Abstract
Salmonella is regarded as the predominant cause of foodborne illnesses worldwide, and the increase of these antimicrobial-resistant strains makes it more difficult to prevent. On this occasion, bacteriophages (phages) stand out as an alternative biocontrol agent with high efficiency and low mutation rates. Salmonella phages have confronted challenges to counteract with more than 2,500 serovars of Salmonella spp. and overcome the universality of antibiotics to different species, and thus, broad-host-range phages infecting Salmonella spp. are urgently required to realize precise poultry treatment or clinical therapy. First, phage STP4-a was screened to have a broad host range through bioinformatics analysis, and then the host range assay proved that phage STP4-a could inhibit 88 out of 91 Salmonella strains. Then, in silico analysis excluded the possibility of phage STP4-a possessing any known lysogeny factors, toxins, pathogen-related genes, or foodborne allergens, and oral toxicity studies further ensured the safety of unknown factors or suspected risks. In addition, strong inhibition effects of phage STP4-a were seen on both single Salmonella strain and multiple Salmonella strains in vitro, reducing 3-5 log in 30 min. Phage STP4-a could survive and keep more than 50% activity in simulated stomach or intestine environments in vitro. In terms of antimicrobial activities in chickens, pretreatment with phage STP4-a was the most efficient approach to Salmonella biocontrol, non-detectable in feces during the 14-day experimental period. Therefore, phage STP4-a was an extremely broad-host-range and safe biocontrol agent, performing its potential as a food additive or therapeutic drug in poultry industry.
Key words: Salmonella bacteriophage, broad-host-range, safety, antimicrobial activity, poultry industry
Introduction
In recent years, multidrug-resistant bacteria have emerged and become more and more serious due to the wide spread of antibiotic resistance genes, becoming a global health crisis. It has been estimated that annual death toll from drug-resistant infections would increase from currently 700,000 to 10 million and would cost US$100 trillion worldwide by 2,050 if no actions were taken (O'Neill, 2014). In particular, China is the largest consumer of antibiotics in the world; for example, a total of 162,000 tons of antibiotics were used in human and animals in 2013 (Zhang et al., 2015, Yassin et al., 2017). Consequently, China has confronted the greatest challenge of drug resistance in the world, and the safety of humans and animals was greatly threatened. To solve the increasing serious problem of antibiotic resistance, substitutes for antibiotics have gained more attention.
Bacteriophages (abbr. phages), that is, viruses to bacteria, are regarded as a good choice in combating drug resistance due to its high efficiency and low mutation rates (Hagens and Loessner, 2007, Zhang and Buckling, 2012). In addition, phages show their great advantages over antibiotics in that they can usually kill just one species or strain, giving a more precise way to target pathogenic bacteria (Domingo-Calap and Delgado-Martinez, 2018). In 2014, phage therapy was listed as one of seven promising solutions in the plan of US National Institute of Allergy and Infectious Diseases to counteract with antibiotic resistance (Reardon, 2014). Then, Grégory Resch of the University of Lausanne reported the Phagoburn Plan to use phage therapy in the clinical treatment for human infections (Reardon, 2014). Therefore, phages are considered to be ideal biocontrol agents in various fields, such as food protection, pathogen therapy, and so on (Atterbury et al., 2007, Shahin and Bouzari, 2018). However, it will be a disadvantage if the specificity of phages is too high for further application. For example, Salmonella enterica, a gram-negative facultative anaerobe, is one of the most frequently reported causes of foodborne illnesses worldwide, leading to 155,000 deaths every year (Eng et al., 2015), and poultry was the main pathogenic vehicle (Heredia and Garcia, 2018). Salmonella contains more than 2,500 serovars (Coburn et al., 2007), which makes it a huge challenge to prevent bacterial infections with such specific phages. Thus, broad-host-range Salmonella phages are in great demands to realize precise poultry treatment or clinical therapy while avoiding antibiotic resistance.
To the best of our knowledge, more than 160 nucleotide sequences of Salmonella phages have been reported in National Center for Biotechnology Information (NCBI) till now. Nonetheless, only about 16% of these phages were proved to have broad lysis spectrum properties, such as Salmonella phages VB-SenS-Ent1 (NC_019539.1) (Turner et al., 2012), SFP10 (Park et al., 2012), and PhiSG-JL2 (Kwon et al., 2008). Hence, broad-host-range phages are valuable resources to be explored because of its low proportion, and their safety and efficacy need to be evaluated ahead of permission in food control or infectious therapy. For instance, ListexP100 (containing phage P100) was firstly considered as GRAS (generally recognized as safe, GRAS Notices GRN000218) among phages by FDA in 2007, which has already been approved as a food additive for the control of Listeria monocytogenes in ready-to-eat foods (Bren, 2007). The safety and efficacy of phage P100 was studied comprehensively to ensure its acceptance. Then some phage products, such as EcoShield (Boyacioglu et al., 2013) and SalmoFresh (Sukumaran et al., 2016), have been granted approval successively by FDA to prepare phage cocktails for specific biocontrol. Kutateladze and Resch believed that more phages should be ready for updating phage products, similar to the seasonal influenza vaccine (Reardon, 2014). Currently reported phages were far from the aim to cover more than 2,500 serovars of Salmonella spp., and thus, broad-host-range phages and their safety and efficacy were indispensably required to be figured out for a thoughtful shield for Salmonella spp.
In this work, we analyzed the host range, safety, and animal therapy of phage STP4-a, which was first isolated in our previous work with the host of Salmonella Typhimurium ATCC 14028. On the one hand, compared with the bioinformatics assessment, we determined the lysis spectrum of phage STP4-a using 91 Salmonella strains, containing different species, subspecies, and serovars. On the other hand, the safety and biocontrol effects of phage STP4-a were taken into consideration in the comprehensive assessment of it as an antimicrobial agent targeting Salmonella spp., which was performed by combining bioinformatics analysis and animal experiments.
Materials and methods
Bacteria and Phages
Phage STP4-a was previously isolated from the sewage treatment plant in Qingdao, China, by using S. Typhimurium ATCC 14028 as the host strain, which was stored at China Center for Type Culture Collection (CCTCC) with the accession number CCTCC M2014145. There were 95 strains, containing 91 Salmonella strains, 2 Escherichia coli (E. coli) strains, and 2 Klebsiella pneumonia (K. pneumonia) strains being used for the host range assay (Table 1). All strains were grown overnight in nutrient broth at 37°C with shaking. Among those strains, all the WFS/E/K series were isolated from the sick chicken and the rest were kindly provided by Dr. Xiuping Jiang (Clemson University, US). The proliferation of phage STP4-a was administrated according to the literature of Li et al. (2016), and the final titer of phage STP4-a could reach up to 1.5 × 1010 PFU/mL.
Table 1.
