Abstract
Aging is a fundamental biological process that is still not fully understood. As many of the most significant human diseases have aging as their greatest risk factor, a better understanding of aging potentially has enormous practical implications in treating these diseases. The nematode C. elegans is an exceptionally useful genetic model organism that had been used with great success to shed light on many genes and pathways that are involved in aging. Many of these pathways and mechanisms have been shown to be conserved through mammals. The standard methods for assaying survival in C. elegans to measure changes in lifespan are tedious and time consuming. This limits the throughput and productivity of C. elegans aging researchers. In recent years, many inroads have been made into automating various facets of the collection and analysis of C. elegans lifespan experimental data. The advances described in this review all work to ameliorate some of the hurdles that come with manual worm lifespan scoring, by automating or eliminating some of the most time consuming aspects of the assay. By greatly increasing the throughput of lifespan assays, these methods will enable types of experiments (e.g., drug library screens) whose scale is currently impractical. These methods have already proved exceptionally useful, and some of them are likely to be the predecessors of even more refined methods that could lead to breakthroughs in the ability to study lifespan in C. elegans. This could in turn potentially revolutionize our understanding of the basic biology of aging, and one day lead to treatments that could offset or delay age-related diseases in humans.
Keywords: Aging, lifespan, C. elegans, automation, microfluidics, healthspan
1. Introduction
Aging is a fundamental biological puzzle of great and longstanding interest [1,2]. Why do we age [3]? Why do closely related species, diverged from a recent common ancestor, have such different natural lifespans [4]? Although much recent progress has been made, there is still much to learn about the basic underlying causes of aging. Practically speaking, aging is the shared strongest risk factor for age-related diseases, including cancer, heart disease, and Alzheimer’s disease. As a result, work to understand aging itself could bear fruit in the form of interventions that could simultaneously postpone the onset, or lessen the severity, of multiple age-related diseases [5]. This practical significance of improvements in our understanding of the basic biology of aging will only increase, as the human population continues demographically to become more aged in the next several decades [6].
The nematode Caenorhabditis elegans [7] has been proven to be an extremely useful model in the study of aging [8–13]. Genetic alterations have extended the lifespan of C. elegans up to 10-fold [14]. Further, many genetic pathways that have been found to affect lifespan in C. elegans have also been shown to have broad conservation of their lifespan-extending phenotypes, in diverse other organisms such as Drosophila and mice [15,16]. Multiple drugs have also been found to greatly increase the lifespan of C. elegans [17–19], and some of these drugs have also been shown to have conserved effects on lifespan in other, distantly-related organisms, including mice [20]. The gold standard for the collection of C. elegans lifespan data has long been manual scoring, by assessing movement of worms on solid media. This process involves manually prodding worms using a worm pick, as they age and begin to move less and become less responsive. It also involves manually transferring worms away from any plates that may have become contaminated with mold or progeny [21,22]. In addition, manually scored lifespans must deal with the problem of separating the adult worms from their own progeny. As newly hatched worms typically become indistinguishable from their own parents in around 48 hours, this must involve either laboriously transferring the adult worms no less than every 48 hours for the first several days of their lives, or sterilization using either fluorodeoxyuridine (FUdR) [23] or temperature-sensitive or other conditionally sterile mutants [24]. FUdR blocks DNA synthesis by inhibiting thymidylate synthetase, and thus prevents the development of C. elegans progeny. Although these interventions do not appear to affect the lifespan of wild-type C. elegans under standard conditions, fluorodeoxyuridine treatment has been shown to affect lifespan in multiple other genotypes, including worms with mutations in tub-1, a modulator of fat storage, and exo-3, an endonuclease known to decrease lifespan. FUdR also has been shown to alter lifespan under certain conditions, such as hypertonic stress conditions [25–28]. As a result, it is likely that either FUdR treatment or conditionally sterile mutants could lead to false positive or false negative outcomes in large-scale screening experiments for lifespan phenotypes.
The process of manually scoring lifespans as described above is an incredibly time consuming and tedious process. Because of this, there is a great need for an automated system to determine worm lifespan and other phenotypes such as movement, size, and fecundity. The manual process also puts a number of limitations on the number of worms and conditions that can be tested at once. Someone manually scoring a lifespan can likely score the worms of one to two thousand worms a day, taking most of the day depending on the lifespan stage. This lack of throughput and time commitment can make performing large-scale screens unfeasible using current methods. With current limitations, researchers may miss dose-specific lifespan extensions of drugs, or may miss subtle effects of an RNAi treatment due to a limited number of worms scored. Advances in automation will not only allow researchers to be able to measure lifespan more efficiently, but to measure additional phenotypes without having to put in additional time or resources. Most importantly, greatly increased throughput often inspires entirely new types of experiments that could not previously be contemplated.
Here we review some current advances in the methods for assaying lifespan in C. elegans. These primarily involve automation, but also include variations in the manual techniques for scoring these assays, and in the software used to analyze these data. Some automated methods are already available to the interested researcher, on either an academic [29] or commercial [30,31] basis, and improvements are rapidly ongoing in all areas. However, for many studies, manual scoring is still the de facto standard at this time in many laboratories. These advances have already provided many new insights into aging in C. elegans, that may shed light on broadly conserved aging biology including human aging biology. In the future, many of these methods may serve as a foundation for subsequent additional advances in lifespan methodology. In conclusion, we consider where this area of research is likely to be headed next, and discuss some considerations of interest to developers and users of the next developments in this exciting field.
2. Current Systems
Pioneering work in C. elegans lifespan automation has focused largely on microfabricated and flatbed scanner approaches, along with some notable examples that do not fall neatly into either of these categories, or that cross these categories. Here we will consider the advantages and disadvantages of the various broad technological approaches outlined, as well as the advantages and disadvantages of several of the specific implementations.
2.1. Microfabricated approaches
Scoring worm lifespans on both solid media plates and liquid media has provided much insight into mechanisms of aging. However, these techniques typically involve scoring worms for movement once per day, or even less frequently than that. Recent advances in small-scale polymer fabrication arising from improvements in soft lithography and photolithography have led to lab-scale production of microfluidic devices for diverse research purposes including cell structure analysis, cell cycle studies, tissue culture, and lifespans [32]. These microfluidic devices provide a means to potentially increase the throughput of lifespan assays relative to manual scoring, and in some cases they also allow continuous observation and recording as opposed to intermittent observation. The general design of microfluidic chip-based devices typically includes inlets to allow for addition of fresh liquid media and food, outlets to allow removal of waste, eggs, and progeny, and and one or more areas for housing and examination of C. elegans (Fig. 1a). Microfluidic devices, and the methods for fabricating them, are under rapid and sustained development for uses far outside of the measurement of lifespan in C. elegans, and for reasons independent of this use [33]. As a result, approaches that rely on these technologies will benefit from the rising tide of outside advances motivated by their many other uses. More generally, modular designs that favor hardware and software components that are under rapid outside development for reasons beyond the scope of nematode aging, are likely to outpace other approaches.
