Abstract
It is generally accepted that domestic ducks are valuable protein sources for humans. The gastrointestinal ecosystem contains enormous and complicated microbes that have a profound effect on the nutrition, immunity, health, and production of domestic ducks. To deeply understand the gastrointestinal microbial composition of domestic ducks, we investigated the microbiomes of 7 different gastrointestinal locations (proventriculus, gizzard, duodenum, jejunum, ileum, cecum, and rectum) and the short-chain fatty acids in 15 healthy muscovy ducks based on 16S rRNA gene sequencing, qPCR, and gas chromatography. As a result, 1 029 735 sequences were identified into 35 phyla and 359 genera. Firmicutes, Proteobacteria, Bacteroidetes, Cyanobacteria, and Actinobacteria were the major phyla, with Bacteroidetes being most abundant in the cecum. The population of the total bacteria and the representatives of the Firmicutes, Bacteroidetes, and Bacteroides groups increased from the proximal to the distal part of the GIT. Bacteroides was the most dominant group in the cecum. Acetate, propionate, and butytrate, as well as gene copies of butyryl-CoA including acetate-CoA transferase and butyrate kinase, were significantly higher in cecum than in other sections. Isobutyrate, valerate, and isovalerate were only found in the cecum. The differences of microbial composition and the short-chain fatty acids of their metabolites among these 7 intestinal locations might be correlated with differences in gut function. All these results provide a reference for the duck gastrointestinal microbiome and a foundation for understanding the types of bacteria that promote health and enhance growth performance and decrease instances of disease in duck breeding.
Key words: duck, gastrointestine, microbiome, short-chain fatty acids
Introduction
As valuable sources of nutritious meat and eggs, high-quality liver fat, and feathers for humans, ducks are an economically important waterfowl in Asia and Europe, especially in China and France, and are now being raised worldwide. Over 24.5 million ducks are reared each year in the United States (Best et al., 2017). The health, disease, and productivity of ducks are closely associated with the microbiome of their gastrointestinal tract (GIT) (Vasaï et al., 2014a, Stanley et al., 2014, Dai et al., 2018). Indeed, the GIT is colonized by a large number of bacterial species, which have proved to play a crucial role in feed digestion, immunomodulation, pathogen exclusion, and endocrine activity (Corrigan et al., 2015, Waite and Taylor, 2015, Best et al., 2017, Li et al., 2017). In addition, the gastrointestinal microbiome ferments and degrades the indigestible carbohydrates to produce short-chain fatty acids (SCFA) as an energy source for the host, the acetate, propionate, and butyrate being the main metabolic products of the microbiome in animals (Wong et al., 2006, Waite and Taylor, 2014). Disturbances of the GIT microbiome in poultry would lead to the increased susceptibility to pathogens and infectious diseases, which may inflict serious loss for farmers, and potentially cause the contamination of the poultry products by foodborne pathogens (Corrigan et al., 2015).
It is well known that avian intestinal contents are significantly different from those of monogastric mammals (Pérez de Rozas, 2004). However, most research is focused on the gut microbiome of galliformes, specifically broiler chickens (Stanley et al., 2014, Xiao et al., 2017) and turkeys (Scupham et al., 2008, Andreano et al., 2017). Recently, some research works were conducted regarding the duck microbiome. Vasaï et al. (2014b) reported that Firmicutes and Bacteroidetes were the dominant phyla in the ileum and cecum of Pekin (Anser platyrhynchos) and Muscovy (Cairina moschata) ducks (>80%), with the microbiome in cecum consisting of ∼65 and ∼50% Bacteroidetes, respectively. Besides, the the cecum in Shaoxing ducks is composed of ∼33% Bacteroidetes and ∼20% Bacteroides (Zhao et al. 2019), and Cherry Valley duck's cecum was composed of ∼33.4% Bacteroidetes and ∼28.8% Bacteroides (Dai et al., 2018). However, Best et al. (2017) reported Bacteriodetes were absent from Pekin duck's cecal microbiome. Proteobacteria dominated, ranging from 77 to 99% of the microbial population in the cecum of Pekin duck's on day 1, and the population had shifted to the dominance by Firmicutes which ranged from 81 to 98% on day 8. The dominance of Firmicutes then extended through the rest of the grow-out period, making-up an average of 96% of the microbial population.
Most of these studies have centered upon the microbiome in part of the GIT or feces, especially in the cecum as the cecum is considered to be crucial in poultry health and as a crucial pathogen reservoir (Qu et al., 2008, Stanley et al., 2014, Vasaï et al., 2014a, Sun et al., 2016, Best et al., 2017). However, little information is available to elucidate the spatial variation of gastrointestinal microbiota in duck. In this study, we aim to perform a comprehensive assessment for the spatial distribution of the microbial community by high-throughput sequencing and SCFAs of their metabolites by gas chromatography in the GIT of common ducks. As the proventriculus, gizzard, duodenum, jejunum, ileum, cecum, and rectum are the main gastrointestinal sections of poultry (Pan and Yu, 2014, Yang et al., 2018a), the microbial community and SCFAs were investigated in these 7 different GIT sections of ducks.
Materials and methods
Ducks and Sample Collection
A total of 2000 one-day-old muscovy (C. moschata) ducks (Lanxi Hewang Breeding Duck Co. Ltd., Jinhua, China) were raised in a rearing pen under the standard commercial conditions. A commercial duck diet (Table 1) was supplied ad libitum for 70 D, and then, 15 male ducks with a body weight of 4.12 ± 0.26 kg close to mean BW were selected and euthanized by cervical dislocation. The GIT was removed from the carcasses immediately, and the luminal contents of the proventriculus, gizzard, duodenum, jejunum, ileum, cecum, and rectum were collected. The contents were then frozen immediately in liquid nitrogen and stored at −80°C until the isolation of microbial genomic DNA.
Table 1.
Composition of the experimental diets (as-fed basis).
| Ingredient (%) | Calculated nutrients levels (%) | ||
|---|---|---|---|
| Corn | 56.5 | Metabolizable energy, MJ/kg | 11.58 |
| Soybean meal | 20 | Crude protein | 16.5 |
| Wheat | 18 | Calcium | 0.95 |
| Soybean oil | 1.85 | Phosphorus | 0.52 |
| Sodium carbonate | 1.16 | Lysine | 0.92 |
| Dicalcium phosphate | 0.64 | Methionine | 0.49 |
| Lysine | 0.315 | Threonine | 0.67 |
| Methionine | 0.235 | Tryptophan | 0.21 |
| Salt | 0.24 | Crude fiber | 4.36 |
| Choline chloride | 0.06 | ||
| Vitamin and trace mineral premix1 | 1 |
The amount supplied per kilogram of total diet: vitamin A, 8,000IU; vitamin D3, 600IU; vitamin E, 8.0 mg; vitamin K3, 1 mg; vitamin B1, 3 mg; vitamin B2, 5 mg; vitamin B6, 2 mg; vitamin B12, 0.2 mg; nicotinic acid, 60 mg; pantothenic acid, 18 mg; folic acid, l mg; Fe, 120 mg; Cu, 20 mg; Mn, 30 mg; Zn, 60 mg; I, 0.45 mg; Se, 0.2 mg.
