus aureus ATCC 25923, Bacillus cereus CICC 23828, and Listeria monocytogenes ATCC 13932) were used to evaluate the antimicrobial activities of these 2 peptides. Briefly, overnight cultured bacteria were subcultured to the mid-log phase at 37°C in Luria broth or tryptone soya broth and then washed by PBS (pH 7.4) and resuspended to 5 × 105 CFU/ml in the same buffer. After 90 μL of bacteria had been dispensed into 96-well cell culture plates, 10 μL of peptides was added in serial 2-fold dilutions duplicate. The minimum inhibitory concentration (MIC) of each peptide was determined as the lowest concentration of peptide that completely inhibited the growth of the microbe.
Results
Genomic Identification of the Goose Cathelicidin Genes
After thoroughly screening different databases of the goose, no target sequence was identified by using the HMM method. Instead, 3 hits with high similarity to the chicken cathelicidin genes were obtained from NW_013185891.1 through using the TBLASTN workflow. By translating the 3 fragments into amino acid sequences, it was found that the 3 sequence segments belonged to 2 different cathelicidin genes. The first hit, which is located in the plus strand, was a signal peptide of cathelicidin. The remaining 2 hits are harbored in the minus strand and could be well aligned with the first and the last exons of chicken CATH2, respectively (Supplementary Figure 1). Current information in NCBI shows that the 3 hits are also located between the KLHL18 and TBRG4 genes, and multiple sequences surrounding the target region are unrecognized (data not shown). Thus, these observations collectively suggest that the goose cathelicidin genes are hidden in the low-quality region of the goose genome.
Molecular Cloning of the Goose Cathelicidins
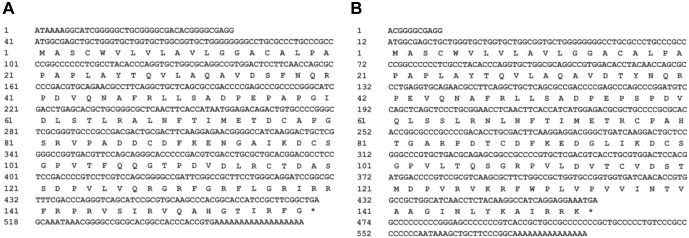
Limited by the quality of the current goose genome, we used RACE technology to obtain the full cDNA fragments of the goose cathelicidin genes. As illustrated in Figure 1, the 2 cathelicidin genes, namely goose CATH2 (GenBank accession number: MH631013.1) and goose CATH3 (GenBank accession number: MH631012.1) were obtained by using the cDNA from the spleen of a 7-day-old goose. The full length of mRNA of the goose CATH2 is 571 bp, containing 477 bp of open reading frame and encoding 158 amino acids, and the 5′ and 3′ untranslated region (UTR) was 40 bp and 54 bp, respectively. The complete cDNA of the goose CATH3 is 573 bp in length, preceded by a 55 bp 5′ UTR and a 103 bp 3′ UTR. By aligning the 2 mRNAs and the goose genome, it was found that the nucleotide sequences in the corresponding regions are almost identical (data not shown), which indicated the 3 hits obtained by homology search of the goose genome belong to these 2 functional cathelicidin genes. However, we failed to obtain any fragment of CATHB1 and other cathelicidin genes from the spleen and even from the bursa of Fabricius from the 7-days-old goose.
Figure 1.
Complementary cDNA and deduced amino acids sequences of goose CATH2 (A) and CATH3 (B). The asterisk represents the termination codon, and the numbers refer to the position of nucleotide or amino acid.
