Abstract
During the process of transmission and spread of avian leukosis virus subgroup J (ALV-J) in chickens worldwide, the viral genome is constantly changing. A comprehensive and systematic study of the evolutionary process of ALV-J in China is needed. In this study, we amplified the full-length viral cDNA sequences of 16 ALV-J isolates of Yellow-chicken origin and analyzed and compared these sequences with another 69 ALV-J strains isolated during the years 1988–2018. These isolates were then sorted into 2 clusters: cluster I included isolates that mainly originated from the layers and White-feather broilers from northern China; cluster II included isolates mainly from the Yellow-chicken, most of them being from southern China. According to the sequence homologies of the whole genome and gag, pol, gp85, and gp37 genes, the ALV-J strains are more likely to randomly change in different directions from the original strain HPRS-103 as time passes. The results of entropy analysis of the sequences of gag, pol, and env revealed that the env gene had the largest variation, and the gag gene nonconserved sites are mainly concentrated in p19, p10, and p12. In addition, 84.71% (72/85) of the isolates had the 205-nucleotide (nt) deletion in the 3′UTR region, and 30.59% (26/85) of the isolates had the 125-nt to 127-nt deletion in the E element. Our study provides evidence for the coexistence of 2 extremely different clusters of ALV-J prevailing in China and in some other countries during the period of 1988–2018 and implies that the clusters are highly dependent on the host genetic background and the geographic location.
Key words: subgroup J avian leukosis viruses, complete genome sequences, genetic variations, genetic background, geographic location
Introduction
Avian leukosis viruses (ALV) are known as a variety of infectious retroviral agents that causes severe economic loss to the poultry industry worldwide (Payne and Nair, 2012). Avian leukosis viruses that infect chickens comprise 7 envelope subgroups designated A–E, J, and K (Mu et al., 2020). Among these subgroups, subgroup J (ALV-J) is the most prevalent in chickens (Zhang et al., 2020). Avian leukosis virus subgroup J was first isolated from meat-type breeder chickens in 1988 and later proved to have a broader neoplastic spectrum (Payne et al., 1991). By the early to mid-1990s, the virus had spread to the USA and to European countries and by the early 2000s to China, spreading rapidly throughout the country (Cui et al., 2003). The host range of ALV-J extended to commercial layers (Lai et al., 2011), broilers (Li et al., 2012), local breeds (Sun and Cui, 2007), and wild birds (Jiang et al., 2014, Shen et al., 2014, Zeng et al., 2014), thus causing great loss to the poultry industry in China (Li et al., 2019b). Owing to the initiation of a Nationwide Eradication Program since 2008 (Wei and Cui, 2015), the infection of the exogenous ALV was significantly reduced among White-feather broilers, layer chickens, and native chickens. However, there was another breakout almost simultaneously in some provinces of China during 2018–2019 (Zhou et al., 2018). In addition, various ALV-J field strains are still circulating in Yellow-chicken breeds where full and strict eradication programs have not been initiated or completed (Li et al., 2019a).
Avian leukosis virus belongs to the genus Alpharetrovirus and the family Retroviridae (Wang et al., 2019). The proviral DNA arrangement of the genome of ALV was 5′LTR-leader-gag-pol-env-3′LTR and the full-length of the genome sequences of ALV was 7,200 bp-7,800 bp (Wang et al., 2018a). The LTR is the nonstructural gene, whereas gag, pol, and env are the structural genes (Meng et al., 2018). The gag gene encodes ALV group–specific antigen and some other proteins (matrix protein, nucleocapsid protein, etc.); pol gene encodes the reverse transcriptase and integrase; env gene encodes the gp37 transmembrane (TM) and the gp85 surface proteins (Li et al., 2018). Although previous studies have been conducted, most have focused on strains from only 1 or 2 genetic backgrounds (Lin et al., 2016, Lin et al., 2017).
To better understand the genomic evolution and variation of the field ALV-J, we analyzed and compared the full-genome sequences of 85 ALV-J isolates, originating from China, UK, USA, and Russia during the years 1988–2018. Our results focused on the genomic characterization of ALV-J isolates and provide evidence to further understand the diversity and complexity of ALV in chickens.
Material and methods
Samples and Virus Isolation
Birds with naturally occurring neoplasms suspected of ALV infection were sent to our laboratory for diagnosis during the period of 2014–2018. A continuous chicken embryo fibroblast cell line DF-1 cells, free of endogenous avian leukosis viruses, was used to isolate ALV (Maas et al., 2006). Briefly, about 2 × 106 DF-1 cells were plated in each well of a 24-well plate. Twenty-four hours later, plasma separated from the blood samples of the birds was inoculated into the monolayer cells for 1h incubation and then replenished with Dulbecco's modified Eagle's medium containing 2% fetal calf serum (Gibco, Australia). Then, the cells were incubated at 37°C in 5% CO2 for 3 serial passages (7 D per passage). Uninfected DF-1 cells served as the negative control.
Virus Identification
After 3 blind passages in DF-1 cells, the viruses were respectively confirmed by indirect immunofluorescence assay using subgroup specific monoclonal antibody JE-9 (Wang et al., 2019). The culture supernatants of the third passage of each sample were harvested for the detection of ALV group-specific antigen p27 with the enzyme-linked immunosorbent assay kit (IDEXX, Westbrook, ME). DNA from the cell samples was prepared using a commercial DNA extraction kit (Tiangen, Beijing, China) and further used for the PCR detection of MDV, REV, and ALV, respectively. The PCR primers used were designed and synthesized according to the sequence published in GenBank and referenced in the literature (Table 1).
Table 1.
Primers for PCR amplifications to detect MDV, REV, and ALV and for amplifying the entire proviral genome of ALV-J.
