Abstract
Human T cell leukemia virus type 1 (HTLV-1) is mainly transmitted vertically through breast milk. The rate of mother-to-child transmission (MTCT) through formula feeding, although significantly lower than through breastfeeding, is approximately 2.4%–3.6%, suggesting the possibility of alternative transmission routes. MTCT of HTLV-1 might occur through the uterus, birth canal, or placental tissues; the latter is known as transplacental transmission. Here, we found that HTLV-1 proviral DNA was present in the placental villous tissues of the fetuses of nearly half of pregnant carriers and in a small number of cord blood samples. An RNA ISH assay showed that HTLV-1–expressing cells were present in nearly all subjects with HTLV-1–positive placental villous tissues, and their frequency was significantly higher in subjects with HTLV-1–positive cord blood samples. Furthermore, placental villous trophoblasts expressed HTLV-1 receptors and showed increased susceptibility to HTLV-1 infection. In addition, HTLV-1–infected trophoblasts expressed high levels of viral antigens and promoted the de novo infection of target T cells in a humanized mouse model. In summary, during pregnancy of HTLV-1 carriers, HTLV-1 was highly expressed in placental villous tissues, and villous trophoblasts showed high HTLV-1 sensitivity, suggesting that MTCT of HTLV-1 occurs through the placenta.
Keywords: Infectious disease, Virology
Keywords: Epidemiology
Introduction
Human T cell leukemia virus type 1 (HTLV-1) infection is endemic in Japan, the southern Unites States, central Australia, the Caribbean, Jamaica, South America, and equatorial Africa (1, 2). At least 5 million to 10 million individuals are infected with HTLV-1 worldwide (2). Although most HTLV-1–infected individuals are asymptomatic (carriers), HTLV-1 can cause related diseases following a persistent latent infection, such as adult T cell leukemia (ATL), HTLV-1–associated myelopathy/tropical spastic paraparesis, and HTLV-1 uveitis in humans (3–5). Presently, there are no curative therapies for lifelong HTLV-1 infection or any prophylactic strategies to avoid the onset of HTLV-1–associated disorders. Thus, preventing infection is crucially important.
A cohort study of HTLV-1 carriers indicated that infection during childhood was a potential risk factor for the development of ATL (6). Furthermore, the most important route of HTLV-1 infection was mother-to-child transmission (MTCT) through breastfeeding (7). Therefore, antenatal screening for HTLV-1 infection to prevent MTCT through avoidance of breastfeeding has been carried out in some endemic areas, such as Nagasaki Prefecture, Japan (8–10). In Japan, which has a high HTLV-1 prevalence, a nationwide MTCT prevention program has been operational since 2010 (11). Formula feeding, short-term breastfeeding, and freeze-thawing of breast milk have conventionally been available as options for pregnant carriers, but formula feeding is now recommended for the prevention of MTCT of HTLV-1 (11).
The rate of HTLV-1 seroconversion is 11.2%–23.7% among children receiving prolonged breastfeeding (>6 months), 5.9%–8.3% among children receiving short-term breastfeeding (<6 months), and 2.4%–3.6% among children receiving formula (12–15). These data demonstrated that breastfeeding was the most likely route for MTCT of HTLV-1. However, as some exclusively formula-fed infants were infected with HTLV-1, other transmission routes may operate during the pregnant and postpartum periods. Thus, elucidation of alternative transmission routes is necessary to establish better prevention programs for MTCT of HTLV-1.
Other potential routes of MTCT during the prenatal, perinatal, and postnatal periods include intrauterine infection by ascending microorganisms from the vagina or transplacental transfusion with contaminated maternal blood (16, 17), perinatal infection by direct exposure to virus-containing maternal blood in the birth canal (18), and postnatal infection by maternal saliva, which is very unlikely (19). However, there is no clear evidence of MTCT without breastfeeding.
HTLV-1 is transmitted efficiently through cell-cell contact with long-term incubation, but not through a cell-free mechanism (20, 21). Furthermore, although the main targets of HTLV-1 are generally thought to be T cells, infection of endothelial cells and other nonlymphocyte cells has been reported (22–24). We hypothesized that transplacental HTLV-1 infection is established during pregnancy and that HTLV-1–infected placental cells are indirectly transmitted to the fetus across the placental barrier.
This study assessed the potential for the transplacental transmission of HTLV-1. We evaluated the presence of HTLV-1 provirus in the maternal blood, cord blood, and placental villous tissues of 254 seropositive pregnant HTLV-1 carriers. We documented abundant infection in placental tissues. We also tested the susceptibility of human primary placental cells to HTLV-1 infection. Notably, villous trophoblasts (VTs) expressed HTLV-1 receptors, and significant viral transcription was observed in the villi of pregnant carriers whose cord blood tested positive for HTLV-1. These findings suggest that HTLV-1 directly targets trophoblasts and that HTLV-1 expression in trophoblasts may be involved in the transplacental transmission of HTLV-1.
Results
Detection and evaluation of HTLV-1 provirus and anti–HTLV-1 antibody in pregnant HTLV-1 carriers.
The present study included 254 pregnant HTLV-1 carriers. The clinical characteristics of the study population are shown in Table 1 and Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/JCI135525DS1). Maternal blood during pregnancy, cord blood, and placental villous tissues at the time of delivery were obtained from subjects to measure the HTLV-1 proviral load (PVL) using real-time PCR. As shown in Figure 1A, HTLV-1 provirus was detected in the maternal blood of 248 of 254 subjects (97.6%), in the placental villous tissues of 140 of 254 subjects (55.1%), and in the cord blood samples of 6 of 254 subjects (2.4%). Overall, 248 women had PVL in the maternal blood, of whom 140 also had PVL in the placenta. Of these 140 women, 6 had PVL in the cord blood. Significant differences in the PVL were observed between the maternal blood, cord blood, and placental villous tissues (Figure 1A).
Table 1. Clinical characteristics of patients with and without HTLV-1–infected placenta among 254 pregnant HTLV-1 carriers.
Figure 1. Comparative analysis of HTLV-1 proviral load in pregnant HTLV-1 carriers.
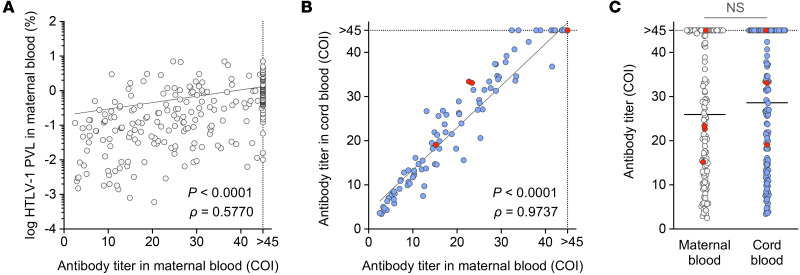
HTLV screening tests for 254 pregnant HTLV-1 carriers were performed during the second trimester of pregnancy. All donors were serologically positive for HTLV-1 infection. Each dot in the dot plots and scatterplots indicates the HTLV-1 proviral load (PVL) of a single specimen. (A) Detection of HTLV-1 in, and differences in PVL between, maternal blood, placental villous tissue, and cord blood in 254 seropositive women. (B) Comparison of maternal blood PVL among 248 pregnant carriers with PVL in the maternal blood, who were distinguished by proviral DNA detection in the placental villous tissue and/or in the cord blood. In A and B, bold lines indicate the median values of PVL. The P values were calculated with Kruskal-Wallis test followed by Dunn’s multiple-comparisons test. (C–E) Correlation analysis of the PVL was performed. Spearman’s rank correlation test was used to identify statistically significant correlations between values. A positive correlation was detected between PVL in the placental villous tissue and PVL in the maternal blood of 140 pregnant HTLV-1 carriers in whom provirus was detected in the placental villous tissue (C). A nonsignificant correlation was observed between cord blood PVL and maternal blood PVL (D) or placental villous PVL (E) for the 6 pregnant HTLV-1 carriers in whom provirus was detected in the cord blood. (F) Representative electrophoretogram of 6 independent experiments of microsatellite genotyping using short tandem repeat (STR) markers. STR loci of the maternal blood were distinct from those of fetal tissues derived from the same specimen. Amelogenin confirmed the presence of the X chromosome–specific allele alone in the maternal blood, and the X and Y chromosome–specific alleles in the placental villous tissue and the cord blood.
The 248 pregnant carriers with PVL in the maternal blood were divided into those with PVL (n = 140) and without PVL (n = 108) in the placental tissue, and their clinical backgrounds were compared. Women with PVL in the placental tissue had a significantly higher peripheral blood PVL, higher antibody titers, and more multiparas compared with women with no PVL in the placental tissue (Table 1 and Figure 1B). These 2 groups did not differ in terms of birth weight and pregnancy complications (Table 1). There was no significant difference in the clinical backgrounds of pregnant women with HTLV-1 in the placenta when divided into those who tested positive versus negative for HTLV-1 in the cord blood (Supplemental Table 1). This was at least in part due to the small number of pregnant women testing positive for HTLV-1 in the cord blood. In addition, there were insufficient numbers of follow-up surveys of cases of MTCT by intrauterine transmission to allow statistical analysis. These issues are subjects for future investigation.
A weak positive correlation between the PVLs in the maternal blood and in the placental villous tissues was observed (Figure 1C), whereas PVL in HTLV-1–positive cord blood samples did not correlate with PVL in the maternal blood or placental villous tissues of the same subject (Figure 1, D and E). To test the possibility that HTLV-1 provirus detected in cord blood was derived from maternal blood contamination of cord blood, microsatellite analysis was performed using short tandem repeat (STR) markers (25). Differences in the patterns of representative STR markers were observed between maternal blood–derived DNA and fetal placental villous tissue– and cord blood–derived DNA (Figure 1F). Similar results were obtained for all 6 samples that tested positive for HTLV-1 provirus in the cord blood. Furthermore, STR analysis and HTLV-1 PVL assay were used to examine how much maternal blood in the cord blood was required to detect a positive signal. A mixing rate of 20% (maternal/fetal cell ratio = 20:80) was the detection limit in the STR analysis, and a mixing rate of 5% (maternal/fetal cell ratio = 5:95) was the detection limit in the HTLV-1 PVL assay (Supplemental Figure 1). A previous study reported that the median rates of maternal blood contamination in the cord blood were 0.27% and 0.10% in HIV-infected women and HIV-uninfected women, respectively (26). These values are markedly lower than the detection limits in the HTLV-1 PVL assay. Although the detection limit may vary depending on the PVL in the maternal blood, comparable median values of maternal blood PVL were observed in women with HTLV-1 detected in the placenta when divided into those testing positive versus negative for HTLV-1 in the cord blood (Supplemental Table 1). This strongly suggests that provirus detected in the cord blood at the time of delivery in this study was derived from cord blood as fetal blood and was not attributable to maternal blood contamination.