Comparison and Evolutionary Analyses of the Goose Cathelicidins
Sequence alignment of the 2 goose cathelicidin genes and the cathelicidin sequences from the represent avian species shows that the amino acid sequences of goose CATH2 (AncyCATH2) and CATH3 (AncyCATH3) are very similar to the other avian cathelicidins (Figure 2). In particular, the deduced mature peptide of goose CATH2 shared high similarity with its counterpart from the duck, while the predicted mature peptide of goose CATH3 was different from the chicken CATH3 (GagaCATH3) only at one amino acid position. Similarly, the estimated net charge of the goose CATH2 and CATH3 mature peptides was 9.1 and 7.0, respectively, which were also identical to some of the other avian cathelicidins. Taken together, these observations of avian cathelicidins strongly indicate that the function of the 2 goose cathelicidin genes may be similar to their avian orthologs.
Figure 2.
Multiple sequence alignment of representative avian cathelicidin sequences. The conserved residues are shaded, and the predicted C-terminal mature peptides are boxed with the lengths and net charges (in parenthesis). The mature peptides of the cathelicidin genes were deduced based on the amino acid alignment of chicken cathelicidin peptides, and the net charge of each mature peptide was estimated by using the online software at https://pepcalc.com/.
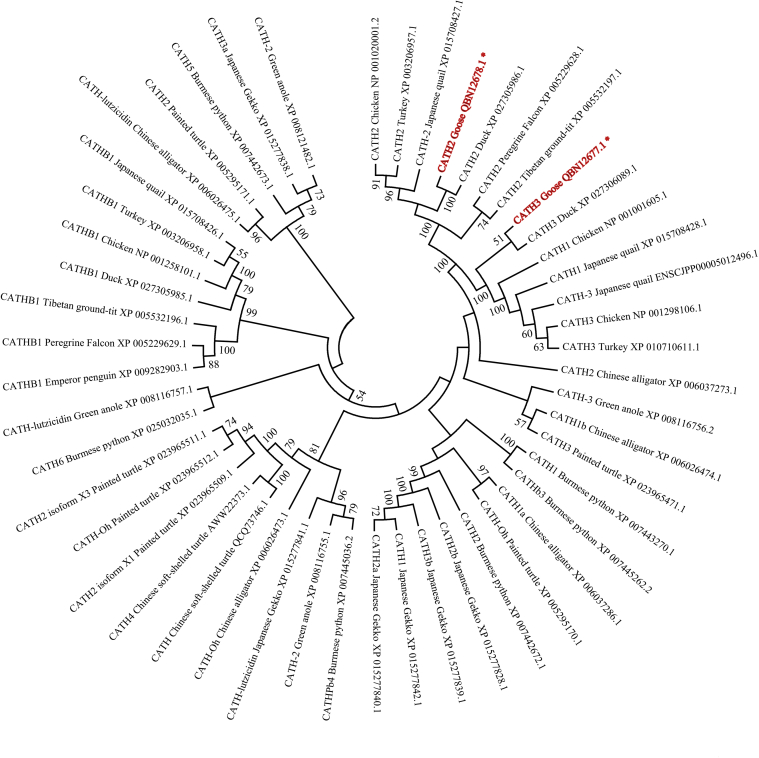
Together with the reported cathelicidins from the Chinese alligator, painted turtle, green anole, and Japanese gecko, the phylogenetic relationships of these representative avian and reptile cathelicidins were analyzed (Figure 3). Supported by bootstrap values of 100 and 51, goose CATH2 and CATH3 exhibited the closest relationships with the sequences of the duck, respectively. Furthermore, the phylogenetic analysis also revealed 2 distinct avian clades, with the CATH2 cluster grouped with the CATH1/3 cluster and the CATHB1s present as an ancient clade. Moreover, the topology of the phylogenetic tree shows none of the reptile cathelicidins grouped within avian clades.
Figure 3.
Phylogenetic analysis of the goose cathelicidin genes and representative cathelicidins from birds and reptiles. The topology was constructed by the amino acid sequences using the Neighbor-Joining method with 1000 bootstrap replicates. Only branches supported by a bootstrap value higher than 50 are shown at branching points.
