Abstract
Chicken plumage color is an important economical trait in poultry breeding, as triple-yellow indigenous broilers are preferred over western commercial broilers in the Chinese market. However, the studies on the pigmentation of plumage coloration are relatively rare at present. Here, we performed a genome-wide mapping study on an F2 intercross, whose 2 founders were one hybrid commercial line “High Quality chicken Line A” that originated from the Anak red chicken and one indigenous line “Huiyang Beard” chicken that is a classical “triple-yellow” Chinese indigenous breed. Moreover, we used an automatic colorimeter that can quantitatively assess the colorations in L∗, a∗, and b∗ values. One major quantitative trait locus (QTL) on chromosome 2 was thus identified by both genome-wide association and linkage analyses, which could explain 10 to 20% of the total phenotypic variance of the b∗ measurements of the back plumage color. Using linkage analysis, 2 additional QTL on chromosome 1 and 20 were also found to be significantly associated with the plumage coloration in this cross. With additional samples from Anak red and Huiyang Beard chickens as well as pooled resequencing data from the 2 founders of this cross, we then further narrowed down the QTL regions and identified several candidate genes, such as CABLES1, CHST11, BCL2L1, and CHD22. As the effects of QTL found in this study were substantial, quantitatively measuring the coloration rather than the descriptive measurements provides stronger statistical power for the analyses. In addition, this major QTL on chromosome 2 that was associated with feather pigmentation at the genome-wide level will facilitate the future chicken breeding for yellow plumage color. In conclusions, we mapped 3 associated QTL on chromosome 1, 2, and 20. The candidate genes identified in this study shed light in the genetic basis of yellow plumage color in chicken.
Key words: yellow plumage color, genome-wide association, linkage analysis
Introduction
In China, chicken plumage color is an important economic trait, as most native breeds have characteristic “triple-yellow” appearance (yellow plumage, beak, and claw) and they are favored by Chinese consumers. Although colorations of plumage were used to be categorized descriptively, its underlying genetic basis is complex, which included the biosynthesis of eumelanin and pheomelanin in melanocytes (Emaresi et al., 2013).
In recent years, genome-wide association studies (GWAS) were widely applied in coat coloration of animals, including Iranian Markhoz goats (Nazari-Ghadikolaei et al., 2018), mice (Miao et al., 2017), horses (Kim et al., 2017), and dogs (Hedan et al., 2019), to screen novel candidate regions. For the black and brown colors of chicken plumage, previous studies adopted GWAS to discover significant SNP on chromosomes 1, 2, 3, and 5 (Yang et al., 2017) and on chromosomes 1, 2, 3, 4, 6, 8, 19, 24, and 33 (Park et al., 2013). For the chocolate color in chicken plumage, recent study using linage analysis (LA) reported that a missense mutation in TYRP1 alters the melanosome structure, which caused the pigment of chocolate color (Li et al., 2019).
So far, many well-known genes including ASIP, TYR, MC1R, TYRP1, SLC24A4, MITF, and KIT have been identified to play vital roles in melanocyte differentiation, proliferation, migration, survival, and morphology (Sulem et al., 2007, Duffy et al., 2010, Minvielle et al., 2010, Roulin and Ducrest, 2013). In feather follicles, the binding of melanin-stimulating hormone α-MSH to Mc1R could increase the intracellular cAMP level, and this promotes MITF expression and further increases the activity of eumelanin-related enzymes such as Tyr and Tyrp1 (Garcia-Borron et al., 2005). Besides, the expression of ASIP in the Japanese quails was higher in the yellow stripes of their back skin than that in the black stripes (Inaba et al., 2019). The binding of ASIP to Mc1R could induce the production of pheomelanin (Sakai et al., 1997, Walker and Gunn, 2010).
Owing to the studies on the pigmentation of pheomelanin (yellow, red) being relatively rare at present, we attempted to quantify the coloration of chicken back plumage. Based on an F2 intercross population, we mapped 3 associated quantitative trait loci (QTL) on chromosome 1, 2, and 20. Then, using genotypes from additional birds and whole genome pooled resequencing data of the founder lines, QTLs were further narrowed down and candidate genes were identified. Our results provide new insights into the genetic basis of yellow plumage color in the chicken.
Materials and methods
Experimental Population
The F2 intercross population and the corresponding genotyping data have been analyzed for its growth traits in the study by Sheng et al. (2013). Detailed descriptions can be found in that study. In brief, the founders were one hybrid commercial line “High Quality chicken Line A” (HQLA) that originated from the Anak red chicken and one indigenous line “Huiyang Beard” (HB) chicken that is a classical “triple-yellow” chicken breed. By reciprocal crossing of the founder lines for 2 generations, a total of 800 F2 birds were produced. Additional samples from Anak red and HB breeds (100 each) were collected from the National Chickens Genetic Resources (Jiangsu Province) (http://www.genebank.org.cn/), which is an off-site conservation base for 29 selected breeds.
Phenotyping and Genotyping
In this study, an automatic colorimeter TCP2 (Beijing Oyke Opto-Electronic Instrument Co., Ltd.) was used to quantitatively assess the back plumage color. The color is quantified using values a∗ (red to green axis; ABPC) and b∗ (yellow to blue axis; BBPC). Light to dark (LBPC) is determined as L∗ on a separate numerical scale (from black = 0 to white = 100). The measurements were performed according to Watson et al. (2018). Birds of the F2 generation were c`onsecutively tested every other week from 8 to 12 wk of age. Four different points on the back plumage of each chicken were measured to obtain an average value.
Blood samples were collected from a total of 585 birds from the F2 cross and 200 birds from the Anak red and HB chickens. Genotyping for all samples were performed using Illumina Chicken 60K SNP Beadchip (Groenen et al., 2011). Quality controls were performed with the following standards: individual data were excluded when their call rates were below 0.9; single-nucleotide polymorphisms (SNP) were removed as a result of call rates < 0.9, minor allele frequency < 0.05, or undetermined positions on the chromosome. SNP on Z chromosome were kept only when their genotypes were homozygous in hens. A final of 44,177 SNPs from the 494 birds of F2 cross were used for the next-step analyses.
Whole-Genome Linkage Analysis
Genetic map was constructed by using CRI-MAP, which was about 3,068 cM in total. Linkage mapping was carried out with least squares regression as described by Haley et al., 1994. With customized R scripts, a multiple linear regression model was used with “sex” and “batch” as the fixed effects. Effects of putative QTL were estimated at each 1-cM interval across the genome, and F statistics were calculated accordingly. One percent and 5% genome-wide thresholds were determined by 1,000 times permutation tests as described by Churchill and Doerge, (1994). To further explore the independent QTL, the most significant QTL would be added to the model as covariate(s) to perform another round of analysis, until no significant result was obtained.
