Abstract
Our long-term goal is to improve chick health and reduce the use of antibiotics in the poultry industry via maternal effects. To link jejunal microbes with chicks' different immune levels and growth performance in our previous research, this study investigated jejunal microbes, jejunal inflammation, and immune responses based on a comparison between different groups. Newly hatched Hy-Line chicks were allotted into 3 groups: a chick control group (cCON), a ciprofloxacin lactate treatment group (Cipro)—the chicks of the cCON and Cipro groups were hatched from laying breeder hens given a basal diet—and a 5-wk β-carotene, curcumin, allicin, and sodium butyrate supplementation group (cCCAB), wherein chicks hatched from laying breeder hens. All groups were fed the same diet for 4 wk; the Cipro group was given ciprofloxacin lactate in drinking water continuously. At the end of the experiment, the results demonstrated that the jejunal microbes of the Cipro group showed significant changes in alpha and beta diversity, and in taxonomy at phylum and genus levels. Statistically, a total of 67 significantly enriched (P < 0.05) taxa were identified between groups by linear discriminant analysis effect size; Firmicutes was significantly enriched (P < 0.05) in the cCCAB group, 65 taxa were significantly enriched (P < 0.05) in the Cipro group, and 32 of the 65 enriched (P < 0.05) taxa were in the Proteobacteria phylum of the Cipro group. Levels of lipopolysaccharide in jejunal content, and nuclear factor kappa-B, and tumor necrosis factor-α in jejunums of the Cipro and cCCAB groups were increased (all P < 0.05) compared to those in the cCON group. There was obvious neutrophil infiltration and upregulated (all P < 0.05) IL-6 mRNA in the Cipro group jejunums compared to the cCON and cCCAB groups. The expression of PSME3 and PSME4 genes was upregulated (all P < 0.05) in the cCCAB group compared to the cCON and Cipro groups. In conclusion, ciprofloxacin lactate administration led to potential hazards in health and growth in chicks via microbial disturbances-induced jejunal inflammation, and laying breeder hens dietary supplementation with β-carotene, curcumin, allicin, and sodium butyrate could enhance jejunal immunity of their offspring via the interaction between host innate immunity selected microbial colonization and microbiota educated adaptive immunity.
Key words: laying breeder hen, β-carotene, antibiotic, microbiota, immune response
Introduction
Massive microbial communities live in the gut, and the microbiota and host immune system interact in induction, education, and function (Belkaid and Harrison, 2017). Immune maturation is likely influenced directly and/or indirectly by the presence of commensal microbes (Gensollen et al., 2016), while the intestines need to avoid developing pathologies and control the colonization of bacteria by innate immunity in early life (Hooper et al., 2012). Although antibiotics were used as an efficient tool to promote growth in the poultry industry, we recognized their negative effects on bacterial resistance (Chattopadhyay, 2014). Besides, there are strong evidences linking gut microbial disruption with antibiotics use in early life with increasing risk of inflammatory diseases (Shaw et al., 2010).
Our pervious study illustrated that laying breeder hens dietary supplementation with β-carotene, curcumin, allicin, and sodium butyrate had a positive effect on hens immunity, and their offspring showed better immunity and growth performance in early life, which may be a result of stronger jejunal immune system development to ensure growth. Moreover, we noticed that chicks supplemented with ciprofloxacin lactate demonstrated enhanced growth performance before 14 D of age, but the promotion effects decreased when ciprofloxacin lactate administration was extended, which was also explained with regard to the changed jejunal immune development and morphology. Hence, we suggest that the variation in growth performance between groups was associated with their intestinal immunity (Gong et al., 2020).
Here, in this study, we will investigate jejunal microbial communities, immunity-related genes, and protein expression in an attempt to explain supplementarily the immune differences and growth performance of each treatment in the previous study.
Methods
This experiment was approved by the Animal Ethics Committee of Jilin Agricultural University.
Bird Management and Experimental Design
The methods of bird management and experimental design have been published in detail in our previous study (Gong et al., 2020). Briefly, a total of 162 Hy-Line Brown laying breeder hens were randomly allotted to 2 groups (a control group [CON] and the β-carotene, curcumin, allicin, and sodium butyrate supplementation group [CCAB]). The CON group was fed a basal diet, while the CCAB group was fed a basal diet supplemented with 60 mg/kg β-carotene, 250 mg/kg curcumin, 250 mg/kg allicin, and 500 mg/kg sodium butyrate (Shaanxi Kingreg Biotech Co. Ltd., China) for 5 wk (including a 1-wk acclimation period). Their breeder eggs were collected respectively for hatching at the end of the hens feeding period. When the offspring chicks were hatched, a total of 60 healthy Hy-Line Brown male chicks with similar body weights hatched from the CON groups' breeder eggs were randomly allotted to 2 treatment groups (a chick control group [cCON] and the ciprofloxacin lactate treatment group [cipro]) with each group comprising 30 chicks each. Thirty other male chicks that hatched from the CCAB group eggs formed a group (cCCAB). All chicks were fed a single basal diet and allowed to eat and drink at will. The Cipro group had 100 mg/L ciprofloxacin lactate (Shaanxi Kingreg Biotech Co. Ltd.) added to their drinking water. The chicks were fed for 4 wk. The detailed chick-brooding management, and basal diet composition of experimental laying breeder hens and chicks are listed in Supplementary Tables 1 and 2, and we declare that these 2 tables have been published in our previous study (Gong et al., 2020).
Sample Collection
At the termination of the experimental period with chicks at 28 D of age, a total of 6 chicks' jejunums of each group were randomly sampled following euthanasia. To standardize the samples for further neutrophil infiltration investigations, the central part with about 2 cm of jejunum of each chick was fixed in 4% paraformaldehyde. We used anatomical features to identify the jejunum: the final intersection point of the duodenum and pancreas was considered as the boundary between the duodenum and jejunum, and the yolk stalk served as the boundary between the jejunum and ileum. The jejunum content samples were collected in germ-free tubes and stored in liquid nitrogen for bacterial genome sequence analysis. The jejunum tissues were collected in RNase-free tubes and frozen at −80°C for quantitative real-time PCR (qRT-PCR) analysis.