Phage STP4-a susceptibilities of different strains used in this study.
| Strain name | Other designation | Origin | Susceptibility | Intensity1 |
|---|---|---|---|---|
| Salmonella Enteritidis | H4639 | UGA-CFS (Dr. Doyle) | + | +++ |
| Salmonella Enteritidis | H2292 | UGA-CFS (Dr. Doyle) | + | +++ |
| Salmonella Enteritidis | H3353 | UGA-CFS (Dr. Doyle) | + | +++ |
| Salmonella Enteritidis | ME18 | UGA-CFS (Dr. Doyle) | + | +++ |
| Salmonella Enteritidis | H4717 | UGA-CFS (Dr. Doyle) | + | +++ |
| Salmonella Enteritidis | Benson | UGA-CFS (Dr. Doyle) | + | +++ |
| Salmonella Enteritidis | H4639(gfp-labeled) | Dr. Xiuping Jiang | + | +++ |
| Salmonella Enteritidis | H2292(gfp-labeled) | Dr. Xiuping Jiang | + | +++ |
| Salmonella Enteritidis | H3353(gfp-labeled) | Dr. Xiuping Jiang | + | +++ |
| Salmonella Enteritidis | ME18(gfp-labeled) | Dr. Xiuping Jiang | + | +++ |
| Salmonella Enteritidis | H4717(gfp-labeled) | Dr. Xiuping Jiang | + | +++ |
| Salmonella Enteritidis | Benson(gfp-labeled) | + | +++ | |
| Salmonella Enteritidis | 15060 | USDA-FSIS | + | +++ |
| Salmonella Enteritidis | 30661 | FDA | + | +++ |
| Salmonella Enteritidis | N9166, chicken breast | Dr. Sonya M. Jones | + | +++ |
| Salmonella Enteritidis | N19847, chicken breast | Dr. Sonya M. Jones | + | +++ |
| Salmonella Enteritidis | N9136, chicken breast | Dr. Sonya M. Jones | + | +++ |
| Salmonella Enteritidis | N16444, chicken breast | Dr. Sonya M. Jones | + | +++ |
| Salmonella Enteritidis | N19890, chicken breast | Dr. Sonya M. Jones | + | +++ |
| Salmonella Enteritidis | WFS-002 | Sick poultry, isolate | + | +++ |
| Salmonella Enteritidis | WFS-003 | Sick poultry, isolate | + | +++ |
| Salmonella Enteritidis | WFS-004 | Sick poultry, isolate | + | +++ |
| Salmonella Enteritidis | WFS-005 | Sick poultry, isolate | + | +++ |
| Salmonella Enteritidis | WFS-006 | Sick poultry, isolate | + | +++ |
| Salmonella Enteritidis | WFS-007 | Sick poultry, isolate | + | +++ |
| Salmonella Enteritidis | WFS-012 | Sick poultry, isolate | + | +++ |
| Salmonella Enteritidis | WFS-013 | Sick poultry, isolate | + | +++ |
| Salmonella Enteritidis | WFS-014 | Sick poultry, isolate | + | +++ |
| Salmonella Enteritidis | WFS-015 | Sick poultry, isolate | + | +++ |
| Salmonella Typhimurium | mutant (Avirulent) | WSU (Dr. Curtis) | + | ++ |
| Salmonella Typhimurium | DT104 97-18-448 | UGA-CFS (Dr. Doyle) | + | +++ |
| Salmonella Typhimurium | var. copenhagen, SS/034, CeftriaxoneR | ISU (Dr. Fey) | + | +++ |
| Salmonella Typhimurium | DT104 ATCC 700408 ISSA GFP | CSU | + | +++ |
| Salmonella Typhimurium | avirulent 8243 (Rifr) | UGA | + | +++ |
| Salmonella Typhimurium | 32463 | FDA | + | +++ |
| Salmonella Typhimurium | ATCC 14028 | ATCC | + | +++ |
| Salmonella Typhimurium | SD11, aroA64 pGFPuv. Avirulent | Dr. Cathy Webb | + | S |
| Salmonella Typhimurium | UK-1 8234# | Dr. Curtiss, WU | + | +++ |
| Salmonella Typhimurium | SL1344 8770# | Dr. Curtiss, WU | + | +++ |
| Salmonella Typhimurium | SL1344 8499# | Dr. Curtiss, WU | + | +++ |
| Salmonella Typhimurium | SL1344 8292# | Dr. Curtiss, WU | + | +++ |
| Salmonella Typhimurium | UK-1 8768# | Dr. Curtiss, WU | + | +++ |
| Salmonella Typhimurium | CMCC (B) 50115 | CMCC | + | +++ |
| Salmonella Chloreasuis | ATCC 8326 | ATCC | + | +++ |
| Salmonella Chloreasuis | ATCC 15480 | ATCC | + | +++ |
| Salmonella Chloreasuis | ATCC 10708 | ATCC | + | ++ |
| Salmonella Senftenberg | ATCC 43845 | ATCC (Roy Curtiss III, Washington University) | - | - |
| Salmonella Newport | H9113 | CDC (Dr. Wu) | + | ++ |
| Salmonella Newport | H9116 | CDC (Dr. Wu) | + | ++ |
| Salmonella Poona | H9301 | CDC (Dr. Wu) | + | ++ |
| Salmonella Poona | H9 G77 | CDC (Dr. Wu) | + | ++ |
| Salmonella Heidleberg | UNL | CDC (Dr. Wu) | + | +++ |
| Salmonella Tenessee | K-4720, peanut butter outbreak | CDC | + | ++ |
| Salmonella Kentucky | N11150 | FDA | - | - |
| Salmonella Anatum | N5396 | FDA | + | ++ |
| Salmonella Newport | N635 | FDA | + | +++ |
| Salmonella Heidelberg | 21380 | FDA | - | - |
| Salmonella Dublin | 23742 | FDA | + | ++ |
| Salmonella St. Paul | 22398 | FDA | + | ++ |
| Salmonella Schwarzengrund | NPAL-1010158-1 | + | ++ | |
| Salmonella Montevideo | NPAL-1010158-2 | + | S | |
| Salmonella Cerro | NPAL-1010274-1 | + | ++ | |
| Salmoenlla Infantis | NPAL-1010651-1 | + | ++ | |
| Salmonella species | Group C1 | NPAL-1011026-1 | + | ++ |
| Salmonella species | Group C1 | NPAL-1011026-2 | + | ++ |
| Salmoenlla Montevideo | NPAL-1011026-3 | + | ++ | |
| Salmonella Winston | NPAL-1011391-1 | + | ++ | |
| Salmonella Winston | NPAL-1011391-2 | + | ++ | |
| Salmonella Minnesota | NPAL-1011391-3 | + | S | |
| Salmonella Tennessee | NPAL-1011664-1 | + | ++ | |
| Salmonella Amager | NPAL-1011664-2 | + | ++ | |
| Salmonella species | Poly F | NPAL-1011687-1 | + | ++ |
| Salmonella species | Group B | NPAL-1011940-1 | + | - |
| Salmonella Orion | NPAL-1012109-1 | + | +++ | |
| Salmonella Infantis | NPAL-1012634-1 | + | ++ | |
| Salmonella species | Livingstone Var. O 14+ | NPAL-1012688-1 | + | ++ |
| Salmonella Rissen | NPAL-1013032-1 | + | ++ | |
| Salmonella Derby | NPAL-1013080-1 | + | ++ | |
| Salmonella species | Group E4 | NPAL-1013471-1 | + | S |
| Salmonella Kentucky | NPAL-1013996-1 | + | ++ | |
| Salmonella species | Orion Var. O 15+ | NPAL-1014374-1 | + | +++ |
| Salmonella Infantis | NPAL-1014374-2 | + | ++ | |
| Salmonella Dabou or Corvallis | NPAL-1014710-1 | + | ++ | |
| Salmonella species | Group C1 | NPAL-1014710-2 | + | + |
| Salmonella Molade | NPAL-1014864-1 | + | ++ | |
| Salmonella Infantis | NPAL-1015324-1 | + | ++ | |
| Salmonella Senftenberg | NPAL-1015386-1 | + | S | |
| Salmonella Montevideo | Factor 14 negative | NPAL-1015543-1 | + | ++ |
| Salmonella species | Group C1 | NPAL-10156268-1 | + | S |
| Salmonella species | Group E4 | NPAL-1016346-1 | + | ++ |
| Salmonella species | Group E1 | NPAL-1016346-2 | + | S |
| Escherichia coli | WFE-001 | Sick poultry, isolate | - | - |
| Escherichia coli | WFE-011 | Sick poultry, isolate | - | - |
| Klebsiella pneumoniae | WFK-008 | Sick poultry, isolate | - | - |
| Klebsiella pneumoniae | WFK-009 | Sick poultry, isolate | - | - |
The intensity results were recorded as follows: +++, confluent lysis; ++, semiconfluent lysis; +, individual plaque; S. shadow lysis; -, no lysis.
