Abstract
ARID1A, a component of the chromatin-remodeling complex SWI/SNF, is one of the most frequently mutated genes in human cancer. We sought to develop rational combination therapy to potentiate the efficacy of immune checkpoint blockade in ARID1A-deficient tumors. In a proteomic analysis of a data set from The Cancer Genomic Atlas, we found enhanced expression of Chk2, a DNA damage checkpoint kinase, in ARID1A-mutated/deficient tumors. Surprisingly, we found that ARID1A targets the nonchromatin substrate Chk2 for ubiquitination. Loss of ARID1A increased the Chk2 level through modulating autoubiquitination of the E3-ligase RNF8 and thereby reducing RNF8-mediated Chk2 degradation. Inhibition of the ATM/Chk2 DNA damage checkpoint axis led to replication stress and accumulation of cytosolic DNA, which subsequently activated the DNA sensor STING-mediated innate immune response in ARID1A-deficient tumors. As expected, tumors with mutation or low expression of both ARID1A and ATM/Chk2 exhibited increased tumor-infiltrating lymphocytes and were associated with longer patient survival. Notably, an ATM inhibitor selectively potentiated the efficacy of immune checkpoint blockade in ARID1A-depleted tumors but not in WT tumors. Together, these results suggest that ARID1A’s targeting of the nonchromatin substrate Chk2 for ubiquitination makes it possible to selectively modulate cancer cell–intrinsic innate immunity to enhance the antitumor activity of immune checkpoint blockade.
Keywords: Cell Biology, Immunology
Keywords: Cancer, Cancer immunotherapy
Introduction
ARID1A is a component of the evolutionarily conserved chromatin-remodeling complex SWI/SNF, which uses the energy from ATP hydrolysis to relax condensed chromatin structure (1). Data sets from the Cancer Genomic Atlas (TCGA) revealed that ARID1A is one of the most frequently mutated genes in a wide spectrum of human cancers (2). Mutations of ARID1A were found in almost 50% of endometrium-related carcinomas, including uterine endometrioid carcinoma, ovarian clear cell carcinoma, and ovarian endometrioid carcinoma (3–5); approximately 30% of gastric cancers (6); and approximately 30% of bladder cancers (7). The majority of ARID1A mutations are inactivating mutations leading to loss of ARID1A expression, which makes ARID1A a poor therapeutic target. In studies conducted to identify druggable molecular changes induced by ARID1A deficiency, our group and others have shown that ARID1A deficiency reduces mismatch repair (MMR) capacity and is enriched in tumors with microsatellite instability (MSI) (6, 8–10). In syngeneic animal models, impaired MMR in ARID1A-deficient tumors increases tumor mutation load and tumor-infiltrating lymphocytes (TILs), and renders the tumors susceptible to immune checkpoint blockade (ICB) (11). However, although tumors with MMR deficiency or MSI exhibit remarkable clinical responses to ICB, largely due to increased neoantigen presentation (12–14), such tumors have also been found to exhibit both primary and secondary resistance to ICB (15, 16). Indeed, we previously reported that not all ARID1A-deficient tumors exhibited remarkable responses to ICB in syngeneic animal models even when relatively molecularly homogeneous ARID1A-depleted tumors were inoculated into highly genetically homogeneous mouse cohorts (8). Therefore, development of a strategy to increase antitumor immune activity in ARID1A-deficient tumors will be critical for achieving extensive and durable responses of such tumors to ICB.
Neoantigens presented by cancer cells with ARID1A deficiency or MMR deficiency can activate cytotoxic T cells and thus elicit an antitumor adaptive immune response (8, 13). In general, adaptive immunity can be primed, stimulated, and refined by innate immunity. It has been shown that activation of the innate immune response, such as the stimulator of interferon genes (STING) DNA-sensing pathway can prime T cells and drive cytotoxic T cell activity (17–20). Antigen-presenting cells (APCs) such as dendritic cells and macrophages detect DNA from pathogens or dying tumor cells. The cytosolic DNA taken up by APCs can be recognized by the pattern-recognition receptor cyclic GMP-AMP synthase (cGAS) and activate the STING pathway (19). Activation of STING can stimulate these APCs to activate T cells and enhance T cell proliferation and trafficking to the tumor microenvironment (17, 21). Activation of STING also leads to the production of proinflammatory cytokines, including type 1 interferons, and thereby enhances antitumor immunity by promoting the function of natural killer cells and cytotoxic T cells and increasing the survival of memory T cells (17, 22, 23). However, it remains largely unknown whether cancer cell–intrinsic STING signaling instead of systemic STING signaling can be selectively increased to invigorate antitumor immunity in the tumor microenvironment and thus potentiate the therapeutic efficacy of ICB.
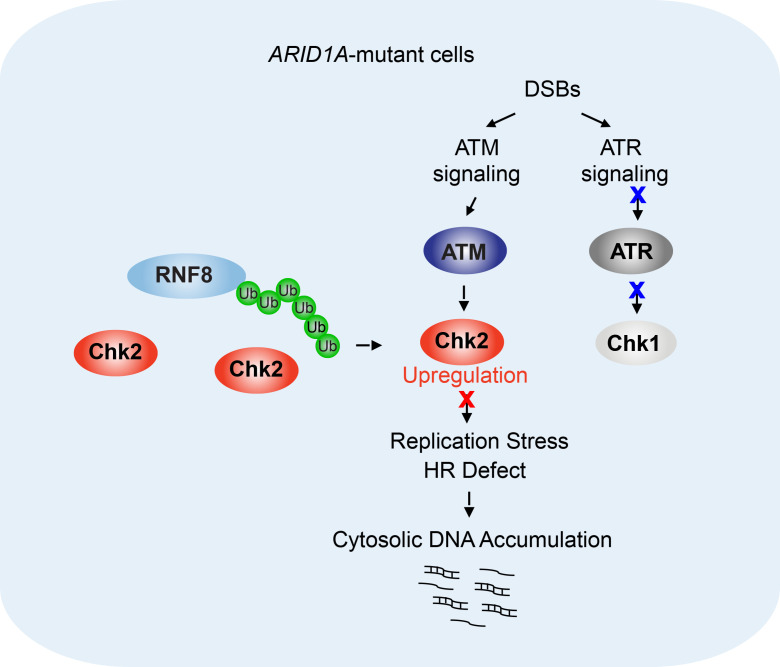
In this study, we identified a chromatin-remodeling–independent function of ARID1A in regulating ubiquitination of CHK2, a key DNA damage checkpoint kinase. Selective inhibition of the DNA damage checkpoint signaling axis ATM/Chk2 triggers cancer cell–intrinsic innate immunity through the STING DNA-sensing pathway and thereby potentiates the therapeutic efficacy of ICB in ARID1A-deficient tumors.
Results
Chk2 expression is increased in ARID1A-deficient cancer cells.
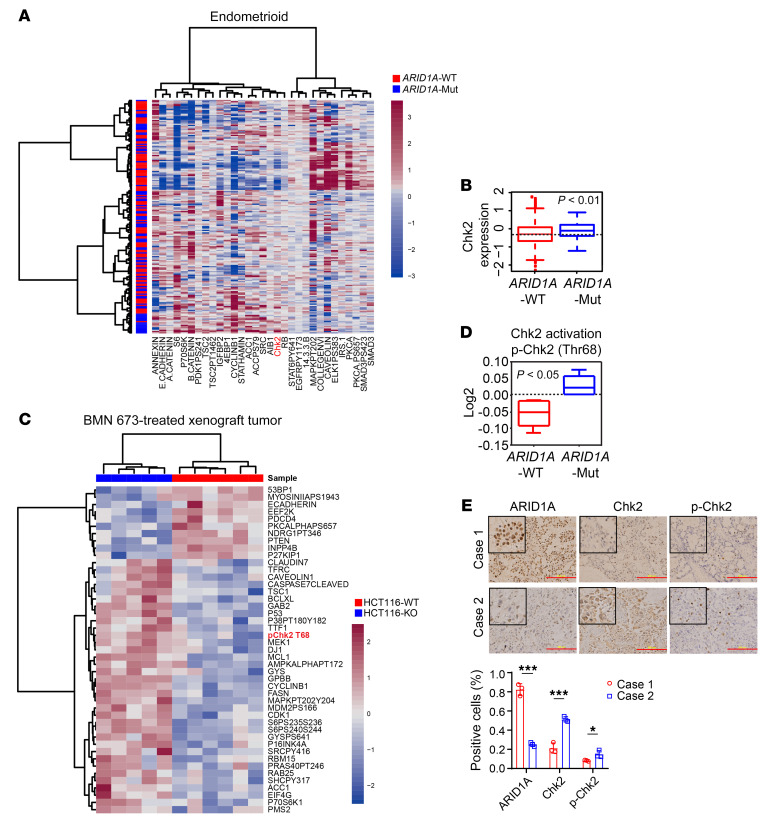
To identify molecules that might be targeted to increase the efficacy of ICB in ARID1A-deficient tumors, we systematically investigated molecular alterations coexisting with ARID1A mutations in tumors. We analyzed the reverse-phase protein array (RPPA) data on endometrioid carcinoma in TCGA databases because of the high frequency of ARID1A mutation in this cancer type. As shown in Figure 1A, we identified a subset of proteins, involved in diverse molecular pathways, whose expression levels differed significantly between ARID1A-WT and ARID1A-mutant tumors. Interestingly, one of these proteins was the DNA damage checkpoint kinase Chk2. The expression level of total Chk2 was significantly higher in ARID1A-mutant tumors than in ARID1A-WT tumors (Figure 1B). Chk2 is phosphorylated and activated in response to DNA damage stimuli and plays a functional role in mediating and regulating DNA damage signaling. To test whether increased expression level of total Chk2 in ARID1A-mutant tumors is associated with a functional increase in Chk2 activation, we conducted RPPA proteomic analysis of WT and ARID1A-depleted xenograft tumors treated with poly (ADP-ribose) polymerase (PARP) inhibitor, a targeted therapeutic drug that induces strong DNA damage signaling (Figure 1C). As expected, the level of phosphorylated Chk2 (Thr68), a marker of Chk2 activation, was remarkably higher in ARID1A-depleted tumors than in the WT control (Figure 1D). This result confirmed that ARID1A-deficient tumors have increased activation of Chk2-mediated signaling in response to DNA damage. Next, to confirm the results from RPPA analysis of TCGA tumors, we performed immunohistochemistry (IHC) staining of specimens from a cohort of patients with clear cell ovarian cancer. Tumor tissues from 8 patients were subdivided into 2 groups on the basis of IHC staining intensity: high ARID1A expression (case 1, at least 50% of cells ARID1A positive) and low ARID1A expression (case 2, less than 50% of cells ARID1A positive). As shown in Figure 1E, tumor samples with low expression of ARID1A exhibited significantly increased expression level of total and phosphorylated Chk2. These data further confirmed the clinical relevance of an enhanced Chk2 signaling pathway in ARID1A-deficient tumors. In addition, we observed increased Chk2 expression in ovarian cancer cell lines with ARID1A mutations that abolished ARID1A expression (Figure 2A). When we restored ARID1A expression in one of these cell lines, Chk2 expression was markedly suppressed, demonstrating that ARID1A regulates Chk2 expression (Figure 2B). Moreover, when we treated ovarian cancer cell lines with ionizing radiation to induce DNA damage, we observed strongly enhanced activation of Chk2 in ARID1A-deficient cancer cells (Figure 2C). Together, these results demonstrated enhanced Chk2 expression and activation in ARID1A-deficient cells.