All animals involved in the experimental procedures were approved by the Institutional Animal Care and Use Committee of Zhejiang Academy of Agricultural Sciences.
DNA Extraction, 16S rRNA Gene Amplification and High-throughput Sequencing
The microbial genomic DNA was extracted from 220 mg of luminal contents of each sample by using a QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA) according to the study by Vasaï et al. (2014a). DNA were quantified by using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA) and stored at −80°C. The 16S rRNA genes of distinct regions (16S V3-V4) were amplified using specific primers (338 F ACTCCTACGGGAGGCAGCA; 806R GGACTACHVGGGTWTCTAAT). With Phusion High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA), PCRs were performed in triplicate using a 20-μL reaction system that contained 5 μM of each primer, 10 ng DNA template, 4 μL 1 × FastPfu Buffer, 2.5 mM deoxyribonucleoside triphosphates, and 0.4 μL FastPfu Polymerase. After visualizing on a 2% (w/v) agarose gel electrophoresis, 400- to 450-bp PCR products were chosen to purify using the Qiagen Gel Extraction Kit (Qiagen, Hilden, Germany). Then, the sequencing libraries were generated by using the TruSeq DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, CA) following manufacturer's recommendations. The library was sequenced on a Illumina HiSeq 2500 platform, and 250-bp paired-end reads were generated after assessing on the Qubit@ 2.0 Fluorometer (Life Technologies, Carlsbad, CA) and Agilent Bioanalyzer 2100 system.
Sequencing Data Analysis
The Quantitative Insights into Microbial Ecology (QIIME) software package, V1.7.0 (Caporaso et al., 2010a, Caporaso et al., 2010b), was used to analyze 16S microbial sequencing data. Paired-end reads were assigned to samples based on their unique barcode and truncated by cutting off the barcode and primer sequences. Paired-end reads were merged using FLASH (V1.2.7) (Magoc and Salzberg, 2011). To obtain clean and effective tags, quality filtering on raw sequences was performed according to the QIIME quality control process (Caporaso et al., 2010a, Caporaso et al., 2010b, Bokulich et al., 2013), and the reference database (Gold database) was used to detect and remove chimera by UCHIME algorithm (UCHIME Algorithm) (Edgar et al., 2011, Haas et al., 2011). Based on GreenGene Database and RDP classifier (version 2.2), sequences with ≥97% identity were assigned to the same operational taxonomic units (OTU), and the representative sequence for each OTU was screened. The PyNAST software (version 1.2, http://pynast.sourceforge.net) was used to study the differences between the dominant species in different samples according to multiple sequence alignment (Caporaso et al., 2010a, Caporaso et al., 2010b). To analyze the complexity of species composition within a sample, α-diversity (Observed-species, Shannon, Simpson, Chao I, abundance-base coverage estimator, Goods coverage) was calculated and displayed by QIIME and R software (version 2.15.3), respectively. The principal coordinate analysis (PCoA) was used to evaluate the differences of GIT microbial community.
The top 35 genera were selected to estimate the correlations from compositional network based on the SparCC algorithm (Wang et al., 2019). The network was presented by using the igraph package in R, with edges connecting nodes. Clusters were made based on the betweenness centrality calculated with the GirvanNewman algorithm (Girvan and Newman, 2002).
qPCR and Primers
The quantification of DNA by qPCR was performed on the Bio-Rad CFX384 (Bio-Rad, Singapore). Amplification and detection were run in 384-well plates using a SYBR Premix Ex Taq II Kit (Takara Bio, Kusatsu, Shiga, Japan). All measurements were done in triplicate in a 15-μL total reaction mixture using 2 μL of 50-ng DNA sample. All qPCR results were expressed as gene copies per g of fresh luminal content. The primers used in this study are listed in Table 2. A melting curve analysis was carried out after amplification to confirm specificity of the reaction. Quantification was done with the standard curves made from known concentrations of genomic bacterial DNA containing the respective amplicon for each primer.
Table 2.
Primers used in the present study.
| Item | Primers (5’→3′) | Reference |
|---|---|---|
| Total bacteria | fwd CGGYCCAGACTCCTACGGG rev TTACCGCGGCTGCTGGCA |
Xu et al. (2017) |
| Firmicutes | FwdGGAGYATGTGGTTTAATTCGAAGCA rev AGCTGACGACAACCATGCAC |
Xu et al. (2017) |
| Bacteroidetes | fwd GGARCATGTGGTTTAATTCGATGAT rev AGCTGACGACAACCATGCAG |
Xu et al. (2017) |
| Bacteroides | fwd GAGAGGAAGGTCCCCCAC rev CGCTACTTGGCTGGTTCAG |
Vasaï et al. (2014a) |
| Butyryl-CoA acetate-CoA transferase | fwd AAGGATCTCGGIRTICAYWSIGARATG rev GAGGTCGTCICKRAAITYIGGRTGNGC |
Xu et al. (2017) |
| Butyrate kinase | fwd TGCTGTWGTTGGWAGAGGYGGA rev GCAACIGCYTTTTGATTTAATGCATGG |
Xu et al. (2017) |
Measurement of SCFA
A total of 100 mg of luminal content of each gastrointestinal section was put into 1.5-mL centrifuge tubes and then suspended in Milli-Q water with 9 volumes of luminal content. After centrifugation at 12,000 rpm for 10 min, 0.5 mL of supernatant was mixed with 0.1 mL of 25% (w/v) solution of crotonic acid (internal standard) and metaphosphoric acid. Finally, the mixture was used to measure the concentrations of SCFAs by capillary GC (GC-2010 plus; Shimadzu, Kyoto, Japan) after filtering with a membrane filter.
Statistical Analysis
Statistics of the microbial diversity and relative abundance, qPCR data, and SCFA concentration were determined by a one-way ANOVA in SPSS software, version 18.0 (IBM, Chicago, IL). Probability values < 0.05 were considered significant. Results are presented as mean ± standard deviation.
Results
Global Sequencing Data
A total of 7,208,163 sequences were obtained from the 105 samples, with the number of sequences ranging from 41,335 to 85,571 per sample after filtering for quality. The sequences were clustered into 511 to 1,400 OTUs for each sample with 97% sequence similarity value (Supplementary Table 1).
Microbial complexity in each gastrointestinal section was estimated according to α-diversity indices (Table 3). The shannon indice of the proventriculus and cecum was higher than that of the gizzard, duodenum, jejunum, ileum, and rectum, which showed that the gizzard and cecum had a higher diversity. Good's coverage was in a range from 0.9944 to 0.9955, which indicates the identification of the majority of bacteria presented in each sample.
Table 3.