Bioinformatics Analysis of Phage STP4-a
The genomic DNA of phage STP4-a was extracted according to the manufacturer's instruction of the TIANamp Virus DNA/RNA Kit obtained from Tiangen Biotech Co., Ltd. (Beijing, China) and then sequenced with the Roche 454 GS FLX + Titanium sequencer in Personalbio (Shanghai, China). After DNA extraction and sequencing, the genomic sequence of phage STP4-a was analyzed to evaluate its host range and safety. The 259 open reading frames (ORF) were compared with known structural or toxic protein sequences in NCBI with Basic Local Alignment Search Tool (BLAST). Computed molecular weights and isoelectric points (pIs) of phage STP4-a gene products were also predicted with proteomic tools from ExPASy (http://www.expasy.org/proteomics). Sequence Search Allergen Database (http://www.allergenonline.org/) was applied to examine the putative food allergens in phage STP4-a encoded proteins. Cleavage sites of phage STP4-a were analyzed with reported restriction enzymes at REBASE (http://rebase.neb.com/rebase/).
Host Range Analysis of Phage STP4-a
The host range of phage STP4-a was tested by the spotting method with 10 μL phages (108-109 PFU/mL) dripped onto the plates, which were spread with different bacterial strains incubated overnight using swabs. Totally 95 strains were used in the host range assay, and the test was repeated 3 times. The appearance of transparent plaques after incubating overnight at 37°C determined the host range of phage STP4-a. In addition, the lysis intensity was evaluated via overlay method. In short, bacterial strains and phages were mixed at an MOI value of 1 (106 CFU/106 PFU) in 5 mL of soft agar [Trypton soya broth/Trypton soya agar, v/v = 1:1] and then poured onto Trypton soya agar plates. Plates were incubated overnight at 37°C. The lysis intensity was classified as confluent lysis, semiconfluent lysis, individual plaque, shadow lysis, or no lysis according to the shape of plaques.
Oral Toxicity Studies of Phage STP4-a With Mice
An acute toxicity study of phage STP4-a was conducted according to OECD Guideline for Testing of Chemicals Acute Oral Toxicity-Fixed Dose Procedure (Organization for Economic Co-operation and Development, 2001). The oral toxicity test was performed with 15 female BALB/c mice (6 wk old, weighing 17.4 ± 0.9 g) and 15 male BALB/c mice (8 wk old, weighing 22.0 ± 0.7 g) (Vital River Laboratories, Beijing, China). Female (F)/male (M) mice were separated randomly into 3 groups, and fresh water and food were provided ad libitum during the experimental period, with a temperature of 22 ± 3°C and humidity of about 30-70%.
Different treatments were shown in Table 2. An aliquot of phage STP4-a (1.5 × 1010 PFU/mL) suspended in the SM buffer (5.8 g/L NaCl, 2 g/L MgSO4•7H2O, and 50 mL/L 1M pH 7.5 Tris-HCl) was prepared to be orally administrated to group F3/M3, which was performed according to body weight (1 mL/100 g body weight) as suggested in the OECD guidelines. The group F1/M1 received SM buffer only and group F2/M2 were treated with inactive phage STP4-a (the same propagation batch) in the same ratio as the control groups. The inactive phage STP4-a was treated by autoclaving at 121°C for 15 min. Mice were provided with water but no food for 3 h before dosing and regained food after a 2-h fastening. During the animal experiment, all processes were conducted in the light of the guidelines of the ethical committee of experimental animal care at Ocean University of China, Qingdao, China.
Table 2.
Different treatments on mice in the bacteriophage STP4-a safety assessment experiment.
| Groups | Number of mice | Treatment |
|---|---|---|
| F1 | 5 | Oral administration with SM buffer |
| F2 | 5 | Oral administration with inactivated bacteriophage STP4-a1 |
| F3 | 5 | Oral administration with bacteriophage STP4-a2 |
| M1 | 5 | Oral administration with SM buffer |
| M2 | 5 | Oral administration with inactivated bacteriophage STP4-a1 |
| M3 | 5 | Oral administration with bacteriophage STP4-a2 |
Five mice were orally administrated with phage STP4-a at the titer of 1.5 × 1010 pfu/mL/mouse.
Five mice were orally administrated with phage STP4-a autoclave-inactivated at the titer of 1.5 × 1010 pfu/mL/mouse.
We gave special attention to changes in weights, pharmacological effects, morbidity, and mortality of animals daily for continuous 14 D. On day 15, all surviving animals were weighed and humanely killed by cervical dislocation, and all test animals were subjected to gross necropsy and dissected. The liver, stomach, spleen, kidneys, heart, and intestines of different groups were preserved in 40% formalin, and their microscopic examination was compared to check their gross pathology. In addition, livers, spleens, kidneys, and thymus of all animals were weighted and the relative organ weights were calculated.
The Antimicrobial Activity of Phage STP4-a In Vitro and Its Tolerance in Simulated Digestion Environments
The antimicrobial activity of phage STP4-a in vitro was determined by using a single bacterial strain and 6 bacterial strains, respectively. The host strain, S. Typhimurium ATCC 14028, was used to test the antimicrobial activity of phage STP4-a for a single strain. Apart from the host strain, another 5 strains, namely S. Typhimurium CMCC 50115, S. Enteritidis CMCC 50041, S. Paratyphi A CMCC50001, S. Paratyphi B CMCC50094, and S. Typhi CMCC50071 were also used in the antimicrobial assay for multibacterial strains. First, 50 mL SM buffer was inoculated with 108 CFU aforementioned Salmonella strains and 1010 PFU phage STP4-a and then incubated at 37°C under rotation. Then, the variation of bacterial counts in different groups was monitored during 6 h incubation.
Simulated chicken digestion environments for phage STP4-a tolerance test were prepared as described by Pizzolitto et al. (2012). Simulated saliva was prepared by dissolving lysozyme in 0.9% NaCl to a final concentration of 2 mg/mL, and adjusting its pH to 6.5 with 5 mol/L NaOH or concentrated 12 mol/L HCl. Next, simulated gastric juice contained 0.125 mol/L NaCl, 0.007 mol/L KCl, and 0.045 mol/L NaHCO3 and its pH was adjusted to 3.0 with concentrated HCl (12 mol/L), and then pepsin (Sigma-Aldrich) was added into the resulting mixture to reach a final concentration of 3 g/L. In addition, simulated intestinal fluid was prepared with trypsin (Sigma-Aldrich) with a final concentration of 1 mg/mL, and its pH was adjusted to 8.0 with 5 mol/L NaOH. Subsequently, the activity of phage STP4-a in vitro was evaluated via measuring titers at different simulated preparations. Accordingly, 1 mL of phage STP4-a was added into 12 mL of simulated digestion preparations (4 mL of simulated saliva and 8 mL of simulated gastric juice), 8 mL of simulated gastric juice, and 12 mL of simulated intestinal fluids. To test different simulated environment tolerance, phage STP4-a was incubated in different simulated preparations at 37°C under agitation (200 rpm). Titers of phage STP4-a were measured at different time intervals in terms of different preparations: 0 and 1 h for simulated digestion preparations; 0, 1, and 2 h for simulated gastric juice; 0 and 3 h for simulated intestinal fluid.
