Abstract
Drivers of sporadic benign pituitary adenoma growth are largely unknown. Whole-exome sequencing of 159 prospectively resected pituitary adenomas showed that somatic copy number alteration (SCNA) rather than mutation is a hallmark of hormone-secreting adenomas and that SCNAs correlate with adenoma phenotype. Using single-gene SCNA pathway analysis, we observed that both cAMP and Fanconi anemia DNA damage repair pathways were affected by SCNAs in growth hormone–secreting (GH-secreting) somatotroph adenomas. As somatotroph differentiation and GH secretion are dependent on cAMP activation and we previously showed DNA damage, aneuploidy, and senescence in somatotroph adenomas, we studied links between cAMP signaling and DNA damage. Stimulation of cAMP in C57BL/6 mouse primary pituitary cultures using forskolin or a long-acting GH-releasing hormone (GHRH) analog increased GH production and DNA damage measured by H2AX phosphorylation and a comet assay. Octreotide, a somatostatin receptor ligand that targets somatotroph adenoma GH secretion in patients with acromegaly, inhibited cAMP and GH and reversed DNA damage induction. In vivo long-acting GHRH treatment also induced pituitary DNA damage in mice. We conclude that cAMP, which induces somatotroph proliferation and GH secretion, may concomitantly induce DNA damage, potentially linking hormone hypersecretion to SCNA and genome instability. These results elucidating somatotroph adenoma pathophysiology identify pathways for targeted treatment.
Keywords: Endocrinology
Keywords: Neuroendocrine regulation
Introduction
Adenomas arising from the anterior pituitary are invariably benign and may either be nonhormone secreting (nonfunctioning) or hormone secreting (functioning) (1, 2). The prevalence of identified pituitary adenomas ranges from a low of 80 to 100 per 100,000 individuals (3, 4) to a high of 1 per 1000 individuals (5), with 9% to 14% of these pituitary adenomas being growth hormone (GH) secreting (3–5). cAMP dysregulation has been implicated in the pathogenesis of GH-secreting somatotroph adenomas seen in rare genetic syndromes such as Carney syndrome, which is associated with a PKRAR1A mutation (6), as well as familial isolated pituitary adenoma (FIPA) associated with AIP gene mutation (7) and X-linked acrogigantism associated with GRP101 gene microduplication/mutation (8–11). Rarely, activating mosaic mutations in cAMP-dependent GNAS, which encodes G protein α subunit, are associated with somatotroph adenomas in McCune-Albright syndrome (9), whereas somatic GNAS mutations are seen in 30% to 40% of nonfamilial somatotroph pituitary adenomas (12). GH-releasing hormone (GHRH) receptor–activating mutations have not been identified to date (13).
We previously showed evidence for pituitary adenoma DNA damage and senescence, particularly in GH-secreting somatotroph adenomas (14). Furthermore, increased expression of the pituitary tumor–transforming gene (PTTG), the index mammalian securin that enables faithful chromatid separation during mitosis (15–17), was also associated with aneuploidy in these adenomas (16). Loss of heterozygosity (18) and somatic copy number alterations (SCNAs) were reported in secreting and more aggressively growing sporadic pituitary adenomas (18–22). De novo SCNAs, commonly seen in tumors, may arise as a consequence of DNA damage and genome instability and can result in cell dysfunction (23, 24).
We prospectively studied sporadic pituitary adenomas using whole-exome sequencing (WES) to evaluate somatic mutations and SCNAs. We then used pathway analysis and determined that both cAMP and Fanconi anemia DNA damage repair pathways are significantly affected in GH-secreting somatotroph adenomas. As somatotrophs are highly sensitive to cAMP, which is important for somatotroph proliferation and GH production (25, 26), we hypothesized that DNA damage is coupled to the striking overproduction of GH from somatotroph pituitary adenomas. We now show increased cAMP signaling leading to DNA damage in WT nontransformed murine pituitary glands, in primary pituitary cultures, and in vivo. Our findings potentially link cAMP to both endocrine hormone hypersecretion and DNA damage and open new avenues for targeting dysregulated cAMP and DNA damage as pathogenetic drivers of sporadic GH-secreting pituitary adenomas.
Results
We performed prospective WES analysis of 159 consecutively resected pituitary adenoma specimens and of buffy coat samples derived from the same patients for use as controls. Samples included nonsecreting gonadotroph (n = 61), null cell (n = 29), and silent corticotroph (n = 8) adenomas, as well as secreting corticotroph (n = 10), lactotroph (n = 16), and somatotroph (n = 35) adenomas. For group analysis, we combined gonadotroph and null cell adenomas into a single category among the nonsecreting adenomas because of their similar clinical phenotype. However, we maintained silent corticotroph adenomas as a distinctive group, as it is yet unclear whether these adenomas behave more aggressively than gonadotroph and null cell adenomas, and because there are reports of rarely occurring silent-to-secreting corticotroph adenoma transformation (27, 28).
Patient demographics, presurgical treatments, adenoma imaging features, and IHC characteristics are detailed in Table 1. Patients with gonadotroph/null cell adenomas were older than those with secreting corticotroph or lactotroph adenomas (P < 0.0001). Nonsecreting adenomas exhibited larger MRI-determined diameters than did hormone-secreting corticotroph and somatotroph adenomas (P < 0.0001) and extended to the cavernous sinus more frequently than did somatotroph and lactotroph adenomas (P < 0.0001). Nonsecreting adenomas also extended to the suprasellar region more commonly than did secreting somatotroph, corticotroph, and lactotroph adenomas (P < 0.0001). We found that Ki67 expression was similar between adenoma subtypes.
Table 1. Characteristics of 159 pituitary adenoma samples included in the WES analysisA.
Somatic mutations are uncommon in pituitary adenomas.
WES revealed a low mutation rate, with 689 nonsynonymous single-nucleotide variations (SNVs) and 31 small insertions/deletions (InDels) in the entire cohort. Nearly all (90.4%) mutated genes appeared once. Approximately 4 nonsynonymous SNVs/InDels per adenoma were detected in both nonsecreting (mean, 4.04 ± 4.9; median, 3 [range, 0–35]) and secreting (mean, 4.7 ± 4.2; median 4, [range, 0–18]) adenomas (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI138540DS1). Among genes rarely mutated in nonfamilial pituitary adenomas, we detected 1 mutation each in USP8 (corticotroph adenoma), AIP (InDel, lactotroph adenoma), and MEN1 (lactotroph adenoma), and none in PRKAR1A, PRKAR2A, CDKN1B, NF1, UPS48, BRAF, HRAS, TP53, DICER1, CABLES1, or SDHx.
However, we observed GNAS mutations in 10 of 35 (29%) somatotroph adenomas; 7 adenomas carried the p.R844C mutation (arginine to cysteine), whereas p.Q870L, p.R844H, and p.R142C mutations were identified in 1 somatotroph adenoma each.
SCNAs are a distinctive genomic hallmark of hormone-secreting adenomas and correlate with clinical phenotype.
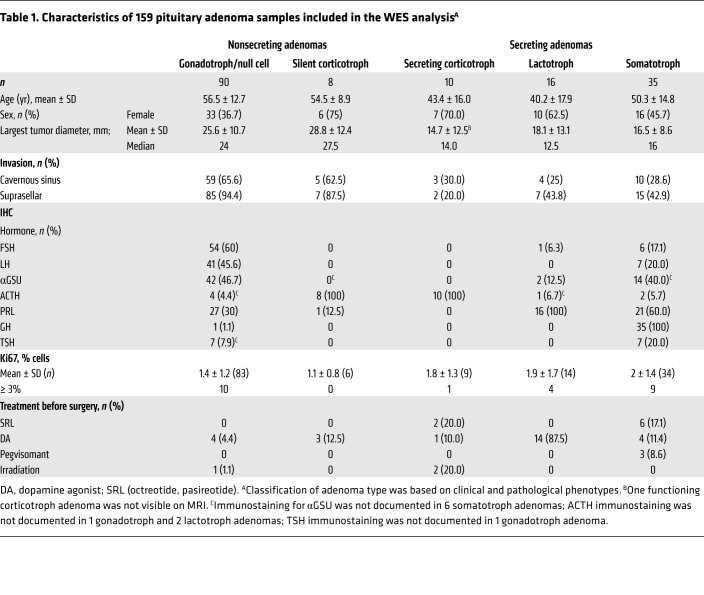
In contrast to the low rates of somatic mutations observed in both hormone-secreting and nonsecreting adenomas, we found SCNAs significantly more frequently in secreting adenomas (Figure 1), with large, often whole chromosomal deletions or amplifications. Somatotroph and lactotroph adenomas expressed the highest SCNA copy ratios. Whole-genome sequencing (WGS) of 14 representative adenomas showed a SCNA copy ratio distribution similar to that obtained by WES for the same adenomas (Supplemental Figure 2).
Figure 1. Heatmap of SCNAs derived from WES of 159 adenomas.
Heatmap of SCNAs obtained from WES of 159 adenomas depicts the SCNA copy ratio as well as adenoma functional status and subtype.
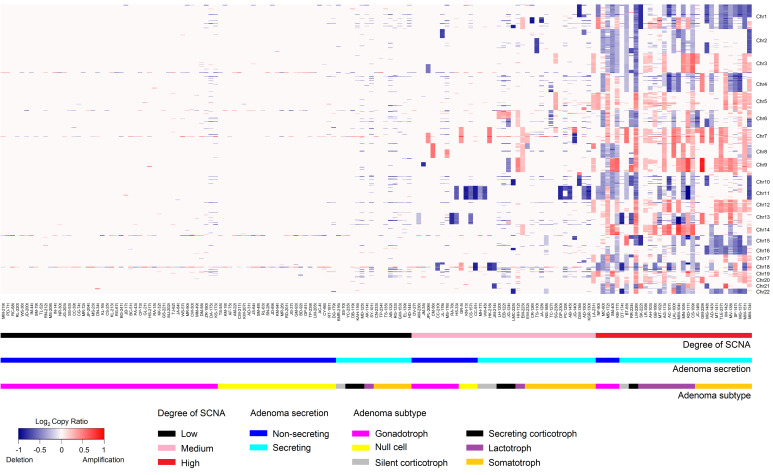
The SCNA degree (low, medium, or high) (Supplemental Figure 3) correlated with IHC staining for prolactin (PRL) (r = 0.55, P = 8.6 × 10–14) and GH (r = 0.36, P = 3.4 × 10–6), and negatively correlated with follicle-stimulating hormone (FSH) (r = –0.29, P = 2.9 × 10–4) and luteinizing hormone (LH) (r = –0.26, P = 8.6 × 10–4) (Figure 2). SCNAs also negatively correlated with suprasellar extension (r = –0.28, P = 5.3 × 10–4) and cavernous sinus extension (r = –0.18, P = 0.03), and we observed a weak correlation between SCNA degree and Ki67 expression (r = 0.19, P = 0.02). Other characteristics, including expression of adrenocorticotrophin (ACTH), thyrotrophin (TSH), and glycoprotein hormone α subunit (αGSU), and the largest tumor diameter did not correlate with SCNA degree (data not shown).