| Primer1 | Sequence (5′-3′) | Position (nt)2 | Size of PCR product (bp) |
|---|---|---|---|
| MDV meq F | CCGTCTAGAAGGCGGGCACGGTAC | 133,351-133,374 | 1,113 |
| MDV meq R | CGGAAGCTTAAACATGGGGCATAGACG | 134,437-134,463 | |
| MDV 132F | TGCGATGAAAGTGCTATGGAGG | 127,553-127,574 | 185, 317 449, 581 |
| MDV 132 R | GAGAATCCCTATGAGAAAGCGC | 127,848-127,869 | |
| REV LTR F | CATACTGGAGCCAATGGT | 335-351 | 291 |
| REV LTR R | AATGTTGTAGCGAAGTACT | 609-627 | |
| ALV-J-F | GGATGAGGTGACTAAGAAAG | 5,258-5,277 | 545 |
| ALV-J-R | CGAACCAAAGGTAACACACG | 5,783-5,802 | |
| ALV-A-F | GGATGAGGTGACTAAGAAAG | 5,026-5,045 | 692 |
| ALV-A-R | AGAGAAAGAGGGGTGTCTAAGGAG | 5,694-5,717 | |
| ALV-B-F | GGATGAGGTGACTAAGAAAG | 5,053-5,072 | 847 |
| ALV-B-R | ATGGACCAATTCTGACTCATT | 5,879-5,899 | |
| ALV-C-F | GGATGAGGTGACTAAGAAAG | 5,267-5,286 | 860 |
| ALV-C-R | GAGGCCAGTACCTCCCACG | 6,108-6,126 | |
| ALV-D-F | GGATGAGGTGACTAAGAAAG | 5,020-5,039 | 797 |
| ALV-D-R | ATCCATACGCACCACAGTATTCG | 5,795-5,817 | |
| ALV-K-F | GATGAGGCGAGCCCTCTCTTTG | 5,273-5,294 | 2,354 |
| ALV-K-R | TGTTGGGAGGTAAAATGGCGT | 7,607-7,627 | |
| J-1F | TGTAGTGTTATGCAATACTCTTATGTAACGATGAAAC | 1-37 | 3,621 |
| J-1R | GGCCATTTTCATGTCTAGATT | 3,600-3,620 | |
| J-2F | GGCGAGGGAATGGAATCT | 3,587-3,604 | 3,069 |
| J-2R | CATCGATTTCTTACTCCTGGCGC | 6,633-6,655 | |
| J-3F | CGAGCAGCCATCGATTTCTTACTC | 6,625-6,648 | 1,216 |
| J-3R | CCTGACGACTACGAGCACCTGCATGAAGCGGATGGCTTCA | 7,802- 7,841 |
Abbreviations: ALV, avian leukosis virus.
F and R denote the forward and reverse primers for a specific fragment, respectively.
Corresponding strain was the standard strain. MDV (GA, AF147806); REV (SNV, DQ003591); ALV-J (HPRS-103, Z46390); ALV-A (RSA, M37980); ALV-B (Schmidt-Ruppin B, AF052428); ALV-C (Prague C, J02342); ALV-D (Schmidt-Ruppin D, D10652); ALV-K (JS11C1, KF746200).
ALV-J Genome Sequencing
The DNA samples were extracted from the DF-1 cells infected with the isolated ALV-J strains by means of a DNA Extraction Kit (Tiangen). Three primer pairs were designed according to the published sequences of HPRS-103 to amplify 3 fragments that overlapped with each other and covered the whole-genome sequences of ALV-J (Table 1). The conditions for PCR were as follows: 95°C, 3 min; 95°C, 15 s; 50–60°C, 15 s; 72°C, 30 s/kb (31 cycles); 72°C, 7 min. The PCR products were analyzed by electrophoresis using 1% agarose in a Tris-EDTA buffer gel containing 0.5 mg/mL ethidium bromide. All the PCR products were isolated from agarose gels and purified with a Universal DNA Purification Kit and then cloned into the pMD18-T vector (Takara, Dalian, China) and transfected into Escherichia coli DH5α competent cells. Clones containing recombinant plasmids were confirmed by the PCR detection, and 3 independent recombinant plasmids were sequenced (BGI, Shenzhen, China) to avoid read errors caused by genetic variations.
Sequence Analysis
The analysis utilized the whole-genome sequences of our ALV-J isolates (n = 16) and the available ALV-J sequences from GenBank (n = 69). Cell-passaged strains and rescued clone strains were excluded from the analysis (Table 2). The nt sequences were retrieved, edited, and analyzed using the EditSeq and MegAlign modules of DNASTAR (ver. 7.1.0). Multiple alignments of ALV-J sequences were generated using MAFFT (ver. 7.311). Phylogenetic analysis of the genomic sequences of ALV-J was performed with the maximum-likelihood and neighbor-joining method using MEGA X (ver.10.1.7). The DAMBE (ver. 5.2.73) was used to test of substitution saturation. Nucleotide and amino acid (aa) entropy analyses were performed using BioEdit (ver.7.2.0) and the online service system Weblogo (http://weblogo.berkeley.edu). The Geneious program (ver.9.1.4) was used for the comparison of the 3′UTR sections of ALV-J strains. Data were analyzed using GraphPad Prism, version 5.0, software and are expressed as means. The one-way ANOVA analysis was used to assess differences among clusters in the Statistical Package for Social Science Windows, version 19, software.
Table 2.
The ALV-J strains used in the study.
| Isolate | Accession no. | Origin | Year | Host | Isolate | Accession no. | Origin | Year | Host |
|---|---|---|---|---|---|---|---|---|---|
| GX14NN01 | MN066154 | Guangxi | 2014 | YL | GX17NN05 | MN066145 | Guangxi | 2017 | YL |
| GX14NN02 | MN066153 | Guangxi | 2014 | YL | GX17NN06 | MN066144 | Guangxi | 2017 | YL |
| GD15MM01 | MN066152 | Guangxi | 2015 | YL | GX17YL05 | MN066142 | Guangxi | 2017 | YL |
| GD15MM02 | MN066151 | Guangxi | 2015 | YL | GX17YL01 | MN066143 | Guangxi | 2017 | YL |
| GX15MM61 | MN066150 | Guangxi | 2015 | YL | GX18NN01 | MN066141 | Guangxi | 2018 | YL |
| GX15MM6-2 | KU934276 | Guangxi | 2015 | YL | GX18NN02 | MN066140 | Guangxi | 2018 | YL |
| GX16MM92 | MN066149 | Guangxi | 2016 | YL | SCDY1 | HQ425636 | Sichuan | 2010 | CL |
| GX16YL01 | MN066148 | Guangxi | 2016 | YL | SCGS-1 | JQ396302 | Sichuan | 2010 | GG |
