Abstract
In neurons, autophagy is crucial to proper axon guidance, vesicular release, dendritic spine architecture, spine pruning and synaptic plasticity and, when dysregulated, is associated with brain disorders, including autism spectrum disorders, and neurodegenerative diseases such as Parkinson’s and Alzheimer’s disease. Once thought to play a housekeeping function of removing misfolded proteins or compromised organelles, neuronal autophagy is now regarded as a finely tuned, real time surveillance and clearance system crucial to synaptic integrity and function. Here we review the role of autophagy in synaptic plasticity and its regulation by epigenetic mechanisms.
Introduction
Autophagy is an evolutionarily conserved cellular mechanism for the degradation and recycling of cellular components via the lysosomal pathway and in neurons, autophagy is increased during periods of low neuronal activity, sensory deprivation, and loss of neurotrophic factors which act indirectly via mTOR signaling, or in response to amino acid starvation [1–4]. Autophagy has three mechanistically distinct forms: chaperone-mediated autophagy, microautophagy, and macroautophagy [5]. In this review, we focus on macroautophagy (hereafter called autophagy), which is the major catabolic mechanism used by neurons to maintain the integrity of synaptic vesicle-dependent transmitter release and quality control of organelles as well as protein homeostasis of synaptic proteins at postsynaptic sites [6]. The neuroanatomical hallmark of autophagy is the presence of autophagosomes. Autophagosomes act via protein adaptors such as p62 to deliver proteins and organelles to lysosomes for degradation. Molecules such as p62, which bind cargo and components of the autophagic machinery, recognize and bind ubiquitinated proteins, enabling their engulfment by autophagosomes targeted for degradation [1]. Reduced autophagy in brains from humans diagnosed with autism is associated with an accumulation of ubiquitinated proteins such as PSD-95 [7••] and an overabundance of spines, presumably due to impaired spine pruning. Reduced autophagy in brains from humans diagnosed with Parkinson’s disease or Alzheimer’s is associated with neurodegeneration [1–4].
The mammalian target of rapamycin (mTOR) is a serine/threonine kinase and plays a central role in cell growth, proliferation, protein synthesis, autophagy, and cell survival [8,9]. In neurons, mTORC1 is localized to presynaptic and postsynaptic sites (or to lysosomes) where it imposes a brake on autophagy [10,11]. The biochemical steps, by which autophagy is activated in neurons under physiological and pathological conditions, are well-delineated. Activated mTOR phosphorylates Unc-51-like autophagy-activating kinase 1 (ULK-1/Atg1) at S757, a target of mTORC1 and well-known anti-autophagy site [12]. This, in turn, sequesters ULK-1 away from AMP kinase (AMPK), inhibiting autophagy. By contrast, in response to cellular stress, including amino acid starvation, loss of neurotrophic factors, endoplasmic reticulum stress, protein aggregates and/or sensory deprivation, AMPK phosphorylates and activates ULK-1 at S317. Upon activation, ULK-1 promotes phosphorylation and activation of Beclin-1 (Atg6) at S14, a crucial step in the ‘nucleation phase’ of autophagy [13] and recruits the PI3K complex, which is essential to elongation of the limiting membrane. Beclin-1 promotes lipidation of LC3-I (Atg8) to generate its lipidated form LC3-II, which localizes to and enables elongation of the limiting membrane and formation of mature LC3-II (+) autophagosomes [1–3]. Autophagosomes act via cargo adaptor proteins such as p62 to deliver the cargo, ubiquitinated proteins, protein aggregates and compromised organelles, to lysosomes for degradation. Ultimately, mature autophagosomes fuse with lysosomes to become autolysosomes, which actively degrade cytosolic cargo via the autosomal/lysosomal pathway.