The proportion of the residual phenotypic variance that was explained by the detected QTL was calculated by the following equation:
where, is the residual mean square of the reduced model (i.e., model that included “sex” and “batch” effects, also other QTL as cofactors), MSF is the residual mean square of the full model (i.e., model that included all covariates, other QTL as well as the additive and dominance effects of the tested position), and MSR is the residual mean square of the reduced model (model that only fit all fixed effects but not any QTL for single-QTL analysis). The methods used for linkage analysis in this study were the same as the study on growth traits of the same population; details can be found in the study by Sheng et al. (2013).
Genome-Wide Association Analysis
A mixed linear model was used to perform association analysis in this study. While “sex” and “batch” were included as the fixed effects, the polygenic effect was also contained in the model as a random effect to correct for population stratifications. The association analyses in this study were all based on this model and were implemented by the R package – GenABEL (Aulchenko et al., 2007). A two-step score test was performed. By using the command “ibs,” the kinship matrix of the experimental population was constructed. Then, the matrix was implemented into the function “polygenic” to obtain the phenotypic residuals that had been accounted for 2 fixed effects and one polygenic (random) effect. The narrow sense heritability was also a direct output of the function “polygenic.” Finally, the obtained residuals were used as dependent traits in a simple regression for single-marker or haplotype-based association tests.
The proportion of the phenotypic variance explained by each of the significant SNP was calculated as follows:
where, p and q are the frequencies of the 2 alleles at the tested locus and Vary is the variance of the tested trait. As a set of significantly associated SNP will be identified for the feather pigmentation at different weeks of age, we only estimated the phenotypic variances explained by the most significant SNP. Thus, is the marker genotype effect of the most significant SNP to the trait.
Phasing and Haplotyping
The selected 3 peak SNP from 3 populations were first phased using fastPHASE 1.2, separately (Scheet and Stephens, 2006), and then, the frequencies were calculated. Haplotypes that had frequencies more than 5% were retained for further analysis. Using a customized R script, the 2 core haplotypes identified were plotted against their corresponding residual phenotypes, which were obtained in the reduced model that only accounted for 2 fixed effects.
FST Calculation Using Whole-Genome Resequencing Data
Whole-genome resequencing on pooled samples from the 2 founder lines (15 HB and 16 HQLA birds) were performed. Four hundred base pair paired-end sequencing libraries were prepared for the next whole-genome resequencing using the Illumina HiSeq 2,000 platform (BGI, Shenzhen). Clean raw reads provided by BGI were mapped onto the chicken reference genome (Gallus_gallus-5.0) using the command “mem” in BWA (Houtgast et al., 2018) and were then sorted by SAMtools (Li et al., 2009); the resulting Binary Alignment/Map files were next processed by Picard 2.8.0 to remove the PCR duplicates; a further recalibration on the base quality score of the aligned data before SNP calling was performed using “BaseRecalibrator” in GATK (Ni and Stoneking, 2016); SAMtools again was used to generate mpileup files for PoPoolation2 to calculate FST over 1 kb sliding window with 50% overlaps. The settings used for PoPoolation2 (Kofler et al., 2011) were minimum count 3, minimum coverage 10, maximum coverage 100, and minimum coverage fraction 1 (Lillie et al., 2017). Threshold was then set to 0.43 as this was the 1% top FST value across the genome in this study. The r2 values of linkage disequilibrium decreased over shorter distances (an average of 3.5 kb) in HB chicken than in Anak red (Peng, 2015). Therefore, we clustered significant regions (FST > 0.43) that were no more than 3.5 kb apart.
Selection of Candidate Genes in the QTL
For GWAS results, the SNP location was referred to the Gallus_gallus-5.0 reference genome, and the genes within the region that 0.35 Mb upstream and downstream of the associated SNP were considered as the candidates. For LA results, genes in the peak regions were considered as candidates.
Results
Phenotypic Summary
The statistical summaries of the L∗, a∗, and b∗ values of back plumage color through 8 to 12 wk of age (ABPC8-12, BBPC8-12, and LBPC8-12) were shown in Supplementary Table 1. The coefficient of variation (CV) values were all at relatively low levels (0.13–0.24), which suggested our measurements were rather stable within this population. Based on the measurements of ABPCs and BBPCs, the back plumage colors were of different degrees of red and yellow, which were in consistent with the fact that they were hybrids of Anak (red) and Huiyang Beard (yellow) chickens.
Pearson's correlation coefficients were calculated between each pair of the recorded phenotypes by “cor.test” in R. ABPC were negatively correlated with BBPC and LBPC (Table 1). The estimated r values between LBPC and ABPC or BBPC were at intermediate to high levels (0.4–0.8), whereas those between ABPC and BBPC were rather low. These are in accordance with the P values; the lower the r values, the higher the P values. In summary, both ABPC and BBPC were highly significantly and closely correlated with LBPC. However, little or low correlations were observed between ABPC and BBPC.
Table 1.
Correlations between pairs of traits recorded in this study.
| ABPC8 | ABPC10 | ABPC12 | BBPC8 | BBPC10 | BBPC12 | LBPC8 | LBPC10 | LBPC12 | |
|---|---|---|---|---|---|---|---|---|---|
| ABPC8 | 1 | 0.50 | 0.36 | −0.03 | −0.04 | −0.16 | −0.52 | −0.44 | −0.35 |
| ABPC10 | 9.69E-32 | 1 | 0.71 | −0.29 | −0.13 | −0.22 | −0.57 | −0.75 | −0.63 |
| ABPC12 | 9.65E-17 | 2.02E-76 | 1 | −0.27 | −0.28 | −0.28 | −0.48 | −0.66 | −0.81 |
| BBPC8 | 5.13E-01 | 3.21E-11 | 7.99E-10 | 1 | 0.64 | 0.59 | 0.66 | 0.58 | 0.52 |
| BBPC10 | 3.72E-01 | 3.92E-03 | 1.73E-10 | 5.26E-58 | 1 | 0.76 | 0.53 | 0.61 | 0.58 |
| BBPC12 | 3.19E-04 | 1.22E-06 | 3.93E-10 | 1.50E-47 | 1.33E-91 | 1 | 0.54 | 0.63 | 0.70 |
| LBPC8 | 4.12E-36 | 1.57E-42 | 1.04E-29 | 5.22E-63 | 1.52E-36 | 1.53E-38 | 1 | 0.76 | 0.67 |
| LBPC10 | 2.87E-24 | 1.03E-90 | 5.57E-62 | 7.69E-46 | 1.19E-50 | 2.36E-55 | 9.40E-92 | 1 | 0.85 |
| LBPC12 | 2.78E-15 | 3.49E-55 | 9.08E-114 | 1.82E-35 | 1.48E-45 | 1.87E-72 | 3.79E-65 | 5.34E-139 | 1 |
Diagonal and upper triangle: Phenotypic correlations. Lower triangle: P values of the phenotypic correlations.
