Abstract
Influenza is a significant cause of morbidity and mortality worldwide. Here we show changes in the abundance and activation states of more than 50 immune cell subsets in 35 individuals over 11 time points during human A/California/2009 (H1N1) virus challenge monitored using mass cytometry along with other clinical assessments. Peak change in monocyte, B cell, and T cell subset frequencies coincided with peak virus shedding, followed by marked activation of T and NK cells. Results led to the identification of CD38 as a critical regulator of plasmacytoid dendritic cell function in response to influenza virus. Machine learning using study-derived clinical parameters and single-cell data effectively classified and predicted susceptibility to infection. The coordinated immune cell dynamics defined in this study provide a framework for identifying novel correlates of protection in the evaluation of future influenza therapeutics.
Keywords: Immunology, Infectious disease
Keywords: Adaptive immunity, Influenza, Innate immunity
Introduction
Influenza virus infection is a global health problem. In the United States in every influenza season since 2010, the US Centers for Disease Control and Prevention estimates that 9 million to 49 million symptomatic infections occurred. These influenza infections led to 4 million to 23 million medical visits, 140,000 to 960,000 hospitalizations, and 12,000 to 79,000 deaths annually (1). Globally, the WHO estimates that seasonal influenza causes 3 million to 5 million cases of severe illness and 290,000 to 650,000 annual deaths (2). In addition to the burden of seasonal influenza epidemics, the last 4 influenza A pandemics, occurring in 1918 (H1N1), 1957 (H2N2), 1968 (H3N2), and 2009 (H1N1), resulted in between 22 million and 58 million fatalities (3).
Countermeasures for seasonal and pandemic influenza outbreaks consist of vaccination, antiviral drugs, physical barriers, hygiene, and physical distancing. Vaccination is an effective measure to control seasonal influenza and mitigate the severity of a pandemic. Unfortunately, currently available vaccines generate strain-specific immunity with limited protection against subtypes not included in the vaccine formulation (3). Therefore, major efforts are under way to develop a universal vaccine that induces broadly neutralizing humoral immunity. One strategy is to direct vaccine immune responses toward the stalk region of influenza hemagglutinin (HA), which is less variable than the globular head region (3). In addition to vaccines, 3 classes of influenza antivirals are approved in the United States and Europe. M2 ion channel blockers (amantadine, rimantadine) have become largely ineffective as a result of resistance (4). Neuraminidase inhibitors (oseltamivir, zanamivir, peramivir) and the cap-dependent endonuclease inhibitor baloxavir marboxil each reduce symptoms by only about 1 day (5–7). Thus, for both seasonal influenza and pandemic preparedness, there is a strong need for more effective influenza therapeutics and for models to study disease and therapeutic responses.
Serum antibodies recognizing HA have long been associated with immunity to influenza infection (8). Newer data suggest that anti-neuraminidase antibodies can also confer immunity (9, 10). Helper and cytotoxic T cell functions both participate in effective influenza immune responses. Preexisting virus-specific CD4+ cells correlated with protection in an experimental setting (11). The presence of CD8+ T cell responses to conserved viral polymerase, matrix protein, and nucleoprotein epitopes may mitigate the severity of illness during ongoing infections (12, 13). Moreover, innate immune cells such as monocytes, dendritic cells (DCs), and plasmacytoid DCs (pDCs) are important for pathogen sensing, triggering cytokine production and the initiation of effective antiviral immune responses (14–17). Whole-blood transcriptomic studies (18–20) offer hints that additional cellular features related to protection and severity of infection remain to be elucidated. However, still lacking is detailed information on the dynamics of many immune cell subsets during human influenza infection, critical knowledge for the development of more effective therapeutics.
Controlled human infection trials, wherein healthy volunteers are challenged with influenza virus, are important settings for the study of disease pathogenesis and rapid testing of novel antivirals and vaccines (21–23). Such studies also provide an extraordinary opportunity to thoroughly characterize the natural history of influenza infection in a highly controlled setting. Here, we used mass cytometry to profile peripheral blood immune cell subsets from 35 healthy individuals challenged with A/California/2009 (H1N1) virus. Cell subsets identified by mass cytometry were highly correlated with complete blood counts (CBCs), and there were dramatic and regularized changes across volunteer subgroups in lymphoid and myeloid subsets over the course of the disease.
Unexpectedly, unsupervised machine learning identified cell clusters closely related to pDCs, characterized by the expression of CD38, that were dramatically increased during infection. In vitro blocking experiments revealed the importance of CD38 for proinflammatory cytokine production in pDCs in response to virus. A multivariate approach incorporating plasma cytokine levels, innate and adaptive immune cell subset abundance, and clinical parameters was used to predict the onset of infection as well as susceptibility to A/California/2009 (H1N1) virus. Together, this study provides an in-depth analysis of the integrated cellular responses that orchestrate infection-acquired immunity upon influenza challenge and will help guide the development of more effective therapeutics.
Results
H1N1 influenza virus challenge in healthy human volunteers.
The objective of this study was to investigate host immune responses in a clinical trial that determined the safety and reactogenicity of an A/California/2009 (H1N1) influenza virus challenge strain (identifier: NCT04106817). Eligible healthy volunteers were inoculated using an atomizer with a single intranasal dose of virus and subjected to extensive clinical assessment while in confinement at a dedicated challenge facility. Before and after inoculation, volunteers were monitored for safety and signs and symptoms of infection, and multiple analyses of biological specimens were performed that included HA antibody titer, virus titer, serology of plasma, CBC, and mass cytometry (Figure 1A).
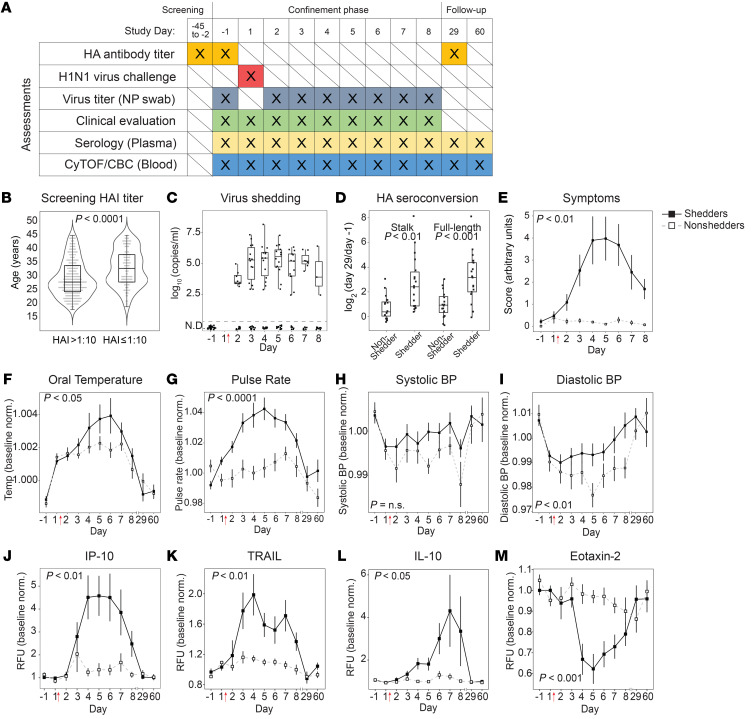
Figure 1. Influenza challenge model in human volunteers.
(A) Schedule of assessments performed throughout the screening, confinement, and follow-up phases of the study. Study days –1 and 1 are consecutive calendar days. Intranasal virus inoculation was performed on study day 1 immediately after blood draw. NP, nasopharyngeal. (B) Of the screened population (n = 437), the distributions of ages for those with HAI titers >1:10 (eligible for inclusion) and with HAI titers ≤1:10 (ineligible). (C) Viral titers as measured by qRT-PCR in nasopharyngeal swabs (n = 19 virus shedders). Viral titers were below the limit of detection for all participants on study day –1, and for 16 individuals throughout the study (nonshedders). (D) Stalk-specific and full-length-HA antibody seroconversion measured by ELISA. Values shown are day 29 relative to day –1. (E) Mean daily symptom score as determined by participant-reported symptom scorecard. (F–I) Baseline-normalized values for maximum daily oral temperature (F), mean pulse rate (G), systolic blood pressure (H), and diastolic blood pressure (I). Vital signs were measured up to 4 times daily. (J–M) Baseline-normalized plasma cytokine and chemokine levels, in relative fluorescence units (RFU), measured by Luminex assay for IP-10 (J), TRAIL (K), IL-10 (L), and eotaxin-2 (M). In E–M, data for virus shedders are indicated with filled squares and solid lines; data for nonshedders are indicated with open squares and dashed lines (mean ± SEM). In F–M, values plotted are normalized to baseline (average of day –1, 1). (C–M) n = 35 volunteers. Welch’s t test (B and D); Bonferroni’s adjusted P value of the time-shedding interaction term (E–M). Red arrows indicate the time point of virus inoculation throughout all figures.
To identify study participants, 427 healthy volunteers between 18 and 44 years of age were screened (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI137265DS1). Individuals with a hemagglutination inhibition (HAI) titer greater than 1:10 were ineligible owing to greater potential for preexisting immunity to the challenge strain (24). In the sampled population, the median age of individuals with HAI titers of ≤1:10 was significantly higher than the median age of individuals with HAI titer >1:10 (Figure 1B and Supplemental Table 1), similar to correlations of age with baseline and post-vaccination HAI titers reported elsewhere (25, 26). Thirty-five volunteers were ultimately enrolled and separated into 3 cohorts receiving escalating doses as indicated in Supplemental Table 2. Overall, 19 of 35 volunteers (54%) developed virus shedding and reported at least 1 symptom consistent with infection. The relatively narrow difference in dose between cohorts did not significantly affect the proportion of volunteers who developed virus shedding, and when subjects were grouped into either virus shedders or nonshedders, clinical exam scores, volunteer-reported symptom scores, and seroconversions were not significantly different between cohorts (Supplemental Tables 2 and 3). Furthermore, when all study data generated for all volunteers were plotted together, shedding status was the major driver of variance (Supplemental Figure 2). Therefore, for the analysis described here, individuals were grouped into virus shedders and nonshedders irrespective of their initial inoculation dose.
