Abstract
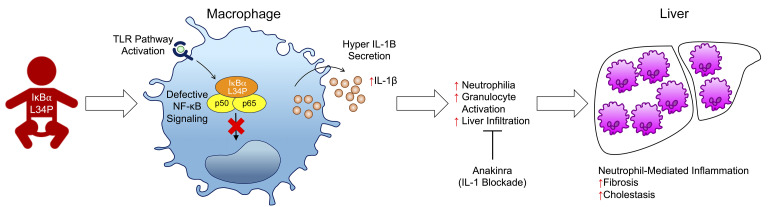
Although IKK-β has previously been shown as a negative regulator of IL-1β secretion in mice, this role has not been proven in humans. Genetic studies of NF-κB signaling in humans with inherited diseases of the immune system have not demonstrated the relevance of the NF-κB pathway in suppressing IL-1β expression. Here, we report an infant with a clinical pathology comprising neutrophil-mediated autoinflammation and recurrent bacterial infections. Whole-exome sequencing revealed a de novo heterozygous missense mutation of NFKBIA, resulting in a L34P IκBα variant that severely repressed NF-κB activation and downstream cytokine production. Paradoxically, IL-1β secretion was elevated in the patient’s stimulated leukocytes, in her induced pluripotent stem cell–derived macrophages, and in murine bone marrow–derived macrophages containing the L34P mutation. The patient’s hypersecretion of IL-1β correlated with activated neutrophilia and liver fibrosis with neutrophil accumulation. Hematopoietic stem cell transplantation reversed neutrophilia, restored a resting state in neutrophils, and normalized IL-1β release from stimulated leukocytes. Additional therapeutic blockade of IL-1 ameliorated liver damage, while decreasing neutrophil activation and associated IL-1β secretion. Our studies reveal a previously unrecognized role of human IκBα as an essential regulator of canonical NF-κB signaling in the prevention of neutrophil-dependent autoinflammatory diseases. These findings also highlight the therapeutic potential of IL-1 inhibitors in treating complications arising from systemic NF-κB inhibition.
Keywords: Immunology, Inflammation
Keywords: Immunotherapy
Introduction
The NF-κB protein complex is integral to the initiation of inflammation and the development of immunity (1–4). NF-κB activation is controlled by inhibitors of κB (IκBα, IκBβ, and IκBε) and by the IκB kinase (IKK) complex, which consists of NEMO, IKK1, and IKK2 (5–12). Patients with genetic defects within the NF-κB signaling pathway, such as in NEMO, IRAK4, MyD88, and NFKBIA, generally display severe immunodeficiencies with impaired cellular responses to immune stimuli such as LPS or TNF-α (13–16).
To date, 16 NFKBIA mutations have been reported in a total of 17 patients (14, 17–28). Thirteen of the patients presented with anhidrotic ectodermal dysplasia (EDA) and showed abnormalities in hair, tooth, and sweat gland development (14, 17–22, 25–28). All of the patients also displayed recurrent infections with impaired inflammation; however, there was heterogeneity in their clinical severity and immunological phenotypes. Nine patients had T cell defects that affected the total count, proliferation, and signaling ability of these cells (14, 17, 19, 20, 22–24). Production of NF-κB–regulated proinflammatory cytokines such as TNF-α or IL-6 was impaired in the majority of the patients’ samples (14, 17–23, 25, 26). However, 1 patient paradoxically showed both a complete absence of immune responses and a suspected autoimmune thyroiditis (21). The molecular mechanisms underlying the differential presentations in these patients were largely unexplored but have been compared in a review (29) and ranked by severity using in vitro fibroblast studies (25).
Here, we report the molecular genetics and cellular studies of an infant with a fatal clinical phenotype combining liver fibrosis, cholestasis, and invasive bacterial and fungal infections. This patient carried a novel heterozygous missense mutation, L34P, in NFKBIA that resulted in impaired NF-κB activation in fibroblasts and macrophages. Functional and immunological studies showed a pronounced alteration in chemokine and cytokine production in the patient’s leukocytes, including hyperproduction of IL-1β. In turn, IL-1β provoked peripheral neutrophil activation and liver damage. Correspondingly, murine bone marrow–derived macrophages carrying the L34P IκBα mutation displayed the hyperproductive IL-1β phenotype. The patient was treated with hematopoietic stem cell transplantation (HSCT) therapy and subsequently with IL-1 blockade. Both treatments led to a reduction in neutrophil activation, IL-1β hyperproduction, and hepatic injury. Our studies of the L34P IκBα mutation revealed that human NF-κB plays an unexpected role as an antiinflammatory agent by regulating IL-1β secretion and preventing myeloid inflammation.
Results
Clinical history of the patient.
The patient was born full term as the first child to nonconsanguineous parents. One week after birth, she presented with a variety of infectious diseases (Figure 1A, Supplemental Figure 3A, and Supplemental Table 3; supplemental material available online with this article; https://doi.org/10.1172/JCI98882DS1), including a disseminated bacille Calmette-Guerin (BCG) infection, with a BCG abscess (Figure 1, C and D) that required hospitalization and prolonged antimicrobial treatment.
Figure 1. Clinical presentation of the patient.
(A and B) Aligned schematic view of infections, liver function tests, and significant procedures before HSCT. (A) Viral (green), fungal (red), bacterial (blue), and mycobacterial (purple) infections are represented as arrows along the timeline. (B) Serum levels of GGT, ALP, ALT, AST, and total and direct (conjugated) bilirubin, with upper healthy limits indicated by corresponding colors in the dashed lines. (C) Wide excision of left buttock abscess at 4 months of age. (D) The abscess was composed of necrotic tissue (black arrow) with large numbers of foamy macrophages (white arrow), inflammatory cells, and poorly formed granulomas. Scale bar: 100 μm. (E) Chest and abdominal CT at 7 months of age showing widespread consolidation in the bilateral lung fields and hepatomegaly. (F) Liver biopsy showing sinusoidal fibrosis surrounding the hepatocytes, demonstrated by Masson’s trichrome staining. Scale bar: 30 μm. (G) Liver biopsy showing hepatocytic cholestasis (black arrows). Scale bar: 30 μm. (H) Lung biopsy showing features of pulmonary alveolar proteinosis, with alveolar spaces filled with amorphous, PAS-positive eosinophilic material (black arrows) and associated florid pneumocytic hyperplasia (white arrow). Scale bar: 100 μm. (I) Milky yield from the first unilateral BAL fluid sample. (J) Papulonodules. (K and L) Skin biopsy showing superficial perivascular dermatitis (K) and absence of sweat glands in deep dermis and superficial subcutaneous fat (L). Scale bars: 100 μm.
Uncontrolled extrahepatic bacterial infections can result in severe liver damage. Accordingly, the patient experienced an early onset of liver inflammation with significant hepatosplenomegaly (Figure 1E). This was accompanied by a persistent and progressive increase in γ-glutamyl transferase (GGT), reaching a peak of 250-fold above normal levels (2525 U/L) (Figure 1B). Bilirubin (total and conjugated), alkaline phosphatase (ALP), and transaminase (alanine aminotransferase [ALT] and aspartate aminotransferase [AST]) levels were also increased (Figure 1B). A liver biopsy showed sinusoidal fibrosis (Figure 1F) and prominent hepatocellular cholestasis (Figure 1G), suggesting an inflammation-based liver injury with intact bile duct function. GGT levels gradually decreased with the control of infections at 5.5 months.
Within the first 6 months of life, the patient had 2 episodes of severe pneumonia requiring prolonged antimicrobial treatment and ventilatory support. At 7 months of age, the patient experienced deterioration in her respiratory status and worsening consolidative changes confirmed by chest CT. Bronchoalveolar lavage (BAL) fluid was negative for pathogens but was milky in appearance, with dense white flocculent material that separated out upon sedimentation (Figure 1I). A lung biopsy showed features consistent with pulmonary alveolar proteinosis (PAP), with alveolar spaces filled by amorphous, periodic acid–Schiff–positive (PAS-positive) eosinophilic material, with florid pneumocyte hyperplasia (Figure 1H and Supplemental Figure 3B).
The patient developed various cutaneous conditions during her stay in the hospital. These ranged in severity from mild symptoms, which included erythematous papulonodules, to more severe and extensive psoriasiform desquamative rashes (Figure 1J and Supplemental Figure 3C). Skin biopsies revealed nonspecific dermatitis with negative microbial cultures (Figure 1K). Notably, however, skin biopsies showed an absence of sweat glands, consistent with a defect in the ectodysplasia receptor (EDA-R) signaling pathway (Figure 1L).
Impaired NF-κB activation in human fibroblasts with NFKBIA mutation.
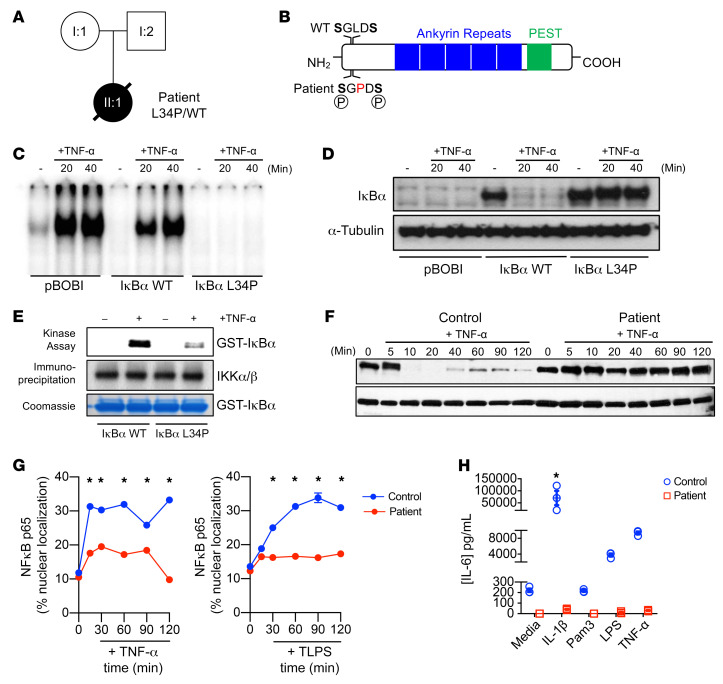
Whole-exome sequencing revealed a novel, de novo, heterozygous T/C mutation in the NFKBIA gene, resulting in a Leu34 to Pro34 (L34P) substitution in the IκBα protein (Figure 2, A and B, and Supplemental Figure 4, A and B). Sanger sequencing confirmed the presence of this mutation. Leu34 in NFKBIA is evolutionarily conserved and lies between 2 phosphorylation sites (Ser32 and Ser36) that are critical for IκBα degradation and NF-κB activation (Supplemental Figure 4C).
