Abstract
African American (AFR) men have the highest mortality rate from prostate cancer (PCa) compared to men of other racial/ancestral groups. Differences in the spectrum of somatic genome alterations in tumors between AFR men and other populations have not been well-characterized due to a lack of inclusion of significant numbers in genomic studies. To identify genomic alterations associated with race, we compared the frequencies of somatic alterations in PCa obtained from four publicly-available datasets containing 250 AFR and 611 European American men (EUR). Mutations in ZFHX3 as well as focal deletions in ETV3 were more frequent in tumors from AFR men. TP53 mutations were associated with increasing Gleason score. Targeted sequencing data from a commercial platform for 436 AFR and 3018 EUR men as also analyzed. MYC amplifications were more frequent in tumors from AFR men with metastatic PCa while deletions in PTEN and rearrangements in TMPRSS2-ERG were less frequent in tumors from AFR men. KMT2D truncations and CCND1 amplifications were more frequent in primary PCa from AFR men. Genomic features that could impact clinical decision making were not significantly different between the two groups including tumor mutation burden, MSI status, and genomic alterations in select DNA repair genes, CDK12, and in AR. While we identified some novel differences in AFR men compared to other populations, the frequencies of genomic alterations in current therapeutic targets for PCa were similar between AFR and EUR men suggesting that existing precision medicine approaches could be equally beneficial if applied equitably.
Keywords: Meta-analysis, Prostate cancer, Race, Genetics, Sequencing
INTRODUCTION
Despite declines in mortality related to cancer in the United States, disparities by race have persisted. African American (AFR) men have a higher incidence, present with more advanced disease at an earlier age, and have increased mortality from prostate cancer (PCa) compared to European Americans (EUR)(1). Differences in outcomes persist even after correcting for socioeconomic covariates(2,3). There is emerging evidence that across some clinical trials and “equal-access” health systems, outcomes between African American men and European American men with prostate cancer are similar(4,5). While these data suggest that disparities can be ameliorated, there is limited knowledge of the genomic alterations that differ between groups and that could impact clinical outcomes. Certain somatic alterations in tumors differ in frequency across ancestral populations and have significant clinical implications. For instance, EGFR mutations are more common in lung adenocarcinoma patients in patients of East Asian ancestry for which targeted therapies are the mainstay of treatment(6). In prostate cancer, TMPRSS2-ERG rearrangements and PTEN deletions are less frequent in prostate cancers from AFR men(1) and lack targeted treatments at this time. Notably, FOXA1 mutations are highly prevalent among Asians with prostate cancer while being less frequent in prostate tumors from men of European ancestry(7).
To date, cancer genomic studies have largely underrepresented racial/ethnic minority groups and have not been powered to detect differences in genomic alterations despite the greater burden of prostate cancer in AFR men(8,9). Larger sample sizes from men of African ancestry are needed to detect significant associations in genes with lower mutational frequencies and to determine whether tumor genomic features associated with benefit from clinical therapies differ between men of African and European ancestry. In this study, we aggregate a large cohort of PCa from AFR men to identify genomic alterations associated with race and investigate tumor genomic features in primary and metastatic disease between these two groups.
METHODS
Mutational analysis and copy number analysis of publicly-available datasets.
Data retrieval and preprocessing.
For the meta-analysis of publicly-available datasets, the selection criteria included studies that fulfilled the following requirements: 1) performed whole exome sequencing (WES) or targeted DNA sequencing of prostate adenocarcinomas, 2) included at least 10 patients with African American ethnicity by self-report, and 3) had mutation calls per tumor that were publicly accessible. For the first dataset we used the MC3 (Multi-Center Mutation Calling in Multiple Cancers) call set from the Pan-Cancer Atlas Project of The Cancer Genome Atlas (TCGA) Network that contain somatic variant calls on TCGA exome sequencing data across 33 tumor types(10). Within the MC3 dataset, variant calls specific to prostate adenocarcinoma (PRAD) were taken for downstream analysis from the “mc3.v0.2.8.PUBLIC.maf” file located at https://api.gdc.cancer.gov/data/1c8cfe5f-e52d-41ba-94da-f15ea1337efc. Somatic variants that were deemed to have not passed all filters denoted within the “FILTER” column were excluded. Clinical annotation information for this cohort was retrieved from the file “nationwidechildrens.org_clinical_patient_prad.txt” available at the GDC Data Portal. Ancestry calls were obtained by running Ethnoseq as previously performed(11).
For the second and third datasets, we utilized somatic mutation calls from the African Ancestry prostate cancer (AAPC) cohort(11) which contained both WES and targeted sequencing data. Mutation calls were obtained from Supplementary Table 5 (WES) and Supplementary Table 10 (Targeted sequencing panel) from the publication. Finally, prostate cancers profiled with the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) panel were included(12). Mutation calls were taken from the tab “Somatic Mutations” and copy number calls from the tab “Amplifications and Deletions” of the Data Supplement file. Race annotation for this cohort was provided within the tab “Clinical Annotation”. Clinical and pathologic data for all cohorts are summarized in Supplementary Tables 1-3. 22 patients from Abida et al and 8 patients from Huang et al did not have annotation for self-reported race or Gleason score and were excluded from subsequent analyses. In total, 250 patients had self-reported African American annotation in the Huang et al and MSK-IMPACT datasets or had confirmed African ancestry in the TCGA dataset (AFR group). 611 patients had self-reported European American annotation in the Abida et al, or had confirmed European ancestry in the TCGA dataset (EUR group).