Overview of sequencing results of samples.
| Item | Proventriculus | Gizzard | Duodenum | Jejunum | Ileum | Cecum | Rectum |
|---|---|---|---|---|---|---|---|
| Sequencing number | 67,922 ± 5,807a,b | 68,534 ± 7,053a,b | 66,680 ± 9,537b | 70,271 ± 6,687a | 70,975 ± 10,530a | 66,350 ± 6,766b | 69,812 ± 7,748a |
| OTU | 903 ± 177 | 787 ± 174 | 767 ± 135 | 812 ± 178 | 843 ± 225 | 857 ± 122 | 739 ± 117 |
| Chao1 | 943 ± 187a | 825 ± 184b | 796 ± 162b | 847 ± 184b | 895 ± 291a,b | 909 ± 144a,b | 792 ± 129b |
| ACE | 948 ± 189a | 842 ± 193a,b | 816 ± 158b | 868 ± 190a,b | 917 ± 304a | 926 ± 153a | 810 ± 127b |
| Shannon | 5.82 ± 0.50a | 4.96 ± 1.17b | 4.65 ± 1.61b | 4.67 ± 1.42b | 4.23 ± 1.39b | 5.90 ± 0.65a | 40.46 ± 0.96b |
| Simpson | 0.94 ± 0.03a | 0.86 ± 0.13a,b | 0.80 ± 0.22a,b | 0.82 ± 0.20a,b | 0.77 ± 0.19b | 0.94 ± 0.04a | 0.83 ± 0.12a,b |
| Good's coverage | 0.9948 | 0.9953 | 0.9955 | 0.9948 | 0.9944 | 0.9949 | 0.9952 |
Means in the same row with different superscript letters differ significantly (P < 0.05).
Abbreviations: ACE, abundance-base coverage estimator; OTU, operational taxonomic units.
Microbial Community Composition and Populations
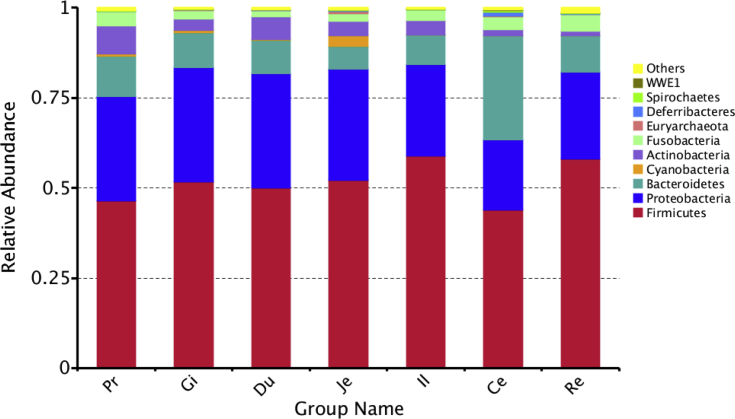
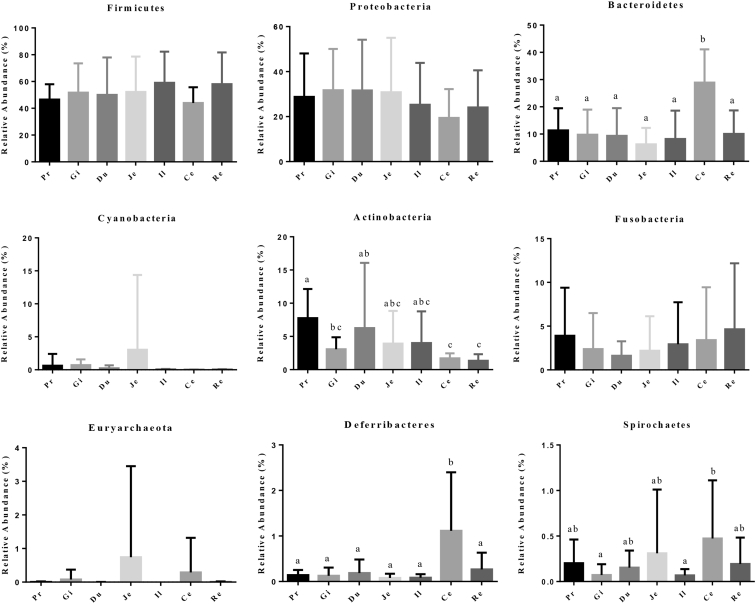
All clean sequences were classified into 35 phyla, among which Firmicutes, Proteobacteria, Bacteroidetes, Cyanobacteria, Actinobacteria, and Fusobacteria were the 6 dominant phyla (Figure 1). Firmicutes, Proteobacteria, and Bacteroidetes were predominant in the 7 gastrointestinal sections, which accounted for more than 80% of the sequences. Bacteroidetes and Deferribacteres were more abundant in the cecum than in other gastrointestinal locations (Figure 2).
Figure 1.
Spatial distribution of the top 10 phyla along the duck gastrointestinal sections. Phyla with abundance <1% were combined into “others”. Abbreviations: Ce, cecum; Du, duodenum; Gi, gizzard; Il, ileum; Je, jejunum; Pr, proventriculus; Re, rectum.
Figure 2.
The differential analysis of the 9 most abundant phyla in 7 gastrointestinal sections of ducks. Different letters represent significant differences. Abbreviations: Ce, cecum; Du, duodenum; Gi, gizzard; Il, ileum; Je, jejunum; Pr, proventriculus; Re, rectum.