Virus shedding was quantified by quantitative reverse transcriptase PCR (qRT-PCR) in nasopharyngeal swabs collected on the day before challenge (study day –1) and daily starting 24 hours after inoculation. Viral titers were below the limit of detection for all participants on study day –1, and for 16 individuals (nonshedders) no shedding was detected throughout the study (Figure 1C). Serum antibodies recognizing either full-length HA or the HA stalk were quantified by 2 separate ELISAs. On day –1, preexisting full-length HA antibody titers, but not preexisting HA stalk antibody titers, were higher among volunteers who did not develop virus shedding (Supplemental Figure 3, A and B). Seroconversion to both full-length HA and HA stalk occurred to a greater extent in shedders compared with nonshedders on day 29 (Figure 1D). Mean daily symptom scores, as reported by volunteers and physician exams, were elevated in shedders compared with nonshedders (Figure 1E and Supplemental Figure 3C). Baseline-normalized oral temperature, pulse rate, and blood pressure were significantly higher in shedders with peak deviations approximately coinciding with days when viral titers and symptom scores were at their maximums (Figure 1, F–I). Sixty-five cytokines and chemokines were measured in plasma, 5 of which changed significantly during infection. In virus shedders, levels of cytokines previously associated with influenza (27, 28), including plasma IP-10 (also known as CXCL10), soluble TRAIL, IL-10, and IL-18, increased. In contrast, levels of eotaxin-2, a factor associated with eosinophil recruitment to the lung and eosinophilic bronchial inflammation (29, 30), decreased (Figure 1, J–M, and Supplemental Figure 4).
These data indicate that the influenza challenge model described here induces measurable virus infection in healthy volunteers including virus shedding, symptoms, vital signs, and serology changes consistent with clinical disease. This model is suitable to investigate the dynamics of cellular immune responses to influenza infection by comparing individuals who do and do not shed virus after challenge.
Mass cytometry recapitulates clinical hematology during influenza infection.
Baseline-normalized values of CBCs evaluated daily showed significant differences between shedders and nonshedders for lymphocyte, monocyte, and neutrophil counts (Supplemental Figure 3, D–K). Consistent with previous reports of experimental and community-acquired influenza A infection (22, 31, 32), lymphocyte counts in virus shedders decreased from baseline, reaching their nadir at day 4, and monocyte counts increased, peaking at day 5. Neutrophil counts first increased and then decreased relative to baseline.
A custom panel of 42 antibodies (Supplemental Tables 4 and 5) recognizing cell surface and intracellular targets (33) was used to profile immune cell subsets in the blood across 11 time points by mass cytometry. We first performed a comparison of mass cytometry data with CBC data across 381 paired samples. Strong correlations were observed for lymphocyte, monocyte, and neutrophil quantifications between the 2 techniques (R = 0.92–0.96, P < 0.001) (Supplemental Figure 3, L–N). Therefore, mass cytometry performed as an ideal tool for in-depth exploration of the differences in cellular immune response dynamics between virus shedders and nonshedders.
Mass cytometry characterizes innate and adaptive immune responses during H1N1 infection.
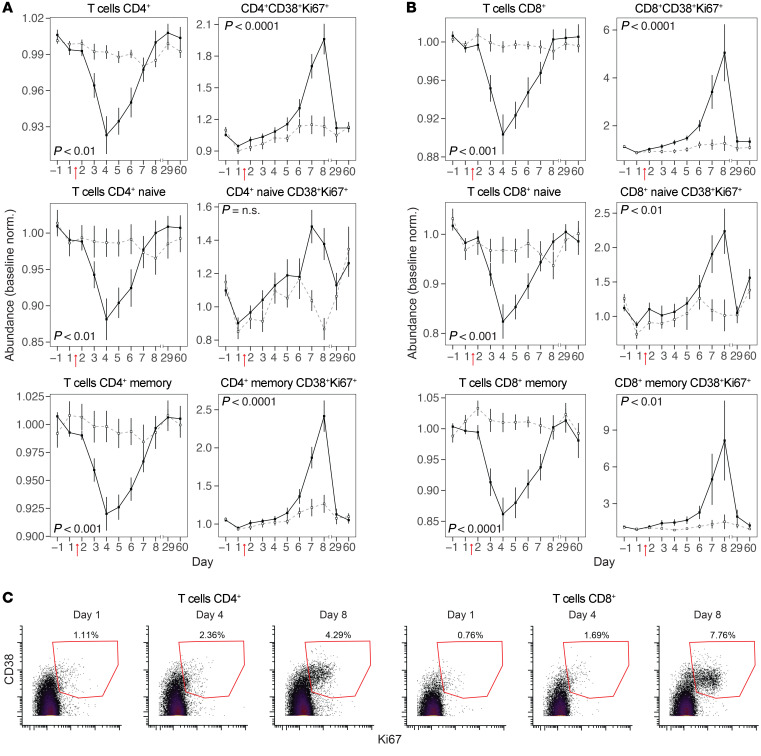
Changes in over 50 innate and adaptive immune cell populations following virus challenge were identified simultaneously in each peripheral blood sample using supervised gating (Figure 2 and Supplemental Figure 5). Relative abundances of total CD4+ and total CD8+ T cells sharply declined following infection, reaching a nadir on day 4 (Figure 3, A and B). Similar changes were observed for naive (CD45RA+) and memory (CD45RA–) subsets. In contrast, for both CD4+ and CD8+ subsets, activated proliferating T cells (CD38+Ki67+) increased in relative abundance to a maximum on day 8 (Figure 3, A–C). Among these activated proliferating CD4+ and CD8+ T cells, memory subsets had considerably larger increases in abundance by day 8 compared with their naive counterparts (Figure 3, A and B).
Figure 2. Simplified gating strategy used to define major immune cell subsets.
Fixed whole blood was stained with a 42-marker metal-conjugated antibody panel and analyzed by mass cytometry. Simplified gating strategy used to define major immune cell subsets is shown for a single representative sample (for complete gating strategy see Supplemental Figure 5).
Figure 3. Kinetics of T cell subsets during H1N1 infection.
(A) Relative abundances of total (left) and activated (CD38+Ki67+, right) CD4+ T cells, naive CD4+ T cells (CD45RA+), and memory CD4+ T cells (CD45RA–). (B) Relative abundances of total (left) and activated (CD38+Ki67+, right) CD8+ T cells, naive CD8+ T cells (CD45RA+CD27+), and memory CD8+ T cells (CD45RA–CD27+). (C) Representative biaxial plots showing the extent of CD38 and Ki67 expression on CD4+ and CD8+ T cells at days 1, 4, and 8 for a representative virus shedder. In B and C, values plotted are relative abundance (percentage of CD66– cells normalized to baseline [average of day –1, 1]) (mean ± SEM); filled squares and solid lines indicate virus shedders, and open squares and dashed lines indicate virus nonshedders. (A and B) n = 35 volunteers. Bonferroni’s adjusted P value of the time-shedding interaction term. For plots of additional gated cell populations, see Supplemental Figure 6.
B cell subsets were defined as total B cells, naive B cells, and memory B cells. Memory B cells were further divided into subsets with and without class switching (CSM and NCSM). Like T cells, most B cell subsets declined after infection, reaching nadirs on day 5 or 6 (Figure 4A). In contrast, the relative abundance of plasmablasts increased steadily from day 5 onward (Figure 4A). Interestingly, the abundance of a previously undescribed subpopulation of plasmablasts that are positive for phosphorylated STAT5 (p-STAT5) rose sharply a week after infection, peaking on study day 8 (Figure 4B).
Figure 4. Kinetics of B cell subsets during H1N1 infection.
(A) Relative abundance of total B cells, non–class-switched memory (NCSM) B cells (CD27+IgM+), naive B cells (CD27–IgM+), and plasmablasts (CD27+IgM–CD38+). (B) Relative abundance of p-STAT5+ plasmablasts (top) and representative histogram of p-STAT5 expression on plasmablasts at days 1 and 8 for a representative virus shedder (bottom). In A and B, values plotted are relative abundance (percentage of CD66– cells normalized to baseline [average of day –1, 1]) (mean ± SEM); filled squares and solid lines indicate virus shedders, and open squares and dashed lines indicate virus nonshedders. (A and B) n = 35 volunteers. Bonferroni’s adjusted P value of the time-shedding interaction term. For plots of additional gated cell populations, see Supplemental Figure 6.
The innate immune response consisting of monocyte, NK cell, DC, and granulocyte subsets was next detailed. Unlike B and T lymphocytes, total NK cell abundance in circulation following infection did not substantially decline (Figure 5A). Notably, activated proliferating NK cells (CD38+Ki67+) steadily increased in relative abundance after infection until at least day 8 for both CD56loCD16+ and CD56+CD16– NK cell subsets (Figure 5, A and E). For DC subsets, we observed differing degrees of reduction in abundance as a function of time after challenge (Figure 5B). Conversely, all monocyte subsets increased strongly in abundance during infection. Classical monocytes (cMCs), intermediate monocytes (intMCs), and nonclassical monocytes (ncMCs) reached peak levels on days 4, 5, and 6, respectively (Figure 5, C and F). Among monocyte subsets, the highest-magnitude increase in abundance was observed for intMCs. Neutrophils (defined as CD66+ cells) had a biphasic response, with abundance increasing soon after challenge and declining late during the study (Figure 5D) in agreement with the CBC measurements (Supplemental Figure 3J). Last, basophils (defined as CD123+HLA-DR– cells) sharply declined in relative abundance until at least day 8 in virus shedders, but not in nonshedders (Figure 5D).
Figure 5. Kinetics of innate immune cell subsets during H1N1 infection.
(A) Relative abundances of total (first and third panels) and activated (CD38+Ki67+, second and fourth panels) populations of CD56+CD16– NK cells and CD56loCD16+ NK cells. (B) Relative abundances of pDCs, CD1c+ myeloid DCs (mDCs), and BDCA3+ mDCs. (C) Relative abundances of classical monocytes (cMCs; CD14+CD16–), intermediate monocytes (intMCs; CD14+CD16+), and nonclassical monocytes (ncMCs; CD14–CD16+). (D) Relative abundances of CD66+ cells and CD123+HLA-DR– cells. (E and F) Biaxial plots of CD38 and Ki67 expression on CD56loCD16+ NK cells (E) and CD14 and CD16 expression on monocytes (F) on days 1–8 and 29. Plots for 1 representative virus shedder are shown. In B–E, values plotted are relative abundance (percentage of CD66– cells normalized to baseline [average of day –1, 1]) (mean ± SEM); data for virus shedders are indicated by filled squares and solid lines, and data for nonshedders are indicated with open squares and dashed lines. (A–F) n = 35 volunteers. Bonferroni’s adjusted P value of the time-shedding interaction term. For plots of additional gated cell populations, see Supplemental Figure 6.
