Abstract
Background
The tumor microenvironment and its interaction with neuroendocrine modulators contribute to prostate carcinogenesis and progression.
Objective
We sought to define the transcriptomic and clinical implications of neuropeptide Y (NPY) expression in prostate cancer progression.
Design, Setting, and Participants
Genome-wide expression profiling of three localized prostate cancer (total n=18,818) and five mCRPC cohorts (total n=495) used to characterize NPY expression. All men underwent radical prostatectomy (RP) for localized prostate cancer.
Outcome measurements and statistical analysis
Patients were grouped into low NPY and high NPY based on NPY expression. Associations between these groups and histologic, genomic, and clinical outcomes including progression (PFS) and metastases-free survival (MFS) were examined. Combining ERG-fusion status with NPY expression, four groups were defined (lowNPY/ERG+, lowNPY/ERG-, highNPY/ERG+, highNPY/ERG-). Cox proportional hazards modeled time to distant metastasis after RP. Genomic-risk scores for metastasis were calculated for prospective samples, based on a 22-gene signature.
Results and limitations
Across cancers, NPY showed the highest expression in prostate cancer in TCGA PAN-cancer cohort (n=9,483, p<0.0001). In 17,967 prospective samples, low NPY expression was associated with aggressive grade group 5 disease and higher genomic risk (p<0.0001). In the retrospective cohort (n=355) and TCGA (n=497), low NPY was associated with shorter MFS and PFS respectively (p=0.001 for both). In mCRPC cohorts, low NPY is associated with neuroendocrine development (p<0.01). NPY was highly correlated to ERG, thus we defined four groups based on NPY expression and ERG fusion. The lowNPY/ERG+ subtype was associated with the highest genomic-risk for metastasis (p<0.0001) and highest rate of metastasis compared to all other subtypes (HR:2.2[1.22–4.03],p=0.008), while the highNPY/ERG- subtype was associated with the lowest genomic-risk for metastasis (p<0.0001) and the lowest rate of metastasis (HR: 0.53[0.35–0.81], p=0.003).
Conclusions
Low NPY expression is associated with adverse genomic features and clinical correlates and outcomes. The lowNPY/ERG+ subtype was associated with the highest risk of developing metastasis. Prognostic subgrouping and tailored treatments by NPY expression and ERG fusion status warrants further study.
Patient summary
The low NPY prostate cancer subtype appears to be aggressive with high risk of developing metastasis.
INTRODUCTION
Human carcinogenesis and progression are influenced by tumor microenvironment [1,2]. Evidence suggests that the interaction between nerve fibers and neuromodulators with the tumor microenvironment has an important role in carcinogenesis and cancer progression through “neoneurogenesis” and other mechanisms [3–6]. Both adrenergic and cholinergic nerves promote prostate cancer development, at least in part, by activating stromal cells via activation of an angiogenic switch through neuromodulators that can lead to exponential tumor growth [4].
Neuroendocrine modulators, such as neuropeptide Y (NPY), are found to be expressed in prostate cancer [7]. In addition to playing a critical role in the regulation of a variety of physiological functions, NPY stimulates cell proliferation and has been implicated as a growth-promoting factor in various malignancies, including prostate cancer [8,9]. NPY has been suggested to be more abundant in cancerous prostate tissue compared to benign[7] and cancerous tissues derived from other organs [10]. Among all neuroendocrine modulators, pro-NPY protein was the only prostate specific prognostic biomarker [7] motivating us to further study its mRNA.
The impact of NPY mRNA on prostate cancer progression and outcomes remains unclear in prostate cancer. We sought to define the transcriptomic and clinical implications of NPY in prostate cancer in over 18,000 patients.
MATERIALS/METHODS
Data and patient cohorts
Genome-wide expression profiles of formalin-fixed paraffin-embedded radical prostatectomy (RP) tumor samples from 18,322 patients (Table S1) were evaluated from two prospective population-based registry cohorts (GRID 5k: n=5,239 and GRID 12k: n=12,728) and a retrospective institutional natural history cohort with long follow-up for the endpoint of metastasis-free survival (MFS) (n=355)[11].