Figure 2. Molecular analysis of the mutant IκBα.
(A) Pedigree of the family with the immunodeficient patient. Circles denote females, square denotes the male; affected individuals are represented by black symbols. (B) Schematic of IκBα protein. The mutation is indicated in red; letters refer to amino acid usage. PEST, domain consisting of a polypeptide sequence rich in proline (P), glutamic acid (E), serine (S), and threonine (T). (C) EMSA assay of HeLa cells transfected with pBOBI-Vec (pBOBI), pBOBI-IκBα (IκBα WT), or pBOBI-mutant IκBα (IκBα L34P) and treated with TNF-α (100 ng/mL) for the indicated durations. (D) Western blotting to detect Flag-IκBα (IκBα WT) and Flag-mutant IκBα (IκBα L34P) in transduced HeLa cells treated with TNF-α (100 ng/mL). α-Tubulin was used as a loading control. (E) Kinase assay of recombinant GST-IκBα WT and mutant constructs exposed to 2 μg IKK complexes from TNF-α–treated HEK293 cells (top panel). Immunoprecipitated IKK complexes are indicated in the middle panels. Coomassie blue (R-250) staining (bottom panel) was used as a loading control. (F) Western blotting of patient and control primary fibroblasts stimulated with TNF-α (100 ng/mL) for the indicated durations. Cells were lysed and blotted for IκBα and the loading control α-tubulin. (G) Quantification of nuclear colocalization of p65 NF-κB with Hoescht DNA staining in patient and control primary fibroblasts stimulated with TNF-α (100 ng/mL) or LPS (1 μg/mL) for the indicated durations. (H) Luminex cytokine assay for IL-6 production in supernatants of patient and control primary fibroblasts stimulated for 18 hours with IL-1β (100 ng/mL), TNF-α (100 ng/mL), or LPS (1 μg/mL), or left unstimulated. Data are representative of 3 experiments unless otherwise indicated and indicate the mean ± SEM. *P < 0.05, by 2-way ANOVA with Dunnett’s multiple-comparisons test.
We assessed the impact of this mutation on NF-κB signaling in HEK293 cells using a NF-κB promoter–luciferase reporter, which was cotransfected with a plasmid encoding for either the patient’s or WT IκBα. Introduction of patient IκBα resulted in more effective inhibition of TNF-α–mediated luciferase expression compared with introduction of the WT IκBα (Supplemental Figure 4D). Moreover, the patient’s IκBα, but not WT IκBα, completely prevented the TNF-α–induced DNA-binding activity of endogenous NF-κB, as assessed by EMSA in a transfected HeLa cell model (Figure 2C). Western blot analysis using the same system showed a severe impairment of the patient’s IκBα degradation (Figure 2D). Kinase assays revealed defective phosphorylation of the patient’s IκBα after TNF-α stimulation (Figure 2E).
To eliminate the possibility that the observed effects were due to overexpression of the variant IκBα, we determined whether this dominant-negative effect was also present in the patient’s primary fibroblasts, which contained only 1 allele of mutated IκBα. In keeping with our earlier findings, stimulation with either TNF-α or LPS showed defective IκBα degradation (Figure 2F and Supplemental Figure 4E), and a severe reduction in NF-κB nuclear translocation (Figure 2G and Supplemental Figure 4F). Furthermore, stimulation with proinflammatory cytokines or TLR agonists resulted in reduced IL-6 and IL-8 production (Figure 2H and Supplemental Figure 4G). Taken together, these data show that the mutation resulted in the persistence of IκBα, with an associated gain of function. This dominant-negative variant thus abolished NF-κB activation, nuclear translocation, and cytokine production.
IκBα mutation causes myeloid hypersecretion of IL-1β and neutrophil accumulation in liver.
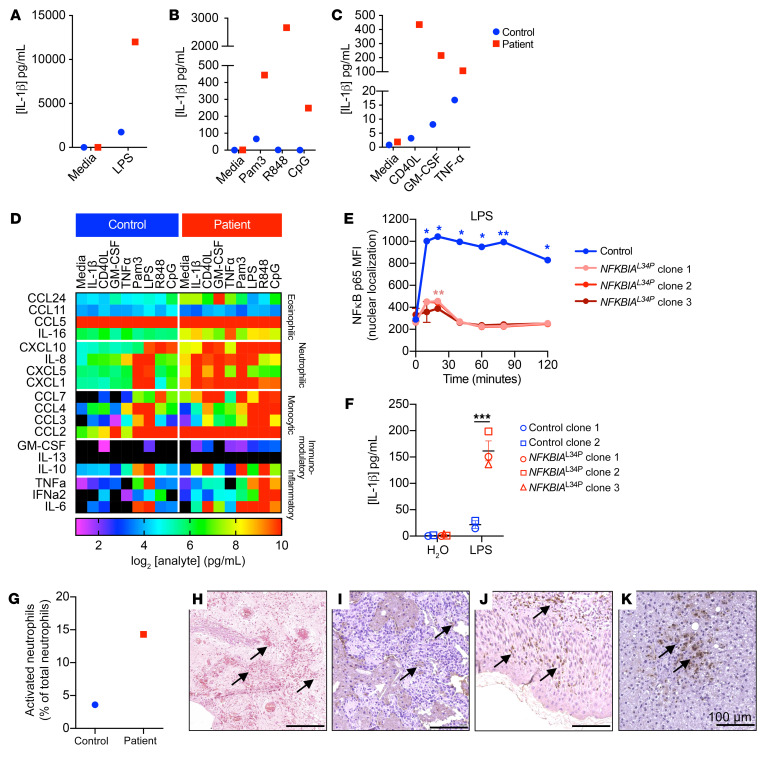
Previous reports showed that endotoxin-induced proinflammatory cytokines such as IL-1β, TNF-α, and IL-6 mediate hepatic damage, including cholestasis and liver fibrosis. LPS stimulation of whole blood in vitro resulted in similar levels of IL-6 and TNF-α in patient and healthy control samples (Supplemental Figure 5, A and B). However, LPS-induced IL-8 and IL-10 production was severely impaired in the patient’s blood (Supplemental Figure 5, C and D). In contrast to the decreased cytokine secretion, the patient’s blood produced 10 times more IL-1β in response to LPS compared with the healthy control’s blood (Figure 3A). Hypersecretion of IL-1β was also evident following stimulation with a variety of TLR and non-TLR agonists, which normally results in low levels of secretion (Figure 3, B and C). Additionally, we found that stimulation of the patient’s whole blood with IL-1β drastically elevated the secretion of chemokines associated with eosinophilic (IL-16 and CCL24), neutrophilic (CXCL1 and CXCL8), and monocytic (CCL2, CCL3, CCL4, and CCL7) inflammation (Figure 3D), suggesting that the patient’s leukocytes were hyperresponsive to IL-1β itself. In keeping with these findings, we noted that several of these cytokines and chemokines were also present in the patient’s peripheral sera (Supplemental Figure 5, E and F).
Figure 3. Activation and infiltration of granulocytes in mucosal tissue and enhanced IL-1β production in the patient.
(A–D) Whole blood from the patient at 8 months of age (red) and a healthy relative (blue) was either left unstimulated or stimulated for 18 hours with the TLR4 agonist E. coli LPS (100 ng/mL) (A), the TLR2 agonist Pam3Csk4 (1 μg/mL), the TLR7/8 agonist R848 (3 μg/mL), the TLR9 agonist CpG (10 μg/mL) (B), CD40L (3 μg/mL), GM-CSF (20 ng/mL), or TNF-α (15 ng/mL) (C). After incubation, supernatants from the cultures were assayed to detect the production of IL-1β (A–C) and chemokines involved in eosinophil, neutrophil, or monocyte chemotaxis, as well as immunomodulatory cytokines and inflammatory cytokines (D). Results are displayed as a heatmap of log2-transformed data, with range bar from no production (in black) to ≥10 log2 pg/mL. (E) Quantification of nuclear colocalization of p65 NF-κB with DNA stain in iMacs stimulated with LPS (100 ng/mL) for the indicated durations. (F) Luminex cytokine assay to detect IL-1β production in supernatants from 2 iMac lines derived from 2 control donors and 3 clonal lines of patient-derived iMacs after 24 hours stimulation with LPS (100 ng/mL). (G) Percentage of activated neutrophils from the patient at 8 months of age (red) and a healthy relative (blue). Graph displays activation as a loss of CD62L. (H) Tissue from the patient was carbol chromatrope-2R stained for eosinophilic infiltrate in the BCG-infected buttock abscess. (I–K) Mucosal tissue was stained for neutrophil elastase and counterstained with hematoxylin in the lung (I), skin (J), and liver (K). Black arrows in H–K indicate granulocyte infiltrate. Scale bars: 100 μm. (A–D and G) Data are from a single experiment. (E and F) Data are representative of 3 experiments and are shown as the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, by 2-way ANOVA with Holm-Šidák’s multiple-comparisons test.
We hypothesized that myeloid cells were the major source of the elevated IL-1β secretion from stimulated whole blood from the patient. Therefore, we generated induced pluripotent stem cell–derived macrophages (iMacs) from patient and healthy control fibroblasts (Supplemental Figure 6). The patient’s iMacs recapitulated defective nuclear translocation of NF-κB following LPS stimulation (Figure 3E and Supplemental Figure 7). Furthermore, stimulation of patient iMacs with LPS or other ligands produced significantly higher IL-1α and IL-1β levels compared with levels detected in control iMacs, whereas IL-6 and TNF-α levels were generally attenuated (Figure 3F and Supplemental Figure 8). These results suggested that inhibition of NF-κB through the prevention of IκBα degradation in macrophages increased IL-1β secretion. We also observed hypersecretion of IL-1β in in vitro experiments with healthy control primary human monocytes treated with the proteasomal inhibitor bortezomib (BTZ). BTZ promotes NF-κB inhibition through the stabilization of IκBα, which we used as a means of replicating the effect of the NFKBIA variant in our patient. Monocytes were primed for 2 hours with LPS to induce expression of IL1B and then treated for 24 hours with BTZ. The addition of BTZ enhanced the release of LPS-induced IL-1β by more than 2-fold in monocytes from 3 different donors. Notably, BTZ alone did not induce IL-1β production without the priming effect of LPS (Supplemental Figure 5G). These experiments confirmed that NF-κB inhibition alone was sufficient to increase IL-1β production.