Figure 2. Correlation between SCNA score and phenotypic adenoma characteristics.
The correlation between SCNA score and phenotypic adenoma characteristics was assessed by immunostaining (hormones and Ki67) or MRI (suprasellar extension [SSE] and cavernous sinus extension [CSE]). Spearman’s rank-order correlation is shown inside the bars; P values are shown above each bar. Dark gray bars indicate a positive correlation; light gray bars indicate a negative correlation. n = 159.
Mutation of GNAS is associated with SCNA degree in somatotroph adenomas.
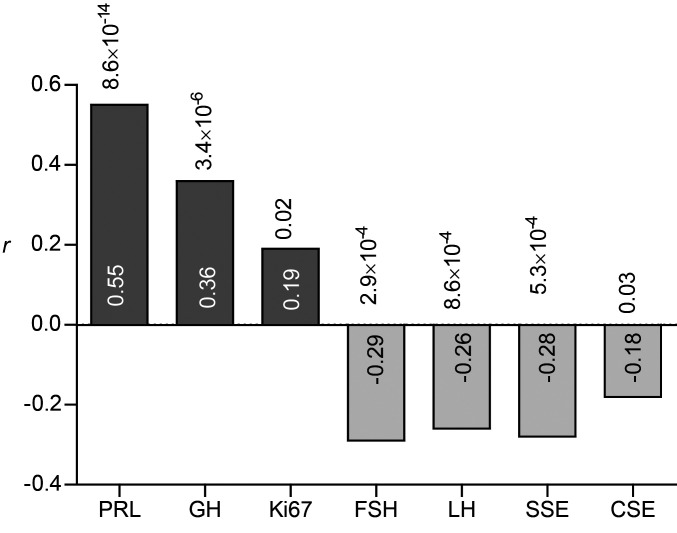
Unlike GNAS mutation–negative somatotroph adenomas, GNAS mutation–positive adenomas mostly harbored medium-degree SCNAs, since of the 10 GNAS-mutated somatotroph adenomas, 9 expressed medium-degree SCNAs and only 1 had low-degree SCNAs. None of the adenomas expressed high-degree SCNAs (Fisher’s exact test, P = 0.001) (Figure 3, A and B). Although 18 of the 25 (72%) somatotroph adenomas negative for the GNAS mutation exhibited medium or high degrees of SCNAs, their distribution across the different SCNA degrees was similar.
Figure 3. Degree of SCNA in GNAS-mutated and nonmutated somatotroph pituitary adenomas.
(A) Adenomas with a low, medium, and high degree of SCNA. The number of adenomas in each group is shown inside the bars. GNAS (+), mutation positive (n = 10); GNAS (–), mutation negative (n = 25). P = 0.001, by Fisher’s exact test. (B) Heatmap of somatotroph adenoma SCNAs (n = 35).
Genes in cAMP and Fanconi anemia pathways are significantly affected by single-gene SCNAs.
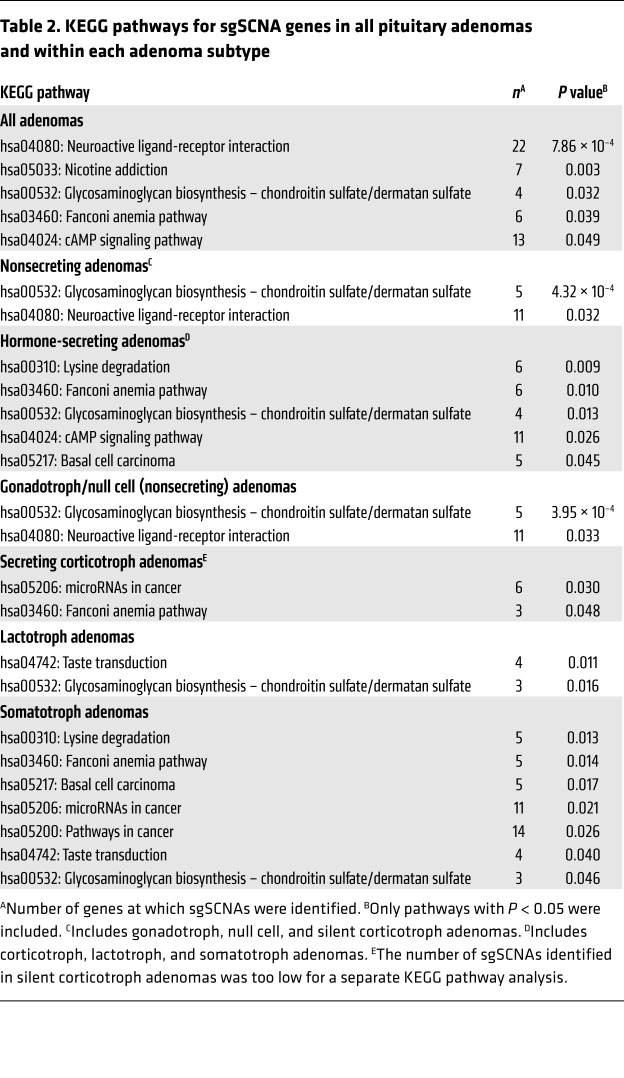
SCNA analysis of the entire cohort revealed 20,938 deleted (69.1%) and 9363 amplified fragments (30.9%). Large chromosomal arm-level SCNAs were observed, each containing large numbers of deleted or amplified genes. In our Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, we focused on SCNAs spanning within a single gene (sgSCNAs) as potential driver candidates and identified 2368 genes across all adenoma samples (1640 deleted and 728 amplified). We further narrowed the list of candidate genes by eliminating sgSCNAs appearing in only 1 adenoma following the criteria described in Methods. Our final analysis set included 853 sgSCNA genes (715 deleted and 138 amplified; Supplemental Table 1). sgSCNAs appeared more frequently in secreting than nonsecreting adenomas (Supplemental Figure 4). The mean number of sgSCNA genes per adenoma was similar in nonsecreting gonadotroph/null cell (25.8 ± 43.5) and silent corticotroph (24.5 ± 50.2) adenomas, and was significantly higher in hormone-secreting corticotroph (79.8 ± 86.2; 2-tailed, unpaired t test with Bonferroni’s correction, P ≤ 0.01), lactotroph (97.4 ± 66.3; P ≤ 0.001), and somatotroph (90.0 ± 67.8; P ≤ 0.001) adenomas compared with gonadotroph/null cell adenomas.
KEGG pathway analysis using these sgSCNA genes identified several pathways significantly affected by SCNAs across all pituitary adenomas as well as when classified by function and subtype (P < 0.05; Table 2). The cAMP pathway was significantly affected by sgSCNA genes in the entire cohort (P = 0.048), and 15 sgSCNA genes in the cAMP pathway, most of which were deletions, appeared 80 times in the entire cohort (Supplemental Figure 5E). Somatotroph adenomas harbored a higher number of cAMP sgSCNAs (55% of identified cAMP pathway sgSCNA genes) than did gonadotroph/null cell (13.8%), lactotroph (18.8%), secreting corticotroph (12.5%), and silent corticotroph (2.5%) (P < 0.0001) adenomas.
Table 2. KEGG pathways for sgSCNA genes in all pituitary adenomas and within each adenoma subtype.
The Fanconi anemia pathway was significantly affected by sgSCNA genes in somatotroph (P = 0.014) and in secreting corticotroph (P = 0.048) adenomas but not in gonadotroph/null cell or lactotroph adenomas. By contrast, we found that the neuroactive ligand-receptor interaction pathway was significantly associated with gonadotroph/null cell adenomas (P = 0.033) but not secreting adenomas (Table 2).
Upon further exploration of the association with the Fanconi anemia pathway, we found that BRCA2, BRCA1, REV3L, HES1, and RMI1 harbored sgSCNAs, all showing deletions, and that Fanconi anemia sgSCNA genes were more frequent in secreting than nonsecreting adenomas (Supplemental Figure 5A). In addition, the sgSCNA gene copy ratio in Fanconi anemia pathway genes correlated with adenoma SCNA scores (Supplemental Figure 5C, r = –0.24, P = 0.006). By contrast, sgSCNAs showing deletions of non-Fanconi anemia pathway genes, which were also more frequently observed in secreting than in nonsecreting adenomas (Supplemental Figure 5B), did not correlate with the SCNA copy ratio (Supplemental Figure 5D), suggesting a unique role for Fanconi anemia gene deletions in pituitary adenoma SCNAs. BRCA1 and BRCA2 mRNA expression levels, which we measured in a cohort of 40 adenomas not previously sequenced but with phenotypic characteristics similar to those of the overall cohort used for WES analysis (Supplemental Table 2), were highly variable and generally did not differ between adenoma types (not shown).