Subsequently in all datasets, variants were filtered to only include nonsynonymous mutations, including as nonsense mutations, missense mutations, and indels. Additionally, mutations with alternate allele less than 0.05 were removed to account for differences in sequencing depth between WES and targeted datasets. For TCGA, 30,869 variants were available in the analysis after filtering. The WES data and the targeted sequencing data from the Huang et al study contained 7,353 and 491 variants after filtering, respectively. 1,470 variants remained after filtering in the MSK-IMPACT cohort (Supplementary Table 4).
18 genes were measured across all four datasets. 15 genes mutated in at least 2 patients were included in the final analysis (Supplementary Tables 5, 6). For each gene, logistic regression was used to determine associations between mutational frequencies and race, age, Gleason score, and mutation rate. Gleason score was treated as a factor with four levels (6, 7, 8, 9+10). A likelihood ratio test was used to compare logistic regression models with and without each term to determine associations with mutation status (car R package). The Benjamini-Hochberg False Discovery Rate (FDR) was applied to the p-values of each term across all genes to correct for multiple hypothesis testing. We also applied the same logistic regression approach while limiting the analysis to truncating mutations for 11 of the aforementioned 15 genes (Supplementary Table 7).
Identification of recurrently altered copy number peaks.
Genome-wide copy number ratios were available from SNP6.0 data in the TCGA cohort and from WES data in Huang et al cohort. GISTICv2.0.23 was applied separately to copy number ratios from AFR men (n=171) or from EUR men (n=626) to identify recurrent deletion peaks (“2 cohort analysis”; Supplementary Tables 8-12). Tumors with a GISTIC thresholded call of −2 were considered deleted. For each gene, logistic regression was used to determine if deletion status for the target gene of 12 recurrently deleted peaks was associated with race, age, or Gleason score (Supplementary Table 13). 8 genes that were a target of a recurrent deletion peak and 15 genes that were a target of a recurrent amplification peak from the GISTIC2.0 analysis mentioned above were also measured in the Abida et al dataset. In the Abida et al dataset, a log2 fold change of less than −1 was considered deleted while a log2 fold change great than 1 was considered amplified. For each gene, logistic regression was used to determine if deletion status (or amplification status) was associated with race, age or Gleason score followed by an FDR correction for each term (“3 cohort analysis”, Supplementary Tables 14-17).
Mutational and copy number analysis from the Foundation cohort.
In total, 251 AFR and 1,940 EUR men with localized PCa, as well as 185 AFR and 1,078 EUR men with metastatic PCa were obtained from Foundation Medicine (Supplementary Tables 18-20). Ancestries were determined for de-identified, consented-for-research samples using a supervised approach. For each platform, Phase 3 1000 Genomes SNPs that overlapped with target regions in the comprehensive genomic profiling assay were projected down to five principal components, and these five features were used to train a random forest classifier that distinguishes between African, admixed American, East Asian, European, and South Asian ancestral groups. Admixture analysis was performed using ADMIXTURE 1g.3.0. Prior to running ADMIXTURE, imputation and phasing were performed using Beagle 5.0. Five ancestral populations were pre-defined by running ADMIXTURE on Phase 3 1000 Genomes data, and the analysis was performed using these projections to determine percent composition for each de-identified sample of these five populations. Five primary categories of alterations were considered including point mutations, truncations, amplifications, deletions, and rearrangements. All of these alterations as well as any other type of variant were included in the “all” category (Supplementary Tables 17-19). Focusing on relatively frequent gene alterations, we counted 26 genes with more than 5 alterations in men with localized PCa and 38 genes with more than 5 alterations in men with metastatic PCa (Supplementary Tables 18-20). For each gene alteration class, the number of patients with this specific alteration was counted and the frequency was computed accordingly. In the Foundation cohort, a Fisher’s exact test was applied to each type of alteration that occurred in more than 5 patients to identify differences between AFR and EUR populations. This test was performed including all men and then separately for men with primary or metastatic disease. Moreover, to analyze the differences between AFR and EUR men PCa at a functional level, we compared the alteration frequencies in gene pathways including gene repair, cell cycle, PI3K, RAS/MAPK, KMT, and WNT pathways(13)(Supplementary Table 21). In this analysis, we included genes in each pathway that were measured in one or more alteration classes reported in the Foundation Cohort for both localized and metastatic PCa patients. Specifically, if the alteration frequency for a gene was 0, it was omitted in the dataset.
For AFR and EUR localized and metastatic PCa patients, tumor mutational burden (TMB) was computed as the percentage of patients with respect to the number of mutations (0 ~ 100). A two-sided T-test was conducted then using the scipy.stats.ttest_ind function in the scipy package (CITE) to compare the means between the two ancestry groups.
All code used for the analysis is available at https://github.com/campbio/Manuscripts.