Prior studies have shown that several cytokines, including IL-1β, promote granulocyte differentiation and neutrophilia in humans and mice (12). In keeping with these previously reported studies, neutrophil counts were markedly increased in the patient, concordant with increased IL-1β production, compared with counts in normal age-matched healthy controls (Supplemental Table 4). Immunophenotyping of the patient’s whole blood revealed striking differences in immune biomarker expression in leukocytes from the patient and a healthy control (Supplemental Figure 9). Moreover, the patient’s neutrophils displayed an activated state indicated by the loss of CD62L (Figure 3G). Staining of tissue sections taken from sites of clinical manifestations showed an infiltration of eosinophils within the buttock abscess (Figure 3H) and of neutrophils in the lung (Figure 3I), skin (Figure 3J), and liver (Figure 3K).
LysM-cre NfkbiaL34P mice replicate the IL-1β hypersecretion phenotype.
In order to clearly demonstrate that this L34P mutation in macrophages is causative of enhanced IL-1β production, we generated heterozygous LysM-cre NfkbiaL34P mice, in which expression of the L34P NFKBIA variant was restricted to myeloid cells (Supplemental Figure 10A). The mice closely mirrored the dominant-negative gain-of-function phenotype observed in the patient. Similar to patient-induced pluripotent stem cell–derived (iPSC-derived) macrophages, LPS-stimulated bone marrow–derived macrophages (BMDMs) from LysM-cre NfkbiaL34P mice exhibited hypersecretion of IL-1β despite reduced Il1b mRNA expression (Figure 4, A and B). An assessment of the NF-κB–regulated genes IL-6 and TNF-α in mutant murine macrophages revealed reduced protein production and mRNA expression, congruent with the findings in the patient’s iPSC-derived macrophages (Figure 4, C and D).
Figure 4. Enhanced IL-1β production by BMDMs from LysM-cre+ NfkbiaL34P conditional-knockin mice.
(A and B) ELISA measurement of IL-1β (A) and IL-6 (C) production in supernatants from WT and LysM-cre NfkbiaL34P BMDMs after 6, 9, 12, and 24 hours of stimulation with LPS (100 ng/mL). qPCR measurement of Il1b (B) and Il6 (D) transcript expression levels in RNA from WT and LysM-cre NfkbiaL34P BMDMs after 6, 12, and 24 hours of stimulation with LPS (100 ng/mL), shown as the 2-ΔΔCt fold change relative to an unstimulated sample (0 hours) and normalized to Actb. (E) Representative confocal microscopic images of NF-κB p65 nuclear translocation, represented by colocalization with DAPI, in BMDMs from WT and LysM-cre NfkbiaL34P mice stimulated for 60 minutes with LPS (100 ng/mL) or left unstimulated. Original magnification, ×20. Scale bars: 10 μm. Data are representative of 3 experiments and indicate the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 1-way ANOVA with a Holm-Šidák multiple-comparisons test.
Similar to the human iMacs, we observed that LPS-induced nuclear translocation of p65 NF-κB was also severely impaired in BMDMs from LysM-cre NfkbiaL34P–mutant mice (Figure 4E and Supplemental Figure 10B). By contrast, exposure of the cells to LPS followed by ATP resulted in comparable IL-1β levels in WT and LysM-cre NfkbiaL34P cells (Supplemental Figure 10C).
Increased susceptibility of the patient to infection is due to an impaired IL-12/IFN-γ axis.
The increased susceptibility of the patient to invasive bacterial infections, including BCG, prompted us to evaluate antibacterial immunity that is highly dependent on IL-12/IFN-γ signaling. Although the patient had unusually high T cell counts (Supplemental Table 5), these T cells showed poor proliferative capacity in response to phytohemagglutinin and concanavalin-A, as well as impaired IFN-γ secretion during BCG stimulation (Supplemental Table 5). Furthermore, addition of exogenous IFN-γ or IL-12 to the patient’s PBMCs cocultured with BCG failed to rescue cytokine production from monocytes and T cells, respectively (Supplemental Table 5).
Taken together, these results show that the IκBα L34P mutation simultaneously caused severe immune deficiency and autoinflammation. The latter, which was characterized by hyperactivation of IL-1β secretion, resulted in neutrophilia and hepatobiliary diseases in the patient.
Restoration of homeostatic immune responses after bone marrow transplantation.
Given the persistence of multiple infections even with antimicrobial therapies, the patient underwent HSCT at 9 months of age, receiving HSCs from an unrelated, fully matched donor. Peripheral blood variable number tandem repeat (VNTR) testing showed full engraftment 28 days after HSCT, with neutrophil numbers reaching normal levels by day +159 (Supplemental Table 6).
The papular skin rash resolved. Serum IgA and IgM levels that were previously undetectable were present within normal ranges in the sera by day +159 after HSCT (0.27 g/L and 0.14 g/L, respectively). By 3 months after HSCT, the patient was noted to have normal hair growth on her scalp and eyebrows, and the first tooth appeared at 18 months of age (9 months after HSCT). Her respiratory support was successfully discontinued on day +53, and the tracheostomy was decannulated on day +102 (4 months after HSCT). Moreover, the patient’s liver function stabilized and showed no signs of further deterioration (Figure 5A).
Figure 5. Partial reduction of immune activation and responses following HSCT.
(A) Serum levels of GGT, ALP, ALT, AST, as well as total and direct bilirubin levels between 8 months of age (1 month before HSCT) and 17 months of age (8 months after HSCT), with upper healthy limits indicated by corresponding colors in the dashed lines. (B) Whole blood was obtained from the patient on day +159 (5 months) after HSCT (purple squares) and assayed for IL-1β secretion after 18 hours of stimulation. Results were compared with those from pre-HSCT stimulation when the patient was 8 months old (red squares). (C) Percentage of activated neutrophils from the patient at 10 months of age (2 months after HSCT) (purple) and a healthy relative (blue). Dot plot displays activation as loss of CD62L. (D) Luminex cytokine assay measurement of cytokine and chemokine concentrations in peripheral sera from the patient at 8 months of age (1 month before HSCT, red) and on day +159 after HSCT (purple). Analytes are categorized by their involvement in neutrophil, monocyte, or eosinophil chemotaxis. Results are displayed as heatmap of log2-transformed data, with the range bar from no production (in black) to ≥10 log2 pg/mL. Absolute values from the cytokine assay are shown in adjacent dot plots in pg/mL. (B and C) Data shown are from a single experiment. (D) Data shown are from a single experiment with 3 technical replicates. **P < 0.01 and ***P < 0.001, by 2-way ANOVA with Bonferroni’s multiple-comparisons test.
Stimulation of the patient’s leukocytes with LPS at 15 months of age (day +159 after HSCT), resulted in reduced IL-1β secretion compared with blood taken before HSCT (Figure 5B). In agreement with these findings, peripheral neutrophils showed reduced activation after transplantation (Figure 5C). Similarly, we observed greatly reduced amounts of eosinophil-, neutrophil-, and monocyte-derived chemokines in the peripheral sera (Figure 5D). This rectification of the many pathological features by HSCT confirmed the essential contribution of the patient’s immune system to her multifaceted disease.
Targeted anti–IL-1 therapy ameliorates hypersecretion of IL-1β and liver damage symptoms.
The patient showed worsening hypertension at 20 months of age (11 months after HSCT), requiring antihypertensive agents and weaning of the oral steroids that had been started for gastrointestinal graft-verus-host disease (GVHD) (diagnosed 8 months after HSCT). An examination of the patient’s chimerism showed that the host VNTR percentage had increased to a high of 12.5% to 25% in both mononuclear cells and granulocytes, indicating the reemergence of the patient’s pathogenic leukocytes (Figure 6A). Congruent with the host cell recurrence, the patient’s blood tests showed increased levels of GGT and ALP (Figure 6, B and C), as well as high levels of conjugated and unconjugated bilirubin (Figure 6D). Repeat liver biopsies revealed cholestasis and portal and perisinusoidal fibrosis, with bile duct damage due to immune cell infiltration (Figure 6E). Immunohistochemical staining of the liver showed neutrophil infiltrate with periportal distribution (Figure 6F). The patient’s clinical deterioration was coincident with a reemergence of activated neutrophils and a novel presence of activated monocytes in the peripheral blood (Figure 6, G and H). These findings were concordant with elevated IL-1β secretion from LPS-stimulated leukocytes (Figure 6I).
Figure 6. Post-HSCT deterioration and disease amelioration by IL-1β–targeting anakinra therapy.
(A) Percentage of patient granulocytic leukocytes and mononuclear leukocytes among the total respective cell population was measured using VNTR analysis between 10 and 25 months of age (1–16 months after HSCT). Dashed lines indicate upper and lower ranges of detection. (B–D) The time course of liver function tests of the patient’s sera before anakinra treatment: GGT and ALP (B), ALT and AST (C), total and direct (conjugated) bilirubin (D). (E) Leukocyte infiltration in liver biopsies and damage to bile ducts at 22 months of age (13 months after HSCT) compared with that observed at 4 months of age (5 months before HSCT). Scale bars: 100 μm. (F) Immunohistochemical staining of neutrophil elastase in liver biopsies from the patient at 16 months of age (7 months after HSCT) and 22 months of age (13 months after HSCT), showing increased granulocytic infiltrate during deterioration. Scale bars: 100 μm. (G–I) Changes in patient’s immune markers during administration and withdrawal of anakinra. Shaded areas with solid lines indicate the dose and duration of anakinra treatment. Time course of activated neutrophils (G) and monocytes (H) from the patient’s peripheral blood, phenotyped immediately ex vivo. (I) Time course of IL-1β production detected in supernatants of patient’s leukocytes stimulated with LPS for 18 hours, when drawn at the indicated ages. (J–L) Time course of liver function tests from the patient’s sera during and after anakinra treatment: GGT and ALP (J), ALT and AST (K), total and direct (conjugated) bilirubin (L).