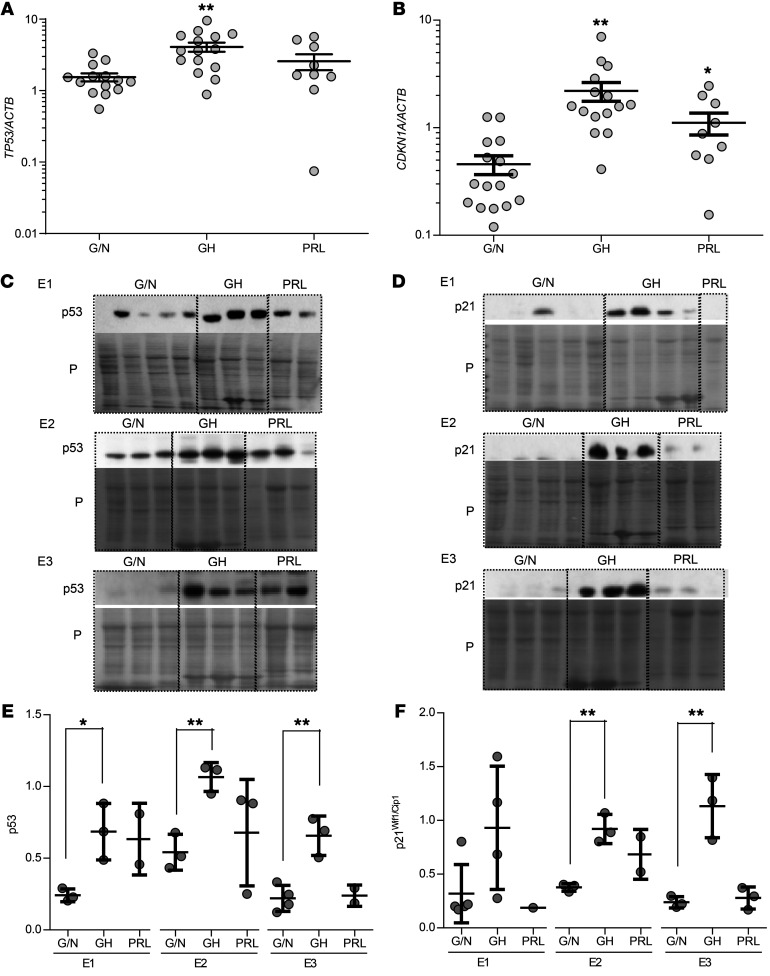
Analysis of this cohort of 40 adenomas not previously sequenced also showed elevated p53 and p21Wif1/Cip1 mRNA and protein levels in somatotroph adenomas but not in gonadotroph/null cell adenomas (Figure 4), further supporting a role for DNA damage response activation specific to somatotroph adenomas.
Figure 4. Expression of p53 and p21Wif1/Cip1 in nonsecreting gonadotroph/null cell, somatotroph, and lactotroph pituitary adenomas.
(A) TP53 (p53) and (B) CDKN1A (p21Wif1/Cip1) mRNA expression in nonsecreting gonadotroph/null cell (G/N), somatotroph (GH), and lactotroph (PRL) pituitary adenomas, normalized to ACTB. Results are presented as the mean ± SEM. *P ≤ 0.05 and **P ≤ 0.01, by 2-tailed, unpaired t test with Bonferroni’s correction. Protein expression of (C) p53 and (D) p21Wif1/Cip1 compared with Ponceau (P) staining in 3 different Western blot membranes with human adenoma sets E1, E2, and E3. (E and F) Quantitative presentation of (E) p53 and (F) p21Wif1/Cip1 in the 3 membranes after normalization to Ponceau, using ImageJ. Each experiment was analyzed separately. Results are presented as the mean ± SD. *P ≤ 0.05 and **P = 0.01, by 2-tailed, unpaired t test with Bonferroni’s correction.
cAMP increases DNA damage in normal mouse primary pituitary cultures.
Several factors constrained our further mechanistic studies in human adenomas. Normal fresh human pituitary tissue is not readily available, there are no normal human pituitary cell lines, and human autopsy pituitary tissue can show early postmortem changes due to stress, inflammation, and apoptosis gene regulation, which could cause DNA damage and influence WES analysis data interpretation (29). We therefore used normal mouse primary pituitary cultures to evaluate DNA damage triggers.
We established normal pituitary cultures derived from 4-month-old C57BL/6 mice not expected to exhibit significant pituitary DNA damage to perform the experiments described in Methods; a total of 710 mice were required to obtain sufficient numbers of cells to derive the reported results.
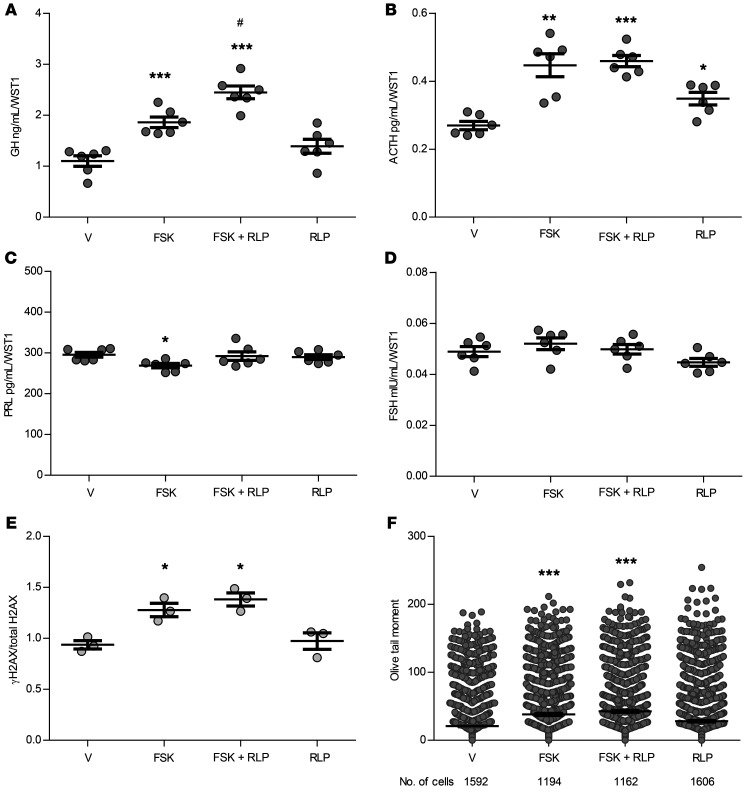
We induced intracellular cAMP levels with 10 μM forskolin to recapitulate signaling of hypothalamus-releasing factors such as GHRH. We also treated cultures with the phosphodiesterase subtype 4 (PDE4) inhibitor rolipram (1 μM) (30) to mimic lower PDE4D mRNA levels observed in somatotroph adenomas compared with levels in gonadotroph/null cell and lactotroph adenomas (Supplemental Figure 6A). As expected, forskolin increased intracellular cAMP levels (Supplemental Figure 7), and forskolin-induced supernatant GH levels (2-tailed, unpaired t test with Bonferroni’s correction P ≤ 0.001 vs. vehicle) were further enhanced with the addition of rolipram (P ≤ 0.001 vs. vehicle, P ≤ 0.05 vs. forskolin alone) (Figure 5A). ACTH levels increased with forskolin alone (P ≤ 0.01) and when combined with rolipram (P ≤ 0.001), as well as with rolipram alone (P ≤ 0.05 vs. vehicle) (Figure 5B). Although forskolin modestly decreased PRL levels (P ≤ 0.05 vs. vehicle) (Figure 5C), neither forskolin alone nor the combination of forskolin and rolipram altered FSH levels (Figure 5D).
Figure 5. Treatment of C57BL/6 normal mouse primary pituitary cultures with forskolin and rolipram.
(A) GH, (B) ACTH, (C) PRL, and (D) FSH concentrations in supernatant after treatment with vehicle (V), 10 μM forskolin (FSK), cotreatment with 10 μM forskolin and 1 μM rolipram (FSK + RLP), or 1 μM rolipram alone, normalized to WST1. n = 6 per treatment group. (E) Quantitative presentation of Western blot results depicting the change in γH2AX levels normalized to total H2AX after 16 hours of treatment as above. Results were derived from 3 separate wells. Band intensities were calculated by ImageJ. The full panel is shown in Supplemental Figure 7B. (F) Results of the comet assay depicting Olive tail moment measurement 16 hours after treatment as above. Results were pooled from 2 experiments. The number of cells analyzed by a blinded observer is indicated. Results are presented as the mean ± SEM (vehicle, 20.8 ± 1.0; forskolin, 38.2 ± 1.4; forskolin plus rolipram, 42.7 ± 1.5; rolipram, 28.1 ± 1.1). *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001 versus vehicle; #P ≤ 0.05 versus forskolin, by 2-tailed, unpaired t test with Bonferroni’s correction. The experiments were performed at least twice.
Phosphorylated H2AX (γH2AX) is considered an early sensitive biomarker for genotoxicity and for monitoring DNA damage (31). Histone H2AX is phosphorylated early upon DNA damage, enabling the accumulation of DNA damage response proteins, and activates cell-cycle checkpoint pathways including p53/p21Wif1/Cip1. γH2AX levels increased within 30 minutes of forskolin treatment and remained elevated at 1 and 3 hours of treatment (Supplemental Figure 8A), suggesting an acute stimulation of the DNA damage pathway, but addition of rolipram did not further enhance forskolin action. We continued to observe an increase in γH2AX after 16 hours of treatment (2-tailed, unpaired t test with Bonferroni’s correction, P ≤ 0.05 with forskolin and P ≤ 0.05 with forskolin and rolipram vs. vehicle) (Figure 5E and Supplemental Figure 8B).
We also performed the comet assay, which is a sensitive method to measure DNA damage in single cells (32, 33). Although γH2AX and comet assay results correlate well, the comet assay detects a larger spectrum of DNA damage than γH2AX and is therefore useful for genotoxicity measurement (34).
The Olive tail moment measured by comet assay similarly increased after 16 hours of forskolin treatment with or without rolipram (P ≤ 0.001) (Figure 5F). Treatment with rolipram alone had no significant effect on either γH2AX levels or comet assay results.