Thus, data from simultaneous examination of abundance and activation status of innate and adaptive immune cell subsets by mass cytometry not only agreed with clinical blood analysis but also provided more granular detail on the coordinated dynamics of the immune system during influenza A infection than has previously been reported.
Unsupervised clustering identifies cell dynamics during influenza infection.
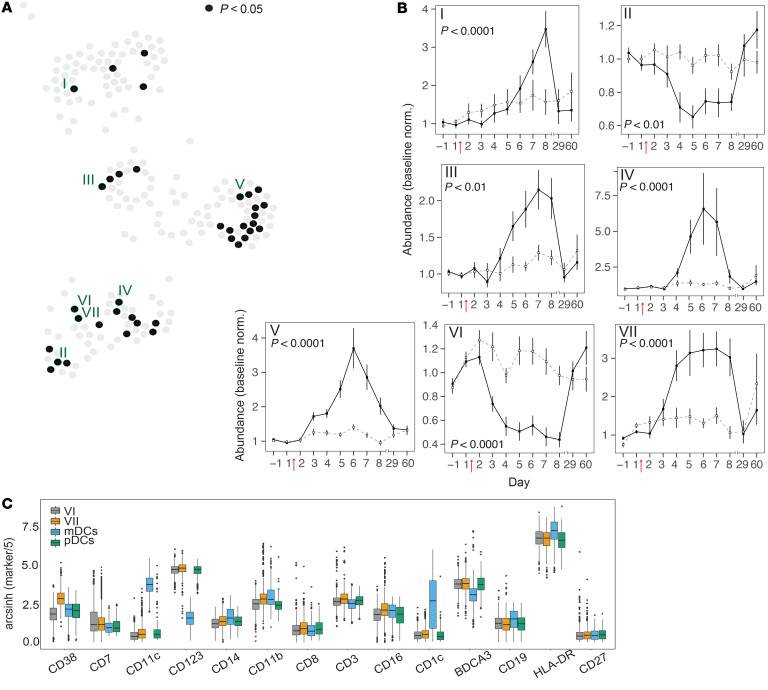
To highlight changes in immune populations that might otherwise have been missed by manual gating, we performed Scaffold analysis (34). Scaffold was chosen because it combines unsupervised clustering of cells (CLARA algorithm [ref. 35]) with ready identification of clusters through comparison with known cellular landmarks and is suitable for combined analysis of a large number of samples. Mass cytometry data from all 381 samples (35 volunteers, 11 time points) were pooled and then grouped based on the similarity of surface marker expression into 200 clusters. Landmark nodes defining major immune cell populations (CD4+ and CD8+ T cells, B cells, NK cells, myeloid and plasmacytoid DCs, cMCs, ncMCs, basophils, and CD66+ cells) were derived by manual gating of a representative sample. The 200 clusters were arranged in 2-dimensional space in relation to the landmarks, with distances proportional to the degree of surface marker similarity to other clusters and landmark nodes (Figure 6). As with the manually gated data, relative abundance kinetics for cell clusters were determined (Supplemental Table 6). To create a single visual representation of the entire data set, the infection-associated changes in the relative abundance of each cluster on study days 2–8 were converted into a color scale and displayed sequentially as wedges of nodes in clockwise orientation (Figure 6). This Scaffold map accurately reflects the global dynamics seen in CBCs and manually gated data, namely decreases from preinfection levels in relative abundances of T and B cell subsets and increases in monocyte subsets (Figure 6). Importantly, this map also allows direct visualization of specific clusters that do not follow bulk trends. Among 37 clusters that differed significantly, 7 clusters of interest with the lowest P values from each map region were chosen for further investigation (Figure 7A and Supplemental Table 6).
Figure 6. Time-based Scaffold analysis of volunteer H1N1 challenge.
Pooled cells from all volunteers and all time points (381 mass cytometry samples) were binned into 1 of 200 nonoverlapping clusters based on the similarity of their expression across all surface markers. Clusters are displayed as nodes of a force-directed Scaffold graph with the length of edges inversely proportional to the similarity of clusters to each other and to landmark nodes defined by manual gating (Supplemental Figure 5). The colored wedges in cluster nodes indicate the degree of increase (red) or decrease (blue) in the mean relative abundance of a given cell cluster for virus shedders on study days 2–8. n = 35 volunteers.
Figure 7. Kinetics of Scaffold cell clusters during H1N1 infection.
(A) Nodes colored black on the Scaffold graph (see Figure 6) indicate clusters with significant differential abundance between shedders and nonshedders (P < 0.05, Bonferroni’s adjusted P value of the time-shedding interaction term). The locations of the 7 Scaffold clusters selected for further analyses are indicated by Roman numerals. (B) Line plots showing abundances (percentage of total clustered cells) (mean ± SEM) normalized to baseline (average of day –1, 1) in Scaffold clusters indicated in A. Data for virus shedders are indicated with filled squares and solid lines, and data for nonshedders are indicated with open squares and dashed lines. (C) Box plots showing expression levels of indicated cell surface markers in Scaffold clusters VI and VII relative to manually gated myeloid and plasmacytoid DC landmarks. Note differential expression of CD38. (A–C) n = 35 volunteers. Bonferroni’s adjusted P value of the time-shedding interaction term. For additional box plots showing all markers for Scaffold clusters I–VII, see Supplemental Figure 7.
The identity of cell populations within each cluster of interest was determined by examination of median marker expression (Supplemental Figure 7). Clusters I, II, III, and IV had marker expression and dynamics like those seen in manual gates for activated memory CD4+ T cells, non–class-switched memory B cells, activated NK cells, and ncMCs, respectively (Figure 7B). Although the mass cytometry panel used in this study was not designed to deeply interrogate granulocytes (as identified here by CD66 expression), 15 of 37 clusters with significant differences between shedders and nonshedders were located near the CD66+ landmark. Of the 200 clusters, cluster V had the largest significant difference between shedders and nonshedders, a differential that peaked on day 6 (Figure 7B). Cells in this cluster express high levels of CD66 and CD16 and low levels of CD11b and CD11c, possibly reflecting the dynamics of a large population of immature neutrophils produced from emergency granulopoiesis (refs. 36, 37, and Supplemental Figure 7).
Clusters VI and VII also differed considerably between shedders and nonshedders. These clusters are located next to each other on the Scaffold map, indicating that they had similar surface marker expression yet had opposite cell abundance kinetics following infection (Figure 7B). This suggests that cluster VI, which drops in abundance during infection, might represent a precursor cell population that gives rise to cluster VII. In terms of marker expression, both clusters aligned closely with the pDC landmark but differed substantially in their expression of CD38 (Figure 7C). The abundance of cells in cluster VII, which has higher CD38 expression than cluster VI, increased sharply during infection, suggesting that elevated CD38 may play a role in pDCs during influenza infection, warranting further investigation.
CD38 is a critical regulator of pDC function in response to influenza virus.
CD38 is a multifunctional protein with both ectoenzymatic activity and adhesion properties and is associated with immune cell activation (38). Based on the Scaffold analysis above, we examined CD38 expression on manually gated pDCs. In virus shedders but not nonshedders, median expression of CD38 on pDCs rose sharply by day 3 after inoculation and remained elevated through at least study day 8 (Figure 8, A and B), confirming findings from unsupervised clustering. CD38 levels were also upregulated in other cell populations (Supplemental Figure 8), consistent with prior reports (38–40).
Figure 8. Analysis of the role of CD38 in pDC activation.
(A) Median CD38 values on manually gated pDCs analyzed by mass cytometry. Baseline-normalized (average of day –1,1) values (mean ± SEM) are plotted for virus shedders (filled squares and solid lines) and nonshedders (open squares and dashed lines). (B) Histograms of CD38 expression on pDCs at days 1, 3, 5, and 8 for a representative virus shedder. (C and D) PBMCs from healthy donors were cultured for 24 hours in the presence or absence of the indicated TLR agonist and analyzed by flow cytometry. (C) Representative histograms for CD38 expression on pDCs (see pDC gating strategy in Supplemental Figure 9). (D) CD38 MFI values from experiments with cells pooled from 4–11 donors in 2–7 independent experiments. (E–K) PBMCs were cultured for 24 hours in the presence (H1N1, blue) or absence (No stim., orange) of heat-inactivated H1N1 virus and, where indicated, in the presence of anti-CD38 blocking antibody or isotype control (Isotype). Flow cytometry data from PBMCs from 3 donors in 2 independent experiments are shown. (E) Representative histograms of CD38 expression on pDCs. (F) Quantification of CD38 MFI on pDCs. (G) Representative biaxial plot showing frequency of TNF-α+ pDCs. (H) Percentage of TNF-α+ pDCs. (I) Representative biaxial plots showing frequency of IFN-α+ pDCs. (J) Percentage of IFN-α+ pDCs. (K) Soluble IFN-α measured in supernatants of PBMC cultures. Bar plots in D, F, H, J, and K show mean ± SEM. Welch’s t test (D, F, and H); Wilcoxon’s signed-rank test (J and K).
pDCs are important early responders to viral pathogens; these cells initiate and orchestrate innate and adaptive immune responses by sensing foreign nucleic acids and producing type I interferons (41). The results raise the question of whether CD38 is involved in the response of pDCs to foreign nucleic acids during influenza exposure. Therefore, we first measured CD38 levels on pDCs in healthy donor PBMCs in response to agonists to Toll-like receptor 7 (TLR7), TLR8, and TLR9 (for gating and antibody panel, see Supplemental Figure 9 and Supplemental Table 7). Levels of CD38 were significantly higher in response to TLR agonists (Figure 8, C and D). Next, we examined CD38 regulation in response to the influenza challenge strain used in this study by incubating PBMCs in the presence or absence of the heat-inactivated A/California/2009 (H1N1) virus. As with the effect of TLR agonists, stimulation with virus resulted in a considerable CD38 increase on pDCs (Figure 8, E and F). Blocking CD38 using an antagonistic mAb (clone AT-1) (42) significantly inhibited the H1N1-induced production of intracellular TNF-α and IFN-α in pDCs and secretion of IFN-α into culture supernatants, indicating that CD38 is a key determinant for pDC functional activation to influenza (Figure 8, G–K). Given that CD38 expression is upregulated on pDCs in response to influenza infection in vivo, this process may trigger the production of proinflammatory cytokines that drive symptomology and host immune responses.