| GX16YL02 | MN066147 | Guangxi | 2016 | YL | SD1005 | KF562375 | Shandong | 2010 | GG |
| GX16ZS01 | MN066146 | Guangxi | 2016 | YL | sdau1001 | JN389517 | Shandong | 2010 | CL |
| SDAU1701 | KY980657 | Shandong | 2017 | CL | SDAU1005 | KT156668 | Shandong | 2010 | CL |
| SDAU1702 | KY980658 | Shandong | 2017 | CL | SDAU1102 | KU159178 | Shandong | 2010 | CL |
| SDAU1703 | KY980659 | Shandong | 2017 | CL | WN100401 | KC711043 | Shandong | 2010 | Wb |
| SDAU1704 | KY980660 | Shandong | 2017 | CL | CAUGX01 | JF931999 | Beijing | 2009 | WB |
| SDAU1705 | KY980661 | Shandong | 2017 | CL | CAUHM01 | JF932000 | Beijing | 2009 | CL |
| SDAU1706 | KY980662 | Shandong | 2017 | CL | CAUSY01 | JF932001 | Beijing | 2009 | CL |
| K243 | KX611833 | Guangdong | 2016 | YL | CAUTS01 | JF932002 | Beijing | 2009 | CL |
| M180 | KX611834 | Guangdong | 2016 | YL | CAUXT01 | JF932003 | Beijing | 2009 | CL |
| GD1406-H | KU500033 | Guangdong | 2014 | YL | CAUYL01 | JF932004 | Beijing | 2009 | CL |
| GD1407 | KU500034 | Guangdong | 2014 | YL | CLB908 M | JX855935 | Russia | 2009 | GG |
| GD1407-L | KU500035 | Guangdong | 2014 | YL | CLB908U | JQ935966 | Russia | 2009 | GG |
| GD1408-1 | KU500036 | Guangdong | 2014 | YL | HAY013 | HM235665 | Jiangsu | 2009 | YL |
| GD1408-2 | KU500037 | Guangdong | 2014 | YL | HLJ09MDJ-1 | LN624880 | Heilongjiang | 2009 | CL |
| GD1411-1 | KU500038 | Guangdong | 2014 | YL | JL09H01 | HQ148554 | Heilongjiang | 2009 | CL |
| GD1411-2 | KU500039 | Guangdong | 2014 | YL | JL09L01 | HQ148555 | Heilongjiang | 2009 | CL |
| GD1411-3 | KU500040 | Guangdong | 2014 | YL | JL093-1 | JN624878 | Jilin | 2009 | CL |
| GD1411-4 | KU500041 | Guangdong | 2014 | YL | JS09GY3 | GU982308 | Jiangsu | 2009 | CL |
| GD14J2 | KU500032 | Guangdong | 2014 | YL | JS09GY6 | GU982310 | Jiangsu | 2009 | CL |
| GDQJ2 | KU156826 | Guangdong | 2014 | YL | MRL905 | JF951728 | Russia | 2009 | GG |
| GX14HG01 | KU997685 | Guangxi | 2014 | YL | SD09DP03 | JN624879 | Shandong | 2009 | CL |
| GX14HG04 | KX058878 | Guangxi | 2014 | YL | SVR807 | HM776937 | Russia | 2008 | GG |
| GX14LT07 | KX034517 | Guangxi | 2014 | YL | NHH | HM235668 | Jiangsu | 2007 | CL |
| GX14ZS14 | KX037423 | Guangxi | 2014 | YL | PDRC-59831 | KP284572 | USA | 2007 | WB |
| GD13GZ | KU500030 | Guangdong | 2013 | YL | SCAU-HN06 | HQ900844 | Guangdong | 2007 | CL |
| GD13HY | KU500031 | Guangdong | 2013 | YL | SD07LK1 | FJ216405 | Shandong | 2007 | CL |
| HLJ13SH01 | KM376510 | Heilongjiang | 2013 | CL | JS-nt | HM235667 | Jiangsu | 2003 | WB |
| BR119 | KF562373 | Shandong | 2012 | YL | NM2002-1 | HM235669 | Inner Mongolia | 2002 | WB |
| GDKP1202 | JX453210 | Guangdong | 2012 | YL | ADOL-7501 | AY027920 | USA | 2001 | WB |
| GDQY1201 | JX423792 | Guangdong | 2012 | YL | NX0101 | DQ115805 | Ningxia | 2001 | WB |
| SCAU11-XG | KC149971 | Guangdong | 2012 | WB | YZ9902 | HM235670 | Jiangsu | 1999 | WB |
| GD1109 | JX254901 | Guangdong | 2011 | CL | HPRS103 | Z46390 | UK | 1988 | WB |
| SCAU11-H | KC149972 | Guangdong | 2011 | WB | WB11098 | JX848322 | Heilongjiang | 2011 | Wb |
| SD110503 | KF562374 | Shandong | 2011 | LC |
Boldface represents the virus strains isolated in this study.
Abbreviations: ALV-J, avian leukosis virus subgroup J; CL, Commercial Layer Chicken; GG, Gallus gallus; WB, White-Feather Broiler; Wb, Wild Bird; YL, Yellow Chicken.
Results
Isolation and Identification of ALV-J
The results showed that a total of 16 DF-1 cell cultures inoculated with plasma samples were positive for ALV p27 antigen (Data not shown) and indirect immunofluorescence assay (Supplementary Figure 1). Furthermore, genomic cDNA was extracted from samples positive for the ALV-p27 antigen and was amplified by PCR with the ALV-specific universal primers and subgroup-specific primers, respectively, and only ALV-J could be amplified; no PCR products were obtained with primers specific for ALV-A, ALV-B, ALV-C, ALV-D, ALV-K, MDV, and REV (Data not shown). The full-length genome sequences of the 16 isolates have been submitted to GenBank with accession numbers MN066140-MN066154 and KU934276, respectively (Table 2).
Summary of ALV-J Virus Strains Used in the Analysis
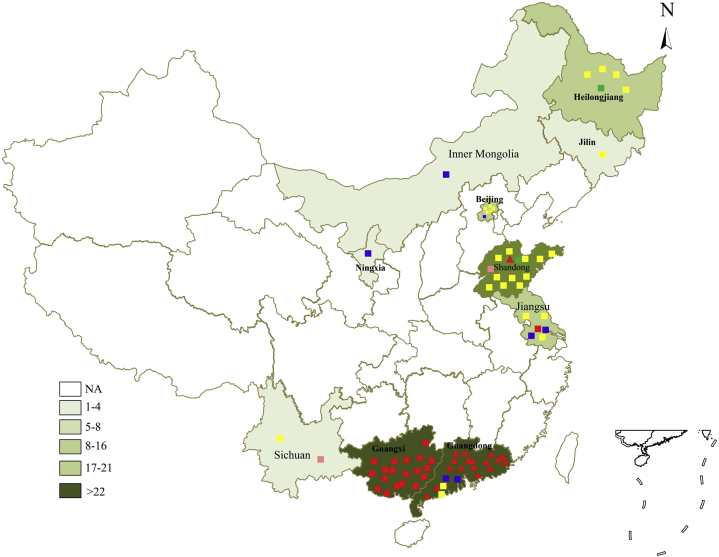
A total of 16 ALV-J strains' complete genomes were sequenced in our laboratory during 2014–2018, as listed in Table 2. A total of 78 strains of ALV-J including our isolates and 62 reference strains were marked on a map of China (Figure 1.). The deeper colors in Figure 1 show the provinces having more strains isolated.
Figure 1.
Geographical distribution of ALV-J isolated in China. The deeper the color suggests the province have more ALV-J strain isolations. ▪ ALV-J isolated from our lab; ▲ ALV-J isolated from Chinese Yellow-chicken; ▪ ALV-J isolated from White-feather broilers; ▪ ALV-J isolated from layer chickens; ▪ ALV-J isolated from Gallus gallus; ▪ ALV-J isolated from wild birds. Abbreviation: ALV-J, avian leukosis virus subgroup J.