Autophagy at the presynaptic synapse
Communication between neurons occurs via action potentials that are propagated down the axon to the presynaptic terminal. Presynaptic terminals (also termed ‘boutons’) are highly specialized compartments packed with synaptic vesicles and the machinery required for fusion of vesicles with the plasma membrane to enable transmitter release together with for autophagy to clear unwanted debris. In neurons, autophagy affords a powerful, highly regulated clearance system, which maintains synapse integrity and function. An essential presynaptic membrane protein Endophilin 1, best known for its role in the endocytosis of synaptic vesicles, localizes to pre-autophagosomal (Atg(+)) membranes and is essential to the initiation of autophagy in axons. Initiation of autophagy starts with nucleation, a step, in which the kinase LRRK2 phosphorylates endophilin (ENDO) 1, inducing a curvature in the bar domain of ENDO1, which initiates biogenesis of an isolation membrane or phagophore, and provides a docking site for Atg3. In the second stage (elongation), LC3 (Atg8) and Atg16 are recruited, and together sequester the autophagic cargo and promote elongation of the phagophore. This stage also depends on Atg3, which drives lipidation of LC3-I to generate its lipidated counterpart LC3-II. Upon lipidation, LC3-II localizes to the phagophore membrane, enabling elongation of the limiting membrane to engulf the substrate/cargo and close to form mature LC3-II (+) autophagosomes (closure) [1–3]. In the final stage, the autophagosome fuses with the lysosome to form autolysosomes (fusion) which ultimately degrade the cargo via the autosomal/lysosomal pathway [2]. In axons, this requires retrograde transport of autophagosomes from the nerve terminal to the cell soma, where degradation occurs. Expression of a constitutively active mutant of LRRK2 or a phosphomimetic mutant of Endo A increases the density of Atg8-positive puncta in presynaptic terminals and increases colocalization of LC3-II (Atg8) with Atg3. Failure of LRRK2 to phosphorylate Endo A at Ser5 is associated with Parkinson’s disease and neurodegeneration.
The lipid phosphatase Synaptojanin-1 is crucial to synaptic vesicle endocytosis. After a synaptic vesicle fuses with the presynaptic membrane and releases transmitter, Synaptojanin-1 retrieves and promotes degradation of the residual vesicular membrane, a step important in ‘resetting’ of the presynaptic cell with new transmitter. This is thought to occur via an attraction between Synaptojanin 1 and clathrin, a protein crucial to endocytosis of synaptic vesicles. A recent study demonstrates that Synaptojanin-1 drives autophagosome biogenesis in the presynaptic compartment independently of its role in endocytosis. Synaptojanin-1 harbors two phosphatase domains, the 5’-phosphatase domain and the SAC1 (N-terminal Sac1-like inositol phosphatase) domains [14]. Whereas the 5’-phosphatase domain drives endocytosis of synaptic vesicles by dephosphorylating PI(4,5)P2, the SAC1 domain drives autophagosome biogenesis by dephosphorylating PI(3)P/PI(3,5)P2 [14]. An R258Q mutation in the SAC1 domain is found in a subset of patients with hereditary, early-onset Parkinson’s disease [14]. The R258Q PD mutation causes a build-up of the PI (3)P/PI(4,5)P2 binding partner Atg18a on pre-formed (nascent) autophagosomes, blocking autophagosome maturation at Drosophila synapses and in neurites of neurons induced from iPSC cells derived from a Parkinson’s disease patients with the R258Q mutation [14]. Moreover, Drosophila carrying the R258Q mutation exhibits neurodegeneration and, specifically, loss of dopaminergic neurons [14].
The cytoskeleton matrix active zone proteins Bassoon and piccolo are thought to keep autophagy in check [15]. The details of how this happens have recently been revealed. Bassoon acts via an Atg5 binding motif in its coiled-coil (CC)2 region to bind Atg5, an E3-like ligase crucial to recognition of ubiquitinated substrates by the autophagy machinery and to recruitment of LC3-I (Atg8) to the phagophore membrane during autophagosome biogenesis [15]. Bassoon inhibits the induction of autophagy, most likely by acting via its own Atg binding motif to block formation of the Atg5/12 complex. Accordingly, expression of a fusion protein containing the Atg5-binding peptide disrupts binding of Bassoon to Atg, rescuing synapses from autophagy [15]. A recent study involving optogenetics showed that autophagy affords a powerful, tightly regulated clearance system capable of responding in 5–10 min to maintain synaptic integrity and function [16]. Thus, the role of synaptic vesicle trafficking proteins in autophagy and the role of autophagy components in presynaptic function are intricately intertwined.
Autophagy at postsynaptic densities
Autophagy and synaptic plasticity
Whereas a great deal of research has focused on autophagy in the presynaptic compartment, much less is known about autophagy in dendrites. One of the first studies to report autophagy at postsynaptic sites focused on GABAergic inhibitory interneurons. Whereas under normal conditions, GABA release from presynaptic sites induces clustering and trafficking of GABAA receptors in postsynaptic cells [17], block of presynaptic GABAergic signaling induces autophagy at postsynaptic sites and accumulation of GABAA receptors in autophagosomes targeted for degradation [17]. Although autophagosomes have been visualized in dendrites, it is as yet unclear as to whether autophagosome biogenesis occurs in dendrites.