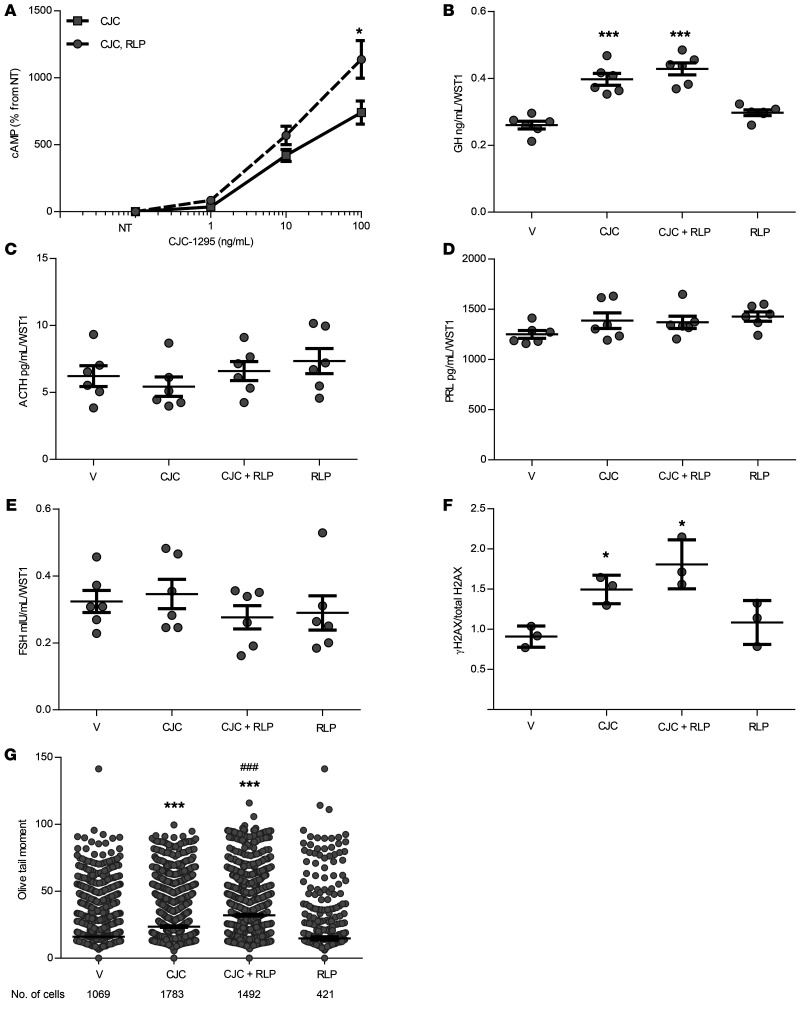
We found that GHRH receptors were more abundantly expressed in human somatotroph adenomas compared with expression in lactotroph or gonadotroph/null cell adenomas (2-tailed, unpaired t test with Bonferroni’s correction, P ≤ 0.001) (Supplemental Figure 6B). As these receptors mediate GHRH induction of somatotroph intracellular cAMP, we treated C57BL/6 mouse primary pituitary cultures with 10 ng/mL CJC-1295, a long-acting GHRH analog, and observed dose-dependent increases in intracellular cAMP levels, which were further enhanced by rolipram (2-way ANOVA with Bonferroni’s correction, P ≤ 0.05 vs. vehicle) (Figure 6A). Importantly, CJC-1295 is 10-fold weaker in stimulating intracellular cAMP production compared with forskolin (Supplemental Figure 7) and therefore would be expected to have a weaker effect on GH stimulation and the extent of DNA damage. We detected increased GH levels with 16 hours of CJC-1295 alone and in combination with rolipram (2-tailed, unpaired t test with Bonferroni’s correction, P ≤ 0.001 vs. vehicle) (Figure 6B). Neither CJC-1295 nor rolipram altered the levels of ACTH (Figure 6C), PRL (Figure 6D), or FSH (Figure 6E).
Figure 6. CJC-1295 and rolipram actions in C57BL/6 normal mouse primary pituitary cultures.
(A) Dose-dependent response of intracellular cAMP to 30 minutes of treatment with CJC-1295 (CJC), with or without 1 μM rolipram. Results are graphed as the percentage of untreated cultures (NT). n = 6/treatment group. *P ≤ 0.05, by 2-way ANOVA with Bonferroni’s correction. (B) GH, (C) ACTH, (D) PRL, and (E) FSH supernatant concentrations after 16 hours of treatment with vehicle, 10 ng/mL CJC-1295, cotreatment with 10 ng/mL CJC-1295 and 1 μM rolipram, or 1 μM rolipram alone, normalized to WST1. n = 6 per treatment group. (F) Quantitative presentation of Western blot depicting the change in γH2AX levels normalized to total H2AX after 16 hours of treatment as above. Results were derived from 3 separate wells. Band intensities were calculated by ImageJ. The full panel is shown in Supplemental Figure 9. (G) Comet assay results depicting Olive tail moment measured after 16 hours of treatment as above. Results were pooled from 2 experiments. The number of cells analyzed by a blinded observer is indicated. Results are presented as the mean ± SEM (vehicle, 16.1 ± 0.7; CJC-1295, 23.6 ± 0.6; CJC-1295 plus rolipram, 32.1 ± 0.7; rolipram, 14.8 ± 1.2). *P ≤ 0.05 versus vehicle and ***P ≤ 0.001 versus vehicle; ###P ≤ 0.001 versus CJC-1295, by 2-tailed, unpaired t test with Bonferroni’s correction. The experiments were performed twice.
As with forskolin treatment, CJC-1295 with and without rolipram increased γH2AX levels at 16 hours (2-tailed, unpaired t test with Bonferroni’s correction, each P ≤ 0.05 vs. vehicle) (Figure 6F and Supplemental Figure 9). A comet assay showed increased Olive tail moment with CJC-1295 compared with vehicle (2-tailed, unpaired t test with Bonferroni’s correction, P ≤ 0.001), which further increased with rolipram cotreatment (P ≤ 0.001 vs. CJC-1295 alone) (Figure 6G).
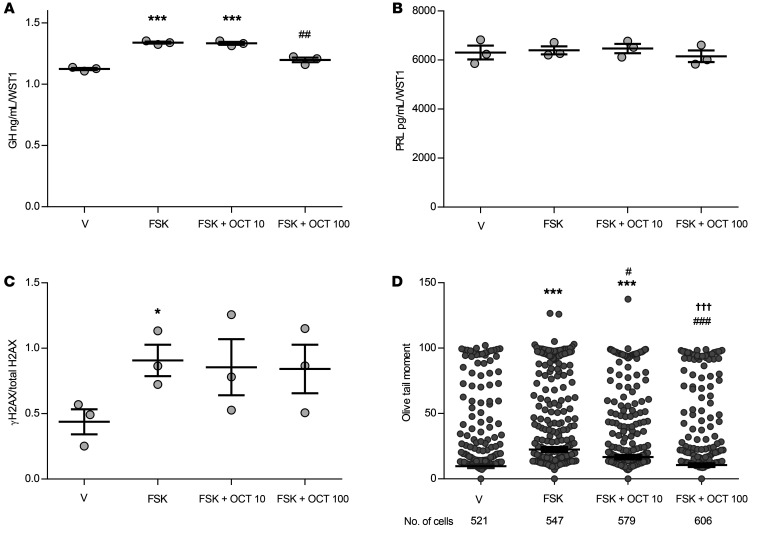
Octreotide, a somatostatin receptor ligand (SRL) that targets somatostatin receptors abundantly expressed on somatotroph adenomas, is used to treat acromegaly caused by a GH-secreting pituitary adenoma. Octreotide reduces adenoma GH secretion by inhibiting adenylate cyclase and reducing cAMP, thereby opposing GHRH action (35, 36). Cotreatment of mouse primary pituitary cultures with forskolin and octreotide (100 nM) reduced forskolin-induced supernatant GH (2-tailed, unpaired t test with Bonferroni’s correction, P ≤ 0.001 forskolin vs. vehicle; P ≤ 0.01 forskolin with 100 nM octreotide vs. forskolin alone) (Figure 7A) but did not alter PRL levels (Figure 7B) or reverse induced γH2AX levels (Figure 7C and Supplemental Figure 10A). Octreotide treatment alone did not change γH2AX levels (Supplemental Figure 10B). However, both 10 nM and 100 nM doses of octreotide attenuated forskolin-induced Olive tail moment (2-tailed, unpaired t test with Bonferroni’s correction, P ≤ 0.01 and P ≤ 0.001, respectively, vs. forskolin), with the 100 nM dose leading to greater attenuation (P ≤ 0.001 vs. forskolin with octreotide 10 nM) (Figure 7D).
Figure 7. Forskolin with and without octreotide actions in C57BL/6 normal mouse primary pituitary cultures.
(A) GH and (B) PRL concentrations in the supernatant after 16 hours of treatment with vehicle, 10 μM forskolin, cotreatment with 10 μM forskolin and 10 nM octreotide (OCT 10), or with 10 μM forskolin and 100 nM octreotide (OCT 100), normalized to WST1. n = 3 per treatment group. (C) Quantitative presentation of Western blot analysis of γH2AX change normalized to total H2AX, derived from 3 wells per treatment group. The full panels are shown in Supplemental Figure 10A. Band intensities were calculated with ImageJ. (D) Comet assay depicting Olive tail moment measured after 16 hours of treatment as above. The number of cells analyzed by a blinded observer is indicated. Results are presented as the mean ± SEM (vehicle, 9.7 ± 1.0; forskolin, 22.5 ± 1.5; forskolin plus 10 nM octreotide, 16.6 ± 1.3; forskolin plus 100 nM octreotide, 10.5 ± 1.0). *P ≤ 0.05 and ***P ≤ 0.001 versus vehicle; #P ≤ 0.05, ##P ≤ 0.01, and ###P ≤ 0.001 versus forskolin; †††P ≤ 0.001 versus forskolin plus 10 nM octreotide, by 2-tailed, unpaired t test with Bonferroni’s correction. The experiment was performed once.