The prospective cohort was comprised of anonymized genome-wide expression profiles from clinical use of the Decipher test, between February 2014 to August 2017, retrieved from the Decipher GRID™ (NCT02609269); the cohort included basic demographic and pathological data, but not longitudinal clinical outcomes. The retrospective cohort was comprised of men treated with RP at Johns Hopkins Medical Institute (JHMI)[11]. TCGA prostate was also used to evaluate associations with outcome.
Additionally, five mCRPC and neuroendocrine (NE) cohorts (Kumar etal[12], Robinson etal[13], Aggarwal etal[14], Beltran etal[15], Tsai etal[16]) were used to evaluate associations of NPY and NE development
Gene expression profiling
Tumor RNA was extracted from FFPE blocks or slides after macrodissection guided by histologic review of the tumor lesion by a GU pathologist. All cases in the retrospective cohort were re-reviewed prior to analysis by expert trained GU pathologists at JHMI[11] Central pathology review was performed for all Prospective cases. All prospective cases had central pathology re-review as samples were ran on a commercial CLIA/CAP platform where every sample is assessed independently prior to analysis. For all cases analyzed, at least 0.5 cm2 of tumor with at least >=60% tumor cellularity was required for the Decipher assay.
Decipher GRID sample specimen selection, RNA extraction, and microarray hybridization were all done in a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory facility (GenomeDx Biosciences, San Diego, CA, USA). Total RNA was extracted and purified using the RNeasy FFPE kit (Qiagen, Valencia, CA). RNA was amplified and labeled using the Ovation WTA FFPE system (NuGen, San Carlos, CA) and hybridized to Human Exon 1.0 ST GeneChips (Thermo-Fisher, Carlsbad, CA). Quality control was performed using Affymetrix Power Tools, and normalization was performed using the Single Channel Array Normalization (SCAN) algorithm[17].
NPY expression in prostate cancer
We characterized the expression of NPY in 9,483 PAN cancer samples across 23 solid tumors including prostate tumor (n=333); data were downloaded from Synapse platform (syn4976369). We further characterized the expression of NPY in prostate tumor vs. normal prostate across three independent cohorts [18–20]. We examined associations between NPY expression and prostate clinicopathological variables including Gleason and CAPRA-S scores. We also looked at correlation between NPY and stromal infiltration based on 141 gene signature[21].
Association of Disease Outcome with NPY
We examined the associations between NPY and clinical outcome using three different cohorts. We examined the association between NPY expression and genomic risk scores for risk of metastasis, using high Decipher score (≥0.6) as the endpoint. Furthermore, in the JHMI retrospective natural history and TCGA cohorts, we examined the association between NPY and MFS and progression-free survival (PFS), respectively.
NPY expression in neuroendocrine development
To study the dynamics of NPY expression under castration and neuroendocrine development, we used expression from the LTL331 patient-derived xenografts system [22] where expression was profiled under several time points (day1, day2, day3, wk1,wk2,wk3,wk8,wk12) until tumor relapsed and neuroendocrine (NE) tumor developed.
Twelve LTL331 patient-derived xenografts were raised in non-obese diabetic (NOD) severe combined immunodeficiency (SCID) mice. After host castration, tissue was harvested, measured, fixed for histopathology, and processed for DNA/RNA analysis. Gene expression microarray profiling was performed using the GE 8×60K Microarray.
Expression data from de novo-small cell neuroendocrine tumors from the Decipher GRID(n=33)[16], and public mCRPC-NE tumors [12–15] was used for analyses characterizing the role of NPY in prostate cancer progression and neuroendocrine development.
ERG fusion determination and NPY group categorization
ERG fusion determined from transcriptomic data as previously described[23]. We examined NPY distribution and used the point splitting the bimodal distribution of expression as the cut-off for high versus low NPY expression (0.75 was used as the cut-off). Four subtypes were defined based on NPY expression and ERG fusion status (lowNPY/ERG+, lowNPY/ERG-, highNPY/ERG+, highNPY/ERG-). Clinical and molecular characterization of these subtypes were elucidated.
Association of Disease Outcome with NPY/ERG subtypes
We examined the associations between NPY/ERG subtypes and pathological variables, genomic risk, and clinical outcome using prospective and retrospective cohorts.