Figure 1. Chk2 signaling is enhanced in tumors with mutant ARID1A or low expression of ARID1A.
(A) Heatmap representing expression profiles of the 31 proteins most differentially expressed between ARID1A-WT and ARID1A-mutant (ARID1A-Mut) endometrioid carcinomas from patients. P < 0.05 (n = 187). (B) Chk2 protein levels in ARID1A-WT and ARID1A-Mut endometrioid carcinomas from patients. P < 0.01 (n = 187). (C) Heatmap representing RPPA expression profiles of the 45 proteins most differentially expressed between HCT116-WT (n = 6) and HCT116–ARID1A-KO (HCT116-KO) (n = 5) xenograft tumors treated with PARP inhibitor BMN 673. P < 0.05. (D) p-Chk2 (Thr68) protein levels in HCT116-WT (n = 6) and HC116–ARID1A-Mut (n = 5) xenograft tumors treated with BMN 673. P < 0.05. (E) Top, representative images of IHC staining of ARID1A, Chk2, and p-Chk2 in ovarian clear cell carcinoma patient specimens (n = 8). Scale bar: 200 μm. Bottom, quantitative results represent the mean ± SD. *P < 0.05; ***P < 0.001. Two-tailed unpaired Student’s t test (A–E).
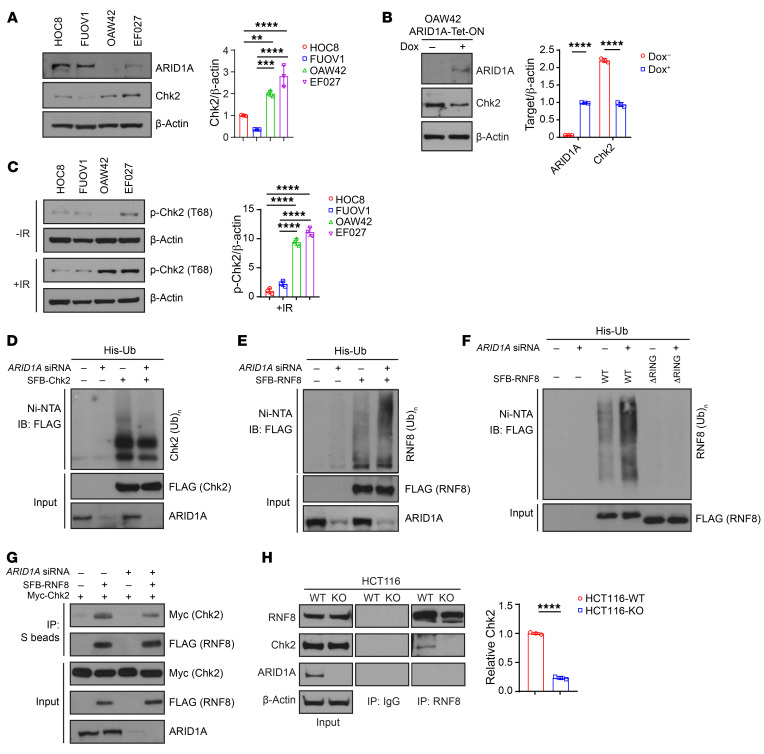
Figure 2. ARID1A regulates E3-ligase RNF8-mediated Chk2 ubiquitination.
(A) Left, Western blots of ARID1A and Chk2 in ARID1A-WT (HOC8 and FUOV1) and -mutant (OAW42 and EF027) ovarian cancer cells. Right, quantitative results represent the mean ± SD from 3 independent experiments. (B) Left, Western blots of ARID1A induction by doxycycline (Dox, 2 μg/mL, 3 days) in ARID1A-null OAW42 cells. Right, quantitative results represent the mean ± SD from 3 independent experiments. (C) Left, Western blots of p-Chk2 (T68) induction by ionizing radiation (IR) (10 Gy) in ARID1A-WT (HOC8 and FUOV1) and -mutant (OAW42 and EF027) ovarian cancer cells. Right, quantitative results represent the mean ± SD from 3 independent experiments. (D) Immunoblot (IB) of U2OS cells transfected with indicated plasmid and siRNA, SFB-tagged (S-tag, Flag epitope tag, and streptavidin-binding peptide tag) Chk2 (SFB-Chk2), si-Nontarget, or siRNA targeting ARID1A along with His-ubiquitin (His-Ub) constructs; Ni–nitrilotriacetic acid (Ni-NTA), nickel bead precipitate. IB, FLAG (immunoblotting by anti-FLAG antibody). (E) Immunoblot of U2OS cells transfected with indicated plasmid and siRNA, SFB-RNF8, si-Nontarget, or siRNA targeting ARID1A along with His-Ub constructs. IB, FLAG. (F) Immunoblot of U2OS cells transfected with indicated plasmid and siRNA, SFB-RNF8, SFB-RNF8 RING domain depletion (ΔRING), si-Nontarget, or siRNA targeting ARID1A along with His-Ub constructs. IB, FLAG. (G) Immunoprecipitation (IP) of SFB-RNF8 with Myc-Chk2 in U2OS cells with si-Nontarget or siRNA targeting ARID1A. (H) Left, coimmunoprecipitation (Co-IP) of endogenous RNF8 and Chk2 in HCT116-WT and ARID1A-KO (HCT116-KO) cells. Right, quantitative analysis from normalization of Chk2 bound by RNF8 represent the mean ± SD from 3 independent experiments. One-way ANOVA with Holm-Šidák’s multiple comparisons test (A and C); 2-tailed unpaired Student’s t test (B and H). **P < 0.01; ***P < 0.001; ****P < 0.0001.
ARID1A regulates RNF8-mediated Chk2 ubiquitination.
We next sought to determine the mechanisms by which ARID1A regulates Chk2. First, we stably knocked down ARID1A in U2OS osteosarcoma cells, a cell model commonly used in DNA damage response and repair studies. Consistent with our observations in ovarian cancer cells, the expression level of total Chk2 was significantly higher in ARID1A-depleted cells (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI130445DS1). Moreover, the induction of p-Chk2 after exposure to ionizing radiation was remarkably higher in ARID1A-depleted cells than in control cells (Supplemental Figure 1A). We then employed quantitative real-time PCR and RNASeq to detect CHK2 mRNA expression in ARID1A-deficient cells. Depletion of ARID1A had no obvious effect on mRNA levels of CHK2, excluding the possibility that CHK2 was regulated at the transcriptional level (Supplemental Figure 1, B and C). Next, we tested whether ARID1A affects the ubiquitination of Chk2, which may contribute to regulation of Chk2 at the protein level. Loss of ARID1A expression reduced the global level of Chk2 ubiquitination (Figure 2D).
To explore how ARID1A regulates Chk2 ubiquitination, we first determined whether ARID1A depletion affects expression of E3-ligase RNF8, which has been reported to specifically induce Chk2 degradation but not Chk1 degradation (24). Neither transient nor stable knockdown of ARID1A changed RNF8 expression (Supplemental Figure 1D). It has been shown that most E3 ligases have the ability to catalyze autoubiquitination, and RNF8 was previously found to autoubiquitinate and degrade itself via its RING domain both in vivo and in vitro (24). Surprisingly, we found that ARID1A knockdown dramatically increased ubiquitination of RNF8 (Figure 2E). Furthermore, loss of the RING domain of RFN8 abolished its increased ubiquitination in ARID1A-depleted cells (Figure 2F), indicating that the increased ubiquitination of RNF8 in ARID1A-depleted cells was mainly dependent on autoubiquitination activity of RNF8 mediated by its RING domain. Depletion of ARID1A did not affect RNF8 protein level (Supplemental Figure 1D), suggesting that ubiquitination of RNF8 did not target itself for degradation. Previous studies demonstrated that linkage of polyubiquitin chain through Lys48 (UbK48) is mainly involved in targeting proteins for degradation. In contrast, linkage through Lys63 (UbK63) plays a role in nonproteolytic signaling, especially in DNA damage response (25, 26). Our data further showed that only K63 ubiquitination of RNF8, not K48 ubiquitination, was increased in ARID1A-deficient cells (Supplemental Figure 1E), confirming the observation that ARID1A deficiency did not affect RNF8 protein level (Supplemental Figure 1D). To further demonstrate the increased Chk2 protein degradation in ARID1A-WT cells, we treated 2 ovarian cancer cell lines (HOC8 and FUOV1, ARID1A-WT cells) with MG132. These 2 cell lines contain proficient ARID1A expression and low Chk2 expression. As expected, the total level of Chk2 was significantly increased by MG132 treatment (Supplemental Figure 2A). We then treated ovarian cancer cell lines HOC8 (ARID1A-WT) and OAW42 (lack of ARID1A expression due to mutation) with cycloheximide (CHX) to assess the half-life of Chk2 protein. As shown in Supplemental Figure 2B, the basal level of Chk2 was higher in ARID1A-deficient OAW42 cells compared with HOC8 cells. In the presence of CHX, ARID1A-deficient OAW42 cells exhibited delayed Chk2 degradation compared with HOC8 cells, indicating a prolonged Chk2 half-life. Collectively, these data showed that ARID1A mutation status has a clear impact on the degradation of Chk2.
Next, we determined how ARID1A might regulate RNF8-mediated Chk2 ubiquitination. We first used 2 full-length constructs, Myc-Chk2 and FLAG-RNF8, to test whether the interaction between RNF8 and Chk2 was affected by loss of ARID1A. As shown in Figure 2G, E3 ligase RNF8 can bind to its substrate Chk2 directly. Interestingly, the interaction between RNF8 and Chk2 was compromised in ARID1A-depleted cells. To further confirm this finding, we used isogenic HCT116 cell lines with ARID1A-WT and ARID1A depletion to test the endogenous interaction between RNF8 and Chk2. Likewise, the capacity of RNF8 to bind Chk2 was partially reduced in ARID1A knockout (KO) cells (Figure 2H). Collectively, these data suggested that the increased K63 ubiquitination of RNF8 in ARID1A-deficient cells may partially block the interaction between RNF8 and Chk2, thereby impairing ubiquitination of Chk2 and preventing Chk2 degradation.
To gain molecular insights into Chk2 ubiquitination, we used a bioinformatic approach to predict the possible protein ubiquitination sites on Chk2 by using Ubpred (27). Three sites (K492, K494, and K520) were identified with medium confidence and one site (K534) was identified with high confidence. Based on the prediction, we conducted site-directed mutagenesis and generated mutant Chk2 construct with these 4 potential sites by replacing K (lysine) with R (arginine) (Supplemental Figure 3A). As shown in Supplemental Figure 3B, the mutant Chk2 construct exhibited significantly impaired polyubiquitination compared with the WT Chk2 construct. This result indicated the requirement of these 4 lysine sites (K492, K494, K520, and K534) for Chk2 polyubiquitination.
Inhibition of the ATM/Chk2 axis augments replication stress and triggers cancer cell–intrinsic innate immune response in ARID1A-deficient cells.