Fig 1:
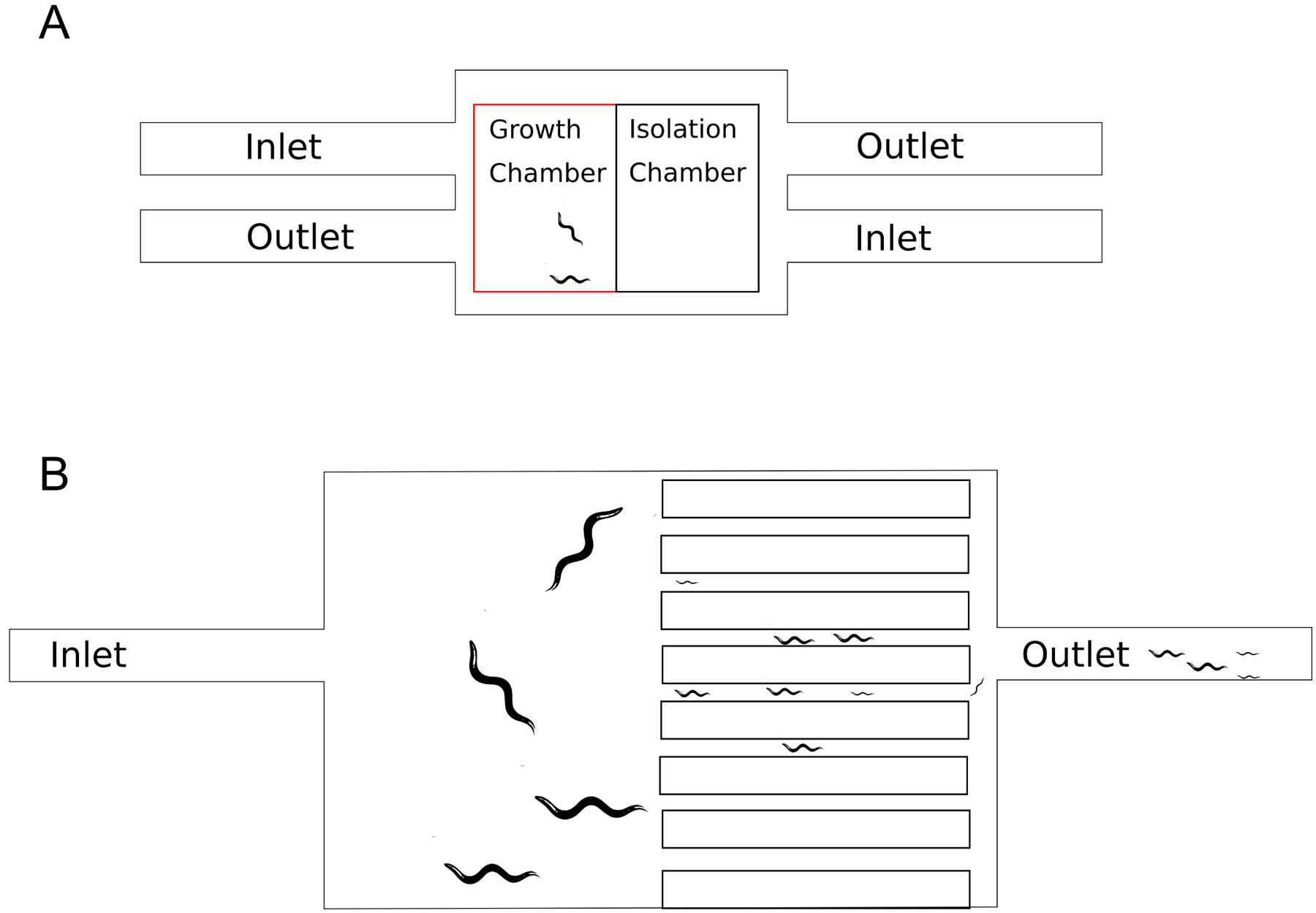
A. General microfluidic design concept demonstrating inlets providing flow into the growth chamber and outlets that allow removal of waste. Worms are kept in the growth chamber unless they need to be moved into the isolation chamber for observation. B. WormFarm design, allowing for removal of larvae and eggs while keeping adults; the original concept uses size restriction and fluid flow to allow eggs and L1 larvae to be washed out while adults remain in the primary growth chamber.
2.1.1. WormFarm for automated measurement of longevity, mean lifespan, body size, and motility.
WormFarm is a microfluidic device that is especially well-suited to lifespan analysis [34]. The design of WormFarm includes three modules. The first module is a food and medium loading module that allows for the introduction of bacterial food in a continuous manner throughout the lifespan. The second module is the WormFarm chip itself, where the worms are housed throughout the duration of the experiment. Finally, the third module is an image acquisition module, that allows the collection of videos and still images for various kinds of further analysis, including automated lifespan scoring.
The food/medium loading module allows for the introduction of not only the standard E. coli OP50 food source, but also dsRNA producing bacteria providing an effective means of doing RNAi knockdown studies for genes of interest [35]. Xian et al. show that using this method, RNAi knockdown of age-1, sptf-3 and the ATP synthesis gene Y82E9BR.3 using clones from the Ahringer RNAi library [35] gave lifespan results that mirrored manual scoring for each of these knockdowns. Additionally, because this method allows the ability to control the food source and input with this module, caloric restriction and other feeding regimes that have previously been shown to affect lifespan can be implemented.
The WormFarm chip where the adult worms are maintained has several size exclusion outlets that allow larvae and eggs to be washed out, while adult worms are retained. This automates the process of progeny removal (Fig. 1b), thus eliminating the need for use of fluorodeoxyuridine or conditionally sterile mutants [36].
The final component of WormFarm is the image acquisition module, which collects videos and photos of the worms within their chambers, and allows for both manual scoring for lifespans as well as the potential to introduce these videos to an automated system. Xian et al. provided one automated system for scoring, but noted the need to maintain small number of worms in each chamber with this automated system because of complications with clusters of worms, and the inability of algorithm to isolate individual worms from within these clusters. For these smaller groups of worms, 40 worms or fewer per device, the automated system was impressively able to show nearly identical results to manual scoring.
2.1.2. WorMotel for tracking behavior and lifespan
WorMotel is a microfabricated device designed with the goal of providing a means of performing longitudinal monitoring of aging C. elegans in a high-throughput manner [37]. In the design of this system, Churgin et al. needed a device that provided clear field of view for each worm, an adequate food source, standardized equipment dimensions, and a method of high-throughput data collection.
One of the bigger challenges with monitoring C. elegans in an automated way is their motility. In a 96 well plate, the worms will climb the sides of the well they are placed in. This makes them difficult to monitor from a microscope or other imaging device. Further, it can lead to aberrant worm deaths due to dehydration from leaving their normal media, and to cross-contamination of different conditions or genotypes if a worm successfully moves from one well to another. To address this issue, Churgin et al. built a device with small concave wells where each well houses only one worm. This concave surface provides a good view of the worms from above in all positions. In addition, the wells are surrounded by moats containing a copper sulfate solution to deter movement outside of these wells (Fig. 2a). Based on current American National Standards Institute (ANSI) standard dimensions, each plate was built to house 240 worms, providing a system that could be monitored by any current technology that works with 96 or 384 plate wells. In this method, each well is filled with nematode growth medium (NGM) agar and bacterial food (Fig. 2b). Because this system is designed to measure changes during the aging process, it also provides a mechanism for generating survival data. From a technical standpoint, this approach is derived in part from considering preceding work that used a polymerized polyethylene glycol layer, overlaid with with droplets of worm media each housing a single worm and capped with polydimethylsiloxane (PDMS) (Fig. 2c) [38].