Twelve months after HSCT (21 months of age), the patient had a hypertensive emergency, with concurrent episodes of fever and worsening of her respiratory symptoms. Analysis of the patient’s BAL revealed the presence of multidrug-resistant bacteria (Supplemental Table 7) and activated neutrophils and T cells, concomitant with elevated levels of IL-1β and chemokines promoting leukocyte recruitment (Supplemental Figure 11). Together, these results suggest that the resurgence of host leukocytes caused a new episode of immunopathology that was further exacerbated by donor cells, resulting in severe liver inflammation, cholestasis, and bile duct damage.
Given the deterioration of the patient’s liver function, the reemergence of activated peripheral neutrophils, and elevated IL-1β levels, we targeted the IL-1β signaling pathway using the recombinant IL-1 antagonist anakinra. Treatment commenced at 24 months of age (15 months after HSCT). Fever then improved over the next 2 weeks. Immunophenotyping showed decreased frequencies of activated neutrophils and monocytes (Figure 6, G and H) during the second administration of anakinra, 15 months after HSCT. In addition, whole blood stimulation assays showed decreased IL-1β production in response to LPS stimulation (Figure 6I). Anakinra treatment was briefly withdrawn, which coincided with an increase in neutrophil and monocyte activation. We restarted therapy at a higher dose when the patient was 25.9 months of age. During treatment with the higher dose of anakinra, the patient’s levels of GGT, ALP, and bilirubin stabilized (Figure 6, J–L). Neutrophil activation also further decreased from the first round of anakinra treatment, while IL-1β production continued to decline (Figure 6, G and I). We found that neutrophil numbers transiently decreased under the higher dose, whereas monocyte and eosinophil numbers were unaffected (Supplemental Figure 12, D–F).
Despite improvements in liver function, fever recurred at 26.8 months of age, concurrent with an extensive rash over the patient’s trunk and limbs that progressed to her face (Supplemental Figure 12, A–C). Anakinra was withdrawn because of the worsening rash, despite this not having been a previously reported adverse event for this therapy. Neutrophil activation rapidly increased in the patient after withdrawal (Figure 6G). Bilirubin levels became 5-fold higher than those detected under anakinra treatment, indicating a worsening cholestasis (Figure 6L), while GGT and ALP amounts decreased (Figure 6J). At 27.5 months of age, he patient experienced consequent acute liver and renal failure concurrent with a severe inflammatory response. We detected multiple pathogens in the BAL, including EBV suggestive of a post-transplant–related lymphoproliferative disorder. The patient succumbed to fungal sepsis with multiple organ failure at 27.5 months of age. Collectively, these observations indicate that dysregulated IL-1β secretion, arising from severe NF-κB inhibition in myeloid cells, played a central role in driving liver immunopathology in the patient. Targeting this specific IL-1 pathway by immunotherapy represents a potential therapeutic intervention to control the progression of liver diseases.
Discussion
In this study, we report an infant with severe immunodeficiency who had a de novo heterozygous IκBα L34P mutation. In the first few months of her life, the patient presented with a multitude of clinical pathologies including cholestasis, liver fibrosis, and recurrent infections. The mutation rendered IκBα resistant to stimuli-induced degradation, causing a severe defect of NF-κB signaling and downstream production of cytokines such as TNF-α and IL-6 in fibroblasts and macrophages. Unexpectedly, the patient’s leukocytes secreted higher amounts of IL-1β after stimulation with LPS or other agonists that normally do not provoke IL-1β release.
Prior studies have shown that the pathogenesis of cholestasis and liver fibrosis is mediated by proinflammatory cytokines such as IL-1β, IL-6, and TNF-α in response to LPS stimulation (30–33). These findings, combined with our own observations, led us to therapeutically target IL-1β secretion in order to ameliorate the patient’s hepatic disease. Anakinra administration confirmed the pathogenic role of IL-1β, with reductions in the amounts of activated neutrophils and hepatic damage markers during the treatment periods, whereas withdrawal coincided with an increase in activated neutrophils and worsening liver condition.
The data in this study show that the cellular source of this IL-1β was experimentally confirmed to originate from the myeloid compartment. First, HSCT reversed hypersecretion of IL-1β and halted the progression of liver damage, confirming that mutant hematopoietic cells were sufficient to confer this pathogenic phenotype. Second, post-transplant disease progressed concurrently with the reemergence of host leukocytes that displayed hypersecretion of IL-1β. Third, patient-derived iPSC macrophages secreted greater amounts of IL-1β after LPS stimulation than did iMacs from control donors. Fourth, murine BMDMs expressing the patient’s NFKBIA variant produced more IL-1β protein following LPS stimulation, despite lower levels of Il1b mRNA. Taken together, these findings demonstrate that the L34P mutation in IκBα caused IL-1β hypersecretion in myeloid cells by enhancing pro–IL-1β processing, thus driving the patient’s fatal hepatic immunopathology.
NF-κB plays a central role in the pathogenesis of many human inflammatory diseases and cancers, which has spurred the development of several small-molecule inhibitors against members of the NF-κB signaling family, such as MLN-0415 (reviewed by Prescott and Cook in ref. 34). Despite promising results in preclinical models of cancer and inflammatory diseases, MLN-0415 failed in phase I clinical trials because of its toxicity (discussed by Verstrepen and Beyaert in ref. 35). While the failure of MLN-0415 in the clinic was widely believed to be caused in part by off-target effects nonspecific to NF-κB, our findings offer an alternative explanation: NF-κB inhibition may be specific, yet inhibition of this pathway also triggers heightened IL-1β responses, which in turn induce toxicity, relapse, or drug resistance. Indeed, neutrophilia, which occurred in our patient, has been reported in patients receiving IKKβ inhibitors (36). These results show that it is imperative to examine the production of IL-1β and its detrimental effects when developing inhibitors of the NF-κB pathway.
The work presented here may also shed light on how to overcome the limitations of other promising cancer treatments. For example, the pharmacologic proteasome inhibitor (PI) BTZ improves clinical outcomes in patients with multiple myeloma (MM) (37–41). However, a subset of these patients develop resistance to this PI after a prolonged period of therapy (42–46). The therapeutic effects of BTZ arise partially from blocking proteasomal degradation of IκBα (38), akin to the mechanism of NF-κB suppression reported here. Thus, it is highly plausible that the toxicity associated with BTZ in these patients with MM is similarly caused by hyper-release of IL-1β. Supporting this hypothesis, McLoed et al. have shown IL-1β overproduction in patients with non–small-cell lung cancer (NSCLC) after treatment with BTZ. In addition, they demonstrated in a murine lung cancer model that IL-1β contributed to resistance against BTZ, which was reversed by coadministration of an IL-1 receptor antagonist (36). Furthermore, increased IL-1β release has recently been demonstrated in in vitro studies of primary human cells. In these experiments, monocytes were first activated by LPS stimulation and subsequently inhibited with BTZ (47), which we replicated in the experiments reported here. Our findings suggest that the increased IL-1β response in patients receiving BTZ or other proteasome inhibitors may result from direct inhibition of NF-κB activation and that anakinra treatment may restore their therapeutic efficacy. Nonetheless, although the precise mechanism for the NF-κB–dependent inhibition of IL-1β hypersecretion in this study remained obscure, its identification may uncover new strategies to overcome NF-κB inhibitor–induced toxicity and resistance.
To the best of our knowledge, our patient’s IL-1β hypersecretion and IL-1β–dependent liver damage, including hyperbilirubinemia, have not been previously reported in other patients with defective NF-κB signaling, including patients with mutations of IRAK4, MyD88, IκBα, NEMO, or IKK2 (48). However, this enhanced IL-1β release has been observed in mice lacking myeloid IKK2 but not in humans with an IKK2 deficiency, suggesting species variation in the regulation of the NF-κB signaling pathway. Single IKK1-, IKK2-, or NEMO-knockout mice did not present liver pathology with hyperbilirubinemia (49, 50). However, severe cholestasis and bile duct damage does occur in murine hepatocytes and biliary epithelial cells with either IKK1 and IKK2 or NEMO and IKK1 combined double-knockouts, which also display lower NF-κB activity (51). Thus, the different presentations among patients with different IκBα mutations may result from distinct effects on NF-κB activity. For instance, 1 patient with a heterozygous mutation at Ser36 in NFKBIA had normal IL-1β production (17), however, in a comparative study of human IκBα mutations, this Ser36 patient possessed a median score for relative disease severity (25). Examination of patients with an increased disease severity score may reveal a similar phenotype in IL-1β processing. To dissect how each IκBα mutation causes its unique pathology, future studies may use iPSC-derived macrophages from each patient to assess differences in NF-κB activity and IL-1β secretion.
Taken together, these results revealed an important role of IκBα in the prevention of uncontrolled IL-1β secretion in myeloid cells. The consequences of this dysregulation promoted granulopoiesis, neutrophilic inflammation, activation, and liver accumulation of neutrophils. Clinically, these studies also imply that the application of NF-kB inhibitors in the treatment of human cancers and inflammatory diseases may require concurrent inhibition of IL-1β production aimed at overcoming IL-1β –induced toxicity due to inhibition of NF-κB.
Methods
Genetic analysis.
Whole-exome sequencing was performed for both parents and the proband (trio). One microgram of high-molecular-weight DNA per sample was used for exome capture with an Ion TargetSeq Exome Kit (Life Technologies, Thermo Fisher Scientific). DNA was sheared using the Covaris M220 Focused Ultrasonicator to target an average fragment size of 200 bp. Shearing was followed by end repair, ligation of adapters, nick repair, purification, size selection, and final amplification before exome capture as per the TargetSeq protocol. The amplified DNA was cleaned with Ampure XP Reagent (Agencourt), and the DNA was eluted in 30 μL low TE buffer. The libraries were quantified using a Qubit 2.0 Fluorometer (Life Technologies, Thermo Fisher Scientific). The exome library was used for emulsion PCR on an Ion OneTouch System or an Ion Chef System (both from Life Technologies, Thermo Fisher Scientific) following the manufacturer’s protocol. Each library was sequenced on an Ion Proton instrument (Life Technologies, Thermo Fisher Scientific) using 1 Ion PI chip.
For this trio, 12.5 Gb, 13.4 Gb, and 14.5 Gb were sequenced per sample, with an average read length of 184 bp, 176 bp, and 177 bp for the father, mother, and affected daughter, respectively. An average coverage of 144× (father), 162× (mother) and 170× (daughter) was achieved over the exome, with 95% of the bases covered at least 20×.