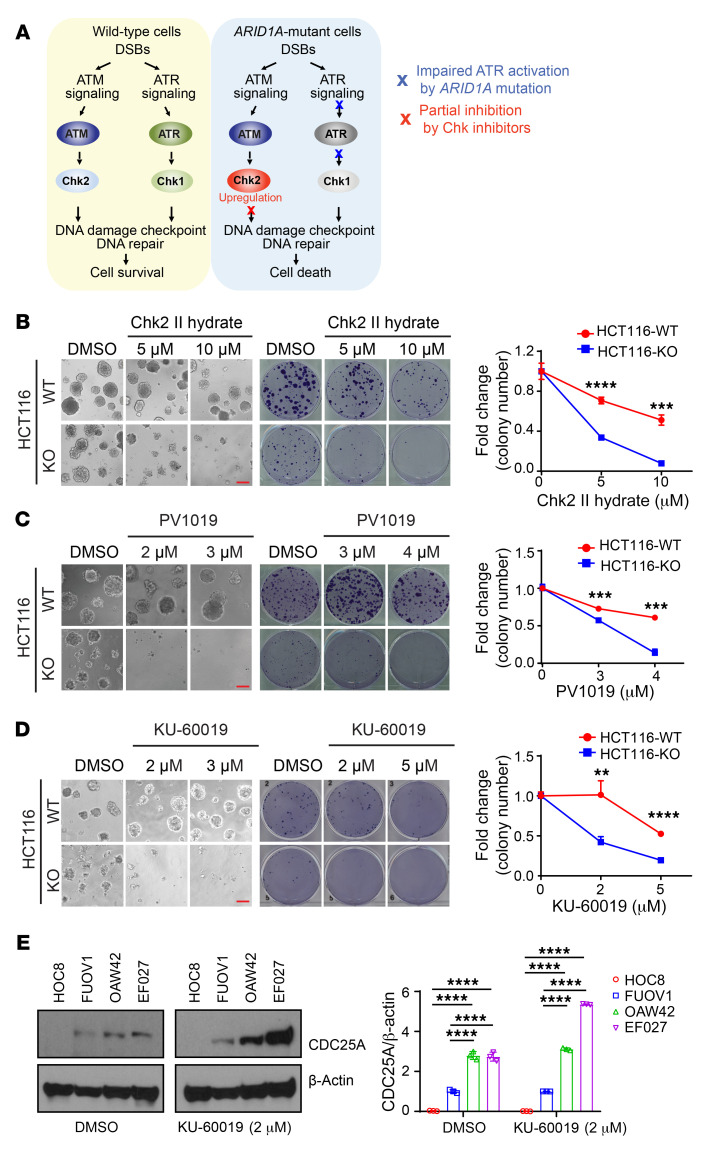
In response to DNA damage, WT cells have 2 coordinated and compensatory pathways, one mediated by the ATM and Chk2 kinases and the other by the ATR and Chk1 kinases, to properly control DNA damage response and DNA repair, which are required for cell survival (refs. 28, 29 and Figure 3A). We previously reported that in ARID1A-deficient cancer cells, activation of ATR in response to DNA damage is impaired, which leads to a weak ATR/Chk1 signaling axis (30). Consistent with this observation, we observed that phosphorylation of Chk1, a downstream kinase activated by ATR, was markedly reduced in ARID1A-deficient ovarian cancer cells (Supplemental Figure 4A). It has also been reported that ARID1A-deficient cancer cells were sensitive to further chemical ablation of ATR kinase activity (31). On the basis of these observations and our observation of enhanced Chk2 signaling in ARID1A-deficient tumors (Figure 1, A–E, and Figure 2, A–C), we reasoned that an enhanced ATM/Chk2 axis may play a crucial role in allowing ARID1A-deficient cells to cope with DNA damage. Findings supporting this hypothesis would suggest that the ATM/Chk2 axis could be a potential target to reduce DNA damage tolerance in ARID1A-deficient tumors.
Figure 3. Inhibition of the ATM/Chk2 axis selectively inhibits ARDI1A-deficient cancer cell growth.
(A) Schematic diagram of DNA damage response signaling in ARID1A-WT and ARID1A-deficient cells. (B–D) ARID1A-WT and ARID1A-KO HCT116 cells treated with the Chk2 inhibitors II hydrate (B) and PV1019 (C) and the ATM inhibitor KU-60019 (D) at indicated concentrations. Clonogenic assays were performed. Left, representative images of 3D culture. Scale bar: 100 μm. Middle, representative images of colony formation. Right, quantitative results represent the mean ± SD from 3 independent experiments from colony formation. **P < 0.01; ***P < 0.001; ****P < 0.0001 by 2-tailed unpaired Student’s t test. (E) Expression of the basal level of CDC25A in ARID1A-WT (HOC8 and FUOV1) and -mutant (OAW42 and EF027) ovarian cancer cells. Left, cells were treated with ATM inhibitor KU-60019 for 48 hours and the whole-cell lysis was subjected to Western blot analysis. Right, quantitative results represent the mean ± SD from 3 independent experiments. ****P < 0.0001 by 1-way ANOVA with Holm-Šidák’s multiple comparisons test.
To test our hypothesis, we treated isogenic parental and ARID1A-KO HCT116 cell lines with 2 different Chk2 inhibitors, II hydrate and PV1019. In 3D culture, ARID1A-deficient cancer cells and ARID1A-WT cells exhibited very similar colony formation capacities in the absence of Chk2 inhibitors (Figure 3, B and C). Interestingly, ARID1A-deficient cancer cells exhibited increased sensitivity to Chk2 inhibition (Figure 3, B and C). In contrast, at the same concentrations, neither Chk2 inhibitor had an obvious inhibitory effect on parental cells. Similarly, both Chk2 inhibitors selectively targeted ARID1A-KO cells by 2D colony formation assay (Figure 3, B and C, and Supplemental Figure 4B). As expected, consistent with the findings with Chk2 inhibitors, ATM inhibitor KU-60019 selectively repressed survival of ARID1A-depleted cells in both 3D and 2D culture (Figure 3D and Supplemental Figure 4C). To further confirm a compensatory requirement for ATM/Chk2 in ARID1A-deficient cells due to the perturbation of ATR/Chk1 signaling, we examined the level of CDC25A protein, a downstream molecule regulated by both ATM/Chk2 and ATR/Chk1 signaling. As shown in Figure 3E, the basal expression level of CDC25A was increased in cells with ARID1A deficiency (OAW42 and EF027 cell lines). This observation is consistent with previous reports that Chk1 regulates CDC25A degradation and the impaired ATR/Chk1 signaling in ARID1A-deficient cells results in changes in CDC25A levels (30, 32). In the absence of exogenous DNA damage stimuli, ATM inhibitor (KU-60019) significantly increased the basal CDC25A protein level in ARID1A-deficient cells, suggesting an increased dependency of ATM/Chk2 signaling in regulating CDC25A protein levels in ARID1A-deficient cells potentially due to the ATR/Chk1 signaling pathway being compromised. Collectively, these data suggested that inhibition of the ATM/Chk2 axis may impair the capacity of ARID1A-deficient cells to control endogenous DNA damage and thereby reduce cell survival.
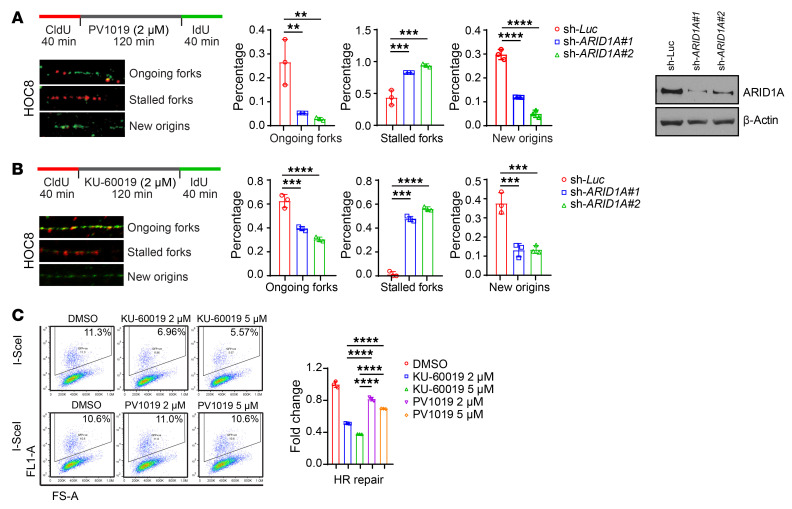
DNA replication is a major generator of endogenous DNA damage. Thus, we visualized DNA fibers to monitor DNA replication perturbation at single-molecule resolution. Both Chk2 inhibitor PV1019 and ATM inhibitor KU-60019 selectively reduced the number of ongoing replication forks and new origins in ARID1A-depleted cells (Figure 4, A and B). Moreover, in the presence of Chk2 or ATM inhibitor the number of stalled replication forks was significantly increased in ARID1A-knockdown cells compared with the control cells (Figure 4, A and B). These data indicated that inhibition of the ATM/Chk2 axis can selectively enhance DNA replication stress in ARID1A-deficient cells. Stalled/collapsed replication forks resulting from replication stress generate endogenous DNA damage, predominantly in the form of replication-associated double-strand DNA breaks. Homologous recombination (HR) is an essential mechanism used by cells to repair and resolve this type of DNA damage. ATM is required for processing double-strand DNA break ends and promoting efficient HR repair, while Chk2 mainly functions in cell-cycle checkpoint. In line with previous findings, we found that ATM inhibitor significantly suppressed HR repair capacity, whereas Chk2 inhibitor modestly reduced HR repair capacity (Figure 4C and Supplemental Figure 4D). These results raised the possibility that inhibition of the ATM/Chk2 axis may lead to accumulation of endogenous DNA damage in ARID1A-deficient cancer cells due to enhanced replication stress and reduced DNA repair capacity. Furthermore, the differential effects of ATM inhibitor and Chk2 inhibitor on HR repair suggested that the 2 types of inhibitor may lead to different levels of accumulation of unresolved endogenous DNA damage in ARID1A-deficient cells.
Figure 4. Inhibition of the ATM/Chk2 axis selectively enhances replication stress in ARID1A-deficient cancer cells.
(A and B) Left, representative images of DNA fiber assay in control (sh-Luc) and ARID1A-depleted (sh-ARID1A#1 and #2) HOC8 cells treated with PV1019 (A) and KU-60019 (B) at indicated concentrations. Middle, quantitative results represent the mean ± SD from 3 independent experiments. **P < 0.01; ***P < 0.001; ****P < 0.0001 by 1-way ANOVA with Dunnett’s multiple comparisons test. Right, Western blots indicate effective ARID1A knockdown. (C) Analysis of homologous recombination (HR) efficiency with the DR-green fluorescent protein (GFP) assay. Left, representative flow cytometry profiles. Right, values are normalized to the percentage of GFP-positive cells in I-SceI–transfected cells without treatment and represent the mean ± SD of 3 independent experiments. ****P < 0.0001 1-way ANOVA with Holm-Šidák’s multiple comparisons test.
When we examined accumulation of unrepaired/unresolved endogenous DNA damage, we were surprised to find higher cytosolic DNA accumulation in ARID1A-knockdown and -KO cells than in control cells (Figure 5, A and B, and Supplemental Figure 5A). Consistent with the differential effects of ATM inhibitor and Chk2 inhibitor on HR repair, ATM inhibitor caused a higher level of cytosolic DNA accumulation compared with Chk2 inhibitor in ARID1A-deficient cancer cells (Figure 5, A and B).