Statistical analyses
MFS for the retrospective cohort was defined by radiographic evidence of metastatic disease with time zero beginning at date of RP. For the prospective cohort, a high genomic classifier score (Decipher ≥ 0.6) was used for genomic high risk patients. For the TCGA cohort, PFS was used[24]. Time to distant metastasis from initial local therapy was modeled using Cox proportional hazards. Spearman’s correlation was used for correlation analysis. Wilcoxon rank sum test was used for continuous statistical analyses, and Chi-square test was used for categorical associations. All tests were performed in R v3.3.1, and all tests were using a 5% significance level.
RESULTS
NPY expression in prostate cancer
Across TCGA PAN-CANCER (n=9,483), NPY showed the highest expression in prostate cancer followed by neuronal solid tumors (Figure 1.A); compared to non-neuronal solid tumors, NPY had a more than 7-fold higher expression in prostate tumors (p<2e−16). In the TCGA cohort and two independent cohorts (DKFZ[18], MSKCC[19]), there was slight differential NPY expression between prostate and non-prostate benign tissues (Supplementary Fig 1).
Figure 1: NPY expression across cancers.
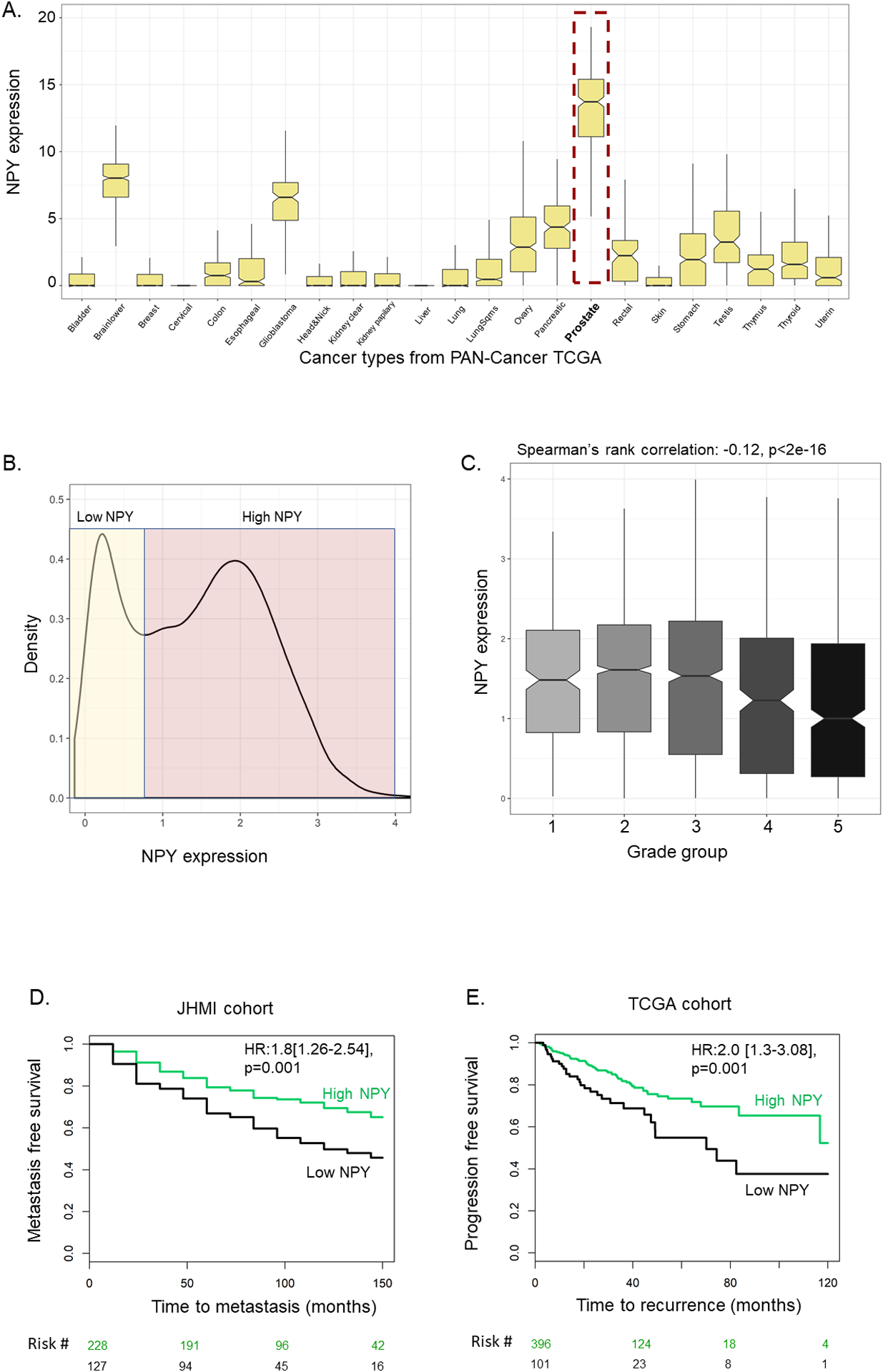
A. Expression of NPY across 23 solid tumors from PAN-CANCER TCGA. B. Density distribution of NPY in 5,239 primary RP prostate tumors from Decipher GRID. C. Expression of NPY across grade groups in the 5,239 RP tumors. D. KM plot of low vs high NPY in JHMI cohort for metastasis endpoint. E. KM plot of low vs high NPY in TCGA-prostate for PFS endpoint.