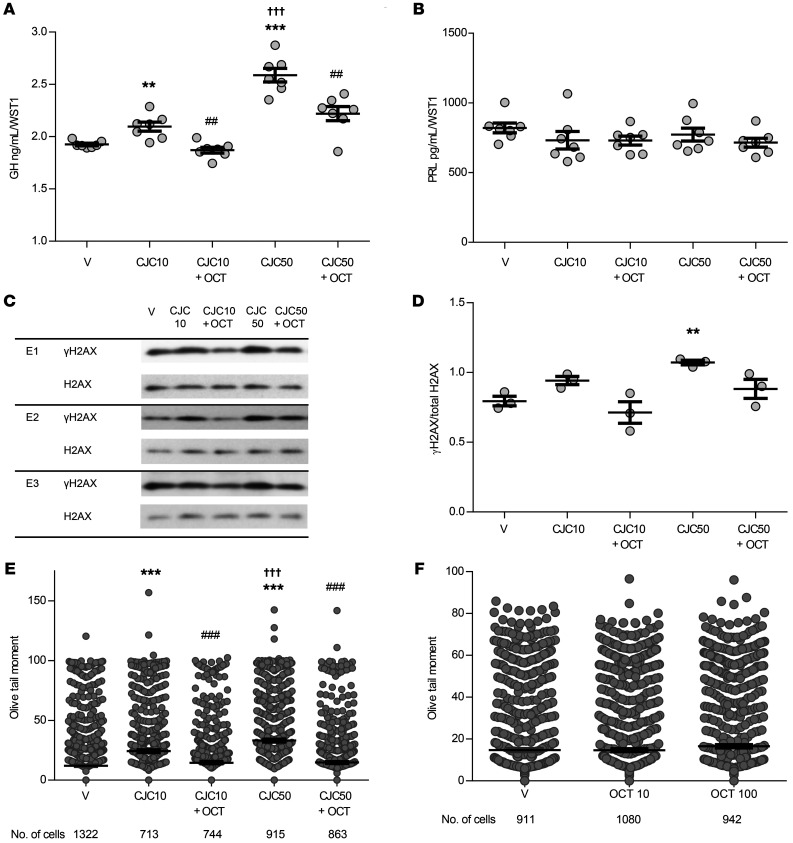
Treatment of normal mouse primary pituitary cultures with 10 and 50 ng/mL CJC-1295 increased GH levels in the supernatant (2-tailed, unpaired t test with Bonferroni’s correction, P ≤ 0.01 and P ≤ 0.001, respectively, vs. vehicle), with greater GH concentrations with the 50 ng/mL dose compared with the 10 ng/mL dose (P ≤ 0.001). The increase in GH we observed with both concentrations of CJC-1295 was attenuated by 100 nM octreotide (P ≤ 0.01 vs. their respective CJC-1295 doses alone) (Figure 8A). PRL levels were unchanged by either CJC-1295 dose, with or without the addition of octreotide (Figure 8B). Although γH2AX induced by CJC-1295 was lower with octreotide treatment (2-tailed, unpaired t test with Bonferroni’s correction, P ≤ 0.01 vs. vehicle), it did not reach significance compared with CJC-1295 alone (Figure 8, C and D). However, Olive tail moment was increased with both doses of CJC-1295 (2-tailed, unpaired t test with Bonferroni’s correction, P ≤ 0.001), more so with the higher CJC-1295 dose (P ≤ 0.001), and cotreatment with 100 nM octreotide attenuated Olive tail moment with both CJC-1295 doses (P ≤ 0.001 vs. their respective CJC-1295 doses alone) (Figure 8E). Neither dose of octreotide alone affected Olive tail moment (Figure 8F).
Figure 8. CJC-1295 with and without octreotide actions in C57BL/6 normal mouse primary pituitary cultures.
(A) GH and (B) PRL concentrations in the supernatant after 16 hours of treatment with vehicle, 10 ng/mL CJC-1295 (CJC 10), cotreatment with 10 ng/mL CJC-1295 and 100 nM octreotide, 50 ng/mL CJC-1295 (CJC 50), or cotreatment with 50 ng/mL CJC-1295 and 100 nM octreotide, normalized to WST1. n = 7 per treatment group. Results are presented as the mean ± SEM. **P ≤ 0.01 and ***P ≤ 0.001 versus vehicle; ##P ≤ 0.01 versus the corresponding CJC-1295 dose; †††P ≤ 0.001 versus 10 ng/mL CJC-1295, by 2-tailed, unpaired t test with Bonferroni’s correction. (C) Western blot showing γH2AX expression compared with total H2AX expression after 16 hours of treatment as above in 3 different experiments: E1, E2, and E3. (D) Quantitative presentation of γH2AX change normalized to total H2AX derived from 2 wells per treatment group. Band intensities were calculated by ImageJ. Results are presented as the mean ± SEM. **P ≤ 0.01, by 2-tailed, unpaired t test with Bonferroni’s correction. (E) Comet assay results depicting Olive tail moment measured after 16 hours of treatment as above. The number of cells analyzed by a blinded observer is indicated. Results are presented as the mean ± SEM (vehicle, 12 ± 0.6; 10 ng/mL CJC-1295, 24.3 ± 1.2; 10 ng/mL CJC-1295 plus octreotide, 14.6 ± 0.9; 50 ng/mL CJC-1295, 33.1 ± 1.2; 50 ng/mL CJC-1295 plus octreotide, 14.8 ± 0.8). ***P ≤ 0.001 versus vehicle; ###P ≤ 0.001 versus the corresponding CJC-1295 dose; †††P ≤ 0.001 versus 50 ng/mL CJC-1295, by 2-tailed, unpaired t test with Bonferroni’s correction. (F) Comet assay results depicting Olive tail moment measured after 16 hours of treatment with 10 nM octreotide and 100 nM octreotide. The number of cells assessed by a blinded observer is indicated. Results are presented as the mean ± SEM (vehicle, 14.7 ± 0.8; 10 nM octreotide, 14.6 ± 0.7; 100 nM octreotide, 16.5 ± 0.8). The experiment was performed once.
cAMP increases DNA damage in normal mouse pituitary in vivo.
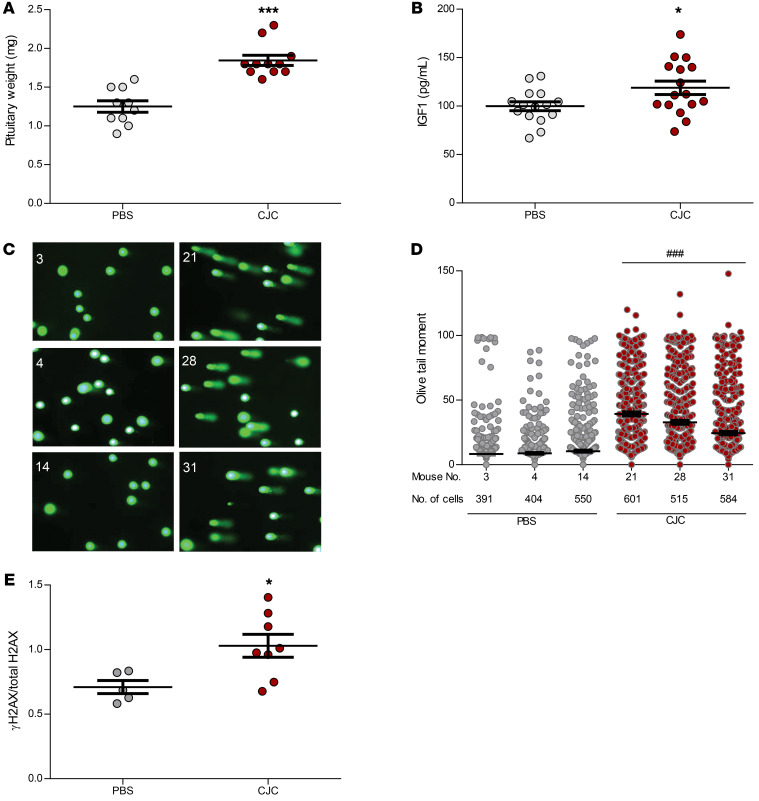
We further evaluated the CJC-1295 long-term effect on pituitary DNA damage in vivo. Four-month-old male C57BL/6 mice were injected s.c. with PBS (PBS mice, n = 15) or 10 μg/kg CJC-1295 (CJC mice, n = 16) 3 times per week for 8 weeks before sacrifice. Total body weights and lengths were similar (Supplemental Figure 11, A and B). Pituitary glands from CJC mice weighed 47% more than pituitary glands from PBS mice (2-tailed, unpaired t test with Bonferroni’s correction, P ≤ 0.001) (Figure 9A), whereas liver (Supplemental Figure 11C) and heart (Supplemental Figure 11D) weights were similar. Although GH is the dominant facilitator of liver IGF1 production (37), clinical diagnosis of GH excess relies mainly on IGF1 levels, as IGF1 has a longer circulating half-life and a more stable secretory pattern than does GH (37). Accordingly, we observed no difference in serum GH levels between the groups (Supplemental Figure 11E) but found higher serum IGF1 levels in CJC mice than in PBS mice (2-tailed, unpaired t test with Bonferroni’s correction, P ≤ 0.05) (Figure 9B). We found that ACTH, PRL, and FSH levels were also similar between the 2 groups (Supplemental Figure 11, F–H). Pituitary DNA damage was induced in CJC mice as evidenced by increased Olive tail moment (2-tailed, unpaired t test with Bonferroni’s correction, P ≤ 0.001) (Figure 9, C and D) and increased γH2AX expression (2-tailed, unpaired t test with Bonferroni’s correction, P ≤ 0.05) (Figure 9E and Supplemental Figure 12). Mouse 32 was excluded from the PBS-treated group because of very low pituitary protein retrieval and undetectable bands on Western blotting. Mouse 7 was also excluded from the PBS group, as, compared with the mean of all PBS and CJC mice, this mouse had a much higher γH2AX/total H2AX ratio (2.8 vs. 0.82 [95% CI: 0.66–0.98]), as well as much higher GH levels (7.6 ng/mL vs. 0.37 ng/mL [95% CI: 0.16–0.57]). After excluding this outlier, the γH2AX/total H2AX ratio correlated with IGF1 levels (r = 0.67, P = 0.01) and with pituitary gland size (r = 0.64, P = 0.02).
Figure 9. CJC-1295 treatment in vivo.