Sequence reads were aligned to the human reference genome (human GRCh37 [hg19]build) using the Torrent Mapping Alignment Program (TMAP) from the Torrent Suite (version 4.2.1). PCR duplicates in the BAM file were identified by the Filter Duplicates plugin (version 4.2) and removed. The variants were called using the Torrent Variant Caller (TVC) plugin (version 4.2.1) and imported into Ion Reporter software (version 4.2, Thermo Fisher Scientific), where each variant was annotated using the “annotate single sample variants” workflow. For each variant, information such as the associated gene, location, quality score, coverage, predicted functional consequences, protein position and amino acid changes, SIFT (52), PolyPhen2 (53), and Grantham prediction scores, phyloP conservation scores (54), and 5000 Genome Project (MLL) minor allele frequencies were provided. Variants were filtered for common SNPs using the NCBI’s “common and no known medical impacts” database (ftp://ftp.ncbi.nih.gov/snp/organisms/human_9606_b141_GRCh37p13/VCF) and the Exome Sequencing Project (http://evs.gs.washington.edu/EVS) common SNP database. Variants that were predicted to be synonymous or that did not have a location on a coding exon, UTR, splice site junction, or flanking intron were filtered off. Variants were next compared with an in-house database of 138 previously sequenced samples, and those that were present in more than 3% of the previously sequenced samples were removed. For the trio, an additional filter was applied to retain variants that were (a) heterozygous in the proband and absent in the parents, (b) homozygous in the proband and heterozygous in both parents, (c) compound heterozygous in the proband and heterozygous in both parents, and (d) X-linked homozygous in the proband and heterozygous in the mother.
A total of 34,738, 34,788, and 34,818 variants were identified in the father, mother, and affected daughter, respectively. A combined unique total of 47,218 variants were identified across protein-coding exons, UTRs, splice sites, and flanking introns. After applying all filters, a final set of 168 variants (5 homozygous, 151 heterozygous, 12 compound heterozygous) were identified. Of these 168 variants, only 33 were protein-changing variants and have not been seen in population or previous in-house exomes. Visualization of each of these variants using the Integrative Genomics Viewer showed that a majority of them were in homopolymer sequences and turned out to be false-positive variants, narrowing the final list of candidate genes to 3. According to the Exome Aggregation Consortium (ExAC), only 1 gene remains, since the heterozygous variant c.565C>T in the YKT6 gene and the compound heterozygous variants c.995A>G and c.13138C>T in the SSPO gene have been reported in the ExAC database. We therefore identified a de novo heterozygous mutation, c.101T>C, in the NFKBIA gene, which leads to the amino acid alteration p.Leu34Pro. Sanger sequencing of the parents and the proband confirmed the variant segregation with the disease.
Lentiviral infection.
Human NFKBIA (1–318 amino acid) and mutant NFKBIA (L34P) were cloned into the pBOBI plasmid with a Flag tag. HEK293T cells (American Type Culture Collection [ATCC] CRL-3216) were used to produce virus as described previously (55), and HeLa cells (ATCC CCL-2) were infected and recovered before use in the experiments described in this work.
EMSA.
EMSA was performed as previously described (55). Briefly, 20 μg total protein was incubated in reaction buffer (50% glycerol, 200 mM HEPES, pH 7.9, 4 mM EDTA, pH 8.0, 4 mM DTT), 2 μg polydI-dC, 1% NP40, and [γ-32P] radio-labeled DNA probe for 30 minutes at room temperature before loading into a nondenaturing polyacrylamide gel. The gel was then dried, exposed to a BAS-IP Image Plate (Fuji), and analyzed with Typhoon FLA 7000 (GE Healthcare).
Kinase assay.
A kinase assay was conducted as previously described (55). GST-IκBα (amino acids 1–54) and mutant GST-IκBα (amino acids 1–54 L34P) constructed in pGEX plasmids were expressed in BL21 and purified using GST Sepharose Beads (Invitrogen, Thermo Fisher Scientific). Total protein (1 mg) from untreated and TNF-α–treated HEK293 cells was used to pull down IKK complexes with anti-NEMO/IKKγ antibody (FL419, sc-8330, rabbit, Santa Cruz Biotechnology) for kinase assays using protein A Sepharose 4B (Invitrogen, Thermo Fisher Scientific). IKK complex–bound beads were washed 3 times with lysis buffer (20 mM HEPES, pH 7.7, 170 mM NaCl, 1 mM EDTA, pH 8.0, 1 mM EGTA, pH 8.0, 20 mM β-glycerophosphate, 10 mM NaF, 0.3 mM Na3VO4) and once with kinase assay buffer (20 mM HEPES, pH 7.7, 10 mM β-glycerophosphate, 10 mM NaF, 2 mM MnCl2, 2 mM MgCl2, and 2 mM DTT with freshly added protease inhibitor). Kinase reaction mixture containing the IKK complex and substrate was incubated in kinase reaction buffer (50 mM HEPES, pH 7.7, 20 mM MgCl2, 2 mM DTT) for 45 minutes at 30°C and then at 95°C for 5 minutes before running in the 12% SDS-polyacrylamide gel. Dried gel was then exposed to a BAS-IP Image Plate for scanning with Typhoon FLA 7000.
IP and Western blot assays.
Immunoblot assays and IP were performed following a previously described standard method (55). Total protein (20 μg) was used for Western blot assays. Anti-Hsp90 (F-8, sc-13119, mouse, Santa Cruz Biotechnology), anti-Flag (F7425, rabbit, MilliporeSigma), anti-IKKα/IKKβ (H-470, sc-7607, rabbit, Santa Cruz Biotechnology), anti-GAPDH (6C5, sc-32233, mouse, Santa Cruz Biotechnology), and normal rabbit IgG (sc-2027, Santa Cruz Biotechnology) were used.
NF-κB luciferase reporter assay.
HEK293 cells were obtained from ATCC (CRL-1573). Transient cotransfected HEK293 cells containing the NF-κB firefly luciferase reporter plasmid pGL4.32 (Promega), and the Renilla luciferase reporter plasmid pGL4.74 (Promega), and pD2509-CMVa vectors containing WT or mutant NFKBIA (DNA 2.0) were created using Lipofectamine 2000 Transfection Reagent (Invitrogen, Thermo Fisher Scientific). The NF-κB firefly luciferase reporter plasmid pGL4.32 (100 ng), the Renilla luciferase reporter plasmid pGL4.74 (1 ng), and a titration of 0.001, 0.01, 0.1, 1, 10, 50, and 100 ng pD2509-CMVa vectors containing WT or mutant NFKBIA were used for transfection.
After an overnight incubation, the transfection media were replaced with DMEM (4.5 g/L d-glucose, l-glutamine) supplemented with 10% FBS (Hyclone, Thermo Fisher Scientific), 2 mM l-glutamine, 0.1 mM MEM-NEAA, 1 mM sodium pyruvate, and 1% penicillin/streptomycin. Cells were then stimulated with 0, 1, 5, 10, 50, and 100 ng/mL TNF-α and incubated for 18 hours before being assayed for NF-κB luciferase activity using the Dual-Glo Luciferase Reporter Assay System (Promega) on an EnVision Multilabel Plate Reader (PerkinElmer).
Western blotting.
Patient and control fibroblasts were seeded in a 6-well plate until full confluence. On the day of the assay, fibroblasts were stimulated with either 100 ng/mL TNF-α or 1 μg/mL LPS (L4391, MilliporeSigma) for 5, 10, 20, 40, or 60 minutes, or they were left unstimulated. Cells were lysed in RIPA Buffer (Life Technologies, Thermo Fisher Scientific) with a HALT Protease and Phosphatase Inhibitor Cocktail (Life Technologies, Thermo Fisher Scientific). Protein amounts were normalized and resuspended in Laemmli buffer (Bio-Rad Laboratories) with 10% β-mercaptoethanol (MilliporeSigma). Samples were then boiled at 99°C for 10 minutes followed by (4%–20%) SDS PAGE and then transferred onto a PVDF membrane using the Trans-Blot Turbo System (Bio-Rad Laboratories). The membrane was then blocked with 5% BSA in 1× TBS with 0.1% Tween-20 (Bio-Rad), followed by blotting with mouse anti-IκBα (4814, Cell Signaling Technology) for 16 hours at 4°C. The following day, the membrane was incubated with HRP-conjugated goat anti–mouse IgG (ab5887, Abcam) for 1 hour at room temperature. Detection was performed using Amersham ECL substrate and hyperfilm (GE Healthcare). In order to normalize protein expression, the membrane was stripped using a mild stripping buffer (1.5% w/v glycine, 0.1% w/v SDS, 1% Tween-20, pH 2.2) at room temperature for 10 minutes. Fresh stripping buffer was reapplied, and stripping occurred for a further 10 minutes. The membrane was then washed twice in PBS for 10 minutes per wash, followed by two 5-minute washes in TBS–Tween-20. Blocking then occurred again as previously stated, followed by incubation with anti–α-tubulin (ab18251, Abcam) or anti–GAPDH-HRP (ab9385, Abcam) for 16 hours at 4°C. Detection of bands occurred as previously described.
Immunofluorescence.
Control and patient fibroblasts were seeded at 1 × 104 cells per well in a 96-well microscopy plate (89626, Ibidi). Twenty-four hours after seeding, media were replaced with 100 μL fresh media. Fibroblasts were then stimulated with either 100 ng/mL TNF-α or 1 μg/mL LPS for 15, 30, 60, 90, or 120 minutes or left unstimulated. Cells were then fixed with 4% paraformaldehyde at room temperature for 20 minutes. Wells were then washed 3 times with PBS and blocked in blocking buffer (1× PBS, 5% normal serum, 0.3% Triton X-100) at room temperature for 1 hour. Blocking buffer was removed, and primary anti–NF-κB p65 (ab32536, Abcam) was diluted in staining buffer (1× PBS, 1% BSA, 0.3% Triton X-100) at 0.148 μg/mL. Cells were stained for 16 hours at 4°C. Subsequently, the cells were washed 3 times with PBS and stained with donkey anti–rabbit DyLight 650 (ab96922 Abcam), Hoechst DNA stain, and phalloidin–Alexa Fluor 647 (A22287, Life Technologies, Thermo Fisher Scientific) at room temperature for 1 hour. Cells were then washed 3 times with PBS and imaged using a Zeiss LSM 700 confocal microscope. Bioinformatics analysis was performed by calculating the total p65 fluorescence on a per-cell basis. The amount of nuclear p65 signal was then calculated as a percentage of the total cellular fluorescence.