Fig 2:
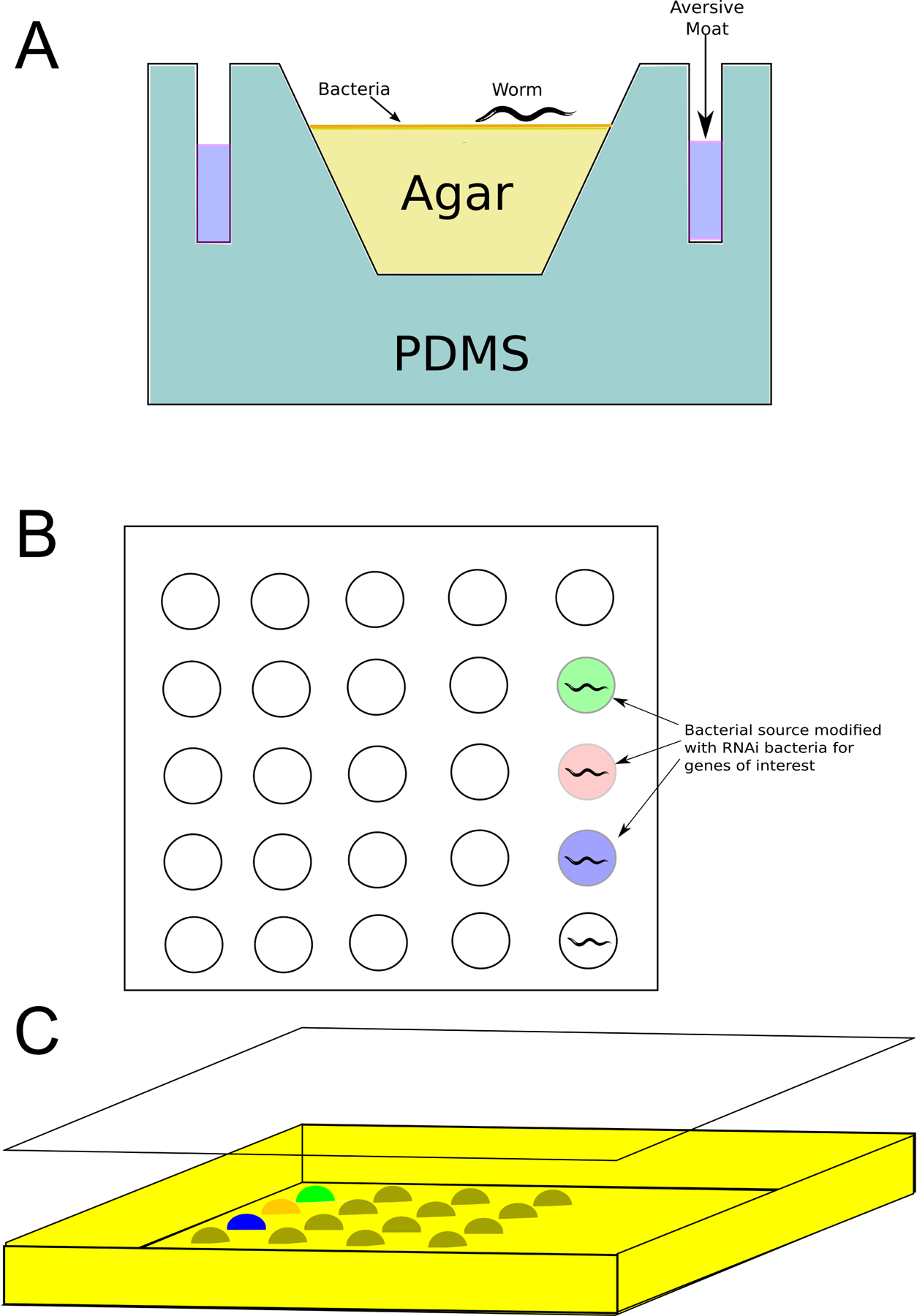
A. WorMotel single location. Aversion moat is filled with copper sulfate to isolate worms, solid NGM agar is placed into each well with a bacterial food source placed on top, to this a single egg is introduced. This system keeps each worm isolated to its single well within a larger 96 or 384 well plate. After Churgin et al. 2017. B. Design by Zhang et al. that placed individual bacteria mounds onto a surface of hygrogel. C. This is then crosslinked with polydimethylsiloxane (PDMS) everywhere this bacteria and hydrogel was absent. PDMS is a clear silicone that allows for easy imaging of each worm in their individual wells. Differing colors represent different bacterial food sources, such as dsRNA-producing bacteria.
For the original design, a digital camera was used to capture videos of the worms throughout their lifespan. These videos captured worm activity for 30 minutes twice a day. At the midpoint of each 30-minute interval, the worms were stimulated with a blue light. This has been previously shown to cause an escape response in worms, that is suspected to be a mechanism for these transparent, soil-dwelling organisms to avoid UV damage due to exposure to sunlight [39]. Here Churgin et al. report results for worms before this blue light stimulation and after in order to assay the worms’ spontaneous movement and their stimulated movement. For aging studies, the potential to more accurately note movement on older, more sedentary worms late in life could offer a great improvement in results during the later stages of a lifespan experiment. In this context, it is worth noting that this method of stimulation by exposure to blue light could be easily incorporated as a possible improvement to many of the other approaches described here.
2.1.3. Lifespan-on-a-chip for measurement of lifespan, behavioral, and physiological phenotypes
Lifespan-on-a-chip is a polydimethylsiloxane (PDMS) based microfluidic device that was designed specifically to score lifespans [40]. Like WormFarm, Lifespan-on-a-chip uses chambers to hold the worms that are fed by an inlet that can carry in fresh media and food, and an outlet that can remove progeny and waste (Fig. 3). In addition, this device can optionally incorporate a previous development by the authors that uses a wedge-shaped channel as a clamp to longitudinally immobilize individual worms [41]. This addition should enable imaging, or laser microdissection surgery [42], to be performed on specific individual worms from an aging cohort at any timepoint during their lifespan. Hulme et al. compare survival curves for wild type worms at 24°C, between the lifespan-on-a-chip device and 96-well plate culture. While the methods do not show perfect correspondence, this proof-of-principle suggests that the method as-is could be used to identify especially long or short lived genotypes or treatments, and further refinements could potentially lead to a closer correlation if desired.