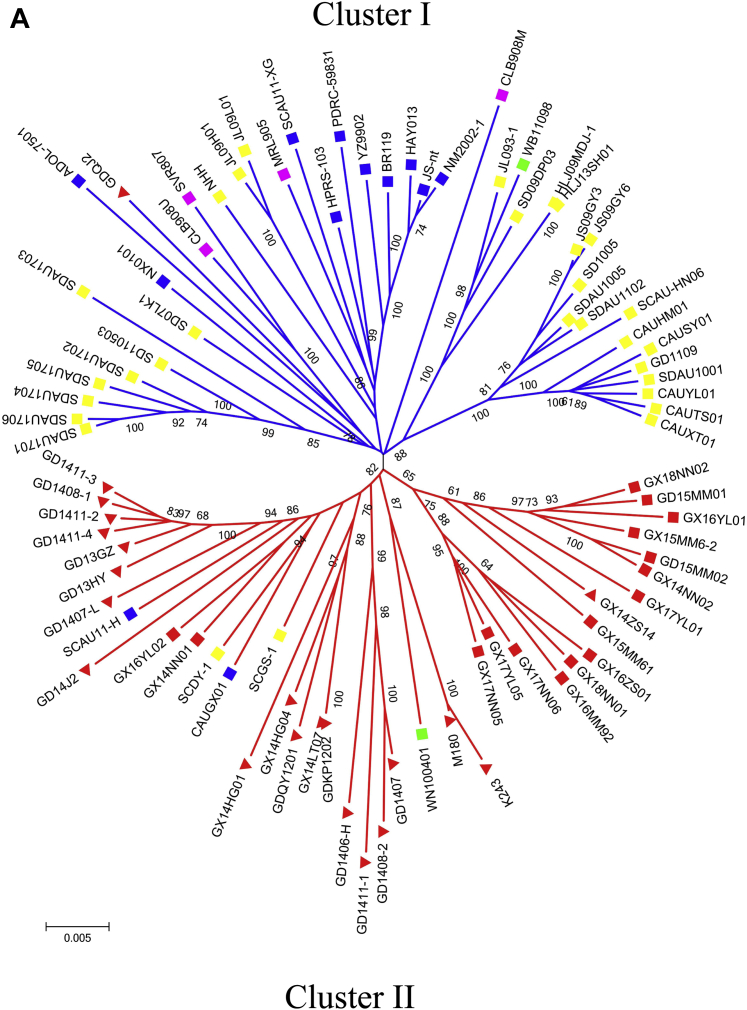
Phylogenetic Analysis
Both the phylogenetic trees constructed with the Neighbor-joining and maximum-likelihood methods showed that they had very similar topography, so only the Neighbor-joining trees are shown here (Figure 2), and the maximum-likelihood trees are shown in Supplementary Figure 2. From the phylogenetic tree analysis of the whole-genome sequence (Figure 2A), we found that 2 extremely different clusters were coexistent among the 85 isolates from birds of different genetic backgrounds. Cluster I includes 28 layer isolates, 10 White-feather broilers' isolates, 4 Gallus' isolates, 1 wild-bird isolate, and 1 Yellow-chicken isolate. Cluster II includes 36 Yellow-chicken isolates, 2 White-feather broilers' isolates, 2 layer isolates, and 1 wild-bird isolate.
Figure 2.
Phylogenetic analysis of the of ALV-J isolates based on nucleotide sequences of the whole-genome and the gag, pol, gp85, and gp37 genes. (A) Whole-genome; (B) GAG; (C) POL; (D) GP85; (E) GP37. ▪ ALV-J isolated from our lab; ▲ ALV-J isolated from Chinese Yellow-chicken;▪ ALV-J isolated from White-feather broilers; ▪ ALV-J isolated from layer chickens; ▪ALV-J isolated from Gallus gallus; ▪ ALV-J isolated from wild birds. The phylogenetic tree was constructed by using the neighbor-joining method based on nucleotide alignment with the Maximum Composite Likelihood Model with 1,000 bootstrap replicates using the software MEGA X 10.1.7. Abbreviation: ALV-J, avian leukosis virus subgroup J.
The phylogenetic trees based on gag and pol gene aa sequences showed that there is no obvious cluster, and the strains isolated from different genetic backgrounds were interlaced (Figures 2B and 2C). The phylogenetic trees of gp85 and gp37 genes showed similar results to the phylogenetic tree based on the whole-genome (Figures 2D and 2E).
Alignment Analysis of nt- and the Deduced aa-Sequences
The whole proviral genomes of the 16 isolates were 7,583-7,634-nt in length. Sequence analysis showed that the maximum divergence throughout the complete genomes was 3.3%, with nt-sequence similarities ranging from 95.2 to 98.5%. The nt-sequence similarities of the gag, pol, and gp37 genes among the 16 isolates were 94.9–99.6% (aa: 95.3–99.7%), 96.6–100% (aa: 95.5-100%), and 97.1–99.6% (aa: 97.9–99.8%), respectively. Further analysis showed that the nt-sequence similarities of gag, pol, and gp37 genes between the 16 isolates and all the reference strains were 94.1–99.6% (aa: 95.1–99.7%), 96.8–99.6% (aa: 96.7–99.8%), and 96.7-100% (aa: 92.0–100%), respectively. These results indicated that the gag, pol, and gp37 genes were highly conserved. Nonetheless, the gp85 gene was found to be much more variable: the 16 isolates shared 92.1–100% nt-sequence similarities and 86.8–100% aa-sequence identities on the gp85 protein with one another, and the nt-sequence similarities of gp85 genes between the 16 isolates and all the reference strains were 78.5–100% (aa: 87.7–100%).
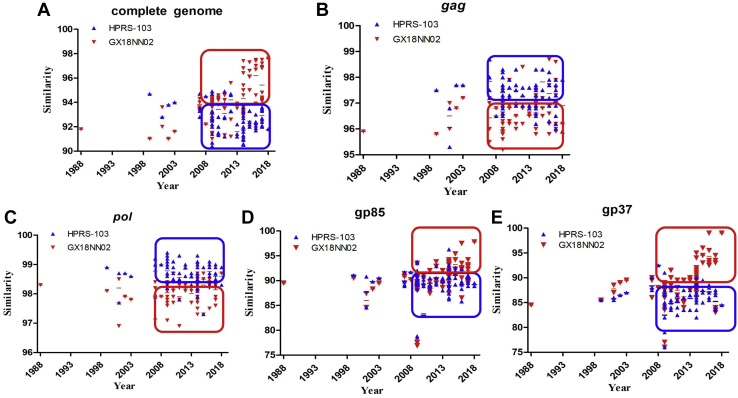
The similarities of ALV-J complete genome, gag, pol, gp85, and gp37 between the prototype HPRS-103, the latest isolate GX18NN02, and the strains isolated in different years are shown in Table 3 and Figure 3. The results of the whole-genome, gag, pol, gp85, and gp37 sequence comparisons indicated that the similarities of ALV-J genome sequences in different strains were irrelevant to the isolated time. According to the similarity analysis of these genes, the ALV-J strains were more likely to randomly change in different directions from the original strains HPRS-103 over time.
Table 3.
Similarity of nucleotides between the isolates with HPRS-103 and GX18NN02.
| Genes | HPRS-103 | GX18NN02 |
|---|---|---|
| Complete genome | 90.2%–94.9% | 90.1%–97.6% |
| gag | 95.3%–98.7% | 95.1%–98.7% |
| pol | 97.3%–99.4% | 96.9%–99.9% |
| gp85 | 78.5%–95.6% | 76.9%–97.8% |
| gp37 | 76.0%–92.5% | 76.0%–99.0% |
Figure 3.