Neutralizing antibodies transported across the placenta are involved in intrauterine viral infection (27, 28). Therefore, we measured anti–HTLV-1 antibody titers in the maternal blood and the cord blood. As previously reported (29), positive correlations between PVLs and antibody titers in the maternal blood were observed (Figure 2A). We then compared antibody titers in the maternal blood and the cord blood, and found a strong correlation and similar titer between the 2 sample types (Figure 2, B and C). We also compared pregnant carriers whose cord blood samples tested negative for HTLV-1 provirus with those whose cord blood samples tested positive and found equivalent anti–HTLV-1 antibody titers (Figure 2C). However, whether the antibodies had qualitative differences is yet to be examined. These results suggest that pregnant carriers have no abnormalities in terms of antibody production and transportation across the placenta, and that other factors play a larger role in MTCT of HTLV-1.
Figure 2. Comparative analysis of anti–HTLV-1 antibody titers in pregnant HTLV-1 carriers.
The anti–HTLV-1 antibody titer was measured in maternal and cord blood samples from 122 subjects using a chemiluminescent enzyme immunoassay (CLEIA). Each dot in the dot plots indicates the HTLV-1 PVL or antibody titer of a single specimen. (A) Relationship between PVL and HTLV-1 antibody titer in the maternal blood measured by CLEIA. (B and C) HTLV-1 antibody titers in maternal blood and cord blood were compared to assess their relationship (B) and their magnitude (C). Bold lines indicate the median values of antibody titers. No significant differences in antibody titer were observed between maternal blood and cord blood (NS by Mann-Whitney U test). Red dots represent pregnant carriers who tested positive for HTLV-1 provirus in the cord blood. In A and B, Spearman’s rank correlation test was used to identify statistically significant correlations between values. COI, cutoff index.
Development of a specific and sensitive HTLV-1 RNA ISH assay and detection of viral RNA in placental villous tissues.
To examine the distribution of HTLV-1–infected cells in the placental villous tissues, we established an RNA ISH assay (RNAscope) (30) targeting HTLV-1 plus- and minus-strand mRNAs with high sensitivity and specificity. RNA probes were designed to target the tax/rex mRNA exon 3, a conserved region in the HTLV-1 plus-strand mRNA (pX probe), as well as a common region in the minus-strand mRNA (HBZ probe). In addition, probes detecting the positive control (hPOLR2A) and negative control (DapB) were used. To analyze infected cells in tissues, a humanized mouse model was established, and mice were infected with HTLV-1 by oral administration as previously reported (31). At 63 days postinfection (dpi), lymphoid organs were collected to quantitate HTLV-1–infected cells (Figure 3A). Characteristics of humanized mice are summarized in Supplemental Table 2. In the spleen, lymph node, and thymus, pX probe–positive foci partially accumulated within the follicles inside tissues (Figure 3B), whereas HBZ probe–positive foci were detected throughout the tissues (Figure 3C). An uninfected mouse showed no signals. DapB was not detected in any mouse tissues, and the expression of hPOLR2A was localized to the same region as HBZ (data not shown).
Figure 3. Development of a plus- and minus-strand HTLV-1 mRNA–specific RNAscope ISH assay using a humanized mouse model.
(A) Schematic drawing of the experimental schedule is shown. All NOJ mice were reconstituted with a human immune system by the intrahepatic transplantation of human CD133+ hematopoietic stem cells into newborn mice. The mice were inoculated orally with HTLV-1–infected MT-2 cells at 0 days postinfection (dpi) and were sacrificed at 63 dpi (arrows). Arrowheads indicate the time points of blood collection. (B and C) RNA ISH with HTLV-1–specific probes targeting plus-strand mRNAs (B) and minus-strand mRNAs (C) was used to stain the spleen and lymph node sections of mock- or HTLV-1–infected humanized mice. Viral mRNAs were detected using DAB chromogen (brown). Boxes indicate regions shown at a higher magnification in adjacent lower panels. Data are representative of at least 3 nonserial tissue sections. Scale bars in lower and higher magnifications represent 500 μm and 100 μm, respectively.
The distribution of HTLV-1–expressing cells in the placental villous tissues was examined using RNA ISH. We prepared 19 placental villous FFPE samples from 19 pregnant carriers, which were divided into groups based on the detection of the HTLV-1 provirus in the villi and/or the cord blood (nos. 1–18), including one ATL patient (no. 19; Table 2). Clinical characteristics of the pregnant carriers are summarized in Table 2. We found that pX probe–positive foci were present in samples from all subjects with HTLV-1–positive placental villous tissues (nos. 7–19), whereas these foci were not identified in subjects with HTLV-1–negative placental villous tissues (nos. 1–6; Figure 4, A and B). In addition, the number of pX probe–positive foci was significantly higher in subjects with HTLV-1–positive cord blood samples compared with those who had HTLV-1–negative cord blood samples (nos. 13–18 vs. nos. 7–12; Figure 4B). No differences were evident between subjects with HTLV-1–negative cord blood and the ATL patient (nos. 7–12 and no. 19; Figure 4B). These results suggested that productive viral expression in the placental villi was maintained during gestation in pregnant carriers.
Table 2. Characteristics of 19 pregnant HTLV-1 carriers for the RNAscope ISH assay.
Figure 4. Detection of HTLV-1 plus-strand mRNAs in pregnant carrier–derived placental villous tissues.
RNA ISH with HTLV-1–specific probes targeting plus-strand mRNAs was used to stain the placental villous tissue sections of pregnant HTLV-1 carriers. The carriers were identical to those listed in Table 2. (A) ISH results are shown for each group of pregnant carriers (nos. 1, 9, and 16), and for a pregnant chronic ATL patient (no. 19). Viral mRNA was detected using DAB chromogen (brown). Boxes indicate regions shown at a higher magnification in adjacent lower panels. Data are representative of at least 5 nonserial tissue sections. Scale bars in lower and higher magnifications represent 200 μm and 50 μm, respectively. (B) Quantitative evaluation of ISH results. The total number of pX probe–positive foci in stained sections was counted by microscopy. Data are expressed as the mean ± SD from 5 nonserial tissue sections. The Mann-Whitney U test was performed to compare statistical differences between groups in subjects with HTLV-1–negative and –positive cord blood samples (nos. 7–12 vs. nos. 13–18).
Evaluation of susceptibility to HTLV-1 infection in primary placental cells.
Next, we examined which types of cells in the placenta were susceptible to HTLV-1 infection. The intervillous space is filled with maternal blood, from which nutrients such as amino acids, fatty acids, and glucose are taken up by the fetus through chorionic villi (32). Maternal blood and fetal blood are completely separated by the villi, referred to as the blood-placental barrier (32, 33). The blood-placental barrier is composed of trophoblasts, mesenchymal fibroblasts, and vascular endothelial cells (from superficial to deep layers within a villus) (34, 35). Trophoblasts are directly in contact with maternal blood and are involved in key placental functions such as nutrient transport (32), and they can be infected by various viruses, including HIV-1, HBV, Zika virus, and Ebola virus (36–39).
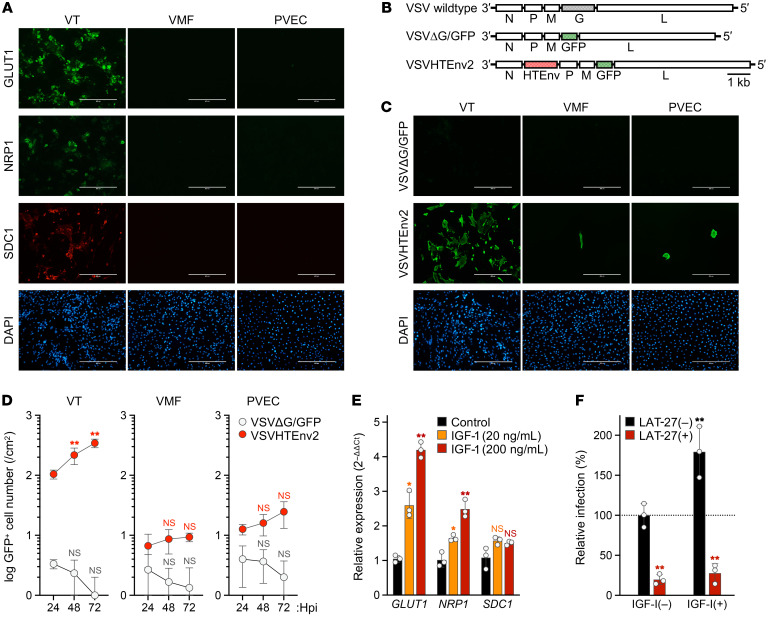
Glucose transporter 1 (GLUT1) plays a key role in glucose transport (32, 40) and is a major receptor for HTLV-1 (41, 42). We therefore examined the expression of HTLV-1 receptors including GLUT1, neuropilin-1 (NRP1) (43), and syndecan-1 (SDC1) (44), a member of the heparan sulfate proteoglycan core protein, in cells isolated from the placentas of HTLV-1–uninfected individuals. We examined the expression of HTLV-1 receptors in placental villous samples from the 18 pregnant carriers in Figure 4B (nos. 1–18) and found no significant differences among these individuals (Supplemental Figure 2). Among the cells making up the blood-placental barrier, VTs had a high expression of GLUT1 and expressed NRP1 and SDC1 (Figure 5A). By contrast, villous mesenchymal fibroblasts (VMFs) and placental vascular endothelial cells (PVECs) had low expression of these receptors (Figure 5A). The relative mRNA expression levels of receptors in VMFs and PVECs were significantly lower than those of receptors in VTs (Supplemental Figure 3). In accordance with the immunostaining results, there was some variability in the expression levels of HTLV-1 receptors on the surface of VTs (Supplemental Figure 4A). The cell purity of VTs, VMFs, and PVECs was 83.8%, 90.8%, and 97.4%, respectively (Supplemental Figure 4B).
Figure 5. Evaluation of the susceptibility of human placental cells to HTLV-1 infection.