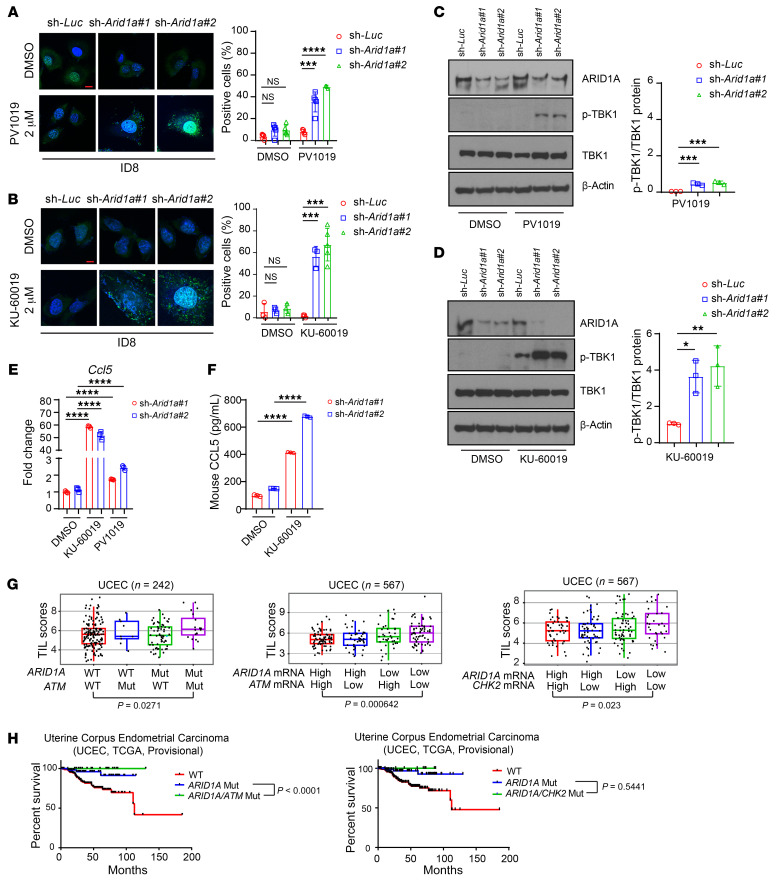
Figure 5. Inhibition of the ATM/Chk2 axis stimulates cytosolic DNA accumulation and promotes immune responsiveness in ARID1A-deficient tumors.
(A and B) Left, representative images of PicoGreen staining in control (sh-Luc) and Arid1a-depleted (sh-Arid1a#1 and #2) ID8 cells treated with DMSO, PV1019 (2 μM) (A), or KU-60019 (2 μM) (B) for 48 hours. DAPI (blue) was used to visualize the nuclei. Scale bar: 10 μm. Right, quantitative results represent the mean ± SD of 3 independent experiments. ***P < 0.001; ****P < 0.0001. (C and D) Left, Western blots of phosphorylated TBK1 (p-TBK1) and total TBK1 (TBK1) in ID8 cells treated with PV1019 (C) or KU-60019 (D) for 48 hours. Right, quantitative data represent the mean ± SD from 3 independent experiments. *P < 0.05; **P < 0.01, ***P < 0.001. (E) qPCR analysis of Ccl5 mRNA expression in ARID1A knockdown ID8 cells under DMSO, KU-60019, or PV1019 treatment. Data represent mean ± SD of 3 independent experiments. ****P < 0.0001. (F) ELISA quantification of mouse CCL5 level in ARID1A knockdown ID8 cells treated with DMSO or KU-60019. Data represent mean ± SD of 3 independent experiments. ****P < 0.0001. (G) Association of TILs with mutation and expression of ARID1A, ATM, and CHK2 in UCEC patient samples as analyzed by TIL signatures. The box plot represents median and quantiles of the data. UCEC mutation data set: n = 242, ARID1A/ATM WT/WT vs. Mut/Mut, P = 0.0271; UCEC expression data set: n = 567, ARID1A/ATM high/high vs. low/low, P = 0.000642; ARID1A/CHK2 high/high vs. low/low, P = 0.023. (H) Survival analysis for UCEC patients with ARID1A, ATM, and CHK2 mutation (Mut). Left, comutation of ARID1A and ATM. Right, comutation of ARID1A and CHK2. UCEC (n = 239): ARID1A/ATM Mut (n = 19) vs. ARID1A Mut (n = 63), P < 0.0001; ARID1A/CHK2 Mut (n = 9) vs. ARID1A Mut (n = 74), P = 0.5441. One-way ANOVA with Dunnett’s multiple comparisons test (A–D); 1-way ANOVA with Holm-Šidák’s multiple comparisons test (E and F); 2-tailed unpaired Student’s t test (G and H).
Previous studies have shown that cytosolic DNA can be recognized by the STING DNA-sensing pathway, which induces phosphorylation and nuclear translocation of interferon transcriptional regulatory factor TANK-binding kinase 1 (TBK1) and leads to production of proinflammatory cytokines, including CCL5 and CXCL10 (20, 33). As expected, we found that ATM inhibitor triggered TBK1 phosphorylation more efficiently than Chk2 inhibitor did (Figure 5, C and D, and Supplemental Figure 5B). Furthermore, KU-60019 treatment significantly increased CCL5 expression, but PV1019 only modestly increased CCL5 expression (Figure 5, E and F).
To further explore the nature of accumulated cytosolic DNA, first we examined whether the nature of cytosolic DNA induced by ATM inhibitor in ARID1A-deficient cells is dsDNA or ssDNA by using specific antibodies for immunostaining. As shown in Supplemental Figure 5, C and D, ATM inhibitor induced the accumulation of both dsDNA and ssDNA in ARID1A-deficient cells. Second, we tested whether the accumulation of cytosolic DNA is dependent on the DNA replication process, as we observed that ATM inhibitor caused elevated replication stress and reduced repair of replication-associated DNA damage. To this end, we used hydroxyurea (HU) as the tool drug, which is known to deplete dNTPs and lead extensively stalled or collapsed replication forks (34). As shown in Supplemental Figure 5, E and F, HU treatment promoted cytosolic DNA and the production of proinflammatory cytokines, including CCL5 and CXCL10, similarly to ATM inhibitor. Furthermore, ATM inhibitor synergized HU to promote higher levels of CCL5 and CXCL10 compared with either single treatment. This result suggested the contribution of replication stress underlying the accumulation of cytosolic DNA induced by ATM inhibitor. Third, we confirmed that the cytosolic DNA arises from stalled or collapsed replication forks by using BrdU-pulsed labeling. BrdU is incorporated into the newly synthesized DNA in S-phase cells. We then costained BrdU and cytosolic DNA in ARID1A-knockdown cells following ATM inhibitor treatment. As we expected, we observed that BrdU-labeled DNA partially overlapped with cytosolic DNA staining (Supplemental Figure 5G). These results indicated that S-phase replication stress is a potential resource of cytosolic DNA in ARID1A-deficient cells treated with ATM inhibitor.
Collectively, these data demonstrated that inhibition of the ATM/Chk2 axis in ARID1A-deficient cells enhances replication stress, which leads to accumulation of cytosolic DNA and in turn activates the STING DNA-sensing pathway, a component of the innate immune response.
Impaired ATM/Chk2 axis increases TILs and predicts better prognosis in ARID1A-deficient tumors.
To determine the clinical relevance of the ATM/Chk2 axis in regulating immune responsiveness in ARID1A-mutated tumors, we analyzed TIL signatures in a TCGA data set. As shown in Supplemental Figure 6A, ovarian tumors with ARID1A mutations or with ATM mutations exhibited enhanced T cell cytotoxic and STING signatures. Interestingly, comutation of ATM with ARID1A significantly promoted TILs in a cohort of 242 uterine corpus endometrial carcinoma (UCEC) tumors (Figure 5G, left). Furthermore, analysis from the TCGA data set containing 567 UCEC tumors showed that tumors with low mRNA expression of both ARID1A and ATM exhibited a significantly increased number of TILs (Figure 5G, middle). As expected given that Chk2 is a substrate of ATM, tumors with low mRNA expression of both ARID1A and CHK2 also exhibited a significantly increased number of TILs (Figure 5G, right). These data from TCGA analysis were in accordance with our observation that inhibition of the ATM/Chk2 axis in ARID1A-deficient cancer cells induces endogenous DNA damage, stimulates the STING DNA-sensing pathway, and thus promotes immune responsiveness. Notably, in cohorts of patients with UCEC and esophagus-stomach cancers, a better prognosis was observed for patients whose tumors had comutations of ARID1A and ATM or comutations of ARID1A and CHK2 than for patients whose tumors had only ARID1A mutation (Figure 5H and Supplemental Figure 6B). On the basis of these results, we next determined whether ATM inhibitor may potentiate the therapeutic efficacy of ICB in ARID1A-deficient tumors.
ATM inhibition enhances the therapeutic efficacy of ICB in ARID1A-deficient tumors.
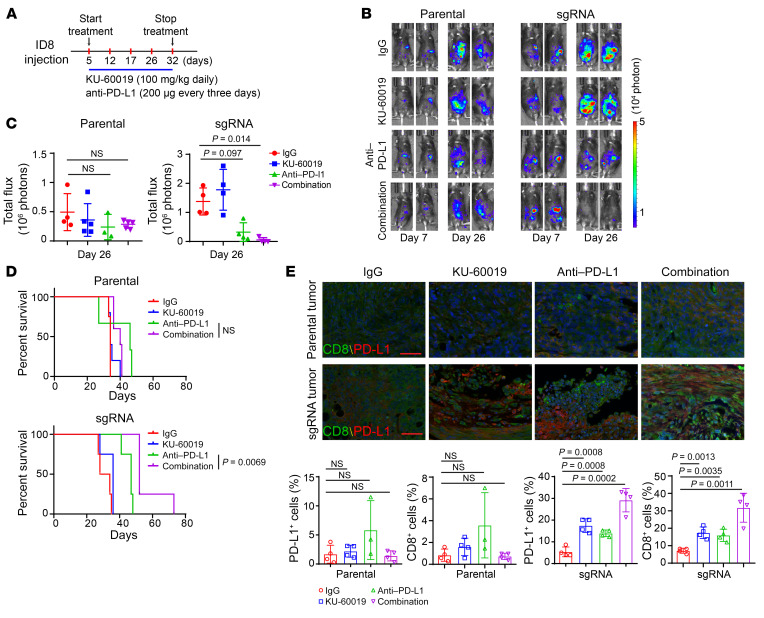
To determine whether ATM inhibitor potentiates the therapeutic efficacy of ICB in ARID1A-deficient tumors, we used luciferase-tagged ID8 ovarian cancer syngeneic mouse models. Beginning 5 days after tumor cell injection, mice were treated with anti–PD-L1 antibody every 3 days and ATM inhibitor daily (Figure 6A). The treatment was given for 28 days. Parental tumors did not respond to either monotherapy or combination therapy (Figure 6, B and C). In contrast, ARID1A-deficient tumors showed increased sensitivity to anti–PD-L1 treatment, as we previously reported (Figure 6, B and C, and ref. 8). As we expected, ATM inhibitor in combination with anti–PD-L1 treatment remarkably reduced tumor burden. More importantly, ATM inhibitor in combination with anti–PD-L1 treatment significantly prolonged the survival of mice bearing ARID1A-deficient tumors but not mice bearing control tumors (Figure 6, B–D). Furthermore, IHC analysis revealed that the combination therapy significantly increased the number of cells positive for CD8, a key cytotoxic T lymphocyte marker, and PD-L1, a key molecule in the immune checkpoint pathway, in ARID1A-depleted tumors compared with either monotherapy (Figure 6E). Collectively, these data indicated that the ATM/Chk2 DNA damage checkpoint axis could be a promising target to activate immune responsiveness and thus potentiate the efficacy of ICB in patients with ARID1A-deficient tumors.