RESULTS
Analysis of 4 publicly-available cohorts
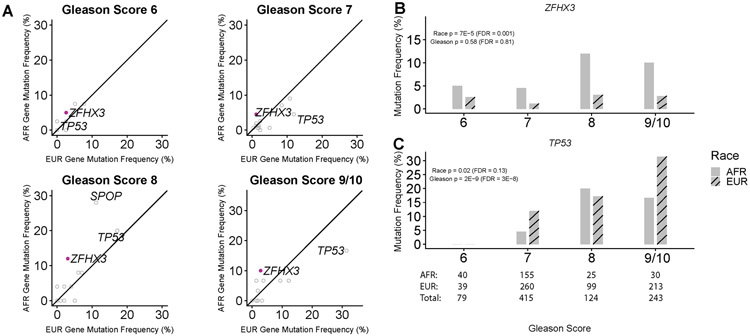
Four sequencing datasets had mutation calls available for analysis including WES data from TCGA, WES and targeted sequencing data from Huang et al(11), and targeted sequencing data from Abida et al(12)(MSK-IMPACT). 861 primary PCa from 250 AFR men (29.0%) and 611 EUR men (71.0%) were included in the analysis across all cohorts after excluding men without sufficient clinical annotation or from Asian background (Methods; Supplementary Tables 1-4). Gleason score was associated with race (p < 0.001; Chi-square test) with higher proportions of lower grade (Gleason 6 and 7) in AFR men. Age was also significantly lower in AFR men (p < 0.001. Wilcoxon rank-sum test). 15 genes were present in assays from all 4 cohorts and were mutated in at least 3 patients. Logistic regression (LR) was used to identify associations with race, Gleason score, age, and mutation rate (Methods; Figure 1A; Supplementary Table 5). The frequencies of somatic mutations in ZFHX3 were significantly higher in tumors from AFRs (6.0% vs. 2.1%; FDR = 0.001; Figure 1B; Supplementary Tables 5,6). The frequency of TP53 mutations was strongly associated with higher Gleason score. (FDR = 3.4 x 10−8; LR; Figure 1C; Supplementary Tables 5,6). While the overall mutation rate of TP53 was not associated with race, truncating mutations were significantly higher in tumors from AFR patients (FDR < 0.05; LR; Supplementary table 7). FOXA1 and SPOP mutations were associated with age (FDR < 0.05; LR).
Figure 1. Mutational frequencies associated with race and Gleason score across 4 prostate cancer cohorts.
(A) Gene mutation frequencies associated with race were identified with a logistic regression model containing race, Gleason score, age, and mutation rate. Solid purple points represent genes significantly associated with race (FDR < 0.05). (B) The frequency of ZFHX3 mutations was associated race. (C) The frequency of TP53 mutations was associated with Gleason score.
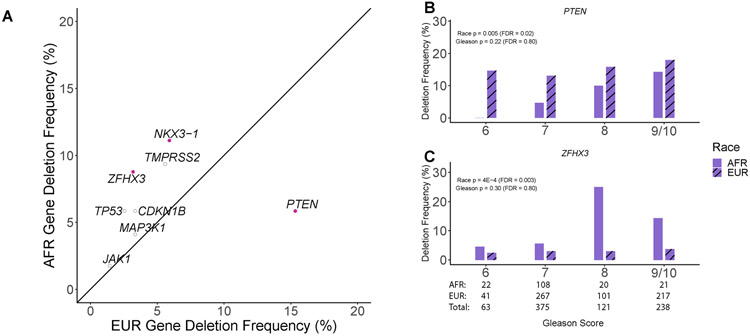
Genome-wide copy number ratios were available for TCGA and Huang et al WES cohorts. Using GISTIC2.0 recurrently altered copy number peaks were identified separately in AFR (n=157) and EUR men (n=392) from these two cohorts (Supplementary Tables 8-12). In AFR men, a novel focal deletion peak was found on chr1q23.1 centered on ETV3 (GISTIC q-value = 0.016; Supplementary Table 9). This peak was not detected by GISTIC2.0 in the much larger cohort of EUR men. The frequency of ETV3 deep deletions was marginally significantly higher in AFR men (6.3% vs. 2.3%; p = 0.021; LR; Supplementary Figure 1; Supplementary Table 13). Although genome-wide copy number ratios were not available in the Abida et al dataset for inclusion in the GISTIC2.0 analysis, copy number calls were accessible for individual genes. 8 target genes of recurrently deleted peaks and 4 target genes of recurrently amplified peaks were measured across Abida et al, Huang et al, and TCGA cohorts. Therefore, we used logistic regression to identify associations between the frequencies of deletions or amplification calls with race, age, and Gleason score. Deletions in ZFHX3 (8.8% vs. 3.2%) and NKX3-1 (11.1% vs. 5.9%) were significantly enriched in tumors from AFR men (FDR < 0.05; LR; Figure 2A; Supplementary Tables 14-15) while deletions in PTEN were significantly decreased in AFR men (FDR < 0.05; LR; 5.9% vs. 15.3%; Figure 2B,C). MYC amplifications were associated with higher Gleason score as previously described(14) (FDR < 0.05; LR; Supplementary Figure 2; Supplementary Table 16). A combined analysis of deletions and mutations further demonstrated associations between race and ZFHX3 or PTEN alterations (Supplementary Figure 3; Supplementary Table 17).