(A) Surface expression of HTLV-1 receptor molecules on primary placental cells. The expressions of GLUT1, NRP1, and SDC1 were confirmed by immunofluorescence (green and red), and cell nuclei were stained with DAPI (blue). (B) Schematic structure of rVSVs expressing GFP, with or without an HTLV-1 envelope (HTEnv), and the WT construct is shown. (C) HTLV-1 envelope–dependent infection of placental cells. Cells were infected with non–G-complemented VSVΔG/GFP or VSVHTEnv2. At 24 hours postinfection (hpi), GFP expression was evaluated to identify rVSV-infected cells (green) among all cells stained with DAPI (blue). In A and C, stained cells were observed by fluorescence microscopy. Scale bars: 400 μm. (D) Evaluation of viral growth kinetics. In rVSV-infected placental cells, the total number of GFP-positive cells was counted every 24 hours until 72 hpi. The results were calculated as the cell number per square centimeter. (E) Relative expression level of HTLV-1 receptor genes in VTs. Cells were stimulated with IGF-1 or mock-stimulated. The normalized expression levels for mock controls were set as 1 as a reference. (F) Increase in HTLV-1 susceptibility in IGF-1–stimulated VTs. Cells were initially stimulated with IGF-1 or mock-stimulated and then infected with non–G-complemented VSVHTEnv2 in the presence or absence of HTLV-1 envelope–specific neutralizing antibody (LAT-27). The total number of GFP-positive cells was counted, and the relative infectivity was calculated. In D–F, asterisks represent significant differences versus the data for 24 hpi, control, and LAT-27(–), respectively. *P < 0.05, **P < 0.01 by 2-way ANOVA followed by Dunnett’s multiple-comparisons test. Data are means ± SD of 3 independent experiments. VT, villous trophoblasts; VMF, villous mesenchymal fibroblasts; PVEC, placental vascular endothelial cells.
Next, we tested the sensitivity of cells making up the blood-placental barrier to HTLV-1 using a primary villous cell culture system. A previously developed viral surrogate for HTLV-1 was used for quantitative evaluation (45). This recombinant vesicular stomatitis virus (VSV) was engineered to express the HTLV-1 envelope gene instead of the endogenous envelope G gene, along with a gene encoding GFP (VSVHTEnv2; Figure 5B). The VSVΔG/GFP virus was used as a negative control. As expected, significant infection was observed in VTs inoculated with VSVHTEnv2 but not in cells inoculated with VSVΔG/GFP (Figure 5C). By contrast, VMFs and PVECs showed limited infection (Figure 5C). In addition, the spread of infection over time was minimal in VMFs and PVECs but was apparent in VTs (Figure 5D). Levels of receptor expression were suggested to be a key factor in HTLV-1 sensitivity, and GLUT1 expression in trophoblasts was reported to be regulated primarily by IGF-1 (46). We therefore added IGF-1 to VTs and observed concentration-dependent increases in GLUT1 expression as well as increases in NRP1 expression (Figure 5E). SDC1 expression was unaffected (Figure 5E). IGF-1–stimulated VTs showed an approximately 2-fold increase in HTLV-1 sensitivity (Figure 5F). An HTLV-1 envelope–specific neutralizing antibody significantly inhibited infection of VTs by the surrogate virus irrespective of IGF-1 stimulation (Figure 5F), suggesting a potential new intervention to prevent MTCT by transferring neutralizing antibodies to the fetus through the placenta.
Characteristics of HTLV-1–infected placental cells in viral transmission.
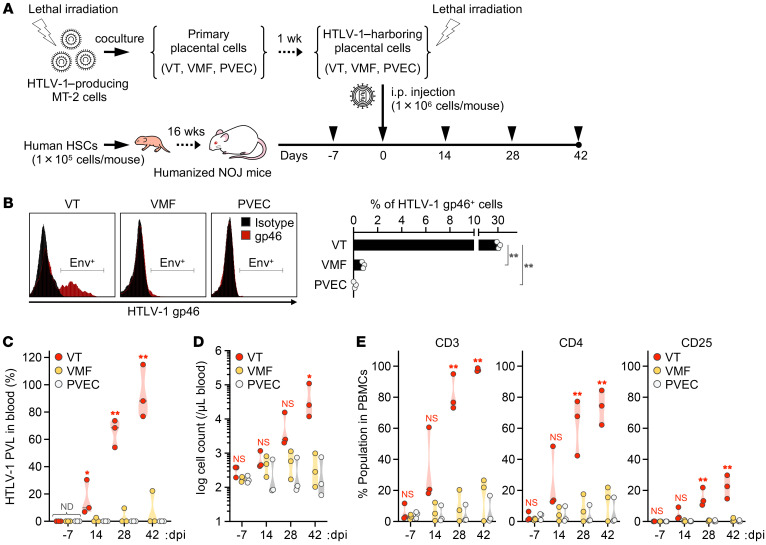
We examined whether placental cells infected with HTLV-1 expressed viral antigens. Considering that HTLV-1 infection is spread by cell-to-cell transmission, a coculture model of HTLV-1–producing cells and target primary cells was employed (47). Target VTs, VMFs, and PVECs were cocultured with HTLV-1–producing cells (Figure 6A), and the expression level of viral envelope glycoprotein on the surface of the target cells was analyzed by flow cytometry. The expression level of envelope glycoprotein was significantly higher in VTs than in the other cell types. The proportions of envelope-expressing cells were 30.7%, 0.8%, and 0.1% for VTs, VMFs, and PVECs, respectively (Figure 6B). We also investigated whether these 3 types of placental cell cocultured with HTLV-1–producing cells could give rise to the de novo infection of target T cells in vivo by using a humanized mouse model. After inoculation of the cells to humanized mice, the PVL in the peripheral blood, the leukocyte cell count, and the T cell phenotype were analyzed every 14 days (Figure 6A). HTLV-1 was detected in the peripheral blood of all 3 humanized mice inoculated with VTs cocultured with HTLV-1–producing cells, and the leukocyte cell count was increased significantly in proportion to the increase in PVL over time (Figure 6, C and D). In contrast, HTLV-1 was detected in the peripheral blood of 1 of 3 humanized mice inoculated with VMFs, and was not detected in any of the humanized mice inoculated with PVECs (Figure 6C). Leukocyte cell counts did not change in these 2 groups (Figure 6D). Furthermore, the percentage of CD3+ T cells among PBMCs was markedly increased in humanized mice inoculated with VTs, which was associated with significant increases in the percentages of CD4+ and CD25+ cells (Figure 6E). No significant differences were found between the VMF-inoculated group and the PVEC-inoculated group in the analysis of PVL, leukocyte cell count, and T cell phenotype (Figure 6, C–E). These results indicate that HTLV-1–infected VTs expressed a high level of viral antigens and induced the de novo infection of target T cells in vivo.
Figure 6. HTLV-1–infected trophoblasts induce de novo infection in humanized mice.
(A) Schematic of the experimental schedule. After humanization, 9 mice were inoculated i.p. with HTLV-1–harboring placental cells at 0 dpi (VT, n = 3; VMF, n = 3; PVEC, n = 3). Arrowheads indicate the time points of blood collection. All mice were observed carefully during the experimental period at 42 dpi. (B) Cell surface expression level of HTLV-1 envelope glycoprotein was analyzed in primary placental cells cocultured with MT-2 cells. The cells were stained with anti–HTLV-1 gp46 mAb or mouse IgG1 mAb as an isotype control. Representative cells (left) and the percentages of HTLV-1 gp46–positive cells (right) are shown. Data are means ± SD of 3 independent experiments. (C) Quantification of HTLV-1 PVL in the peripheral blood of inoculated mice. HTLV-1 PVL was determined by real-time PCR at 14, 28, and 42 dpi. One dot represents the result of an individual mouse. Undetected samples were given an arbitrary value of 100. (D and E) Human CD45+ leucocytes, CD3+ lymphocytes, total CD4+ T cells, and CD25+ T cells were routinely analyzed by flow cytometry. One dot represents the result of an individual mouse. The absolute numbers of human CD45+ leucocytes are shown in D. Frequencies of lymphocytes positive for the indicated marker are shown in E. CD3+ lymphocytes were gated to analyze the populations of CD4+ and CD25+ T cells. Asterisks represent significant differences versus the data for VTs in B, or PVEC-inoculated mice in C–E. *P < 0.05, **P < 0.01 by 1-way ANOVA followed by Dunnett’s multiple-comparisons test. ND, not detected.
Detection of HTLV-1–infected trophoblasts in the placenta of pregnant carriers.
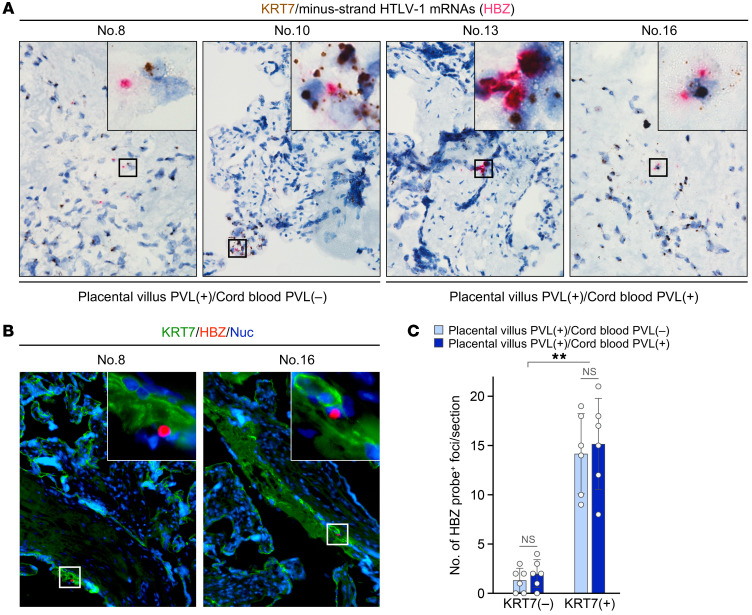
Last, we tested whether trophoblasts in the placental villous tissues were infected by HTLV-1. Using RNA ISH, we examined the mRNA expression of KRT7, a trophoblast marker (48), and HTLV-1 HBZ in the same tissue section. We found that some cells expressed both RNAs irrespective of the presence of HTLV-1 provirus in the cord blood (Figure 7A). Similar results were obtained using the HTLV-1 provirus–positive placental villous FFPE samples listed in Table 2. Costaining of HTLV-1 HBZ mRNA with KRT7 protein was performed using the same placental villous FFPE samples (Figure 7B). KRT7-positive cells had a significantly higher number of HBZ probe–positive foci compared with KRT7-negative cells (Figure 7C). In addition, there was no difference in the number of HBZ probe–positive foci in placental villi when pregnant carriers testing positive for HTLV-1 in the cord blood were compared with those negative for HTLV-1 in the cord blood (Figure 7C). This clearly demonstrated the presence of HTLV-1–infected trophoblasts in the placenta. Taken together, these results suggested that trophoblasts in the placenta are also major targets of HTLV-1 infection.
Figure 7. Detection of HTLV-1–infected placental trophoblasts in vivo.