Predictive modeling of H1N1 virus shedding by machine learning.
In line with other influenza challenge studies (21, 22), 16 of 35 volunteers (46%) did not shed virus after challenge (Supplemental Table 2). This provided an opportunity to investigate factors other than HAI or HA antibodies in the blood that are relevant to protection. We therefore used our data set to computationally define an immune correlate of protection that could predict whether an individual would go on to become a viral shedder versus a nonshedder.
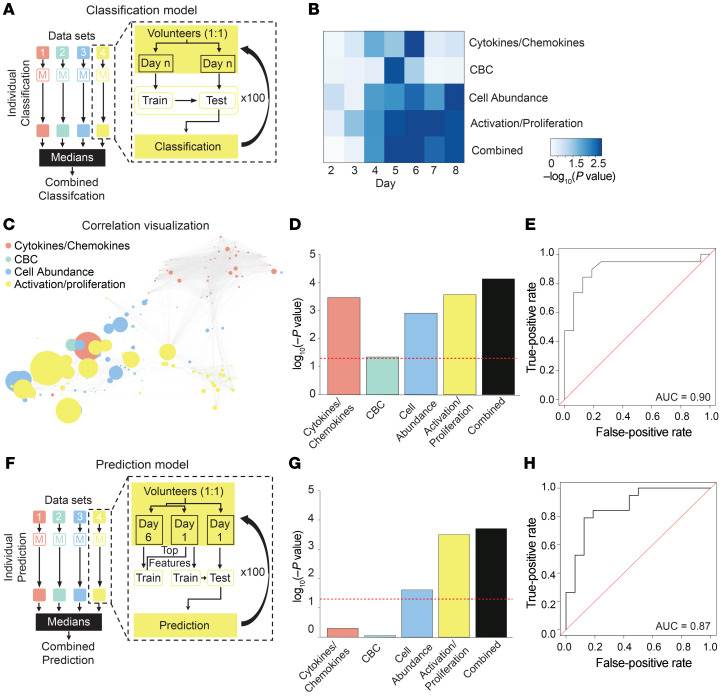
We first developed a bootstrapping-based random forest (43) classifier to diagnose viral shedding after challenge. We used this approach rather than a more traditional cross-validation to reduce variance in the reported results given the relatively small sample size (44–47). To test the importance of different types of data collected in this study (excluding symptomology or viral titer) for accurate diagnosis of infection, 4 individual random forest models were generated using 4 data sets: cytokine/chemokine data, CBC counts, the relative abundance of immune cell populations, and cellular activation and proliferation marker expression (the latter 2 sets determined by mass cytometry) (Supplemental Table 8). Individual classifier models based on each data set were trained and tested iteratively for their ability to distinguish virus shedders from nonshedders. After training and testing of each data set classifier model independently, models were stacked to create a single combined classifier for each day (Figure 9A). The performance of both individual and combined classifiers was assessed at each day of the post-challenge confinement phase by calculation of the area under the receiver operating characteristic curve (AUC) and performance of a Wilcoxon’s signed-rank test between the actual and the predicted data (Figure 9B). The ability to accurately classify shedders versus nonshedders was greatest on study day 6 (P < 0.001), when the stacked classifier outperformed all individual models (Figure 9, C–E). Using the combined classifier on day 6, the AUC indicates that the probability that a randomly chosen subject is correctly classified as a virus shedder or nonshedder is 0.90 (Figure 9E). To address the potential for lack of robustness in tree-based analysis in high-dimensional data sets (48), we implemented a rigorous 100-iteration cross-validation strategy. The performance of the classification model, measured by the AUC, stabilized after approximately 50 iterations with only minor changes resulting from random subsampling thereafter (Supplemental Figure 10A).
Figure 9. Machine learning–based classification and prediction of H1N1 virus shedding status.
(A) Schematic of models used to classify virus shedding after challenge. Four models were generated from 4 data sets: cytokines/chemokines; CBC; cell abundance; and cell activation and proliferation (Supplemental Table 8). For each data set, the model was iteratively trained and tested on 50% held-out data 100 times. The performance for each individual model was the median value from 100 iterations. For the stacked model, individual models were combined. (B) Heatmap of P values for classification of virus shedding status on days 2–8. (C) Correlation network displays the relationship between features used in the combined model. Node sizes are proportional to the univariate correlation between a given feature and virus shedding. (D) The P values for each model on the day with the best overall classification power (day 6). (E) Receiver operating characteristic (ROC) curve evaluating the performance of the combined classification model on day 6. (F) Schematic of model used to predict virus shedding before challenge. The 4 data sets from A were used to generate a predictive model that was trained and tested iteratively on 50% held-out data 100 times. In contrast to A, only the top features associated with classification of virus shedding on day 6 from each data set were used to train models on day 1 data. Models were subsequently evaluated on the excluded day 1 test set. The performance for each individual model was the median value from 100 iterations. For the final model, individual models were combined. (G) The P values for each model on day 1. (H) ROC curve evaluating the performance of the combined predictive model on data collected on day 1. Wilcoxon’s signed-rank test (A–H). n = 35 volunteers.
Next, using a similar approach, a bootstrapping-based random forest predictive model was devised to predict susceptibility to virus shedding before challenge. This was accomplished using distinct training and test groups and a subset of the features from the 4 data sets described above that were important for classifying virus shedding after challenge (Figure 9F). The resulting model was able to predict whether a volunteer was likely to later develop virus shedding using data from the prechallenge period (P < 0.01) with a probability of 0.87 (Figure 9, G and H). Like above, a 100-iteration cross-validation of the performance of the predictor, measured by the AUC, stabilized after approximately 70 iterations with only minor changes resulting from random subsampling thereafter (Supplemental Figure 10C). For additional details on the tuning of the classifier and predictor model and parameter selection, see Methods.
Among the top features that synergistically contributed to powering the classification and prediction models (Supplemental Table 9) were plasma levels of IP-10 and abundances of CD38+ pDCs, CD38+ CSM B cells, and monocytes, features already examined in detail above. This aligns with prior literature that associates IP-10 with influenza infection (49, 50), and CD38 as a component of a multicohort gene metasignature for influenza infection symptomatology and vaccine response (19). The success of this approach suggests the existence of cellular and humoral correlates of protection and infection beyond those currently exploited in the clinic, warranting further investigation in future studies.
Discussion
Here we characterized host responses to influenza virus infection in a controlled challenge study in human volunteers. H1N1 challenge resulted in virus shedding at high copy numbers, with an attack rate and symptomology similar to those in historical challenge studies (21, 22). Preexisting ELISA titers for full-length HA protein were significantly higher in nonshedders compared with shedders, while there was no difference in HA “stalk-only” titers between groups. This is consistent with recent reports of higher effectiveness of HA head-targeted antibodies compared with HA stalk–targeted antibodies (51, 52).
Prior studies of experimental and community-acquired human influenza A reported decreases in circulating bulk lymphocytes with concomitant increases in bulk monocytes (22, 31, 32). Our study confirms and greatly extends the findings of these previous studies, adding increased granularity and depth. Consistent with Wilkinson et al. (11), we observed in virus shedders that total activated and proliferating CD4+ and CD8+ T cells steadily increased until at least study day 8 and returned to baseline by day 29. The magnitude of the changes observed was larger for memory than for naive T cells.
In virus shedders, abundances of naive and non–class-switched memory B cells declined after infection, and remained depressed until at least day 8, while plasmablasts increased over the same period. Interestingly, a population of p-STAT5+ plasmablasts emerged 7 days after challenge. STAT3 and STAT5 phosphorylation in B cells can be activated by cytokines, including IL-2, IL-4, IL-7, and IL-21, which support B cell differentiation (53, 54). In previous in vitro studies, STAT3-dependent BLIMP-1 expression was associated with plasmablast differentiation, whereas STAT5-dependent BCL-6 induction promoted memory B cell formation (55, 56). Plasmablasts with sustained high levels of STAT5 phosphorylation had not been previously observed during infection in vivo. The role(s) played during influenza infection by these subpopulations of plasmablasts expressing high and low levels of p-STAT5 merits investigation in future studies.
Monocyte subsets increased dramatically after infection, peaking at days 4 (cMCs), 5 (intMCs), and 6 (ncMCs), consistent with the presumed stepwise differentiation trajectory of these subsets (57). The largest magnitude of elevations was seen for intMCs, consistent with a prior report of severe pandemic H1N1 infection (15). IntMCs and ncMCs are also a known source of IP-10 (58); in our study, the changes in plasma levels of IP-10 had similar kinetics to those of these subsets.
We observed a biphasic curve for neutrophil kinetics, a sharp decline in CD123+HLA-DR– basophils, and significant differences in CD66+ Scaffold clusters. These data point toward possible functions for granulocyte subsets in influenza A infection that warrant further investigation.
Prior studies have reported transient declines in circulating total NK cells (particularly CD56+ NK cells) during pandemic H1N1 2009 infection (59); similar declines were observed in virus shedders in our study. Conversely, sizeable increases were observed in activated, proliferating CD56+CD16– and CD56loCD16+ NK cell subsets. Upregulation of CD38 on NK cells has been reported previously in individuals with acute influenza infection; expression of CD38 by NK cells is linked to cytolytic function and immune synapse formation with infected cells (39).
pDCs are resistant to influenza infection and have a high capacity to activate influenza-specific T cells and drive Th1 polarization (14, 41, 60). Their importance was demonstrated in a patient with life-threatening influenza who had IRF7 deficiency resulting in poor production of type I and III interferons by pDCs (61). Inspection of the Scaffold map generated from our data pointed to a sharp upregulation of CD38 on pDCs in individuals shedding virus. This was recapitulated in vitro by exposure of PBMCs from healthy donors to TLR agonists or heat-inactivated H1N1, consistent with a report of herpes simplex virus–induced CD38 upregulation on cultured pDCs (62). Importantly, experiments with CD38-blocking antibodies shown here support the conclusion that CD38 is a regulator of type I interferon production in pDCs. CD38 is a multifunctional protein with both ectoenzymatic activity and adhesion properties mediated through interaction with CD31 on endothelial cells (38). While CD38 is well known for involvement in lymphocyte activation (38), a recent report highlights its role in inflammatory cytokine production in macrophages and monocytes (40), and similar mechanisms may operate in pDCs.