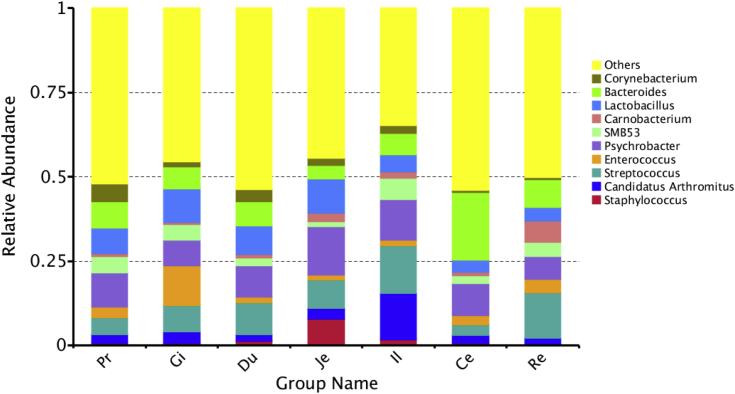
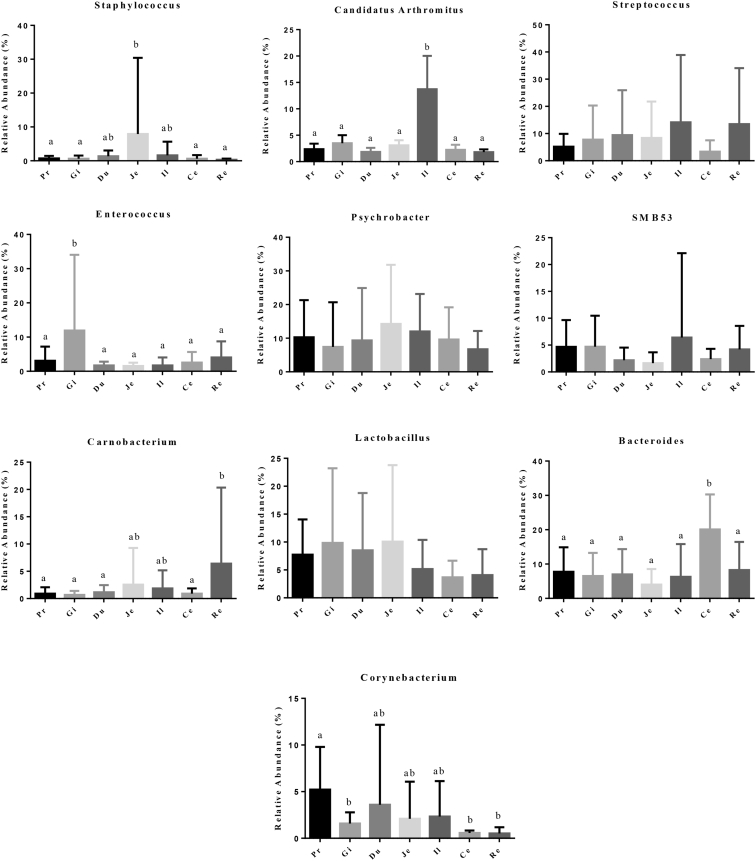
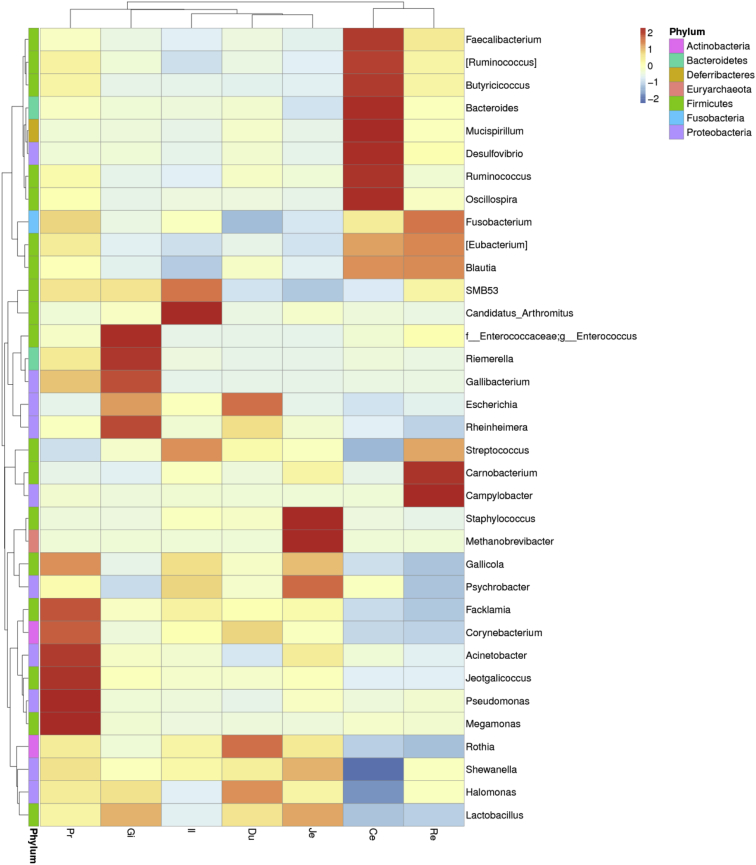
At the genus level, all sequences from 105 samples were identified into 359 genera. The top 10 genera were Staphylococcus, Candidatus Arthromitus, Streptococcus, Enterococcus, Psychrobacter, SMB53, Carnobacterium, Lactobacillus, Bacteroides, and Corynebacterium (Figure 3). Staphylococcus and Candidatus Arthromitus were most abundant in the jejunum and ileum, respectively (Figure 4). Bacteroides was the most dominant group in the cecum, accounting for approximately 20%; however, this genus had a dramatically lower presence in other sections (less than 8%). The distribution of other dominant genera in the GIT of duck is shown in Figure 5. It was observed that the relative abundance of Faecalibacterium, Butyricicoccus, Bacteroides, Mucispirillum, and Desulfovibrio was also higher in the cecum than in the other GIT sections.
Figure 3.
Spatial distribution of the top 10 genera along the duck gastrointestinal sections. Abbreviations: Ce, cecum; Du, duodenum; Gi, gizzard; Il, ileum; Je, jejunum; Pr, proventriculus; Re, rectum.
Figure 4.
The differential analysis of the 10 most abundant genera in 7 gastrointestinal sections of ducks. Different letters represent significant differences. Abbreviations: Ce, cecum; Du, duodenum; Gi, gizzard; Il, ileum; Je, jejunum; Pr, proventriculus; Re, rectum.
Figure 5.
Hierarchically clustered heat map for 7 gastrointestinal sections of ducks. Rows indicate the 35 predominant bacterial genera. Abbreviations: Ce, cecum; Du, duodenum; Gi, gizzard; Il, ileum; Je, jejunum; Pr, proventriculus; Re, rectum.
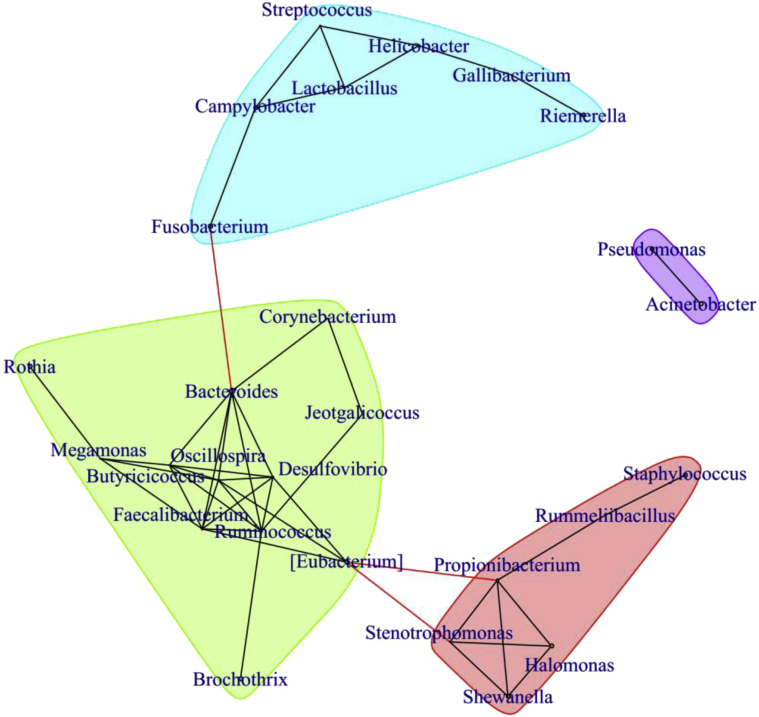
A network analysis using the SparCC algorithm also demostrated the interactions between genera, and 3 large clusters within the network were observed (Figure 6). It is interesting that Bacteroides, Oscillospira, Butyricicoccus, Faecalibacterium, Ruminococcus, Desulfovibrio, and Eubacterium showed a high level of connectivity.