Figure 2. Copy number alterations associated with race and Gleason score across 4 prostate cancer cohorts.
(A) Gene deletion frequencies associated with race were identified with a logistic regression model containing race, Gleason score and age (AFR: 171, EUR: 626). Solid purple points represent genes significantly associated with race (FDR < 0.05). (B) The frequency of PTEN deletions was significantly lower in AFR men (FDR < 0.05). Differences between groups were most pronounced in Gleason 6 and 7 tumors. (C) The frequency of ZFHX3 deletions was significantly higher in AFR men (FDR < 0.05).
Analysis of the Foundation cohort
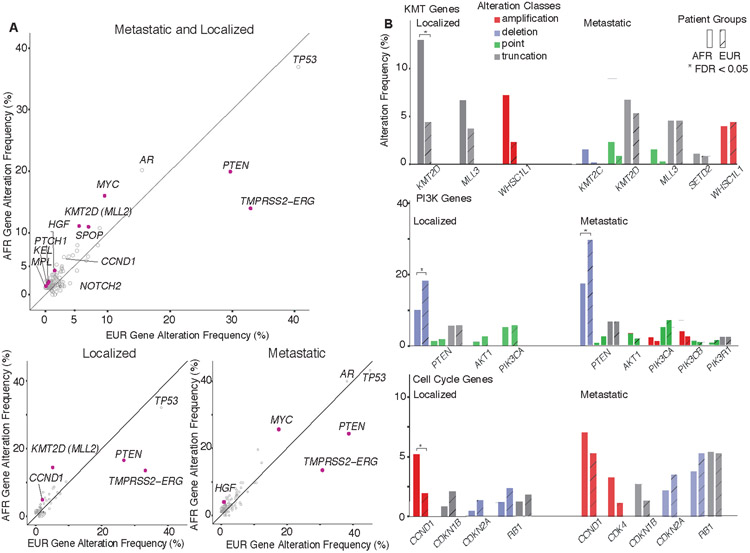
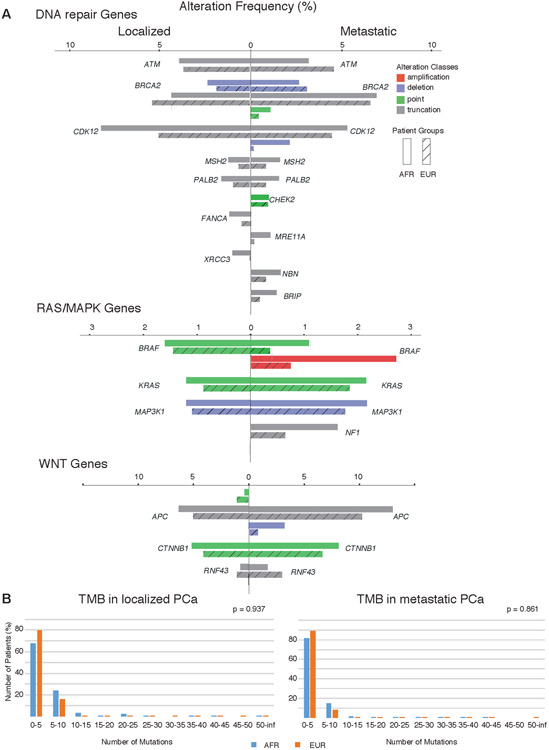
We next examined a large cohort of localized and metastatic PCa prostate cancer patients (n=3,454) sequenced with a comprehensive genomic profiling assay (Supplementary Tables 18-20). European (EUR) and African (AFR) ancestry was determined using principal components analysis (Methods). Fisher’s exact tests (FET) were used to compute the statistical significance of gene alteration frequencies followed by correction for multiple hypothesis testing. When examining all tumors, genomic alterations significantly higher in AFR men (n=436) included CCND1 amplification (6.0% vs. 3.1%), HGF amplification (3.4% vs. 1.4%), KMT2D truncation (10.1% vs. 4.7%), MYC amplification (15.8% vs. 9.4%), SPOP point mutation (10.8% vs. 6.9%), and KEL (2.1% vs. 0.5%), NOTCH2 (1.8% vs. 0.5%), PTCH1 (1.8% vs. 0.4%) overall alterations (FDR < 0.05; FET; Figure 3A). In contrast, PTEN deletions (13.1% vs. 22.2%) and TMPRSS2-ERG rearrangements (13.8% vs. 32.9%) were significantly lower in AFR men, consistent with previous studies (FDR < 0.001; FET; Figure 3B)(1,11). When examining localized PCa, KMT2D truncation (13% vs. 4.3%) and CCND1 amplification (5.2% vs. 1.9%) events were significantly higher in AFR men (FDR < 0.05; FET)(15), whereas no significant differences were detected in selected genes in DNA repair, RAS/MAPK or WNT pathways (Figure 4A). In the case of KMT2D, a significant difference in alteration frequency was detected only for truncation events but not point mutations.
Figure 3. Association of mutations with race and metastasis in the Foundation cohort.