Cross-species Analysis of CATHB1 Genes in Anseriformes
To further investigate whether the CATHB1 gene is present in the goose, we conducted cross-species genomic gene identification in Anseriformes by using the same workflow. Within the other 3 species with complete genome sequence in Anseriformes, fragments of cathelicidins were obtained from the pink-footed goose (GCA_002592135.1) and the Eastern spot-billed duck (GCA_002224875.1) but not from the bar-headed goose (GCA_006229135.1). In addition, hits of CATHB1 were only identified from the scaffold NOIK01005096.1 of the Eastern spot-billed duck, together with the sequences of 2 other cathelicidins (Supplementary Figure 2). After carefully checking the surrounding regions of cathelicidin fragments in the pink-footed goose genome, we found that the cathelicidin segments in the pink-footed goose genome belong to the first 3 exons of CATH2, and the adjacent gaps might have led to the absence of CATHB1 fragments in the pink-footed goose genome (Supplementary Figure 3).
Expression Patterns of Goose CATH2 and CATH3
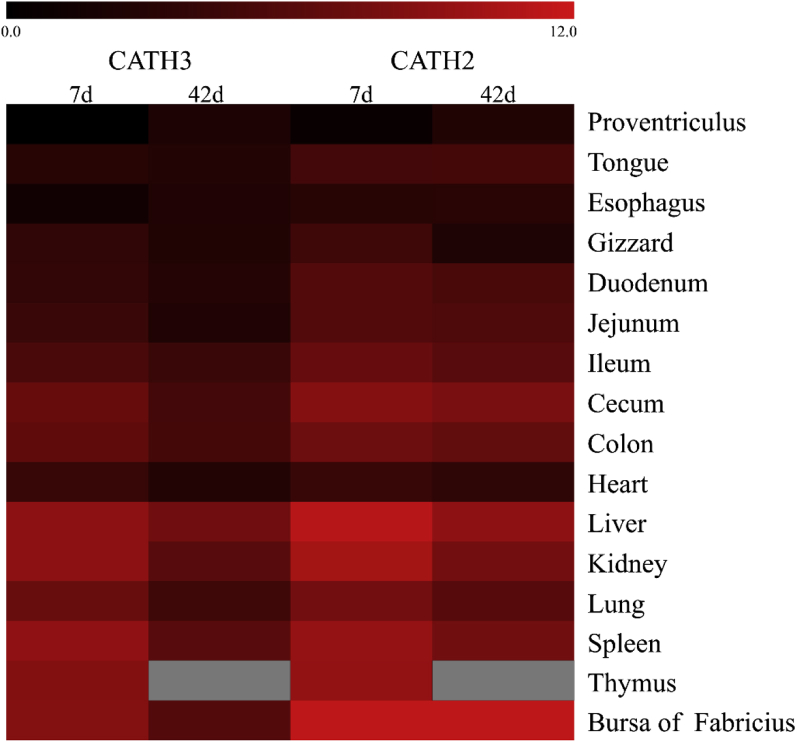
To investigate the expression patterns of goose CATH2 and CATH3, we performed real-time PCR to reveal their mRNA abundance in a total of 16 tissues from both 7-day-old and 42-day-old geese. As summarized in Figure 4, the distributions of the 2 cathelicidin mRNA were ubiquitously detected in all the examined tissues. Similar to the observations in chickens, the goose CATH2 and CATH3 presented a very similar tissue expression pattern in the 16 examined tissues at the 2 time points, which also suggested a close relationship in phylogenetics. The mRNA transcripts were highly expressed in the spleen, thymus, and bursa of Fabricius and moderately appear in the intestinal tract. However, a negligible amount of mRNA was found in tissues such as proventriculus, esophagus, and heart, which indicated a broad expression of goose cathelicidins in lymphoid tissues. Compared with the tissue distribution at 7 D, further augmentation of goose CATH2 and CATH3 mRNA levels in the proventriculus, tongue, and esophagus was observed at 42 D. However, the mRNA amount of each goose cathelicidin gene trend to be reduced in most of the other parts, including the spleen, lung, liver, cecum, and colon. The changes of cathelicidin expression in these tissues between 7 D and 42 D might be associated with the maturation of adaptive immunity.