Neutrophil Infiltration Observation
Jejunum samples (n = 6 in each group) fixed in 4% paraformaldehyde were embedded in paraffin according to conventional methods. Briefly, 5-μm thick sections were cut onto gelatinized slides, and then these sections were stained using hematoxylin and eosin and photographed under 40× and 100× magnification using an Axio Examiner Zeiss microscope (Zeiss, Germany).
RNA Extraction and qRT-PCR
Expression of the mRNAs of inflammatory cytokines genes and immunoproteasome genes in jejunums was determined by qRT-PCR and the primers are listed in Supplementary Table 3. The jejunal total RNA of each sample was extracted using an RNAiso Plus Kit (TaKaRa Bio Inc., Shiga, Japan); the quantity and quality of the harvested RNAs were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively, and the total RNA of each sample was considered to be valid in the range of 1.9 < OD260/OD280 < 2.0 and 1.8 < 28S/18S < 2.0 simultaneously; cDNA was synthesized for qRT-PCR using a PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa Bio Inc.). Quantitative real-time PCR was performed with RealStar Green Fast Mixture (GenStar Biosolutions, Beijing, China) in a CFX Connect Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The total volume of the PCR reactions was 20 μL comprising 10 μL RealStar Green Fast Mixture, 6 μL RNase-free H2O, 2 μL cDNA, and 1 μL of each primer. The cycling conditions were as follows: 95°C for 2 min, then 40 cycles of 95°C for 5 s, and then 60°C for 30 s. Data analysis was conducted using the 2−△△Ct method, and the β-actin gene was chosen as the internal standard.
Determination of Inflammation-Related Factors in the Jejunums
To determine the levels of lipopolysaccharide (LPS) in jejunal content, nuclear factor kappa-B (NF-κB) in jejunums, and tumor necrosis factor-α (TNF-α) in jejunums, jejunum tissues (or content) of each sample of about 0.1 g with 1 mL PBS were grinded and centrifuged at 3,500 × g for 15 min, and then the total protein was quantified by using an Enhanced BCA Protein Assay Kit (Beyotime Biotech, Shanghai, China); the quantitative analysis was conducted using commercial ELISA kits (MeiMian Industrial Co., Ltd, Yancheng, China) according to the manufacturer's instructions. The results were expressed as ng/g total protein.
DNA Extraction of Jejunal Bacterial Genome
Total jejunal bacterial genomic DNA samples were extracted using FastDNA SPIN extraction kits (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturer's instructions. The quantity and quality of harvested DNAs were measured using a NanoDrop ND-1000 spectrophotometer and 0.8% agarose gel electrophoresis, respectively.
16S rDNA Amplicon Pyrosequencing
PCR amplification of the bacterial 16S rRNA genes V3 to V4 region was performed using the forward primer 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and the reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′). Sample-specific 7-bp barcodes were incorporated into the primers for multiplex sequencing. The PCR system contained 5 μL of Q5 reaction buffer (5×), 5 μL of Q5 High-Fidelity GC buffer (5×), 0.25 μL of High-Fidelity DNA Polymerase (5U/μL), 2 μL (2.5 mmol) of deoxyribonucleotide triphosphates, 1 μL (10 umol) of each forward and reverse primer, 2 μL of DNA template, and 8.75 μL of ddH2O. Thermal cycling was composed of initial denaturation at 98°C for 2 min, followed by 25 cycles of denaturation at 98°C for 15 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, with a final extension of 5 min at 72°C. PCR amplicons were purified with Agencourt AMPure Beads (Beckman Coulter, Indianapolis, IN, USA) and quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). After the individual quantification step, amplicons were pooled in equal amounts, and paired-end 2 × 300 bp sequencing was performed using the Illumina MiSeq platform with MiSeq PE250 at Shanghai Personal Biotechnology Co. Ltd. (Shanghai, China).
Sequence Analysis
The Quantitative Insights Into Microbial Ecology (QIIME, v1.8.0) pipeline was employed to process the sequencing data, as previously described (Caporaso et al., 2010). Briefly, raw sequencing reads with exact matches to the barcodes were assigned to respective samples and identified as valid sequences. The low-quality sequences were filtered using the following criteria (Gill et al., 2006, Chen and Jiang, 2014): sequences that had a length of <150 bp, sequences that had average Phred scores of <20, sequences that contained ambiguous bases, and sequences that contained mononucleotide repeats of >8 bp. Paired-end reads were assembled using FLASH (Magoc and Salzberg, 2011). After chimera detection, the remaining high-quality sequences were clustered into operational taxonomic units (OTU) at 97% sequence identity by UCLUST (Edgar, 2010). A representative sequence was selected from each OTU using default parameters. OTU taxonomic classification was conducted by BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi; NCBI, Bethesda, MD) searching the representative sequences set against the Greengenes Database (DeSantis et al., 2006) using the best hit (Altschul et al., 1997). An OTU table was further generated to record the abundance of each OTU in each sample and the taxonomy of these OTUs. OTUs containing less than 0.001% of the total sequences across all samples were discarded. To minimize the difference of sequencing depth across samples, an averaged, rounded rarefied OTU table was generated by averaging 100 evenly resampled OTU subsets under 90% of the minimum sequencing depth for further analysis.
Bioinformatics and Statistical Analyses
Sequence data analyses were mainly performed using QIIME and R package (v 3.2.0; Lucent Technologies, Inc., Murray Hill, NJ) for MacOS. QIIME (http://qiime.org) is an open-source bioinformatics pipeline for performing microbiome analysis from raw DNA sequencing data. In QIIME, raw sequencing data generated on the Illumina or other platforms can be designed into scientific graphics and statistics, including OTU picking, taxonomic assignment, and diversity analyses and visualizations. OTU-level alpha diversity indices (including Chao1 richness estimator, Abundance-based Coverage Estimator [ACE] metric, Shannon diversity index, and Simpson index) were calculated using the OTU table in QIIME. Beta diversity analysis was also performed in QIIME to investigate the structural variation of microbial communities across samples using UniFrac distance metrics (Lozupone and Knight, 2005, Lozupone et al., 2007) and visualized via nonmetric multidimensional scaling (NMDS) (Ramette, 2007). Linear discriminant analysis (LDA) effect size (LEfSe) was used to detect differentially abundant taxa across groups using the default parameters (Segata et al., 2011), and significant enrichment was considered when log10 LDA score >2 and P < 0.05 at the same time. A relative abundance matrix at the genus level for LEfSe analysis was submitted via the Galaxy online analysis platform (http://huttenhower.sph.harvard.edu/galaxy/). Then, LEfSe can automatically perform statistical analysis on the composition of each classification level, and visualize the analysis results. Other statistical analyses were conducted by one-way ANOVA using SPSS 23.0 (IBM Corporation, Armonk, NY, USA) for MacOS, and the results are expressed as the mean ± SEM. A value of P < 0.05 was considered statistically significant.