Three Independent QTL Were Found to be Significantly Associated With the L∗ and b∗ Values of Back Plumage Color in This Population
Given the recorded pedigree of this population, genetic map was constructed and both GWAS and LA were performed (all results were listed in Supplementary Table 1 significant results were summarized in Table 2). For the ABPC measurements, only a single significant SNP was found that was associated with ABPC10 in GWAS. However, based on the increasing order of the corresponding P values, the following SNP were identified on chromosome 20. In addition, this locus was not significantly associated with ABPC10 using LA, and the locus with the highest F value in LA was on the other chromosome – GGA10 (Supplementary Table 1). Therefore, no significant results were found for ABPC measurements in this study.
Table 2.
Loci that were significantly associated with the plumage color on birds' back in this population.
| Traits (h2) | QTL | Chromosome | Peak positions | Test statistics (F/P values) | Peak regions | Variance explained | Covariates | Methods |
|---|---|---|---|---|---|---|---|---|
| LBPC8 (43.7%) | QTL2 | 2 | 103.2 Mb (rs14228181) | 1.25E-09∗∗ | 103.2-106.7 Mb | 7.85% | / | GWAS |
| QTL2 | 2 | 102.4 Mb (222 cM) | 18.56∗∗ | 88.9-108.7 Mb (201-235 cM) | 6.73% | / | LA | |
| LBPC10 (52.9%) | QTL2 | 2 | 103.2 Mb (rs14228181) | 2.91E-08∗∗ | 103.2-106.7 Mb | 6.65% | / | GWAS |
| QTL2 | 2 | 101.7 Mb (221 cM) | 24.63∗∗ | 85.5-111.9 Mb (199-240 cM) | 8.83% | / | LA | |
| QTL1 | 1 | 54.1 Mb (139 cM) | 13.64∗∗ | 53.6-56.1 Mb | 4.51% | chr2:221 cM | LA | |
| LBPC12 (46.2%) | QTL2 | 2 | 106.7 Mb (rs317554548) | 3.19E-07∗ | / | 5.59% | / | GWAS |
| QTL2 | 2 | 101.1 Mb (220 cM) | 17.95∗∗ | 91.3-108.1 Mb (207-233 cM) | 6.48% | / | LA | |
| QTL5 | 5 | 53.9 Mb (149 cM) | 11.04∗∗ | / | 3.70% | chr2:220 cM | LA | |
| BBPC8 (44.0%) | QTL2 | 2 | 103.2 Mb (rs14228181) | 8.06E-13∗∗ | 93.8-106.7 Mb | 12.08% | / | GWAS |
| QTL2 | 2 | 101.7 Mb (221 cM) | 36.91∗∗ | 83.1-113.2 Mb (195-242 cM) | 12.85% | / | LA | |
| QTL20 | 20 | 10.2 (63 cM) | 11.96∗∗ | 9.9-10.4 Mb (61-65 cM) | 3.77% | chr2:221 cM | LA | |
| BBPC10 (56.7%) | QTL2 | 2 | 103.2 Mb (rs14228181) | 1.27E-11∗∗ | 93.8-107.6 Mb | 11.50% | / | GWAS |
| QTL2 | 2 | 102.9 Mb (223 cM) | 57.86∗∗ | 76.1-116.4 Mb (189-249 cM) | 19.02% | / | LA | |
| QTL20 | 20 | 10.3 Mb (64 cM) | 20.01∗∗ | 9.3-13.3 Mb (56-75 cM) | 5.92% | chr2:223 cM | LA | |
| QTL1 | 1 | 50.5 Mb (126 cM) | 13/31∗∗ | 49.5-53 Mb (123-135 cM) | 3.66% | chr2:223 cM; chr20:64 cM | LA | |
| BBPC12 (55.7%) | QTL2 | 2 | 103.2 Mb (rs14228181) | 4.61E-11∗∗ | 100.1-108.3 | 10.28% | / | GWAS |
| QTL2 | 2 | 102.4 Mb (222 cM) | 45.62∗∗ | 81.3-112.3 Mb (194-241 cM) | 15.54% | / | LA | |
| QTL20 | 20 | 10.8 Mb (67 cM) | 10.98∗ | 10.6-10.9 Mb (66-67 cM) | 3.35% | chr2:222 cM | LA |
∗Denotes 5% genome-wide significance, ∗∗denotes 1% genome-wide significance.
Abbreviations: GWAS, genome-wide association studies; LA, linage analysis; QTL, quantitative trait locus.
Using GWAS, 6 QTLs were identified, and each was significantly associated with one measurement of BBPC and LBPC. Twelve QTLs were found to be associated with the 6 traits of BBPC and LBPC by LA. As shown in Table 2, these detected QTLs clustered to 4 locations on 4 different chromosomes in the genome. We then named each QTL by combining “QTL” and the number of chromosome where it was located.
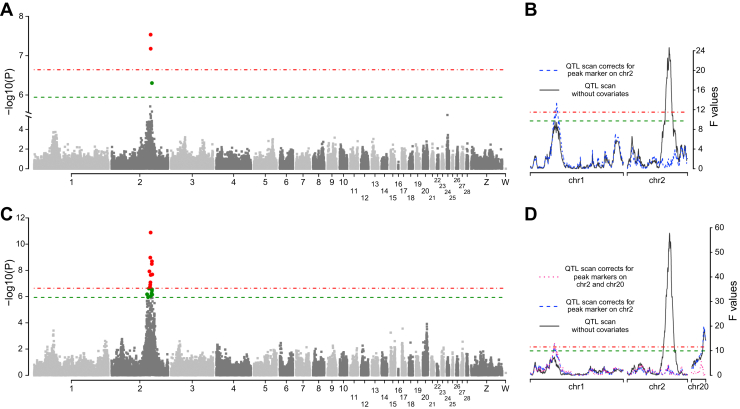
One major QTL on chromosome 2 (QTL2) were not only identified by both methods but were also associated with LBPC and BBPC across 8 to 12 wk of age. For LBPC, QTL2 explained a large proportion of phenotypic variances (5.5–8.8%). An even larger proportion of phenotypic variances of BBPC were explained by QTL2 (10–19%; Table 2, Figures 1A and 1C). After correcting for the effect(s) of the most significant SNP as covariate(s) in the model, additional 3 QTLs on chromosomes 1, 5, and 20 were found by LA, whereas no significant results were detected by GWAS. As QTL5 was identified to be associated with LBPC12 alone and it spanned only one centimorgan, we excluded it as an unreliable result from this study. Therefore, another 2 independent QTL20 and QTL1 were found to be associated with BBPC by LA (Figures 1B and 1D).