Figure 4.
Tissues expression pattern of goose CATH2 and CATH3. Tissues were collected from six 7-day-old goose and 42-day-old geese, respectively. The expression levels of the cathelicidins were calculated relative to that of CATH3 in proventriculus from 7-day-old goose using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the endogenous control gene. The color elements represent the average log2 ratios of fold change from 6 samples. The gray areas mean the absence of data due to the unavailability of the thymus from 42-day-old geese.
Antibacterial Activity of the Goose Cathelicidins
To further measure and compare the antibacterial properties of the goose CATH2 and CATH3, the putative mature peptides of the 2 cathelicidins were chemically synthesized, and the antibacterial activity was tested against 6 bacterial strains, respectively. The MIC values of the goose CATH2 and CATH3 against each bacteria strain are listed in Table 2. Both of the mature peptides exhibited a broadly antibacterial spectrum against all the 6 bacteria stains, with MICs ranging from 1 to 16 μM. In addition, the antibacterial efficiency of the CATH3 was generally higher than that of the CATH2.
Table 2.
Minimum inhibitory concentrations (μM) of goose CATH2 and CATH3 mature peptides.
| Bacteria | MIC(μM) |
|
|---|---|---|
| CATH2 | CATH3 | |
| Gram-negative | ||
| Escherichia coli CICC 10389 | 4–8 | 1 |
| Salmonella typhimurium ATCC 14028 | 8–16 | 4 |
| Shigella flexneri ATCC12022 | 2 | 2 |
| Gram-positive | ||
| Staphylococcus aureus ATCC25923 | 8 | 1 |
| Bacillus cereus CICC 23828 | 4 | 4 |
| Listeria monocytogenes ATCC 13932 | 2 | 1 |
Abbreviation: MIC, minimal inhibitory concentration.
These concentrations represent the ranges of MIC values of 3 independent experiments performed in duplicate.
Discussion
Using a combination of bioinformatics tools and PCR methods, we reported 2 cathelicidin genes in the goose for the first time. The discovered and confirmed goose cathelicidin sequences in the present study indicate that the absence of cathelicidin gene annotation in the current database results from low-quality sequences in the corresponding regions of the goose genome. Sequence comparisons and net charge prediction of the mature peptide, as well as phylogenetic analysis and antimicrobial assay in this research, show that the 2 goose cathelicidin genes share high similarities with other avian cathelicidins.
Although we could not rule out the possibility that the absence of the CATHB1 gene segment in the genome of the 3 anserid species (goose, pink-footed goose, and bar-headed goose) might be an artifact from a misassemble of the genome, the current observations trend to imply that CATHB1 may have been lost in Anserinae because we also failed to obtain the CATHB1 segment from geese by molecular cloning. Different from the gene structure in other species, both the annotated duck CATHB1 gene in the NCBI database (LOC113842527) and our predicted CATHB1 sequence from the Eastern spot-billed duck (Supplementary Figure 2) consisted of 3 exons instead of the canonical 4 exons. In addition, only 2 of the 4 cysteines of the duck CATHB1 are conserved in the cathelin domain, and multiple amino acid sequences are missing or show large diversity at the amino acid level (Figure 2). Furthermore, only the orthologs of CATH2 and CATH3 have been confirmed in the duck (Gao et al., 2015), and no evidence has shown the presence of CATHB1 mRNA in ducks. These observations collectively implied a possibility that the CATHB1 in the duck and Eastern spot-billed duck was a pseudogene. Thus, the fragments of the CATHB1 gene in Anserinae might either be deleted from the genome or have become diverged from its counterparts because of sequence degeneration followed by pseudogenization. The availability of additional genome and transcriptome sequences with higher quality will help to shed light on these hypotheses.