Efficacy of Post-treatments and Pretreatments With Phage STP4-a in Chickens
To demonstrate the efficacy of phage STP4-a in controlling S. Typhimurium in vivo, 2-week-old commercial layer chickens (n = 60) were divided into 6 experimental groups (Table 3), and each group was composed of 10 birds. Groups A, B, C, and D were bred for 7 D without any pretreatments. By contrast, groups E and F were pretreated with 109 PFU/g feed of phage STP4-a, which started 7 D before bacterial challenge. All birds in groups A, C, and E were challenged with 1 × 108 CFU of S. Typhimurium, and all birds in groups B, D, and F were set as negative controls without any challenges. At the same time, birds in groups C and D were treated with 109 PFU/g feed of phage STP4-a after bacterial challenge for 14 D. Thus, group C/D and group E/F were used for evaluating the efficacy of post-treatment and pretreatment with phages therapy on Salmonella in poultry. The counts of Salmonella and phage STP4-a in chicken fecal samples were determined. Three fecal samples were collected stochastically from each group. These samples were diluted 10-fold in 0.85% NaCl, and 1 mL of each serial dilution was dispensed and spread on Xylose-Lysine Desoxycholate agar. Samples that were undetectable for Salmonella through direct plating method were preenriched in Universal Preenrichment Broth (Hopebio, Qingdao, China) at 37°C for 24 h, and a secondary enrichment was conducted subsequently in Rappaport Vassiliadis Broth (Hopebio, Qingdao, China) at 42°C for 24 h. The enriched cultures were then plated onto Xylose-Lysine Desoxycholate plates after enrichment. Each agar plate was incubated at 37°C for 24 h before the typical Salmonella colonies were counted.
Table 3.
Different treatments on chickens in the phage STP4-a efficacy evaluation experiment.
| Groups | Number of chickens | Pretreatment | Challenge | Treatment |
|---|---|---|---|---|
| A | 10 | None | S. Typhimurium ATCC 14028 challenged1 | Untreated |
| B | 10 | None | Unchallenged | Untreated |
| C | 10 | None | S. Typhimurium ATCC 14028 challenged1 | STP4-a treated2 |
| D | 10 | None | Unchallenged | STP4-a treated2 |
| E | 10 | STP4-a pre-treated2 | S. Typhimurium ATCC 14028 challenged1 | Untreated |
| F | 10 | STP4-a pre-treated2 | Unchallenged | Untreated |
Ten chickens were orally challenged with S. Typhimurium (ATCC 14028) at the concentration of 1 × 108 CFU/bird.
Ten chickens were treated with bacteriophages as feed additives at a concentration of 1 × 109 pfu/g feed.
Statistics Analysis
In the mice experiment, body weights of 5 animals and organ weights of 3 samples were recorded. Results were then expressed in mean values, with error bars indicating the standard deviations. The data in tables were analyzed by using Duncan test with SPSS 17.0 (Chicago, IL, USA). Student's t-test was used to compare significant differences between test groups and negative control groups or blank groups, with P<0.05 considered as statistically significant difference. Figures were drawn with OriginPro 8.5 (OriginLab, Northampton, MA, USA).
Genome Accession Number
The sequence of phage STP4-a genome was available in GenBank, with the accession number of KJ000058.2.
Results and discussion
Genomic Sequence and the Host Range Analysis of Phage STP4-a
Phage STP4-a genomic sequence consisted of 159,914 bp, with a G + C content of 36.86%. A total of 259 ORF, accounting for 95.1% of STP4-a sequence, were predicted in its genome. All hypothetical functions, pIs, and molecular weights of 259 proteins were listed in Supplementary Table 1. Bioinformatics analysis of all 259 gene products of phage STP4-a revealed that 128 ORF shared a high similarity to predicted genes with known functions, and 131 ORF showed homology to hypothetical proteins.
The host range of phage STP4-a was first estimated through bioinformatics analysis. After genomic sequence comparison, phage STP4-a shared great similarity with broad-host-range Salmonella phages S16 (GeneBank accession number HQ331142.1) (Marti et al., 2013) and STML-198 (GeneBank accession number JX181825.1). Phage STP4-a was proved to belong to T4 family in the previous study (Li et al., 2016). Then, key genes or proteins of phage STP4-a in the infection cycle were analyzed to further estimate the host range of phage STP4-a as follows, including both attachment and synthesis of nucleic acids processes.
First, in attachment process, Schwarzer et al. (2012) and Santos et al. (2011) verified that tail fiber proteins could determine the threshold to get into the hosts through recognizing outer membrane receptors. For example, phage phi92 with a multivalent adsorption sites could recognize distinct O-antigens of different Salmonella serovars, contributing to its wide host range (Schwarzer et al., 2012). In addition, phage PVP-SE1 took the inner lipopolysaccharide (LPS) core as the receptor, which was conserved and held a similar structure in S. Typhimurium and some E. coli strains. Thus, phage PVP-SE1 had a broader host range than Felix O1, whose receptor was a less conserved region, outer LPS core (Santos et al., 2011). Consequently, broad-host-range phages usually had a conserved receptor structure or multivalent adsorption sites. In phage STP4-a genome, gp237-gp241 shared great homology with gp34-gp38 (tail fiber proteins) in phage S16. In particular, gp240 (long tail fiber distal subunit) and gp241 (receptor recognition protein) of phage STP4-a were similar to gp169 (87%) and gp170 (62%) of phage S16. The 2 proteins of phage S16 determined its broad host range, which were contributed by 2 receptors of Salmonella strains, Omp C and outer LPS core. According to its possible multiple adsorption sites, there was a possibility that phage STP4-a had a broad-host-range.
Next, the synthesis of nucleic acids for phages can be stopped by restriction endonucleases in bacterial restriction-modification (R-M) system (Stern et al., 2011). Broad-host-range phages usually escape restriction endonucleases due to a lack of cleavage sites or production of antiresitriction proteins (Santos et al., 2011). After cleavage sites analysis, phage STP4-a DNA had only one cleavage site for Salmonella type II restriction enzyme SshAI (CCTNAGG) and 5 for SthI (GGTACC), but it possessed 126 sites for SinI (GGWCC). Similarly, a broad-host-range phage PVP-SE1 had no cleavage sites for the Salmonella type II restriction enzymes SbaI and SthBI but that it possessed 141 sites for SblAI (Santos et al., 2011). Furthermore, gp247 of phage STP4-a showed 75% homology to anti-restriction nuclease of Klebsiella phage KPV15. Then the putative anti-restriction nuclease of phage STP4-a may be resistant to the host R-M system, preventing the host endonuclease from cleaving specific DNA sequences of phages (Evdokimov et al., 2007), and then widen the host range in a sense. Such conclusions were also found in phage PVP-SE1 that a putative DNA cytosine methyltransferase (gp9) may thus methylate cytosine resides at endonuclease sites, which could protect phage DNA from being cut by its hosts' restriction endonucleases in the R-M system (Santos et al., 2011). Hence, phage STP4-a possessed such 2 strategies to escape bacterial R-M system, potentially broadening its host range.