To verify the pluripotency of the iPSCs used in this study, staining was performed as previously described (56) using the primary antibodies anti-OCT4, anti-SOX2, anti–SSEA-4, and anti–TRA-1-60. Cells were imaged on a FluoView 1000 microscope (Olympus).
Induced macrophages (iMacs) were seeded at 1 × 104 cells per well in a 384-well microscopy plate (6057300, PerkinElmer). One hour after seeding, media were replaced with 100 μL fresh media to remove nonadherent cells. iMacs were stimulated with 100 ng/mL LPS for 40 minutes or left unstimulated, followed by fixation with buffer containing 4% paraformaldehyde (557870, BD) at 37°C for 10 minutes and permeabilization in buffer containing methanol (558050, BD) on ice for 30 minutes. Cells were washed 3 times with PBS before staining with primary anti–NF-κB p65 antibody (8242, Cell Signaling Technology), as with fibroblasts. Subsequently, the cells were washed twice with PBS and stained with donkey anti–rabbit DyLight 650 and Hoechst DNA stain at room temperature for 1 hour. Cells were then washed 3 times with PBS and imaged using an Opera Phenix confocal microscope (PerkinElmer). Bioinformatics analysis was performed using Columbus software (PerkinElmer) by calculating the total p65 fluorescence per nucleus indicated by Hoescht staining.
BMDMs were seeded at 1.2 × 106 cells per well in 6-well plates with coverslips and stimulated for 60 minutes with LPS (100 ng/mL). After stimulations, cells were washed with PBS, fixed with 4% PFA (sc-281692, Santa Cruz Biotechnology), and permeabilized with 0.2% Triton X-100 (H5142, Promega), with both steps performed for 20 minutes at room temperature. Cells were then washed twice in PBS and blocked for 1 hour with PBS containing 5% FBS (FBS–HI-12A, Capricorn Scientific) and 5% BSA (P6154, Biowest). Next, cells were incubated with anti-p65 antibody (8242, Cell Signaling Technology) at 1:400 for 1 hour. Subsequently, the cells were washed twice in PBS before secondary staining with goat anti–rabbit IgG Alexa Fluor 546 (A-11035, Invitrogen, Thermo Fisher Scientific) at 1:200 for 1 hour. Finally, the cells were washed 4 times with PBS, counterstained with DAPI (D1306, Life Technologies, Thermo Fisher Scientific) according to the manufacturer’s protocol, and then imaged on a Zeiss LSM 510 Meta confocal microscope.
Flow cytometry.
Whole blood samples from the patient and a healthy control were stained with 4 different fluorescently labeled monoclonal antibody panels to determine cell lineage and activation status. T cells were identified with CD38-FITC (560982, BD Biosciences), CTLA4/CD152-phycoerythrin (CTLA4/CD152-PE) (555853, BD Biosciences), CD3-ECD (IM2705U, Beckman Coulter), CD4-PerCP-Cy5.5 (347324, BD Biosciences), CCR7-PE CY7 (557648, BD Biosciences), PD1-APC (329907, BioLegend), CD8-APC7 (557834, BD Biosciences), HLADR–Pacific blue (83-9956, eBioscience), CD45–Pacific orange (563204, BD Biosciences), CD127-BV 711 (351327, BioLegend), and CD25–BUV 395 (BD Biosciences). B cell lineages were identified with IGD-FITC (555778, BD Biosciences), CD24-PE (555428, BD Biosciences), CD19–PE ECD (IM2708U, Beckman Coulter), CD20–PE CY5 (555624, BD Biosciences), CD5–PerCP Cy5.5 (300617, BioLegend), CD38–PE Cy7 (335790, BD Biosciences), CD138-APC (347193BD, Biosciences), CD27–APC CY7 (302816, BioLegend), CD86–Pacific blue (305418, BioLegend), CD45–Pacific orange (563204, BD Biosciences), CD3 SINK-BUV395 (563546, BD Biosciences), CD56 SINK-BUV395 (BD Biosciences), CD14 SINK-BUV395 (563562, BD Biosciences). Granulocyte subsets and NK cells were identified by HLADR-FITC (555811 BioLegend), CD56-PE (130-090-755 Macs, Miltenyi Biotec), CD62L–PE Tx Rd (IM2713U, Beckman Coulter), CD66b-PerCP (305108, BioLegend), CD3–PE CY5 (555341, BD Biosciences), CD11b–PE CY7 (557743, BD Biosciences), CD69–APC CY7 (557834, BD Biosciences), CD34–Pacific blue (562577, BD Biosciences), CD45–Pacific orange (563204, BD Biosciences), CD16–Qdot 605 (93-0168, eBioscience), and CD15–BUV395 (563872, BD Biosciences).
Whole blood was then lysed with BD Lyse/Fix Buffer and acquired using an LSR II (BD Biosciences). Data were analyzed on a FACSDiva (BD Biosciences) with Flowjo software.
Macrophages differentiated from iPSCs were checked for purity by flow cytometry on the day of harvesting using CD45–APC Cy7 (368516, BioLegend), CD11b–PE-Cy7 (25-0118-42, eBioscience), CD163-PE (FAB1607P-100, R&D Systems), and CD14-FITC (325604, BioLegend). Cells were acquired using a 5-laser LSR II System (BD Biosciences), and data were analyzed with Flowjo software.
Whole blood stimulation.
Whole blood was obtained from the patient and a healthy relative and subsequently stimulated with TNF-α (15 ng/mL), IL-1β (10 ng/mL), CD40L (3 μg/mL), granulocyte macrophage CSF (GM-CSF) (20 ng/mL), TLR2 agonist Pam3Csk4 (1 μg/mL), TLR3 agonist PolyI:C (1 μg/mL), TLR4 agonist E. coli LPS (100 ng/mL), TLR7/8 agonist R848 (3 μg/mL), or TLR9 agonist CpG (10 μg/mL), or left unstimulated at 37°C in 5% CO2 for 18 hours. The cells were then pelleted and the supernatant collected and stored at –80°C.
Luminex assay.
The following kits were used: (a) human cytokine panel 1 measuring EGF, eotaxin, basic FGF (FGF-2), Fms-like tyrosine kinase 3 (Flt-3) ligand, fractalkine, granulocyte CSF (G-CSF), granulocyte macrophage CSF (GM-CSF), growth-regulated oncogene (GRO), IL-6, IL-8, macrophage inflammatory protein-1a (MIP-1a), MIP-1b, VEGF, soluble CD40 ligand (sCD40L); (b) human cytokine panel 2 measuring chemokine (C-C) motif ligand-21 (CCL21/6Ckine), B lymphocyte chemoattractant-1 (BCA-1, also known as CXCL13), cutaneous T cell–attracting chemokine (CTACK, also known as CCL27), epithelium-derived neutrophil-activating peptide-78 (ENA-78, also known as CXCL5), eotaxin-2, eotaxin-3, I-309/CCL1, IL-16, IL-20, IL-21, IL-23, IL-28a, IL-33, leukemia inhibitory factor (LIF), monocyte chemoattractant protein-2 (MCP-2), MCP-4, macrophage inflammatory protein-1d (MIP-1d), stem cell factor (SCF), stromal cell–derived factor (SDF) 1A+β, thymus and activation-regulated chemokine (TARC, also known as CCL17), thrombopoietin (TPO), TRAIL, thymic stromal lymphopoietin (TSLP); and a human soluble cytokine receptor panel measuring soluble EGFR (sEGFR), sIL-1RII (sCD121b), sIL-2Rα (sCD25), sIL-4R (sCD124), sIL-6R (sCD126), soluble receptor for advanced glycation endproducts (sRAGE), sTNFRI (sTNFRSF1A), sTNFRII (sTNFRSF1B), sVEGFR2 (sFlk-1), sVEGFR3 (sFlt-4), and soluble glycoprotein 130 (sGp130). Plates were washed using a BioTek ELx405 washer (BioTek Instruments) and read with the FLEXMAP 3D System (Luminex) following the manufacturer’s instructions. Data were analyzed using Bio-Plex Manager 6.0 Software (Bio-Rad) with a 5-parameter curve-fitting algorithm applied for standard curve calculations.
Histological analysis.
Formalin-fixed tissue samples were paraffin embedded and sections taken and prepared for staining by standard histological techniques. Sections were stained with carbol chromatrope 2R for assessment of eosinophils. Immunohistochemical analysis of neutrophil elastase and NF-κB p65 was performed using the EXPOSE Rabbit-Specific HRP/DAB Detection Kit (Abcam) following the manufacturer’s instructions. Briefly, samples were quenched of endogenous peroxidase activity before heat-mediated antigen retrieval at 95°C in target retrieval solution (Dako) and protein blocking. Primary antibodies for either anti–rabbit neutrophil elastase or anti–rabbit NF-κB p65 (both from Abcam) were added at a dilution of 1:500 and 1:100, respectively, and sections were left to incubate overnight at 4°C before HRP/DAB detection was performed. Slides were then counterstained with H&E and mounted with DPX mountant (MilliporeSigma). Assessment of acid-fast bacilli in tissue sections was performed by Ziehl-Neelsen staining following standard procedures.
Generation of iMacs from patient fibroblasts and stimulation for cytokine expression.
Skin fibroblasts from the patient were obtained by punch biopsy. Control neonatal dermal fibroblasts were purchased from ATCC (PCS-201-010). Upon isolation, fibroblasts were cultured in high-glucose DMEM (Gibco, Thermo Fisher Scientific) containing 10% (v/v) heat-inactivated FBS (Gibco, Thermo Fisher Scientific), 1× MEM nonessential amino acids (NEAAs) (Invitrogen, Thermo Fisher Scientific), GlutaMAX Supplement (Gibco, Thermo Fisher Scientific) sodium pyruvate, and 1% penicillin/streptomycin.