Figure 6. ATM inhibition enhances the therapeutic efficacy of ICB in ARID1A-deficient tumors.
(A) Schematic of isotype control IgG, KU-60019, and anti–PD-L1 antibody treatment. Treatments were started on day 5 after inoculation and stopped on day 32. (B) Representative images for bioluminescence of mice with i.p. ID8 tumors on day 7 and day 26. Left, parental ID8 tumors. Right, ARID1A-depleted (sgRNA) ID8 i.p. tumors. (C) Endpoint of bioluminescence in mice bearing parental and ARID1A-depleted (sgRNA) ID8 i.p. tumors. Parental: IgG vs. anti–PD-L1, not significant; IgG vs. combination, not significant. sgRNA: IgG vs. anti–PD-L1, P = 0.097; IgG vs. combination, P = 0.014. Data represent mean ± SD (n = 3–5). (D) Survival curves of mice with ID8 i.p. tumors. Top, parental ID8 tumors. Bottom, ARID1A-depleted (sgRNA) ID8 tumors. Parental: anti–PD-L1 vs. combination, not significant; sgRNA: anti–PD-L1 vs. combination, P = 0.0069. (E) Top, CD8 and PD-L1 fluorescence-based IHC staining in parental and ARID1A-depleted ID8 i.p. tumors (n = 3 or 4). Bottom, quantitative analysis represent mean ± SD with indicated P value. +, positive. Scale bar: 50 μm. Two-tailed unpaired Student’s t test (C and E); log-rank (Mantel-Cox) test (D).
Discussion
Our findings from this study support the model outlined in Supplemental Figure 7. We found that loss of ARID1A leads to an increased level of Chk2 and activation due to disrupted RNF8-mediated Chk2 ubiquitination and degradation. Furthermore, inhibition of the ATM/Chk2 axis in the DNA damage response pathway selectively promotes the accumulation of cytosolic DNA due to elevated replication stress and reduced repair of replication-associated DNA damage in ARID1A-deficient tumors, potentially due to impaired ATR/Chk1 axis. As a consequence, the STING DNA-sensing pathway is activated and stimulates type I interferon signaling. Consistent with these observations, tumors from cancer patients with ARID1A and ATM mutations or low expression exhibited an increased number of TILs and were associated with a better prognosis compared with tumors with only ARID1A mutations or low expression. Notably, treatment with the combination of an ATM inhibitor and anti–PD-L1 antibody reduced tumor burden and prolonged survival compared with either monotherapy in mice bearing ARID1A-deficient ovarian tumors but not in mice bearing Arid1a-WT ovarian tumors.
ARID1A is a component of the chromatin-remodeling complex SWI/SNF, a large protein complex containing 12 subunits (35). The SWI/SNF complex has an evolutionarily conserved role in chromatin remodeling from yeast to mouse and human and regulates chromatin transaction activities, including gene transcription, DNA replication, and DNA damage response/repair (36). KO of different SWI/SNF subunits in mouse models produces different phenotypes (37). Consistent with these observations, mutations in subunits of the SWI/SNF complex have been found to occur in approximately 20% of all human cancers (38). Mutations of different subunits are enriched in distinct cancer types and are associated with unique pathological changes. For example, in ovarian cancers, mutations of ARID1A (BAF250) are found in almost 50% of endometrium-related carcinomas, including uterine endometrioid carcinoma, ovarian clear cell carcinoma, and ovarian endometrioid carcinoma (3–5). In contrast, mutations of BRG1 (SMARCA4), a catalytic subunit of the SWI/SNF complex, are found in almost 100% of small cell carcinomas of the ovary (39, 40). It remains largely unknown how the impaired function of individual subunits of the SWI/SNF complex leads to such discrete phenotypes in mouse models and human cancers, even though impaired function in any subunit can disrupt the chromatin-remodeling activity of SWI/SNF. One explanation is that different subunits of SWI/SNF are essential for the chromatin-remodeling function of the SWI/SNF complex in different cellular and molecular contexts. To our surprise, we identified a novel role of ARID1A in regulating ubiquitination of Chk2, a checkpoint kinase, which is not one of the conventional chromatin substrates of the SWI/SNF complex, which include DNA, histones, and nucleosomes. Studies in yeast have shown that the chromatin-remodeling complexes SWI/SNF and INO80 can target nonchromatin substrates to stimulate activity of DNA damage checkpoint kinases Mec1 (ATR in mammals) and Rad53 (Chk2 in mammals), respectively (41, 42). In line with these findings, our study suggested that targeting nonchromatin substrates such as Chk2 and nonchromatin transactions such as protein ubiquitination may provide a new mechanistic basis to explain the diverse molecular and pathological changes caused by mutations in subunits of the SWI/SNF complex in human cancers. It is worth noting that the increased levels of Chk2 in the HCT116-derived ARID1A-KO xenografts (Figure 1C) did not seem apparent when the cells were grown in culture (Figure 2H). This could reflect a dependence of Chk2 on growth rate. In our previous study, we observed that HCT116 ARID1A-KO cells grew slower than ARID1A-WT cells in culture dish. In contrast, in vivo HCT116-derived ARID1A-KO xenografts showed larger tumors when compared with HCT116-WT xenografts (30). Chk2 plays an essential role in regulating cell-cycle G1-S-G2/M phases checkpoints (43), which underlies the regulation between Chk2 and the growth rate. HCT116 ARID1A-KO cells in culture exhibited a much slower growth rate. It is possible that additional mechanisms might be activated in ARID1A-deficient cells to reduce the Chk2 level and thus allow cells to progress in cell cycles.
A previous study showed that E3-ligase RNF8 can induce K48-linked ubiquitination chains on chromatin, which promote substrate degradation and regulate the abundance of DNA damage response and DNA repair proteins at DNA damage sites (24). However, it remains unclear how RNF8-mediated ubiquitination and protein degradation is regulated. Our study showed that ARID1A may regulate the function of RNF8 through RNF8 autoubiquitination and thus alter RNF8’s substrate targeting capacity. Our data suggest that chromatin-remodeling proteins residing on chromatin, such as the ARID1A-SWI/SNF complex, may, in addition to their well-established role in regulating DNA-histone contact, have additional functions that impact protein-protein interactions, such as the interaction between E3-ligase RNF8 and its substrate Chk2.
There are several possible mechanisms underlying how ARID1A mediates RNF8 autoubiquitination. First, the ubiquitin ligase RNF8 recruits ubiquitin-conjugating enzymes such as the heterodimeric E2 enzyme UBC13-UEV, which promotes K63-linked polyubiquitination (44, 45). It is possible that ARID1A interacts with the RNF8-UBC13-UEV ubiquitination complex and inhibits RNF8 autoubiquitination by shielding RNF8 from the UBC13-UEV heterodimeric E2 enzyme. Loss of ARID1A thus increases RNF8 autoubiquitination. Second, loss of ARID1A may cause protein conformational changes of the RNF8-UBC13-UEV ubiquitination complex. These conformational changes may localize the protein domains of RNF8 to the proximity of the E2 enzyme and promote RNF8 autoubiquitination. Third, autoubiquitination may occur intramolecularly or intermolecularly. It is possible that loss of ARID1A may facilitate accessibility of intramolecular and/or intermolecular RNF8 protein domains to the RNF8-UBC13-UEV ubiquitination complex. Finally, a study in the budding yeasts showed that the chromatin-remodeling complex SWI/SNF, of which ARID1A is a component, regulates activation of the DNA damage checkpoint kinase Mec1 (ATR in mammals) through direct interaction between Snf2 ATPase, the catalytic core component of SWI/SNF, and Mec1 (42). The ATP-dependent SWI/SN complex had previously been proposed to use nucleosomes as substrates to alter DNA-protein interactions. This study suggested a new role of the chromatin-remodeling complex in regulating protein-protein interactions in the absence of chromatin substrates (DNA-protein). It also remains to be determined whether the subunit requirement of SWI/SNF-mediated ubiquitination regulation differs from that of SWI/SNF-mediated chromatin remodeling or whether the chromatin-remodeling activity is required for the ubiquitination regulation by the SWI/SNF complex.
Our study revealed that inhibition of the ATM/Chk2 axis stimulates cancer cell–intrinsic innate immune response mediated by the SING pathway in ARID1A-deficient tumors. This finding serves as a mechanistic basis for combining ATM inhibitor with anti–PD-L1 to enhance therapeutic responses in ARID1A-deficient tumors. Recent studies have shown that the therapeutic efficacy of IBC requires cytotoxic T cell activation and infiltration into tumors (46). Preclinical studies have shown that activation of the STING pathway increases TILs and thereby leads to increased T cell priming, activation, and survival of memory T cells and an inflamed tumor microenvironment (18, 47, 48). As expected, STING agonists have recently been shown to elicit strong antitumor activity (49). Furthermore, STING activation and ICB have also exhibited synergistic promotion of antitumor immunity in mouse models (50–52). However, these studies using STING agonists aim to amplify the signaling of the STING pathway by boosting its general activation. This approach to STING activation lacks selectivity against tumor cells. Uncontrolled or excessive activation of the STING pathway can induce autoimmune responses in normal cells and result in tissue damage and autoimmune disease (33). To gain targeting specificity, STING agonists are commonly administrated to tumors by localized intratumor injections, which may not be easily applied in the clinic (53, 54). In addition to being activated by administration of STING agonists, STING can be activated by endogenous DNA damage generated from radiation therapy and mitotic progression (17–20, 55–59).
In our study, instead of directly inducing DNA damage, we selectively activated a cancer cell–intrinsic STING response in tumors by targeting their molecular vulnerabilities conferred by altered DNA damage checkpoint signaling. ARID1A-deficient cancer cells exhibit an enhanced ATM/Chk2 axis in DNA damage response, which may provide an adaptive mechanism to compensate for the impaired signaling from the ATR/Chk1 axis. As expected, in this specific molecular context, we found that inhibition of ATM/Chk2 selectively led to the accumulation of cytosolic DNA and increased immune responsiveness, and subsequently enhanced the therapeutic efficacy of ICB in ARID1A-deficient tumors. In contrast, fewer effects were observed in control tumors. Our study provides a proof-of-principle example of genetic context–dependent targeting of DNA damage responses as a means to selectively promote innate immune response. Thus, results from our study elucidate a mechanistic basis for developing rational combination therapy to promote the efficacy of ICB beyond increased mutation burden and antigenicity in ARID1A-deficient tumors, or more broadly in MMR-deficient tumors, by modulating complementary innate immune responses.