The dramatic response of CD38 upregulation in pDCs may be part of an early immune program activated by the presence of the virus, causally important to eventual control of viral load in an infected individual. Interestingly, a study detailing acute Ebola infection of humans similarly found that CD38 upregulation on pDCs was correlated to declining viral load (48), suggesting that pDC CD38’s role in viral infections may be broader than just influenza. The relevance of CD38 on pDCs is further underscored by a report that the CD38-targeted therapeutic daratumumab (63), approved for the treatment of multiple myeloma, depletes circulating pDCs (64). Strategies for the modulation of pDC CD38 may warrant investigation as potential therapies for viral infections or autoimmune diseases associated with dysregulated overaction of pDC IFN-I production.
We applied machine learning approaches to accurately classify and predict virus shedding based on amalgamated data from this study. This adds to knowledge from previous whole-blood transcriptomic studies (18–20) that correlates of protection from influenza A infection beyond previously described serum antibodies and IFN-γ+CD4+ T cells exist in the blood (11). However, it is important to recognize that the modeling approach used here considers all features in a coordinated manner for classification and prediction of virus shedding; therefore the performance of individual features requires validation in future independent cohorts. Furthermore, protection in challenge studies may not completely reflect protection from community exposures that could differ in strain, particle size of exposure, or dose.
This study provides a detailed view of the landscape of coordinated immune cell dynamics during influenza A infection and can serve as a framework on which to evaluate the impact of investigational influenza therapeutics on host immune responses in future human challenge studies. This work may also provide knowledge to guide the design of next-generation clinical tools for influenza diagnosis and susceptibility prediction.
Methods
Clinical evaluation
Vital signs including oral temperature, pulse rate, and systolic and diastolic blood pressure were collected 4 times daily. Mean daily values were calculated for all values except oral temperature, which was reported as maximum daily value. Vital signs are reported in figures normalized to each individual’s baseline values (day –1, 1). Symptoms were scored twice daily by participants using a symptom scorecard as absent (0), mild (1), moderate (2), or severe (3) for 17 symptoms: abdominal pain, breathing difficulty, cough, diarrhea, earache, facial or eye pain/tenderness, fatigue, hot/feverish/chills/rigor, headache, hoarseness, musculoskeletal ache, nasal stuffiness/congestion, nausea/vomiting, runny nose, sneezing, sore throat, and wheezy chest. Symptom scores were summed across all categories and reported as the mean of 2 daily scorecards. A targeted physical exam was also conducted twice daily by a physician investigator to evaluate nasal discharge, otitis, pharyngitis, sinus tenderness, new wheezes and crackles, percussion, and abdominal tenderness. Numerical scores were summed across all categories and reported as the mean of 2 daily exams.
Virus challenge
The challenge virus strain, A/California/2009 (H1N1), was isolated from a 3-year-old child in 2009 and manufactured under good manufacturing practices as previously described (65). This strain has reported greater than 99.4% sequence identity with A/California/04/2009 (H1N1) in HA and neuraminidase regions (65) and has been used in additional challenge studies (65, 66). Volunteers were given a single intranasal dose of the challenge virus delivered by a mucosal atomizer while seated in a semirecumbent position. See Supplemental Table 2 for dosing information. The 50% tissue culture infectious dose (TCID50) of the viral stock was determined by standard methods by Viroclinics Biosciences Inc.
Phlebotomy
Phlebotomy occurred at consistent times daily throughout the confinement phase. Routine CBC and blood chemistry were performed by Consolidated Medical Bio-Analysis Inc. Serum for HAI titer and plasma for Luminex assay were isolated on-site and stored at –80°C until analysis. For mass cytometry, 750 μL of heparinized blood was mixed with 1050 μL of Smart Tube Proteomic Stabilizer, incubated for 11 minutes at ambient temperature, then flash-frozen on dry ice and stored at –80°C until analysis. Blood processing in Smart Tube Proteomic Stabilizer occurred within approximately 10 minutes of phlebotomy.
Hemagglutination inhibition assay
HAI titer was determined by analysis of serum samples using A/California/7/2009 (H1N1) (obtained from the National Institute for Biological Standards and Control, Ridge, United Kingdom) and turkey red blood cells by Focus Diagnostics Inc. as previously described (67).
HA ELISA
ELISAs were performed as described previously (51). In brief, ELISAs were performed on 96-well Immulon 4HBX flat-bottomed microtiter plates (Thermo Fisher Scientific) coated with 0.5 μg/well streptavidin (MilliporeSigma). Biotinylated headless H1 HA stalk or biotinylated full-length H1 HA protein (secreted A/California/07/2009 HA) was diluted in biotinylation buffer to 0.25 μg/mL and 2 μg/mL, respectively; 50 μL/well was added and incubated for 1 hour. Each well was subsequently blocked for 1 hour with biotinylation blocking buffer. Each serum sample was serially diluted in biotinylation buffer starting at 1:100 dilution, added to the ELISA plates, and incubated for 1 hour. As a control, the human HA stalk–specific mAb CR9114 was serially diluted starting at 0.03 μg/mL to verify equal coating between plates and to determine relative serum titers. Peroxidase-conjugated goat anti-human IgG (Jackson ImmunoResearch Laboratories Inc.) was diluted in biotinylation buffer at 1:5000, added to each plate, then incubated for 1 hour. Finally, KPL SureBlue TMB peroxidase substrate (5120-0077, SeraCare) was added to each well for 5 minutes, then the reaction was quenched with 250 mM HCl solution. All incubation steps were performed on a rocker at room temperature. Plates were washed between each incubation step with PBS-Tween using a BioTek 405 LS microplate washer. Relative titers were determined using a consistent concentration of CR9114 mAb for each plate and reported as the corresponding inverse of the serum dilution that generated equivalent optical densities. Each type of ELISA was performed twice. Plasmids to produce headless H1 HA stalk protein were provided by Adrian McDermott and Barney Graham (Vaccine Research Center, NIH), and plasmids to produce HA stalk–specific mAb CR9114 were provided by Patrick Wilson (University of Chicago, Chicago, Illinois, USA).
Virus shedding analysis
Virus shedding data were obtained from ClincialTrials.gov (NCT04106817) with assay details provided by WCCT Global. Assays were performed by Matthew Memoli at the National Institute of Allergy and Infectious Diseases, NIH, as previously described (68). Briefly, nasopharyngeal swabs were collected daily for viral shedding assessment using a BD Universal Viral Transport Kit with a 3 mL vial containing transport medium and a flocked swab (product 220528, Becton Dickinson). Virus shedding was determined by a qRT-PCR assay targeting the influenza matrix 1 (M1) RNA gene segment (69). An external standard was used to construct a standard curve for copy number estimation. Virus shedders were defined as volunteers with at least 1 positive qRT-PCR test during the post-challenge period.
Luminex cytokine assay
Circulating cytokines and chemokines were measured in plasma samples by Eve Technologies using a multiplexed Luminex-based assay (Human Cytokine/Chemokine 65-Plex Panel). Targets examined were EGF, eotaxin, FGF-2, Flt-3 ligand, fractalkine, G-CSF, GM-CSF, GRO, IFN-α2, IFN-γ, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IL-17A, IL-18, IL-1ra, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IP-10, MCP-1, MCP-3, MDC (CCL22), MIP-1α, MIP-1β, PDGF-AA, PDGF-AB/BB, RANTES, TGF-α, TNF-α, TNF-β, VEGF, sCD40L, eotaxin-2, MCP-2, BCA-1, MCP-4, I-309, IL-16, TARC, 6CKine, eotaxin-3, LIF, TPO, SCF, TSLP, IL-33, IL-20, IL-21, IL-23, TRAIL, CTACK, SDF-1a+B, ENA-78, MIP-1d, and IL-28A. Luminex data are reported as bead count and fluorescence intensity. Fluorescence values with corresponding bead count lower than 35 were considered unreliable and eliminated from further analysis. Data reported are fluorescence intensities normalized to each individual’s baseline values (day –1, 1) (70).
Mass cytometry
Whole-blood samples treated with Smart Tube Proteomic Stabilizer were thawed, treated to lyse red blood cells, barcoded, and stained with a panel of metal-conjugated antibodies (Supplemental Tables 4 and 5) before analysis using a Fluidigm CyTOF 2 mass cytometer (71). A total of 381 samples were processed and analyzed by mass cytometry (35 volunteers × 11 time points, minus 4 unavailable samples [volunteer 112 day 29, volunteer 112 day 60, volunteer 304 day 29, volunteer 305 day 60]). Samples were processed in 6 large batches with complete sample sets (samples from all time points) for 11 or 12 volunteers thawed simultaneously. Each volunteer sample set was thawed alongside a control aliquot created from a single blood draw from a healthy volunteer for normalization of plate-specific batch effects. After red blood cell lysis with 1X Thaw-Lyse buffer (Smart Tube Inc.), cells were counted, and 1.1 × 106 cells were manually arranged in a 96-well block. Subsequent steps were performed on a previously described robotics platform (71). Each sample set along with 1 control sample was barcoded with palladium metal (72) and combined into a single well. Barcoded samples were treated with Fc block (Human TruStain FcX, BioLegend) for 10 minutes before surface antibody staining for 30 minutes in cell staining medium (PBS with 0.5% BSA and 0.02% sodium azide). After surface staining, cells were permeabilized with ice-cold 100% methanol, washed, and stained for 60 minutes with intracellular antibodies in cell staining medium. Following intracellular staining, cells were washed and resuspended in an iridium intercalator (Fluidigm) solution containing 1.6% paraformaldehyde. Finally, samples were washed, resuspended in 1× five-element normalization beads (La, Pr, Tb, Tm, Lu) (Fluidigm), and analyzed on a freshly cleaned and tuned cytometry by time-of-flight (CyTOF) instrument. Mass cytometry data were bead-normalized across all runs and debarcoded as previously described (73). For normalization of plate-specific batch effects, first, all markers from all shared controls were linearly transformed using a smoothing spline to match the mean and standard deviation of the control in the first plate. Next, the same plate-specific transformations were applied to all other samples in the respective plates (74).
Scaffold analysis
For Scaffold analysis, FCS files were first created from CD61–CD235– gated mass cytometry events. Events were randomly subsampled to 50,000 events per file, and events from all volunteers and time points (~19 million events) were pooled and assigned into 1 of 200 nonoverlapping clusters based on the similarity of surface marker expression using the CLARA function in R (35) as part of the Scaffold R package (see Supplemental Table 4 for list of surface markers) (34). Scaffold code can be found at https://github.com/nolanlab/scaffold Gated populations from a single representative sample were used to define landmark nodes.