Four-month-old male C57BL/6 mice were injected s.c. with PBS (n = 15) or 10 μg/kg CJC-1295 (n = 16) 3 times per week for 8 weeks. (A) Pituitary weight (mg) in PBS-treated mice (n = 9) and CJC-1295–treated mice (n = 10). (B) Serum IGF1 levels (pg/mL) in PBS-treated mice (n = 15) and CJC-1295–treated mice (n = 16). Results are presented as the mean ± SEM. *P ≤ 0.05 and ***P ≤ 0.001 versus PBS, by 2-tailed, unpaired t test with Bonferroni’s correction. (C) Comet assay and tail appearance in representative mice. Images on the left are of PBS-treated mice (mice 3, 4, and 14); images on the right are of CJC-1295–treated mice (mice 21, 28, and 31). One image from each mouse is presented. Cells from each mouse pituitary gland were plated on 3 slides, and 61–89 images were taken for each mouse (original magnification, ×20), depending on the number of cells visualized. (D) Comet assay results depicting Olive tail moment for the 6 mice presented in C. The number of cells analyzed by a blinded observer is indicated. Results are presented as the mean ± SEM (mouse 3, 8.3 ± 0.9; mouse 4, 8.8 ± 0.7; mouse 14, 10.5 ± 0.8; mouse 21, 39.3 ± 1.5; mouse 28, 32.9 ± 1.5; mouse 31, 24.4 ± 1.2). ###P ≤ 0.001 for each CJC-1295–treated mouse versus each PBS-treated mouse, by 1-way ANOVA. (E) Quantitative analysis of γH2AX change normalized to total H2AX in PBS-treated mice (n = 5/7) versus CJC-1295–treated mice (n = 8/8), as presented in Supplemental Figure 11. Two mice were excluded from the PBS-treated group, as described in Results. Band intensities were calculated with ImageJ. Results are presented as the mean ± SEM. *P ≤ 0.05 versus PBS, by 2-tailed, unpaired t test with Bonferroni’s correction. The experiment was performed twice.
Discussion
Pituitary adenoma hormone hypersecretion is determined by lineage-specific gene expression as well as by the autonomously growing mass of differentiated adenoma cells (37). Our results support a distinct pituitary adenoma genotype-phenotype for cAMP-dependent, GH-secreting somatotroph adenomas versus nonsecreting adenomas.
We show by WES of 159 pituitary adenomas that SCNAs, often with amplification or deletion of an entire chromosome, are a genomic hallmark of benign somatotroph pituitary adenomas. In distinction, nonsecreting adenomas have been shown to harbor fewer SCNAs despite their larger size and more frequent extrasellar expansion. Furthermore, we did not detect many somatic mutations in secreting or nonsecreting pituitary tumors, supporting the findings of prior reports (22, 38, 39).
SCNAs reflect DNA damage and genomic instability and occur in normal tissue and cancerous tumors (23, 40). Our observation of genome instability with sparse SNV/InDel mutations was previously described in nonproliferating, terminally differentiated cells, which are highly metabolically active (41, 42). Similar to our observations in benign pituitary adenomas, these nonreplicating cells were shown to not accumulate mutations despite dampened DNA repair mechanisms that lead to DNA damage accumulation (41).
Our analysis of sgSCNAs identified 853 genes that could be used in KEGG pathway analysis and highlighted several pathways that are most affected, particularly cAMP and Fanconi anemia DNA damage repair pathways (43, 44).
Of all differentiated hormone-expressing pituitary cells, somatotrophs are the most sensitive to cAMP action (26). Somatotroph cAMP production is induced by GHRH, a GPCR coupled to Gαs and encoded by GNAS. Several lines of evidence support a role for cAMP activation in somatotroph proliferation, differentiation, and hormone secretion as well as in somatotroph adenoma pathogenesis (43–47). For example, human diseases associated with cAMP activation are commonly associated with somatotroph hyperplasia and GH hypersecretion (7–12, 48, 49), and ectopic GHRH secretion from neuroendocrine tumors leads to pituitary somatotroph hyperplasia in humans and the development of pituitary adenomas in some patients (50–52), whereas transgenic mice overexpressing human GHRH develop pituitary hyperplasia (53) and later somatomammotroph tumors that secrete both GH and PRL (46). Somatomammotroph hyperplasia also develops in mice overexpressing cholera toxin, which recapitulates the pathology observed with GNAS mutation (45). GHRH action is counteracted by hypothalamic somatostatin, which activates Gαi/o coupling to adenylate cyclase, mainly via somatostatin receptors SST2 and SST5, to inhibit cAMP production and reduce somatotroph cell secretory activity (36, 54). Furthermore, specific PDE enzymes hydrolyze and deactivate cAMP, thereby attenuating its accumulation and, in turn, probably affecting somatotroph adenoma function (11). We observed lower PDE4D gene expression in somatotroph adenomas compared with expression in gonadotroph and lactotroph adenomas, further fueling cAMP sensitivity in the somatotroph adenomas.
Concomitantly, sgSCNA KEGG analysis also showed that canonical DNA damage response genes of the Fanconi anemia pathway harbor significant sgSCNA deletions in somatotroph adenomas. Fanconi anemia proteins repair dsDNA breaks, preventing replication failure and genomic instability, and also act to maintain proper chromosome segregation (55, 56). Mutations in DNA damage repair genes were identified in pituitary adenomas using a next-generation sequencing panel of 300 cancer-related genes (57), and our KEGG analysis showed that BRCA1, BRCA2, and REV3L sgSCNA deletions were frequently associated with somatotroph and corticotroph adenomas and correlated with the pituitary adenoma SCNA score.
The results of mRNA expression of these genes were difficult to interpret, given the extremely large variation and small sample size. Moreover, our attempts to analyze a CRISPR/Cas9 BRCA2-knockout GH3 rat somatotroph tumor cell line were not successful because of early cell death and an inability to propagate cells for experiments (not shown).
DNA damage repair pathway sgSCNAs in somatotroph adenomas caught our attention, as we previously showed upregulated p53/p21Wif1/Cip1 in somatotroph adenomas in association with DNA damage, aneuploidy, and cellular senescence (14). p53 is activated with cellular stress such as DNA damage, resulting in the regulation of cell-cycle arrest, chromosome segregation, DNA damage repair, apoptosis, and senescence (58, 59). We now confirm that, unlike nonsecreting gonadotroph/null cell adenomas, which showed little SCNA, and lactotroph adenomas, which showed high SCNA, only somatotroph adenomas had high expression levels of p53 and p21Wif1/Cip1, which are known to downregulate Fanconi anemia pathway genes such as FANCD2 and BRCA2, further inhibiting DNA repair (59–61). These observations are bolstered by our demonstration that GH suppresses DNA damage repair in epithelial colon cells (62). Thus, our findings indicate that abundant intracellular somatotroph GH produced upon cAMP stimulation also increases expression of p53 and p21Wif1/Cip1, further linking cAMP to the SCNA and DNA damage observed in somatotroph adenomas.
Building on these results, we explored links between cAMP activation and the DNA damage response. In both ex vivo normal primary pituitary cultures and in vivo, we demonstrate that increased cAMP induced by forskolin, which universally stimulates cAMP, or by CJC-1295, which selectively stimulates somatotroph cAMP, led to somatotroph DNA damage. We also show that octreotide counteracts GHRH action, suppressing cAMP and leading to attenuated DNA damage as well as GH production.
The extent of CJC-1295–induced DNA damage we observed was dose dependent, consistent with the observation that GHRH-overexpressing transgenic mice developed pituitary somatotroph adenomas only at 1 year of age (46), suggesting that the intensity and chronicity of intracellular cAMP exposure likely determine the degree of cAMP-induced DNA damage. Further, genetic mutations associated with dysfunctional cAMP pathways also manifest as somatotroph adenomas at different ages. For example, familial isolated somatotroph adenomas associated with AIP mutations manifest at older ages, whereas somatotroph adenomas associated with GPR101, specifically mosaic microduplications of Xq26.3, manifest before 2 years of age (63). Although cAMP-associated mutations have not been identified in most sporadic somatotroph adenomas, several lines of evidence support the presence of cAMP pathway hyperactivation. For example, the low PDE4D levels in somatotroph adenomas compared with the levels in lactotroph and gonadotroph/null cell adenomas observed in our cohort may have led to higher levels of intracellular cAMP due to attenuated hydrolysis and deactivation of cAMP. Indeed, both GNAS mutation–positive and GNAS mutation–negative somatotroph adenomas were previously shown to express similarly enhanced CREB phosphorylation as compared with nonfunctioning pituitary adenomas (64), further supporting an association for these adenomas with cAMP pathway activity.
Insights into the molecular mechanisms underlying cAMP-induced DNA damage and SCNA in somatotroph adenomas can be derived from several observations. cAMP promotes somatotroph proliferation and terminal differentiation via GHRH and its receptor (25, 26, 44–47, 65), and replication stress in terminally differentiated cells (akin to somatotrophs) causes replication fork stalling and chromosomal instability without significant mutation accumulation (41). Moreover, well-differentiated cells exhibit dampened DNA repair mechanisms, further enhancing DNA damage accumulation (41). These observations support our conclusion that GHRH-induced replication stress in terminally differentiated somatotroph cells may result in genomic instability and DNA damage with low mutation rates.
Constitutive cAMP elevation, as occurs in GNAS-mutated somatotroph adenomas, may also protect somatotroph adenomas from a high degree of DNA damage, as none of these tumors harbored high-level SCNAs, consistent with cAMP attenuation of DNA damage in cells previously induced by UV light or toxins (66, 67). Thus, our observations support the notion that, although cAMP promotes DNA damage, it concomitantly protects against further extensive DNA damage. Our observed p53/p21Wif1/Cip1 induction is also consistent with this hypothesis, which may explain the invariably benign nature of these senescent adenomas. Indeed, GH carcinomas are extremely rare, and malignant transformation of GNAS mutation–positive somatotroph adenomas has not been described to our knowledge.