Figure 6.
Network analysis of the interactions between genera. SparCC was used to calculate the relationships between bacterial taxa.
qPCR Data indicated a significant increase in total bacteria and representatives of the Firmicutes and Bacteroidetes including the Bacteroides group from the proximal to the distal part of the GIT. The populations of total bacteria and Firmicutes were the highest in the cecum and rectum, while those of Bacteroidetes and Bacteroides were the highest in the cecum (Table 4).
Table 4.
The abundance of different bacterial groups and functional genes in different gastrointestinal sections of ducks.
| Item | Proventriculus | Gizzard | Duodenum | Jejunum | Ileum | Cecum | Rectum |
|---|---|---|---|---|---|---|---|
| Total bacteria | 4.67 ± 0.53a | 4.39 ± 0.39a | 6.02 ± 0.41b | 7.08 ± 0.44b,c | 8.95 ± 0.68c | 11.89 ± 0.82d | 10.92 ± 1.06d |
| Firmicutes | 5.07 ± 0.57a,b | 4.38 ± 0.50a | 5.89 ± 0.24b | 6.64 ± 0.59b | 8.86 ± 0.79c | 11.05 ± 0.61d | 10.54 ± 0.78d |
| Bacteroidetes | 4.08 ± 0.46a | 4.12 ± 0.28a | 5.92 ± 0.61b | 5.78 ± 0.73b | 6.27 ± 0.63b | 9.40 ± 0.83c | 8.06 ± 0.45d |
| Bacteroides | 3.99 ± 0.47a | 4.20 ± 0.56a | 5.23 ± 0.20b | 5.54 ± 0.59b | 5.81 ± 0.49b | 9.25 ± 0.67c | 7.78 ± 0.77d |
| Butyryl-CoA:acetate-CoA transferase | 2.54 ± 0.16a | 2.19 ± 0.31a | 3.66 ± 0.43b | 3.57 ± 0.19b | 4.61 ± 0.38b,c | 6.69 ± 0.70d | 5.16 ± 0.47c |
| Butyrate kinase | 3.10 ± 0.47a | 1.99 ± 0.15b | 3.90 ± 0.29a | 3.81 ± 0.46a | 5.02 ± 0.59c | 6.36 ± 0.51d | 5.73 ± 0.30c,d |
The abundance of bacterial groups was expressed as log10 16S rRNA gene copies/g of fresh feces, and functional genes were expressed as log10 gene copies of total DNA/g of fresh feces.
Means in the same row with different superscript letters differ significantly (P < 0.05).
Similarity of Microbial Composition in Different GIT Sections
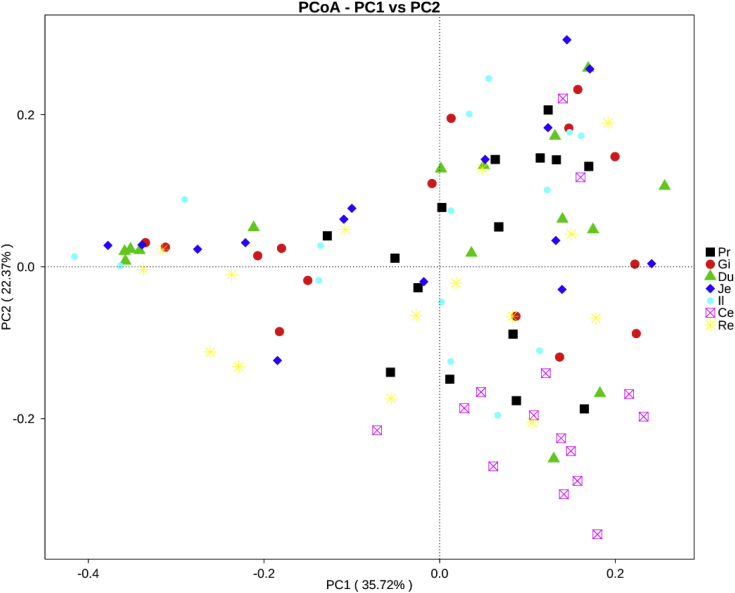
The similarities and differences of microbial community composition in the 105 samples taken from the 7 gastrointestinal sections of 15 ducks are shown in the PCoA plot (Figure 7); PC1 and PC2 accounted for 35.72 and 22.37% of the total variation, respectively. The microbial community of the cecum formed a distinct cluster that was separated from those of other gut sections. To understand the similarity, richness, and diversity of the bacterial community in the GIT of ducks, the relative abundance of microbial composition was visualized with a hierarchically clustered heat map (Figure 5). The microbial composition of the cecum and rectum shared a cluster, while that of duodenum, jejunum, and ileum formed another cluster with proventriculus and gizzard. The hierarchical cluster of the bacterial community in different GIT sections is consistent with the anatomy of the GIT in ducks.
Figure 7.
Principal coordinates analysis (PCoA) of the difference for microbial community composition of 7 gastrointestinal sections in ducks with weighted unifrac distance. The percentages present the relative contribution of the 2 principal coordinates (PC1-PC2). Abbreviations: Ce, cecum; Du, duodenum; Gi, gizzard; Il, ileum; Je, jejunum; Pr, proventriculus; Re, rectum.