In comparison with the tissue expression pattern in chickens (Achanta et al., 2012), abundant expressions of cathelicidins were found in multiple lymphoid organs and the gastrointestinal tract of geese as well. However, some species-specific tissue expression patterns were also observed in some surveyed tissues of this study. For instance, the expression of all 4 chicken cathelicidin genes was found to be high in the heart (Achanta et al., 2012), whereas relatively low magnitudes of the goose cathelicidin transcripts were observed in the heart. On the other hand, a different expression profile was also found between chickens and geese. A gradual increase of cathelicidin gene expressions was shown in the lung and cecum in chickens (Achanta et al., 2012), while opposite results were observed in the same 2 tissues between the 7-day and 42-day geese in the present study. We speculated that some of these results might be due to the gene number variation of the cathelicidin family because the chicken genome harbors 4 cathelicidin genes and current evidence supports only 2 cathelicidins coded by the goose genome. Besides, some discrepancies in mRNA expressions might be due to the different living environments and raising conditions of birds because the expression of avian cathelicidins can be modulated by microbe-associated molecular patterns and immune factors (Abdel-Mageed et al., 2016, Abdel-Mageed et al., 2017, Mohammed et al., 2017). Importantly, different from the other known avian cathelicidin genes, CATHB1 belongs to a distant family member, especially in tissue distribution. Northern blot and situ hybridization analyses exhibit that chicken CATHB1 is expressed exclusively in the secretory epithelial cell neighbors of the M cells (Goitsuka et al., 2007). In this regard, it would be particularly interesting to investigate whether alternatives could be generated in the epithelium of the bursa of Fabricius if CATHB1 was absent in the goose.
Consistent with the earlier studies (Xiao et al., 2006, Yu et al., 2015, Lee et al., 2016), the goose CATH2 and CATH3 mature peptides proved to possess potent antimicrobial activities against both gram-negative and gram-positive bacteria at micromolar concentrations, with generally higher MIC values of CATH2 than those of CATH3. The overall antibacterial efficiency of the goose CATH3 was higher than that of CATH2, also implying conserved antimicrobial activity across the avian species. It is known that net charge, helicity, amphipathicity, and hydrophobicity are significantly involved in the antimicrobial potency of HDPs (Takahashi et al., 2010, Yang et al., 2016). Coupled with the results of our amino acid sequence analyses, we speculated that these conservations in antibacterial property might be because most amino acid substations in these peptides did not change the essential structural and physicochemical parameters of avian cathelicidins. Accumulating evidence suggests potential applications of avian cathelicidins for both therapeutic and prophylactic use, albeit limited by natural cytotoxicity and some other external factors. For instance, chicken CATH1 shows strong antibacterial efficacy in vivo against methicillin-resistant Staphylococcu saureus (Bommineni et al., 2010), and pretreating mice with this peptide before a lethal methicillin-resistant Staphylococcu saureus infection can save them from death (Bommineni et al., 2014). In addition, the immunomodulatory effects of avian cathelicidins exhibit the potential to be developed as adjuvants or components of an adjuvant complex (Zhang and Sunkara, 2014, van Dijk et al., 2016, Kraaij et al., 2017). On the account of the cost and stability for the direct administration of HDPs, indirect instigation of synthesis of endogenous HDPs by dietary compounds seems to be a convenient and cost-effective strategy in animal production (Deng et al., 2018, Lyu et al., 2018). Because these HDP-inducing compounds have no direct interactions with pathogens, they can boost the synthesis of HDPs without triggering resistance and provoking inflammation (Zhang and Sunkara, 2014, Robinson et al., 2018). The discovery of the 2 cathelicidin genes in the present study will help explore novel antimicrobial strategy in the future.
In summary, we identified and cloned the first 2 cathelicidins from the goose. Sequence and phylogenetic analyses indicated that avian cathelicidins evolved after the split of birds and reptiles, and avian cathelicidins present high conservations in amino acid sequence and net charge. The results of cross-species genome analysis indicated that CATHB1 underwent changes in gene structure in Anatidae and was lost in Anserinae. Tissue expression patterns of the 2 cathelicidin genes showed similar transcript abundance in multiple, but not all, tissues. Similar to other avian cathelicidins, the chemically synthesized goose cathelicidin peptides have similar bactericidal activity against a broad range of bacteria strains. This work enriches our knowledge in avian cathelicidin genes, and additional evolutionary analyses, as well as functional characterization, are still needed to understand this gene family better.