Fig. 3:
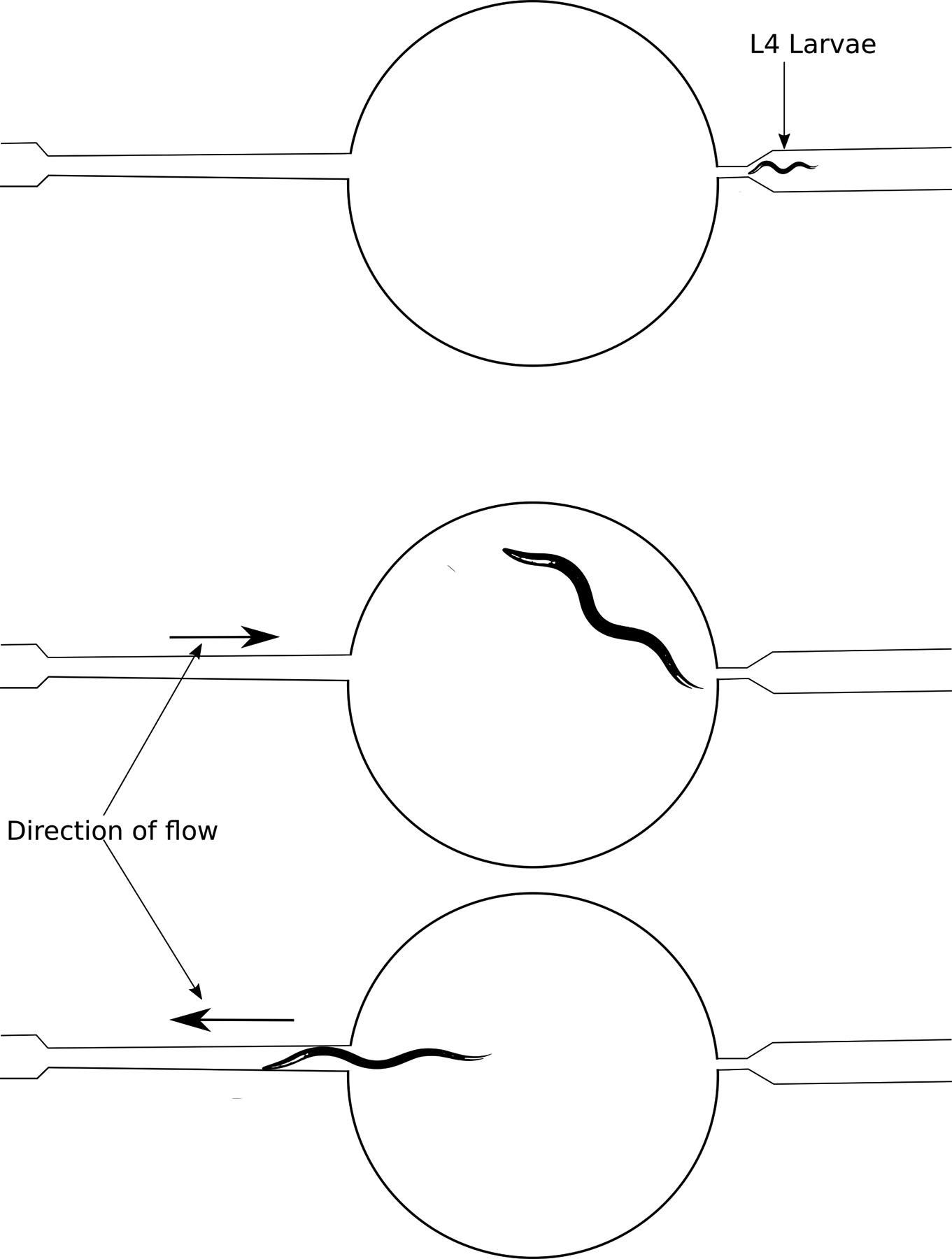
Lifespan-on-a-chip has control valves that allow movement of fluids in either direction, as indicated. This movement allows for temporary immobilization of worms for study. The right side of the system is just large enough for an L4 larvae to be pushed through, but too small for adult worms to exit.
More recently, a device using similar wedge-shaped worm clamps for non-lifespan studies has been developed including a recent addition that provides input from multiple sources, and individual outlets for waste removal [43]. Through this newer design, it is possible to monitor phenotypes such as motility, total length of worms, and number of progeny. This design allows for multiple different food sources through individual inlets, allowing for the introduction of different RNAi bacteria to individual worms, or a means of doing caloric restriction studies on a per-worm basis. Applying these changes to lifespan-focused studies could increase the potential longitudinal study options offered by this type of approach.
2.1.4. Automated measurement of reproductive aging
While we focus here primarily on methods that measure organismal survival, reproductive aging is another, closely related phenotype that is of great interest. In studying reproductive aging, a very successful approach has been reported that uses a microfluidic device coupled to a relatively sophisticated analysis pipeline. The analysis pipeline uses the LabView [44] graphical programming environment as well as MATLAB [45]. Using this approach [46], Li et al. were able to very accurately quantify the fecundity over time of C. elegans, in an automated fashion.
2.2. Flatbed scanner based approaches
Several early approaches to cost-effective automated lifespan analysis in C. elegans involved pioneering use of commercially available flatbed scanners (Fig. 4). These devices have been used in a multitude of biomedical assays, from colorimetric detection to the scanning of rat brain sections [47]. With a large visual field, and the possible use of multiple relatively inexpensive scanners, these systems can allow for increased throughput. Like microfluidic approaches, flatbed scanner-based methods are also capable of simultaneously assaying a number of other phenotypes during the course of the lifespan analysis. The evolution of flatbed scanner based methods has allowed for a number of modifications that require varying degrees of technical and computational expertise for their successful implementation, including a recent approach, Automated WormScan, that is designed to be reasonably easy for an interested experimenter to implement. As flatbed scanners themselves have become a somewhat specialized niche tool that is not undergoing intensive development, this technology does not necessarily benefit from the rapid improvements and implementations microfluidic devices enjoy due to their use in several differing research and commercial applications. Nevertheless, these approaches offer many advantages of their own. This is evidenced by the fact that they are still under active development, and that some of the largest single-experiment worm lifespan datasets to date have been generated using flatbed scanner-based approaches.
Fig. 4:
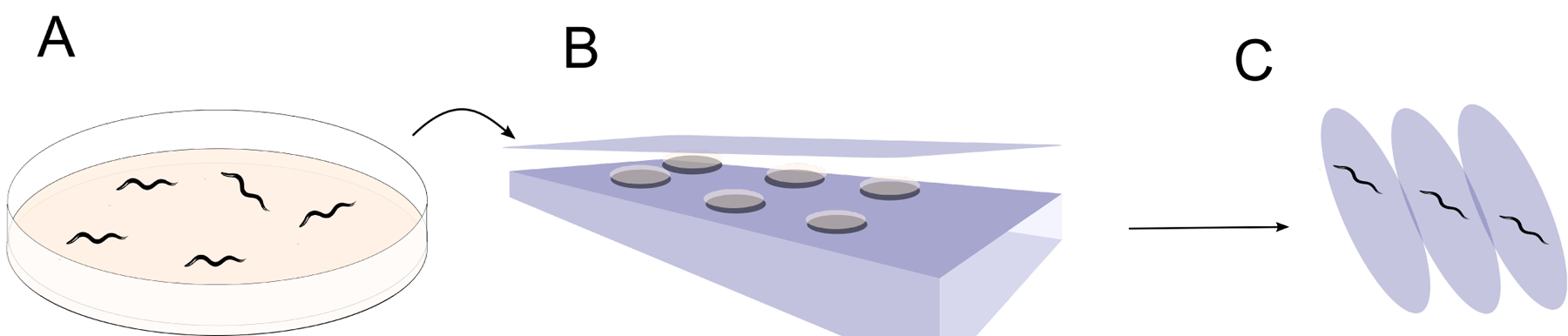
A. Representative worm solid media agar plate. B. Agar plate is flipped upside-down and placed on scanner to allow for image capture for specified time duration. C. Time lapse images depicting worm movement between images, as one method of assaying which worms are alive. After Stroustrup et al. 2013.