Evaluation of Common Anseriforme Bacterial Pathogens
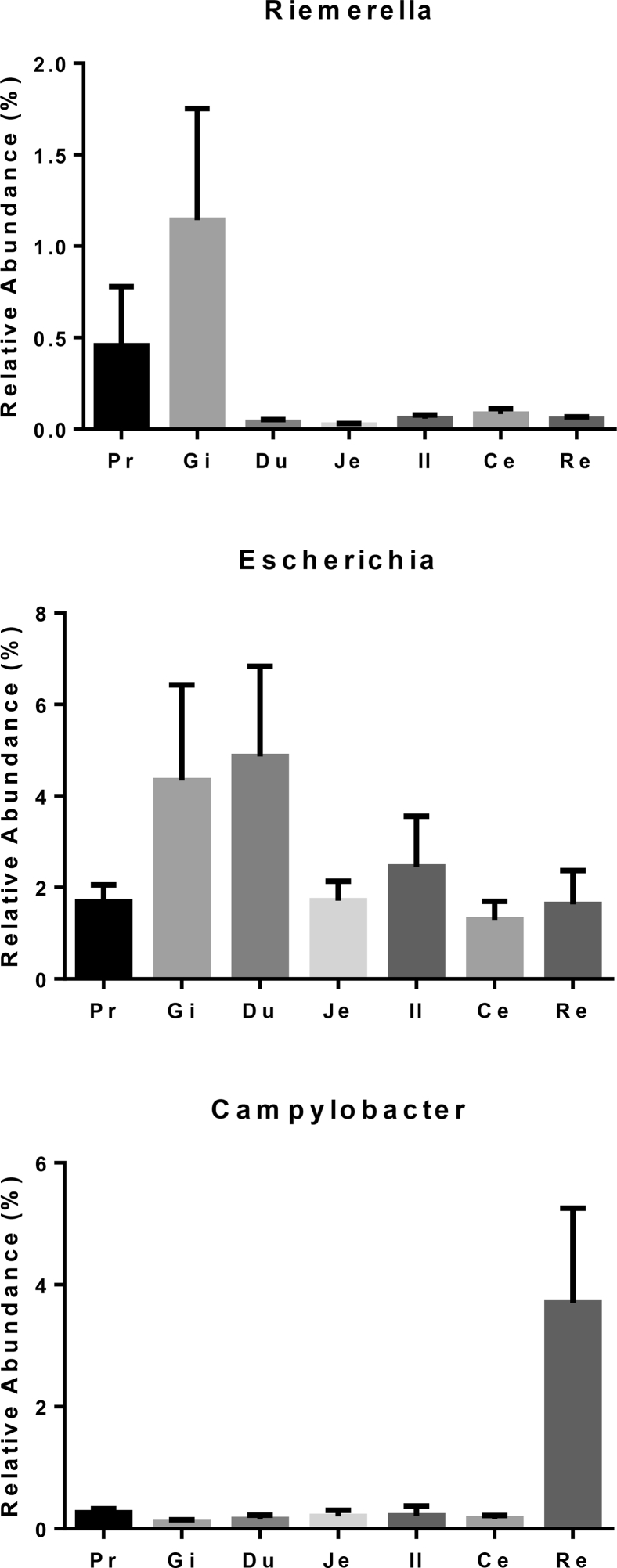
The 5 most common bacterial pathogens associated with duck health include Riemerella anatipestifer, Escherichia coli, Salmonella, Streptococcus, and Enterococcus. None of the assigned OTUs were classified into Salmonella. Assignments to the genus level for Riemerella, Escherichia, Streptococcus, and Enterococcus were present (Figures 4 and 8).
Figure 8.
The relative abundance of Riemerella, Escherichia, and Campylobacter in 7 gastrointestinal sections of ducks.
SCFAs in Different GIT Sections
The concentrations of SCFAs varied significantly in the 7 sections of the GIT in ducks. Acetate existed in all gastrointestinal sections; propionate and butyrate were found in the ileum, cecum, and rectum; and isobutyrate, valerate, and isovalerate were only observed in the cecum (Table 5). In addition, the concentrations of acetate, propionate, and butyrate in the cecum were significantly higher than those in other sections (P < 0.05). Futhermore, the relative abundance of butyryl-CoA acetate-CoA transferase and butyrate kinase was higher in cecum (Table 4), as these 2 function genes are involved in the regulation of butyrate-producing metabolism of the microbiome.
Table 5.
Concentrations of SCFAs in different gastrointestinal sections of ducks (mg/g).
| Item | Proventriculus | Gizzard | Duodenum | Jejunum | Ileum | Cecum | Rectum |
|---|---|---|---|---|---|---|---|
| Acetate | 0.0176 ± 0.0042a | 0.0216 ± 0.0076a | 0.0281 ± 0.0071a | 0.0288 ± 0.0151a | 0.0767 ± 0.1060b | 0.9965 ± 0.6369c | 0.2216 ± 0.1764a |
| Propionate | — | — | — | — | 0.0158 ± 0.0166 | 0.5907 ± 0.3947 | 0.0183 ± 0.0153 |
| Butyrate | — | — | — | — | 0.0122 ± 0.0173 | 0.3323 ± 0.3164 | 0.0162 ± 0.0283 |
| Isobutyrate | — | — | — | — | — | 0.0234 ± 0.0124 | — |
| Valerate | — | — | — | — | — | 0.0237 ± 0.0191 | — |
| Isovalerate | — | — | — | — | — | 0.0149 ± 0.0080 | — |
Means in the same row with different superscript letters differ significantly (P < 0.05). “—“ Indicates that the SCFA is undetected in the GIT section.
Abbreviations: GIT, gastrointestinal tract; SCFA, short-chain fatty acid.
Discussion
As a popular dish for Chinese, ducks are widely distributed in China because of its standing as the largest duck producer and consumer in the world. Meanwhile, more than 24.5 million ducks are raised for consumption each year in the United States (Best et al., 2017). The rapid development of duck renders it an important source for liver fat, meat, eggs, and feather for humans (Vasaï et al., 2014a, Pan and Yu, 2014). The comprehensive characterization of the microbial community and SCFAs in the GIT of ducks is vital in understanding the healthy state and in predicting the variations in microbiome related to disease and feed changes. Different gastrointestinal sections have different microbial community structures and play different roles in the health and growth of poultry (Lee and Pang, 1992, Pan and Yu, 2014). Although the microbial community of the duck GIT has already been reported, previous studies have mainly focused on the part of intestinal sections and feces (Sun et al., 2016, Best et al., 2017, Dai et al., 2018, Wang et al., 2018). In the present study, we have investigated the microbial composition in the GIT of duck, including the proventriculus, gizzard, duodenum, jejunum, ileum, cecum, and rectum through 16S rRNA gene sequencing.
Abundance-base coverage estimator and Chao1 indices indicated that the microbiome in the proventriculus was the richest, while the microbiome in the rectum was the poorest in ducks. Shannon and Simpson indices suggested that the microbiome in the proventriculus and cecum were the most diverse, while the microbiome in the ileum and rectum were the least diverse. The community composition analysis showed different dominant bacterial communities present in different gastrointestinal sections, which was in accordance with our previous results in chickens and geese (Xiao et al., 2017, Yang et al., 2018a). The microbiome in the cecum was notably different from that in the other gastrointestinal sections by PCoA score plot. The causal factors for these differences were that different gut compartments possess different functions and physiochemical characteristics (Stanley et al., 2014, Nakao et al., 2015) and that each section is inhabited by a specific microbial community (Dethlefsen et al., 2007, Rinttilä and Apajalahti, 2013).
In the present study, Firmicutes, Proteobacteria, Bacteroidetes, Cyanobacteria, and Actinobacteria were the major phyla in the GIT of duck, which suggested that the dominant phyla in the duck GIT are similar to those in other poultry, including turkeys, geese, and chickens (Qu et al., 2008, Scupham et al., 2008, Waite and Taylor, 2015, Xiao et al., 2017, Yang et al., 2018a). Bacteroidetes in the cecum of duck was considerably more abundant than that in other sections, which is similar to the findings of several previous studies (Vasaï et al., 2014a, Vasaï et al., 2014b, Wang et al., 2018). However, Best et al. (2017) reported that Bacteriodetes was absent in the Pekin duck's cecal microbiome while Proteobacteria dominated in early stages and then Firmicutes dominated from 8 D of age to the rest of the grow-out period. Given that diet and genetics play a fundamental role in determining the composition of gastrointestinal microbiome, the discrepancy in these studies might be due to the different diets or breeds of ducks used in the experiments.