In primary prostate tumors, NPY showed a bimodal distribution in both the GRID 5K prospective and the JHMI retrospective cohorts (Figure 1.B, Supplementary Fig 2). We found this signal is not influenced by the stromal infiltration score (r=−0.1). This motivated us to further study the biological and clinical factors associated with NPY expression. Across grade groups and CAPRA-S groups, lower NPY expression was associated with more aggressive groups (GG5, high CAPRA-S) in the GRID 5K prospective cohort (Spearman’s correlation p<0.0001 for both) (Figure 1.C, supplementary Fig 3.A). These findings were validated within the GRID 12k cohort (supplementary Fig 4). There was no significant association between NPY and perineural invasion (Wilcoxon rank sum test, p=0.37) (supplementary Fig 3.B).
Association of Disease Outcome with NPY expression
When splitting patients into deciles based on NPY expression, patients with low NPY in the GRID 5K prospective cohort were more enriched with high genomic risk scores, where the lowest decile of NPY expression had 41% high genomics risk compared to average of 25% for remaining deciles (supplementary Fig 5.A). Similarly, in the JHMI cohort, tumors with lowest decile of NPY expression had high absolute rates of metastases (51% for low decile vs 33% for remaining deciles (Chi -square, p=0.02)(supplementary Fig 5.B). In retrospective cohorts, low NPY was associated with shorter MFS (10-yr MFS: 45% vs 68%, HR:1.8[1.26–2.54], p=0.001), and shorter PFS in TCGA (5-yr PFS: 54% vs 73%, HR:2.0 [1.3–3.08], p=0.001) (Figure 1D–E). In MVA adjusting for pathological grade groups, SVI, LNI, EPE and SM, low NPY remained independent prognostic biomarker of MFS in JHMI cohort (HR:1.5[1.05–2.15],p=0.02) and PFS in TCGA cohort (HR:1.7[1.04–3.03,p=0.04) (Table S2).
NPY expression in mCRPC and neuroendocrine development
Characterizing the expression of NPY in mCRPC cohorts revealed that NPY has also a bimodal distribution (Figure 2.A). Low NPY in these cohorts was associated with neuroendocrine differentiation. NPY was significantly lower in mCRPC-NE compared to adenocarcinoma mCRPC (p<0.01 for all) (Figure 2.B–C). NPY expression was also significantly reduced in de novo SC/NE compared to high grade (GG5) adenocarcinoma and mCRPC compared to adenocarcinoma (Figure 2.D–E). When characterizing NPY expression in patient-derived xenograft model upon castration [22], there was a gradual reduction in NPY expression upon castration and neuroendocrine development(Figure 2.F).
Figure 2: Expression of NPY in neuroendocrine development.