2.2.1. WormScan
One of the first uses of flatbed scanners for C. elegans survival scoring automation was the WormScan system [48]. This system utilizes a modified Epson Perfection V700 Photo Scanner, and measures worm movement in order to determine whether a worm is alive or dead. The scanner takes a few images of a field of plates over the course of a few minutes, and compares the consecutive images to note worms that have moved. For image analysis, the Advanced WEKA Segmentation [49] and AnalyzeSkeleton [50] plugins of the widely available open source FIJI package [51] were used in order to determine the size and movement of assayed worms. With continuous scanning, this system can precisely pinpoint the time of cessation of movement. This potential increase in time resolution offers another possible advantage of this system over once-a-day manual scoring, particularly for tests shorter assays involving acute stress survival analysis where all worms are dead within several hours. Assays like this would require scoring at least once per hour.
It is well known that aging worms often require stimulation in order to determine if they remain alive, generally in the form of a manual poke from a researcher. Here, the bright white light from the scanning bed itself was shown to stimulate the worms, in much the same manner as manual stimulation using a worm pick. Because the manual stimulation is not necessary when using this method, plates are able to remain closed. This greatly reduces the risk of any contamination that might necessitate transfer of worms to fresh plates, or lead to loss of data or reduction of overall cohort numbers due to censoring of worms lost to contamination. Although both increased temperature and high intensity light can affect lifespan in C. elegans, Mathew et al. note that they did not see any decreased survival over 18 hours of exposure to the light source in the Epson Perfection V700 scanner. Mathew et al. also note that these scanners also provide enough resolution to clearly distinguish L2 stage larvae from adult worms. This ability allows the software to be used for assays of fecundity, without the need to manually score progeny.
Most importantly, when used for survival analysis, there was no significant difference between manual worm scoring and the results of the flatbed scanner analysis. WormScan demonstrated accurate survival analysis for worms exposed to phosphine gas, as well as accurate analysis of worm size, and of worm fecundity. This system as well as other flatbed scanner methods utilize FUdR in order to eliminate growth of the progeny from the adult worms that being scored for lifespan. However, if treatment is not timed properly, then worms may still need to be manually transferred away from any progeny. Mathew et al. did note that they began with 30–150 individual worms per seeded 5.5cm plate in some of these experiments. This range of worm densities could easily lead to starvation on a plate of this size under normal worm culture conditions, without supplemental additions of food, over the length of time of a typical worm lifespan. This problem could be avoided by simply lowering the number of worms on each plate, although this would come at a cost of lowered overall throughput per flatbed scanner.
2.2.2. The Caenorhabditis elegans Lifespan Machine
Another flatbed scanner-based approach, the C. elegans Lifespan Machine [52], also used the Epson Perfection V700 Photo Scanner. Here, Stroustrup et al. heavily modified the stock Epson V700 photo scanner in order to extensively optimize their approach. To each flatbed scanner, they attached seven direct current (DC) powered fans of the type commonly used in desktop computer chassis cooling, along with a suitable DC power supply to drive the fans. This set of modifications allows regulation of the heat produced by the internal components of the scanner. In addition to this, in this approach the entire apparatus is also intended to be housed in a temperature-controlled room or incubator, in order to provide the greatest possible temperature control and stability. These efforts are significant due to the exquisite sensitivity of C. elegans lifespan to temperature [24,53,54]. Stroustrup et al. also made a modification to the optics of each scanner, manually adjusting the position of the flatbed scanner’s lens. This was done in order to move the scanner focal plane from the manufactured plane that is coincident with the surface of the scanner bed glass, to a new position above the glass that coincides with the plane of the agar surface on which the worms will live out their lives. In a truly impressive effort, Stroustrup et al. also developed their own very sophisticated analysis software from scratch for this platform. This software includes a component with its own custom interactive graphical user interface, the Worm Browser. Using a clever and relatively computationally intensive process that begins with machine-learning classification of worms from the scanner-derived images, this overall approach was able to provide extremely accurate and precise survival curves, that corresponded very well with hand-scored lifespan data generated using traditional manual manipulation [52,55].
2.2.3. Automated WormScan
One of the most recent adaptations of the flatbed scanner technology to the scoring of C. elegans lifespans is Automated WormScan [56]. This method iterates on the WormScan method [48], and again takes advantage of the Epson V700 photo scanner hardware platform, or alternatively may also use the newer upgraded V800 model scanner. Puckering et al. here thoroughly automate the image analysis and data acquisition aspect of lifespan and survival experiments in C. elegans. Automated WormScan utilizes a well-documented free and open source FIJI [51] plugin for image analysis, as well as a Microsoft Excel macro that is capable of automatically populating spreadsheets with the data extracted from the FIJI analysis. This analysis measures the number of moving worms by comparing the images of various consecutive scans. Further simplifying the use of this method, an automated installer for the software is available for the Windows platform, under the free and open source GNU General Public License. The provided software package requires little computational expertise to run, and is capable of greatly decreasing the time spent by experimenters on image analysis and data acquisition. This protocol also shifts worms from a temperature regulated environment to scanners left at ambient conditions, once per day, then back to the temperature-regulated environment. In this regard this approach is very similar to manual lifespan analysis, where small groups of plates are typically moved from a temperature controlled incubator to a lab bench for manual scoring once per day, then returned to the incubator. Here each batch of plates was only exposed to room temperature for 60 minutes or less for each time the plates were scored, which is intended to minimize the effect of temperature fluctuations on lifespan, or other phenotypes of interest. This approach eliminates the need to house the flatbed scanners inside a temperature-controlled room. Puckering et al. show that there is no significant difference between manual worm counting and that of the Automated WormScan. Although Automated WormScan was not used for lifespan analysis, it can easily be used for this assay by comparing daily scan results.
2.3. Other approaches: LFASS
Label-free automated survival scoring, or LFASS, takes advantage of an unusual phenotype previously observed by Coburn et al. and dubbed “death-associated fluorescence”. The intestines of C. elegans contain gut granules, which themselves exhibit blue fluorescence caused by a glycosylated form of anthranilic acid [57]. Diffuse spreading of this compound and its associated fluorescence is a reliable indicator of imminent death in C. elegans [58], and it is this phenotype that Coburn et al. exploit in the LFASS method. Using a standard 96- or 384-well plate reader (here a Tecan Infinite 200, Tecan Group Ltd., Switzerland), Coburn et al. are able to measure death-associated fluorescence of worms on standard NGM / E. coli OP50 media [59]. Unlike many of the other methods described here, the output is not a complete survival curve, but a very repeatable and accurate estimate of the median time of death. Although Benedetto et al. focus here on survival under acutely stressful conditions, such as heat shock, E. faecalis infection, and oxidative stress to to tert-butyl hydroperoxide (t-BHP), this method should in principle be easily adaptable to the longer time periods involved in normal C. elegans aging.