Human iPSCs were grown on feeder CF-1 mouse embryonic fibroblasts (MEFs) and generated according to the protocol described in Teo et al. (56). After the iPSC lines were validated by confocal staining for pluripotency markers, they were differentiated into induced macrophages as in Takata et al. (54) to obtain iMacs. iMacs were stimulated with LPS (100 ng/mL), Pam3Csk4 (1 μg/mL), and flagellin from Salmonella typhimurium (1 μg/mL, tlrl-stfla, InvivoGen) for 24 hours, and supernatant was collected for Luminex cytokine analysis.
NfkbiaL34P mice.
Mice were bred at the Biological Resource Centre, A*STAR in Singapore or at the Cyagen Transgenic Animal Center (Jiang Su, China). Breeding pairs for lysozyme 2 cre recombinase mice [LysMcre, B6.129P2-Lyz2tm1(cre)Ifo/J/004781] were obtained from The Jackson Laboratory (57). The modified nkfb1 bacterial artificial chromosome (BAC) transgene was introduced into C57BL/6 ES cells at the Cyagene Animal Center.
Generation of the NFKBIA vector.
A C57/BL6J-derived bacterial artificial chromosome, RP23-214K21, was purchased from the BACPAC Resources Center (Oakland, California, USA). It contained the full genomic sequence for nfkbia and psma6 and served as the template to amplify the arms and exons of nfkbia, which were then cloned into the KI238-fv vector. To create the nfkbia-knockin switch allele, exon 1 was cloned to incorporate the CTG-to-CCG mutation at location L34P. This was downstream of the WT sequence and was in the reverse orientation. The 50 bp homology primer sequences for nfkbia are as follows and are listed in Supplemental Table 1. The WT and mutant exons 1 were flanked by loxP sites, and the mutant by lox2272 sites. The loxP sequence consisted of a [core] flanked by palindrome sequences: 5′-ATAACTTCGTATA[GCATACAT]TATACGAAGTTAT-3′ and 3′-TATTGAAGCATAT[CGTATGTA]ATATGCTTCAATA-5′. Lox2272 sequences consisted of two 13 bp inverted repeats flanking an 8 bp asymmetric spacer sequence (5′-ATAACTTCGTATAaagtatccTATACGAAGTTAT-3′). The loxP or lox2272 paired sites were each in opposite orientations to enable inversion (and reversal) of the DNA sequence on recombination with cre. Following this inversion, the loxP or lox2272 sites would then be in the same orientation to enable deletion of the WT sequence for effective translation of the point mutation (Supplemental Figure 1).
Efficient modification was verified using genomic sequencing and multiple enzyme digestion with pulse field gel analyses. The targeting vector was incorporated into embryonic stem cell (ESC) clones using electroporation. Correctly targeted ESC clones were validated using Southern blotting and selected for blastocyst microinjection, followed by chimera production. Founders were confirmed as germline following crossbreeding with B6 WT mice and effective PCR and sequencing using forward TTACGAGTCTCCGTCCTCCGTGA and reverse CTGTACTTCCCCTCGCGGATAAC primers (Supplemental Figure 2).
DNA isolation and genotyping for the knockin switch allele.
DNA was isolated from a tail snip using the TaKaRa MiniBEST Universal Genomic DNA Extraction Kit Version 5.0 (Takara 9765). To Genotype the mice, a custom TaqMan SNP Genotyping Assay was generated, which detected the point mutation at L34P. The CTG SNP was identified using a VIC probe and the CCG SNP with a FAM probe. The assay was completed according to the manufacturer’s instructions.
Generation and stimulation of BMDMs.
Cells were differentiated in culture for 7 days with 20 ng/mL M-CSF (416-ML-50, R&D Systems) in RPMI 1640 media (L0500, Biowest) supplemented with 10% FBS (FBS-HI-12A, Capricorn Scientific) and 1% penicillin/streptomycin (15140122, Gibco, Thermo Fisher Scientific). Media were topped up on days 3 and 5, and the cells were visualized with a Nikon Eclipse TS100 microscope. On day 7, the adherent cells were harvested using 10 mM EDTA in PBS and seeded in 6-well plates (Greiner Bio-One).
For cytokine production, cells were seeded at 1.5 × 106 cells/mL overnight in 6-well plates. The cells were then stimulated with LPS (100 ng/mL), ATP (5 mM, A2383, MilliporeSigma), and nigericin (5 μM, 7143, MilliporeSigma).
Cytokine concentrations in supernatants were measured by ELISA. Murine IL-1β was measured using a precoated kit (432601, BioLegend). For the murine IL-6 ELISA, IL-6 was detected by purified mouse IL-6 (1 μg/mL, 554400, BD Biosciences) and detected by mouse IL-6 biotin (0.5 μg/mL, 554402, BD Biosciences);
TNF-α was captured by purified hamster anti–mouse/rat TNF (1 μg/mL, 557516, BD Biosciences) and detected using biotin human anti–mouse/rat TNF (0.5 μg/mL, 558415, BD Biosciences). Chemiluminescence assays were performed with HRP-conjugated avidin (405103, BioLegend). Absorbance was recorded on a BioTek Synergy H1 Hybrid Reader.
Quantitative PCR.
To measure cytokine transcript levels in BMDMs, cells were seeded at 1.5 × 106 cells/mL overnight in 6-well plates and then stimulated with LPS (100 ng/mL) for 6, 12, or 24 hours, or left unstimulated. Cells were harvested using TRIzol (15596018, Invitrogen, Thermo Fisher Scientific) and isopropanol, and the RNA pellet was washed with 75% ethanol before reconstitution in diethyl-pyrocarbonate (DEPC) RNase-free water. RNA samples were converted to cDNA using ImProm-II Reverse Transcriptase Enzyme (A3803, Promega) and dNTP (U1515m, Promega), according to the manufacturer’s instructions, in a T100 Thermal Cycler (Bio-Rad). Quantitative PCR (qPCR) was run on cDNA using GoTaq(R) qPCR Master Mix (A6002, Promega) with a CFX connect RT-PCR machine (Bio-Rad). The qPCR probe sequences are provided in Supplemental Table 2.
Statistics.
Experiments using the patient’s whole blood before HSCT were only performed once because of severely limited volumes. All other experiments were repeated 3 times. Significance was calculated with GraphPad Prism 7 (GraphPad Software) using a 1- or 2-way ANOVA.
Study approval.
The care and use of the laboratory animals conformed to NIH guidelines, and all experimental procedures conformed to an animal use protocol approved by the BRC committee in Singapore (no. 191489). All experiments using human samples were approved by SingHealth, Singapore (protocol CIRB: 2013/1029/E). Written informed consent was obtained from all participants in the study.
Author contributions
EEKT and RAH are co–first authors and were responsible for writing the initial draft of the manuscript. EEKT appears as first author, because she was the patient’s primary care physician, made the initial observations of the patient, and introduced the case to the rest of the group. AMT, WKL, EEKT, S.S. Jamuar, CO, KCT, MJAK, DWQL, EST, and BT were involved in the clinical care of the patient and contributed to the interpretation of the data. In addition, MJAK, DWQL, RAH, CKL, and S.S. Jamuar contributed to the provision and editing of tables and figures. RAH, CKL, VBA, EC, EMS, LAJ, NK, JK, CZWL, LXW, YLZ, KHO, WY, MW, AAPS, CSL, AKKT, S.S. James, BV, WY, FG, VT, BR, LH, and KCC contributed to in vitro experiments and data analysis and interpretation. AAPS, CSL, AKKT, HYL, AMF contributed to in vivo experiments and data analysis. S.S. James, CO, AMF, KCC, JEC and WKL were responsible for critical appraisal of the final manuscript. All coauthors reviewed and approved the final version of the submitted manuscript.
Supplementary Material
Acknowledgments
We thank the patient and her family for participating in this research. We would also like to thank Tan Ene Choo at KK Women and Children’s Hospital Research Centre (KKH KKRC) for assistance with sample handling. We thank Alvin Yu-Jin Ng and Sumanty Tohari for their help with exome sequencing and the identification of variants.
Version 1. 08/04/2020
In-Press Preview
Version 2. 09/28/2020
Electronic publication
Version 3. 11/02/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(11):5817–5832.https://doi.org/10.1172/JCI98882.
Contributor Information
Chrissie K. Lim, Email: chlim@imcb.a-star.edu.sg.
Christina Ong, Email: c1ong.dr@gmail.com.
Mark J.A. Koh, Email: mark.koh.ja@kkh.com.sg.
Madhushanee Weerasooriya, Email: micmw@nus.edu.sg.
Chang Siang Lim, Email: ephlcs@nus.edu.sg.
Hui Yin Lee, Email: leehy@imcb.a-star.edu.sg.
Leigh Ann Jones, Email: leighannjones@hotmail.com.
Nivashini Kaliaperumal, Email: nivashinik@imcb.a-star.edu.sg.
Ee Shien Tan, Email: tan.ee.shien@kkh.com.sg.
Biju Thomas, Email: biju.thomas@singhealth.com.sg.
Lena Ho, Email: lena@ho-lab.org.
Florent Ginhoux, Email: Florent_Ginhoux@immunol.a-star.edu.sg.
Adrian K.K. Teo, Email: ateo@imcb.a-star.edu.sg.
Weimiao Yu, Email: wmyu@imcb.a-star.edu.sg.
Byrappa Venkatesh, Email: mcbbv@imcb.a-star.edu.sg.
Vinay Tergaonkar, Email: vinayt@imcb.a-star.edu.sg.
Bruno Reversade, Email: bruno.reversade@imb.a-star.edu.sg.
Ah Moy Tan, Email: tan.ah.moy@kkh.com.sg.
Woei Kang Liew, Email: liew.woei.kang@kkh.com.sg.
John E. Connolly, Email: jeconnolly@imcb.a-star.edu.sg.