(A) Simultaneous detection of 2 target genes within the same placental villous sections using RNA ISH duplex assay. Probes targeting HTLV-1 minus-strand mRNAs (HBZ) and KRT7, a trophoblast marker, were used to stain the tissue sections of pregnant carriers. The carriers with placental villous PVL (placental villous PVL–positive, and cord blood PVL–negative or –positive) were identical to those in Figure 4 and listed in Table 2. KRT7 was detected using DAB chromogen (brown), and HBZ was detected using fast red chromogen (red). ISH results are shown in the groups of subjects with HTLV-1–negative and –positive cord blood samples (nos. 8 and 10 vs. nos. 13 and 16). (B) RNA ISH combined with immunofluorescence (IF) assay was performed as a confirmatory test. KRT7 protein was detected using FITC (green), HBZ was detected using fast red as a fluorochrome (red), and cell nuclei were stained with NucBlue (blue). Black boxes in A and white boxes in B represent 25 μm squares and 50 μm squares, respectively, and indicate the regions shown at a higher magnification in the same panels. In A and B, data are representative of at least 2 nonserial tissue sections. (C) Quantitative evaluation of RNA ISH combined with IF results. The total number of HBZ probe–positive foci in KRT7-positive or -negative cells of each section was counted by fluorescence microscopy. Data are mean ± SD values from 6 tissue sections of each group. The multiple t test was performed to compare statistical differences between groups in subjects with HTLV-1–negative and –positive cord blood samples (nos. 7–12 vs. nos. 13–18). Asterisks in C represent significant differences between KRT7-negative and KRT7-positive cells; **P < 0.01 by Mann-Whitney U-test.
Discussion
The best-studied route for MTCT of HTLV-1 is breastfeeding; other routes, including intrauterine transmission, are only partially understood. Limited studies have focused on HTLV-1 infection during pregnancy (49–54). However, a subpopulation of infants who were formula-fed acquired HTLV-1 infection (8, 14), which prompted us to examine the potential routes of transmission before delivery. Currently, a major challenge is that it is difficult to follow all newborns of pregnant carriers for a long period and no follow-up survey has been carried out to determine whether newborns were infected with HTLV-1. In 1992, Kawase et al. reported that 18 of 717 infants born to pregnant carriers tested positive for HTLV-1 in the cord blood, but only 5 of these infants were followed up (51). They also showed that the 5 infants with cord blood HTLV-1 provirus detected by PCR were not HTLV-1 carriers based on the results of an antibody screening test (51). Conversely, all infants (4 of 4) who were identified as HTLV-1 carriers at 1 year of age or older tested negative for HTLV-1 provirus in the cord blood (51). Therefore, perinatal transmission was considered the potential route of HTLV-1 transmission to formula-fed infants rather than intrauterine transmission (51, 52). However, we found that HTLV-1 provirus was detected in the cord blood and placental villous tissues. We also tested sensitivity to HTLV-1 in the cells of the blood-placental barrier and found increased susceptibility to HTLV-1 infection in the VTs in accordance with previous reports (55–57). In addition, our study showed that HTLV-1 directly infected VTs in pregnant carriers and that HTLV-1 RNA was expressed at a high level in some of these infected cells.
Interestingly, the frequency of HTLV-1–positive cord blood samples among pregnant carriers was comparable to the frequency of HTLV-1 infection among exclusively formula-fed infants as previously reported (12–15). The presence of proviral DNA in the cord blood is highly suggestive of intrauterine transmission, but further investigation is warranted. Comparable PVLs in the placental and maternal blood were observed in women with HTLV-1 detected in the placenta when divided into those testing positive versus negative for HTLV-1 in the cord blood (Supplemental Table 1). It is difficult to predict the rate of HTLV-1 transmission to cord blood based on the status of placental infection. However, pregnant women with HTLV-1 detected in the placenta had significantly higher maternal blood PVL than those without HTLV-1 in the placenta (Table 1 and Figure 1B). This suggests that pregnant women with high maternal blood PVL are at high risk of placental infection. In the future, we plan to investigate the route of MTCT of HTLV-1 by long-term follow-up of infants born to women with HTLV-1 detected in the placenta, including those testing positive for HTLV-1 in the cord blood, according to the mode of nutrition.
Different viruses can be transmitted to the fetus through the infection of placental tissues including the villi (58, 59), and HTLV-1 may exploit the same route of transmission. For example, bovine leukemia virus (BLV), which belongs to the same genus as HTLV-1, Deltaretrovirus, was transmitted to the fetus through the placenta and birth canal in infected cattle (60, 61). It was also reported that maternal viral load was significantly correlated with the frequency of prenatal and perinatal transmission of BLV (61). Unfortunately, however, we did not observe such relationships in pregnant HTLV-1 carriers in this study, but the PVL in maternal blood was positively correlated with that in the placental villous tissue. Further investigation using nonhuman primate animal models is anticipated, as well as a prospective follow-up study of infants testing positive for HTLV-1 provirus in the cord blood and/or the placental villous tissues to examine whether HTLV-1 infection is established. Because natural infection with STLV-1, a model virus of HTLV-1, occurs in Japanese macaques, it is possible to analyze the kinetics of the virus in the placenta in the early, middle, and late stages of pregnancy and to follow up the offspring in detail (62). With significant advances in testing technologies in recent years, including standardized real-time PCR (63), digital PCR (64), chemiluminescence enzyme immunoassays (65), and line immunoassays (66), the use of higher-sensitivity testing is expected to lead to a more accurate understanding of the transplacental transmission of HTLV-1.
Previous reports revealed that GLUT1, NRP1, and SDC1 played a significant role in cellular susceptibility to HTLV-1 as coreceptors (41, 43, 44). Analysis of human primary placental cells revealed that trophoblasts expressed GLUT1 at a high level and correspondingly expressed NRP1 and SDC1. In accordance with the receptor expression, we demonstrated the higher susceptibility of primary trophoblasts to HTLV-1 infection. GLUT1 is a key molecule regulating glucose transport from mother to fetus (32, 40), and NRP1 functions as a receptor for placental growth factor in trophoblasts (67). SDC1 was reported to be expressed in syncytiotrophoblasts, which are involved in placental angiogenesis and inflammation (68). It is reasonable to assume that HTLV-1 can directly target and infect VTs in contact with the maternal blood of the intervillous space. Furthermore, in clinical practice, IGF-1, GLUT1, and NRP1 were reported to have important roles in fetal and placental development (69). Placental expression of GLUT1 was decreased in hypertensive disorders of pregnancy (HDP) and increased in gestational diabetes mellitus (GDM) (70, 71). In addition, placental expression of NRP1 was inhibited in preeclampsia and fetal growth deficiency (72, 73). When we compared 3 groups of pregnant HTLV-1 carriers (those who tested positive for HTLV-1 provirus in the maternal blood, in the placental villous tissue, and in the cord blood), no differences were observed in their clinical backgrounds including maternal age at birth, birth history, length of gestation, mode of delivery, and the child’s sex (Table 1 and Supplemental Table 1). We also compared birth weight and incidence of pregnancy-related complications, as fetal development is known to correlate with GLUT1 expression, but again no differences were observed (Table 1 and Supplemental Table 1). Concordantly, no differences were noted in the expression levels of HTLV-1 receptors in the placental villi of pregnant carriers regardless of their clinical background (Supplemental Figure 2). Furthermore, as mentioned above, HDP and GDM were reported to affect GLUT1 expression levels, but these conditions were not observed in pregnant women testing positive for HTLV-1 in the cord blood in the present study (Supplemental Table 1). Most of the pregnant women with pregnancy complications had mild conditions. Further accumulation of cases is needed to investigate the effect of HTLV-1 infection on pregnancy complications. The expression of GLUT1 and growth factors inside the placenta can change dynamically during fetal development, and there may be specific periods or conditions during pregnancy in which HTLV-1 infection is more easily established.
Interestingly, HTLV-1 provirus was detected in the placental villous tissues of 55.1% of pregnant carriers, but in the cord blood from only 2.4% of carriers. This discrepancy suggests that the placenta functions as a physical and biological barrier against HTLV-1 infection, although the mechanism involved remains unknown. Of the cells that make up the blood-placental barrier, VTs are highly susceptible to HTLV-1, whereas VMFs and PVECs are less susceptible (Figure 5, C and D, and Figure 6B). This may be critical for the placental barrier function. Furthermore, blood antibody titers were significantly higher in pregnant women with HTLV-1 detected in the placenta than in those without HTLV-1 in the placenta (Table 1). HTLV-1 was detected in the placenta of more than half of pregnant carriers, whereas HTLV-1 was detected in the cord blood of a limited number of women, suggesting that such high antibody titers in the blood may contribute to the prevention of intrauterine transmission.
The placental barrier was reported to deteriorate because of disease and infections (28). Indeed, HTLV-1 infection induced apoptosis in the placental cells of HTLV-1–seropositive pregnant women (74). HTLV-1 and EBV coinfection of trophoblasts was reported to significantly increase HTLV-1 antigen levels, possibly increasing the risk of de novo infection (75). Furthermore, coinfection of trophoblasts with HIV-1 and CMV or human herpesvirus resulted in high production of infectious HIV-1 (76, 77). These studies indicated that the transient activation of retroviral production by coinfecting viruses in trophoblasts may be involved in the mechanism of transplacental transmission of HTLV-1. These data were derived from limited studies, and further examination of the association of HTLV-1 and/or other viral infections with fetal development and pregnancy-related complications is required.
During transplacental infection by viruses such as HIV-1, which cause viremia and spread infection through infectious viral particles, maternal-fetal microtransfusion during delivery is a key route of MTCT (78) in addition to transmission through directly infected placental cells (79, 80). In contrast, HTLV-1 produces a limited number of infectious viral particles and establishes infection by cell-to-cell transmission (20, 21); therefore, the virus is more likely to exploit the direct infection of the placenta, including trophoblasts, as a route of transplacental infection instead of microtransfusion.
Notably, the expression of HTLV-1 antigens is thought to be strongly suppressed, but the behavior and localization of virus-expressing cells in tissues are not fully understood (81). In addition, multiple viral genes are encoded in the sense and antisense strands of the HTLV-1 provirus, resulting in complex expression patterns (81). Recently, a new technique with high sensitivity and specificity was established for detecting mRNA in tissue sections and was applied to the analysis of viral gene–expressing cells in tissues (82, 83). Using this technique, we developed a humanized mouse model to study the infection of human placental tissues by HTLV-1 in situ. As mentioned above, we successfully established a new method to detect viral mRNA in tissues using RNAscope (30). This method is highly sensitive for the detection and identification of cells expressing the plus- and minus-strand mRNAs of HTLV-1 and is useful for analyzing the behavior of HTLV-1 and identifying tissues harboring the virus in the dormant state.