Results
Sequencing Results and Quality Control
A total of 299,418 valid reads with length distribution at about 430 bp were harvested from raw sequencing reads (Supplementary Figure 1), and 1,329 OTUs were identified from chicks' jejunum content samples in this experiment (Supplementary Figure 2). A rarefaction curve shows that experimental samples with sufficient sequencing depth were representative of the sampled population (Supplementary Figure 3).
Alpha and Beta Diversity of Jejunal Microbial Communities
To investigate the alpha diversity in each sample, the Chao1 estimator, ACE, Simpson index, and Shannon diversity index were measured (Table 1), and these 4 indices in the Cipro group were significantly higher (all P < 0.05) than those in the cCON group. The Chao1 estimator, ACE, and Shannon diversity indices in the Cipro group were significantly increased (all P < 0.05) compared to those in the cCCAB group, and there was no statistical difference (P > 0.05) between cCON and cCCAB groups in these 4 indices.
Table 1.
Alpha diversity indices in jejunal microbiota of each group.
| Chao1 | ACE | Simpson | Shannon | |
|---|---|---|---|---|
| cCON | 406.27 ± 37.93a | 410.33 ± 40.65a | 0.75 ± 0.06a | 3.94 ± 0.31a |
| Cipro | 591.41 ± 49.42b | 598.14 ± 56.15b | 0.95 ± 0.02b | 6.14 ± 0.34b |
| cCCAB | 423.97 ± 30.80a | 425.55 ± 32.17a | 0.79 ± 0.06a,b | 4.50 ± 0.53a |
a,bMeans with no common superscripts within a column differ significantly (P < 0.05) by Duncan's multiple range test, n = 3.
Abbreviations: cCON, chick control group; cCCAB, chicks from the β-carotene, curcumin, allicin, and sodium butyrate supplementation group; Cipro, ciprofloxacin lactate treatment group.
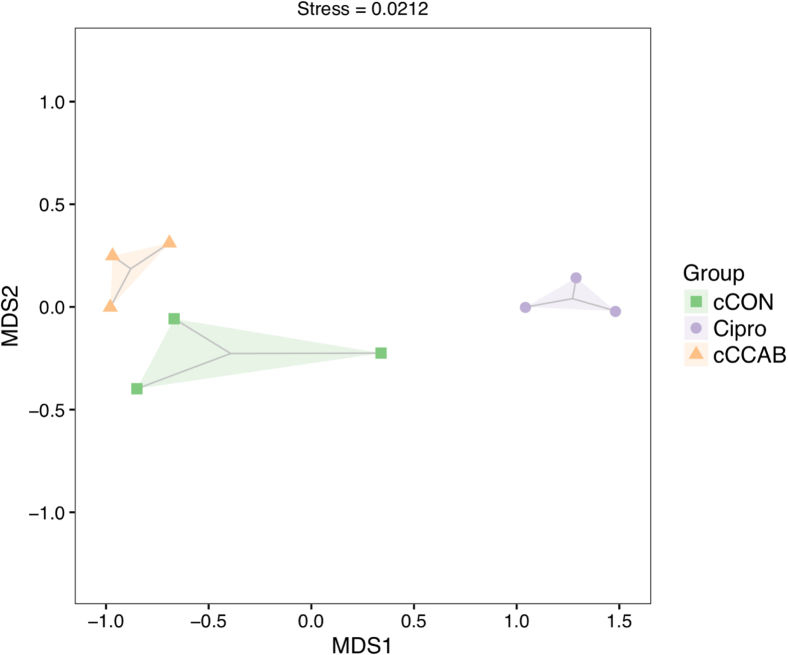
Beta diversity of microbiota in the jejunal content was evaluated by using NMDS as shown in Figure 1, which illustrates that replicates were clustered in the interior of different groups, but the Cipro group was clearly separated away from the cCON and cCCAB groups (stress = 0.0212).
Figure 1.
Beta diversity in jejunal microbiota of each group. n = 3. Abbreviations: cCON, chick control group; Cipro, ciprofloxacin lactate treatment group; cCCAB, chicks from the β-carotene, curcumin, allicin, and sodium butyrate supplementation group; MDS, multidimensional scaling.
Changes in Jejunal Microbial Communities
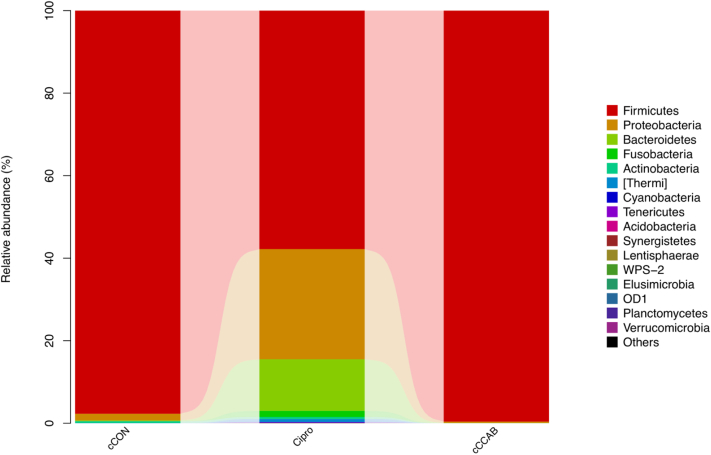
The bacterial constituents at the phylum level in the jejunal content of each group (Figure 2) show that the average relative abundance of Firmicutes at 57.80% in the chicks with ciprofloxacin lactate supplementation was lower than that in cCON (97.70%), whereas Proteobacteria (26.70%), Bacteroidetes (12.50%), and Fusobacteria (1.47%) in the Cipro group were increased compared to the corresponding values in the cCON group (1.63, 0.20, and 0.0%, respectively). In addition, except for the rare occurrence (relative abundance < 1%) of Actinobacteria and Thermi in the cCON and Cipro groups, Cyanobacteria, Tenericutes, and other phyla (unclassified) appeared uniquely in the Cipro group in very small proportions (<1%). However, Firmicutes at 99.60% in the cCCAB group was higher than that in the cCON group (97.70%), and Proteobacteria (0.33%) and Bacteroidetes (0.07%) were lower than the corresponding values in the cCON group (1.63 and 0.20%, respectively).