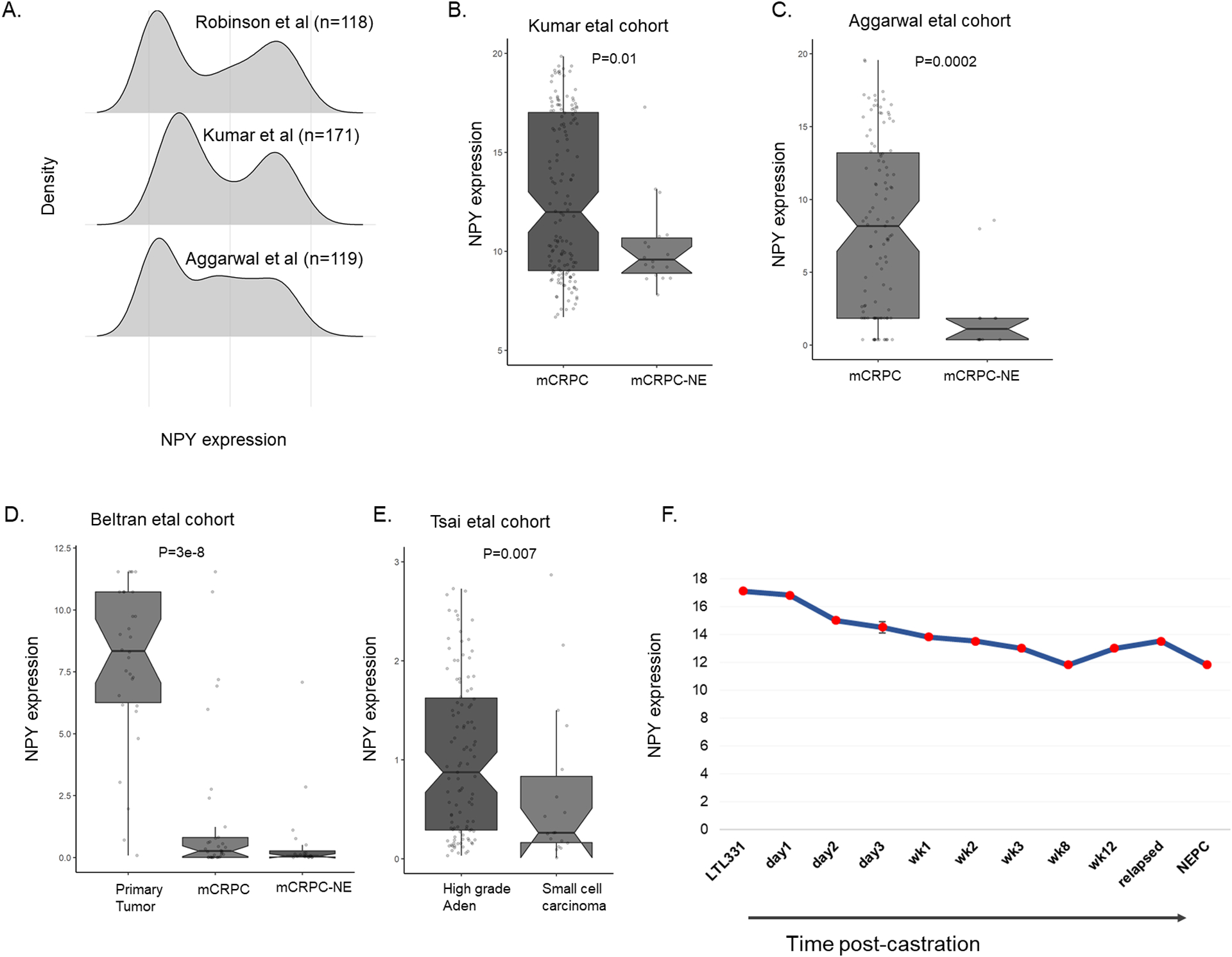
A. Distribution of NPY in mCRPC cohorts. B-C. Expression of NPY is down in mCRPC-NE compared to mCRPC in Kumar etal and Aggarwal etal. D. Expression of NPY is down in mCRPC compared to primary in Beltran etal. E. Expression of NPY is down in prostate tumors with denovo-small cell neuroendocrine. F. Expression of NPY in xenograft model under castration and NE development in LTL331R system (data downloaded from GSE 59986).
Combining NPY and ERG for subtype identification
To further characterize the biology of NPY, we correlated NPY to 46K genes and found ERG to be among the most correlated (r=0.43) genes in adenocarcinoma cohort (GRID5K) (Table S3). When plotting NPY vs ERG, both genes showed bimodal distribution, where 91% of ERG+ had high NPY (>0.75), while only 53% of high NPY was ERG+ (Figure 3.A), suggesting a potential biological cross-talk between the two genes. When associating NPY with molecular subtypes defined by [25], the ERG+ subtype (41.5% of total) was associated with the highest expression of NPY in the GRID 5K prospective cohort (Figure 3.B). These findings were validated within the GRID 12k cohort as well (supplementary Fig 6). Based on these results, four groups were defined based on NPY expression and ERG fusion status (lowNPY/ERG+, lowNPY/ERG-, highNPY/ERG+, highNPY/ERG-). We further characterised the clinical implications of these subtypes.