Figure 1.
Genome-wide association and linkage analysis for LBPC10 and BBPC10 traits. (A) Manhattan plot for the genome-wide association study for LBPC10 traits. (B) The identification of QTL related with LBPC10 trait by linkage analysis. (C) Manhattan plot for the genome-wide association study for BBPC10 traits. (D) The identification of QTLs related with BBPC10 trait by linkage analysis. The Manhattan plot indicates the −log10 (observed P values) for genome-wide SNP (y-axis) plotted against the irrespective positions on each chromosome (x-axis).
Identifying the Core Haplotypes Underlying the Major QTL on Chromosome 2
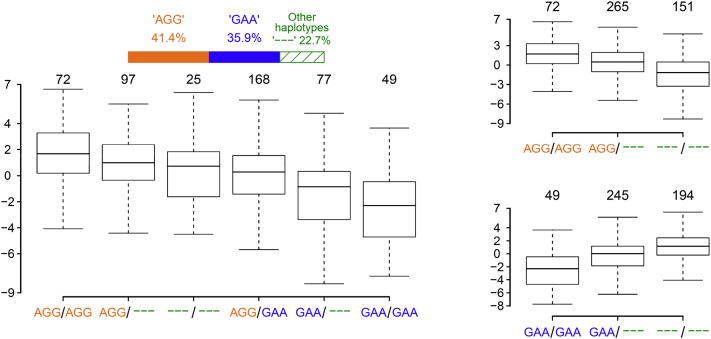
Compared with the estimated heritability of each BBPC traits, the peak SNP of QTL2 accounted for about 20% of the additive genetic variances, which suggested QTL2 was a major QTL that was associated with measurements of BBPC in this population. We then further define the haplotypes using the top 3 peak SNPs found by GWAS for trait LBPC10. Two main haplotypes “AGG” and “GAA” were identified, whose frequencies took up nearly 80% of all the haplotypes found. By using residual phenotypes that were obtained by corrected for sex and batch effects, “AGG” homozygous individuals had the highest b values, whereas “GAA” homozygous individuals had the lowest b values (Figure 2). We further examined the frequency distributions of “AGG” and “GAA” in additional 200 Anak red and HB birds (100 birds each; Table 3). “AGG” that increased the extent of yellowness was not found in the Anak chicken, whereas the frequency of “GAA” was substantial (0.78). Moreover, “GAA” took up significantly smaller proportion in the HB chicken.
Figure 2.
The distribution of LBPC10 measurements on the different core haplotypes. “AGG” and “GAA” were the 2 main haplotypes. Letters in yellow mean the corresponding haplotypes have an effect to increase L∗ values, while letters in blue denote their effects in decreasing the L∗ values.
Table 3.
Frequency distributions of core haplotypes from QTL2 in 3 populations.
| Populations | AGG | GAA | others1 |
|---|---|---|---|
| F2 population | 0.41 | 0.36 | 0.23 (3) |
| Anak red | 0 | 0.78 | 0.22 (2) |
| HB | 0.07 | 0.24 | 0.69 (4) |
Numbers of haplotypes that had frequencies more than 5% were shown in brackets.
QTL Were Further Narrowed Down Using Whole-Genome Pooled Resequencing Data of the Founder Lines
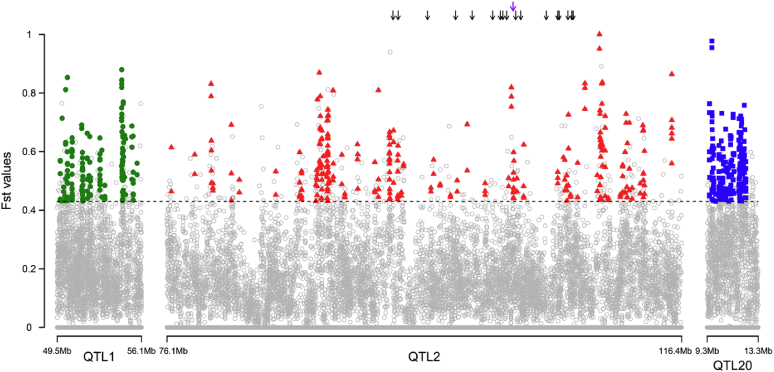
The candidate genes listed in Supplementary Table 1 were determined based on the position of the associated SNP identified by GWAS and the promising regions identified by LA. Whole-genome FST values were obtained by comparing the HB and HQLA pooled sequencing data, and the FST profiles of the QTL regions were shown in Figure 3. Almost 200 significant FST clusters were defined within the 3 identified QTLs (Supplementary Table 1). On QTL1, the candidate genes including MIEF1, MGAT3, TNRC6B, and CHST11 were overlapped with FST clusters. The CHST11 is a key enzyme in the biosynthesis of chondroitin sulfates. On the core haplotypes of the QTL2, the genes that contain the associated SNP were CABLES1, TAF4B, GREB1L, ROCK1, THOC1, COLEC12, and ANKRD29, besides, 4 candidate genes including ZNF521, LAMA3, TMEM241, and CABLES1 were overlapped with the significant FST clusters. Furthermore, there were 104 candidate genes within the region of the QTL20. Of these candidates, BCL2L1 and CDH22 were overlapped with the significant FST clusters. Combined with the implication of the relevant published references, the promising genes were emphasized and summarized in Table 4.
Figure 3.
The distribution of the whole-genome FST clusters within the QTL. The arrows represent FST clusters in the core haplotypes. 1% top FST clusters across the genome was defined as the threshold set to 0.43.
Table 4.