PBMC culture and stimulation
Blood was collected using EDTA-coated tubes (BD Biosciences), and cells were processed within 2 hours of blood draw. PBMCs were isolated by density gradient centrifugation using Ficoll-Paque PLUS (GE Healthcare). Cells were washed with PBS and 2 × 106 cells were cultured in 500 μL complete RPMI (Corning) containing 10% FBS (GIBCO), 2 mM l-glutamine (Corning), 25 mM HEPES (Corning), 1 mM sodium pyruvate (Corning), 100 μM nonessential amino acids (Corning), and 55 μM 2-mercaptoethanol (GIBCO) plus 10 ng/mL recombinant human IL-3 (R&D Systems) for 24 hours at 37°C in 5 mL polypropylene tubes. Cells were stimulated with 5 μg/mL imiquimod (Invivogen), resiquimod (Invivogen), CpG-A (2216, Invivogen), or CpG-B (2006-G5, Invivogen) or with heat-inactivated challenge strain at MOI of 2. Virus was heat-inactivated for 30 minutes at 56°C. For intracellular cytokine staining, Brefeldin A (10 μg/mL) was added 10 hours after stimulation with virus. Where indicated, PBMCs were preincubated with anti-CD38 (5 μg per 1 × 106 cells) clone B307 (AT-1) (Abcam) to block CD38 (42), or Rb isotype control (R&D Systems) for 30 minutes at 37°C before addition of stimulus. After 24 hours, supernatants were isolated and stored at –80°C until analysis. For IFN-α ELISA, culture supernatants were analyzed in triplicate according to the manufacturer’s instructions using a Human IFN Alpha Multi-Subtype ELISA Kit (TCM [tissue culture media]) (PBL Assay Science).
Flow cytometry
Before staining, cells were incubated with human IgG (Invitrogen) for 10 minutes at room temperature to block nonspecific binding. Cells were stained for surface markers for 20 minutes at room temperature. Cells were fixed and permeabilized for 20 minutes at room temperature using BD Cytofix/Cytoperm (BD Biosciences), then stained for IFN-α and TNF-α for 30 minutes at room temperature in permeabilization buffer. Data were acquired on a 5-laser LSRFortessa X20 (BD Biosciences). pDCs were gated as singlet, live Lin– (CD3–CD19–CD20–CD335–CD66b–CD14–CD16–CD11c–) HLA-DR+CD123+BDCA4+.
Statistics
Longitudinal models.
To investigate the differences between shedders and nonshedders for a given response variable, a mixed-effect model was used. For baseline normalized data, values were first log2(y+1)-transformed to unskew the data, followed by baseline normalization using the average value from the 2 baseline days (days –1, 1). A longitudinal linear random intercept model was developed through the day of the maximum difference between shedders and nonshedders. Shedding and time were covariates, and a subject-specific random intercept term, uj, was used to address correlation among multiple measurements. The P value for the interaction term between time and shedding is reported. The longitudinal regression model is described as follows: yij = β0 + β1sheddingj × timei + uj, where yij denotes the response variable at the ith day for subject j.
P value adjustment.
All P values were adjusted for multiple hypothesis correction using Bonferroni’s method. A P value less than 0.05 was considered significant.
Missing and excluded values.
Day 29 samples were not available for analysis from volunteer 112. Day 60 samples were not available for analysis from volunteers 112, 304, and 305. All available samples from the confinement phase (days –1 through 8) were included in the analysis. For the follow-up period only (days 29, 60), samples were excluded from analysis for any volunteer who reported new symptoms immediately before a follow-up visit that could be suggestive of unrelated community-acquired infections (e.g., sore throat). This included day 29 samples from volunteers 110, 304, and 306 and day 60 samples from volunteers 104, 105, and 202.
Correlation analysis.
Correlation analysis was performed using the nonparametric Spearman’s correlation test, and the Spearman ρ value of correlation is reported.
Receiver operating characteristic.
The receiver operating characteristic curve was drawn for each category of the population. True-positive and false-positive rates were computed as previously described (75). The area under the receiver operating characteristic curve was used to evaluate the performance of the predictor and classifier.
Classification model.
For each day a matrix X of all features from a given data set and a vector of viral shedding, Y, were used to generate a decision tree–based random forest classifier. A random forest classifier consists of a collection of tree-structured classifiers {t(X, Θb), b = 1, …, B}, where {Θb} are independent identically distributed random vectors, and each tree casts a unit vote for the most popular class at input X (43). Breiman’s implementation of the random forest (43, 76) was used in this study because it is a well-established method that has had excellent predictive power in prior studies with a similarly large number of features relative to sample n (77, 78).
For each day, 4 individual models were generated based on 4 data sets: cytokine and chemokine data; CBC counts; relative abundance of immune cell populations (determined by mass cytometry); and cellular activation (CD38) and proliferation (Ki67) marker expression (determined by mass cytometry) (Supplemental Table 8). These 4 models were trained and tested for their ability to classify viral shedding. For the final multivariate modeling step, individual models were combined as previously described (79, 80). The median values across all 4 data sets were reported as the final classification (81).
Predictive model.
For the predictive model, data were first split into separate training and test sets. Using training data only, a random forest model was generated using a matrix X of all features from day 6 for each data set. Features with importance greater than 0.09 (Supplemental Table 9) were selected to create a reduced feature set that was used to retrain the model on day 1 training data using a gradient-boosted decision tree system (82). The model was then tested on the excluded test set data from day 1. This step was repeated 100 times.
The performance for the predictor was the median value from 100 repetitions. The results from the analysis of single data sets were then combined in a final multivariate modeling step, where the median value across all 4 data sets was reported as the final prediction.
Robustness of classification and prediction models.
For both classification and prediction models, the performance is reported as the median value from 100 repetitions. Bootstrapping was implemented by splitting of the data set into 2 equal parts: a training set and a testing set. Models were trained on the training set and tested using the test set. This step was repeated 100 times. To ensure that 100 bootstrapping repetitions was a suitable number, a range of 2 to 200 iterations were examined, finding the performance of the model, indicated by AUC, to stabilize, and variance to decline, after approximately 50 iterations for the classifier (Supplemental Figure 10A), or approximately 70 iterations for the predictor (Supplemental Figure 10C). Furthermore, to explore the potential impact of different train-test splits of the data set on model performance, 2:1, 4:1, and 9:1 train-test splits were tested, resulting in classifier AUCs of 0.92, 0.92, and 0.91, respectively (Supplemental Figure 10B), and predictor AUCs of 0.77, 0.78, and 0.80, respectively. These results are comparable to the AUC of 0.90 for the classifier and AUC of 0.87 for the predictor using the 1:1 split of the data set shown in Figure 9, E and H.
Box plots.
In box plots, the center line represents the median; the box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; and points, all data points (Figures 1 and 8) or only outliers (Figure 7 and Supplemental Figure 7). Where indicated, Welch’s t tests (2-tailed) were performed.
Software
Gating and processing of both mass cytometry and flow cytometry data and the creation of biaxial plots and histograms were accomplished using CellEngine (Primity Bio). Other plots were generated with Prism 8 (GraphPad Software Inc.) or R 3.5.2 (R Core Team, 2018). R packages used in the computational modeling included randomForest (83), pROC (75), ROCR (84), xgboost (85), matrixStats (86), ColorBrewer (87), and Rtsne (88). The full reproducible code is made available to readers.
Publicly available data and code
The code and data generated during and/or analyzed during the current study are available to the public in the Flow Repository (https://flowrepository.org/id/FR-FCM-Z2NZ) (89).
Study approval
The clinical study was conducted at WCCT Global’s challenge facility in Costa Mesa, California, between January 2015 and August 2015 with informed consent obtained from all study subjects. Volunteers were recruited from the greater Los Angeles, California, metropolitan area. The study was listed under ClinicalTrials.gov identifier NCT04106817, was approved by an independent institutional review board (Aspire IRB, Santee, California, protocol WCCTG 14-01), and was monitored by a data and safety monitoring board. An investigational drug application was filed for the challenge virus with the US FDA (IND 16268). The study was conducted in accordance with the ethical principles that have their origin in the Declaration of Helsinki and clinical research guidelines established by the Code of Federal Regulations (Title 21, CFR Parts 50, 56, and 312) and International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH) guidelines.
Control blood for mass cytometry and in vitro experiments involving PBMCs was obtained from healthy adult volunteers following the guidelines of the Environmental Health and Safety Biosafety program of Stanford University. Donors provided informed consent in accordance with IRB protocols approved by the Stanford University Administrative Panel on Human Subjects Research.
Author contributions
DRM, GPN, KK, CMS, and ZBH designed the study, analyzed data, and wrote the manuscript with contributions from all authors. BB, JY, KK, and MA conducted the clinical study. SEH and SRC contributed to HA ELISA experiments. RL and JI executed and analyzed flow cytometry experiments. DRM, HC, GKF, MHS, NK, NM, SJ, BG, and ZBH conducted and analyzed mass cytometry experiments. ZR, PFG, HC, NA, and SSB designed and completed computational modeling and statistical analysis of all data.
Supplementary Material
Acknowledgments
We thank Adrian McDermott, Barney Graham, and Patrick Wilson for plasmids; and Matthew Memoli and Allison Han for virus and HAI titer quantification. This work was supported by the US NIH (grants and sub-awards: 2U19AI057229-16, 5P01HL108797-07, 5R01GM109836-04, 5R33CA183654-03, 5U01AI101984-07, 5UH2AR067676-04, 5R01CA196657-03, 5U54CA209971-03, 5F99CA212231-02, 1F32CA233203-01, 5U01AI140498-02, 1U54HG010426-01, 5U19AI100627-07, 1R01HL120724-01A1, R33CA183692, R01HL128173-04, 5P01AI131374-02, 5UG3DK114937-02, 1U19AI135976-01, IDIQ17X149, 1U2CCA233238-01, 1U2CCA233195-01, 1R01AI113047 [to SEH], 1R01AI108686 [to SEH], CEIRS HHSN272201400005C [to SEH], 1DP2AR069953 [to JI], and K23GM111657 [to BG]); an NIH Cooperative Centers for Translational Research in Human Immunology and Biodefense Opportunity Fund seed grant; the Department of Defense (W81XWH-14-1-0180 and W81XWH-12-1-0591); the FDA (HHSF223201610018C); Cancer Research UK (C27165/A29073); the Bill and Melinda Gates Foundation (OPP1113682); the Cancer Research Institute; the Parker Institute for Cancer Immunotherapy; the Kenneth Rainin Foundation (2018-575); the Silicon Valley Community Foundation (2017-175329 and 2017-177799-5022); the Doris Duke Charitable Foundation (to BG); the Beckman Center for Molecular and Genetic Medicine; Juno Therapeutics Inc. (122401); Pfizer Inc. (123214); Celgene Inc. (133826 and 134073); Vaxart Inc. (137364); and the Rachford and Carlota A. Harris Endowed Chair (to GPN). DRM was supported by a Canadian Institutes of Health Research post-PhD fellowship. CMS was supported by an Advanced Postdoc Mobility Fellowship from the Swiss National Science Foundation (P300PB_171189 and P400PM_183915). RL is the recipient of a National Science Foundation Graduate Research Fellowship Program fellowship (DGE-1147470). SSB was supported by a Stanford Interdisciplinary Graduate Fellowship and Stanford Bio-X. SEH holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. This article reflects the views of the authors and should not be construed as representing the views or policies of the US FDA, NIH, or other institutions and companies providing funding or affiliated with the authors.