Bioinformatics analysis above predicted phage STP4-a to have a broad host range due to its multivalent receptor recognition sites of tail fibers, a lack of cleavage sites for restriction endonucleases and possessing anti-restriction endonucleases. In host range assay, phage STP4-a proved to have a broad lysis spectrum in infecting Salmonella strains, with 88 out of 91 Salmonella strains being susceptible, consistent with predicted conclusion, while phage STP4-a could not inhibit E. coli or K. pneumonia, indicating a high host specificity (Table 1). Therefore, phage STP4-a took both merits in owning a broad host range, as well as high specificity to Salmonella strains, getting rid of microbiota collateral damage. For example, ListexP100, which was a single broad-host-range phage, was identified as GRAS by FDA, giving us a clear direction for the development of phage commercialization in the future. Currently, most phages were prone to have relatively narrow lysis spectrum (Mathur et al., 2003), so they were less effective when used alone. Therefore, exploring “multivalent” phages or phage cocktails was difficult but necessary to ensure that they were lytic to most bacterial strains within a given species of pathogens (Carlton, 1999).
Toxicity Analysis of Phage STP4-a In Silico and in Mice
In silico analysis, all 259 gene products of phage STP4-a had no relevance to any known virulence, toxin, or pathogen-associated protein family or gene products of Salmonella strains or any other pathogens. With a 0.01 E-value cutoff, none of those 259 gene products had any relationship with any polypeptides or protein sequences contained in the food allergenic protein sequence database except gp231. For gp231, an 8-amino-acid short-sequence of phage STP4-a (DDEDEDED) was identical with the 8-amino-acid short-sequence of glycinin in Glycine max. Two epitopes of glycinin in Gly m glycinin G1 and 11 epitopes of glycinin in Gly m glycinin G2 were reported in Structural Database of Allergenic Proteins. The identical 8-amino-acids had no relation with those epitopes, which could exclude the possibility that this gene product or phage STP4-a had the cross-reactivity with IgE. In addition, when comparing this protein sequence in NCBI database, it showed that this sequence was common among different species, such as Drosophila grimshawi, Atropa belladonna, A. belladonna, and so on. To the current knowledge of those species above in Structural Database of Allergenic Proteins, they could not cause allergy to animals or human beings, suggesting that the gene products had no side effects on humans. Therefore, it can be concluded that phage STP4-a posed no risks in silico.
After bioinformatics analysis of phage STP4-a, functions of 131 gene products were still unknown, and thus, further studies were necessary to clarify the safety of phage STP4-a. In this work, animal experiments were conducted with single-dose oral administration of phage STP4-a in the 14-day period, which was corresponded with Salmonella phage wksl3 (Kang et al., 2013). The oral administration of phage STP4-a (1011 PFU/kg body weight) in the 14-day experimental period produced no death in both sexes of mice, indicating that mice could survive with a single dose of 1011 PFU/kg body weight administrated. No development of abnormal behavior, changes in physical appearance (such as skin, hair, eyes, or mucous membrane), or any other toxicological effects, including inflammation, allergy or diarrheal symptoms, and no strange phenomena in breathing, circulatory, vegetative nerve, or nervous centralis, were observed in any mice during the experimental period.
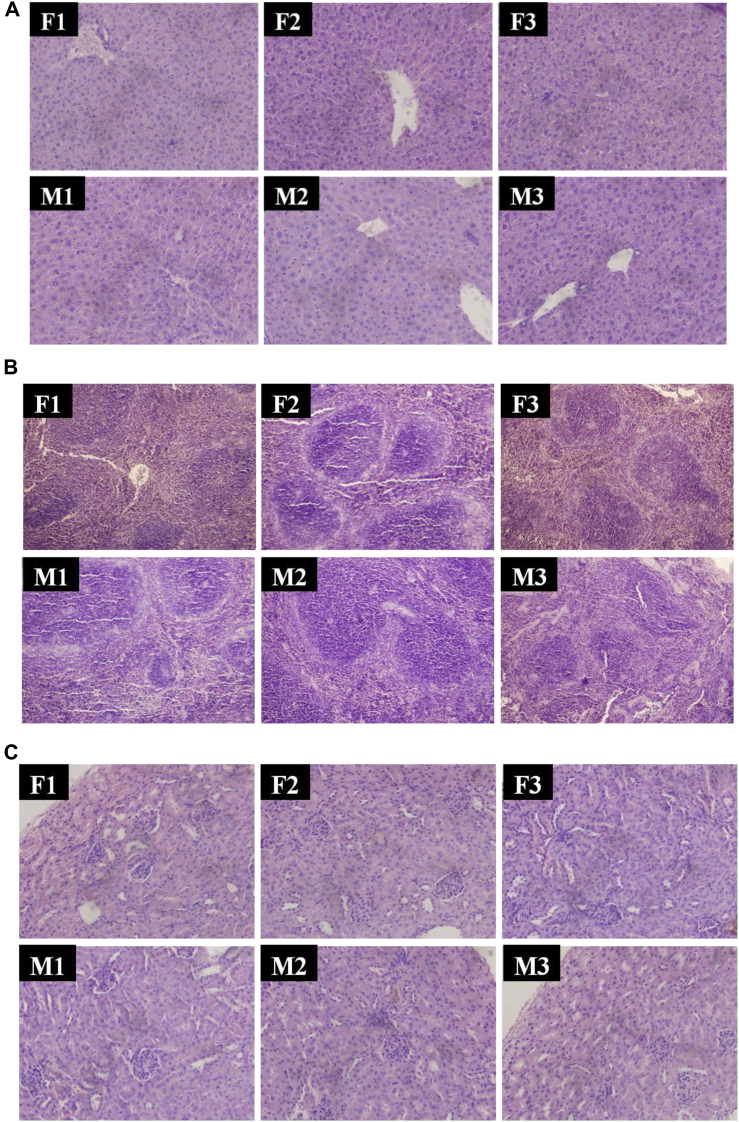
The data of body weight, organ weight, and relative organ weight were shown as follows. Body weight (Supplementary Table 2A) in group F2/M2 and group F3/M3 showed no significant difference (P > 0.05) from group F1/M1. The weight and relative weight of kidney, spleen, and thymus in group F2/M2 and group F3/M3 showed no significant difference (P > 0.05) from group F1/M1. Although the liver weight and relative weight in group F2 were higher than group F1 significantly (P < 0.05), Group F3 showed no difference from group F1 (Supplementary Table 2B and C). Usually, weights of organs, the index for toxicity, enzyme induction, physiological perturbations, and acute injury were correlated with histopathological changes (Michael et al., 2007). Furthermore, relative organ weights were also used to indicate the essential organ growth, such as liver, kidney, spleen, and thymus. In this study, no significant changes were observed in weights of organs among active or inactive phage-treated groups when compared with SM buffer-treated groups, manifesting that phage STP4-a with its residues was not harmful. However, the liver weights and the relative liver weights in group F2 were a little higher (P < 0.05) than those in group F1, within the normal organ weights and coefficients range (Zhang et al., 2011), whereas the minor differences of liver weights and relative liver weights were considered significant just from statistics rather than its biology. Then, phage STP4-a with its residues was further proved to have no adverse effects on liver organ growth. In addition, gross anatomy of the vital organs of mice in active or inactive phage-treated groups showed no significant changes in color or texture compared with SM buffer-treated groups. Therefore, it could be concluded that phage STP4-a with its residues was not harmful to organs at the dose of 1011 PFU/kg body weight. Macroscopic examination of organs of all phage-treated mice revealed normal color and texture compared with SM buffer-treated or inactive phage-treated animals. Histological sections of the liver, spleen, kidney, stomach, intestine, and heart in different groups showed no significant differences in Figure 1. For those organs, no congestion, ischemia, edema, necrosis, fibrosis, and envelope were developed. Therefore, it could be inferred that phage STP4-a had no negative effects on different organs in mice through the 14-day oral toxicity study.