Figure 3: Associations of NPY and ERG.

A. Expression of NPY vs ERG in GRID cohort showing tumors with ERG-fusion have high NPY. B. Expression of NPY across molecular subtypes in the GRID cohort. C.Genomic risk scores (based on 22 gene signature across NPY/ERG subtypes in the GRID 5K prospective cohort. D. Percentage of high genomic risk and metastasis events across NPY/ERG subtypes showing lowNPY/ERG+ have highest percentage of high genomic risk and metastasis events.
Association of Disease Outcome with NPY/ERG subtypes
When examining genomic risk across the NPY/ERG subtypes. The genomic risk of metastasis was highest in the lowNPY/ERG+ subtype and lowest in the highNPY/ERG- subtype (Wilcoxon rank sum test, p<0.0001) (Figure 3.C); These findings were validated in the GRID 12K prospective cohort (supplementary Fig 7). The lowNPY/ERG+ subtype was associated with the highest rate of the high genomic risk class in the GRID 5K prospective cohort (51%) compared to 20% in the highNPY/ERG- subtype (Figure 3.D, Chi-square test for trend, P<0.0001). When associating NPY/ERG subtypes with clinicopathological variables, lowNPY/ERG+ was significantly associated with GG5, EPE, SVI, LNI (p<0.0001 for all) in the GRID 5k cohort(Table S4).
The lowNPY/ERG+ subtype (5% of total) in the JHMI cohort was associated with a higher absolute rate of metastases (63%) compared to 24% for the highNPY/ERG- subtype (Figure 3.D, Chi-square for trend, P=0.006).
In JHMI patients, when adjusting for ERG-fusion status, both low NPY and ERG+ were significantly association with MFS [NPY, HR:2.11[1.44–3.09],p=0.0001; ERG, HR: 1.53[1.04–2.26],p=0.02). LowNPY/ERG+ group was associated with the highest risk of metastasis compared to the other three groups (HR:2.2[1.22–4.03],p=0.008), and highNPY/ERG- subtype was associated with the most favorable genomic features and the lowest risk of metastasis (HR: 0.53[0.35–0.81], p=0.003). LowNPY/ERG+ was associated with the lowest 10-year MFS (34%) compared to 53%, 62%, 77% for lowNPY/ERG-, highNPY/ERG+ and highNPY/ERG-, respectively (supplementary Fig 8).
Molecular characterization of the lowNPY/ERG+ subtype
To further understand the molecular biology behind the aggressive nature of the lowNPY/ERG+ subtype, we conducted differential gene expression analysis between lowNPY/ERG+ versus highNPY/ERG+ and lowNPY/ERG- versus highNPY/ERG- (Table S5). Using t-SNE dimensionality reduction method, lowNPY/ERG+ and highNPY/ERG+ were distinct from the other groups (supplementary Fig 9.A). lowNPY/ERG+ was distinct from highNPY/ERG+ and highNPY/ERG- was distinct from lowNPY/ERG- (supplementary Fig 9.B–C, Hotelling’s T^2 test p<2e-16 for both). These results reveal that NPY and ERG defined transcriptomically distinct groups. Analyzing the differentially expressed genes showed that genes downregulated in the lowNPY/ERG+ are enriched with AR-targets and the overexpressed genes are enriched with genes related to neuronal development and focal adhesion (Table S6).
DISCUSSION
We found that though NPY is highly expressed in prostate cancers relative to other cancers, lower NPY expression is associated with aggressive high-grade disease and progression to neuroendocrine disease. Utilizing 18,322 transcriptomes we discovered and validated a subclass of low NPY expression coupled with ERG fusion, that had high risk of metastasis and decreased AR-activity. Our findings from multiple independent cohorts demonstrate that low NPY expression coupled with ERG fusion is associated with the least favorable genomic risk scores and the poorest PFS and MFS compared to other subtypes, while the high NPY/ERG- subtype is associated with the most favorable genomic risk scores and best PFS and MFS. Furthermore, our findings highlight that genes downregulated in the lowNPY/ERG+ subgroup are enriched with AR-targets.