In rat hippocampal neurons, treatment with NMDA at a dose that induces chemical long-term depression (LTD), increased LC3-II and the number of LC3(+) autophagosomes at postsynaptic sites, indicative of increased autophagy, and enhanced degradation of the AMPA receptor subunit GluA1 [18]. The inhibitor of autophagy wortmannin or shRNA to Atg7 blocked NMDA-induced autophagy and partially recovered GluA1 levels, consistent with the concept that AMPA receptors are degraded, a least in part, via the autophagy/lysosomal pathway [18]. In a recent study, induction of autophagy in the amygdala or hippocampus was reported to destabilize synapses and contextual fear memory in an activity-dependent manner. Destabilization of synapses coincided with autophagy-dependent degradation of AMPARs in dendritic spines in brain regions associated with contextual fear memory [19]. These findings are significant in that they indicate that autophagy is regulated in an activity-dependent manner and can recognize and degrade postsynaptic proteins with precision and specificity.
A recent study provides evidence that autophagy can regulate synaptic plasticity at CA1 synapses [20]. Fasting in rats was shown to increase BDNF and suppress autophagy in the hippocampus. This is significant in that theta-burst stimulation induced long-term potentiation (TBS-LTP), a major form of synaptic plasticity, at Schaffer collateral to CA1 pyramidal cell synapses requires BDNF and suppression of autophagy [21]. Additionally, fasting markedly enhanced freezing behavior in a contextual fear conditioning assay, indicative of enhanced memory, and increased the number of dendritic spines on CA1 and CA3 neurons, presumably as a consequence of impaired autophagy. The investigators identified postsynaptic scaffolding/signaling proteins (PSD-95, SHANK3, PICK1) that are degraded by autophagy, offering a likely mechanism for the role of BDNF and suppressed autophagy in TBS-LTP [21]. These findings are in keeping with recent observations that inhibition of neuronal autophagy results in a buildup of synaptic proteins such as Arc and PSD-95, consistent with the concept that autophagy is involved in degradation of these and other synaptic proteins under physiological conditions [22••]. Thus, autophagy is pivotal to the homeostasis of several key synaptic proteins and serves as a significant regulator of structural plasticity, synaptic strength, and memory consolidation [2,18].
Synapse elimination and spine pruning
Synaptic activity influences the number and strength of synapses that form between neurons. During early postnatal development, the rate of synapse formation exceeds that of elimination, resulting in a greater number of immature excitatory synapses. At more mature ages, destabilization and spine pruning reduce synapse number. thus refining neural circuits that underlie behavior and cognition [23]. Two papers provide evidence for a role for impaired autophagy as a mechanism underlying compromised synapse elimination (an overabundance of spines) in autism spectrum disorders (ASDs). In layer V of the neocortex of Tsc2+/− mice [7••] and the hippocampus of Fmr1 KO mice, mTOR signaling is aberrantly activated and autophagy is constrained during postnatal development [22••]. Moreover, reduced autophagy is causally linked to impaired spine pruning in cortical layer V pyramidal neurons of Tsc2+/− mice [7••] and hippocampal neurons in Fmr1 KO mice [22••]. Rapamycin, an inhibitor of mTORC1, corrects deficits in spine pruning and social interactions in Tsc2+/− mice, but not in Atg7cKO neuronal autophagy-deficient mice or Tsc2+/−: Atg7cKO double mutants [7••]. Interestingly, activation of autophagy in neurons enables spine elimination, with little or no effect on spine biogenesis. Moreover Atg7 KO mouse exhibit ASD phenotypes including impaired social interactions [7••].