Figure 1.
Histological observations of different organs in different groups. (A) Liver (H.E × 400), (B) spleen (H.E × 200), (C) kidney (H.E × 400), (D) stomach (H.E × 200), (E) intestine (H.E × 200), (F) heart (H.E × 200).
Aside from phage STP4-a, the safety of other 3 lytic phages was evaluated through both bioinformatics analysis and animal experiments to our current knowledge. Carlton et al. (2005) and Kang et al. (2013) studied the safety of lytic phages treatments on Listeria and Salmonella, respectively, and concluded that they showed no harmful factors in silico and no abnormal phenomena in animal experiments. Furthermore, FDA considered phage P100 as GRAS in 2007 and granted its approval as a food additive for the control of L. monocytogenes in ready-to-eat foods (Bren, 2007). However, Mccallin et al. (2013) identified 2 undesired genes (dps gene and lambda bor-like gene) in the genome of Microgen phage cocktail and then they excluded the risks of Microgen phage cocktail to humans. Therefore, it was essential to evaluate the safety of phages through bioinformatics analysis and animal experiments simultaneously. Herein, phage STP4-a was proved to be safe through bioinformatics analysis and animal experiments.
The Antimicrobial Activities of Phage STP4-a In Vitro and Its Stability in Simulated Environments
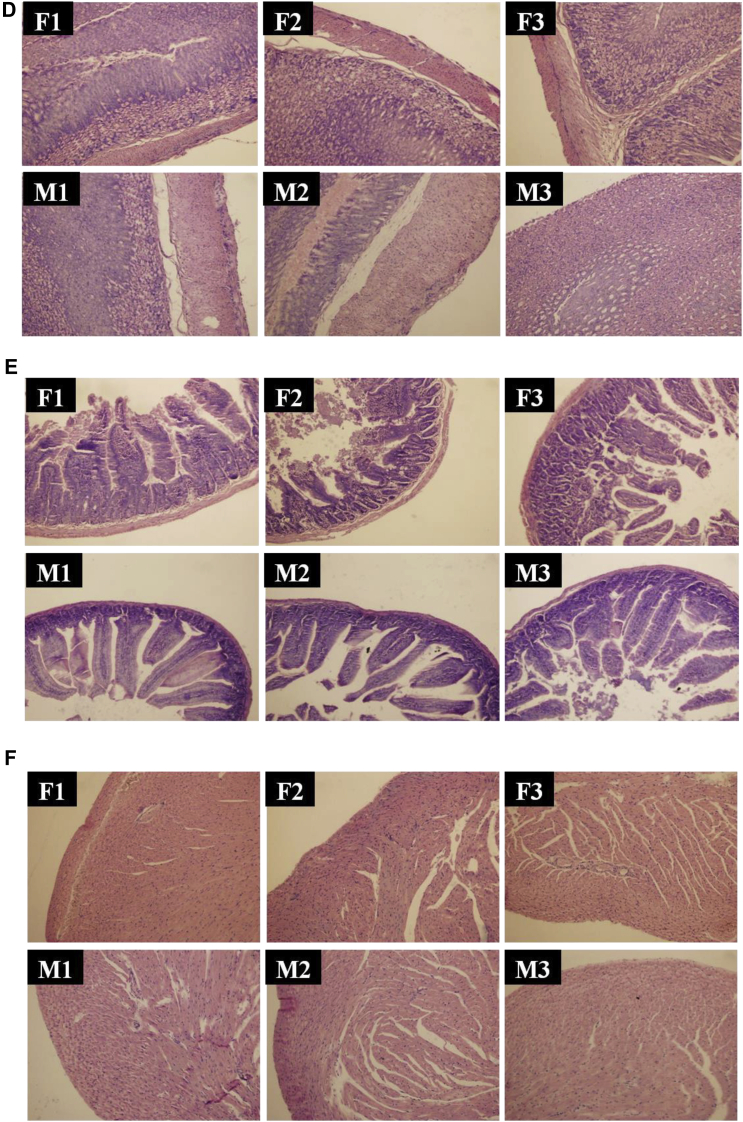
Before in vivo therapy, the antimicrobial activities of phage STP4-a in vitro needed to be evaluated both on a single Salmonella strain and a mixture of 6 Salmonella strains. As shown in Figure 2A, counts of S. Typhimurium ATCC 14028 decreased from 106 CFU/mL to less than 10 CFU/mL sharply in 30 min under phage STP4-a treatment. For phage STP4-a treatment on multiple Salmonella strains (Figure 2B), total bacterial number dropped from 106 CFU/mL to 103 CFU/mL in 30 min and then reached below 100 CFU/mL after 1 h treatment. Therefore, phage STP4-a could efficiently reduce counts of a single or multiple Salmonella strains by 4-5 log in vitro.
Figure 2.
The in vitro antimicrobial activities of phage STP4-a and its stability in simulated environments for different time intervals. (A) In vitro antimicrobial activity for a single strain, Salmonella Typhimurium ATCC 14028. (B) In vitro antimicrobial activity for 6 Salmonella strains. (C) The stability of phage STP4-a in simulated environments. Borders with blue color represented titers of phage in 12 mL simulated digestion preparations (4 mL simulated saliva and 8 mL simulated gastric juice, abbr. “SA + GA”) for 0, 1 h; Borders with red color represented titers of phage 8 mL simulated gastric juice (abbr. “GA”) for 0, 1, 2 h. Borders with green color represented titers of phage 12 mL simulated intestinal fluid (abbr. “IN”) for 0, 3 h.
Then, the stability of phage STP4-a in the digestion process was estimated in different simulated environments (Figure 2C). Since saliva content would pass into the stomach, the activity of phage STP4-a was measured in the mixture of simulated saliva and gastric juice (SA + GA) with the ratio of 1:2. The titer of phage STP4-a was 1.29 × 109 PFU/mL at initial and decreased to 0.99 × 109 PFU/mL after 1-h digestion. Similarly, the titer of phage STP4-a in 8 mL gastric juice (GA) was 2.11 × 109 PFU/mL at initial, and then reduced to 1.20 × 109 PFU/mL and 1.09 × 109 PFU/mL after 1- and 2-h incubation, respectively. The titer of phage STP4-a had no significant change after 3-h incubation in simulated intestinal fluid (IN). In general, more than 50% of phage STP4-a survived through all simulated digestion processes, including saliva, gastric juice, and intestinal fluid, which could be prepared for efficient in vivo therapy on Salmonella.