Distinct microorganisms reside in different gastrointestinal sections of animals (Xiao et al., 2017, Yang et al., 2018a, Zhang et al., 2018). The relative abundance of Lactobacillus, Staphylococcus, Candidatus Arthromitus, Escherichia, and Rothia was commonly higher in the proventriculus, gizzard, and small intestine of duck, while Faecalibacterium, Ruminococcus, Butyricicoccus, Bacteroides, Mucispirillum, Desulfovibrio, Oscillospira, [Eubacterium], and Blautia were enriched in the large intestine, especially in cecum. In addition, the amounts of total bacteria, Firmicutes, Bacteroidetes, and Bacteroides measured by qPCR were found to be increased from the proximal to the distal part of the GIT in this study. Given that a strong pH gradient and a huge oxygen gradient occur from the proximal small intestine to the large intestine (Albenberg et al., 2014), the bulk of bacterial growth presents itself in the large intestine where host nutrient absorption is minimal, luminal pH is relatively neutral, and oxygen level is in the submicromolar range (Wexler and Goodman, 2017).
The GIT are a complex ecosystem, where the symbiosis between the host and different resident microorganisms occurs. For example, Bacteroides was dominant in the cecum and had a higher population there than that in other sections of ducks. It has been reported that Bacteroides could promote the development of the immune system, play distinguishing roles in immunomodulatory effects, and improve the mucosal barrier function in animals (Wexler and Goodman, 2017). The microbiome is also involved in metabolic reactions, which produce the microbial metabolism substrates, such as SCFA (Nicholson et al., 2012, Yang et al., 2018a). The concentrations of SCFAs were higher in cecum than in the other sections in duck, which suggests that the cecum is the core location of fiber fermentation. In addition, the SCFAs' production in the cecum might be closely correlated with the microbial community structure of duck. Bacteroides, Faecalibacterium, Butyricicoccus, Ruminococcus, Oscillospira, and mucispirillum were enriched in the cecum and were closely connected to one another based on the network analysis. It is well known that these genera are the critical bacteria to produce SCFAs (den Besten et al., 2013, Zhang et al., 2015, Yang et al., 2018b). Futhermore, the relative abundance of butyryl-CoA acetate-CoA transferase and butyrate kinase was highest in the cecum. The biosynthesis of butyrate occurs commonly via the butyryl CoA:acetate CoA transferase pathway or via the butyrate kinase pathway (Louis et al., 2004). This illustrates the cause of more SCFAs present in the cecum than in the other GIT sections from the aspect of SCFAs-producing function genes in microbiomes.
The most common bacterial pathogens related to duck health are R. anatipestifer, E. coli, Salmonella, Streptococcus, and Enterococcus (Best et al., 2017). Riemerella was observed in the contents of different GIT sections in this study, similar to the result of the study by Dai et al. (2018). Sequences of rDNA of nonserotypable R. anatipestifer-like strains isolated from the pharyngeal flora of healthy Pekin ducks were found to be 99% identical to those of R. anatipestifer (Ryll et al., 2008). Both Enterococcus and Streptococcus appear to be part of the microbiome of the duck GIT, comprising up to 15-50% of the population (Vasaï et al., 2014a). The ducks in this study harboring Riemerella, Enterococcus, and Streptococcus were healthy, which highlights the difference between presence of a potential pathogen and an actual instance of disease (Casadevall and Pirofski, 2014).
In conclusion, we characterized the microbial community structure and SCFAs in 7 different gastrointestinal locations of duck. We found that Firmicutes, Proteobacteria, Bacteroidetes, Cyanobacteria, and Actinobacteria were the major phyla in the GIT of duck. The total bacteria and the representatives of the Firmicutes and Bacteroidetes group from the proximal to the distal of GIT were increased. Bacteroides was the most dominant group in the cecum. Acetate, propionate, and butytrate, as well as gene copies of butyryl-CoA:acetate-CoA transferase and butyrate kinase, were significantly higher in the cecum than in other sections. Isobutyrate, valerate, and isovalerate were only found in the cecum. Collectively, these findings could provide useful information for the future study of the relationship between the GIT microbiome and domestic duck growth performance and health.
Acknowledgements
This work was supported by the National Waterfowl Industry Technology System of China (CARS-42-27); the State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Zhejiang Academy of Agricultural Sciences (2010DS700124-ZZ1905).
Conflict of Interest Statement: The authors did not provide a conflict of interest statement.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.03.040.
Contributor Information
Wen Wang, Email: ww_hi1018@163.com.
Yingping Xiao, Email: ypxiaozj@hotmail.com.
Supplementary Data
References
- Albenberg L., Esipova T.V., Judge C.P., Bittinger K. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano S.D., Bonastre A.S., Francino O., MartÍ A.C., Lecchi C., Grilli G., Giovanardi D., Ceciliani F. Gastrointestinal microbial population of Turkey (Meleagris gallopavo) affected by hemorrhagic enteritis virus. Poult. Sci. 2017;96:3550–3558. doi: 10.3382/ps/pex139. [DOI] [PubMed] [Google Scholar]
- Best A.A., Porter A.L., Fraley S.M., Fraley G.S. Characterization of gut microbiome dynamics in developing pekin ducks and impact of management system. Front Microbiol. 2017;7:2125. doi: 10.3389/fmicb.2016.02125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N.A., Subramanian S., Faith J.J., Gevers D., Gordon J.I., Knight R., Mills D.A., Caporaso J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Bittinger K., Bushman F.D., DeSantis T.Z., Andersen G.L., Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L.A. Ditch the term pathogen. Nature. 2014;516:165–166. doi: 10.1038/516165a. [DOI] [PubMed] [Google Scholar]
- Corrigan A., de Leeuw M., Penaud-Frézet S., Dimova D., Murphy R.A. Phylogenetic and functional alterations in bacterial community compositions in broiler ceca as a result of mannan oligosaccharide supplementation. Appl. Environ. Microb. 2015;81:3460–3470. doi: 10.1128/AEM.04194-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten G., van Eunen K., Groen A.K., Venema K., Reijngoud D.J., M Bakker B. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipld. Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S.J., Zhang K.Y., Ding X.M., Bai S.P., Luo Y.H., Wang J.P., Zeng Q.F. Effect of dietary non-phytate phosphorus levels on the diversity and structure of cecal microbiota in meat duck from 1 to 21 d of age. Poult. Sci. 2018;97:2441–2450. doi: 10.3382/ps/pey090. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L., McFall-Ngai M., Relman D.A. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girvan M., Newman M.E. Community structure in social and biological networks. Proc. Natl. Acad. Sci. U S A. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Gevers D., Earl A.M., Feldgarden M., Ward D.V., Giannoukos G., Ciulla D., Tabbaa D., Highlander S.K., Sodergren E., Methé B., DeSantis T.Z., Petrosino J.F., Knight R., Birren B.W., The Human Microbiome Consortium Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.P., Pang S.F. Identification and characterization of melatonin binding sites in the gastrointestinal tract of ducks. Life Sci. 1992;50:117–125. doi: 10.1016/0024-3205(92)90293-x. [DOI] [PubMed] [Google Scholar]