In the hippocampus of Fmr1 KO mice, acute knockdown of Raptor, a defining component of mTOR complex 1 (mTORC1) in the CA1 of Fmr1 KO mice corrects mTORC1 signaling, activates autophagy, and restores spine density to near that of WT mice. Because suppression of mTORC1 not only activates autophagy, but also limits protein translation, which may independently affect spine density [24–26] and can potentially inhibit cell growth, proliferation and ribosome biogenesis, the investigators next delivered lentivirus carrying shRaptor together with shRNA directed to Atg7 [7••,27], an E1-like ligase essential for autophagosome formation, to block autophagyt [28]. Expression of shAtg7 together with shRaptor reversed, at least in part, the rescue of spine density by shRaptor, consistent with the concept that rescue occurs, at least partially, via activation of autophagy [22••]. These findings are consistent with a model whereby mTOR-dependent autophagy is essential for spine pruning in late development, and activation of neuronal autophagy corrects synaptic pathology and social behavioral deficits in ASDs with hyperactivated mTOR, and suggest that impaired autophagy and its link to impaired spine elimination is likely to be more widespread in a number of ASDs [7••].
Epigenetic and transcriptional control of autophagy
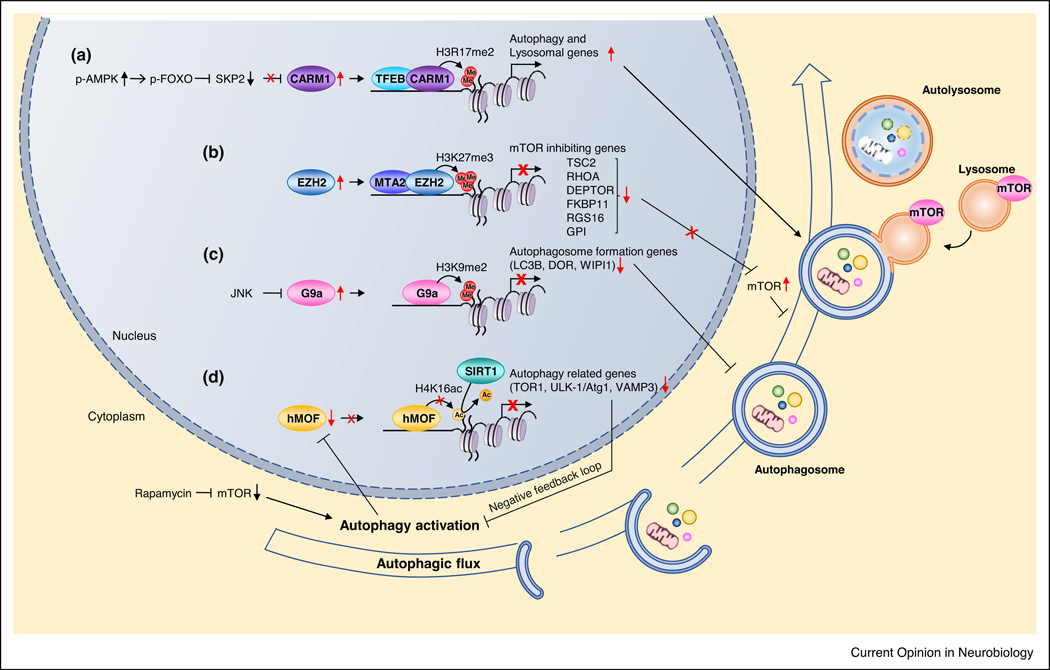
Whereas the role of autophagy in neurons has been well studied, epigenetic and transcriptional regulation of autophagy is a new, emerging topic. Considerable evidence implicates chromatin remodeling in the fine-tuning of the autophagy pathway in response to environmental cues such as starvation. Several chromatin remodeling enzymes are ‘master regulators’ of autophagy genes (reviewed in Refs. [29,30]). These include co-activator-associated arginine methyl-transferase 1 (CARM1), the H3K9 methyltransferase G9a [31••], the polycomb protein EZH2 (enhancer of zeste 2 polycomb repressive complex-2, which confers a trimethylation mark on H3 at lysine 27 [32], hMOF (human ortholog of the Drosophila MOF gene (males absent on the first)) H4K16 acetyltransferase, and its counterpart SIRT1 (Sirtuin 1) H4K16 deacetylase [33], which confer epigenetic marks of gene activation or repression on core histones (Figure 1).
Figure 1.