3. Other worm tracking and automation
Here we consider some approaches to tracking and automation in C. elegans that are not currently primarily applied to collection of survival or lifespan data, but that may serve as useful resources in the development and improvement of future C. elegans lifespan automation approaches.
3.1. The Movement Tracker and The Parallel Worm Tracker
The Movement Tracker [60], originally named as the Parallel Worm Tracker [61], is a system designed to automate behavioral analysis in C. elegans. Although initially described for measuring average speed of worms and drug-induced paralysis, it has broad applications for measuring organism movement and lifespan. Intended to track the movement of a single C. elegans over time, this open source, freely available software has been used for measurements in both C. elegans and Drosophila. Derivatives of these open source programs could potentially lend themselves to other uses in the future, specifically perhaps to the automation of C. elegans lifespan experiments.
3.2. QuantWorm
QuantWorm is a software package that has the capacity to look at both time-lapse images and videos, in order to computationally understand events including lifespan and mobility in C. elegans [62]. This package contains a data acquisition module called WormScanner, as well as four analysis modules called WormLocomotion, WormEgg, WormLength, and WormLifespan. For lifespan analysis, the WormLifespan software module uses a method that takes the differences between two images from two time points t0 and t1. The process looks at the changes between these two images, and highlights pixels that have changed between t0 and t1, and then measures those pixels to compare them to an established metric of typical size for adult worms. In this way, WormLifespan is able to identify a worm that has moved between the two frames collected at t0 and t1 respectively. If the measured change is within an acceptable range, the worm is scored as alive, and otherwise it is scored as dead. In order to validate this method for lifespan experiments, Jung et al. used measurements of worms grown on standard NGM media in a 6-well plate format. These were scanned with WormScanner once daily, and processed with WormLifespan. In a side-by-side comparison of manual scoring and automated scoring, Jung et al. reported that for n between 0 and 100 worms on each plate, the software was accurate to within 3.7+/−5.12 worms. This software has a number of advantages over manual lifespan scoring. By using individual pixels to determine if a worm is living or dead, worms may be determined alive where to they eye they could look as though they are not moving, giving more accurate lifespan results. The time between the two points t0 and t1 could also be increased in order to more accurately identify death which will otherwise be time consuming in manual scoring.
3.3. Wide Field-of-View Nematode Tracking Platform (WF-NTP)
The Wide Field-of-View Nematode Tracking Platform, or WF-NTP [63], uses a Python [64] program released under the open-source GNU General Public License to analyze data from a high-resolution USB camera. This software has been shown to accurately measure paralysis in worms, by quantifying worms’ bends-per-minute. The software allows for modifications to parameters controlling e.g. background subtraction, so that users can fine tune the output to their specific recording environment depending on the camera, light source, etc. that are available to them. This kind of tune-ability is exceptionally useful not only to users of the software as-is, but to developers hoping to extend the capabilities of this open source program in the future. Although Perni et al. did not perform a comparison for lifespan measurements against the gold standard of manual scoring, they propose that this software could be used for automated lifespan measurements.
4. Conclusions and future directions in the field
In the past several years, rapid advances have been made in the automation of survival assays for C. elegans. Diverse approaches have been used, from flatbed scanners to microfabricated devices, and all of these lines of development appear to be both promising, and currently ongoing. In parallel, software that can be used to analyze C. elegans survival data has also been developed using several relatively independent approaches. Some of these analysis pipelines are restricted in that they will work only with proprietary software or file formats. For a subset of these proprietary tools, such as MATLAB [45], free and open source alternatives such as GNU Octave [65] exist that may facilitate more rapid development of improved versions in the future. In several other cases, the analysis software is already freely available, open source, or both. In considering a platform to further develop or customize, there are extraordinary benefits to researchers that plan ahead to attempt to avoid lock-in to non-standard or highly proprietary hardware or software components. As automation-based approaches by their inherent nature strive for constantly increasing throughput, these methods may scale by multiple orders of magnitude over the course of their development. As a result, things that may at first seem to be small inconveniences may grow to become significant obstacles as development continues. A forward-looking approach, taking this type of scalability into account from the outset, can help thoughtful researchers avoid painting themselves into a corner [66].
As the kinds of high throughput approaches outlined here continue to improve, the larger and higher-resolution survival datasets generated by these methods will become more amenable to more advanced types of analysis by, quantitative methods such as systems biology [67] and network-based approaches [68]. These approaches themselves are currently undergoing rapid development. As a result, the methods described here and their successors and derivatives are likely to play an extremely important role in our understanding of aging in C. elegans, and perhaps in our understanding of the conserved biology of aging more generally.
Acknowledgments
This work was supported by National Institutes of Health - National Institute of General Medical Sciences P20GM121176 to M.M., and a Glenn Foundation for Medical Research - American Federation for Aging Research Junior Investigator Grant to M.M.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest to disclose.