In conclusion, to our knowledge this is the first report to demonstrate that HTLV-1 can directly infect and be expressed in placental villous tissues. Our results also highlight high viral expression levels in the villi of fetuses with HTLV-1–positive cord blood. Assuming that HTLV-1 is transmitted via the placenta, the expression of viral antigens in placental villi may be involved. In the humanized mouse model, HTLV-1–infected VTs caused the de novo infection of target T cells with HTLV-1. Similarly, HTLV-1–infected VTs expressed viral antigens in placental villous tissues of pregnant carriers, indicating they might be a potential source of the spread of HTLV-1 infection. Some VMFs also expressed HTLV-1 antigens and caused the de novo infection of target cells in vivo. Accordingly, it is of interest to elucidate whether the blood-placental barrier is disrupted by the HTLV-1 infection of VTs and VMFs and the mechanism by which intrauterine transmission to the fetus occurs. The route for MTCT of HTLV-1 other than breast milk has long been controversial. Although our findings do not substantiate the transplacental route of HTLV-1 transmission, the present study should aid future research to elucidate the mechanism by which intrauterine HTLV-1 transmission occurs.
Methods
Primary human cells.
Human primary placental cells, including villous trophoblasts (VTs), villous mesenchymal fibroblasts (VMFs), and placental vascular endothelial cells (PVECs), were purchased from Sciencell Research Laboratories (SCR) and cultured under the manufacturer’s recommended conditions. After normal delivery, placental cells were isolated from human placental villi and were cryopreserved at passage 1. Cells from 3 different lots (one lot reflected a single healthy donor) were used for downstream experiments. Optimized culture medium for each cell type was used as follows: Trophoblast Medium (SCR) for VTs, Fibroblast Medium (SCR) for VMFs, and Endothelial Cell Medium (SCR) for PVECs. All cells were cultured under an atmosphere containing 5% CO2 at 37°C. To evaluate the impact of IGF-1 stimulation on the expression levels of HTLV-1 receptor genes, VTs were plated in 12-well culture plates and incubated in Trophoblast Medium until the cultures were 70%–80% confluent. Cells were then switched to serum-free Trophoblast Medium for 24 hours before incubation in the same medium with or without human recombinant IGF-1 (PeproTech) (20 or 200 ng/mL) for 24 hours, as previously described (46). To infect placental cells with HTLV-1, VTs, VMFs, and PVECs were cocultured with lethally irradiated HTLV-1–producing MT-2 cells as previously reported (47). On the next day of cocultivation, the cells were thoroughly washed with PBS to remove residual MT-2 cells. Coculture medium was replaced with fresh cell type–specific medium every 24 hours, and the cells were further cultured for 1 week.
Donor selection and sample preparation.
The collection of donor samples was conducted from June 2012 to February 2016 at the Nagasaki University Hospital and regional clinics in the Nagasaki Prefecture. Based on the results of HTLV-1 screening tests (detailed below) conducted during the second trimester of pregnancy, 391 of 41,863 pregnant women (0.93%) were diagnosed as HTLV-1 carriers. During this period, 254 eligible pregnant seropositive women were included in this study when a triad of maternal blood, cord blood, and placental tissue were collected after receipt of written informed consent (Table 1 and Supplemental Table 1). The maternal blood was collected before delivery from each pregnant participant, and placental villous tissue and cord blood were collected after delivery. Third-trimester placentas were collected after vaginal delivery or cesarean section, and a first-trimester placenta was collected from a chronic ATL patient after an induced abortion at 15 weeks gestation (84). Collected placental tissues were immediately stored in PBS at 4°C. Following the mechanical removal of the amnion and blood vessels by extensive washing in Dulbecco’s PBS, placental villous tissues were cut into small pieces. Freshly chopped placental tissues were used for DNA extraction, and other tissue specimens were fixed in 10% formalin, embedded in paraffin, and sectioned for histological analysis (detailed below). PBMCs were isolated from the maternal and cord blood by Ficoll-Paque density gradient centrifugation (Lymphoprep; Alere Technologies AS) according to the manufacturer’s instructions. DNA was extracted from PBMCs (maternal and cord blood) and fresh placental tissues using a QIAamp DNA Mini Kit (QIAGEN) according to the manufacturer’s instructions. DNA samples were stored at −80°C until measurement of HTLV-1 proviral DNA levels (detailed below). This study was approved by the Institutional Review Board for Ethical, Legal, and Social Issues of Nagasaki University. All clinical samples were obtained from participants with informed consent.
Antibody tests.
Serological tests for HTLV-1 antibody in the maternal blood samples were performed using a chemiluminescent enzyme immunoassay (CLEIA; Lumipulse HTLV-I, Fujirebio), a particle agglutination assay (PA; Serodia-HTLV-I, Fujirebio), and Western blotting (WB; Problot HTLV-I; Fujirebio), according to the manufacturer’s instructions. HTLV-1 antibody titers in cord blood samples were determined by CLEIA. Antibody titers determined by CLEIA were represented as a cutoff index (COI), and samples with a COI greater than 45 were assigned a COI of 45. Nagasaki University defined HTLV-1 seropositivity as a titer ≥ 8 on PA or a COI ≥ 1.0 on CLEIA according to the manufacturer’s recommendations. Screening tests for HTLV-1 infection were primarily performed using CLEIA or PA. WB was performed afterward as a confirmatory test for samples with positive screening results (29, 85).
Quantitation of HTLV-1 PVL.
To quantitate HTLV-1 infection, the copy number of proviral DNA was measured using a real-time PCR method as previously reported (54, 63, 86). In brief, genomic DNA from human or mouse samples was used as a PCR template (100 ng per reaction). Quantitative real-time PCR was performed in triplicate to measure the copy numbers of the pX region of the HTLV-1 provirus and of the human β-globin (HBB) gene as an internal control. The primers and probe for HTLV-1 pX were as follows: 5′-CCCACTTCCCAGGGTTTGGA-3′ (forward), 5′-GGCCAGTAGGGCGTGA-3′ (reverse), and FAM-5′-CCAGTCTACGTGTTTGGAGACTGTGTACA-3′-BHQ (probe). The primers and probe for HBB were as follows: 5′-GTGCACCTGACTCCTGAGGAGA-3′ (forward), 5′-CCTTGATACCAACCTGCCCAG-3′ (reverse), and FAM-5′-AAGGTGAACGTGGATGAAGTTGGTGG-3′-BHQ (probe). The PVL was calculated as [(copy number of pX)/(copy number of HBB/2)] × 100, and expressed as pX copies per 100 cells (%).
STR genotyping analysis.
Genomic DNA derived from maternal blood, placental villous tissue, and cord blood samples of seropositive pregnant carriers was subjected to microsatellite analysis as previously reported (87). Briefly, genotyping was performed using the Powerplex 16 system (Promega), which amplifies 15 tetranucleotide STR loci and the sex-determining marker amelogenin. STR loci were analyzed using an ABI PRISM 3500 Genetic Analyzer (Applied Biosystems) and GeneMapper software (Applied Biosystems) according to the manufacturer’s instructions.
Establishment of humanized mice and HTLV-1 infection.
NOD/SCID Jak3-knockout (NOJ) mice were purchased from Kyudo Co. These mice were maintained under specific pathogen–free conditions and were handled in accordance with the institutional guidelines for animal experimentation at the National Institute of Infectious Diseases. Humanized mice were generated using an established method (31, 88). CD133+ hematopoietic stem cells were purified from human cord blood (provided by the Japanese Red Cross Kanto-Koshinetsu Cord Blood Bank, Tokyo, Japan) with the CD133 MicroBead Kit for Hematopoietic Tissue (Miltenyi Biotec) according to the manufacturer’s instructions. Cells with a purity greater than 95% were immediately transplanted intrahepatically into newborn NOJ mice aged 0–2 days (1 × 105 cells per mouse) or stored at −80°C until use. Twenty weeks after transplantation, 3 humanized mice were inoculated with irradiated MT-2 cells by oral administration (1 × 107 cells per mouse) or HTLV-1–harboring placental cells by i.p. injection (1 × 106 cells per mouse) to establish HTLV-1 infection.
Flow cytometry.
To evaluate the populations of human lymphocytes in humanized mice, flow cytometric analysis was performed as previously reported (31, 88). In brief, PBMCs were collected from mice, and then anti-human CD45–phycoerythrin-Cy7 (clone HI30), CD3-FITC (clone OKT3), CD4-phycoerythrin (clone OKT4), and CD25-allophycocyanin (clone BC96) mouse mAbs (Bay bioscience Co. Ltd.) were used to stain cell surface markers according to the manufacturer’s recommendations. For the staining of HTLV-1 envelope glycoprotein, anti–HTLV-1 gp46 mouse mAb (clone 67/5.5.13.1, Abcam), mouse IgG1 isotype control mAb (clone 15-6E10A7, Abcam), and a phycoerythrin-conjugated anti-mouse IgG goat polyclonal antibody (pAb) (Abcam) were used according to the manufacturer’s recommendations (31). To evaluate the purity of primary placental cells, anti-human KRT7-FITC (clone LP5K), CD31-FITC (clone WM-59), and CD90-FITC (clone 5E10) mouse mAbs (MilliporeSigma) were used to stain cell surface markers according to the manufacturer’s recommendations. Similarly, anti-human GLUT1-phycoerythrin (clone 202915, R&D Systems), NRP1-phycoerythrin (clone 446921, R&D Systems), and SDC1-phycoerythrin (clone B-A38, EXBIO) mouse mAbs were used to stain cell surface HTLV-1 receptors. Stained cells were immediately analyzed using a BD Accuri C6 flow cytometer with CFlow software (Becton Dickinson).
Immunofluorescence assay.
To assess the surface expression of GLUT1, NRP1, and SDC1, an immunofluorescence (IF) assay was performed as previously reported (31). In brief, for GLUT1 and NRP1 staining, VTs, VMFs, and PVECs were incubated with the relevant mouse mAb (R&D Systems), followed by incubation with a FITC-conjugated anti-mouse IgG goat pAb (Jackson ImmunoResearch Laboratories Inc.). For SDC1 staining, a phycoerythrin-conjugated mouse mAb (EXBIO) was used for the direct staining of cell surface proteins. Stained cells were observed and photographed with an EVOS FL Auto Imaging System (Thermo Fisher Scientific).
Infection and titration of recombinant VSV.
A recombinant vesicular stomatitis virus (rVSV) expressing HTLV-1 envelope glycoprotein was developed as previously reported (45). Briefly, the HTLV-1 envelope (HTEnv) gene was amplified from an HTLV-1 molecular clone, K30. The amplified product was excised using restriction enzymes and ligated into pVSVΔG/GFP in which the VSV G gene was replaced with the GFP gene (pVSVHTEnv) (45). Infectious rVSVs were recovered from pVSVHTEnv and pVSVΔG/GFP using an established method (89). These non–G-complemented virus stocks were then harvested and immediately used for downstream experiments.