The functions of candidate genes identified in this study.
| QTLs | Gene | Position | Function | Reference |
|---|---|---|---|---|
| 1 | CHST111 | 54433922-54593321 | Catalyze the transfer of sulfate to position 4 of the GalNAc residue of chondroitin. | Zhou et al., 2016 |
| 2 | DSG4 | 106052985-106078954 | Haplotypes related with the color of the goat coat. | E et al., 2016 |
| 2 | CABLES11 | 102881201-102953764 | An adapter protein that links cyclin-dependent kinase with nonreceptor tyrosine kinases and regulates the activity of Cdks by enhancing their Y15 phosphorylation. | Huang et al., 2017 |
| 2 | GATA6 | 102636810-102654409 | Promotes hair follicle progenitor cell renewal. | Wang et al., 2017 |
| 2 | GREB1L | 102280644-102409696 | A coactivator for retinoic acid receptors. | Brophy et al., 2017 |
| 2 | YES1 | 101881440-101930073 | Mediates the biological process of tyrosine. | Patel et al., 2013, Ta et al., 2018 |
| 20 | EDN3 | 10799441-10816861 | Variation in EDN3 resulted in dermal hyperpigmentation phenotype or fibromelanosis | Dorshorst et al., 2011, Lillie et al., 2017, Shinomiya et al., 2012 |
| 20 | BCL2L11 | 10015061-10033228 | Homologous to BCL2 gene related with gray hair in mouse | Seiberg et al., 2013 |
| 20 | CDH221 | 10647698-10705460 | The expression was associated with melanoma metastasis | Piche et al., 2011 |
Genes overlapped with the significant FST clusters.
Discussion
Over the last few decades, researchers attempted to develop different methodologies and approaches to measure the coloration of animal appearance, such as Munsell tables (Peiponen, 1992), photographic analysis (Fitze and Richner, 2002, Figuerola and Senar, 2010), digital colorimeters (Figuerola et al., 1999, Juan Carlos et al., 2003), or reflection spectrometers (Bennett et al., 1996, Hunt et al., 1997, Alonsoalvarez et al., 2004). In our study, we quantified the plumage colorations using an automatic colorimeter. The measurements of plumage coloration can be conducted in 2 ways: one way was measuring the coloration directly on the bird (Siitari and Huhta, 2002, Eaton and Lanyon, 2003, Alonsoalvarez et al., 2004) and the other way was collecting feathers for measurement in the laboratory (Zuk and Decruyenaere, 1994, Bennett et al., 1997, Keyser and Hill, 1999, Siefferman and Hill, 2003). Earlier studies have demonstrated that the measured values obtained directly on the animal or from collected feathers, were nicely correlated (Quesada and Senar, 2006).
Traditionally, the plumage color is regarded as a qualitative trait; in this study, we developed the measuring method to obtain quantified values of the plumage color for GWAS and LA, and the results showed the QTL distributing on chromosome 1 (49.5–56.1 Mb), 2 (76.1–116.4 Mb), and 20 (9.3–13.3 Mb). Besides, it is noticeable that the proportion of phenotypic variance explained by QTL2 was substantial, which we speculated was attributed to the improvement of the measurement method.
Our results indicated that the major genes were likely located in QTL2 and very likely in the core haplotypes. The core haplotypes constructed by using the 3 most significant SNPs (rs14228181, rs14227093, and rs14230934) were ranging from 102.3 Mb to 106.4 Mb where many candidate genes were located. Previous studies demonstrated that DSG4 was related with the goat white and black color (E et al., 2016), and GATA6 promotes hair follicle progenitor cell renewal (Wang et al., 2017). Besides, the dietary intake of tyrosine could influence the color expression of the hair coat of adult dogs (Watson et al., 2018), and the YES1 as a candidate of interested is a member of the Src family tyrosine kinases. Network results from geneMANIA with human database showed YES1 have the physical interaction with KIT. The KIT gene is confirmed to be responsible for several common colors in pigs and horses, such as Dominant White and spotting (Marklund et al., 1998, Fontanesi et al., 2010, Durig et al., 2017, Hoban et al., 2018). In addition, we found 2 associated SNPs were located in CABLES1 that was also overlapped with the significant FST clusters. It was reported that CABLES1 was a critical regulator of corticotrope proliferation (Roussel-Gervais et al., 2016), linked cyclin-dependent kinase with nonreceptor tyrosine kinases (Huang et al., 2017). In emydid turtles or male painted turtles, carotenoids promoted lutein access to increased yellow and red chroma, as well as reduced ultraviolet chroma and brightness (Steffen et al., 2019). The carotenoids in vivo can be transformed to retinoic acid by the metabolic reaction (Weaver et al., 2018). In this study, we found one associated SNP (rs315171337) located in the intron of the GREB1L, and this gene implicated in activation of retinoic acid receptor targets (Brophy et al., 2017).
Considering that the feather pigmentation possibly effected by multiple genes, the candidates on QTL1 and QTL20 require attention as well. As the candidates on QTL1, CACNA1I belong to the family members of voltage-gated calcium channels and involved in the potential role of neuronal calcium signaling in schizophrenia (Xie et al., 2018). CHST11 can catalyze the transfer of sulfate to position 4 of the GalNAc residue of chondroitin (Zhou et al., 2016). As the candidates on QTL20, EDN3 affects melanocyte proliferation and differentiation (Baynash et al., 1994). Studies on chickens have illustrated that the structural variation of EDN3 resulted in dermal hyperpigmentation phenotype or fibromelanosis (Dorshorst et al., 2011, Shinomiya et al., 2012, Li et al., 2017). Besides, BCL2L1 and CDH22 were overlapped with the significant FST clusters. BCL2L1 is homologous to BCL2, which was incorporated in the Color Genes database (http://www.espcr.org/micemut/) for gray hair in mice (Seiberg, 2013), and CDH22 expression was associated with melanoma metastasis (Piche et al., 2011).
SLC24A5 is a putative cation transporter belonging to the potassium-dependent sodium–calcium ion exchanger family. Association of the SLC24A5 with pigmentation has been established in zebrafish, mice, humans, and chickens (Lamason et al., 2005, Vogel et al., 2008, Liu et al., 2011).This gene was downregulated in yellow quail and negatively regulated by ASIP (Nadeau et al., 2008). In this study, we found many of its homologous genes in the candidates, including SLC41A2 (chr1), SLC35C2 (chr20), SLC52A3 (chr20), SLC12A5 (chr20), and SLC38A6 (chr5). However, their functions on pigment synthesis were rarely reported.
In summary, no significant QTL was found for ABPC measurements. For the BBPC and LBPC measurements, QTL2 was associated with LBPC and BBPC across 8 to 12 wk of age; QTL20 and QTL1 were 2 additional independent QTLs that were found to be associated with BBPC. Moreover, we constructed the core haplotypes using the 3 most significant SNPs, which narrowed the region to 102.3–106.4 Mb. Combined with the whole-genome FST values, we further narrowed down the QTL regions and found several candidate genes. Further analyses and experiments are required to validate their functions in the feather pigmentation in chickens.