Acknowledgment
This research was funded by the Department of Science and Technology of Sichuan Province (grant number 2019YJ0338), the Doctoral Scientific Research Funds of China West Normal University (grant number 17E077), the Fundamental Research Funds of China West Normal University (grant number 18B030), and the State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Zhejiang Academy of Agricultural Sciences (grant number 2010DS700124-ZZ1905).
Conflict of Interest Statement: The authors did not provide a conflict of interest statement.
Footnotes
Supplementary data associated with this article can be found in the online version at http://doi.org/10.1016/j.psj.2020.03.021.
Supplementary data
Reference
- Abdel-Mageed A.M., Isobe N., Yoshimura Y. Effects of virus-associated molecular patterns on the expression of cathelicidins in the hen vagina. J. Poult. Sci. 2016;53:240–247. doi: 10.2141/jpsa.0150180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Mageed A.M., Nii T., Isobe N., Yoshimura Y. Modulatory roles of proinflammatory cytokines on the expression of cathelicidins in the lower regions of the oviduct of laying hens. Cytokine. 2017;99:66–72. doi: 10.1016/j.cyto.2017.07.008. [DOI] [PubMed] [Google Scholar]
- Achanta M., Sunkara L.T., Dai G., Bommineni Y.R., Jiang W., Zhang G. Tissue expression and developmental regulation of chicken cathelicidin antimicrobial peptides. J. Anim. Sci. Biotechno. 2012;3:15. doi: 10.1186/2049-1891-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bechinger B., Gorr S.U. Antimicrobial peptides: mechanisms of action and resistance. J. Dent. Res. 2017;96:254–260. doi: 10.1177/0022034516679973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommineni Y.R., Achanta M., Alexander J., Sunkara L.T., Ritchey J.W., Zhang G. A fowlicidin-1 analog protects mice from lethal infections induced by methicillin-resistant Staphylococcus aureus. Peptides. 2010;31:1225–1230. doi: 10.1016/j.peptides.2010.03.037. [DOI] [PubMed] [Google Scholar]
- Bommineni Y.R., Pham G.H., Sunkara L.T., Achanta M., Zhang G. Immune regulatory activities of fowlicidin-1, a cathelicidin host defense peptide. Mol. Immunol. 2014;59:55–63. doi: 10.1016/j.molimm.2014.01.004. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Prickett M.D., Gutowska W., Kuo R., Belov K., Burt D.W. Evolution of the avian β-defensin and cathelicidin genes. BMC Evol. Biol. 2015;15:188. doi: 10.1186/s12862-015-0465-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z., Wang J., Lyu W., Wieneke X., Matts R., Ma X., Zhang G. Development of a cell-based high-throughput screening assay to identify porcine host defense peptide-inducing compounds. J. Immunol. Res. 2018;2018:5492941. doi: 10.1155/2018/5492941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng F., Chen C., Zhu W., He W., Guang H., Li Z., Wang D., Liu J., Chen M., Wang Y., Yu H. Gene cloning, expression and characterization of avian cathelicidin orthologs, Cc-CATHs, from Coturnix coturnix. FEBS J. 2011;278:1573–1584. doi: 10.1111/j.1742-4658.2011.08080.x. [DOI] [PubMed] [Google Scholar]
- Gao W., Xing L., Qu P., Tan T., Yang N., Li D., Chen H., Feng X. Identification of a novel cathelicidin antimicrobial peptide from ducks and determination of its functional activity and antibacterial mechanism. Sci. Rep. 2015;5:17260. doi: 10.1038/srep17260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goitsuka R., Chen C.L., Benyon L., Asano Y., Kitamura D., Cooper M.D. Chicken cathelicidin-B1, an antimicrobial guardian at the mucosal M cell gateway. Proc. Natl. Acad. Sci. 2007;104:15063–15068. doi: 10.1073/pnas.0707037104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R.E., Sahl H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotech. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- Haney E.F., Mansour S.C., Hancock R.E. Antimicrobial peptides: an introduction. Methods Molecular Biology (Clifton, N.J.) 2017;1548:3–22. doi: 10.1007/978-1-4939-6737-7_1. [DOI] [PubMed] [Google Scholar]