For the titration of rVSVs, primary placental cells were inoculated with aliquots of viral stocks (0.1 mL per well) as described above. The cells were fixed every 24 hours until 72 hours after VSV inoculation, and the total number of fluorescent (GFP-positive) cells was counted. In addition, the virus stock of VSVHTEnv2 was preincubated with or without the HTLV-1 envelope glycoprotein–neutralizing mAb LAT-27 (90) (10 μg/mL) at 37°C for 1 hour and then used to inoculate primary trophoblasts. At 24 hours after inoculation, VSVHTEnv2-infected cells were assessed by counting of the number of GFP-positive cells. The cells were photographed using an EVOS FL Auto Imaging System, and the total number of GFP-positive cells in each well was counted by fluorescence microscopy using an Olympus IX70 microscope to calculate the relative infectivity of rVSVs.
Gene expression analysis.
Total RNA was extracted from VT, VMF, and PVEC cell suspensions using an RNeasy Mini Kit according to the manufacturer’s instructions (QIAGEN). Placental villous FFPE samples were treated with a deparaffinization solution, followed by the extraction of total RNA using an RNeasy FFPE Kit (QIAGEN). To analyze gene expression levels, a SYBR Green–based 1-step reverse transcriptase PCR was performed as described previously (31, 88). Human hypoxanthine phosphoribosyltransferase 1 (HPRT1) was used as an internal control gene. The sequences of the forward and reverse primers were as follows: GLUT1: 5′-CTGCTCATCAACCGCAAC-3′ and 5′-CTTCTTCTCCCGCATCATCT-3′; NRP1: 5′-CAGGTGATGACTTCCAGCTCA-3′ and 5′-CCCAGTGGCAGAAGGTCTTG-3′; SDC1: 5′-GCCGCAAATTGTGGCTACT-3′ and 5′-GCTGCGTGTCCTTCCAAGT-3′; HPRT1: 5′-CCTTGGTCAGGCAGTATAATCCA-3′ and 5′-CCAACAAAGTCTGGCTTATATCCAA-3′. Data were collected using the 7500 Fast real-time PCR System (Thermo Fisher Scientific), and relative expression levels were calculated using the 2–ΔΔCt method.
RNAscope ISH assays.
RNA ISH was performed using RNAscope 2.5 HD kits (Advanced Cell Diagnostics [ACD]) according to the manufacturer’s instructions. For manual single-plex RNAscope assays (30), FFPE tissue sections were baked for 1 hour at 60°C and deparaffinized, and endogenous peroxidases were quenched with hydrogen peroxide for 10 minutes at room temperature. Slides were then boiled for 30 minutes in RNAscope Target Retrieval Reagents and incubated with RNAscope Protease Plus for 30 minutes at 40°C before probe hybridization. Probes were then hybridized for 2 hours at 40°C followed by RNAscope amplification and DAB chromogenic detection. The C1 probes targeting HTLV-1 plus-strand (pX) and minus-strand (HBZ) mRNAs were designed and synthesized by ACD (catalog 529981 and 432901, respectively). The probes targeting the human POLR2A and bacterial DapB genes were purchased from ACD and used as positive and negative controls (catalog 310451 and 310043, respectively). For the statistical analysis of pX probe–stained samples, positive foci (most likely resembling single cells) were counted manually for each slide as previously reported (91).
For the manual duplex RNAscope assays (92), the RNAscope 2.5 HD Duplex Reagent Kit was used (catalog 322430). As described above, FFPE tissue sections were pretreated with reagents and hybridized with probes. Fast red chromogenic detection for the C2 probe was performed first, followed by DAB chromogenic detection of the C1 probe. C1 and C2 probes targeting human KRT7 and HBZ mRNAs were designed and synthesized by ACD (catalog 550151 and 432901-C2, respectively). Duplex positive and negative control probes were purchased from ACD (catalog 321641 and 320751, respectively). Slides were counterstained with hematoxylin solution and visualized by standard bright-field microscopy.
For the simultaneous visualization of viral RNA and cell surface markers, an assay combining RNA ISH with IF was developed (83). Anti–human KRT7 mouse mAb (clone OV-TL 12/30, MilliporeSigma) and FITC-conjugated anti-mouse IgG goat pAb (Jackson ImmunoResearch Laboratories Inc.) were used for fluorescent detection. RNAscope was performed using a 2.5 HD Reagent Kit–RED kit with the C1 probe targeting HTLV-1 HBZ mRNA, followed by a standard IF assay according to the manufacturer’s instructions. Slides were mounted with ProLong Glass Antifade Mountant with NucBlue Stain and visualized with an EVOS FL Auto Imaging System (Thermo Fisher Scientific).
Statistics.
Differences between groups were assessed using the Mann-Whitney U test, 2-tailed multiple t tests, the Kruskal-Wallis test followed by Dunn’s multiple-comparisons test, 1-way or 2-way ANOVA followed by Dunnett’s multiple-comparisons test, and the χ2 test. The strength of associations between variables was evaluated using Spearman’s rank correlation coefficient (ρ). All statistical analyses were performed using GraphPad Prism 8 (GraphPad Software) or SPSS Statistics v23 (IBM). P values less than 0.05 were considered statistically significant.
Study approval.
All protocols involving human subjects and animal experiments were reviewed and approved by the Institutional Review Board of Nagasaki University and the National Institute of Infectious Diseases (approval numbers 12052814 and 117113, respectively).
Author contributions
KT and NF performed experiments, analyzed and interpreted data, designed figures, wrote the manuscript, and contributed to study design. KT visualized the figures. KO performed and supervised virological experiments. TT, S Miura, YH, AN, and NK collected and provided clinical specimens. HH, DS, and KY performed and supervised the evaluation of HTLV-1 PVL and serological tests. ES and TM performed and supervised histological experiments. MK and S Matsuoka contributed to data interpretation. KM and IH designed and supervised the study, contributed to data interpretation, and wrote the manuscript. All authors contributed to the discussion of the project and reviewed and approved the manuscript. The authorship order of the co–first authors was assigned on the basis of the relative contributions of each person and was agreed upon by all authors.
Supplementary Material
Acknowledgments
We gratefully acknowledge the technical assistance of Keiko Imai and Keiko Furuhata of the National Institute of Infectious Diseases and Yuko Sakakida, Maasa Miyamoto, Ayako Ueyama, and Yasuko Noguchi of Nagasaki University. We also thank the Edanz Group for editing a draft of the manuscript. This work was supported in part by Grants-in-Aid for Scientific Research (C) (grants 18K09265 to S Miura, 26462495 and 17K11241 to KM, and 15K09462 and 18K08378 to IH) and Grants-in-Aid for Young Scientists (grants 19K16032 to KT and 16K20198 and 18K16805 to NF) from the Japan Society for the Promotion of Science, and a Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development (JP19fk0108037 to IH).
Version 1. 10/19/2020
Electronic publication
Version 2. 11/02/2020
Print issue publication
Funding Statement
Grant-in-Aid for Scientific Research (C)
Grant-in-Aid for Young Scientists
Research Program on Emerging and Re-emerging Infectious Diseases
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(11):6171–6186.https://doi.org/10.1172/JCI135525.
Contributor Information
Kenta Tezuka, Email: tezukakn@niid.go.jp.
Naoki Fuchi, Email: naoki@fuchi-ladies.or.jp.
Kazu Okuma, Email: kokuma@niid.go.jp.
Takashi Tsukiyama, Email: t.tsukky@mac.com.
Shoko Miura, Email: shokom@nagasaki-u.ac.jp.
Yuri Hasegawa, Email: haseyuri@nagasaki-u.ac.jp.
Ai Nagata, Email: aimelody@nagasaki-u.ac.jp.
Nahoko Komatsu, Email: nh22a.ilme@gmail.com.
Hiroo Hasegawa, Email: hhase@nagasaki-u.ac.jp.
Daisuke Sasaki, Email: d-sasaki@nagasaki-u.ac.jp.
Eita Sasaki, Email: esasaki@niid.go.jp.
Takuo Mizukami, Email: tmiz@niid.go.jp.
Madoka Kuramitsu, Email: mkura@nih.go.jp.
Sahoko Matsuoka, Email: sahoko@niid.go.jp.
Katsunori Yanagihara, Email: k-yanagi@nagasaki-u.ac.jp.
Kiyonori Miura, Email: kiyonori@nagasaki-u.ac.jp.
Isao Hamaguchi, Email: 130hama@niid.go.jp.