References
- [1].Medawar PB, An Unsolved Problem of Biology, 1st ed, Lewis HK and Co., London, London. [Google Scholar]
- [2].Haldane JBS, New Paths in Genetics, 1st ed., Harper, London, 1942. [Google Scholar]
- [3].Kirkwood TBL, Austad SN, Why do we age?, Nature. 408 (2000) 233–238. [DOI] [PubMed] [Google Scholar]
- [4].Austad SN, Comparative Biology of Aging, J Gerontol A Biol Sci Med Sci. 64A (2009) 199–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kaeberlein M, Longevity and aging, F1000Prime Rep. 5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nikolich-Žugich J, Goldman DP, Cohen PR, Cortese D, Fontana L, Kennedy BK, Mohler MJ, Olshansky SJ, Perls T, Perry D, Richardson A, Ritchie C, Wertheimer AM, Faragher RGA, Fain MJ, Preparing for an aging world: Engaging biogerontologists, geriatricians, and the society, The Journals of Gerontology. Series A, Biological Sciences and Medical Sciences. 71 (2016) 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kenyon C, The nematode Caenorhabditis elegans, Science. 240 (1988) 1448–1453. [DOI] [PubMed] [Google Scholar]
- [8].Klass MR, A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results, Mech. Ageing Dev 22 (1983) 279–286. [DOI] [PubMed] [Google Scholar]
- [9].Friedman DB, Johnson TE, A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility, Genetics. 118 (1988) 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R, A C. elegans mutant that lives twice as long as wild type, Nature. 366 (1993) 461–464. [DOI] [PubMed] [Google Scholar]
- [11].McCormick M, Chen K, Ramaswamy P, Kenyon C, New genes that extend Caenorhabditis elegans’ lifespan in response to reproductive signals, Aging Cell. 11 (2012) 192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hansen M, Taubert S, Crawford D, Libina N, Lee S-J, Kenyon C, Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans, Aging Cell. 6 (2007) 95–110. [DOI] [PubMed] [Google Scholar]
- [13].Kaeberlein TL, Smith ED, Tsuchiya M, Welton KL, Thomas JH, Fields S, Kennedy BK, Kaeberlein M, Lifespan extension in Caenorhabditis elegans by complete removal of food, Aging Cell. 5 (2006) 487–494. [DOI] [PubMed] [Google Scholar]
- [14].Ayyadevara S, Alla R, Thaden JJ, Shmookler Reis RJ, Remarkable longevity and stress resistance of nematode PI3K-null mutants, Aging Cell. 7 (2008) 13–22. [DOI] [PubMed] [Google Scholar]
- [15].Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, Johnston ED, Tchao BN, Pak DN, Welton KL, Promislow DEL, Thomas JH, Kaeberlein M, Kennedy BK, Quantitative evidence for conserved longevity pathways between divergent eukaryotic species, Genome Res. 18 (2008) 564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].McCormick MA, Delaney JR, Tsuchiya M, Tsuchiyama S, Shemorry A, Sim S, Chou AC-Z, Ahmed U, Carr D, Murakami CJ, Schleit J, Sutphin GL, Wasko BM, Bennett CF, Wang AM, Olsen B, Beyer RP, Bammler TK, Prunkard D, Johnson SC, Pennypacker JK, An E, Anies A, Castanza AS, Choi E, Dang N, Enerio S, Fletcher M, Fox L, Goswami S, Higgins SA, Holmberg MA, Hu D, Hui J, Jelic M, Jeong K-S, Johnston E, Kerr EO, Kim J, Kim D, Kirkland K, Klum S, Kotireddy S, Liao E, Lim M, Lin MS, Lo WC, Lockshon D, Miller HA, Moller RM, Muller B, Oakes J, Pak DN, Peng ZJ, Pham KM, Pollard TG, Pradeep P, Pruett D, Rai D, Robison B, Rodriguez AA, Ros B, Sage M, Singh MK, Smith ED, Snead K, Solanky A, Spector BL, Steffen KK, Tchao BN, Ting MK, Wende HV, Wang D, Welton KL, Westman EA, Brem RB, Liu X, Suh Y, Zhou Z, Kaeberlein M, Kennedy BK, A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging, Cell Metab. 22 (2015) 895– 906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Solis GM, Kardakaris R, Valentine ER, Bar-Peled L, Chen AL, Blewett MM, McCormick MA, Williamson JR, Kennedy B, Cravatt BF, Petrascheck M, Translation attenuation by minocycline enhances longevity and proteostasis in old post-stress-responsive organisms, Elife. 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cabreiro F, Au C, Leung K-Y, Vergara-Irigaray N, Cochemé HM, Noori T, Weinkove D, Schuster E, Greene NDE, Gems D, Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism, Cell. 153 (2013) 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen J, Ou Y, Li Y, Hu S, Shao L-W, Liu Y, Metformin extends C elegans lifespan through lysosomal pathway, Elife. 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin M-J, Schwab M, Pollak M, Zhang Y, Yu Y, Becker KG, Bohr VA, Ingram DK, Sinclair DA, Wolf NS, Spindler SR, Bernier M, de Cabo R, Metformin improves healthspan and lifespan in mice, Nat Commun. 4 (2013) 2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sutphin GL, Kaeberlein M, Measuring Caenorhabditis elegans Life Span on Solid Media, J Vis Exp. (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Park H-EH, Jung Y, Lee S-JV, Survival assays using Caenorhabditis elegans, Mol. Cells 40 (2017) 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hosono R, Sterilization and growth inhibition of Caenorhabditis elegans by 5-fluorodeoxyuridine, Exp. Gerontol 13 (1978) 369–374. [DOI] [PubMed] [Google Scholar]
- [24].Hosono R, Mitsui Y, Sato Y, Aizawa S, Miwa J, Life span of the wild and mutant nematode Caenorhabditis elegans. Effects of sex, sterilization, and temperature, Exp. Gerontol 17 (1982) 163– 172. [DOI] [PubMed] [Google Scholar]
- [25].Aitlhadj L, Stürzenbaum SR, The use of FUdR can cause prolonged longevity in mutant nematodes, Mech. Ageing Dev 131 (2010) 364–365. [DOI] [PubMed] [Google Scholar]
- [26].Anderson EN, Corkins ME, Li J-C, Singh K, Parsons S, Tucey TM, Sorkaç A, Huang H, Dimitriadi M, Sinclair DA, Hart AC, elegans C lifespan extension by osmotic stress requires FUdR, base excision repair, FOXO, and sirtuins, Mech. Ageing Dev 154 (2016) 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kato Y, Miyaji M, Zhang-Akiyama Q-M, FUdR extends the lifespan of the short-lived AP endonuclease mutant in Caenorhabditis elegans in a fertility-dependent manner, Genes Genet. Syst 91 (2017) 201–207. [DOI] [PubMed] [Google Scholar]
- [28].Van Raamsdonk JM, Hekimi S, FUdR causes a twofold increase in the lifespan of the mitochondrial mutant gas-1, Mech. Ageing Dev 132 (2011) 519–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Costs of Services - Biology of Aging at UW, (n.d.). http://www.uwaging.org/shock-center/costs (accessed September 2, 2019).
- [30].Services Our, (n.d.). https://magnitudebiosciences.com/services (accessed September 2, 2019).
- [31].Services | NemaLife Inc. | United States, NemaLife Inc (n.d.). https://www.nemalifeinc.com/services (accessed September 2, 2019).