References
- 1.Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 2.DiDonato JA, Mercurio F, Karin M. NF-κB and the link between inflammation and cancer. Immunol Rev. 2012;246(1):379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 3.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5(10):749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 4.Xi L, et al. Expressions of IL-6, TNF-α and NF-κB in the skin of Chinese brown frog (Rana dybowskii) Eur J Histochem. 2017;61(4):2834. doi: 10.4081/ejh.2017.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109 Suppl:S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 6.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 7.Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1(4):a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 9.Baldwin AS. Series introduction: the transcription factor NF-kappaB and human disease. J Clin Invest. 2001;107(1):3–6. doi: 10.1172/JCI11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Häcker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;2006(357):re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, Lenardo MJ, Baltimore D. 30 years of NF-κB: a blossoming of relevance to human pathobiology. Cell. 2017;168(1-2):37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stancovski I, Baltimore D. NF-kappaB activation: the I kappaB kinase revealed? Cell. 1997;91(3):299–302. doi: 10.1016/S0092-8674(00)80413-4. [DOI] [PubMed] [Google Scholar]
- 13.Picard C, Casanova JL, Puel A. Infectious diseases in patients with IRAK-4, MyD88, NEMO, or IκBα deficiency. Clin Microbiol Rev. 2011;24(3):490–497. doi: 10.1128/CMR.00001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Courtois G, et al. A hypermorphic IkappaBalpha mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J Clin Invest. 2003;112(7):1108–1115. doi: 10.1172/JCI18714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Bernuth H, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008;321(5889):691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picard C, et al. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine (Baltimore) 2010;89(6):403–425. doi: 10.1097/MD.0b013e3181fd8ec3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssen R, et al. The same IkappaBalpha mutation in two related individuals leads to completely different clinical syndromes. J Exp Med. 2004;200(5):559–568. doi: 10.1084/jem.20040773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald DR, Mooster JL, Reddy M, Bawle E, Secord E, Geha RS. Heterozygous N-terminal deletion of IkappaBalpha results in functional nuclear factor kappaB haploinsufficiency, ectodermal dysplasia, and immune deficiency. J Allergy Clin Immunol. 2007;120(4):900–907. doi: 10.1016/j.jaci.2007.08.035. [DOI] [PubMed] [Google Scholar]
- 19.Lopez-Granados E, et al. A novel mutation in NFKBIA/IKBA results in a degradation-resistant N-truncated protein and is associated with ectodermal dysplasia with immunodeficiency. Hum Mutat. 2008;29(6):861–868. doi: 10.1002/humu.20740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohnishi H, et al. A rapid screening method to detect autosomal-dominant ectodermal dysplasia with immune deficiency syndrome. J Allergy Clin Immunol. 2012;129(2):578–580. doi: 10.1016/j.jaci.2011.09.042. [DOI] [PubMed] [Google Scholar]
- 21.Schimke LF, et al. A novel gain-of-function IKBA mutation underlies ectodermal dysplasia with immunodeficiency and polyendocrinopathy. J Clin Immunol. 2013;33(6):1088–1099. doi: 10.1007/s10875-013-9906-1. [DOI] [PubMed] [Google Scholar]
- 22.Yoshioka T, et al. Autosomal dominant anhidrotic ectodermal dysplasia with immunodeficiency caused by a novel NFKBIA mutation, p.Ser36Tyr, presents with mild ectodermal dysplasia and non-infectious systemic inflammation. J Clin Immunol. 2013;33(7):1165–1174. doi: 10.1007/s10875-013-9924-z. [DOI] [PubMed] [Google Scholar]
- 23.Lee AJ, et al. Severe mycobacterial diseases in a patient with GOF IκBα mutation without EDA. J Clin Immunol. 2016;36(1):12–15. doi: 10.1007/s10875-015-0223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sogkas G, et al. A novel NFKBIA variant substituting serine 36 of IκBα causes immunodeficiency with warts, bronchiectasis and juvenile rheumatoid arthritis in the absence of ectodermal dysplasia. Clin Immunol. 2020;210:108269. doi: 10.1016/j.clim.2019.108269. [DOI] [PubMed] [Google Scholar]
- 25.Petersheim D, et al. Mechanisms of genotype-phenotype correlation in autosomal dominant anhidrotic ectodermal dysplasia with immune deficiency. J Allergy Clin Immunol. 2018;141(3):1060–1073.e3. doi: 10.1016/j.jaci.2017.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staples E, et al. Disseminated Mycobacterium malmoense and Salmonella infections associated with a novel variant in NFKBIA. J Clin Immunol. 2017;37(5):415–418. doi: 10.1007/s10875-017-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giancane G, Ferrari S, Carsetti R, Papoff P, Iacobini M, Duse M. Anhidrotic ectodermal dysplasia: a new mutation. J Allergy Clin Immunol. 2013;132(6):1451–1453. doi: 10.1016/j.jaci.2013.05.034. [DOI] [PubMed] [Google Scholar]
- 28.Moriya K, Sasahara Y, Ohnishi H, Kawai T, Kanegane H. IKBA S32 mutations underlie ectodermal dysplasia with immunodeficiency and severe noninfectious systemic inflammation. J Clin Immunol. 2018;38(5):543–545. doi: 10.1007/s10875-018-0522-y. [DOI] [PubMed] [Google Scholar]
- 29.Boisson B, Puel A, Picard C, Casanova JL. Human IκBα gain of function: a severe and syndromic immunodeficiency. J Clin Immunol. 2017;37(5):397–412. doi: 10.1007/s10875-017-0400-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosters A, Karpen SJ. The role of inflammation in cholestasis: clinical and basic aspects. Semin Liver Dis. 2010;30(2):186–194. doi: 10.1055/s-0030-1253227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szabo G, Csak T. Inflammasomes in liver diseases. J Hepatol. 2012;57(3):642–654. doi: 10.1016/j.jhep.2012.03.035. [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez-Garay EA. Cholestasis: human disease and experimental animal models. Ann Hepatol. 2003;2(4):150–158. doi: 10.1016/S1665-2681(19)32126-X. [DOI] [PubMed] [Google Scholar]
- 33.Geier A, Fickert P, Trauner M. Mechanisms of disease: mechanisms and clinical implications of cholestasis in sepsis. Nat Clin Pract Gastroenterol Hepatol. 2006;3(10):574–585. doi: 10.1038/ncpgasthep0602. [DOI] [PubMed] [Google Scholar]
- 34.Prescott JA, Cook SJ. Targeting IKKβ in cancer: challenges and opportunities for the therapeutic utilisation of IKKβ inhibitors. Cells. 2018;7(9):E115. doi: 10.3390/cells7090115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verstrepen L, Beyaert R. Receptor proximal kinases in NF-κB signaling as potential therapeutic targets in cancer and inflammation. Biochem Pharmacol. 2014;92(4):519–529. doi: 10.1016/j.bcp.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 36.McLoed AG, et al. Neutrophil-derived IL-1β impairs the efficacy of NF-κB inhibitors against lung cancer. Cell Rep. 2016;16(1):120–132. doi: 10.1016/j.celrep.2016.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park JE, Miller Z, Jun Y, Lee W, Kim KB. Next-generation proteasome inhibitors for cancer therapy. Transl Res. 2018;198:1–16. doi: 10.1016/j.trsl.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paramore A, Frantz S. Bortezomib. Nat Rev Drug Discov. 2003;2(8):611–612. doi: 10.1038/nrd1159. [DOI] [PubMed] [Google Scholar]
- 39.Richardson PG, et al. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med. 2005;352(24):2487–2498. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- 40.Richardson PG, et al. A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med. 2003;348(26):2609–2617. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- 41.Palumbo A, et al. Daratumumab, bortezomib, and dexamethasone for multiple myeloma. N Engl J Med. 2016;375(8):754–766. doi: 10.1056/NEJMoa1606038. [DOI] [PubMed] [Google Scholar]
- 42.Ruschak AM, Slassi M, Kay LE, Schimmer AD. Novel proteasome inhibitors to overcome bortezomib resistance. J Natl Cancer Inst. 2011;103(13):1007–1017. doi: 10.1093/jnci/djr160. [DOI] [PubMed] [Google Scholar]
- 43.Robak P, Robak T. Bortezomib for the treatment of hematologic malignancies: 15 years later. Drugs R D. 2019;19(2):73–92. doi: 10.1007/s40268-019-0269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lichter DI, et al. Sequence analysis of β-subunit genes of the 20S proteasome in patients with relapsed multiple myeloma treated with bortezomib or dexamethasone. Blood. 2012;120(23):4513–4516. doi: 10.1182/blood-2012-05-426924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Politou M, Karadimitris A, Terpos E, Kotsianidis I, Apperley JF, Rahemtulla A. No evidence of mutations of the PSMB5 (beta-5 subunit of proteasome) in a case of myeloma with clinical resistance to Bortezomib. Leuk Res. 2006;30(2):240–241. doi: 10.1016/j.leukres.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Franke NE, et al. Impaired bortezomib binding to mutant β5 subunit of the proteasome is the underlying basis for bortezomib resistance in leukemia cells. Leukemia. 2012;26(4):757–768. doi: 10.1038/leu.2011.256. [DOI] [PubMed] [Google Scholar]
- 47.Tang AC, et al. Combination therapy with proteasome inhibitors and TLR agonists enhances tumour cell death and IL-1β production. Cell Death Dis. 2018;9(2):162. doi: 10.1038/s41419-017-0194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pannicke U, et al. Deficiency of innate and acquired immunity caused by an IKBKB mutation. N Engl J Med. 2013;369(26):2504–2514. doi: 10.1056/NEJMoa1309199. [DOI] [PubMed] [Google Scholar]
- 49.Hsu LC, et al. IL-1β-driven neutrophilia preserves antibacterial defense in the absence of the kinase IKKβ. Nat Immunol. 2011;12(2):144–150. doi: 10.1038/ni.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greten FR, et al. NF-kappaB is a negative regulator of IL-1beta secretion as revealed by genetic and pharmacological inhibition of IKKbeta. Cell. 2007;130(5):918–931. doi: 10.1016/j.cell.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luedde T, Heinrichsdorff J, de Lorenzi R, De Vos R, Roskams T, Pasparakis M. IKK1 and IKK2 cooperate to maintain bile duct integrity in the liver. Proc Natl Acad Sci USA. 2008;105(28):9733–9738. doi: 10.1073/pnas.0800198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 53.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takata K, et al. Induced-pluripotent-stem-cell-derived primitive macrophages provide a platform for modeling tissue-resident macrophage differentiation and function. Immunity. 2017;47(1):183–198.e6. doi: 10.1016/j.immuni.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 55.Shin EM, et al. DEAD-box helicase DP103 defines metastatic potential of human breast cancers. J Clin Invest. 2014;124(9):3807–3824. doi: 10.1172/JCI73451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teo AK, et al. Derivation of human induced pluripotent stem cells from patients with maturity onset diabetes of the young. J Biol Chem. 2013;288(8):5353–5356. doi: 10.1074/jbc.C112.428979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8(4):265–277. doi: 10.1023/A:1008942828960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.