Epigenetic regulators of autophagy genes. (a) CARM1 confers a dimethylation mark on H3 at arginine 17, which activates transcription of autophagy genes (ULK-1/Atg1, Atg11, Atg12) via the AMPK-SKP2-CARM1 signaling axis. (b) EZH-2 promotes trimethylation of H3 at lysine 27, and thereby represses genes that inhibit mTOR signaling (DEPTOR, TSC2) which in turn, activates mTOR signaling and suppresses autophagy. (c) G9a promotes demethylation of H3 at lysine 9, and thereby represses genes related to autophagosome formation. (d) Control of H4K16 acetylation status by hMOF and SIRT1. In response to cellular stress, mTOR (and Raptor) translocates to the surface of lysosomes, where mTOR is strategically positioned to be phosphorylated by AKT and to phosphorylate and inhibit its downstream target Transcription factor EB (TFEB), which drives transcription of genes crucial to autophagy and lysosomal degradation. Upon conditions of high nutrients, inhibition of mTOR signaling leads to activation of autophagy; under these conditions, hMOF and H4K16 acetylation are downregulated which in turn, represses transcription of autophagy related genes (TOR1, ULK-1/Atg1, VAMP3). Thus, a negative feedback regulatory loop occurs which might prevent overly high stimulation of autophagic flux.
CARM1, a histone methyltransferase that confers dimethylation on histone H3 at arginine 17 (H3R17), a mark of active gene transcription, is a crucial regulator of autophagy [29]. CARM1 serves as a transcriptional coactivator of the transcription factor TFEB, a master regulator of autophagy-related and lysosomal genes, in non-neuronal cells [34••]. CARM1 is regulated at the level of protein stability by the SKP2-SCF complex, a E3 ubiquitin ligase, which targets CARM1 for degradation via the ubiquitin-based, proteasomal pathway [35]. In response to glucose starvation, AMPK is activated in the nucleus, where it phosphorylates FOXO3A, a transcription factor that negatively regulates the SKP2/SCF complex by inhibiting SKP2 gene expression transcription and by binding SKP2 directly and disrupting the SKP2/SCF complex [35] (Figure 1a). Loss of SKP2 increases CARM1 stability (and abundance) [35]. Genome-wide association studies reveal a global increase in CARM1 and its functional readout, dimethylation of H3 at arginine 17 at the promoter of TFEB target genes, which include a vast number of autophagy-related and lysosomal genes (ULK1, Atg12, Atg13, Atg14, Sirt1) [34••] (Figure 1a). These findings identify the AMPK-SKP2-CARM1 signaling cascade as an important mechanism in the activation of autophagy in response to environmental cues.
EZH2 and G9a suppress autophagy [31••,32]. EZH2 represses transcription of several negative regulators of the mTOR signaling pathway — TSC2, RHOA, DEPTOR, FKBP11, RGS16 and GPI, thereby suppressing autophagy [32]. EZH2 is recruited to the promoter of these genes via a chromatin-binding protein MTA2, where it (EZH2) increases trimethylation of H3K27 (Figure 1b). As an example, EZH2-dependent repression of TSC2 (tuberous sclerosis 2) releasing the brake on mTOR and thereby inhibiting autophagy. By contrast, G9a represses genes associated with autophagosome formation [31••] (Figure 1c). Upon activation of autophagy, G9a binding (and dimethylation of H3K9) are diminished at promoters of target genes LC3B, DOR and WIPI1, which participate in autophagosome formation. This process is prevented by pharmacological inhibition of JNK (c-Jun N-terminal kinase) as well as G9. These findings demonstrate that EZH2 and G9a suppress autophagy and underscore EZH2 and G9a as potential therapeutic targets for intervention in the neurodegeneration associated with Parkinson’s disease. Two additional epigenetic regulators of autophagy are hMOF and its counterpart SIRT1 (Figure 1d). hMOF catalyzes acetylation of histone H4 lysine 16 (H4K16), while SIRT1 removes it. Genome wide control of the acetylation status on H4K16 by these enzymes has been suggested as a determinant in the process of death and survival decision in autophagic cells [36]. Autophagy gene targets include TOR1, ULK1 and VAMP3.
Autophagy is activated in selectively vulnerable CA1 neurons in response to induction of global ischemia [37••]. Immediately following ischemia, mTOR protein is decreased, coinciding with an increase in biochemical markers of autophagy and autophagic flux in hippocampal neurons. The mTORC1 inhibitor rapamycin or acute knockdown of mTOR promotes autophagy and attenuates ischemia-induced neuronal death, indicating an inverse causal relation between mTOR, autophagy, and neuronal death. However, the mechanism by which ischemia regulates mTOR in hippocampal neurons and how this signaling impacts on synaptic function and plasticity is, as yet, unclear. A possible scenario is that an endogenous inhibitor of mTOR (DEPTOR, TSC2) is upregulated in response to loss of EZH2.