Dual and antagonistic effects of cAMP were shown to occur via intracellular compartmentalization regulated by PDE4D and PDE4B expression, which enables differential effects of EPAC and PKA (68). These 2 downstream effectors opposingly regulate the trafficking and activity of DNA-dependent protein kinase catalytic subunit (DNA-PKcs) in and out of the nucleus. As this sensor of DNA damage is required for DNA repair and maintenance of chromosomal integrity (68), our finding of lower PDE4D levels in human somatotroph adenomas may reflect unopposed EPAC/Rap2 removal of nuclear DNA-PKcs and prevention of chromosomal stability maintenance. Interestingly, cAMP was also shown to inhibit DNA damage repair proteins in lung cells via EPAC signaling (69).
Of note, our findings identify a possible mechanism to explain several known somatotroph adenoma phenotypes and treatment responses. Our observations that intracellular cAMP stimulation in somatotroph cells, either via GHRH activation or constitutive activation of Gαs due to an activating GNAS mutation, concomitantly increased GH production and DNA damage may explain distinctive somatotroph adenoma features. GNAS mutation–positive somatotroph adenomas are usually smaller (70, 71) and more frequently present as microadenomas (diameter ≤1 cm). Yet, compared with somatotroph adenomas that do not harbor GNAS mutations, GNAS mutation–positive adenomas secrete more GH (70, 71) and are less responsive to GHRH (70) but are highly responsive to octreotide (71–73). These same features are common to densely granulated somatotroph adenomas (74), which tend to be smaller in size, produce more GH per unit volume, and respond better to octreotide compared with sparsely granulated adenomas (75). We also observed higher expression of somatotroph adenoma p53 and p21Wif1/Cip1, which are known to induce cell-cycle arrest upon DNA damage (76). This finding may explain why GNAS mutation–positive and densely granulated somatotroph adenomas, both of which secrete higher GH levels, are phenotypically small, show more restricted tumor expansion, and have an enhanced response to cAMP-suppressing octreotide. Likewise, clinical resistance of nonsecreting adenomas to octreotide may be explained by their preferred proliferative properties over cAMP-dependent hormone induction. In fact, in nonsecreting adenomas, differentiated hormone production appears to be repressed, suggesting a neoplastic switch by more rapidly proliferating pituitary cell populations (77). Overall, these results also offer a rationale for further subclassification of somatotroph adenomas based on the degree of DNA damage. Whether this approach would supplement current phenotype-genotype classifications (73) requires a prospective study of clinical outcomes.
In summary, using WES, we show that SCNAs, but not SNV/InDel mutations, are dominant genomic alterations in hormone-secreting pituitary adenomas. Using sgSCNA genes in KEGG analysis, we observed frequent copy number deletions in cAMP and DNA damage repair genes in GH-secreting somatotroph adenomas. We show that p53 and p21Wif1/Cip1 are selectively highly expressed in somatotroph adenomas. Using ex vivo normal primary pituitary cultures and in vivo GHRH analog treatment in mice, we show a dual role for somatotroph cAMP action in promoting GH secretion while concomitantly causing DNA damage. These findings help explain the smaller size and less aggressive behavior of densely granulated somatotroph adenomas and of those harboring GNAS mutations, both of which secrete higher GH levels. Our observations support a distinct pituitary adenoma phenotype for cAMP-dependent, GH-secreting adenomas versus nonsecreting adenomas, which are less sensitive to cAMP effects, have lower levels of DNA damage, and continue to proliferate, generating larger tumors. Elucidation of driver mechanisms for cAMP-stimulated DNA damage in GH-secreting pituitary adenomas will offer new approaches for targeted therapy.
Methods
Sample collection from patients.
Patients diagnosed at our Pituitary Center were scheduled to undergo surgical resection of a pituitary adenoma by a single neurosurgeon and provided written informed consent before inclusion in the study. On the day of surgery, a blood sample (5 mL) was collected, buffy coat separated, and stored at –80°C. Immediately after resection, adenoma fragments were placed in RNAlater solution (QIAGEN) for 48 hours at 4°C, and then removed from the solution and stored at –80°C.
Gene sequencing and analysis.
Whole-exome and genome capture, library preparation, and sequencing of patient genomic DNA samples followed by sequence alignment and marking duplicate reads were conducted at the Beijing Genomics Institute (BGI) (Cambridge, Massachusetts, USA). We performed all downstream analyses, including: (a) detection and annotation of SNVs, InDels, and SCNAs; (b) calculation, comparison, and visualization of SCNAs and SNVs; and (c) sgSCNA isolation, pathway enrichment, and other statistical analyses.
Exome capture, library preparation, and WES.
Frozen blood and tissue samples were batch shipped to BGI for WES. DNA was extracted from adenoma tissue and buffy coats using the DNeasy Blood & Tissue Kit (QIAGEN), and total DNA concentration was measured by Qubit dsDNA BR Assay (Thermo Fisher Scientific). Qualified genomic DNA samples were randomly fragmented with peaks of 150 to 200 bp, and adapters were ligated to both ends of the resulting fragments. Adapter-ligated templates were purified by Agincourt AMPure SPRI beads (Beckman Coulter), and fragments of approximately 200 bp in length were excised. Extracted DNA was amplified by ligation-mediated PCR (LM-PCR), purified, and hybridized for enrichment to the SureSelect Biotinylated RNA Library (Applied Biosystems), and then bound to streptavidin beads. Nonhybridized fragments were washed out after 24 hours. LM-PCR products were subjected to the Agilent 2100 Bioanalyzer (Agilent Technologies) to estimate enrichment magnitude, and exome-enriched shotgun libraries were sequenced using the Illumina HiSeq 2000 platform (Illumina). Paired-end reads with an average size of 100 bp were generated with 100X average on-target coverage. Image analysis and base calling were performed with Illumina Genome Analyzer Pipeline, version 1.3 (Illumina), using default parameters.
WES data processing and analysis.
Reads were aligned to a human reference genome (hg19) using Burrows-Wheeler Alignment (BWA), version 0.5.9-r16 (78). Duplicate reads were marked using Picard, version 1.54 (http://broadinstitute.github.io/picard/), by our Biostatistics and Bioinformatics Core, which also analyzed raw data and alignment files provided by BGI.
Analysis workflows for variant discovery followed best practices (79, 80), as described on the Genome Analysis Toolkit (GATK) website (https://software.broadinstitute.org/gatk/best-practices).
SNV/InDels were identified using MuTect2 (81) in GATK, version 3.6 (82). HapMap 3.3 (83) was used to estimate cross-sample contamination. Both dbSNP, version 147 (84), and COSMIC, version 78 (85), were applied to filter variants. ANNOVAR (86) was applied to annotate identified somatic variants.
SCNAs were called using copy number analysis tools in GATK, version 4.0 Beta (82). To detect small regions of SCNAs, the length of exon interval bins was set to 200 bp. To eliminate copy ratio noise, SCNAs were identified after filtering out low-coverage exon interval bins with fewer than 100 read counts. Adenoma sample copy ratios were calculated using normalized read counts of coverage against a control panel created by collecting proportional coverage from respective whole blood genomic DNA samples in our cohort. A circular binary segmentation algorithm was applied to segment copy ratios to identify genomic regions with deletion/amplification of SCNAs by default parameters. ANNOVAR (86) was applied to annotate identified SCNAs.
SCNA analysis using WES data.
As large chromosomal arm-level SCNAs were observed on chromosomes 1 to 22, but not on sex chromosomes, we adjusted for possible overall shifts by calculating segmentation copy ratios by the GATK framework divided by the mean of the copy ratio of segmentation across intervals on chromosome X for each adenoma (the copy ratio mean for chromosome X is 0.95–1.02). The SCNA heatmap was plotted according to chromosomal gain or loss defined by |log2(adjusted copy ratio)| >0.2. The SCNA score for each adenoma represented the overall SCNA disruption and was calculated by summating the absolute gain or loss on chromosomes 1 to 22.
On the basis of the SCNA scores, adenoma samples were further separated into groups to indicate low (≤0.2), medium (>0.2 and <2.0), and high (≥2.0) degree of SCNA disruption; representative adenoma SCNAs for each score are shown in Supplemental Figure 3. Spearman’s rank-order correlation was applied to check correlations between the degree of SCNA and clinical adenoma phenotypic characteristics. The coverage plot of SCNAs was generated using Bedtools, version 2.21.0 (87), and visualized using Integrative Genomics Viewer, version 2.3.34 (88, 89).
sgSCNA isolation.
sgSCNAs were identified by filtering out genes that met the following criteria: (a) absolute SCNA deletion or amplification of log2-adjusted copy ratio of less than 0.2; (b) known to be readily altered in cancers per Lawrence et al. (90); (c) identified as chromosomal instability genes (CIN70) per Carter et al. (91); (d) occurred only once in our cohort; (e) consistently occurred as either deletion or amplification at a frequency of 90% or less.
Pathway enrichment analysis.
We used KEGG pathway enrichment analysis of sgSCNA genes for all adenomas and per adenoma subtype using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (92).
WGS data processing and analysis.
WGS libraries of 14 tumors selected from the WES cohort were prepared using TruSeq Nano DNA HT (Illumina), and a quantified adapter was added to each sample to allow for DNA pooling. After clustering a HiSeq X flow cell, libraries were sequenced on the HiSeq Xten platform (Illumina). Raw image files were processed by the Illumina pipeline for base calling with default parameters, and sequences of each individual sample were generated as 150 bp paired-end reads. The adapter sequence in the raw data was removed, and low-quality reads and bases were discarded according to the default parameters. Raw data and BWA alignment results were provided by BGI, and coverage visualization was performed by our Biostatistics and Bioinformatics Core.
Mouse primary pituitary cell cultures.
Four-month-old male C57BL/6 mice purchased from The Jackson Laboratory were sacrificed using isoflurane and CO2, the pituitary glands were harvested and placed in DMEM, and the cells were immediately dispersed using the Neural Tissue Dissociation Kit (Miltenyi Biotec). Primary pituitary cells were counted before plating using the TC10 Automated Cell Counter (Bio-Rad) and then cultured in NeuroCult Basal Medium with NeuroCult Differentiation Supplement (STEMCELL Technologies) in 6% CO2 in a 37°C humidified incubator.