- Jin L., Yu J.P., Yang Z.J., Merilä J., Liao W.B. Modulation of gene expression in liver of Hibernating Asiatic Toads (Bufo gargarizans) Int. J. Mol. Sci. 2018;19:2363. doi: 10.3390/ijms19082363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kościuczuk E.M., Lisowski P., Jarczak J., Strzałkowska N., Jóźwik A., Horbańczuk J., Krzyżewski J., Zwierzchowski L., Bagnicka E. Cathelicidins: family of antimicrobial peptides. A review. Mol. Biol. Rep. 2012;39:10957–10970. doi: 10.1007/s11033-012-1997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraaij M.D., van Dijk A., Haagsman H.P. CATH-2 and LL-37 increase mannose receptor expression, antigen presentation and the endocytic capacity of chicken mononuclear phagocytes. Mol. Immunol. 2017;90:118–125. doi: 10.1016/j.molimm.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.O., Jang H.J., Rengaraj D., Yang S.Y., Han J.Y., Lamont S.J., Womack J.E. Tissue expression and antibacterial activity of host defense peptides in chicken. BMC Vet. Res. 2016;12:231. doi: 10.1186/s12917-016-0866-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu L., Chen Y., Wang Z., Li X., Chen W., Tao Z., Shen J., Tian Y., Wang D., Li G., Chen L., Chen F., Fang D., Yu L., Sun Y., Ma Y., Li J., Wang J. The goose genome sequence leads to insights into the evolution of waterfowl and susceptibility to fatty liver. Genome Biol. 2015;16:89. doi: 10.1186/s13059-015-0652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu W., Deng Z., Sunkara L.T., Becker S., Robinson K., Matts R., Zhang G. High throughput screening for natural host defense peptide-inducing compounds as novel alternatives to antibiotics. Front. Cell. Infect Microbiol. 2018;8:191. doi: 10.3389/fcimb.2018.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J., Finn R.D., Eddy S.R., Bateman A., Punta M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013;41:e121. doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed E.S.I., Isobe N., Yoshimura Y. Effects of probiotics on the expression of cathelicidins in response to stimulation by Salmonella Minnesota lipopolysaccharides in the proventriculus and cecum of broiler chicks. J. Poult. Sci. 2017;53:298–304. doi: 10.2141/jpsa.0160064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson K., Ma X., Liu Y., Qiao S., Hou Y., Zhang G. Dietary modulation of endogenous host defense peptide synthesis as an alternative approach to in-feed antibiotics. Anim. Nutr. 2018;4:160–169. doi: 10.1016/j.aninu.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafee T.M., Lay F.T., Hulett M.D., Anderson M.A. The defensins consist of two independent, convergent protein superfamilies. Mol. Biol. Evol. 2016;33:2345–2356. doi: 10.1093/molbev/msw106. [DOI] [PubMed] [Google Scholar]
- Takahashi D., Shukla S.K., Prakash O., Zhang G. Structural determinants of host defense peptides for antimicrobial activity and target cell selectivity. Biochimie. 2010;92:1236–1241. doi: 10.1016/j.biochi.2010.02.023. [DOI] [PubMed] [Google Scholar]