- [32].Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE, Soft lithography in biology and biochemistry, Annu Rev Biomed Eng. 3 (2001) 335–373. [DOI] [PubMed] [Google Scholar]
- [33].Tokeshi M, Applications of microfluidic systems in biology and medicine, Springer, 2019. [DOI] [PubMed] [Google Scholar]
- [34].Xian B, Shen J, Chen W, Sun N, Qiao N, Jiang D, Yu T, Men Y, Han Z, Pang Y, Kaeberlein M, Huang Y, Han J-DJ, WormFarm: a quantitative control and measurement device toward automated Caenorhabditis elegans aging analysis, Aging Cell. 12 (2013) 398–409. [DOI] [PubMed] [Google Scholar]
- [35].Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J, Systematic functional analysis of the Caenorhabditis elegans genome using RNAi, Nature. 421 (2003) 231–237. [DOI] [PubMed] [Google Scholar]
- [36].Mitchell DH, Stiles JW, Santelli J, Sanadi DR, Synchronous Growth and Aging of Caenorhabditis elegans in the Presence of Fluorodeoxyuridine, J Gerontol. 34 (1979) 28–36. [DOI] [PubMed] [Google Scholar]
- [37].Churgin MA, Jung S-K, Yu C-C, Chen X, Raizen DM, Fang-Yen C, Longitudinal imaging of Caenorhabditis elegans in a microfabricated device reveals variation in behavioral decline during aging, ELife. 6 (2017) e26652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang WB, Sinha DB, Pittman WE, Hvatum E, Stroustrup N, Pincus Z, Extended Twilight among Isogenic C. elegans Causes a Disproportionate Scaling between Lifespan and Health, Cell Systems. 3 (2016) 333–345.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Edwards SL, Charlie NK, Milfort MC, Brown BS, Gravlin CN, Knecht JE, Miller KG, A Novel Molecular Solution for Ultraviolet Light Detection in Caenorhabditis elegans, PLOS Biology. 6 (2008) e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Elizabeth Hulme S, Shevkoplyas SS, McGuigan AP, Apfeld J, Fontana W, Whitesides GM, Lifespan-on-a-chip: microfluidic chambers for performing lifelong observation of C . elegans, Lab on a Chip. 10 (2010) 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hulme SE, Shevkoplyas SS, Apfeld J, Fontana W, Whitesides GM, A microfabricated array of clamps for immobilizing and imaging C. elegans, Lab Chip. 7 (2007) 1515–1523. [DOI] [PubMed] [Google Scholar]
- [42].Allen PB, Sgro AE, Chao DL, Doepker BE, Scott Edgar J, Shen K, Chiu DT, Single-synapse ablation and long-term imaging in live C. elegans, J. Neurosci. Methods 173 (2008) 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Baris Atakan H, Cornaglia M, Mouchiroud L, Auwerx J, Gijs MAM, Automated high-content phenotyping from the first larval stage till the onset of adulthood of the nematode Caenorhabditis elegans, Lab on a Chip. 19 (2019) 120–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yang Y, LabVIEW graphical programming cookbook: 69 recipes to help you build, debug, and deploy modular applications using LabVIEW, Packt Pub, Birmingham, 2014. [Google Scholar]
- [45].Attaway S, MATLAB: a practical introduction to programming and problem solving, 5th ed., Butterworth-Heinemann, 2019. [Google Scholar]
- [46].Li S, Stone HA, Murphy CT, A microfluidic device and automatic counting system for the study of C. elegans reproductive aging, Lab Chip. 15 (2015) 524–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Göröcs Z, Ozcan A, Biomedical imaging and sensing using flatbed scanners, Lab Chip. 14 (2014) 3248–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mathew MD, Mathew ND, Ebert PR, WormScan: A Technique for High-Throughput Phenotypic Analysis of Caenorhabditis elegans, PLoS One. 7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Witten IH, Frank E, Hall MA, Pal CJ, Data Mining, Fourth Edition: Practical Machine Learning Tools and Techniques, 4th ed., Morgan Kaufmann Publishers Inc., San Francisco, CA, USA, 2016. [Google Scholar]
- [50].Arganda-Carreras I, Fernández-González R, Muñoz-Barrutia A, Ortiz-De-Solorzano C, 3D reconstruction of histological sections: Application to mammary gland tissue, Microsc. Res. Tech 73 (2010) 1019–1029. [DOI] [PubMed] [Google Scholar]
- [51].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis, Nat. Methods 9 (2012) 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Stroustrup N, Ulmschneider BE, Nash ZM, López Moyado IF, Apfeld J, Fontana W, The C elegans Lifespan Machine, Nat Methods. 10 (2013) 665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Miller H, Fletcher M, Primitivo M, Leonard A, Sutphin GL, Rintala N, Kaeberlein M, Leiser SF, Genetic interaction with temperature is an important determinant of nematode longevity, Aging Cell. 16 (2017) 1425–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lee S-J, Kenyon C, Regulation of the longevity response to temperature by thermosensory neurons in Caenorhabditis elegans, Curr. Biol 19 (2009) 715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Stroustrup N, Anthony WE, Nash ZM, Gowda V, Gomez A, López-Moyado IF, Apfeld J, Fontana W, The temporal scaling of Caenorhabditis elegans ageing, Nature. 530 (2016) 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Puckering T, Thompson J, Sathyamurthy S, Sukumar S, Shapira T, Ebert P, Automated Wormscan, F1000Res. 6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Coburn C, Allman E, Mahanti P, Benedetto A, Cabreiro F, Pincus Z, Matthijssens F, Araiz C, Mandel A, Vlachos M, Edwards S-A, Fischer G, Davidson A, Pryor RE, Stevens A, Slack FJ, Tavernarakis N, Braeckman BP, Schroeder FC, Nehrke K, Gems D, Anthranilate fluorescence marks a calcium-propagated necrotic wave that promotes organismal death in C. elegans, PLoS Biol. 11 (2013) e1001613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Coburn C, Gems D, The mysterious case of the C. elegans gut granule: death fluorescence, anthranilic acid and the kynurenine pathway, Front Genet. 4 (2013) 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Benedetto A, Bambade T, Au C, Tullet JMA, Monkhouse J, Dang H, Cetnar K, Cabreiro F, Chan B, Gems D, New label-free automated survival assays reveal unexpected stress resistance patterns during C. elegans aging, Aging Cell. 0 (2019) e12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Mouchiroud L, Sorrentino V, Williams EG, Cornaglia M, Frochaux MV, Lin T, Nicolet-Dit-Félix AA, Krishnamani G, Ouhmad T, Gijs MAM, Deplancke B, Auwerx J, The Movement Tracker: A Flexible System for Automated Movement Analysis in Invertebrate Model Organisms, Curr Protoc Neurosci. 77 (2016) 8.37.1–8.37.21. [DOI] [PubMed] [Google Scholar]
- [61].Ramot D, Johnson BE, Jr TLB, Carnell L, Goodman MB, The Parallel Worm Tracker: A Platform for Measuring Average Speed and Drug-Induced Paralysis in Nematodes, PLOS ONE. 3 (2008) e2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jung S-K, Aleman-Meza B, Riepe C, Zhong W, QuantWorm: A Comprehensive Software Package for Caenorhabditis elegans Phenotypic Assays, PLOS ONE. 9 (2014) e84830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Perni M, Challa PK, Kirkegaard JB, Limbocker R, Koopman M, Hardenberg MC, Sormanni P, Müller T, Saar KL, Roode LWY, Habchi J, Vecchi G, Fernando N, Casford S, Nollen EAA, Vendruscolo M, Dobson CM, Knowles TPJ, Massively parallel C elegans tracking provides multi-dimensional fingerprints for phenotypic discovery, Journal of Neuroscience Methods. 306 (2018) 57–67. [DOI] [PubMed] [Google Scholar]
- [64].Lutz M, Mark, Learning Python, 5th ed., O’ Reilly, n.d. [Google Scholar]
- [65].Schmidt Hansen J, GNU Octave: beginner’s guide : become a proficient Octave user by learning this high-level scientific numerical tool from the ground up, Packt Pub, Birmingham, 2011. [Google Scholar]
- [66].Stallman R, Free community science and the free development of science, PLoS Medicine. 2 (2005) e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].McCormick MA, Promislow DEL, Recent Advances in the Systems Biology of Aging, Antioxid. Redox Signal 29 (2018) 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].McCormick MA, Promislow DE, Networks in the biology of aging, Annual Review of Gerontology and Geriatrics. 34 (2013). [Google Scholar]