Acknowledgments
This work was supported financially by the National Natural Science Foundation of China (31702108 and 31472083) and the Fundamental Research Funds for the Central Universities (2662016QD018).
Ethics Statement: All animals used in the current study were cared for and used in accordance with the requirements of the Institute of Animal Science, Guangdong Academy of Agricultural Sciences (No. GAAS-IAS-2009-73) and the guidance (HZAUMU2013-0005) approved by the Ethics Committee of Huazhong Agricultural University.
Conflict of Interest Statement: The authors did not provide a conflict of interest statement.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.01.030.
Contributor Information
Zheya Sheng, Email: zheya.sheng@mail.hzau.edu.cn.
Xiaoxiang Hu, Email: huxx@cau.edu.cn.
Supplementary data
References
- Alonsoalvarez C., Doutrelant C., Sorci G. Ultraviolet reflectance affects male-male interactions in the blue tit (Parus caeruleus ultramarinus) Behav. Ecol. 2004;15:805–809. [Google Scholar]
- Aulchenko Y.S., Ripke S., Isaacs A., van Duijn C.M. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- Baynash A.G., Hosoda K., Giaid A., Richardson J.A., Emoto N., Hammer R.E., Yanagisawa M. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994;79:1277–1285. doi: 10.1016/0092-8674(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Bennett A.T., Cuthill I.C., Partridge J.C., Lunau K. Ultraviolet plumage colors predict mate preferences in starlings. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8618–8621. doi: 10.1073/pnas.94.16.8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett A.T.D., Cuthill I.C., Partridge J.C., Maier E.J. Ultraviolet vision and mate choice in zebra finches. Nature. 1996;380:433–435. [Google Scholar]
- Brophy P.D., Rasmussen M., Parida M., Bonde G., Darbro B.W., Hong X., Clarke J.C., Peterson K.A., Denegre J., Schneider M., Sussman C.R., Sunde L., Lildballe D.L., Hertz J.M., Cornell R.A., Murray S.A., Manak J.R. A gene implicated in activation of retinoic acid receptor targets is a novel renal Agenesis gene in humans. Genetics. 2017;207:215–228. doi: 10.1534/genetics.117.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G.A., Doerge R.W. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorshorst B., Molin A.M., Rubin C.J., Johansson A.M., Stromstedt L., Pham M.H., Chen C.F., Hallbook F., Ashwell C., Andersson L. A complex genomic rearrangement involving the endothelin 3 locus causes dermal hyperpigmentation in the chicken. PLoS Genet. 2011;7:e1002412. doi: 10.1371/journal.pgen.1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy D.L., Zhao Z.Z., Sturm R.A., Hayward N.K., Martin N.G., Montgomery G.W. Multiple pigmentation gene polymorphisms account for a substantial proportion of risk of cutaneous malignant melanoma. J. Invest. Dermatol. 2010;130:520–528. doi: 10.1038/jid.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durig N., Jude R., Holl H., Brooks S.A., Lafayette C., Jagannathan V., Leeb T. Whole genome sequencing reveals a novel deletion variant in the KIT gene in horses with white spotted coat colour phenotypes. Anim. Genet. 2017;48:483–485. doi: 10.1111/age.12556. [DOI] [PubMed] [Google Scholar]
- E G.X., Zhao Y.J., Ma Y.H., Cao G.L., He J.N., Na R.S., Zhao Z.Q., Jiang C.D., Zhang J.H., Arlvd S., Chen L.P., Qiu X.Y., Hu W., Huang Y.F. Desmoglein 4 diversity and correlation analysis with coat color in goat. Genet. Mol. Res. 2016;15:15017814. doi: 10.4238/gmr.15017814. [DOI] [PubMed] [Google Scholar]
- Eaton M.D., Lanyon S.M. The ubiquity of avian ultraviolet plumage reflectance. Proc. Biol. Sci. 2003;270:1721–1726. doi: 10.1098/rspb.2003.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emaresi G., Ducrest A.L., Bize P., Richter H., Simon C., Roulin A. Pleiotropy in the melanocortin system: expression levels of this system are associated with melanogenesis and pigmentation in the tawny owl (Strix aluco) Mol. Ecol. 2013;22:4915–4930. doi: 10.1111/mec.12438. [DOI] [PubMed] [Google Scholar]
- Figuerola J., Pascual J., Senar J.C. The use of a colorimeter in field studies of Blue Tit Parus caeruleus coloration. Ardea. 1999;87:269–275. [Google Scholar]
- Figuerola J., Senar J.C. Measurement of plumage badges: an evaluation of methods used in the Great Tit Parus major. Ibis. 2010;142:482–484. [Google Scholar]
- Fitze P.S., Richner H. Differential effects of a parasite on ornamental structures based on melanins and carotenoids. Behav. Ecol. 2002;13:401–407. [Google Scholar]
- Fontanesi L., D'Alessandro E., Scotti E., Liotta L., Crovetti A., Chiofalo V., Russo V. Genetic heterogeneity and selection signature at the KIT gene in pigs showing different coat colours and patterns. Anim. Genet. 2010;41:478–492. doi: 10.1111/j.1365-2052.2010.02054.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Borron J.C., Sanchez-Laorden B.L., Jimenez-Cervantes C. Melanocortin-1 receptor structure and functional regulation. Pigm. Cell Res. 2005;18:393–410. doi: 10.1111/j.1600-0749.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- Groenen M.A., Megens H.J., Zare Y., Warren W.C., Hillier L.W., Crooijmans R.P., Vereijken A., Okimoto R., Muir W.M., Cheng H.H. The development and characterization of a 60K SNP chip for chicken. BMC Genom. 2011;12:274. doi: 10.1186/1471-2164-12-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley C.S., Knott S.A., Elsen J.M. Mapping quantitative trait loci in crosses between outbred lines using least squares. Genetics. 1994;136:1195–1207. doi: 10.1093/genetics/136.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedan B., Cadieu E., Botherel N., Dufaure de Citres C., Letko A., Rimbault M., Drogemuller C., Jagannathan V., Derrien T., Schmutz S., Leeb T., Andre C. Identification of a missense variant in MFSD12 involved in Dilution of Phaeomelanin Leading to white or Cream coat color in dogs. Genes. (Basel). 2019;10:356. doi: 10.3390/genes10050386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban R., Castle K., Hamilton N., Haase B. Novel KIT variants for dominant white in the Australian horse population. Anim. Genet. 2018;49:99–100. doi: 10.1111/age.12627. [DOI] [PubMed] [Google Scholar]