A correlation comparison between the ALV-J isolates and HPRS-103, GX18NN02 on the sequences of the whole-genome and the gag, pol, gp85, and gp37 genes. (A) Whole genome; (B) GAG; (C) POL; (D) GP85; (E) GP37. Data shown represent means. Abbreviation: ALV-J, avian leukosis virus subgroup J.
Further analysis of the sequences of subunits of these genes revealed some interesting findings. The homologies of ALV-J whole-genome, gag, pol, gp85, and gp37 among the prototype HPRS-103, the latest isolate GX18NN02, and the strains isolated after 2008 showed extremely significant differences (P < 0.01). The sequence similarities of the whole-genome, gp85, and gp37 between the prototype HPRS-103 and strains isolated after 2008 were much higher than those between GX18NN02 and strains isolated after 2008 (P < 0.01), but that of the pol and gag genes are just the opposite (Figure 3.).
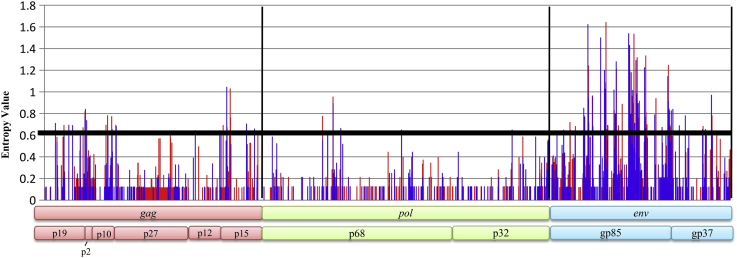
An entropy value greater than 0.6 indicates the corresponding aa-site in the gene sequence is not conserved (Mo et al., 2013). The higher the peak, the greater the entropy is, indicating the higher variation frequency of aa sites. The results of analysis of entropy of gag, pol, and env genes on aa-sequences are shown in Figure 4 and Table 4. The percentage entropy greater than 0.6 and the average entropy of each cluster's structure genes are shown in Table 4. By comparing the average entropy of each structural gene and the number of aa-sites that are not conserved, we found that viral structural genes in the cluster II strains are more variable than those in the cluster I strains. Further analysis found that the gag gene nonconserved sites are mainly concentrated in p19, p10, and p12, the pol gene nonconserved sites are mainly concentrated in p68, and the env gene nonconserved sites are mainly concentrated in gp85.
Figure 4.
Amino acid entropy analyses of the viral structure genes. The X-axis represents position, whereas the Y-axis represents entropy level. Black line represents the variance threshold of 0.6. Blue line represents entropy level of ALV-J in Cluster I; blue line represents entropy level of ALV-J in Cluster II. Abbreviation: ALV-J, avian leukosis virus subgroup J.
Table 4.
Amino acid entropy analysis of viral structure genes.
| Gene | Type | Cluster I | Cluster II |
|---|---|---|---|
| gag | Entropy bigger than 0.6 | 2.42% (17/702) | 1.71% (12/702) |
| Average of entropy | 0.0478661 | 0.0501436 | |
| pol | Entropy bigger than 0.6 | 0.57% (5/874) | 0.57% (5/874) |
| Average of entropy | 0.031243 | 0.035644 | |
| env | Entropy bigger than 0.6 | 8.51% (48/564) | 7.09% (40/564) |
| Average of entropy | 0.147965 | 0.129868 |
Also we found other interesting aa-mutations (Table 5). Unique aa-substitutions were similar in the majority of sequences from the same clusters, such as 194G, 659V, and 194E, 659L of the gag gene in cluster I and cluster II strains, along with 208H, 404R, and 208S, 404K of the env gene in cluster I and cluster II.
Table 5.
Mutation sites in gag, pol, and env genes of the isolates compared with HPRS-103.
| Item | gag | pol | env | |||||
|---|---|---|---|---|---|---|---|---|
| Position | 77V | 122I | 133V | 194 G | 550K | 659 L | 217V | 63 |
| Cluster I | I (50%) | I (97%) | V (66%) | G (89%) | R (66%) | V (69%) | I (38%) | L (97%) |
| V (50%) | A (32%) | K (34%) | L (37%) | V (62%) | ||||
| Cluster II | V (93%) | A (63%) | A (73%) | E (50%) | K (55%) | L (70%) | I (63%) | L (68%) |
| I (37) | V (28%) | G (50%) | R (45%) | V (30%) | V (34%) | M(30%) | ||
| HPRS-103 | Env | |||||||
| Position | 64 | 106 | 107 | 121 | 159 | 206 | 208 | 247 |
| Cluster I | Q (97%) | N (53%) | T (58%) | D (64%) | T (72%) | D (86%) | H (58%) | R (42%) |
| S (42%) | A (42%) | N (22%) | _(33%) | L (14%) | G (19%) | |||
| Cluster II | Q (80%) | N (100%) | T (80%) | N (63%) | T (97%) | C (53%) | S (53%) | R (40%) |
| R (15%) | A (20%) | R (13) | D (45) | P (18%) | K (20%) | |||
| HPRS-103 | Env | |||||||
| Position | 250 | 258 | 404 | 423 | 434 | 489 | 506 | |
| Cluster I | L (36%) | G (81%) | R (58%) | I (58%) | I (78%) | R (64%) | L (61%) | |
| F (30%) | R (19%) | K(41%) | V (42%) | V (20%) | Q (36%) | F (39%) | ||
| Cluster II | F (40%) | G (50%) | K(95%) | I (88%) | I (100%) | Q (97%) | L (78%) | |
| L (45%) | R (45%) | W (23%) | ||||||
Molecular Characterization of 3′UTR
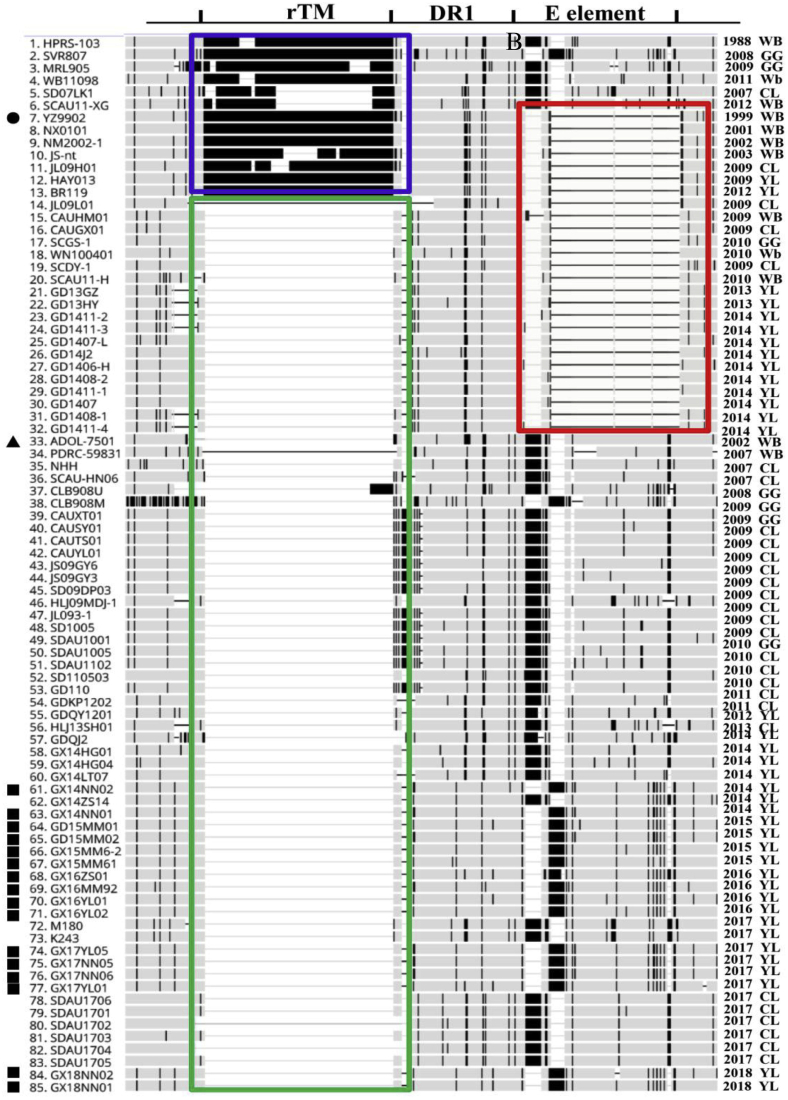
The 3′UTR sequences from 85 ALV-J isolates were analyzed and compared (Figure 5). A total of 205 nt, respectively in the rTM (175-nt deletion) and DR-1 (30-nt deletion) regions, were deleted in 84.71% (72/85) of the isolates. And, this deletion first appeared in the American ALV-J isolate ADOL-7501. In addition, 30.59% (26/85) of the isolates were found to have a 125- to 127-nt deletion in the E element, and this first appeared in the Chinese White-feather broilers' isolate YZ9902.