Conclusions and future perspectives
An emerging theme is the specificity of autophagy in degradation of synaptic proteins and organelles in an activity-dependent manner and the precision by which it is regulated. Despite a wealth of information concerning epigenetic regulation of autophagy genes, none of the research was performed in neurons. Thus, an important future direction would be to identify epigenetic mechanisms by which autophagy genes are regulated in neurons. CARM1 is not only expressed in neurons, but localizes to postsynaptic densities in hippocampal neurons where it negatively regulates spine maturation and dendritic arborization [38]. EZH2 is expressed in neurons where it plays a role in reconsolidation of fear memory [39]. Future studies are warranted to examine epigenetic and transcriptional mechanisms that could potentially regulate autophagy in neurons.
Acknowledgements
We would like to acknowledge all the authors whose valuable work we could not include due to space limitations and the limited number of citations allowed. This work was supported by; National Institutes of HealthNS046742, HD083828, NS100047, MH092877 and a generous grant from the F. M. Kirby Foundation to RSZ; National Institutes of Health NS100047, AHA Scientist Development Grant, NARSAD Young Investigator Award and LB692 Nebraska Tobacco Settlement Biomedical Research Development Funds to JYH. RSZ is the F.M. Kirby Chair in Neural Repair and Protection.
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
•• of outstanding interest
- 1.Harris H, Rubinsztein DC: Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol 2011, 8:108–117. [DOI] [PubMed] [Google Scholar]
- 2.Nixon RA: The role of autophagy in neurodegenerative disease. Nat Med 2013, 19:983–997. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ: Autophagy fights disease through cellular self-digestion. Nature 2008, 451:1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong E, Cuervo AM: Autophagy gone awry in neurodegenerative diseases. Nat Neurosci 2010, 13:805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider JL, Cuervo AM: Autophagy and human disease: emerging themes. Curr Opin Genet Dev 2014, 26:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine B, Kroemer G: Biological functions of autophagy genes: a disease perspective. Cell 2019, 176:11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A et al. : Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 2014, 83:1131–1143•• This paper was the first to show that autophagy is impaired in a model of autism spectrum disorders and causally related to impaired spine pruning.
- 8.Laplante M, Sabatini DM: mTOR signaling in growth control and disease. Cell 2012, 149:274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hay N, Sonenberg N: Upstream and downstream of mTOR. Genes Dev 2004, 18:1926–1945. [DOI] [PubMed] [Google Scholar]
- 10.Hoeffer CA, Klann E: mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci 2010, 33:6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N: Translational control of long-lasting synaptic plasticity and memory. Neuron 2009, 61:10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH: ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009, 20:1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL: ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013, 15:741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanhauwaert R, Kuenen S, Masius R, Bademosi A, Manetsberger J, Schoovaerts N, Bounti L, Gontcharenko S, Swerts J, Vilain S et al. : The SAC1 domain in synaptojanin is required for autophagosome maturation at presynaptic terminals. EMBO J 2017, 36:1392–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okerlund ND, Schneider K, Leal-Ortiz S, Montenegro-Venegas C, Kim SA, Garner LC, Waites CL, Gundelfinger ED, Reimer RJ, Garner CC: Bassoon controls presynaptic autophagy through Atg5. Neuron 2017, 93:897–913 e897. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann S, Orlando M, Andrzejak E, Bruns C, Trimbuch T, Rosenmund C, Garner CC, Ackermann F: Light-activated ROS production induces synaptic autophagy. J Neurosci 2019, 39:2163–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowland AM, Richmond JE, Olsen JG, Hall DH, Bamber BA: Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. J Neurosci 2006, 26:1711–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shehata M, Matsumura H, Okubo-Suzuki R, Ohkawa N, Inokuchi K: Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J Neurosci 2012, 32:10413–10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shehata M, Abdou K, Choko K, Matsuo M, Nishizono H, Inokuchi K: Autophagy enhances memory erasure through synaptic destabilization. J Neurosci 2018, 38:3809–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikoletopoulou V, Tavernarakis N: Regulation and roles of autophagy at synapses. Trends Cell Biol 2018, 28:646–661. [DOI] [PubMed] [Google Scholar]
- 21.Nikoletopoulou V, Sidiropoulou K, Kallergi E, Dalezios Y, Tavernarakis N: Modulation of autophagy by BDNF underlies synaptic plasticity. Cell Metab 2017, 26:230–242 e235. [DOI] [PubMed] [Google Scholar]
- 22.Yan J, Porch MW, Court-Vazquez B, Bennett MVL, Zukin RS: Activation of autophagy rescues synaptic and cognitive deficits in fragile X mice. Proc Natl Acad Sci U S A 2018, 115: E9707–E9716•• This paper was the first to show that autophagy is impaired in Fragile X syndrome and causally related to synaptic and cognitive deficits including impaired spine pruning.