These findings have several potentially critical clinical implications. It appears that though NPY expression is higher in prostate cancer relative to other solid tumors, low NPY expression may be an adverse feature in prostate cancer that predicts for aggressive disease and progression. Notably, there was no association between NPY expression and perineural invasion suggesting NPY predicts for adverse prostate cancer features and outcome independent of perineural invasion. We found that though NPY was highly expressed in the ERG+ subgroup, high NPY expression was not mutually exclusive to ERG fusion. Specifically, ERG+ represented only a subset (53%) of high NPY group, suggesting that NPY may actually influences the microenvironment to modulate ERG-fusions.
Furthermore, subgroupings by NPY expression and ERG fusion status may have significant prognostic implications for prostate cancer, and our findings highlight a potentially novel approach to prostate cancer subgroupings. The interaction between NPY and ERG has been previously studied, but there are conflicting results about the prognostic role of NPY alone or in combination with ERG-rearrangements [7,10,26]. In a previous study, combination of pro-NPY and ERG expression did not show association with risk of BF, castration-based treatment, CRPC, and PCa-specific death following RP[26]. Here, we found in multiple independent large cohorts that low NPY expression with ERG fusions predict for adverse genomic features and poor clinical outcomes, while high NPY expression without ERG fusions predict for more favorable genomic markers and clinical outcome. Ultimately, whether subgroupings by NPY expression and ERG fusion status improve upon currently available risk prediction schemas and nomograms warrants further investigation.
Lastly, given that genes downregulated in the lowNPY/ERG+ are enriched with AR-targets, the low NPY/ERG+ subtype may represent a group potentially at risk of ADT resistance. This subgroup could be more likely to benefit from the addition of chemotherapy or novel systemic agents. Ultimately, prospective trials may need to investigate the relationship between the lowNPY/ERG+ subtype and response to ADT. The underlying driving mechanisms behind the observed aggressiveness of the low NPY/ERG+ subtype will need further exploration to best tailor treatment and study design for this subgroup.
The findings of our study must be viewed within the limitations. First, defining the groups is based on whole genome transcriptomic data from FFPE tissue sample, making its clinical adoption challenging given the high price of genome-wide chip arrays. Second, these samples were all RP samples and thus do not reflect the complete clinical spectrum of prostate tumors. Additionally, these analyses are based on RNA expression and not protein level which may provide additional clinical validation. Further testing the clinical utility of NPY/ERG subtypes in other tissues and sample types (i.e. blood, urine) needs further exploration. Further experimental validation of the impact of NPY-knockdown in prostate cells with and without ERG-fusion will shed more light on the interactions between these two genes.
Despite these potential limitations, this study highlights the novel findings that though NPY is highly expressed in prostate cancers relative to other cancers, low NPY expression is associated with adverse genomic and histologic features, disease progression, and poor clinical outcomes. Furthermore, patients with low NPY and ERG fusions are at a high risk of developing metastasis and may be at risk of ADT resistance. Whether prognostic subgroupings and tailored treatments should be made by NPY expression and ERG fusion status warrants further careful examination.