Figure 5.
The sequence comparison on the 3′UTR of the ALV-J strains. Blue box represents ALV isolates without 205-nt deletion; red box represents ALV isolates which include 127 bp deletions; green box represents isolates that include the 205-nt deletion; ● YZ9902, the stains in which 125 bp deletions were first appeared; ▲ ADOL-7501, the strains in which 205-nt deletions first appeared; ▪ ALV-J isolated from our lab. Abbreviation: ALV-J, avian leukosis virus subgroup J.
Discussion
ALV-J was first isolated from a case of myeloid leukosis in meat-type breeder chickens in 1988 and later proved to have a broader neoplastic spectrum (Payne and Nair, 2012). In 1999, ALV-J was first isolated in China from White-feather boilers (Yang et al., 2003), and since then, the host range of ALV-J has gradually expanded to layer flocks, Yellow-chickens, indigenous chicken breeds, and wild birds (Li et al, 2019b). This may be because of the instability and diversity of the Retroviridae (Garcia-Montojo et al., 2018) and the rapid evolution of the virus to help it escape the immune protection mechanisms of the host by the gene mutations and reorganization of the genome (Zhang et al., 2018). During the process of ALV-J transmission and spread all over the world, the ALV-J genome is constantly changing (Ji et al., 2012, Li et al., 2016).
In the present study, the whole-genome sequences of 85 ALV-J isolates from chickens of different genetic backgrounds were analyzed. Phylogenetic analysis of 85 ALV-J isolates originally from birds of different genetic backgrounds based on their whole-genome sequences indicated 2 different clusters. Cluster I consists mainly the strains isolated from layer chickens and White-feather broilers. Interestingly, most of these strains are originally from northern China. Cluster II consists mainly the strains isolated from Yellow-chickens, and most of these strains are originally from southern China (mainly Guangdong and Guangxi provinces, which produced nearly 40% of the Yellow-chickens in China). We know that the provinces in the northeast of China mainly have White-feather broilers and layers and had a yearly production of 4.5 billion birds, whereas the provinces in southern China mainly have Yellow-chickens and had a yearly production of 4.2 billion birds in 2018 (Wei, 2018). These geographical areas are separated by the Yellow River in the middle of mainland China, and these different bird lines barely have crossbreeding in commercial production. So, we speculate that the strains classified in cluster I might have the same origin, and the strains classified in cluster II have another origin.
The phylogenetic trees based on gag and pol gene aa-sequences showed that there is no obvious cluster, and the phylogenetic trees of the gp85 and gp37 genes showed similar results based on the whole-genome sequences. The results indicate that the gag and pol genes are relatively conserved during the transmission process of the virus, but the env gene was constantly changing in the same process (Meng et al., 2016). The author's previous studies indicated that the gp85 genes of ALV-J isolates from chickens of different genetic backgrounds were sorted into 5 clusters (Wang et al., 2018b). We speculate that the difference between the 2 results is because of the number of ALV-J strains. In our previous studies, the number of gp85 gene was 198 (compared to the present study, n = 85), which means that more detailed clusters can be made. On comprehensive consideration of the author's and some other researcher's results (Wang et al., 2018b, Li et al., 2018), the ALV-J strain based on gp85 gene can be divided into 2 large clusters as in this article, and the 2 big clusters could be further subdivided into 5 clusters, according to the author's previous studies.
An entropy value bigger than 0.6 indicates the corresponding aa-site was not conserved (Fan et al., 2019). The result of analysis of entropy of gag, pol, and env genes on aa-sequences showed that the env gene had the largest variation, and this is consistent with previous studies (Wang et al., 2018b). It is worth noting that in the gag and pol genes, we found areas with higher entropy values, such as the p19, p10, and p12 in gag gene and the p68 in pol gene (Table 5). A detailed statistical analysis of the aa-composition of each cluster found that unique aa-substitutions were present. The p19, p12, and p68 genes encode matrix protein, nucleocapsid protein, and reverse transcriptase, respectively. Previous studies showed that ALV with mutations in the pol gene show competitive replication advantages both in vivo and in vitro (Su et al., 2018). The effect of the mutations we found on the biological properties of the virus remains to be further studied.
The 3′UTR is prone to nt-substitutions and deletions, which influences chromosomal and viral gene expressions and is important in viral pathogenesis and replication (Han et al., 2015). A total 205-nt deletion was observed in this region; it contains a 175-nt-deletion at the 3′end of the rTM region and a 30-nt-deletion at the 5′end of the DR-1 region. Previous studies showed that this 205-nt deletion favors viral replication and enhances pathogenicity in layers and broilers (Wang et al., 2012). In the present study, we found that the 205-nt deletion first appeared in the American ALV-J isolates ADOL-7501, and 84.71% (72/85) of the strains isolated from birds of different genetic backgrounds contained this deletion (Figure 5). Based on this evidence, we believe this deletion was one of mechanisms of ALV-J induced oncogenicity, consistent with the presumption of previous studies (Wang et al., 2012).