- van Dijk A., van Eldik M., Veldhuizen E.J., Tjeerdsma-van Bokhoven H.L., de Zoete M.R., Bikker F.J., Haagsman H.P. Immunomodulatory and anti-Inflammatory activities of chicken cathelicidin-2 derived peptides. PloS One. 2016;11:e0147919. doi: 10.1371/journal.pone.0147919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lu Z., Feng F., Zhu W., Guang H., Liu J., He W., Chi L., Li Z., Yu H. Molecular cloning and characterization of novel cathelicidin-derived myeloid antimicrobial peptide from Phasianus colchicus. Dev. Comp. Immunol. 2011;35:314–322. doi: 10.1016/j.dci.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Cai Y., Bommineni Y.R., Fernando S.C., Prakash O., Gilliland S.E., Zhang G. Identification and functional characterization of three chicken cathelicidins with potent antimicrobial activity. J. Biol. Chem. 2006;281:2858–2867. doi: 10.1074/jbc.M507180200. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Yu L., Gui G., Gong Y., Wen X., Xia W., Yang H. Molecular cloning and expression analysis of interleukin-8 and -10 in yellow catfish and in response to bacterial pathogen infection. Biomed. Res. Int. 2019;2019:9617659. doi: 10.1155/2019/9617659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Zhou P., Zhang Y., Zhang Z., Liu J., Zhang H. Short-chain fructo-oligosaccharides alleviates oxidized oil-induced intestinal dysfunction in piglets associated with the modulation of gut microbiota. J. Funct. Foods. 2019;2019:103661. [Google Scholar]
- Yang M., Zhang C., Zhang X., Zhang M.Z., Rottinghaus G.E., Zhang S. Structure-function analysis of Avian beta-defensin-6 and beta-defensin-12: role of charge and disulfide bridges. BMC Microbiol. 2016;16:210. doi: 10.1186/s12866-016-0828-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young-Speirs M., Drouin D., Cavalcante P.A., Barkema H.W., Cobo E.R. Host defense cathelicidins in cattle: types, production, bioactive functions and potential therapeutic and diagnostic applications. Int. J. Antimicrob. Ag. 2018;51:813–821. doi: 10.1016/j.ijantimicag.2018.02.006. [DOI] [PubMed] [Google Scholar]
- Yu H., Lu Y., Qiao X., Wei L., Fu T., Cai S., Wang C., Liu X., Zhong S., Wang Y. Novel cathelicidins from pigeon highlights evolutionary convergence in Avain cathelicidins and cunctions in modulation of innate immunity. Sci. Rep. 2015;5:11082. doi: 10.1038/srep11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Xiao Y., Li J., Ran J., Yin L., Liu Y., Zhang L. Molecular characterization of a novel ovodefensin gene in chickens. Gene. 2018;678:233–240. doi: 10.1016/j.gene.2018.08.029. [DOI] [PubMed] [Google Scholar]
- Yu L., Zhang L., Yang H., Gui G., Liu Y., Xiao Y. Identification and characterization of the myeloid differentiation factor 88 gene in yellow catfish. 3 Biotech. 2018;8:430. doi: 10.1007/s13205-018-1448-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti M. Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 2004;75:39–48. doi: 10.1189/jlb.0403147. [DOI] [PubMed] [Google Scholar]
- Zhang G., Sunkara L.T. Avian antimicrobial host defense peptides: from biology to therapeutic applications. Pharmaceuticals. 2014;7:220–247. doi: 10.3390/ph7030220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen D., Yu L., Wei Y., Li J., Zhou C. Genome-wide analysis of the ovodefensin gene family: monophyletic origin, independent gene duplication and presence of different selection patterns. Infect. Genet. Evol. 2019;68:265–272. doi: 10.1016/j.meegid.2019.01.001. [DOI] [PubMed] [Google Scholar]
- Zhang L., Jie H., Xiao Y., Zhou C., Lyu W., Bai W. Genomic identification and expression analysis of the cathelicidin gene family of the forest musk deer. Animals. 2019;9:481. doi: 10.3390/ani9080481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Gao B. A fossil antibacterial peptide gives clues to structural diversity of cathelicidin-derived host defense peptides. FASEB J. 2009;23:13–20. doi: 10.1096/fj.08-114579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.