Supplementary Material
References:
- [1].Wang M, Zhao J, Zhang L, Wei F, Lian Y, Wu Y, et al. Role of tumor microenvironment in tumorigenesis. J Cancer 2017;8:761–73. doi: 10.7150/jca.17648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gao F, Liang B, Reddy S, Farias-Eisner R, Su X. Role of Inflammation-Associated Microenvironment in Tumorigenesis and Metastasis. Curr Cancer Drug Targets 2014;14:30–45. doi: 10.2174/15680096113136660107. [DOI] [PubMed] [Google Scholar]
- [3].Magnon C, Hall S, Lin J, Xue X, Gerber L, Freedland S, et al. Autonomic Nerve Development Cancer Progression. Science (80-) 2013;341:1–10. doi: 10.1038/aja.2013.113. [DOI] [PubMed] [Google Scholar]
- [4].Zahalka AH, Arnal-Estapé A, Maryanovich M, Nakahara F, Cruz CD, Finley LWS, et al. Adrenergic nerves activate an angio-metabolic switch in prostate cancer. Science (80-) 2017;358:321–6. doi: 10.1126/science.aah5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hayakawa Y, Wang TC. Nerves switch on angiogenic metabolism. Science (80-) 2017;358:305–6. doi: 10.1126/science.aaq0365. [DOI] [PubMed] [Google Scholar]
- [6].Hondermarck H, Jobling P. The Sympathetic Nervous System Drives Tumor Angiogenesis. Trends in Cancer 2018;4:93–4. doi: 10.1016/j.trecan.2017.11.008. [DOI] [PubMed] [Google Scholar]
- [7].Iglesias-Gato D, Wikström P, Tyanova S, Lavallee C, Thysell E, Carlsson J, et al. The Proteome of Primary Prostate Cancer. Eur Urol 2016;69:942–52. doi: 10.1016/j.eururo.2015.10.053. [DOI] [PubMed] [Google Scholar]
- [8].Tilan J, Kitlinska J. Neuropeptide Y (NPY) in tumor growth and progression: Lessons learned from pediatric oncology. Neuropeptides 2016;55:55–66. doi: 10.1016/j.npep.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li J, Tian Y, Wu A. Neuropeptide Y receptors: a promising target for cancer imaging and therapy. Regen Biomater 2015;2:215–9. doi: 10.1093/rb/rbv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Massoner P, Kugler KG, Unterberger K, Kuner R, Mueller LAJ, Fälth M, et al. Characterization of Transcriptional Changes in ERG Rearrangement-Positive Prostate Cancer Identifies the Regulation of Metabolic Sensors Such as Neuropeptide Y. PLoS One 2013;8. doi: 10.1371/journal.pone.0055207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ross AE, Johnson MH, Yousefi K, Davicioni E, Netto GJ, Marchionni L, et al. Tissue-based Genomics Augments Post-prostatectomy Risk Stratification in a Natural History Cohort of Intermediate- and High-Risk Men. Eur Urol 2016;69:157–65. doi: 10.1016/j.eururo.2015.05.042. [DOI] [PubMed] [Google Scholar]
- [12].Kumar A, Coleman I, Morrissey C, Zhang X, True L, Gulati R, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 2016;22:369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas GV., et al. Clinical and Genomic Characterization of TreatmentEmergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J Clin Oncol 2018;36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tsai H, Lehrer J, Alshalalfa M, Erho N, Davicioni E, Lotan T. Gene expression signatures of neuroendocrine prostate cancer and primary small cell prostatic carcinoma. BMC Cancer 2017;17:759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Piccolo SR, Sun Y, Campbell JD, Lenburg ME, Bild AH, Johnson WE. A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics 2012;100:337–44. doi: 10.1016/j.ygeno.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Boormans JL, Korsten H, Ziel-Van Der Made AJC, Van Leenders GJLH, De Vos CV., Jenster G, et al. Identification of TDRD1 as a direct target gene of ERG in primary prostate cancer. Int J Cancer 2013;133:335–45. doi: 10.1002/ijc.28025. [DOI] [PubMed] [Google Scholar]
- [19].Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative Genomic Profiling of Human Prostate Cancer. Cancer Cell 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry CD, et al. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015;163:1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Akamatsu S, Wyatt AW, Lin D, Lysakowski S, Zhang F, Kim S, et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep 2015;12:922–36. doi: 10.1016/j.celrep.2015.07.012. [DOI] [PubMed] [Google Scholar]
- [23].Torres A, Alshalalfa M, Tomlins S, Erho N, Gibb E, Chelliserry J, et al. Comprehensive Determination of Prostate Tumor ETS Gene Status in Clinical Samples Using the CLIA Decipher Assay. J Mol Diagn 2017;19:475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tomlins SA, Alshalalfa M, Davicioni E, Erho N, Yousefi K, Zhao S, et al. Characterization of 1577 primary prostate cancers reveals novel biological and clinicopathologic insights into molecular subtypes. Eur Urol 2015;68:555–67. doi: 10.1016/j.eururo.2015.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kristensen G, Røder MA, Berg KD, Elversang J, Iglesias-Gato D, Moreira J, et al. Predictive value of combined analysis of pro-NPY and ERG in localized prostate cancer. APMIS 2018;126:804–13. doi: 10.1111/apm.12886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