Figure 2.
The bacterial constituents at the phylum level in jejunal microbiota of each group. [Bacteria] refers to the official classification information for the Greengenes database, but these recommendations have not been included in Berger's Manual. n = 3. Abbreviations: cCON, chick control group; Cipro, ciprofloxacin lactate treatment group; cCCAB, chicks from the β-carotene, curcumin, allicin, and sodium butyrate supplementation group.
As shown in Table 2, at the genus level of jejunal microbial communities, the average relative abundance of the top 10 phyla in the Cipro group differed from the cCON group. The main changes were that Lactobacillus at 26.73% in the Cipro group was decreased compared to the cCON group (93.83%) and cCCAB group (87.6%), and only Lactobacillus, Unclassified Comamonadaceae, and Megamonas were present in the Cipro and cCON groups. In the cCCAB group, the main changes were that Lactobacillus at 87.6% was lower compared to the cCON group at 93.83%, but Enterococcus at 7.5% was higher than the corresponding value in the cCON group at 0.93%.
Table 2.
The top 10 average relative abundance at the genus level in jejuna microbial communities of each group.
| Abundance rank | cCON (relative abundance %) | Cipro (relative abundance %) | cCCAB (relative abundance %) |
|---|---|---|---|
| 1 | Lactobacillus (93.83) | Lactobacillus (26.73) | Lactobacillus (87.60) |
| 2 | Enterococcus (0.93) | Unclassified_Comamonadaceae (11.93) | Enterococcus (7.50) |
| 3 | Unclassified_Peptostreptococcaceae (0.63) | Megamonas (8.67) | Unclassified_Lactobacillales (2.30) |
| 4 | Unclassified_Comamonadaceae (0.60) | Bacteroides (5.70) | Unclassified_Peptostreptococcaceae (1.03) |
| 5 | Unclassified_Lactobacillales (0.57) | Unclassified_Ruminococcaceae (4.40) | Unclassified_Enterobacteriaceae (0.30) |
| 6 | Megamonas (0.30) | Unclassified_Clostridiales (3.43) | Unclassified_[Mogibacteriaceae] (0.30) |
| 7 | Unclassified_Enterobacteriaceae (0.30) | [Ruminococcus] (2.87) | Unclassified_Clostridiales (0.20) |
| 8 | Corynebacterium (0.30) | Unclassified_Burkholderiales (2.47) | Unclassified_Ruminococcaceae (0.17) |
| 9 | Unclassified_Leuconostocaceae (0.30) | Sediminibacterium (2.40) | Megamonas (0.07) |
| 10 | Unclassified_Planococcaceae (0.30) | Lachnospiraceae (2.30) | [Ruminococcus] (0.07) |
[Bacteria] means the official classification information for the Greengenes database, but these recommendations have not been included in Berger's Manual. n = 3.
Abbreviations: cCON, chick control group; cCCAB, chicks from the β-carotene, curcumin, allicin, and sodium butyrate supplementation group; Cipro, ciprofloxacin lactate treatment group.
Statistical Differences of Jejunal Microbiota
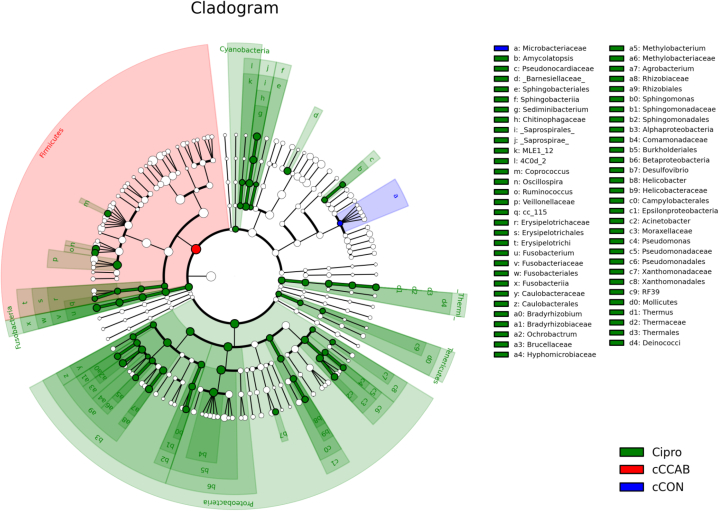
Linear discriminant analysis effect size was used to identify significantly enriched microbiota from the phylum to genus levels (Figure 3). The results show that there were 67 taxa with significant differences (log10 LDA score >2 and P < 0.05) between groups. Firmicutes in the jejunal content of the cCCAB group was significantly enriched (P < 0.05) than that in the cCON and Cipro groups, and Microbacteriaceae in the cCON group was higher (P < 0.05) than the other groups, but the other 65 taxa with statistical differences were enriched (P < 0.05) in the Cipro group. And these significantly enriched (P < 0.05) taxa in the Cipro group were centralized in Proteobacteria, Cyanobacteria, Fusobacteria, [Thermi], and Tenericutes phyla. In addition, 32 of the 65 enriched (P < 0.05) taxa were in the Proteobacteria phylum of the Cipro group, including 3 classes (AlphaProteobacteria, BetaProteobacteria, and EpsilonProteobacteria), 7 orders (Caulobacterales, Rhizobiales, Sphingomonadales, Burkholderiales, Campylobacterales, Pseudomonadales, and Xanthomonadales), 12 families (Caulobacteraceae, Bradyrhizobiaceae, Brucellaceae, Hyphomicrobiaceae, Methylobacteriaceae, Rhizobiaceae, Sphingomonadaceae, Comamonadaceae, Helicobacteraceae, Moraxellaceae, Pseudomonadaceae, and Xanthomonadaceae), and 9 genera (Bradyrhizobium, Ochrobactrum, Methylobacterium, Agrobacterium, Sphingomonas, Desulfovibrio, Helicobacter, Acinetobacter, and Pseudomonas). The other 33 significantly enriched taxa of the Cipro group were mainly distributed in the Cyanobacteria, Fusobacteria, [Thermi], and Tenericutes phyla.