The ATM/Chk2 kinase signaling axis is the central regulator of the DNA damage response, particularly in response to double-strand DNA breaks (60). Thus, ATM and Chk2 are logical therapeutic targets for developing inhibitors to block DNA damage response when given as monotherapies or in combination with genotoxic agents (61). However, owing to low therapeutic efficacy or toxic effects, no inhibitors of ATM or Chk2 have progressed in the clinic (61). It has been proposed that the genetic context of the tumor is important for the efficacy of DNA damage response inhibitors. Cancer cells with p53 deficiency showed increased sensitivity to the inhibition of CHK2 (62). One patient whose small cell lung cancer exhibited RAD50 mutation and ATM deficiency achieved a complete response to the combination of an inhibitor targeting both Chk1 and Chk2 and irinotecan (63). These data suggest that inhibition of the ATM/Chk2 axis, or more broadly inhibitors of DNA damage response, may require synergistic association with defects in DNA damage checkpoint and repair pathways to achieve optimal immune-modulating effects. Indeed, our study demonstrated that ARID1A-deficient cancer cells were more sensitive than ARID1A-WT cancer cells were to ATM or Chk2 inhibitors in the in vitro experiments and exhibited a stronger innate immune response, possibly because of their increased dependence on the ATM/Chk2 axis resulting from weakened ATR/Chk1 signaling. Notably, in our in vivo syngeneic mouse models, ATM inhibitor did not significantly cause cytotoxic effects to inhibit tumor growth. It is possible that the dosage we used for our in vitro experiment was not achieved in vivo at the concentration of ATM inhibitor we gave to mice. Interestingly, the addition of ATM inhibitor at the current dosage to anti–PD-L1 treatment enhanced therapeutic responses to anti–PD-L1. These data raise an important consideration regarding the clinical applications of ATM/Chk2 inhibitors or DNA damage response inhibitors: the dosages needed for these drugs to modulate immune responses in tumors may be less than the doses needed to inhibit or kill tumor cells. Our data suggest that the combination of ICB with these inhibitors at their suboptimal dosages, which do not cause significant cytotoxic effects, may be sufficient to elicit cancer cell–intrinsic innate immunity and thus enhance the antitumor efficacy of ICB. In summary, our study may reveal a new strategy for using inhibitors of the ATM/Chk2 axis or more broadly DNA damage response as immune-modulating agents in tumors with a defined genetic background, such as ARID1A-deficient tumors, to overcome the current clinical challenges of ICB, including toxic effects and low efficacy.
Methods
Cell culture.
U2OS cell was purchased from the American Type Culture Collection (ATCC). Colon cancer HCT116 WT and ARID1A-KO (Q456*/Q456*) cell lines were purchased from Horizon Discovery Ltd. and maintained according to the manufacturer’s instructions. Ovarian cancer HOC8, FUOV1, OAW42, and EF027 cells were provided by Gordon Mills’s laboratory (MD Anderson Cancer Center, Houston, Texas, USA). U2OS and HCT116 cells were cultured in McCoy’s 5A medium (Cellgro). Ovarian cancer cells were cultured in RPMI 1640 medium (Cellgro). All media were supplemented with 10% FBS with glutamine, penicillin, and streptomycin. ID8 mouse ovarian surface epithelial cells were provided by Vahid Afshar-Kharghan’s laboratory (MD Anderson Cancer Center, Houston, Texas, USA) and maintained in DMEM (high glucose, Cellgro) supplemented with 4% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 5 μg/mL insulin, 5 μg/mL transferrin, and 5 ng/mL sodium selenite. Cells were incubated in a 37°C humidified incubator with 5% CO2.
Antibodies, reagents, and plasmids.
Anti-Myc (catalog sc-40), -RNF8 (catalog sc-271462) (for IP), and -CD8 (catalog sc-7970) antibodies were purchased from Santa Cruz. Anti-FLAG (catalog F1804) and –β-Actin (catalog A1978) antibodies were purchased from MilliporeSigma. Anti-human ARID1A (catalog A301-040A) antibodies were purchased from Bethyl Laboratories. Anti-Chk1 (catalog 2360), -Chk2 (catalog 2662), –p-Chk2 (Thr68) (catalog 2661), –p-Chk1 (s345) (catalog 2348), -CDC25A (catalog 3652), -mouse ARID1A (catalog 12354), –PD-L1 (mouse specific; IHC specific catalog 64988), –phospho-TBK1/NAK (Ser172) (catalog 5483), and -TBK1/NAK (catalog 3013) antibodies were purchased from Cell Signaling Technology. Anti-BrdU (catalog ab6326) antibody was purchased from Abcam. Anti-BrdU (catalog 347580) antibody was purchased from BD Biosciences. Anti-DNA antibody single stranded specific (catalog MAB3299) and anti-DNA antibody double stranded (catalog MAB1293) were purchased from MilliporeSigma. Isotype control IgG and anti–PD-L1 (catalog BE0101, clone 10F.9G2) antibodies were purchased from Bio X Cell. RNF8 antibody used in Western blotting was a gift from MSY Huen (The University of Hong Kong, Hong Kong, China).
Doxycycline was purchased from MilliporeSigma. D-luciferin potassium (catalog 217531G) and Pico-Green dsDNA reagent and kits (catalog P7581) were purchased from Thermo Fisher Scientific. The ATM inhibitor KU-60019 (catalog S1570) was purchased from Selleck Chemicals. Chk2 inhibitors II hydrate (catalog C3742) and PV1019 (catalog 220488) were purchased from MilliporeSigma. Vectastain Elite ABC-Peroxidase kits (catalog PK-6200) were purchased from Vector Laboratories. An Opal 4-color manual IHC kit (catalog NEL810001KT) was purchased from PerkinElmer Health Sciences. A mouse/rat CCL5/RANTES Quantikine ELISA kit (catalog MMR00) was purchased from R&D Systems.
Plasmids encoding histidine-tagged (his-tagged) WT ubiquitin and the ubiquitin mutants containing only Lys48 and Lys63 were gifts from Hui-Kuan Lin (Wake Forest School of Medicine, Winston-Salem, North Carolina, USA). SFB-tagged (S-tag, Flag epitope tag, and streptavidin-binding peptide tag) Chk2, Myc-Chk2, WT RNF8, and RNF8 RING domain depletion plasmid were gifts from Junjie Chen (MD Anderson Cancer Center, Houston, Texas, USA). SFB-tagged Chk2 mutation construct (K492, K494, K520, and K534) was generated and purchased from Epoch Life Science. pLenti-3-TR and pLenti6-ARID1A were described previously (8).
RNA interference and CRISPR/Cas9 gene editing.
ARID1A knockdown was achieved by RNA interference using a lentiviral vector–based MISSION shRNA or siRNA (MilliporeSigma). Transfection of siRNA duplex was performed by using oligofectamine (Life Technology) according to the manufacturer’s instructions. The siRNA sequences were as follows: ARID1A CCAACAACAUGGCGGACAA and UGAAGAACUCGAACGGGAA. The shRNA sequences were as follows: ARID1A: V3LHS 410041(#1), TAAATAGCTGTGTCTCGCT; V2LHS_71866 (#2), TCTTGAGATAGCTCCTGCG; mouse Arid1a: TRCN0000238304 (#1), CTAGGCAGCCTAACTATAAT; TRCN0000238306 (#2), TTTATAGTATGGCGAGTTAA. For CRISPR/Cas9 gene knockout, the mouse Arid1a sgRNAs were purchased from GeneCopoeia (MCP225840-SG01-3). Specificity and efficacy of ARID1A knockdown or -KO were evaluated by Western blotting.
Colony-forming assay and 3D culture.
For colony-forming assay, cells were seeded in a 6-well plate at a low density (500 cells/well), and the drug was added the next day. The cells were cultured for 2 weeks to allow colony formation. Colonies were fixed with 4% buffered paraformaldehyde and stained with 0.01% Crystal violet. Colonies containing 50 or more cells were scored as positive for statistical analysis.
For 3D culture, briefly, 50 μL Matrigel was coated to each well of a pre-cooled 8-chamber slide (BD Falcon), which was placed immediately in a 37°C incubator and left there for 30 minutes to allow reconstituted basement membrane to solidify. Then 4000 cells were seeded in 400 μL medium in each well of the 8-chamber slide. After 2 days, drug was added at the indicated concentration and 3 days later cells were subjected to microscopy.
Tet-ON expression of ARID1A.
ARID1A-deficient OAW42 ovarian cancer cells were infected by the lentivirus vectors pLenti-3-TR and pLenti6-ARID1A in sequence, and after selection by puromycin (1 μg/mL), cells were treated with doxycycline (1 μg/mL) for 48 hours to induce the expression of ARID1A.
Immunohistochemistry.
Patient tissue specimens were obtained from Tongji Hospital, The University of Huazhong Science & Technology. IHC was completed by using Vectastain Elite ABC-peroxidase kits according to the manufacturer’s instructions. Briefly, paraffin-embedded tissue slides were baked at 60°C for 1 hour, deparaffinized, rehydrated with serial passage through changes of xylene and graded alcohol, and washed in water. Antigen retrieval was performed by citrate buffer. Endogenous peroxidase in tissues was blocked by incubation of slides in 3% hydrogen peroxide solution before incubation with primary antibody. Stained slides were counterstained with hematoxylin and coverslipped for review. The multiplexed immunofluorescence analysis was performed according to the manufacturer’s instructions (PerkinElmer). Stained slides were counterstained with DAPI and coverslipped for review by Vectra Polaris Automated Quantitative Pathology Imaging System (at the Flow Cytometry and Cellular Imaging Core Facility, MD Anderson Cancer Center). Positivity was defined as at least 5% of cells staining, or the percentage of positive cells per slide was calculated. The following antibodies were used for IHC: human anti-ARID1A (1:100), human anti-Chk2 (1:100), human anti–p-Chk2 (1:100), anti-CD8 (1:200), and anti–PD-L1 (1:100).
Immunoblotting and immunoprecipitation.
Cell pellets were washed in PBS and total proteins were extracted using 8 mol/L urea lysis buffer containing protease and phosphatase inhibitors (GenDEPOT) for 30 minutes at 4°C. Lysates were cleared by centrifugation, and then proteins were separated by SDS-PAGE. PVDF membranes (Bio-Rad) were blocked using 5% (wt/vol) nonfat dried milk in PBST (PBS containing 0.1% Tween-20) for 1 hour at room temperature (20°C–25°C) and then incubated with primary antibodies at 4°C overnight. Subsequently, membranes were washed with PBST 3 times and incubated with horseradish peroxidase secondary antibody (1:2000; Santa Cruz). After extensive washing with PBST, bound antibody was detected by enhanced chemiluminescence according to the manufacturer’s instructions (Bio-Rad).
For immunoprecipitation, whole-cell extracts (1–2 mg) were prepared with RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% Nonidet P40, 150 mM NaCl, 1 mM EDTA, 10% sodium deoxycholate, freshly added with 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, and 1 mM NaF) on ice and were incubated with corresponding antibody (1:100) at 4°C overnight, followed by addition of 30 μL protein G magnetic beads (Cell Signaling Technology). The precipitates were washed 4 times with ice-cold lysis buffer, resuspended in 5 times SDS loading buffer, boiled at 95°C for 5 minutes, resolved by SDS-PAGE, and then immunoblotted. Blotting was quantitated by ImageJ software, and statistics were calculated using Prism software.
Protein stability assay.