Efficacy of Phage Therapy on Chicken Infection In Vivo
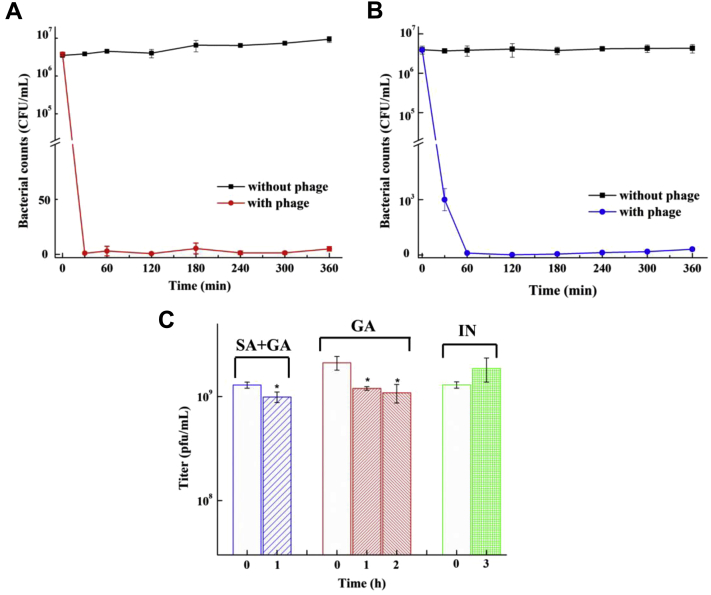
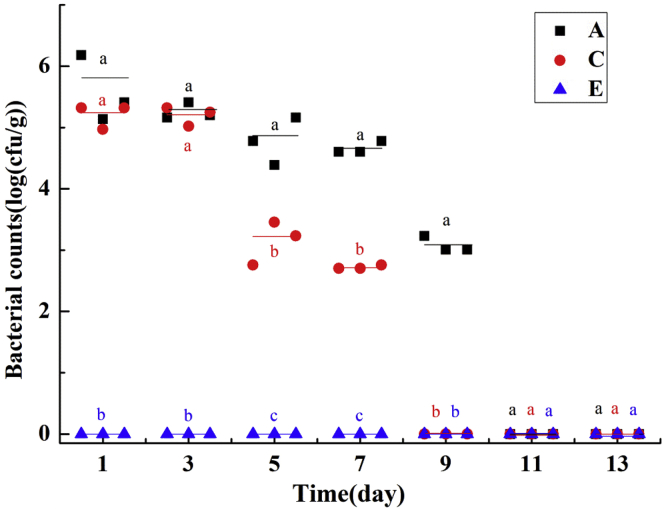
During this experiment, no clinical signs or mortality was observed in those groups and the Salmonella counts [CFU/g (lg)] in feces among Salmonella-infected groups decreased in different degrees (Figure 3). Control groups B, D, and F were absent of Salmonella (data not shown), and Salmonella-infected groups showed different variations under different treatments. In group E, Salmonella was not detectable during the 2-week experiment, showing more significant and better antimicrobial activity than groups A and C. In addition, Salmonella counts in feces of group C decreased by 2 log than group A from the fifth day, and Salmonella became non-detectable on the ninth day, which was earlier than 11th day in group A. Hence, we found that phage treatment could inhibit bacterial colonization in chickens, and pretreatment with phages could prevent and eliminate Salmonella absolutely.
Figure 3.
Bacteria counts in feces among Salmonella-infected groups during the 2-week experiments. Data in the same day with different letters (a, b, c) are significantly different (P < 0.05).
Pretreatment with phage STP4-a could not only work as the “vaccination” but also further test its safety in poultry. On the one hand, Fujiwara et al. (2011) clarified that pretreatment of tomato seedling with phages could limit activities of bacterial cells drastically, which was consistent with our results. Phage STP4-a was distributed evenly through the digestion glands, including stomachs and intestines. Once bacterial cells went into digestion system, phages would instantly catch and kill them to stop bacteria cells colonization and get itself proliferation at the same time. Therefore, pretreatment with phages could build a “firewall” against multiple bacterial invasions. On the other hand, all chickens in experimental groups and control groups treated with phage STP4-a all survived and thus further proved its safety under phage therapy.
Conclusion
It is our purpose to provide broad-host-range Salmonella phages to realize precise poultry or clinical treatments. Instead of tedious and complicated laboratory work, we have successfully screened possible broad-host-range phage STP4-a under bioinformatics analysis and then proved that phage STP4-a could inhibit 88 out of 91 Salmonella strains. For the sake of phage STP4-a application, safety and antimicrobial activities were both evaluated. In silicon analysis and animal experiments for phage STP4-a excluded any risks from any known lysogeny factors, toxins, pathogen-related genes, or foodborne allergens, as well as unknown factors or suspected risks. Furthermore, strong antimicrobial activities of phage STP4-a were seen in vitro and in vivo. Phage STP4-a could survive and keep more than 50% activity in simulated digestion environments in vitro. As for antimicrobial activities in chickens, groups pretreated with phage STP4-a showed non-detectable Salmonella counts in their feces during the 14-day experimental period. Therefore, phage STP4-a was a safe antimicrobial agent with an extremely broad-host-range and could be used in poultry industry.
Acknowledgments
This work was financially supported by the Natural Science Foundation of China, China (31870166) and the Natural Science Foundation of Shandong Province, China (2017GNC13108). The authors thank Dr. Richard Goodman (University of Nebraska at Lincoln) for discussing putative allergenic proteins in phage STP4-a when 8-amino-acid short-sequence is identical with glycinin in Glycine max.
Conflicts of Interest Statement: There are no conflicts to declare.
Footnotes
Supplementary data associated with this article can be found in the online version at http://doi.org/10.1016/j.psj.2020.03.051.