(A) Comparison of alteration frequencies for each gene between AFR and EUR populations in combined samples (AFR: N = 436, EUR: N = 3,018, upper panel), localized PCa samples (AFR: N = 251, EUR: 1,940, lower left panel) and metastatic PCa samples (AFR: N = 185, EUR: 1,078, lower right panel). Alteration frequencies are overall frequencies of all classes. Solid black lines represent a 1:1 relationship between AFR and EUR patients. Open grey circles are genes with no significant differences (FDR >= 0.05) and solid purple circles are genes with significantly different alteration rates (FDR < 0.05). Gene names are labeled beside their corresponding markers in the scatter plots. (B) Frequency of major pathway alterations with significant differences between AFR (solid) and EUR (shadow) in localized or metastatic PCa samples, including KMT (lysine methyltransferase) genes, PI3K pathway genes and cell cycle pathway genes in which alterations were detected in the Foundation cohort. Gene alteration rates are shown as side-by-side bar charts consisting of a total of four color-coded alteration classes. Genes with significantly different alteration rates (FDR < 0.05) were highlighted with asterisks.
Figure 4. Frequency of major pathway alterations and tumor mutational burdens.
(A) Frequency of major pathway alterations with no significant differences between AFR (solid) and EUR (shadow) in localized or metastatic PCa samples, including DNA repair pathway genes, RAS/MAPK pathway genes and WNT pathway genes in which alterations were detected in the Foundation cohort. Gene alteration rates are shown as side-by-side bar charts consisting of a total of four color-coded alteration classes. If the alteration frequency for a gene was 0, it was omitted in the dataset. (B) Tumor mutational burdens (TMB) in both localized and metastatic PCa (localized: n_AFR = 124, n_EUR = 852; metastatic: n_AFR = 202, n_EUR = 1695). TMBs were computed as the percentage of patients within the cohort with respect to the number of mutations detected (0~100). No significant difference was found between AFR (blue) and EUR (orange) populations.
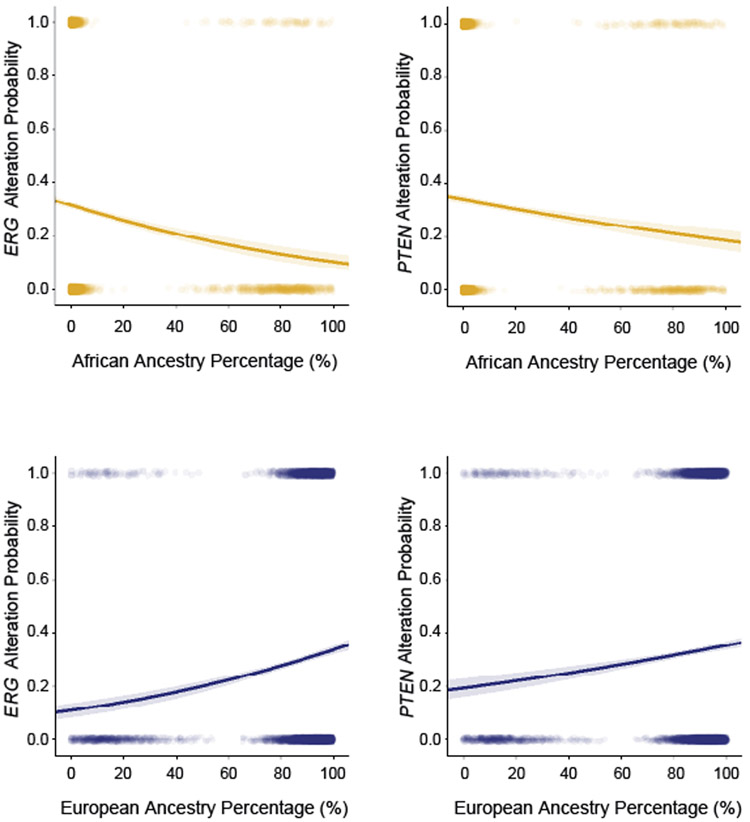
Among metastatic PCa, TMPRSS2-ERG rearrangement (AFR: 14.1% vs. EUR: 31.6%) and PTEN deletion (AFR: 24.3% vs. EUR: 37.5%) rates were significantly lower in AFR men (FDR<0.05; Fisher’s Exact Test) while MYC amplifications were more frequent (25.4% vs. 16.4%; Fisher’s Exact Test; FDR = 0.052). Among AFR men, RB1, MYC and AR were more frequently altered in metastatic compared to localized tumors (FDR<0.05; Fisher’s Exact Test; Supplementary Figure 4). Among EUR men, RB1, MYC, AR, TP53, PTEN, APC and CCND1 were more frequently altered in metastatic samples (FDR<0.05; Fisher’s Exact Test). Assessments of tumor mutation burden and MSI status showed no statistically significant difference between AFR and EUR prostate cancers (Figure 4B, Supplementary Table 22). Logistic regression analysis using continuous measures of AFR and EUR ancestry percentages showed that the frequency of TMPRSS2-ERG rearrangements and PTEN deletions were positively correlated to EUR ancestry percentages and negatively correlated to AFR ancestry percentages (p<0.001; LR; Figure 5).
Figure 5. Association of ERG and PTEN alteration frequencies to ancestry percentage.