Figure 3.
Significantly enriched microbiota from the phylum to genus levels in jejunal microbiota of each group. _Bacteria_ refers to the official classification information for the Greengenes database, but these recommendations have not been included in Berger's Manual. n = 3. Abbreviations: cCON, chick control group; Cipro, ciprofloxacin lactate treatment group; cCCAB, chicks from the β-carotene, curcumin, allicin, and sodium butyrate supplementation group.
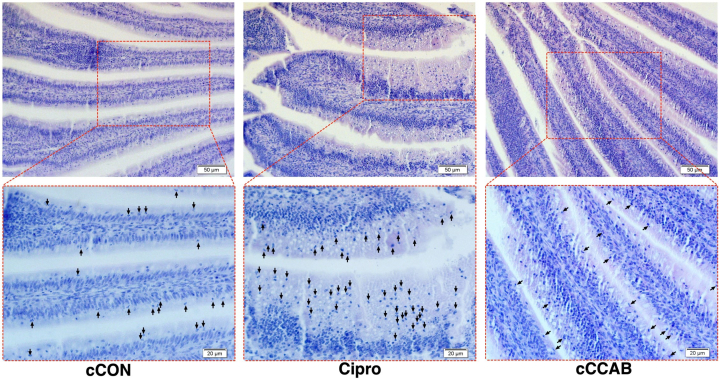
Neutrophil Infiltration in Jejunums
Neutrophils were identified using hematoxylin and eosin staining (Figure 4). Increased neutrophils (arrows) were observed at the jejunal submucosa area of chicks supplemented with ciprofloxacin lactate, but neutrophil infiltration in the jejunums of chicks hatched from the breeder hens with dietary β-carotene, curcumin, allicin, and sodium butyrate supplementation was at a similar level compared to the cCON group.
Figure 4.
Neutrophil infiltration (arrow) in jejunal mucosa of each group. Bar = 50 μm (top row) and 20 μm (bottom row). Abbreviations: cCON, chick control group; Cipro, ciprofloxacin lactate treatment group; cCCAB, chicks from the β-carotene, curcumin, allicin, and sodium butyrate supplementation group.
Expression of Inflammation-Related Factors and Genes in Jejunums
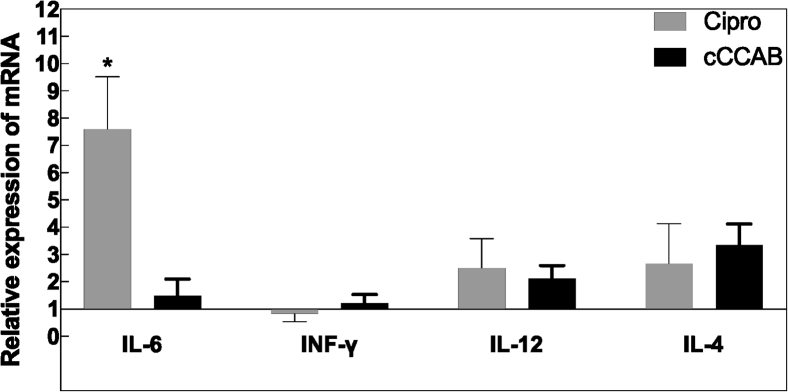
Regarding inflammation-related factors, the values of LPS in jejunal content, NF-κB in jejunums, and TNF-α in jejunums of the Cipro and cCCAB groups were significantly increased (all P < 0.05) compared with the cCON group (Table 3). The expression of jejunal IL-6 gene in the Cipro group was significantly upregulated compared to the cCON (P < 0.05) and cCCAB (P < 0.05) groups, and the expression of jejunal IL-12 and IL-4 genes was also upregulated (all P > 0.05) in the Cipro and cCCAB groups compared to those in the cCON group (Figure 5).
Table 3.
Inflammatory factors in jejunums (or jejunal content) of each group.
| LPS (ng/gprot) | NF-κB (ng/gprot) | TNF-α (ng/gprot) | |
|---|---|---|---|
| cCON | 9.72 ± 2.75a | 72.44 ± 6.32a | 4.65 ± 0.44a |
| Cipro | 25.65 ± 4.05b | 113.97 ± 7.29b | 6.82 ± 0.51b |
| cCCAB | 20.55 ± 1.75b | 112.02 ± 2.41b | 6.45 ± 0.20b |
a,bMeans with no common superscripts within a column differ significantly (P < 0.05) by Duncan's multiple range test, n = 6.
Abbreviations: cCON, chick control group; cCCAB, chicks from the β-carotene, curcumin, allicin, and sodium butyrate supplementation group; Cipro, ciprofloxacin lactate treatment group; LPS, lipopolysaccharide; NF-κB, nuclear factor kappa-B; TNF-α, tumor necrosis factor-α.
Figure 5.
Expression of inflammation-related genes in jejunums of each group. The cCON group is hidden with mean = 1. Significant differences compared with the cCON group are expressed as *P < 0.05 by Duncan’s multiple range test, n = 6. Abbreviations: cCON, chick control group; Cipro, ciprofloxacin lactate treatment group; cCCAB, chicks from the β-carotene, curcumin, allicin, and sodium butyrate supplementation group; INF-γ, interferon gamma.