In the present study, 30.59% (26/85) of ALV-J isolates had a 125 to 127-nt-deletion in the E element. Although a previous study showed that the 125 to 127-nt-deletion had no effect on viral replication and tumor induction (Han et al., 2015), this deletion does have its own significance. The same mutations (205-nt deletion or 125 to 127-nt-deletion) occurred in the isolates of different breeds of birds from different provinces, indicating that the viruses spread in the birds across different provinces. This requires us to test the ALV in advance of the trans-province transportation of the birds to prevent the spread of the disease from 1 location to another.
In conclusion, the whole-genome sequences from 85 ALV-J isolates were sorted into 2 clusters and that was mainly determined by the sequences of the env gene. Cluster I included isolates that mainly originated from the layers and White-feather broilers from northern China, and cluster II included isolates mainly from the Yellow-chicken, and most of them were from southern China. Through sequence analysis of ALV-J, it is implied that there is high host genetic background and geographic location dependency. This study focused on the characterization of the whole genome of ALV-J, and this might help us to further understand the diversity and complexity of ALV-J in local chickens. Importantly, it has informed us that more efforts should be made toward the eradication of ALV-J in the Yellow-chicken lines.
Acknowledgments
The manuscript was kindly reviewed by Dr. Richard Roberts, Aurora, CO 80014, USA.
This work was supported by the Guangxi Special Funding on Science and Technology Research [AA17204057], the Guangxi Program for Modern Agricultural Industry Technical System Construction-Chicken Industry [nycytxgxcxtd-19-03], the Shandong Provincial Natural Science Foundation Project [ZR201807070202].
Conflicts of Interest Statement: The authors declare no conflict of interest.
Footnotes
Supplementary data associated with this article can be found in the online version at http://doi.org/10.1016/j.psj.2020.04.023.
Supplementary data
References
- Cui Z.Z., Du Y., Zhang Z., Silva R.F. Comparison of Chinese field strains of avian leukosis subgroup J viruses with prototype strain HPRS-103 and United States strains. Avian Dis. 2003;47:1321–1330. doi: 10.1637/6085. [DOI] [PubMed] [Google Scholar]
- Fan W.S., Tang N., Dong Z.H., Chen J.M., Zhang W., Zhao C.R., He Y.N., Li M., Wu C.L., Wei T.C., Huang T., Mo M.L., Wei P. Genetic analysis of avian Coronavirus infectious Bronchitis virus in yellow chickens in southern China over the Past decade: Revealing the changes of genetic diversity, dominant Genotypes, and Selection Pressure. Viruses. 2019;11:898. doi: 10.3390/v11100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Montojo M., Doucet-O'Hare T., Henderson L., Nath A. Human endogenous retrovirus-K (HML-2): a comprehensive review. Crit. Rev. Microbiol. 2018;44:715–738. doi: 10.1080/1040841X.2018.1501345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C.Y., Hao R.J., Liu L.L., Zeng X.W. Molecular characterization of 3′UTRs of J subgroup avian leukosis virus in passerine birds in China. Arch. Virol. 2015;160:845–849. doi: 10.1007/s00705-014-2321-y. [DOI] [PubMed] [Google Scholar]
- Ji J., Li H.X., Zhang H.M., Xie Q.M., Chang S., Shang H.Q., Ma J.Y., Bi Y.Z. Complete genome sequence of an avian leukosis virus isolate associated with hemangioma and myeloid leukosis in egg-type and meat-type chickens. J. Virol. 2012;86:10907–10908. doi: 10.1128/JVI.01894-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Zeng X.W., Hua Y.P., Gao Q., Fan Z.B., Chai H.L., Wang Q., Qi X.L., Wang Y.Q., Gao H.L., Gao Y.L. Genetic diversity and phylogenetic analysis of glycoprotein gp85 of avian leukosis virus subgroup J wild-bird isolates from Northeast China. Arch. Virol. 2014;159:1821–1826. doi: 10.1007/s00705-014-2004-8. [DOI] [PubMed] [Google Scholar]
- Lai H.Z., Zhang H.N., Ning Z.Y., Chen R.A., Zhang W.Y., Qing A.J., Xin C.A., Yu K.Z., Cao W.S., Liao M. Isolation and characterization of emerging subgroup J avian leukosis virus associated with hemangioma in egg-type chickens. Vet. Microbiol. 2011;151:275–283. doi: 10.1016/j.vetmic.2011.03.037. [DOI] [PubMed] [Google Scholar]
- Li H.J., Wang P.K., Lin L.L., Shi M.Y., Gu Z.M., Huang T., Mo M.L., Wei T.C., Zhang H.M., Wei P. The emergence of the infection of subgroup J avian leucosis virus escalated the tumor incidence in commercial Yellow chickens in Southern China in recent years. Transbound. Emerg. Dis. 2019;66:312–316. doi: 10.1111/tbed.13023. [DOI] [PubMed] [Google Scholar]
- Li T.F., Xie J., Liang G.C., Ren D., Sun S., Lv L., Xie Q., Shao H.X., Gao W., Qin A.J., Ye J.Q. Co-infection of vvMDV with multiple subgroups of avian leukosis viruses in indigenous chicken flocks in China. BMC Vet. Res. 2019;15:288. doi: 10.1186/s12917-019-2041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.X., Xue C.Y., Ji J., Chang S., Shang H.Q., Zhang L.J., Ma J.Y., Bi Y.Z., Xie Q.M. Complete genome sequence of a J subgroup avian leukosis virus isolated from local commercial broilers. J. Virol. 2012;86:11937–11938. doi: 10.1128/JVI.02009-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.L., Meng F.F., Li W.H., Wang Y.X., Chang S., Zhao P., Cui Z.Z. Characterization of avian leukosis virus subgroup J isolated between 1999 and 2013 in China. Poult. Sci. 2018;97:3532–3539. doi: 10.3382/ps/pey241. [DOI] [PubMed] [Google Scholar]
- Li Y., Fu J.Y., Cui S., Meng F.F., Cui Z.Z., Fan J.H., Chang S., Zhao P. Gp85 genetic diversity of avian leukosis virus subgroup J among different individual chickens from a native flock. Poult. Sci. 2016;96:1100–1107. doi: 10.3382/ps/pew407. [DOI] [PubMed] [Google Scholar]
- Lin L.L., Wang P.K., Yang Y.L., Li H.J., Teng H., Ping W. Full-length genome sequence analysis of four subgroup J avian leukosis virus strains isolated from chickens with clinical hemangioma. Virus Genes. 2017;53:868–875. doi: 10.1007/s11262-017-1490-7. [DOI] [PubMed] [Google Scholar]