Logistic regression was used to identify associations of ERG and PTEN alteration status with percentage of AFR or EUR ancestry within each patient (N = 5,624). Solid lines represent the logistic regression curves between alteration status and ancestry percentage. The semi-transparent band around the solid lines represent the 95% confidence interval of the logistic regression curves.
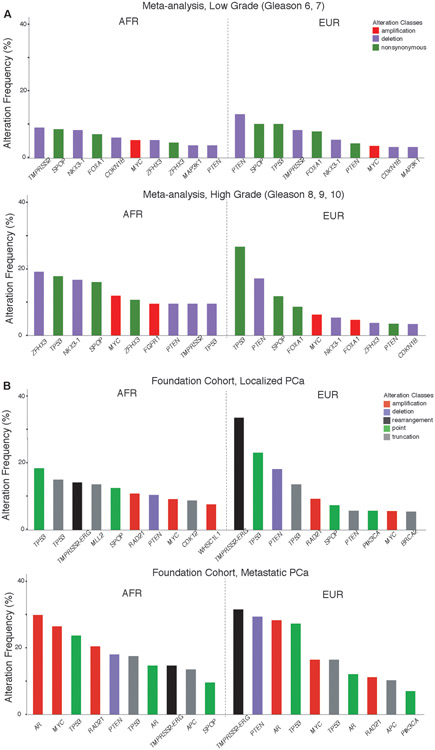
Examining common associations across both the meta-analysis and Foundation cohorts, a higher frequency of MYC amplifications was observed in tumors with more severe disease (higher Gleason grade or metastatic disease) and PTEN deletions were enriched in tumors from EUR patients (Figure 6). Alterations in the AR gene were observed at higher frequencies in metastatic tumors, likely reflecting resistance mechanisms to androgen-directed therapies of metastatic PCa. TP53 had the highest frequency of all non-synonymous mutations or point mutations in all cohorts except for the low-grade tumors in the meta-analysis cohort. The next most commonly mutated gene was SPOP, which was found among the most frequently altered genes in all cohorts except metastatic tumors from EUR patients. Overall, no significant differences were observed in currently clinically-actionable genes between AFR and EUR men across all cohorts.
Figure 6. Top ten most frequently altered genes in AFR and EUR men.
(A) The ten most frequent alterations are shown from AFR and EUR patients in the meta-analysis cohort for tumors with low-grade disease (top; Gleason scores 6, 7) and high-grade disease (bottom; Gleason scores 8, 9, and 10). Gene alteration classes considered include non-synonymous mutations, amplifications, and deletions. (B) The ten most frequent alterations in AFR and EUR patients in the Foundation Medicine cohort for tumors with localized PCa (top) and metastatic PCa (bottom). Gene alteration classes considered include amplifications, deletions, rearrangements, point (missense) mutations and truncations.
DISCUSSION
Given the paucity of African American men in sequencing cohorts, the goal of this study was to identify mutational events associated with race or ancestry in PCa. We first performed a meta-analysis across four different publicly available datasets and identified 2 putative prostate cancer tumor suppressors, ETV3 and ZFHX3, that were more frequently altered in AFR men. Loss of ZFHX3 has been shown to cause neoplastic lesions in the prostate in mice(16). ETV3, an ETS transcription factor, was recurrently deleted in AFR men and has not previously been implicated in PCa. We have previously shown similar to loss-of-function mutations in another ETS repressor, ERF, which were enriched in PCa from AFR men. Together these results suggest that while ERG rearrangements are less common in AFR men, additional mechanisms may still converge upon dysregulation of ETS factors(11,17). We also observed that mutations in TP53 were associated with an increase in Gleason score across all racial groups. Associations between TP53 mutations or TP53 expression and metastatic disease have been previously reported(18,19). These results suggest that if TP53 mutations are found in low-grade disease, they may potentially indicate a more aggressive clinical trajectory. PTEN has been reported to have a lower mutation frequency in African Americans in endometrial cancer patients(20). A major limitation of the meta-analysis is that some cohorts lacked matched tumors from EUR men, limiting our ability to control for cohort-specific effects that could arise due to differences in region, clinical setting, or sequencing assay. Another limitation was that the meta-analysis was limited to 15 genes since 2 of the 4 cohorts relied on smaller targeted sequencing panels for profiling. Studies examining larger numbers of tumors form AFR men profiled with whole-exome or whole-genome platforms may reveal additional genomic alterations associated with ancestry.
In our large cohort analysis of PCa profiled with the Foundation Medicine assay, we found higher rates of CCND1 amplifications and KMT2D truncations in African-ancestry tumors but fewer PTEN deletions and TMPRSS2-ERG rearrangements. Significantly more frequent CCND1 amplifications and KMT2D truncation events were identified within AFR localized prostate cancer patients. Higher expression of CCND1 has been implicated with perineural invasion in prostate cancer(21), an aggressive histological feature in prostate cancer. Truncating mutations in KMT2D have been reported in both localized and metastatic prostate cancer patients with unclear clinical significance(22). As a tumor suppressor, KMT2D alterations are more frequently detected in metastatic than primary tumors(23), which is consistent with the higher truncation and overall alteration frequencies in EUR metastatic tumors. MYC amplifications were more frequent in AFR metastatic samples. Germline susceptibility variants at 8q24 have been found more frequently in AFR men and may exert their effect via an impact on MYC expression (24). Our data raises the possibility that MYC amplifications may also contribute to high-risk disease in this population. The major limitation of this cohort was the unavailability of the clinical covariates such as age, tumor stage, and Gleason grade were not available and thus could not be included in statistical analyses.