Expression of Immunoproteasome Genes in Jejunums
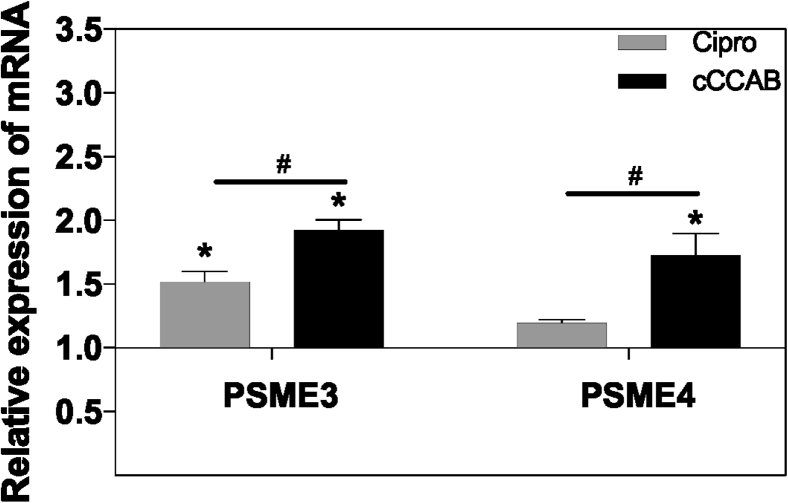
As shown in Figure 6, the expression of jejunal proteasome activator subunit 3 and 4 (PSME3 and PSME4) genes was significantly upregulated in the cCCAB group compared to the cCON and Cipro (both P < 0.05) groups, and the PSME3 gene was upregulated (P < 0.05) in the Cipro group compared to the cCON group.
Figure 6.
Expression of immunoproteasome genes in jejunums of each group. The cCON group is hidden with mean = 1. Significant differences compared with the cCON group are expressed as ∗P < 0.05, and significant differences between the Cipro group and the cCCAB group are expressed as #P < 0.05 by Duncan's multiple range test, n = 6. Abbreviations: cCON, chick control group; Cipro, ciprofloxacin lactate treatment group; cCCAB, chicks from the β-carotene, curcumin, allicin, and sodium butyrate supplementation group; PSME3 and PSME4, proteasome activator subunit 3 and 4.
Discussion
There are multiple indices to access alpha diversity of microbial communities, such as the Chao1 estimator and the ACE estimator that illustrate the microbial richness by evaluating the number of OTUs (Chao, 1984, Chao and Yang, 1993), while the Simpson index and Shannon diversity index emphasize evenness on the microbiota by calculating the differences in abundance of each microbial community (Shannon, 1948, Simpson, 1949). In this study, we found that the jejunal microbiota of chicks supplemented with ciprofloxacin lactate had higher richness and evenness compared to the cCON and cCCAB groups according to the alpha diversity results (Table 1), and the distribution of the Cipro group individuals in NMDS was distant from that of the cCON and cCCAB groups for beta diversity analysis. Therefore, we suggest that the chicks with ciprofloxacin lactate supplementation might lead to higher richness, evenness, and varied microbial structures in jejunums, similar to the results of a recent study reporting that broiler chickens supplemented with an antibiotic (chlortetracycline) showed significantly increased Chao1 and Shannon diversity indices in cecal microbiota after 14 D of treatment (Hong et al., 2019), and a previous study considered antibiotic therapy as a factor that influenced the intestinal microbial composition (Cisek and Binek, 2014).
We also observed that there were obvious changes in jejunal microbial communities of the chicks supplemented with ciprofloxacin lactate in taxonomy and statistics. At the phylum level in taxonomy, the jejunal communities in the cCCAN group were similar to the cCON group in general, but the Cipro group had decreased Firmicutes and increased Proteobacteria compared to the cCON and cCCAB groups. As for the genus level, the primary changes in the jejunal microbiome between the cCON and cCCAB groups were that the cCCAB groups showed slightly lower levels of Lactobacillus of relative abundance and slightly higher Enterococcus. However, Lactobacillus in the jejunal microbial communities of the Cipro group decreased largely compared to the cCON and cCCAB groups, and the total relative abundance of the top 10 genera in the Cipro group was 70.90% only (cCON 98.06%, cCCAB 99.54%), which also supported the results of alpha diversity. Xiao et al. (2017) reported that Firmicutes is the major phylum, and Lactobacillus, widely reported in probiotics, is the most predominant genus in jejunal microbiota of broiler chicken. Therefore, we suggest that ciprofloxacin lactate supplementation could change the jejunal microbial communities from the phylum to genus levels according to the comparison between groups.
In statistical differences of microbial communities, a total of 67 significantly enriched taxa were identified from phyla to genera levels; 65 of the 67 taxa belonged to the Cipro group, and 32 of the 65 taxa were classified in Proteobacteria. Although Proteobacteria is a minor constituent in the intestines of poultry, this group includes many pathogenic bacteria in this phylum, such as Escherichia coli, Salmonella, Vibrio cholerae, and Helicobacter, etc. (Shin et al., 2015). Moreover, Bradyrhizobium, Ochrobactrum, Methylobacterium, Agrobacterium, Sphingomonas, Desulfovibrio, Helicobacter, Acinetobacter, and Pseudomonas genera (classified in the Proteobacteria phylum) in the Cipro group were significantly enriched. Therefore, we investigated the relationship between these genera and hosts from limited studies. Ochrobactrum species are Gram-negative rods belonging to the family Brucellaceae; in particular, Ochrobactrum anthropic and Ochrobactrum intermedium have been increasingly reported as emerging pathogens with infecting capacity in animals (Daxboeck et al., 2002, Teyssier et al., 2005, Hong et al., 2016). Green (2006) suggested that Methylobacterium species also are Gram-negative potential opportunistic pathogens, because they are also found in a variety of contaminants. The genus Desulfovibrio with the function of dissimilatory sulfate reduction may play an important role in the development of ulcerative colitis in humans (Gibson et al., 1993). Helicobacter species are increasingly recognized as microbial pathogens in humans and animals, and Helicobacter pullorum is associated with enteritis and hepatitis in broiler chickens and laying hens, and diarrhea, gastroenteritis, and liver disease in humans (Young et al., 2000, Ceelen et al., 2005). Acinetobacter species are emerging as common pathogens involving the urinary and respiratory tracts in humans (Wise and Tosolini, 1990). And Pseudomonas aeruginosa is the most common cause of Gram-negative pneumonia in humans (Dominguez et al., 2012). However, Firmicutes in the cCCAB group was significantly enriched in our study, and a previous study reported that Firmicutes is the most predominant phylum in Hy-Line layer chicks at 28 D of age (Ballou et al., 2016), with fermentation capacity, enabling the fermentation of more volatile fatty acids and thereby promoting growth performance (Ley et al., 2005). Therefore, our study suggests that 1) chicks with ciprofloxacin lactate administration might lead jejunal microbiota communities toward disorder and abnormality and 2) good growth performance in chicks hatched from laying breeder hens with dietary β-carotene, curcumin, allicin, and sodium butyrate supplementation might depend on the enriched Firmicutes in jejunum.