Supplementary data
References
- Atterbury R.J., Van Bergen M.A.P., Ortiz F., Lovell M.A., Harris J.A., De Boer A., Wagenaar J.A., Allen V.M., Barrow P.A. Bacteriophage therapy to reduce Salmonella colonization of broiler chickens. Appl. Environ. Microb. 2007;73:4543–4549. doi: 10.1128/AEM.00049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyacioglu O., Sharma M., Sulakvelidze A., Goktepe I. Biocontrol of Escherichia coli O157: H7 on fresh-cut leafy greens. Bacteriophage. 2013;3:e24620. doi: 10.4161/bact.24620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bren L. Bacteria-eating virus approved as food additive. FDA Consum. 2007;41:20–22. [PubMed] [Google Scholar]
- Carlton R.M. Phage therapy: past history and future prospects. Arch. Immunol. Ther. Ex. 1999;47:267. [PubMed] [Google Scholar]
- Carlton R.M., Noordman W.H., Biswas B., de Meester E.D., Loessner M.J. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharmacol. 2005;43:301–312. doi: 10.1016/j.yrtph.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Coburn B., Grassl G.A., Finlay B.B. Salmonella, the host and disease: a brief review. Immunol. Cell Biol. 2007;85:112–118. doi: 10.1038/sj.icb.7100007. [DOI] [PubMed] [Google Scholar]
- Domingo-Calap P., Delgado-Martinez J. Bacteriophages: protagonists of a post-antibiotic era. Antibiotics. 2018;7:66. doi: 10.3390/antibiotics7030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng S.K., Pusparajah P., Mutalib N.S.A., Ser H.L., Lee L.H. Salmonella: a review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015;8:1–10. [Google Scholar]
- Evdokimov A.A., Sclavi B., Zinoviev V.V., Malygin E.G., Hattman S., Buckle M. Study of bacteriophage T4-encoded Dam DNA (adenine-N6)-methyltransferase binding with substrates by rapid laser UV cross-linking. J. Biol. Chem. 2007;282:26067–26076. doi: 10.1074/jbc.M700866200. [DOI] [PubMed] [Google Scholar]
- Fujiwara A., Fujisawa M., Hamasaki R., Kawasaki T., Fujie M., Yamada T. Biocontrol of Ralstonia solanacearum by treatment with lytic bacteriophages. Appl. Environ. Microb. 2011;77:4155–4162. doi: 10.1128/AEM.02847-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagens S., Loessner M.J. Application of bacteriophages for detection and control of foodborne pathogens. Appl. Microbiol. Biotechnol. 2007;76:513–519. doi: 10.1007/s00253-007-1031-8. [DOI] [PubMed] [Google Scholar]
- Heredia N., Garcia S. Animals as sources of food-borne pathogens: a review. Anim. Nutr. 2018;4:250–255. doi: 10.1016/j.aninu.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Kim J., Jung T., Woo G. wksl3, a new biocontrol agent for Salmonella enterica serovars Enteritidis and Typhimurium in foods: characterization, application, sequence analysis, and oral acute toxicity study. Appl. Environ. Microb. 2013;79:1956–1968. doi: 10.1128/AEM.02793-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H.J., Cho S.H., Kim T.E., Won Y.J., Jeong J., Park S.C., Kim J.H., Yoo H.S., Park Y.H., Kim S.J. Characterization of a T7-like lytic bacteriophage (phiSG-JL2) of Salmonella enterica serovar Gallinarum biovar Gallinarum. Appl. Environ. Microbiol. 2008;74:6970–6979. doi: 10.1128/AEM.01088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Li M., Lin H., Wang J., Jin Y., Han F. Characterization of the novel T4-like Salmonella enterica bacteriophage STP4-a and its endolysin. Arch. Virol. 2016;161:377–384. doi: 10.1007/s00705-015-2647-0. [DOI] [PubMed] [Google Scholar]
- Marti R., Zurfluh K., Hagens S., Pianezzi J., Klumpp J., Loessner M.J. Long tail fibres of the novel broad-host-range T-even bacteriophage S16 specifically recognize Salmonella OmpC. Mol. Microbiol. 2013;87:818–834. doi: 10.1111/mmi.12134. [DOI] [PubMed] [Google Scholar]
- Mathur M.D., Vidhani S., Mehndiratta P.L. Bacteriophage therapy: an alternative to conventional antibiotics. J. Assoc. Physicians India. 2003;51:593. [PubMed] [Google Scholar]
- McCallin S., Alam S.S., Barretto C., Sultana S., Berger B., Huq S., Krause L., Bibiloni R., Schmitt B., Reuteler G., Brussow H. Safety analysis of a Russian phage cocktail: from metagenomic analysis to oral application in healthy human subjects. Virology. 2013;443:187–196. doi: 10.1016/j.virol.2013.05.022. [DOI] [PubMed] [Google Scholar]
- Michael B., Yano B., Sellers R.S., Perry R., Morton D., Roome N., Johnson J.K., Schafer K., Pitsch S. Evaluation of organ weights for rodent and non-rodent toxicity studies: a review of regulatory guidelines and a survey of current practices. Toxicol. Pathol. 2007;35:742–750. doi: 10.1080/01926230701595292. [DOI] [PubMed] [Google Scholar]
- Organization for Economic Co-operation and Development. 2001. OECD guidelines for acute toxicity of chemicals, no. 420. Organization for Economic Co-operation and Development, Paris, France. [Google Scholar]
- O'Neill J. Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist. 2014;20:1–16. [Google Scholar]
- Park M., Lee J.H., Shin H., Kim M., Choi J., Kang D.H., Heu S., Ryu S. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7. Appl. Environ. Microbiol. 2012;78:58–69. doi: 10.1128/AEM.06231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzolitto R.P., Armando M.R., Combina M., Cavaglieri L.R., Dalcero A.M., Salvano M.A. Evaluation of Saccharomyces cerevisiae strains as probiotic agent with aflatoxin B1 adsorption ability for use in poultry feedstuffs. J. Environ. Sci. Heal. B. 2012;47:933–941. doi: 10.1080/03601234.2012.706558. [DOI] [PubMed] [Google Scholar]
- Reardon S. Phage therapy gets revitalized. Nature. 2014;510:15–16. doi: 10.1038/510015a. [DOI] [PubMed] [Google Scholar]
- Santos S.B., Kropinski A.M., Ceyssens P.J., Ackermann H.W., Villegas A., Lavigne R., Krylov V.N., Carvalho C.M., Ferreira E.C., Azeredo J. Genomic and proteomic characterization of the broad-host-range Salmonella phage PVP-SE1: creation of a new phage genus. J. Virol. 2011;85:11265–11273. doi: 10.1128/JVI.01769-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer D., Buettner F.F., Browning C., Nazarov S., Rabsch W., Bethe A., Oberbeck A., Bowman V.D., Stummeyer K., Muhlenhoff M., Leiman P.G., Gerardy-Schahn R. A multivalent adsorption apparatus explains the broad host range of phage phi92: a comprehensive genomic and structural analysis. J. Virol. 2012;86:10384–10398. doi: 10.1128/JVI.00801-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahin K., Bouzari M. Bacteriophage application for biocontrolling Shigella flexneri in contaminated foods. J. Food Sci. Tech. 2018;55:550–559. doi: 10.1007/s13197-017-2964-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A., Sorek R. The phage-host arms race: shaping the evolution of microbes. Bioessays. 2011;33:43–51. doi: 10.1002/bies.201000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran A.T., Nannapaneni R., Kiess A., Sharma C.S. Reduction of Salmonella on chicken breast fillets stored under aerobic or modified atmosphere packaging by the application of lytic bacteriophage preparation SalmoFreshTM. Poult. Sci. 2016;95:668–675. doi: 10.3382/ps/pev332. [DOI] [PubMed] [Google Scholar]
- Turner D., Hezwani M., Nelson S., Salisbury V., Reynolds D. Characterization of the Salmonella bacteriophage vB_SenS-Ent1. J. Gen. Virol. 2012;93:2046–2056. doi: 10.1099/vir.0.043331-0. [DOI] [PubMed] [Google Scholar]
- Yassin A.K., Gong J., Kelly P., Lu G., Guardabassi L., Wei L., Han X., Qiu H., Price S., Cheng D., Wang C. Antimicrobial resistance in clinical Escherichia coli isolates from poultry and livestock, China. Plos One. 2017;12:e185326. doi: 10.1371/journal.pone.0185326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.G., Buckling A. Phages limit the evolution of bacterial antibiotic resistance in experimental microcosms. Evol. Appl. 2012;5:575–582. doi: 10.1111/j.1752-4571.2011.00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.Q., Ying G.G., Pan C.G., Liu Y.S., Zhao J.L. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol. 2015;49:6772–6782. doi: 10.1021/acs.est.5b00729. [DOI] [PubMed] [Google Scholar]
- Zhang X.Y., Zhan C.L., Xiao Y.H. Measurement and comparisons of organ weight,organ coefficient,hematological parameters and hematological biochemical parameters of specific pathogen free Balb/c mice. Chin. J. Clin. Rehab. Tissue Eng. Res. 2011;15:7734–7737. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.