Between the Foundation Medicine and the meta-analysis cohorts, the mutation frequencies of a subset of genes were tested in both assays including AR, BRAF, BRCA2, BRIP1, CDKN1B, CDK6, CTNNB1, FGFR1, FLCN, JAK1, KDM6A, MAP3K1, MCL1, MED12, PIK3CA, PTEN, SPOP, TMPRSS2, and TP53. Of these genes, PTEN deletion was lower in the AFR population in both analyses of localized PCa. All other genes were not significantly associated with race, suggesting an overall concordance between the two analyses. We note that MYC amplifications were found to be associated with Gleason score and not race in the meta-analysis while these amplifications were associated with race in the Foundation Medicine cohort. The lower alteration frequency observed in the meta-analysis cohorts may be partially explained by the focus on primary localized samples for the analysis. Alternatively, the lack of ability to control for clinical covariates in the Foundation Medicine cohort could produce an association if the underlying Gleason score was confounded with race.
Based on our analysis of the largest number of PCa from AFR men to our knowledge, a single gene alteration is unlikely to account for the observed prostate cancer disparities. Furthermore, no significant differences were seen in clinically-actionable DNA repair genes, MSI-high status, and tumor mutation burden, suggesting that current therapeutic strategies may be equally applied in both populations(25). While recent studies by Dess et al and Halabi et al may be interpreted to mean that biological differences driving disparities do not exist, we believe that it is important to understand that significant differences exist in tumor biology, genomics, and proteomics that underlie the molecular etiology of prostate cancer in African Americans. These differences occur in the context of structural social and economic determinants that can drive and amplify disparities.
Additional studies that profile large numbers of well-matched tumors from AFR and non-AFR men from the same clinical setting will be needed to confirm the novel associations reported in this study and to understand the clinical significance. The genomic differences seen in genes such as CCND1, ZFHX3, PTEN, and TMPRSS2-ERG suggest that different pathways of carcinogenesis may be active in AFR men, which could lead to further disparities if targeted therapies for some of these alterations become available. Understanding how outcomes are influenced by the genomic and biological features of prostate cancers in AFR men with the interaction of social and environmental factors remains an understudied area of cancer disparities research. Determining a comprehensive understanding of the genomic features of African American prostate cancers and how they relate to adverse features or contribute to poorer outcomes overall for AFR men could inform our strategies to improve precision medicine for these patients. These studies will remain important to understand when certain therapies may preferentially benefit African American patients, who remain underrepresented in clinical trials(26). Examining additional features such as the noncoding genome, epigenome and tumor microenvironment will be needed to fully understand the biological contribution to the observed disparities in AFR men.
Supplementary Material
Statement of Translational Relevance:
African American men have the highest incidence and mortality from prostate cancer. It is not known whether currently targetable genomic alterations in prostate cancer differ between men of European ancestry and men of African ancestry. Analyzing a large prostate cancer cohort, we identify certain differences and show that the frequency of alterations in AR, CDK12, DNA repair genes, tumor mutation burden, and MSI-high status are similar, suggesting that therapeutic approaches based on these features can be beneficial in both populations.
Acknowledgments
Financial Support:
The results here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/ as outlined in the TCGA publications guidelines http://cancergenome.nih.gov/publications/publicationguidelines. This work was funded by the Department of Defense W81XWH-14-1-0514 (F.W.H.) and W81XWH-17-PCRP-HD (F.W.H. and J.D.C.), the National Institutes of Health/National Cancer Institute P20 CA233255-01 (F.W.H. and J.D.C.) U19 CA214253 (F.W.H.), and the Prostate Cancer Foundation (F.W.H.).
Footnotes
Conflict of interest disclosure statement: Newberg and Frampton are employees at Foundation Medicine, Inc. No other potential conflict of interest was reported by the authors.