- Houtgast E.J., Sima V.M., Bertels K., Al-Ars Z. Hardware acceleration of BWA-MEM genomic short read mapping for longer read lengths. Comput. Biol. Chem. 2018;75:54–64. doi: 10.1016/j.compbiolchem.2018.03.024. [DOI] [PubMed] [Google Scholar]
- Huang J.R., Tan G.M., Li Y., Shi Z. The Emerging role of Cables1 in Cancer and other Diseases. Mol. Pharmacol. 2017;92:240–245. doi: 10.1124/mol.116.107730. [DOI] [PubMed] [Google Scholar]
- Hunt S., Cuthill I.C., Swaddle J.P., Bennett A.T.D. Ultraviolet vision and band-colour preferences in female zebra finches, Taeniopygia guttata. Anim. Behav. 1997;54:1383–1392. doi: 10.1006/anbe.1997.0540. [DOI] [PubMed] [Google Scholar]
- Inaba M., Jiang T.X., Liang Y.C., Tsai S., Lai Y.C., Widelitz R.B., Chuong C.M. Instructive role of melanocytes during pigment pattern formation of the avian skin. Proc. Natl. Acad. Sci. U. S. A. 2019;116:6884–6890. doi: 10.1073/pnas.1816107116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan Carlos S., Jordi F., Jordi D. Plumage coloration and nutritional condition in the great tit Parus major: the roles of carotenoids and melanins differ. Die Naturwissenschaften. 2003;90:234–237. doi: 10.1007/s00114-003-0414-7. [DOI] [PubMed] [Google Scholar]
- Keyser A.J., Hill G.E. Condition-dependent variation in the blue-ultraviolet coloration of a structurally based plumage Ornament. Proc. R. Soc. B. Biol. Sci. 1999;266:771–777. [Google Scholar]
- Kim N.Y., Bhuiyan M.S.A., Chae H.S., Baek K.S., Son J.K., Shin S.M., Woo J.H., Park S.H., Lee S.H. Genome-wide association study for tobiano spotting coat color in Korean Jeju x Thoroughbred horse population. Anim. Genet. 2017;48:728–729. doi: 10.1111/age.12596. [DOI] [PubMed] [Google Scholar]
- Kofler R., Pandey R.V., Schlotterer C. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq) Bioinformatics. 2011;27:3435–3436. doi: 10.1093/bioinformatics/btr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason R.L., Mohideen M.A., Mest J.R., Wong A.C., Norton H.L., Aros M.C., Jurynec M.J., Mao X., Humphreville V.R., Humbert J.E., Sinha S., Moore J.L., Jagadeeswaran P., Zhao W., Ning G., Makalowska I., McKeigue P.M., O'Donnell D., Kittles R., Parra E.J., Mangini N.J., Grunwald D.J., Shriver M.D., Canfield V.A., Cheng K.C. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- Li J., Bed'hom B., Marthey S., Valade M., Dureux A., Moroldo M., Pechoux C., Coville J.L., Gourichon D., Vieaud A., Dorshorst B., Andersson L., Tixier-Boichard M. A missense mutation in TYRP1 causes the chocolate plumage color in chicken and alters melanosome structure. Pigment Cell Melanoma Res. 2019;32:381–390. doi: 10.1111/pcmr.12753. [DOI] [PubMed] [Google Scholar]
- Li H., Fan L., Zhu S., Shin M.K., Lu F., Qu J., Hou L. Epilation induces hair and skin pigmentation through an EDN3/EDNRB-dependent regenerative response of melanocyte stem cells. Sci. Rep. 2017;7:7272. doi: 10.1038/s41598-017-07683-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., S. Genome Project Data Processing The sequence Alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie M., Sheng Z., Honaker C.F., Dorshorst B.J., Ashwell C.M., Siegel P.B., Carlborg O. Genome-wide standing variation facilitates long-term response to bidirectional selection for antibody response in chickens. BMC Genom. 2017;18:99. doi: 10.1186/s12864-016-3414-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.F., Luo J., Hu X.X., Yang H., Lv X.Q., Feng C.G., Tong J., Wang Y.Q., Wang S.H., Liu X.J., Lin T.H., Fei J., Liu Y., Li N. Repression of Slc24a5 can reduce pigmentation in chicken. Front. Biosci. 2011;3:158–165. doi: 10.2741/e229. [DOI] [PubMed] [Google Scholar]
- Marklund S., Kijas J., Rodriguez-Martinez H., Ronnstrand L., Funa K., Moller M., Lange D., Edfors-Lilja I., Andersson L. Molecular basis for the dominant white phenotype in the domestic pig. Genome Res. 1998;8:826–833. doi: 10.1101/gr.8.8.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Soudy F., Xu Z., Liao M., Zhao S., Li X. Candidate gene identification of feed Efficiency and coat color traits in a C57BL/6J x Kunming F2 mice population using genome-wide association study. Biomed. Res. Int. 2017;2017:7132941. doi: 10.1155/2017/7132941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle F., Bed'hom B., Coville J.L., Ito S., Inoue-Murayama M., Gourichon D. The "silver" Japanese quail and the MITF gene: causal mutation, associated traits and homology with the "blue" chicken plumage. BMC Genet. 2010;11:15. doi: 10.1186/1471-2156-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau N.J., Minvielle F., Ito S., Inoue-Murayama M., Gourichon D., Follett S.A., Burke T., Mundy N.I. Characterization of Japanese quail yellow as a genomic deletion upstream of the avian homolog of the mammalian ASIP (agouti) gene. Genetics. 2008;178:777–786. doi: 10.1534/genetics.107.077073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazari-Ghadikolaei A., Mehrabani-Yeganeh H., Miarei-Aashtiani S.R., Staiger E.A., Rashidi A., Huson H.J. Genome-wide association studies identify candidate genes for coat color and Mohair traits in the Iranian Markhoz goat. Front. Genet. 2018;9:105. doi: 10.3389/fgene.2018.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni S., Stoneking M. Improvement in detection of minor alleles in next generation sequencing by base quality recalibration. BMC Genom. 2016;17:139. doi: 10.1186/s12864-016-2463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.N., Choi J.A., Lee K.T., Lee H.J., Choi B.H., Kim H., Kim T.H., Cho S., Lee T. Genome-wide association study of chicken plumage pigmentation. Asian-Australasian J. Anim. Sci. 2013;26:1523–1528. doi: 10.5713/ajas.2013.13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P.R., Sun H., Li S.Q., Shen M., Khan J., Thomas C.J., Davis M.I. Identification of potent Yes1 kinase inhibitors using a library screening approach. Bioorg. Med. Chem. Lett. 2013;23:4398–4403. doi: 10.1016/j.bmcl.2013.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiponen V.A. Colour Discrimination of two Passerine bird Species in the Munsell system. Ornis Scand. 1992;23:143–151. [Google Scholar]