Ovarian cancer cells were treated with 25 μg/mL CHX for 0, 3, 6, 12, and 24 hours. Ovarian cancer cells were treated with 1 μM proteasome inhibitor MG132 for 6 hours before the cells were harvested for Western blots.
In vivo ubiquitination assay.
In vivo ubiquitination assays were performed as described elsewhere (64). U2OS or 293T cells were transfected with the indicated plasmids. At 60 hours after transfection, cells were trypsinized and lysed in denaturing buffer (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, pH 8.0, 10 mM imidazole). This step was followed by affinity purification with nickel-nitrilotriacetic acid resins (MilliporeSigma) and immunoblotting analysis.
Quantitative real-time PCR.
Total RNAs (1–2 μg), which were purified by using PureLink RNA mini kit (Life Technologies), were used in a reverse transcriptase reaction with the high-capacity RNA-to-cDNA kit (Thermo Fisher Scientific). Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) was used for the thermocycling reaction in an ABI-VIIA7 real-time PCR system (Thermo Fisher Scientific).
The PCR analysis was carried out in triplicate with the following primer sets: mouse Ccl5 (forward: 5′-ATATGGCTCGGACACCACTC-3′; reverse: 5′-TCCTTCGAGTGACAAACACG-3′); mouse β-Actin (forward: 5′-TGTCCACCTTCCAGCAGATGT-3′; reverse: 5′-AGCTCAGTAACAGTCCGCCTAG-3′); human CHK2 (forward: 5′-GCAGACCCAGCTCTCAATGT-3′; reverse: 5′-CCCTTCCATCAATTCCAAAA-3′); and human β-Actin (forward: 5′-GAGCACAGAGCCTCGCCTTT-3′; reverse: 5′-TCATCATCCATGGTGAGCTG-3′).
DNA fiber assay.
DNA fiber assays were performed as described previously (65). In brief, HOC8 cells were labeled with CldU (100 μM) for 40 minutes, followed by exposure to KU-60019 (2 μM) or PV1019 (2 μM) for 2 hours, and chased with IdU (250 μM) for 40 minutes. Labeled DNA fibers were then diluted by unlabeled ones and were essentially spread as described (66) before standard detection of CldU and IdU tracts (primary antibodies: anti-IdU, anti-BrdU from BD Biosciences, catalog 347580; anti-CldU, anti-BrdU from Abcam ab6326; secondary antibodies: Alexa Fluor 488 and Alexa Fluor 594 from Thermo Fisher Scientific). Fibers were imaged with an FV1000 Olympus Laser Confocal Microscope at the Flow Cytometry and Cellular Imaging Core Facility at MD Anderson Cancer Center and analyzed using ImageJ software. Statistics were calculated using Prism software.
HR repair assays.
The HR repair assays were performed as previously described (67).
PicoGreen staining and BrdU incorporation.
PicoGreen staining was performed as we previously described (68). Briefly, PicoGreen staining was performed using Quant-iT Pico-Green dsDNA reagent and kits (Thermo Fisher Scientific). PicoGreen was diluted into cell culture medium with a final concentration of 3 μL/mL, and cells were cultured in a 37°C incubator for 1 hour. The cells were then washed and fixed for confocal microscopy with an FV1000 Olympus Laser Confocal Microscope at the Flow Cytometry and Cellular Imaging Core Facility at MD Anderson Cancer Center with DAPI counterstaining. For BrdU incorporation, cells were labeled with 10 μM BrdU for 30 minutes, fixed by 4% paraformaldehyde for 30 minutes at room temperature. Cells were then treated with 2M HCl for 20 minutes and neutralized with 0.1 M sodium borate buffer pH 8.5 for 2 minutes. Cells were stained with anti-BrdU antibody, followed by secondary antibody Alexa Fluor 594 from Thermo Fisher Scientific.
Cytosolic ssDNA and dsDNA staining.
ssDNA and dsDNA stainings were performed as previously described (69). Briefly, cells were fixed according to manufacturer’s instructions for dsDNA antibody and treated with 2 mg/mL RNase A for 30 minutes at 37°C. Cells were stained with ssDNA or dsDNA antibody, followed by secondary antibody Alexa Fluor 594 from Thermo Fisher Scientific. The cells were then washed and fixed for confocal microscopy with an FV1000 Olympus Laser Confocal Microscope at the Flow Cytometry and Cellular Imaging Core Facility at MD Anderson Cancer Center with DAPI counterstaining.
ELISA.
Following a 2-day KU-60019 treatment, the cell culture supernatants were collected and processed using a mouse/rat CCL5/RANTES Quantikine ELISA Kit (R&D Systems) according to the manufacturer’s instructions.
RPPA and statistical analysis.
RPPA assays were performed at the Functional Proteomics RPPA Core Facility at MD Anderson Cancer Center as previously described (70–72). BMN 673–treated xenografts have been previously described (30). Briefly, the protein lysates of xenograft or patient tumor tissues were serially diluted and spotted onto FAST slides (Schleicher & Schuell BioSciences) using a robotic GeneTAC arrayer (Genomic Solutions). After printing, the slides were probed with primary antibodies and then biotin-conjugated secondary antibodies, and the signals were captured using a DakoCytomation-catalyzed system and DAB colorimetric reaction (DakoCytomation). The processed slides were scanned with a flatbed scanner, and spot intensities were analyzed and quantified using MicroVigene software (VigeneTech). Slide quality control was performed as described (73), and SuperCurve software (available at https://bioinformatics.mdanderson.org/public-software/supercurve/) was used to process the spot intensities to calculate protein expression levels (74). The protein expression measurements were corrected and normalized for loading using a median centering approach as previously described (71, 72, 75, 76).
Two-tailed Student’s t test was used to compare protein expression between the ARID1A-mutant and ARID1A-WT groups, and the differentially expressed proteins were selected using a false discovery rate adjusted P value of less than 0.05 as the cutoff. The expression patterns of the significant proteins were visualized by 2-way unsupervised hierarchical clustering heatmaps for which Pearson’s correlation coefficient was used as distance matrix and Ward was used as linkage rule.
TIL scoring analysis.
As we previously described (8), we analyzed gene expression and mutation of UCEC tumors and gene expression of ovarian cancer (OV) tumors from TCGA. RNA-Seq data were downloaded from the TCGA Data Portal and were log-transformed. TIL scores were determined by an 8-gene signature from the study that profiled the mRNA expression landscape of immune cells (77). The TIL scores were analyzed in OV and UCEC patients stratified by (a) mutations and mRNA levels of ARID1A and ATM and (b) mRNA levels of ARID1A and CHK2. Differential expression of TIL scores in subsets of patients were examined by using Student’s t test.
Survival analysis.
Patients survival data were obtained from CBioPortal for Cancer Genomics (78, 79). Two data cohorts, one of patients with UCEC (TCGA Firehose Legacy, previously known as TCGA Provisional) and one of patients with esophagus-stomach cancer (TCGA, ref. 80), were selected. Gene expression profiles were categorized by 4 mutation statuses: ARID1A mutation, ATM/ARID1A double mutation, CHK2/ARID1A double mutation, and WT. A total of 239 patients with UCEC and 518 patients with esophagus-stomach cancer were divided into subgroups as described above. Overall survival of various treatment groups was analyzed using the Cox regression model unless otherwise noted.
In vivo mouse models.
Animals were randomly assigned to different groups. For the i.p. model, ID8 cells (5 × 106) were injected into the peritoneal cavity of C57BL/6 mice (female, 6–8 weeks old, CRL/NCI). Tumor progression was monitored once a week by using a Xenogen IVIS Spectrum in vivo bioluminescence imaging system (Small Animal Imaging Facility, MD Anderson Cancer Center). Tumor volume was determined on the basis of the total flux (photons per second). Tumor-bearing mice were treated with isotype control IgG (i.p. injection) or anti–PD-L1 antibody (i.p. injection, 200 μg/mouse, Bio X Cell) every 3 days, KU-60019 (100 mg/kg, oral delivery) daily, or combination treatment. Mice reaching a humane endpoint or weighing more than 32 g as a result of tumor burden and/or ascites were euthanized. There were 3–5 mice in each treatment group.
Statistics.
All statistical analyses were done using GraphPad Prism 8 software. Overall survival of various treatment groups was analyzed using the Cox regression model. For other analyses, unpaired t tests were used to generate 2-tailed P values. Otherwise, 1-way ANOVA was used to generate P values.
Study approval.
All animal studies were conducted in compliance with protocols approved by the MD Anderson Cancer Center Institutional Animal Care and Use Committee (Houston, Texas, USA).
Author contributions
LW designed the project, carried out the experimental work, analyzed the data, and wrote the manuscript. LY carried out the in vivo ubiquitination assay. W Zhao carried out the TIL scoring analysis. YP performed the survival analysis of patients. ZJ carried out RPPA and RPPA-related statistical analysis. JS carried out RNA interference and CRISPR/Cas9 gene editing. W Zhang carried out knockdown ARID1A in U2OS cells. CW conducted the IHC staining of tumor tissue in ovarian cancer patients. XS, CA, and YTL provided suggestions for this project. GP, SS, TAY, and JAA revised the manuscript.
Supplementary Material
Acknowledgments
We thank Shiaw-Yih Lin for critically reviewing our manuscript. This research was supported by NCI Cancer Center Support grant CA016672 to The University of Texas MD Anderson Cancer Center; NIH R01 grant CA181663 to GP; Cancer Prevention and Research Institute of Texas grant RP160242 to XS and GP; Ovarian Cancer Research Foundation grants 5P50 CA217685-02, 7U01 CA217842-03, 5P50 CA098258-13, and 545152; Adelson Medical Research Foundation grant 04-7023433 to GBM; and a Developmental Research Program grant to GP under the MD Anderson Uterine Cancer Specialized Programs of Research Excellence (SPORE) in Uterine Cancer (NCI grant P50CA098258).
Version 1. 10/05/2020
Electronic publication
Version 2. 10/12/2020
Grant added to Acknowledgments section
Version 3. 11/02/2020
Print issue publication
Footnotes
LY’s and CW’s present address is: Department of Medical Oncology, Tongji Hospital, University of Huazhong Science and Technology, Wuhan, China.
JS’s present address is: Department of Ophthalmology, Ninth People’s Hospital, Shanghai JiaoTong University School of Medicine, Shanghai, China. Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai, China.
Conflict of interest: GBM has received sponsored research support from Nanostring Center of Excellence, Ionis (provision of tool compounds) and clinical trials support from AstraZeneca, Genentech, GSK, and Eli Lilly; he has ownership interest in Catena Pharmaceuticals, ImmunoMet, SignalChem, and Tarveda; he is a consultant/advisory board member of AstraZeneca, Chrysallis Biotechnology, ImmunoMet, Ionis, Lilly,PDX Pharmaceuticals, SignalChem Lifesciences, Symphogen, Tarveda, Turbine, and Zentalis Pharmaceuticals; he has licensed technology for a homologous recombination deficiency assay to Myriad Genetics and for digital spatial profiling to Nanostring. TAY has received sponsored research support from AstraZeneca, Bayer, Clovis, Constellation, Cyteir, Eli Lilly, EMD Serono, Forbius/Formation Biologics, F-Star, GlaxoSmithKline, Genentech, ImmuneSensor, Ipsen, Jounce, Karyopharm, Kyowa, Novartis, Pfizer, Ribon Therapeutics, Regeneron, Sanofi, Seattle Genetics, Tesaro, and Vertex Pharmaceuticals; he is a consultant/advisory board member of Almac, Aduro, AstraZeneca, Atrin, Axiom, Bayer, Calithera, Clovis, Cybrexa, EMD Serono, F-Star, Guidepoint, Ignyta, I-Mab, Jansen, Kyn Therapeutics, Merck, Pfizer, Roche, Seattle Genetics, and Zai Labs. GP has received sponsored research support from Pfizer.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(11):5951–5966.https://doi.org/10.1172/JCI130445.