References
- 1.Khani F, Mosquera JM, Park K, Blattner M, O'Reilly C, MacDonald TY, et al. Evidence for molecular differences in prostate cancer between African American and Caucasian men. Clin Cancer Res 2014;20(18):4925–34 doi 10.1158/1078-0432.CCR-13-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman RM, Gilliland FD, Eley JW, Harlan LC, Stephenson RA, Stanford JL, et al. Racial and ethnic differences in advanced-stage prostate cancer: the Prostate Cancer Outcomes Study. J Natl Cancer Inst 2001;93(5):388–95 doi 10.1093/jnci/93.5.388. [DOI] [PubMed] [Google Scholar]
- 3.Mahal BA, Berman RA, Taplin ME, Huang FW. Prostate Cancer-Specific Mortality Across Gleason Scores in Black vs Nonblack Men. Jama 2018;320(23):2479–81 doi 10.1001/jama.2018.11716. [DOI] [PubMed] [Google Scholar]
- 4.Dess RT, Hartman HE, Mahal BA, Soni PD, Jackson WC, Cooperberg MR, et al. Association of Black Race With Prostate Cancer-Specific and Other-Cause Mortality. JAMA Oncol 2019;5(7):975–83 doi 10.1001/jamaoncol.2019.0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halabi S, Dutta S, Tangen CM, Rosenthal M, Petrylak DP, Thompson IM, et al. Overall Survival of Black and White Men With Metastatic Castration-Resistant Prostate Cancer Treated With Docetaxel. J Clin Oncol 2019;37(5):403–10 doi 10.1200/JCO.18.01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi Y, Li J, Zhang S, Wang M, Yang S, Li N, et al. Molecular Epidemiology of EGFR Mutations in Asian Patients with Advanced Non-Small-Cell Lung Cancer of Adenocarcinoma Histology - Mainland China Subset Analysis of the PIONEER study. PLoS One 2015;10(11):e0143515 doi 10.1371/journal.pone.0143515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Xu C, Lee HJ, Ren S, Zi X, Zhang Z, et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature 2020. doi 10.1038/s41586-020-2135-x. [DOI] [PubMed] [Google Scholar]
- 8.Spratt DE, Chan T, Waldron L, Speers C, Feng FY, Ogunwobi OO, et al. Racial/Ethnic Disparities in Genomic Sequencing. JAMA Oncol 2016;2(8):1070–4 doi 10.1001/jamaoncol.2016.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan J, Hu Z, Mahal BA, Zhao SD, Kensler KH, Pi J, et al. Integrated Analysis of Genetic Ancestry and Genomic Alterations across Cancers. Cancer Cell 2018;34(4):549–60.e9 doi 10.1016/j.ccell.2018.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst 2018;6(3):271–81.e7 doi 10.1016/j.cels.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang FW, Mosquera JM, Garofalo A, Oh C, Baco M, Amin-Mansour A, et al. Exome Sequencing of African-American Prostate Cancer Reveals Loss-of-Function. Cancer Discov 2017;7(9):973–83 doi 10.1158/2159-8290.CD-16-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abida W, Armenia J, Gopalan A, Brennan R, Walsh M, Barron D, et al. Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. JCO Precis Oncol 2017;2017 doi 10.1200/PO.17.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161(5):1215–28 doi 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fromont G, Godet J, Peyret A, Irani J, Celhay O, Rozet F, et al. 8q24 amplification is associated with Myc expression and prostate cancer progression and is an independent predictor of recurrence after radical prostatectomy. Hum Pathol 2013;44(8):1617–23 doi 10.1016/j.humpath.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol 2015;16(1):25–35 doi 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 16.Sun X, Fu X, Li J, Xing C, Frierson HF, Wu H, et al. Deletion of atbf1/zfhx3 in mouse prostate causes neoplastic lesions, likely by attenuation of membrane and secretory proteins and multiple signaling pathways. Neoplasia 2014;16(5):377–89 doi 10.1016/j.neo.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bose R, Karthaus WR, Armenia J, Abida W, Iaquinta PJ, Zhang Z, et al. ERF mutations reveal a balance of ETS factors controlling prostate oncogenesis. Nature 2017;546(7660):671–5 doi 10.1038/nature22820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powell E, Piwnica-Worms D, Piwnica-Worms H. Contribution of p53 to metastasis. Cancer Discov 2014;4(4):405–14 doi 10.1158/2159-8290.Cd-13-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grignon DJ, Caplan R, Sarkar FH, Lawton CA, Hammond EH, Pilepich MV, et al. p53 status and prognosis of locally advanced prostatic adenocarcinoma: a study based on RTOG 8610. J Natl Cancer Inst 1997;89(2):158–65 doi 10.1093/jnci/89.2.158. [DOI] [PubMed] [Google Scholar]
- 20.Althubiti MA. Mutation Frequencies in Endometrial Cancer Patients of Different Ethnicities and Tumor Grades: An Analytical Study. Saudi J Med Med Sci 2019;7(1):16–21 doi 10.4103/sjmms.sjmms_154_18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura Y, Felizola SJ, Kurotaki Y, Fujishima F, McNamara KM, Suzuki T, et al. Cyclin D1 (CCND1) expression is involved in estrogen receptor beta (ERbeta) in human prostate cancer. Prostate 2013;73(6):590–5 doi 10.1002/pros.22599. [DOI] [PubMed] [Google Scholar]
- 22.Network CGAR. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015;163(4):1011–25 doi 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Testa U, Castelli G, Pelosi E. Cellular and Molecular Mechanisms Underlying Prostate Cancer Development: Therapeutic Implications. Medicines (Basel) 2019;6(3) doi 10.3390/medicines6030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proceedings of the National Academy of Sciences of the United States of America 2006;103(38):14068–73 doi 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med 2015;373(18):1697–708 doi 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sartor O, Armstrong AJ, Ahaghotu C, McLeod DG, Cooperberg MR, Penson DF, et al. Survival of African-American and Caucasian men after sipuleucel-T immunotherapy: outcomes from the PROCEED registry. Prostate cancer and prostatic diseases 2020. doi 10.1038/s41391-020-0213-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.