- Peng D.Z. China Agricultural University; Beijing: 2015. Study of Genetic Diversity and Genome-wide Positive Selection in Chinese Indigenous and Commercial Chicken Breeds. phD Diss. [Google Scholar]
- Piche B., Khosravi S., Martinka M., Ho V., Li G. CDH22 expression is reduced in metastatic melanoma. Am. J. Cancer Res. 2011;1:233–239. [PMC free article] [PubMed] [Google Scholar]
- Quesada J., Senar J.C. Comparing plumage colour measurements obtained directly from live birds and from collected feathers: the case of the great tit Parus major. J. Avian Biol. 2006;37:609–6016. [Google Scholar]
- Roulin A., Ducrest A.L. Genetics of colouration in birds. Semin. Cell Dev. Biol. 2013;24:594–608. doi: 10.1016/j.semcdb.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Roussel-Gervais A., Couture C., Langlais D., Takayasu S., Balsalobre A., Rueda B.R., Zukerberg L.R., Figarella-Branger D., Brue T., Drouin J. The Cables1 gene in Glucocorticoid regulation of Pituitary corticotrope growth and Cushing Disease. J. Clin. Endocrinol. Metab. 2016;101:513–522. doi: 10.1210/jc.2015-3324. [DOI] [PubMed] [Google Scholar]
- Sakai C., Ollmann M., Kobayashi T., Abdel-Malek Z., Muller J., Vieira W.D., Imokawa G., Barsh G.S., Hearing V.J. Modulation of murine melanocyte function in vitro by agouti signal protein. EMBO J. 1997;16:3544–3552. doi: 10.1093/emboj/16.12.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P., Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiberg M. Age-induced hair greying - the multiple effects of oxidative stress. Int. J. Cosmet. Sci. 2013;35:532–538. doi: 10.1111/ics.12090. [DOI] [PubMed] [Google Scholar]
- Sheng Z., Pettersson M.E., Hu X., Luo C., Qu H., Shu D., Shen X., Carlborg O., Li N. Genetic dissection of growth traits in a Chinese indigenous x commercial broiler chicken cross. BMC Genom. 2013;14:151. doi: 10.1186/1471-2164-14-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomiya A., Kayashima Y., Kinoshita K., Mizutani M., Namikawa T., Matsuda Y., Akiyama T. Gene duplication of endothelin 3 is closely correlated with the hyperpigmentation of the internal organs (Fibromelanosis) in silky chickens. Genetics. 2012;190:627–638. doi: 10.1534/genetics.111.136705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefferman L., Hill G. Structural and melanin coloration indicate parental effort and reproductive success in male Eastern Bluebirds. Behav. Ecol. 2003;14:855–861. [Google Scholar]
- Siitari H., Huhta E. Individual color variation and male quality in pied flycatchers (Ficedula hypoleuca): a role of ultraviolet reflectance. Behav. Ecol. 2002;13:737–741. [Google Scholar]
- Steffen J.E., Hultberg J., Drozda S. The effect of dietary carotenoid increase on painted turtle spot and stripe color. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2019;229:10–17. doi: 10.1016/j.cbpb.2018.12.002. [DOI] [PubMed] [Google Scholar]
- Sulem P., Gudbjartsson D.F., Stacey S.N., Helgason A., Rafnar T., Magnusson K.P., Manolescu A., Karason A., Palsson A., Thorleifsson G., Jakobsdottir M., Steinberg S., Palsson S., Jonasson F., Sigurgeirsson B., Thorisdottir K., Ragnarsson R., Benediktsdottir K.R., Aben K.K., Kiemeney L.A., Olafsson J.H., Gulcher J., Kong A., Thorsteinsdottir U., Stefansson K. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- Ta N.L., Chakrabandhu K., Huault S., Hueber A.-O. The tyrosine phosphorylated pro-survival form of Fas intensifies the EGF-induced signal in colorectal cancer cells through the nuclear EGFR/STAT3-mediated pathway. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-30804-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel P., Read R.W., Vance R.B., Platt K.A., Troughton K., Rice D.S. Ocular albinism and hypopigmentation defects in Slc24a5-/- mice. Vet. Pathol. 2008;45:264–279. doi: 10.1354/vp.45-2-264. [DOI] [PubMed] [Google Scholar]
- Walker W.P., Gunn T.M. Shades of meaning: the pigment-type switching system as a tool for discovery. Pigment Cell Melanoma Res.s. 2010;23:485–495. doi: 10.1111/j.1755-148X.2010.00721.x. [DOI] [PubMed] [Google Scholar]
- Wang A.B., Zhang Y.V., Tumbar T. Gata6 promotes hair follicle progenitor cell renewal by genome maintenance during proliferation. EMBO J. 2017;36:61–78. doi: 10.15252/embj.201694572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A., Wayman J., Kelley R., Feugier A., Biourge V. Increased dietary intake of tyrosine upregulates melanin deposition in the hair of adult black-coated dogs. Anim. Nutr. 2018;4:422–428. doi: 10.1016/j.aninu.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R.J., Santos E.S.A., Tucker A.M., Wilson A.E., Hill G.E. Carotenoid metabolism strengthens the link between feather coloration and individual quality. Nat. Commun. 2018;9:73. doi: 10.1038/s41467-017-02649-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Huang D., Wei L., Luo X.J. Further evidence for the genetic association between CACNA1I and schizophrenia. Hereditas. 2018;155:16. doi: 10.1186/s41065-017-0054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Du X., Wei S., Gu L., Li N., Gong Y., Li S. Genome-wide association analysis identifies potential regulatory genes for eumelanin pigmentation in chicken plumage. Anim. Genet. 2017;48:611–614. doi: 10.1111/age.12573. [DOI] [PubMed] [Google Scholar]
- Zhou H., Li Y., Song X., Zhao Y., Cheng L., Zhao L., Jia L. CHST11/13 regulate the metastasis and Chemosensitivity of human Hepatocellular Carcinoma cells via Mitogen-Activated protein kinase Pathway. Dig. Dis. Sci. 2016;61:1972–1985. doi: 10.1007/s10620-016-4114-5. [DOI] [PubMed] [Google Scholar]
- Zuk M., Decruyenaere J.G. Measuring individual variation in colour: a comparison of two techniques. Biol. J. Linn. Soc. 1994;53:165–173. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.