Contributor Information
Lulu Wang, Email: lwang14@mdanderson.org.
Lin Yang, Email: bjxhyl@sina.com.
Chen Wang, Email: l367142303@aliyun.com.
Wei Zhao, Email: wei.zhao2@nih.gov.
Zhenlin Ju, Email: zju@mdanderson.org.
Wei Zhang, Email: weizhangncbi@163.com.
Jianfeng Shen, Email: darkingfeng@hotmail.com.
Yang Peng, Email: ypeng2@mdanderson.org.
Clemens An, Email: ca336@cornell.edu.
Shumei Song, Email: ssong@mdanderson.org.
Xuetong Shen, Email: xshen@mdanderson.org.
Guang Peng, Email: gpeng@mdanderson.org.
References
- 1.Hodges C, Kirkland JG, Crabtree GR. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb Perspect Med. 2016;6(8):a026930. doi: 10.1101/cshperspect.a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3(1):35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363(16):1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330(6001):228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang H, et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012;22(11):2120–2129. doi: 10.1101/gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43(12):1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 7.Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43(9):875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen J, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24(5):556–562. doi: 10.1038/s41591-018-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allo G, et al. ARID1A loss correlates with mismatch repair deficiency and intact p53 expression in high-grade endometrial carcinomas. Mod Pathol. 2014;27(2):255–261. doi: 10.1038/modpathol.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou A, et al. Loss of ARID1A expression in colorectal carcinoma is strongly associated with mismatch repair deficiency. Hum Pathol. 2014;45(8):1697–1703. doi: 10.1016/j.humpath.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Kim MS, Je EM, Yoo NJ, Lee SH. Loss of ARID1A expression is uncommon in gastric, colorectal, and prostate cancers. APMIS. 2012;120(12):1020–1022. doi: 10.1111/j.1600-0463.2012.02930.x. [DOI] [PubMed] [Google Scholar]
- 12.Le DT, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Germano G, et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature. 2017;552(7683):116–120. doi: 10.1038/nature24673. [DOI] [PubMed] [Google Scholar]
- 14.Le DT, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mouw KW, D’Andrea AD. DNA repair deficiency and immunotherapy response. J Clin Oncol. 2018;36(17):1710–1713. doi: 10.1200/JCO.2018.78.2425. [DOI] [PubMed] [Google Scholar]
- 16.Lee V, Murphy A, Le DT, Diaz LA. Mismatch repair deficiency and response to immune checkpoint blockade. Oncologist. 2016;21(10):1200–1211. doi: 10.1634/theoncologist.2016-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corrales L, McWhirter SM, Dubensky TW, Gajewski TF. The host STING pathway at the interface of cancer and immunity. J Clin Invest. 2016;126(7):2404–2411. doi: 10.1172/JCI86892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo SR, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41(5):830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ablasser A, et al. cGAS produces a 2’-5’-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498(7454):380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barber GN. STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 2014;35(2):88–93. doi: 10.1016/j.it.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 22.Swann JB, et al. Type I IFN contributes to NK cell homeostasis, activation, and antitumor function. J Immunol. 2007;178(12):7540–7549. doi: 10.4049/jimmunol.178.12.7540. [DOI] [PubMed] [Google Scholar]
- 23.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nat Rev Immunol. 2015;15(7):405–414. doi: 10.1038/nri3845. [DOI] [PubMed] [Google Scholar]
- 24.Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol. 2012;19(2):201–206. doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315(5809):201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 26.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 27.Radivojac P, et al. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 2010;78(2):365–380. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14(3):278–288. [PMC free article] [PubMed] [Google Scholar]
- 29.Hirao A, et al. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287(5459):1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 30.Shen J, et al. ARID1A deficiency impairs the DNA damage checkpoint and sensitizes cells to PARP inhibitors. Cancer Discov. 2015;5(7):752–767. doi: 10.1158/2159-8290.CD-14-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williamson CT, et al. ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nat Commun. 2016;7:13837. doi: 10.1038/ncomms13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao Z, et al. Chk1 mediates S and G2 arrests through Cdc25A degradation in response to DNA-damaging agents. J Biol Chem. 2003;278(24):21767–21773. doi: 10.1074/jbc.M300229200. [DOI] [PubMed] [Google Scholar]
- 33.Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell. 2014;54(2):289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 34.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37(4):492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21(3):396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11(7):481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 37.Tang L, Nogales E, Ciferri C. Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog Biophys Mol Biol. 2010;102(2–3):122–128. doi: 10.1016/j.pbiomolbio.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 2015;1(5):e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karnezis AN, et al. Dual loss of the SWI/SNF complex ATPases SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type. J Pathol. 2016;238(3):389–400. doi: 10.1002/path.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jelinic P, et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet. 2014;46(5):424–426. doi: 10.1038/ng.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapoor P, et al. Phosphorylation-dependent enhancement of Rad53 kinase activity through the INO80 chromatin remodeling complex. Mol Cell. 2015;58(5):863–869. doi: 10.1016/j.molcel.2015.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapoor P, et al. Regulation of Mec1 kinase activity by the SWI/SNF chromatin remodeling complex. Genes Dev. 2015;29(6):591–602. doi: 10.1101/gad.257626.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou BB, Bartek J. Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat Rev Cancer. 2004;4(3):216–225. doi: 10.1038/nrc1296. [DOI] [PubMed] [Google Scholar]
- 44.Plans V, Scheper J, Soler M, Loukili N, Okano Y, Thomson TM. The RING finger protein RNF8 recruits UBC13 for lysine 63-based self polyubiquitylation. J Cell Biochem. 2006;97(3):572–582. doi: 10.1002/jcb.20587. [DOI] [PubMed] [Google Scholar]
- 45.Ito K, et al. N-Terminally extended human ubiquitin-conjugating enzymes (E2s) mediate the ubiquitination of RING-finger proteins, ARA54 and RNF8. Eur J Biochem. 2001;268(9):2725–2732. doi: 10.1046/j.1432-1327.2001.02169.x. [DOI] [PubMed] [Google Scholar]
- 46.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corrales L, et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 2015;11(7):1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohkuri T, et al. Intratumoral administration of cGAMP transiently accumulates potent macrophages for anti-tumor immunity at a mouse tumor site. Cancer Immunol Immunother. 2017;66(6):705–716. doi: 10.1007/s00262-017-1975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramanjulu JM, et al. Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature. 2018;564(7736):439–443. doi: 10.1038/s41586-018-0705-y. [DOI] [PubMed] [Google Scholar]
- 50.Luo M, et al. A STING-activating nanovaccine for cancer immunotherapy. Nat Nanotechnol. 2017;12(7):648–654. doi: 10.1038/nnano.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghaffari A, et al. STING agonist therapy in combination with PD-1 immune checkpoint blockade enhances response to carboplatin chemotherapy in high-grade serous ovarian cancer. Br J Cancer. 2018;119(4):440–449. doi: 10.1038/s41416-018-0188-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H, et al. cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc Natl Acad Sci USA. 2017;114(7):1637–1642. doi: 10.1073/pnas.1621363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ager CR, Reilley MJ, Nicholas C, Bartkowiak T, Jaiswal AR, Curran MA. Intratumoral STING activation with T-cell checkpoint modulation generates systemic antitumor immunity. Cancer Immunol Res. 2017;5(8):676–684. doi: 10.1158/2326-6066.CIR-17-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sivick KE, et al. Magnitude of therapeutic STING activation determines CD8+ T cell-mediated anti-tumor immunity. Cell Rep. 2018;25(11):3074–3085.e5. doi: 10.1016/j.celrep.2018.11.047. [DOI] [PubMed] [Google Scholar]
- 55.Erdal E, Haider S, Rehwinkel J, Harris AL, McHugh PJ. A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev. 2017;31(4):353–369. doi: 10.1101/gad.289769.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 2017;548(7668):466–470. doi: 10.1038/nature23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mackenzie KJ, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548(7668):461–465. doi: 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parkes EE, et al. Activation of STING-dependent innate immune signaling by S-phase-specific DNA damage in breast cancer. J Natl Cancer Inst. 2016;109(1):djw199. doi: 10.1093/jnci/djw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vanpouille-Box C, et al. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat Commun. 2017;8:15618. doi: 10.1038/ncomms15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pilié PG, Tang C, Mills GB, Yap TA. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol. 2019;16(2):81–104. doi: 10.1038/s41571-018-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Antoni L, Sodha N, Collins I, Garrett MD. CHK2 kinase: cancer susceptibility and cancer therapy - two sides of the same coin? Nat Rev Cancer. 2007;7(12):925–936. doi: 10.1038/nrc2251. [DOI] [PubMed] [Google Scholar]
- 63.Al-Ahmadie H, et al. Synthetic lethality in ATM-deficient RAD50-mutant tumors underlies outlier response to cancer therapy. Cancer Discov. 2014;4(9):1014–1021. doi: 10.1158/2159-8290.CD-14-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang WL, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325(5944):1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145(4):529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140(6):1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peng G, et al. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol. 2009;11(7):865–872. doi: 10.1038/ncb1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen J, et al. PARPi triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCAness. Cancer Res. 2019;79(2):311–319. doi: 10.1158/0008-5472.CAN-18-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen YJ, et al. Genome-derived cytosolic DNA mediates type I interferon-dependent rejection of B cell lymphoma cells. Cell Rep. 2015;11(3):460–473. doi: 10.1016/j.celrep.2015.03.041. [DOI] [PubMed] [Google Scholar]
- 70.Tibes R, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5(10):2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 71.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ju Z, et al. Development of a robust classifier for quality control of reverse-phase protein arrays. Bioinformatics. 2015;31(6):912–918. doi: 10.1093/bioinformatics/btu736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu J, He X, Baggerly KA, Coombes KR, Hennessy BT, Mills GB. Non-parametric quantification of protein lysate arrays. Bioinformatics. 2007;23(15):1986–1994. doi: 10.1093/bioinformatics/btm283. [DOI] [PubMed] [Google Scholar]
- 75.Hennessy BT, et al. Pharmacodynamic markers of perifosine efficacy. Clin Cancer Res. 2007;13(24):7421–7431. doi: 10.1158/1078-0432.CCR-07-0760. [DOI] [PubMed] [Google Scholar]
- 76.Gonzalez-Angulo AM, et al. Functional proteomics can define prognosis and predict pathologic complete response in patients with breast cancer. Clin Proteomics. 2011;8(1):11. doi: 10.1186/1559-0275-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.West NR, Milne K, Truong PT, Macpherson N, Nelson BH, Watson PH. Tumor-infiltrating lymphocytes predict response to anthracycline-based chemotherapy in estrogen receptor-negative breast cancer. Breast Cancer Res. 2011;13(6):R126. doi: 10.1186/bcr3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cancer Genome Atlas Research Network, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.