Version 1. 10/12/2020
Electronic publication
Version 2. 11/02/2020
Print issue publication
Footnotes
Conflict of interest: MA, KK, BB, and JY are current or former employees of WCCT Global. DRM is an unpaid advisor for WCCT Global. ZBH is an employee of Primity Bio. PFG is an employee of the Parker Institute for Cancer Immunotherapy. KK and JY are employees of ARK Clinical Research. NA is an advisor for January Inc. and Vasognosis and a consultant for MaraBio, and has received research funding support from Alkahest. CMS has equity in and is a scientific advisor for Enable Medicine LLC. SEH has received consultancy fees from Sanofi Pasteur, Lumen, Novavax, and Merck for work unrelated to this report. GPN and DRM have received research funding support from Vaxart Inc. GPN holds patents (US8003312B2 and US8679858B2) related to CyTOF and associated reagents that were used in this study and receives royalties derived from those patents in accordance with Stanford’s publicly available distribution agreement.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(11):5800–5816.https://doi.org/10.1172/JCI137265.
Contributor Information
Zainab Rahil, Email: zrahil@stanford.edu.
Rebecca Leylek, Email: rleylek@stanford.edu.
Christian M. Schürch, Email: schuerch@stanford.edu.
Han Chen, Email: hchen95@stanford.edu.
Zach Bjornson-Hooper, Email: bjornson@stanford.edu.
Pier Federico Gherardini, Email: pfgherardini@parkerici.org.
Matthew H. Spitzer, Email: matthew.spitzer@ucsf.edu.
Gabriela K. Fragiadakis, Email: Gabriela.Fragiadakis@ucsf.edu.
Nilanjan Mukherjee, Email: nilanjan.mpb@gmail.com.
Nelson Kim, Email: nelsonkimster@gmail.com.
Sizun Jiang, Email: sizunj@stanford.edu.
Jennifer Yo, Email: jyo@arkclinicalresearch.com.
Brice Gaudilliere, Email: gbrice@stanford.edu.
Scott E. Hensley, Email: hensley@pennmedicine.upenn.edu.
Juliana Idoyaga, Email: jidoyaga@stanford.edu.
Nima Aghaeepour, Email: naghaeep@stanford.edu.
Kenneth Kim, Email: allergydoc81@gmail.com.
David R. McIlwain, Email: mcilwain@stanford.edu.
References
- 1.Xu X, et al. Update: Influenza activity in the United States during the 2018-19 season and composition of the 2019-20 influenza vaccine. MMWR Morb Mortal Wkly Rep. 2019;68(24):544–551. doi: 10.15585/mmwr.mm6824a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. WHO. Influenza (Seasonal) 2020. https://www.who.int/en/news-room/fact-sheets/detail/influenza-(seasonal) Updated November 5, 2018. Accessed August 27, 2020.
- 3. WHO. Pandemic Influenza Risk Management Guidance. https://www.who.int/influenza/preparedness/pandemic/PIRM_update_052017.pdf Updated May 2017. Accessed August 27, 2020.
- 4.Fiore AE, et al. Antiviral agents for the treatment and chemoprophylaxis of influenza — recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep. 2011;60(1):1–24. [PubMed] [Google Scholar]
- 5.Jefferson T, Jones M, Doshi P, Spencer EA, Onakpoya I, Heneghan CJ. Oseltamivir for influenza in adults and children: systematic review of clinical study reports and summary of regulatory comments. BMJ. 2014;348:g2545. doi: 10.1136/bmj.g2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayden FG, et al. Baloxavir marboxil for uncomplicated influenza in adults and adolescents. N Engl J Med. 2018;379(10):913–923. doi: 10.1056/NEJMoa1716197. [DOI] [PubMed] [Google Scholar]
- 7.De Clercq E, Li G. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev. 2016;29(3):695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hobson D, Curry RL, Beare AS, Ward-Gardner A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J Hyg (Lond) 1972;70(4):767–777. doi: 10.1017/s0022172400022610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stadlbauer D, et al. Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science. 2019;366(6464):499–504. doi: 10.1126/science.aay0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng S, et al. Novel correlates of protection against pandemic H1N1 influenza A virus infection. Nat Med. 2019;25(6):962–967. doi: 10.1038/s41591-019-0463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilkinson TM, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18(2):274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 12.Krammer F, et al. Influenza. Nat Rev Dis Primers. 2018;4(1):3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sridhar S, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med. 2013;19(10):1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- 14.Cella M, Facchetti F, Lanzavecchia A, Colonna M. Plasmacytoid dendritic cells activated by influenza virus and CD40L drive a potent TH1 polarization. Nat Immunol. 2000;1(4):305–310. doi: 10.1038/79747. [DOI] [PubMed] [Google Scholar]
- 15.Cole SL, et al. M1-like monocytes are a major immunological determinant of severity in previously healthy adults with life-threatening influenza. JCI Insight. 2017;2(7):e91868. doi: 10.1172/jci.insight.91868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Duin D, et al. Prevaccine determination of the expression of costimulatory B7 molecules in activated monocytes predicts influenza vaccine responses in young and older adults. J Infect Dis. 2007;195(11):1590–1597. doi: 10.1086/516788. [DOI] [PubMed] [Google Scholar]
- 17.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12(8):786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walters KA, et al. Differential effects of influenza virus NA, HA head, and HA stalk antibodies on peripheral blood leukocyte gene expression during human infection. mBio. 2019;10(3):e00760-19. doi: 10.1128/mBio.00760-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andres-Terre M, et al. Integrated, multi-cohort analysis identifies conserved transcriptional signatures across multiple respiratory viruses. Immunity. 2015;43(6):1199–1211. doi: 10.1016/j.immuni.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davenport EE, Antrobus RD, Lillie PJ, Gilbert S, Knight JC. Transcriptomic profiling facilitates classification of response to influenza challenge. J Mol Med. 2015;93(1):105–114. doi: 10.1007/s00109-014-1212-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balasingam S, Wilder-Smith A. Randomized controlled trials for influenza drugs and vaccines: a review of controlled human infection studies. Int J Infect Dis. 2016;49:18–29. doi: 10.1016/j.ijid.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Sherman AC, Mehta A, Dickert NW, Anderson EJ, Rouphael N. The future of flu: a review of the human challenge model and systems biology for advancement of influenza vaccinology. Front Cell Infect Microbiol. 2019;9:107. doi: 10.3389/fcimb.2019.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrat F, et al. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol. 2008;167(7):775–785. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
- 24.Potter CW, Oxford JS. Determinants of immunity to influenza infection in man. Br Med Bull. 1979;35(1):69–75. doi: 10.1093/oxfordjournals.bmb.a071545. [DOI] [PubMed] [Google Scholar]
- 25.Bahadoran A, et al. Immune responses to influenza virus and its correlation to age and inherited factors. Front Microbiol. 2016;7:1841. doi: 10.3389/fmicb.2016.01841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broberg E, Nicoll A, Amato-Gauci A. Seroprevalence to influenza A(H1N1) 2009 virus—where are we? Clin Vaccine Immunol. 2011;18(8):1205–1212. doi: 10.1128/CVI.05072-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davey RT, et al. The association between serum biomarkers and disease outcome in influenza A(H1N1)pdm09 virus infection: results of two international observational cohort studies. PLoS One. 2013;8(2):e57121. doi: 10.1371/journal.pone.0057121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peteranderl C, Herold S. The impact of the interferon/TNF-related apoptosis-inducing ligand signaling axis on disease progression in respiratory viral infection and beyond. Front Immunol. 2017;8:313. doi: 10.3389/fimmu.2017.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ackerman SJ, et al. Ex vivo models for the study of eosinophils. In: Eosinophils in Health and Disease. Elsevier Inc.; 2013:39–71. [Google Scholar]
- 30.Ravensberg AJ, et al. Eotaxin-2 and eotaxin-3 expression is associated with persistent eosinophilic bronchial inflammation in patients with asthma after allergen challenge. J Allergy Clin Immunol. 2005;115(4):779–785. doi: 10.1016/j.jaci.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 31.McClain MT, et al. Longitudinal analysis of leukocyte differentials in peripheral blood of patients with acute respiratory viral infections. J Clin Virol. 2013;58(4):689–695. doi: 10.1016/j.jcv.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 32.Coşkun O, et al. Relative lymphopenia and monocytosis may be considered as a surrogate marker of pandemic influenza a (H1N1) J Clin Virol. 2010;47(4):388–389. doi: 10.1016/j.jcv.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 33. doi: 10.1101/577759. Bjornson-Hooper ZB, et al. Cell type-specific monoclonal antibody cross-reactivity screening in non-human primates and development of comparative immunophenotyping panels for CyTOF [preprint]. Posted on bioRxiv March 14, 2019. [DOI]
- 34.Spitzer MH, et al. IMMUNOLOGY. An interactive reference framework for modeling a dynamic immune system. Science. 2015;349(6244):1259425. doi: 10.1126/science.1259425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. Wiley; 2005. [Google Scholar]
- 36.Manz MG, Boettcher S. Emergency granulopoiesis. Nat Rev Immunol. 2014;14(5):302–314. doi: 10.1038/nri3660. [DOI] [PubMed] [Google Scholar]
- 37.Elghetany MT. Surface antigen changes during normal neutrophilic development: a critical review. Blood Cells Mol Dis. 2002;28(2):260–274. doi: 10.1006/bcmd.2002.0513. [DOI] [PubMed] [Google Scholar]
- 38.Quarona V, et al. CD38 and CD157: a long journey from activation markers to multifunctional molecules. Cytometry B Clin Cytom. 2013;84(4):207–217. doi: 10.1002/cyto.b.21092. [DOI] [PubMed] [Google Scholar]
- 39. doi: 10.1101/349084. Gars LM, et al. CD38 contributes to human natural killer cell responses through a role in immune synapse formation [preprint]. Posted on bioRxiv May 24, 2019. [DOI]