References
- 1.Watanabe T. Current status of HTLV-1 infection. Int J Hematol. 2011;94(5):430–434. doi: 10.1007/s12185-011-0934-4. [DOI] [PubMed] [Google Scholar]
- 2.Gessain A, Cassar O. Epidemiological aspects and world distribution of HTLV-1 infection. Front Microbiol. 2012;3:388. doi: 10.3389/fmicb.2012.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc Natl Acad Sci U S A. 1982;79(6):2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osame M, et al. HTLV-I associated myelopathy, a new clinical entity. Lancet. 1986;1(8488):1031–1032. doi: 10.1016/s0140-6736(86)91298-5. [DOI] [PubMed] [Google Scholar]
- 5.Mochizuki M, et al. HTLV-I uveitis: a distinct clinical entity caused by HTLV-I. Jpn J Cancer Res. 1992;83(3):236–239. doi: 10.1111/j.1349-7006.1992.tb00092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy EL, et al. Modelling the risk of adult T-cell leukemia/lymphoma in persons infected with human T-lymphotropic virus type I. Int J Cancer. 1989;43(2):250–253. doi: 10.1002/ijc.2910430214. [DOI] [PubMed] [Google Scholar]
- 7.Hino S, et al. Breaking the cycle of HTLV-I transmission via carrier mothers’ milk. Lancet. 1987;2(8551):158–159. doi: 10.1016/s0140-6736(87)92358-0. [DOI] [PubMed] [Google Scholar]
- 8.Hino S. Establishment of the milk-borne transmission as a key factor for the peculiar endemicity of human T-lymphotropic virus type 1 (HTLV-1): the ATL Prevention Program Nagasaki. Proc Jpn Acad Ser B Phys Biol Sci. 2011;87(4):152–166. doi: 10.2183/pjab.87.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribeiro MA, et al. Blocking vertical transmission of human T cell lymphotropic virus type 1 and 2 through breastfeeding interruption. Pediatr Infect Dis J. 2012;31(11):1139–1143. doi: 10.1097/INF.0b013e318263215e. [DOI] [PubMed] [Google Scholar]
- 10.Rosadas C, Malik B, Taylor GP, Puccioni-Sohler M. Estimation of HTLV-1 vertical transmission cases in Brazil per annum. PLoS Negl Trop Dis. 2018;12(11):e0006913. doi: 10.1371/journal.pntd.0006913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishijima T, Shimada S, Noda H, Miyake K. Towards the elimination of HTLV-1 infection in Japan. Lancet Infect Dis. 2019;19(1):15–16. doi: 10.1016/S1473-3099(18)30735-7. [DOI] [PubMed] [Google Scholar]
- 12.Hino S, Katamine S, Miyata H, Tsuji Y, Yamabe T, Miyamoto T. Primary prevention of HTLV-1 in Japan. Leukemia. 1997;11(suppl 3):57–59. [PubMed] [Google Scholar]
- 13.Ureta-Vidal A, et al. Mother-to-child transmission of human T-cell-leukemia/lymphoma virus type I: implication of high antiviral antibody titer and high proviral load in carrier mothers. Int J Cancer. 1999;82(6):832–836. doi: 10.1002/(SICI)1097-0215(19990909)82:6<832::AID-IJC11>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 14.Moriuchi H, Masuzaki H, Doi H, Katamine S. Mother-to-child transmission of human T-cell lymphotropic virus type 1. Pediatr Infect Dis J. 2013;32(2):175–177. doi: 10.1097/INF.0b013e31827efc39. [DOI] [PubMed] [Google Scholar]
- 15.Paiva AM, et al. Risk factors associated with HTLV-1 vertical transmission in Brazil: longer breastfeeding, higher maternal proviral load and previous HTLV-1-infected offspring. Sci Rep. 2018;8(1):7742. doi: 10.1038/s41598-018-25939-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujino T, Fujiyoshi T, Yashiki S, Sonoda S, Otsuka H, Nagata Y. HTLV-I transmission from mother to fetus via placenta. Lancet. 1992;340(8828):1157. doi: 10.1016/0140-6736(92)93181-l. [DOI] [PubMed] [Google Scholar]
- 17.Bittencourt AL, Sabino EC, Costa MC, Pedroso C, Moreira L. No evidence of vertical transmission of HTLV-I in bottle-fed children. Rev Inst Med Trop Sao Paulo. 2002;44(2):63–65. doi: 10.1590/S0036-46652002000200002. [DOI] [PubMed] [Google Scholar]
- 18.Sawada T, Taguchi T, Miyoshi I, Nakachi H, Nagayama T. HTLV-1 proviral DNA in oral aspirates of newborns born to seropositive mothers. JAMA. 1995;273(4):284. doi: 10.1001/jama.1995.03520280028023. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto T, et al. Inhibitory activity in saliva of cell-to-cell transmission of human T-cell lymphotropic virus type 1 in vitro: evaluation of saliva as an alternative source of transmission. J Clin Microbiol. 1995;33(6):1510–1515. doi: 10.1128/JCM.33.6.1510-1515.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igakura T, et al. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science. 2003;299(5613):1713–1716. doi: 10.1126/science.1080115. [DOI] [PubMed] [Google Scholar]
- 21.Pais-Correia AM, et al. Biofilm-like extracellular viral assemblies mediate HTLV-1 cell-to-cell transmission at virological synapses. Nat Med. 2010;16(1):83–89. doi: 10.1038/nm.2065. [DOI] [PubMed] [Google Scholar]
- 22.Hoxie JA, Matthews DM, Cines DB. Infection of human endothelial cells by human T-cell leukemia virus type I. Proc Natl Acad Sci U S A. 1984;81(23):7591–7595. doi: 10.1073/pnas.81.23.7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakai M, et al. Infection of human synovial cells by human T cell lymphotropic virus type I. Proliferation and granulocyte/macrophage colony-stimulating factor production by synovial cells. J Clin Invest. 1993;92(4):1957–1966. doi: 10.1172/JCI116789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura H, et al. Direct infection of primary salivary gland epithelial cells by human T lymphotropic virus type I in patients with Sjögren’s syndrome. Arthritis Rheumatol. 2015;67(4):1096–1106. doi: 10.1002/art.39009. [DOI] [PubMed] [Google Scholar]
- 25.Masuzaki H, et al. Labor increases maternal DNA contamination in cord blood. Clin Chem. 2004;50(9):1709–1711. doi: 10.1373/clinchem.2004.036517. [DOI] [PubMed] [Google Scholar]
- 26.Lee TH, Chafets DM, Biggar RJ, McCune JM, Busch MP. The role of transplacental microtransfusions of maternal lymphocytes in in utero HIV transmission. J Acquir Immune Defic Syndr. 2010;55(2):143–147. doi: 10.1097/QAI.0b013e3181eb301e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maidji E, McDonagh S, Genbacev O, Tabata T, Pereira L. Maternal antibodies enhance or prevent cytomegalovirus infection in the placenta by neonatal Fc receptor-mediated transcytosis. Am J Pathol. 2006;168(4):1210–1226. doi: 10.2353/ajpath.2006.050482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arora N, Sadovsky Y, Dermody TS, Coyne CB. Microbial vertical transmission during human pregnancy. Cell Host Microbe. 2017;21(5):561–567. doi: 10.1016/j.chom.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsumoto C, et al. Analysis of HTLV-1 proviral load (PVL) and antibody detected with various kinds of tests in Japanese blood donors to understand the relationship between PVL and antibody level and to gain insights toward better antibody testing. J Med Virol. 2017;89(8):1469–1476. doi: 10.1002/jmv.24802. [DOI] [PubMed] [Google Scholar]
- 30.Wang F, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14(1):22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tezuka K, et al. Control of human T-cell leukemia virus type 1 (HTLV-1) infection by eliminating envelope protein-positive cells with recombinant vesicular stomatitis viruses encoding HTLV-1 primary receptor. J Virol. 2018;92(4):e01885-17. doi: 10.1128/JVI.01885-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jansson T, Myatt L, Powell TL. The role of trophoblast nutrient and ion transporters in the development of pregnancy complications and adult disease. Curr Vasc Pharmacol. 2009;7(4):521–533. doi: 10.2174/157016109789043982. [DOI] [PubMed] [Google Scholar]
- 33.Kitano T, et al. Conditionally immortalized syncytiotrophoblast cell lines as new tools for study of the blood-placenta barrier. Biol Pharm Bull. 2004;27(6):753–759. doi: 10.1248/bpb.27.753. [DOI] [PubMed] [Google Scholar]
- 34.Hemmings DG, Kilani R, Nykiforuk C, Preiksaitis J, Guilbert LJ. Permissive cytomegalovirus infection of primary villous term and first trimester trophoblasts. J Virol. 1998;72(6):4970–4979. doi: 10.1128/JVI.72.6.4970-4979.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novakovic B, Saffery R. DNA methylation profiling highlights the unique nature of the human placental epigenome. Epigenomics. 2010;2(5):627–638. doi: 10.2217/epi.10.45. [DOI] [PubMed] [Google Scholar]
- 36.Zachar V, et al. Genetic analysis reveals ongoing HIV type 1 evolution in infected human placental trophoblast. AIDS Res Hum Retroviruses. 1999;15(18):1673–1683. doi: 10.1089/088922299309711. [DOI] [PubMed] [Google Scholar]
- 37.Bhat P, Anderson DA. Hepatitis B virus translocates across a trophoblastic barrier. J Virol. 2007;81(13):7200–7207. doi: 10.1128/JVI.02371-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheridan MA, et al. Vulnerability of primitive human placental trophoblast to Zika virus. Proc Natl Acad Sci USA. 2017;114(9):E1587–E1596. doi: 10.1073/pnas.1616097114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muehlenbachs A, et al. Ebola virus disease in pregnancy: clinical, histopathologic, and immunohistochemical findings. J Infect Dis. 2017;215(1):64–69. doi: 10.1093/infdis/jiw206. [DOI] [PubMed] [Google Scholar]
- 40.Takata K, Hirano H, Kasahara M. Transport of glucose across the blood-tissue barriers. Int Rev Cytol. 1997;172:1–53. doi: 10.1016/s0074-7696(08)62357-8. [DOI] [PubMed] [Google Scholar]
- 41.Manel N, Kim FJ, Kinet S, Taylor N, Sitbon M, Battini JL. The ubiquitous glucose transporter GLUT-1 is a receptor for HTLV. Cell. 2003;115(4):449–459. doi: 10.1016/S0092-8674(03)00881-X. [DOI] [PubMed] [Google Scholar]
- 42.Jin Q, Agrawal L, VanHorn-Ali Z, Alkhatib G. Infection of CD4+ T lymphocytes by the human T cell leukemia virus type 1 is mediated by the glucose transporter GLUT-1: evidence using antibodies specific to the receptor’s large extracellular domain. Virology. 2006;349(1):184–196. doi: 10.1016/j.virol.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 43.Ghez D, et al. Neuropilin-1 is involved in human T-cell lymphotropic virus type 1 entry. J Virol. 2006;80(14):6844–6854. doi: 10.1128/JVI.02719-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka A, et al. Entry of human T-cell leukemia virus type 1 is augmented by heparin sulfate proteoglycans bearing short heparin-like structures. J Virol. 2012;86(6):2959–2969. doi: 10.1128/JVI.05783-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okuma K, Dalton KP, Buonocore L, Ramsburg E, Rose JK. Development of a novel surrogate virus for human T-cell leukemia virus type 1: inhibition of infection by osteoprotegerin. J Virol. 2003;77(15):8562–8569. doi: 10.1128/JVI.77.15.8562-8569.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baumann MU, et al. Regulation of human trophoblast GLUT1 glucose transporter by insulin-like growth factor I (IGF-I) PLoS One. 2014;9(8):e106037. doi: 10.1371/journal.pone.0106037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma G, Yasunaga JI, Ohshima K, Matsumoto T, Matsuoka M. Pentosan polysulfate demonstrates anti-human T-cell leukemia virus type 1 activities in vitro and in vivo. J Virol. 2019;93(16):e00413–19. doi: 10.1128/JVI.00413-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee CQ, et al. What is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Reports. 2016;6(2):257–272. doi: 10.1016/j.stemcr.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saito S, et al. Identification of HTLV-I sequence in cord blood mononuclear cells of neonates born to HTLV-I antigen/antibody-positive mothers by polymerase chain reaction. Jpn J Cancer Res. 1990;81(9):890–895. doi: 10.1111/j.1349-7006.1990.tb02663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Satow Y, et al. Detection of HTLV-I antigen in peripheral and cord blood lymphocytes from carrier mothers. Lancet. 1991;338(8772):915–916. doi: 10.1016/0140-6736(91)91775-P. [DOI] [PubMed] [Google Scholar]