- Lin W.C., Li X.J., Dai Z.K., Zhang X.H., Shuang C., Peng Z., Zhang H.M., Feng C., Xie Q.M. Molecular epidemiology of J-subgroup avian leukosis virus isolated from meat-type chickens in southern China between 2013 and 2014. Arch. Virol. 2016;161:3039–3049. doi: 10.1007/s00705-016-3003-8. [DOI] [PubMed] [Google Scholar]
- Maas R., Van Z.D., Oei H., Claassen I. Replacement of primary chicken embryonic fibroblasts (CEF) by the DF-1 cell line for detection of avian leucosis viruses. Biologicals. 2006;34:177–181. doi: 10.1016/j.biologicals.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Meng F.F., Li Q.C., Zhang Y.B., Cui Z.Z., Chang S., Zhao P. Isolation and characterization of subgroup J Avian Leukosis virus associated with hemangioma in commercial Hy-Line chickens. Poult. Sci. 2018;97:2667–2674. doi: 10.3382/ps/pey121. [DOI] [PubMed] [Google Scholar]
- Meng F.F., Xuan D., Tao H., Shuang C., Fan J.H., Peng Z., Cui Z.Z. A deep sequencing reveals significant diversity among dominant variants and evolutionary dynamics of avian leukosis viruses in two infectious ecosystems. BMC Vet. Res. 2016;12:287. doi: 10.1186/s12917-016-0902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo M.L., Li M., Huang B.C., Fan W.S., Wei P., Wei T.C., Cheng Q.Y., Wei Z.J., Lang Y.H. Molecular characterization of major structural protein genes of avian Coronavirus infectious Bronchitis virus isolates in southern China. Viruses. 2013;5:3007–3020. doi: 10.3390/v5123007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X.H., Xu M.R., Zhu S.Y., Xiao W.H., Shen X., Qin A.J. Geese not susceptible to virulent subgroup J avian leukosis virus isolated from chickens. Avian Pathol. 2020;49:29–35. doi: 10.1080/03079457.2019.1657559. [DOI] [PubMed] [Google Scholar]
- Payne L.N., Brown S.R., Bumstead N., Howes K., Frazier J.A., Thouless M.E. A novel subgroup of exogenous avian leukosis virus in chickens. J. Gen. Virol. 1991;72:801–807. doi: 10.1099/0022-1317-72-4-801. [DOI] [PubMed] [Google Scholar]
- Payne L.N., Nair V. The long view: 40 years of avian leukosis research. Avian Pathol. 2012;41:11–19. doi: 10.1080/03079457.2011.646237. [DOI] [PubMed] [Google Scholar]
- Shen Y., Cai L., Wang Y., Wei R., He M., Wang S., Wang G., Cheng Z. Genetic mutations of avian leukosis virus subgroup J strains extended their host range. J. Gen. Virol. 2014;95:691–699. doi: 10.1099/vir.0.059915-0. [DOI] [PubMed] [Google Scholar]
- Su Q., Li Y., Cui Z.Z., Chang S., Zhao P. The emerging novel avian leukosis virus with mutations in the pol gene shows competitive replication advantages both in vivo and in vitro. Emerg. Microbes Infect. 2018;7:117. doi: 10.1038/s41426-018-0111-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.H., Cui Z.Z. Epidemiological and pathological studies of subgroup J avian leukosis virus infections in Chinese local "yellow" chickens. Avian Pathol. 2007;36:221–226. doi: 10.1080/03079450701332345. [DOI] [PubMed] [Google Scholar]
- Wang P.K., Lin L.L., Li H.J., Shi M.Y., Gu Z.M., Wei P. Full-length genome sequence analysis of an avian leukosis virus subgroup J (ALV-J) as contaminant in live poultry vaccine: the commercial live vaccines might be a potential route for ALV-J transmission. Transbound. Emerg. Dis. 2018;65:1103–1106. doi: 10.1111/tbed.12841. [DOI] [PubMed] [Google Scholar]
- Wang P.K., Lin L.L., Li H.J., Yang Y.L., Huang T., Wei P. Diversity and evolution analysis of glycoprotein GP85 from avian leukosis virus subgroup J isolates from chickens of different genetic backgrounds during 1989-2016: coexistence of five extremely different clusters. Arch. Virol. 2018;163:377–389. doi: 10.1007/s00705-017-3601-0. [DOI] [PubMed] [Google Scholar]
- Wang P.K., Shi M.Y., He C.W., Lin L.L., Li H.J., Gu Z.M., Li M., Gao Y.L., Huang T., Mo M.L., Wei T.C., Wei P. A novel recombinant avian leukosis virus isolated from gamecocks induced pathogenicity in Three-Yellow chickens: a potential infection source of avian leukosis virus to the commercial chickens. Poult. Sci. 2019;98:6497–6504. doi: 10.3382/ps/pez548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Gao Y.L., Wang Y.Q., Qin L.T., Qi X.L., Qu Y., Gao H.L., Wang X.M. A 205-nucleotide deletion in the 3′ Untranslated region of avian leukosis virus subgroup J, Currently Emergent in China, contributes to its pathogenicity. J. Virol. 2012;86:12849–12860. doi: 10.1128/JVI.01113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P. Challenge and Development Opportunity of Quality chicken industry in China: a review about 9 Issues on the topic. China Poult. 2018;41:1–6. [Google Scholar]
- Wei P., Cui Z.Z. Avian leukosis and pullorum disease: threats to Chinese native breeder chickens and preventive measures for elimination. China Poult. 2015;37:1–4. [Google Scholar]
- Yang Y.Y., Ye J.Q., Zhao Z.H., Qin A.J., Gu Y.F. Isolation and identification of Inner Mongolia strain of ALV subgroup. J. Virologica Sinica. 2003;18:454–458. [Google Scholar]
- Zeng X.W., Gao Y.L., Li D.L., Hao R.J., Liu W.S., Han C.Y., Gao H.L., Qi X.L., Wang Y.Q., Liu L.L., Wang X.M. Molecular characteristics of the complete genome of a J-subgroup avian leukosis virus strain isolated from Eurasian teal in China. Virus Genes. 2014;49:250–258. doi: 10.1007/s11262-014-1081-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Guan X.l., Chen Z.W., Cao D.G., Kang Z.F., Shen Q.C., Lei Q.X., Li F.W., Li H.Q., Leghari M.F., Wang Y.Q., Qi X.L., Wang X.M., Gao Y.L. The high conserved cellular receptors of avian leukosis virus subgroup J in Chinese local chickens contributes to its wide host range. Poult. Sci. 2018;97:4187–4192. doi: 10.3382/ps/pey331. [DOI] [PubMed] [Google Scholar]
- Zhang Y.W., Su Q., Zhang Z.H., Cui Z.Z., Chang S., Zhao P. Molecular characteristics of the re-emerged avian leukosis virus in China, 2018-2019. Transbound. Emerg. Dis. 2020;67:1141–1151. doi: 10.1111/tbed.13440. [DOI] [PubMed] [Google Scholar]
- Zhou D.F., Xue J.W., Zhang Y., Wang G.H., Feng Y.S., Hu L.P., Shang Y.L., Cheng Z.Q. Outbreak of myelocytomatosis caused by mutational avian leukosis virus subgroup J in China, 2018. Transbound Emerg. Dis. 2018;66:622–626. doi: 10.1111/tbed.13096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.