- 23.Rakic P, Bourgeois JP, Eckenhoff MF, Zecevic N, Goldman-Rakic PS: Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science 1986, 232:232–235. [DOI] [PubMed] [Google Scholar]
- 24.Bagni C, Greenough WT: From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci 2005, 6:376–387. [DOI] [PubMed] [Google Scholar]
- 25.Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, Kaphzan H, Klann E: Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature 2013, 493:411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darnell JC, Klann E: The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci 2013, 16:1530–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai Y, Yang L, Hu G, Chen X, Niu F, Yuan L, Liu H, Xiong H, Arikkath J, Buch S: Regulation of morphine-induced synaptic alterations: role of oxidative stress, ER stress, and autophagy. J Cell Biol 2016, 215:245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ: Autophagy regulates adipose mass and differentiation in mice. J Clin Invest 2009, 119:3329–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fullgrabe J, Klionsky DJ, Joseph B: The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol 2014, 15:65–74. [DOI] [PubMed] [Google Scholar]
- 30.Baek SH, Kim KI: Epigenetic control of autophagy: nuclear events gain more attention. Mol Cell 2017, 65:781–785. [DOI] [PubMed] [Google Scholar]
- 31.Artal-Martinez de Narvajas A, Gomez TS, Zhang JS, Mann AO, Taoda Y, Gorman JA, Herreros-Villanueva M, Gress TM, Ellenrieder V, Bujanda L et al. : Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol 2013, 33:3983–3993•• This was the first study to show that EZH2 suppresses autophagy by repressing transcription of one or more negative regulators of mTOR signaling such as TSC2.
- 32.Wei FZ, Cao Z, Wang X, Wang H, Cai MY, Li T, Hattori N, Wang D, Du Y, Song B et al. : Epigenetic regulation of autophagy by the methyltransferase EZH2 through an MTOR-dependent pathway. Autophagy 2015, 11:2309–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fullgrabe J, Klionsky DJ, Joseph B: Histone post-translational modifications regulate autophagy flux and outcome. Autophagy 2013, 9:1621–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin HJ, Kim H, Oh S, Lee JG, Kee M, Ko HJ, Kweon MN, Won KJ, Baek SH: AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature 2016, 534:553–557•• This study was the first to identify the AMPK-SKP2-CARM1 signaling cascade as an epigenetic mechanism important to activation of autophagy in response to environmental stimuli.
- 35.Wu J, Lee SW, Zhang X, Han F, Kwan SY, Yuan X, Yang WL, Jeong YS, Rezaeian AH, Gao Y et al. : Foxo3a transcription factor is a negative regulator of Skp2 and Skp2 SCF complex. Oncogene 2013, 32:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fullgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, Joseph B: The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 2013, 500:468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang JY, Gertner M, Pontarelli F, Court-Vazquez B, Bennett MV, Ofengeim D, Zukin RS: Global ischemia induces lysosomalmediated degradation of mTOR and activation of autophagy in hippocampal neurons destined to die. Cell Death Differ 2017, 24:317–329•• The first evidence showing that loss of mTOR promotes autophagy and attenuates neuronal death in a clinically relevant model of global ischemia. This study represents an advance over previous studies in that it delineates mechanisms underlying ischemia-induced activation of autophagy and reveals a novel mechanism, by which mTOR self-regulates its own degradation in insulted hippocampal neurons.
- 38.Lim CS, Alkon DL: Inhibition of coactivator-associated arginine methyltransferase 1 modulates dendritic arborization and spine maturation of cultured hippocampal neurons. J Biol Chem 2017, 292:6402–6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jarome TJ, Perez GA, Hauser RM, Hatch KM, Lubin FD: EZH2 methyltransferase activity controls Pten expression and mTOR signaling during fear memory reconsolidation. J Neurosci 2018, 38:7635–7648. [DOI] [PMC free article] [PubMed] [Google Scholar]