- 40.Amici SA, et al. CD38 is robustly induced in human macrophages and monocytes in inflammatory conditions. Front Immunol. 2018;9:1593. doi: 10.3389/fimmu.2018.01593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reizis B. Plasmacytoid dendritic cells: development, regulation, and function. Immunity. 2019;50(1):37–50. doi: 10.1016/j.immuni.2018.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaisitti T, et al. CD38 increases CXCL12-mediated signals and homing of chronic lymphocytic leukemia cells. Leukemia. 2010;24(5):958–969. doi: 10.1038/leu.2010.36. [DOI] [PubMed] [Google Scholar]
- 43.Breiman L. Random forests. Machine Learning. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 44.Braga-Neto UM, Dougherty ER. Is cross-validation valid for small-sample microarray classification? Bioinformatics. 2004;20(3):374–380. doi: 10.1093/bioinformatics/btg419. [DOI] [PubMed] [Google Scholar]
- 45.Kim JH. Estimating classification error rate: repeated cross-validation, repeated hold-out and bootstrap. Comput Stat Data Anal. 2009;53(11):3735–3745. doi: 10.1016/j.csda.2009.04.009. [DOI] [Google Scholar]
- 46.Efron B, Tibshirani R. Improvements on cross-validation: the.632+ bootstrap method. J Am Stat Assoc. 1997;92(438):548–560. [Google Scholar]
- 47.Efron B. Estimating the error rate of a prediction rule: improvement on cross-validation. J Am Stat Assoc. 1983;78(382):316–331. doi: 10.1080/01621459.1983.10477973. [DOI] [Google Scholar]
- 48.McElroy AK, et al. Immunologic timeline of Ebola virus disease and recovery in humans. JCI Insight. 2020;5(10):137260. doi: 10.1172/jci.insight.137260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheung CY, et al. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet. 2002;360(9348):1831–1837. doi: 10.1016/S0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- 50.Lu X, Masic A, Liu Q, Zhou Y. Regulation of influenza A virus induced CXCL-10 gene expression requires PI3K/Akt pathway and IRF3 transcription factor. Mol Immunol. 2011;48(12-13):1417–1423. doi: 10.1016/j.molimm.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 51.Christensen SR, et al. Assessing the protective potential of H1N1 influenza virus hemagglutinin head and stalk antibodies in humans. J Virol. 2019;93(8):e02134-18. doi: 10.1128/JVI.02134-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park JK, et al. Evaluation of preexisting anti-hemagglutinin stalk antibody as a correlate of protection in a healthy volunteer challenge with influenza A/H1N1pdm virus. mBio. 2018;9(1):e02284-17. doi: 10.1128/mBio.02284-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berglund LJ, et al. IL-21 signalling via STAT3 primes human naive B cells to respond to IL-2 to enhance their differentiation into plasmablasts. Blood. 2013;122(24):3940–3950. doi: 10.1182/blood-2013-06-506865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwakkenbos MJ, van Helden PM, Beaumont T, Spits H. Stable long-term cultures of self-renewing B cells and their applications. Immunol Rev. 2016;270(1):65–77. doi: 10.1111/imr.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diehl SA, et al. STAT3-mediated up-regulation of BLIMP1 is coordinated with BCL6 down-regulation to control human plasma cell differentiation. J Immunol. 2008;180(7):4805–4815. doi: 10.4049/jimmunol.180.7.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheeren FA, et al. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat Immunol. 2005;6(3):303–313. doi: 10.1038/ni1172. [DOI] [PubMed] [Google Scholar]
- 57.Ziegler-Heitbrock L, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116(16):e74–e80. doi: 10.1182/blood-2010-02-258558. [DOI] [PubMed] [Google Scholar]
- 58.Cros J, et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 2010;33(3):375–386. doi: 10.1016/j.immuni.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pulendran B, Maddur MS. Innate immune sensing and response to influenza. Curr Top Microbiol Immunol. 2015;386:23–71. doi: 10.1007/82_2014_405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fonteneau JF, et al. Activation of influenza virus-specific CD4+ and CD8+ T cells: a new role for plasmacytoid dendritic cells in adaptive immunity. Blood. 2003;101(9):3520–3526. doi: 10.1182/blood-2002-10-3063. [DOI] [PubMed] [Google Scholar]
- 61.Ciancanelli MJ, et al. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348(6233):448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schuster P, et al. Co-ordinated regulation of plasmacytoid dendritic cell surface receptors upon stimulation with herpes simplex virus type 1. Immunology. 2010;129(2):234–247. doi: 10.1111/j.1365-2567.2009.03176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van de Donk NWCJ, Richardson PG, Malavasi F. CD38 antibodies in multiple myeloma: back to the future. Blood. 2018;131(1):13–29. doi: 10.1182/blood-2017-06-740944. [DOI] [PubMed] [Google Scholar]
- 64.Stocker N, et al. Daratumumab prevents programmed death ligand-1 expression on antigen-presenting cells in de novo multiple myeloma. Cancer Med. 2020;9(6):2077–2084. doi: 10.1002/cam4.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watson JM, et al. Characterisation of a wild-type influenza (A/H1N1) virus strain as an experimental challenge agent in humans. Virol J. 2015;12:13. doi: 10.1186/s12985-015-0240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liebowitz D, et al. Efficacy, immunogenicity, and safety of an oral influenza vaccine: a placebo-controlled and active-controlled phase 2 human challenge study. Lancet Infect Dis. 2020;20(4):435–444. doi: 10.1016/S1473-3099(19)30584-5. [DOI] [PubMed] [Google Scholar]
- 67.Greenberg ME, et al. Response to a monovalent 2009 influenza A (H1N1) vaccine. N Engl J Med. 2009;361(25):2405–2413. doi: 10.1056/NEJMoa0907413. [DOI] [PubMed] [Google Scholar]
- 68.Memoli MJ, et al. Validation of the wild-type influenza A human challenge model H1N1pdMIST: an A(H1N1)pdm09 dose-finding investigational new drug study. Clin Infect Dis. 2015;60(5):693–702. doi: 10.1093/cid/ciu924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krafft AE, et al. Evaluation of PCR testing of ethanol-fixed nasal swab specimens as an augmented surveillance strategy for influenza virus and adenovirus identification. J Clin Microbiol. 2005;43(4):1768–1775. doi: 10.1128/JCM.43.4.1768-1775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Breen EJ, Tan W, Khan A. The statistical value of raw fluorescence signal in Luminex xMAP based multiplex immunoassays. Sci Rep. 2016;6:26996. doi: 10.1038/srep26996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. doi: 10.1101/574160. Bjornson-Hooper ZB, et al. A comprehensive atlas of immunological differences between humans, mice and non-human primates [preprint]. Posted on bioRxiv March 11, 2019. [DOI] [PMC free article] [PubMed]
- 72.Zunder ER, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc. 2015;10(2):316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Finck R, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83(5):483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Green PJ, Silverman BW. Nonparametric Regression and Generalized Linear Models: A Roughness Penalty Approach. Chapman & Hall; 1994. [Google Scholar]
- 75.Robin X, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pretorius A, Bierman S, Steel SJ. A meta-analysis of research in random forests for classification. Paper presented at: 2016 Pattern Recognition Association of South Africa and Robotics and Mechatronics International Conference (PRASA-RobMech); November 30–December 2, 2016; Stellenbosch, South Africa. https://ieeexplore.ieee.org/document/7813171 Accessed August 27, 2020.
- 77.Díaz-Uriarte R, Alvarez de Andrés S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Okun O, Priisalu H. Random forest for gene expression based cancer classification: overlooked issues. In: Marti J, Benedi MJ, Mendonça AM, Serrat J, eds. Pattern Recognition and Image Analysis: Third Iberian Conference, IbPRIA 2007, Girona, Spain, June 6-8, 2007, Proceedings, Part II. Springer; 2007:483–490. Lecture Notes in Computer Science; 4478. [Google Scholar]
- 79.Ge G, Wong GW. Classification of premalignant pancreatic cancer mass-spectrometry data using decision tree ensembles. BMC Bioinformatics. 2008;9:275. doi: 10.1186/1471-2105-9-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He L, Yang Z, Zhao Z, Lin H, Li Y. Extracting drug-drug interaction from the biomedical literature using a stacked generalization-based approach. PLoS One. 2013;8(6):e65814. doi: 10.1371/journal.pone.0065814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ghaemi MS, et al. Multiomics modeling of the immunome, transcriptome, microbiome, proteome and metabolome adaptations during human pregnancy. Bioinformatics. 2019;35(1):95–103. doi: 10.1093/bioinformatics/bty537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Vanwinckelen G, Blockeel H. On estimating model accuracy with repeated cross-validation. In: Proceedings of the 21st Belgian-Dutch Conference on Machine Learning. Benelearn; 2012:39–44. [Google Scholar]
- 83.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2/3:18–22. [Google Scholar]
- 84.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21(20):3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 85. Chen T, Guestrin C. XGBoost: a scalable tree boosting system. In: Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. Association for Computing Machinery; 2016:785–794. [Google Scholar]
- 86. Bengtsson H. Functions that Apply to Rows and Columns of Matrices (and to Vectors). https://cran.rstudio.com/web/packages/matrixStats/index.html Accessed August 27, 2020.
- 87.Zeileis A, Hornik K, Murrell P. Escaping RGBland: selecting colors for statistical graphics. Comput Stat Data Anal. 2009;53(9):3259–3270. doi: 10.1016/j.csda.2008.11.033. [DOI] [Google Scholar]
- 88. Krijthe JH. Rtsne: T-Distributed Stochastic Neighbor Embedding using a Barnes-Hut Implementation. https://github.com/jkrijthe/Rtsne Accessed August 27, 2020.
- 89.Spidlen J, Breuer K, Rosenberg C, Kotecha N, Brinkman RR. FlowRepository: a resource of annotated flow cytometry datasets associated with peer-reviewed publications. Cytometry A. 2012;81(9):727–731. doi: 10.1002/cyto.a.22106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.