- Li M., Zhou H., Pan X., Xu T., Zhang Z., Zi X., Jiang Y. Cassava foliage affects the microbial diversity of Chinese indigenous geese caecum using 16S rRNA sequencing. Sci. Rep. 2017;7:45697. doi: 10.1038/srep45697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P., Duncan S.H., McCrae S.I., Millar J., Jackson M.S., Flint H.J. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J. Bacteriol. 2004;186:2099–2106. doi: 10.1128/JB.186.7.2099-2106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao N., Kaneda H., Tsushima N., Ohta Y., Tanaka M. Characterization of primary structure and tissue expression profile of the chicken apical sodium-dependent bile acid transporter mRNA. Poult. Sci. 2015;94:722–727. doi: 10.3382/ps/pev027. [DOI] [PubMed] [Google Scholar]
- Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Pan D., Yu Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes. 2014;5:108–119. doi: 10.4161/gmic.26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez de Rozas A.M. A comparative study of intestinal microbial diversity from birds, pigs and rabbits by restriction fragment length polymorphism analysis. Reprod Nutr Develop. 2004;44:S4. [Google Scholar]
- Qu A., Brulc J.M., Wilson M.K., Law B.F., Theoret J.R., Joens L.A., Konkel M.E., Angly F., Dinsdale E.A., Edwards R.A., Nelson K.E., White B.A. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS One. 2008;3:e2945. doi: 10.1371/journal.pone.0002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinttilä T., Apajalahti J. Intestinal microbiota and metabolites-implications for broiler chicken health and performance. J. Appl. Poult. Res. 2013;22:647–658. [Google Scholar]
- Ryll M., Christensen H., Bisgaard M., Christensen J.P., Hinz K.H., Köhler B. Studies on the prevalence of riemerella anatipestifer in the upper respiratory tract of clinically healthy ducklings and characterization of untypable strains. J. Vet. Med. Ser. B. 2008;48:537–546. doi: 10.1046/j.1439-0450.2001.00471.x. [DOI] [PubMed] [Google Scholar]
- Scupham A.J., Patton T.G., Bent E., Bayles D.O. Comparison of the cecal microbiota of domestic and wild turkeys. Microb. Ecol. 2008;56:322–331. doi: 10.1007/s00248-007-9349-4. [DOI] [PubMed] [Google Scholar]
- Stanley D., Hughes R.J., Moore R.J. Microbiota of the chicken gastrointestinal tract: influence on health, productivity and disease. Appl. Microbiol. Biot. 2014;98:4301–4310. doi: 10.1007/s00253-014-5646-2. [DOI] [PubMed] [Google Scholar]
- Sun D., Duan C., Shang Y., Ma Y., Tan L., Zhai J., Gao X., Guo J., Wang G. Application of Faecalibacterium 16S rDNA genetic marker for accurate identification of duck faeces. Environ. Sci. Pollut. Res. Int. 2016;23:7639–7647. doi: 10.1007/s11356-015-6024-z. [DOI] [PubMed] [Google Scholar]
- Vasaï F., Ricaud K.B., Cauquil L., Daniel P., Peillod C., Gontier K., Tizaoui A., Bouchez O., Combes S., Davail S. Lactobacillus sakei modulates mule duck microbiota in ileum and ceca during overfeeding. Poult. Sci. 2014;93:916–925. doi: 10.3382/ps.2013-03497. [DOI] [PubMed] [Google Scholar]
- Vasaï F., Ricaud K.B., Bernadet M.D., Cauquil L., Bouchez O., Combes S., Davail S. Overfeeding and genetics affect the composition of intestinal microbiota in Anas platyrhynchos (Pekin) and Cairina moschata (Muscovy) ducks. FEMS Microbiol. Ecol. 2014;87:204–216. doi: 10.1111/1574-6941.12217. [DOI] [PubMed] [Google Scholar]
- Wang S., Chen L., He M., Shen J., Li G., Tao Z., Wu R., Lu L. Diferent rearing conditions alter gut microbiota composition and host physiology in Shaoxing ducks. Sci. Rep. 2018;8:7387. doi: 10.1038/s41598-018-25760-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Tsai T., Deng F., Wei X., Chai J., Knapp J., Apple J., Maxwell C.V., Lee J.A., Li Y., Zhao J. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome. 2019;7:109. doi: 10.1186/s40168-019-0721-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.M., de Souza R., Kendall C.W., Emam A., Jenkins D.J. Colonic health: Fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- Waite D.W., Taylor M.W. Characterizing the avian gut microbiota: membership, driving influences, and potential function. Front Microbiol. 2014;5:223. doi: 10.3389/fmicb.2014.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite D.W., Taylor M.W. Exploring the avian gut microbiota: current trends and future directions. Front Microbiol. 2015;6:673. doi: 10.3389/fmicb.2015.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler A.G., Goodman A.L. An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2017;2:17026. doi: 10.1038/nmicrobiol.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Xiang Y., Zhou W., Chen J., Li K., Yang H. Microbial community mapping in intestinal tract of broiler chicken. Poult. Sci. 2017;96:1387–1393. doi: 10.3382/ps/pew372. [DOI] [PubMed] [Google Scholar]
- Xu J., Verbrugghe A., Lourenço M., Cools A., Liu D.J.X., Wiele T.V., Marzorati M., Eeckhaut V., Immerseel F.V., Vanhaecke L., Campos M., Hesta M. The response of canine faecal microbiota to increased dietary protein is influenced by body condition[J] BMC Vet. Res. 2017;13:374. doi: 10.1186/s12917-017-1276-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Xiao Y., Gui G., Li J., Wang J., Li D. Microbial community and short-chain fatty acid profile in gastrointestinal tract of goose. Poult. Sci. 2018;97:1420–1428. doi: 10.3382/ps/pex438. [DOI] [PubMed] [Google Scholar]
- Yang H., Xiao Y., Wang J., Xiang Y., Gong Y., Wen X., Li D. Core gut microbiota in Jinhua pigs and its correlation with strain, farm and weaning age. J. Microbiol. 2018;56:346–355. doi: 10.1007/s12275-018-7486-8. [DOI] [PubMed] [Google Scholar]
- Zhang J., Guo Z., Xue Z., Sun Z., Zhang M., Wang L., Wang G., Wang F., Xu J., Cao H., Xu H., Lv Q., Zhong Z., Chen Y., Qimuge S., Menghe B., Zheng Y., Zhao L., Chen W., Zhang H. A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J. 2015;9:1979–1990. doi: 10.1038/ismej.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wu W., Lee Y.K., Xie J., Zhang H. Spatial heterogeneity and co-occurrence of mucosal and luminal microbiome across swine intestinal tract. Front. Microbiol. 2018;9:48. doi: 10.3389/fmicb.2018.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Li K., Luo H., Duan L., Wei C., Wang M., Jin J., Liu S., Mehmood K., Shahzad M. Comparison of the intestinal microbial community in ducks reared differently through high-throughput sequencing. Biomed. Res. Int. 2019;2019:14. doi: 10.1155/2019/9015054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.