Microbiota play a fundamental role in induction, education, and function of the host immune system (Belkaid and Harrison, 2017), and disruption of the microbiota by antibiotics use early in life causes an increased risk of inflammation (Scott et al., 2018); so the inflammation-related and immunity-related parameters were investigated in this study. In histological observations, we found that there were more neutrophils in the jejunal mucosa of the Cipro group; the LPS in jejunal content, and NF-κB and TNF-α in jejunums were increased significantly in the Cipro group; and the jejunal IL-6 gene was significantly upregulated in the Cipro group. During the process of inflammation, LPS stimulation is transferred into the intestinal epithelium by Toll-like receptor (Cario et al., 2000), and then the NF-κB transcription family is activated to induce transcription of proinflammatory factors (Tak and Firestein, 2001), such as TNF-α, IL-6, and IL-12, with IL-6 regulating the recruitment of neutrophils to remove inflammation via STAT3 (Fielding et al., 2008). In our study, the increased LPS in jejunal content of the Cipro group might be caused by the disrupted microbiota and significantly enriched Gram-negative bacteria, such as Bradyrhizobium, Ochrobactrum, Methylobacterium, Agrobacterium, Sphingomonas, Helicobacter, Acinetobacter, and Pseudomonas genera, etc. (information source: MicrobeWiki, https://microbewiki.kenyon.edu/index.php/MicrobeWiki). Consequently, our study showed that chicks with ciprofloxacin lactate administration caused inflammatory response in jejunums via ciprofloxacin lactate-induced microbial disturbances to interfere with intestinal immunity and growth. However, we noticed that the levels of LPS, NF-κB, and TNF-α in the cCCAB group were also significantly increased, but expression of IL-6 gene and neutrophils infiltration were at a common level, and PSME3 and PSME4 genes were upregulated significantly. The increased LPS in the jejunal content of the cCCAB group might be caused by the significantly enriched Firmicutes (Figure 3) and the higher richness of microbiota in jejunums (Table 1), because there are Gram-negative stains partly in Firmicutes (Vesth et al., 2013). TNF-α not only plays a key role in inflammatory response, but also controls the immunoproteasome expression for regulating T cell differentiation and survival (Zaiss et al., 2008). Also, immunoproteasome with identified functions in antigen processing for major histocompatibility complex class I helps to diversify the antigenic peptide repertoire facilitating an improved adaptive immune response (Kloetzel and Ossendorp, 2004). Thus, we suggest that the improved jejunal immunity might be contributed by jejunal microbial communities upregulating PSME3 and PSME4 indirectly in the chicks hatched from laying breeder hens with dietary β-carotene, curcumin, allicin, and sodium butyrate supplementation.
In summary, ciprofloxacin lactate administration led to potential hazards in health and growth in chicks via microbial disturbances-induced jejunal inflammation, while the chicks hatched from laying breeder hens with dietary β-carotene, curcumin, allicin, and sodium butyrate supplementation maintained enhanced jejunal immunity via the interaction between the host innate immunity selected microbial colonization and microbiota educated adaptive immunity, possibly. Our study also suggests that 1) long-term antibiotics treatment in the chicks' early life disrupted jejunal microbial communities to impact the development of jejunal adaptive immunity, even inducing inflammation to harm health and growth and 2) development of the offspring chicks in early life benefited from the enhanced maternal immunity by nutritional interventions, and the better jejunal immunity of their offspring chicks might depend on the chick’s innate immune foundation, as well as the evolving adaptive immunity generated by the constant struggle between the host and intestinal microbiota. Moreover, this study indicates that nutritional interventions in laying breeder hens have natural advantages for offspring chicks in early life, and maternal effects may be a potential way to improve chick health and reduce antibiotic use in the poultry industry. We also suggest that strategies should be explored for promoting the health of breeder hens, and that antibiotics as medicines or additives must be used carefully in chicks.
Acknowledgments
This study was supported by the Science and Technology Development Plan Program of Jilin Province (No. 20170204043NY) and the Agricultural Science and Technology Innovation Program for Outstanding Youth of Jilin Province (No. CXGC2017JQ009). The authors thank Shanghai Personal Biotechnology Co. Ltd. (Shanghai, China) for assistance on sequencing and bioinformatics analyses. Finally, the authors thank Yuanyuan Fan (College of Foreign Languages, Jilin Agricultural University, Changchun, China) for a language check of this manuscript.
Conflict of Interest Statement: The authors did not provide a conflict of interest statement.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.03.065.