- 51.Kawase K, et al. Maternal transmission of HTLV-1 other than through breast milk: discrepancy between the polymerase chain reaction positivity of cord blood samples for HTLV-1 and the subsequent seropositivity of individuals. Jpn J Cancer Res. 1992;83(9):968–977. doi: 10.1111/j.1349-7006.1992.tb02009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katamine S, et al. HTLV-I proviral DNA in umbilical cord blood of babies born to carrier mothers. Lancet. 1994;343(8909):1326–1327. doi: 10.1016/S0140-6736(94)92469-4. [DOI] [PubMed] [Google Scholar]
- 53.Kazi A, Miyata H, Kamahora T, Kurokawa K, Katamine S, Hino S. Deleted HTLV-1 provirus in cord-blood samples of babies born to HTLV-1-carrier mothers. Int J Cancer. 1998;77(5):701–704. doi: 10.1002/(SICI)1097-0215(19980831)77:5<701::AID-IJC7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 54.Fuchi N, et al. Natural course of human T-cell leukemia virus type 1 proviral DNA levels in carriers during pregnancy. J Infect Dis. 2018;217(9):1383–1389. doi: 10.1093/infdis/jiy017. [DOI] [PubMed] [Google Scholar]
- 55.Liu X, et al. Cell-mediated transmission of human T cell lymphotropic virus type I to human malignant trophoblastic cells results in long-term persistent infection. J Gen Virol. 1995;76(pt 1):167–173. doi: 10.1099/0022-1317-76-1-167. [DOI] [PubMed] [Google Scholar]
- 56.Liu X, Zachar V, Hager H, Koppelhus U, Ebbesen P. Transfer of human T cell lymphotropic virus type I to human term trophoblast cells in vitro. J Gen Virol. 1996;77(pt 2):369–374. doi: 10.1099/0022-1317-77-2-369. [DOI] [PubMed] [Google Scholar]
- 57.Liu X, Zachar V, Hager H, Ebbesen P. Clonal analysis suggests provirus expression in a subpopulation of human malignant trophoblast cells harbouring the human T cell lymphotropic virus type-I genome. Res Virol. 1996;147(1):45–51. doi: 10.1016/0923-2516(96)80239-1. [DOI] [PubMed] [Google Scholar]
- 58.Tabata T, et al. Zika virus targets different primary human placental cells, suggesting two routes for vertical transmission. Cell Host Microbe. 2016;20(2):155–166. doi: 10.1016/j.chom.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Platt DJ, Smith AM, Arora N, Diamond MS, Coyne CB, Miner JJ. Zika virus-related neurotropic flaviviruses infect human placental explants and cause fetal demise in mice. Sci Transl Med. 2018;10(426):eaao7090. doi: 10.1126/scitranslmed.aao7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meas S, Usui T, Ohashi K, Sugimoto C, Onuma M. Vertical transmission of bovine leukemia virus and bovine immunodeficiency virus in dairy cattle herds. Vet Microbiol. 2002;84(3):275–282. doi: 10.1016/S0378-1135(01)00458-8. [DOI] [PubMed] [Google Scholar]
- 61.Mekata H, et al. Evaluation of the natural perinatal transmission of bovine leukaemia virus. Vet Rec. 2015;176(10):254. doi: 10.1136/vr.102464. [DOI] [PubMed] [Google Scholar]
- 62.Miura M, et al. Characterization of simian T-cell leukemia virus type 1 in naturally infected Japanese macaques as a model of HTLV-1 infection. Retrovirology. 2013;10:118. doi: 10.1186/1742-4690-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuramitsu M, et al. Standardization of quantitative PCR for human T-cell leukemia virus type 1 in Japan: a collaborative study. J Clin Microbiol. 2015;53(11):3485–3491. doi: 10.1128/JCM.01628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuramitsu M, et al. Development of reference material with assigned value for human T-cell leukemia virus type 1 quantitative PCR in Japan. Microbiol Immunol. 2018;62(10):673–676. doi: 10.1111/1348-0421.12644. [DOI] [PubMed] [Google Scholar]
- 65.da Silva Brito V, et al. Performance of commercially available serological screening tests for human T-cell lymphotropic virus infection in Brazil. J Clin Microbiol. 2018;56(12):e00961-18. doi: 10.1128/JCM.00961-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campos KR, et al. Line immunoassay for confirmation and discrimination of human T-cell lymphotropic virus infections in inconclusive Western blot serum samples from Brazil. J Clin Microbiol. 2019;58(1):e01384-19. doi: 10.1128/JCM.01384-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chau K, Hennessy A, Makris A. Placental growth factor and pre-eclampsia. J Hum Hypertens. 2017;31(12):782–786. doi: 10.1038/jhh.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szabo S, et al. Changes of placental syndecan-1 expression in preeclampsia and HELLP syndrome. Virchows Arch. 2013;463(3):445–458. doi: 10.1007/s00428-013-1426-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Acosta O, et al. Increased glucose and placental GLUT-1 in large infants of obese nondiabetic mothers. Am J Obstet Gynecol. 2015;212(2):227.e1–227.e7. doi: 10.1016/j.ajog.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 70.Lüscher BP, et al. Placental glucose transporter (GLUT)-1 is down-regulated in preeclampsia. Placenta. 2017;55:94–99. doi: 10.1016/j.placenta.2017.04.023. [DOI] [PubMed] [Google Scholar]
- 71.Stanirowski PJ, Szukiewicz D, Pyzlak M, Abdalla N, Sawicki W, Cendrowski K. Impact of pre-gestational and gestational diabetes mellitus on the expression of glucose transporters GLUT-1, GLUT-4 and GLUT-9 in human term placenta. Endocrine. 2017;55(3):799–808. doi: 10.1007/s12020-016-1202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arad A, Nammouz S, Nov Y, Ohel G, Bejar J, Vadasz Z. The expression of neuropilin-1 in human placentas from normal and preeclamptic pregnancies. Int J Gynecol Pathol. 2017;36(1):42–49. doi: 10.1097/PGP.0000000000000283. [DOI] [PubMed] [Google Scholar]
- 73.Maulik D, et al. Down-regulation of placental neuropilin-1 in fetal growth restriction. Am J Obstet Gynecol. 2016;214(2):279.e1–279.e9. doi: 10.1016/j.ajog.2015.09.068. [DOI] [PubMed] [Google Scholar]
- 74.Fujino T, Iwamoto I, Otsuka H, Ikeda T, Takesako S, Nagata Y. Apoptosis in placentas from human T-lymphotropic virus type I-seropositive pregnant women: a possible defense mechanism against transmission from mother to fetus. Obstet Gynecol. 1999;94(2):279–283. doi: 10.1016/s0029-7844(99)00322-1. [DOI] [PubMed] [Google Scholar]
- 75.Tóth FD, et al. Epstein-Barr virus permissively infects human syncytiotrophoblasts in vitro and induces replication of human T cell leukemia-lymphoma virus type I in dually infected cells. Virology. 1997;229(2):400–414. doi: 10.1006/viro.1997.8449. [DOI] [PubMed] [Google Scholar]
- 76.Tóth FD, et al. Interactions between human immunodeficiency virus type 1 and human cytomegalovirus in human term syncytiotrophoblast cells coinfected with both viruses. J Virol. 1995;69(4):2223–2232. doi: 10.1128/JVI.69.4.2223-2232.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Csoma E, et al. Human herpesvirus 6 variant a infects human term syncytiotrophoblasts in vitro and induces replication of human immunodeficiency virus type 1 in dually infected cells. J Med Virol. 2002;67(1):67–87. doi: 10.1002/jmv.2194. [DOI] [PubMed] [Google Scholar]
- 78.Kwiek JJ, et al. Maternal-fetal DNA admixture is associated with intrapartum mother-to-child transmission of HIV-1 in Blantyre, Malawi. J Infect Dis. 2008;197(10):1378–1381. doi: 10.1086/587646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langston C, et al. Excess intrauterine fetal demise associated with maternal human immunodeficiency virus infection. J Infect Dis. 1995;172(6):1451–1460. doi: 10.1093/infdis/172.6.1451. [DOI] [PubMed] [Google Scholar]
- 80.Fazeley F, Hu J, Thirkill TL, Douglas GC. Infection of primary human placental fibroblasts with HIV-1, HIV-2, and SIV. Arch Virol. 1997;142(11):2237–2248. doi: 10.1007/s007050050238. [DOI] [PubMed] [Google Scholar]
- 81.Bangham CRM, Matsuoka M. Human T-cell leukaemia virus type 1: parasitism and pathogenesis. Philos Trans R Soc Lond, B, Biol Sci. 2017;372(1732):20160272. doi: 10.1098/rstb.2016.0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deleage C, et al. Defining HIV and SIV reservoirs in lymphoid tissues. Pathog Immun. 2016;1(1):68–106. doi: 10.20411/pai.v1i1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maidji E, Somsouk M, Rivera JM, Hunt PW, Stoddart CA. Replication of CMV in the gut of HIV-infected individuals and epithelial barrier dysfunction. PLoS Pathog. 2017;13(2):e1006202. doi: 10.1371/journal.ppat.1006202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fuchi N, et al. Adult T-cell leukemia-lymphoma in a pregnant woman diagnosed as a human T-cell lymphotropic virus type 1 carrier. J Obstet Gynaecol Res. 2016;42(3):336–340. doi: 10.1111/jog.12904. [DOI] [PubMed] [Google Scholar]
- 85.Ishihara K, et al. Relevance of molecular tests for HTLV-1 infection as confirmatory tests after the first sero-screening. J Immunoassay Immunochem. 2014;35(1):74–82. doi: 10.1080/15321819.2013.792832. [DOI] [PubMed] [Google Scholar]
- 86.Kamihira S, et al. Intra- and inter-laboratory variability in human T-cell leukemia virus type-1 proviral load quantification using real-time polymerase chain reaction assays: a multi-center study. Cancer Sci. 2010;101(11):2361–2367. doi: 10.1111/j.1349-7006.2010.01720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fuchi N, Miura K, Doi H, Li TS, Masuzaki H. Feasibility of placenta-derived mesenchymal stem cells as a tool for studying pregnancy-related disorders. Sci Rep. 2017;7:46220. doi: 10.1038/srep46220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tezuka K, et al. An animal model of adult T-cell leukemia: humanized mice with HTLV-1-specific immunity. Blood. 2014;123(3):346–355. doi: 10.1182/blood-2013-06-508861. [DOI] [PubMed] [Google Scholar]
- 89.Lawson ND, Stillman EA, Whitt MA, Rose JK. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci U S A. 1995;92(10):4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tanaka Y, Zeng L, Shiraki H, Shida H, Tozawa H. Identification of a neutralization epitope on the envelope gp46 antigen of human T cell leukemia virus type I and induction of neutralizing antibody by peptide immunization. J Immunol. 1991;147(1):354–360. [PubMed] [Google Scholar]
- 91.Yú SQ, et al. Specific detection of two divergent simian arteriviruses using RNAscope in situ hybridization. PLoS One. 2016;11(3):e0151313. doi: 10.1371/journal.pone.0151313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Anderson CM, et al. Fully automated RNAscope in situ hybridization assays for formalin-fixed paraffin-embedded cells and tissues. J Cell Biochem. 2016;117(10):2201–2208. doi: 10.1002/jcb.25606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.