Contributor Information
X. Zheng, Email: zhengxin@jlau.edu.cn.
M. Wu, Email: 365531954@qq.com.
Supplementary data
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou A.L., Ali R.A., Mendoza M.A., Ellis J.C., Hassan H.M., Croom W.J., Koci M.D. Development of the chick microbiome: how early exposure influences future microbial diversity. Front. Vet. Sci. 2016;3:2. doi: 10.3389/fvets.2016.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y., Harrison O.J. Homeostatic immunity and the microbiota. Immunity. 2017;46:562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cario E., Rosenberg I.M., Brandwein S.L., Beck P.L., Reinecker H.-C., Podolsky D.K. Lipopolysaccharide activates distinct signaling pathways in intestinal epithelial cell lines expressing Toll-like receptors. J. Immunol. 2000;164:966–972. doi: 10.4049/jimmunol.164.2.966. [DOI] [PubMed] [Google Scholar]
- Ceelen L., Decostere A., Verschraegen G., Ducatelle R., Haesebrouck F. Prevalence of Helicobacter pullorum among patients with gastrointestinal disease and clinically healthy persons. J. Clin. Microbiol. 2005;43:2984–2986. doi: 10.1128/JCM.43.6.2984-2986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984;11:265–270. [Google Scholar]
- Chao A., Yang M.C.K. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika. 1993;80:193–201. [Google Scholar]
- Chattopadhyay M.K. Use of antibiotics as feed additives: a burning question. Front. Microbiol. 2014;5:334. doi: 10.3389/fmicb.2014.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Jiang W. Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Fron. Microbiol. 2014;5:508. doi: 10.3389/fmicb.2014.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisek A.A., Binek M. Chicken intestinal microbiota function with a special emphasis on the role of probiotic bacteria. Pol. J. Vet. Sci. 2014;17:385–394. doi: 10.2478/pjvs-2014-0057. [DOI] [PubMed] [Google Scholar]
- Daxboeck F., Zitta S., Assadian O., Krause R., Wenisch C., Kovarik J. Ochrobactrum anthropi bloodstream infection complicating hemodialysis. Am. J. Kidney Dis. 2002;40:E17. doi: 10.1053/ajkd.2002.35759. [DOI] [PubMed] [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez J.A., Xie Y., Dunne W.M., Yoseph B.P., Burd E.M., Coopersmith C.M., Davidson N.O. Intestine-specific Mttp deletion decreases mortality and prevents sepsis-induced intestinal injury in a murine model of Pseudomonas aeruginosa pneumonia. PLoS One. 2012;7:e49159. doi: 10.1371/journal.pone.0049159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Fielding C.A., McLoughlin R.M., McLeod L., Colmont C.S., Najdovska M., Grail D., Ernst M., Jones S.A., Topley N., Jenkins B.J. IL-6 regulates neutrophil trafficking during acute inflammation via STAT3. J. Immunol. 2008;181:2189–2195. doi: 10.4049/jimmunol.181.3.2189. [DOI] [PubMed] [Google Scholar]
- Gensollen T., Iyer S.S., Kasper D.L., Blumberg R.S. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G.R., Macfarlane G.T., Cummings J.H. Sulphate reducing bacteria and hydrogen metabolism in the human large intestine. Gut. 1993;34:437. doi: 10.1136/gut.34.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S.R., Pop M., Deboy R.T., Eckburg P.B., Turnbaugh P.J., Samuel B.S., Gordon J.I., Relman D.A., Fraser-Liggett C.M., Nelson K.E. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H.Z., Wu M., Lang W.Y., Yang M., Wang J.H., Wang Y.Q., Zhang Y., Zheng X. Effects of laying breeder hens dietary beta-carotene, curcumin, allicin, and sodium butyrate supplementation on the growth performance, immunity, and jejunum morphology of their offspring chicks. Poult. Sci. 2020;99:151–162. doi: 10.3382/ps/pez584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P.N. Methylobacterium. The Prokaryotes. 2006;5:257–265. [Google Scholar]
- Hong Y., Cheng Y., Li Y., Li X., Zhou Z., Shi D., Li Z., Xiao Y. Preliminary study on the effect of Bacillus amyloliquefaciens TL on cecal bacterial community structure of broiler chickens. Biomed. Res. Int. 2019;2019:5431354. doi: 10.1155/2019/5431354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D.J., Kim K.H., Kim J.O., Hong J.S., Jeong S.H., Lee K. First Case Report of human infection with Ochrobactrum tritici causing bacteremia and Cholecystitis. Ann. Lab. Med. 2016;36:278–280. doi: 10.3343/alm.2016.36.3.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L.V., Littman D.R., Macpherson A.J. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloetzel P.-M., Ossendorp F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr. Opin. Immunol. 2004;16:76–81. doi: 10.1016/j.coi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Ley R.E., Bäckhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Eniron. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C.A., Hamady M., Kelley S.T., Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 2007;62:142–160. doi: 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott N.A., Andrusaite A., Andersen P., Lawson M., Alcon-Giner C., Leclaire C., Caim S., Le Gall G., Shaw T., Connolly J.P.R. Antibiotics induce sustained dysregulation of intestinal T cell immunity by perturbing macrophage homeostasis. Sci. Transl. Med. 2018;10:eaao4755. doi: 10.1126/scitranslmed.aao4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423. [Google Scholar]
- Shaw S.Y., Blanchard J.F., Bernstein C.N. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am. J. Gastroenterol. 2010;105:2687. doi: 10.1038/ajg.2010.398. [DOI] [PubMed] [Google Scholar]
- Shin N.R., Whon T.W., Bae J.W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Simpson E.H. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- Tak P.P., Firestein G.S. NF-κB: a key role in inflammatory diseases. J. Clin. Invest. 2001;107:7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyssier C., Marchandin H., Jean-Pierre H., Diego I., Darbas H., Jeannot J.L., Gouby A., Jumas-Bilak E. Molecular and phenotypic features for identification of the opportunistic pathogens Ochrobactrum spp. J. Med. Microbiol. 2005;54:945–953. doi: 10.1099/jmm.0.46116-0. [DOI] [PubMed] [Google Scholar]
- Vesth T., Ozen A., Andersen S.C., Kaas R.S., Lukjancenko O., Bohlin J., Nookaew I., Wassenaar T.M., Ussery D.W. Veillonella, firmicutes: microbes disguised as gram negatives. Stand. Genomic Sci. 2013;9:431. doi: 10.4056/sigs.2981345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise K.A., Tosolini F.A. Epidemiological surveillance of Acinetobacter species. J. Hosp. Infect. 1990;16:319–329. doi: 10.1016/0195-6701(90)90004-8. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Xiang Y., Zhou W., Chen J., Li K., Yang H. Microbial community mapping in intestinal tract of broiler chicken. Poult. Sci. 2017;96:1387–1393. doi: 10.3382/ps/pew372. [DOI] [PubMed] [Google Scholar]
- Young V.B., Chien C.-C., Knox K.A., Taylor N.S., Schauer D.B., Fox J.G. Cytolethal distending toxin in avian and human isolates of Helicobacter pullorum. J. Infect. Dis. 2000;182:620–623. doi: 10.1086/315705. [DOI] [PubMed] [Google Scholar]
- Zaiss D.M.W., de Graaf N., Sijts A.J.A.M. The proteasome immunosubunit multicatalytic endopeptidase complex-like 1 is a T-cell-intrinsic factor influencing homeostatic expansion. Infect. Immun. 2008;76:1207–1213. doi: 10.1128/IAI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.