A total of 710 mice were sacrificed to establish, validate, and reproduce the experiments. Each pituitary gland yielded approximately 1.5 million viable cells. A total of 2.5 million cells were plated per well in 12-well plates (for Western blot analysis and ELISA), 1.5 million cells were plated per well in 24-well plates (for ELISA, quantitative real-time PCR [qRT-PCR], and comet assay), or 0.3 million cells were plated per well in 48- and 96-well plates (for cAMP and WST1 assays). Only adherent cells were studied; floating cells were discarded.
Treatment durations were up to 16 hours after overnight incubation. Forskolin (MilliporeSigma) was resuspended in DMSO (MilliporeSigma). CJC-1295, a long-acting GHRH analog (t1/2 = 8 days in humans), was synthesized by ANASpec on the basis of the published molecular structure (93, 94). Octreotide (Phoenix Pharmaceuticals) and rolipram (MilliporeSigma) were suspended in sterile water. PBS was purchased from MilliporeSigma. Compounds were stocked at –20°C until use.
Mouse in vivo studies.
Four-month-old male C57BL/6 mice (The Jackson Laboratory) were injected s.c. with PBS (n = 15) or 10 μg/kg CJC-1295 (n = 16) 3 times per week (Monday, Wednesday, Friday) for 8 weeks, for a total of 24 injections. No side effects were observed. Mouse weight and length (tip of nose to base of tail) were measured on the first day before injection and weekly before each Monday injection thereafter. After sacrifice, heart blood was collected and serum separated, and heart and liver weights were measured in all mice. Mouse pituitary gland weight was measured in 9 mice treated with PBS and 10 mice treated with CJC-1295 that were randomly selected from the overall cohort. Western blotting of single pituitary glands was performed for 6 mice treated with PBS and 8 mice treated with CJC-1295 that were randomly selected from the overall cohort. One pituitary gland from the group of mice treated with PBS was excluded as an outlier on the basis of excessive elevation of γH2AX levels on Western blot analysis. Hormone measurements were performed for all mice, whereas the comet assay was performed for 6 randomly selected mice (3 treated with PBS and 3 with CJC-1295) as described below.
cAMP assay.
Primary pituitary cells were seeded in 48-well plates, with 6 wells per treatment group, 16 hours before treatment. Cells were treated in DMEM containing 0.3% BSA and 1 mM 3-isobutyl-1-methylxanthine (IBMX) (MilliporeSigma) for 30 minutes in a humidified incubator at 37°C and 6% CO2. Intracellular cAMP was assayed in duplicate using the LANCE cAMP Kit (PerkinElmer), modified for measurement of intracellular cAMP content (54). Results were extrapolated from a standard curve and read by a Victor 3 1420_015 spectrophotometer (PerkinElmer).
Western blot analysis.
Human adenoma tissue was stored in RNAlater (QIAGEN) (95), primary cultured cells were immediately processed for protein extraction, and harvested fresh pituitary glands were snap-frozen and then kept at –80°C until analysis. Samples were placed on ice suspended in RIPA buffer (Cell Signaling Technology) containing protease and phosphatase inhibitors (Thermo Fisher Scientific). Protein concentration was measured (Bio-Rad), and samples of equal concentration were run in Bolt 4% to 12% Bis-Tris Plus gels (Thermo Fisher Scientific) and transferred onto nitrocellulose membranes (Thermo Fisher Scientific). Protein bands were detected using Precision Plus Protein Western C (Bio-Rad), and blots were scanned using ChemiDoc XRS (Bio-Rad). The antibodies used are listed in Supplemental Table 3. Band intensities were measured with ImageJ. Ponceau was used for normalization of proteins from human pituitary adenomas (96, 97) and total H2AX for proteins derived from mouse primary pituitary cultures.
Comet assay.
For in vitro experiments, single-cell nuclear DNA damage was measured by a blinded observer using the OxiSelect Comet Assay Kit (STA350, Cell Biolabs). Single cells were encapsulated in a low-melting-point agarose suspension and lysis buffer for 60 minutes and then under alkaline (pH >13) conditions for an additional 30 minutes, followed by electrophoresis at 18 volt/cm (32, 33). ImageJ was used to measure Olive tail moment, which reflects the percentage of damaged DNA in the tail of total cell DNA, multiplied by tail length. Each experiment was repeated at least twice.
For in vivo experiments, fresh pituitary glands were collected immediately after sacrifice and washed with cold PBS, and then cells were dispersed, passed through a 40 μm cell strainer, and resuspended in 10 mL PBS. After cell dilution in PBS with 0.01% BSA, 1000 cells/μL were subjected to a blinded comet assay as described above.
ELISA.
Medium (in vitro) or serum (in vivo) hormone levels were measured by rat and mouse GH ELISA (MilliporeSigma, catalog EZRMGH-45K; range 0–50 ng/mL, 1:6000 to 1:10,000 dilution), mouse prolactin ELISA (Thermo Fisher Scientific, catalog EMPRL; range 0–20000 pg/mL, 1:10 to 1:100 dilution), mouse ACTH competitive enzyme immunoassay (EIA) (LifeSpan BioSciences, catalog LS-F5354; range 0–1000 pg/mL, 1:5 dilution), and mouse FSH BioAssay ELISA (US Biological Life Sciences, catalog 355754; range 0–150 mIU/mL, 1:1 dilution). In vitro supernatants were collected and centrifuged at 1500 rpm at 4°C for 5 minutes. Cell suspensions were separated from debris and stored at –80°C for analysis. Experiments were repeated at least 3 times and samples assayed in duplicate or triplicate. In vivo heart blood was collected after sacrifice, and blood was allowed to clot at room temperature for 2 hours. Next, the blood was centrifuged (2000 rpm at 4°C for 15 minutes), the serum was separated, and hormone levels were measured. IGF1 was measured by mouse IGF1 ELISA (Thermo Fisher Scientific, catalog EMIGF1, range 0–100 ng/mL, dilution 1:300). GH (dilution 1:200), PRL (dilution 1:10), ACTH (dilution 1:5), and FSH (dilution 1:5) levels were measured using the assays described above for primary pituitary cultures.
WST1.
WST1 is a soluble tetrazolium salt that can only be cleaved by viable cells and is therefore used to measure the relative number of viable cells in cultures. We used WST1 for normalization of supernatant hormone levels. Cells were plated in a 96-well plate, with 8 wells for each treatment group. Following 16 hours of the indicated treatment, WST1 (Takara Bio) was added to the wells, the cells were incubated at 37°C at 6% CO2 for an additional 3 hours, and absorbance was measured at a wavelength of 450 nm.
RNA extraction and qRT-PCR.
RNA was collected using the RNeasy Mini Kit (QIAGEN), and cDNA was synthesized from 1 μg purified RNA using iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad). Commercially available primers were purchased from Bio-Rad. RT-qPCR was performed in 10 μL reactions using SsoAdvanced Universal SYBR Green Supermix in a Bio-Rad CFX96 or 384 instrument (Bio-Rad).
Statistics.
R Project for Statistical Computing, version 3.5.1 (http://www.r-project.org/) was used for statistical analyses of WES and WGS data. Spearman’s rank-order correlation was applied to check correlations between SCNA degree and adenoma phenotypic characteristics. Pearson’s correlation coefficient was used for sgSCNA versus SCNA analysis. Fisher’s exact test was applied to test whether GNAS mutation was associated with SCNA degree in somatotroph adenomas. Comparisons of patient and adenoma phenotypes, protein expression levels in Western blots and ELISA, mRNA expression in qRT-PCR, and comet assays were analyzed using a 2-tailed, unpaired t test. A 2-tailed, unpaired t test with Bonferroni’s correction was used for multiple comparisons. A 1-way ANOVA was used for in vivo comet assay and cAMP sgSCNA distribution in human adenomas. Multiple comparisons were adjusted by Bonferroni’s correction. A 2-way ANOVA with Bonferroni’s analysis was used for cAMP level comparisons. P values of 0.05 or less were considered statistically significant.
Study approval.
Prospective collection of adenoma samples and patient information was approved by the IRB of Cedars-Sinai (Pro00034122) and conducted according to Declaration of Helsinki principles. Written informed consent was received from participants before inclusion in the study. Animal care and handling was approved by the IACUC of Cedars-Sinai (protocol 3392) and was conducted in accordance with the NIH’s Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Author contributions
ABS and SM developed the hypothesis and designed the study. ABS, AM, and AL collected the specimens and provided clinical follow-up data. ED and MY conducted the experiments, and ND performed bioinformatics and statistical analysis. ABS, ND, VC, and SM analyzed, discussed, and interpreted the data. ABS and SM coordinated and directed the project and wrote the manuscript. All authors approved the submitted manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grant DK113998 and the Doris Factor Molecular Endocrinology Laboratory at Cedars-Sinai. The funding sources had no role in the study design, data analysis, or decision to publish. The authors thank Shira Berman for assistance with manuscript editing; Ramtin Khalafi for assistance with biobanking; Yunguang Tong for assistance with data collection; Vivian Hwe for assistance with IRB approvals; and David Raygoza for assistance with adenoma and blood collection. We thank the faculty members of the Cedars-Sinai Pituitary Center, Vivien Bonert, Odelia Cooper, and Ning-Ai Liu, who participated in the patients’ care.
Version 1. 07/16/2020
In-Press Preview
Version 2. 09/28/2020
Electronic publication
Version 3. 11/02/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(11):5738–5755.https://doi.org/10.1172/JCI138540.
See the related Commentary at Oncogene-induced DNA damage: cyclic AMP steps into the ring.
Contributor Information
Anat Ben-Shlomo, Email: anat.ben-shlomo@cshs.org.
Nan Deng, Email: nandeng1213@gmail.com.
Evelyn Ding, Email: EvelynYan.Ding@cshs.org.
Masaaki Yamamoto, Email: mazeran1978@me.com.
Adam Mamelak, Email: adam.mamelak@cshs.org.
Vera Chesnokova, Email: chesnokovav@cshs.org.
Artak Labadzhyan, Email: Artak.Labadzhyan@cshs.org.
Shlomo Melmed, Email: melmed@csmc.edu.
References
- 1.Melmed S. Pituitary-Tumor Endocrinopathies. N Engl J Med. 2020;382(10):937–950. doi: 10.1056/NEJMra1810772. [DOI] [PubMed] [Google Scholar]
- 2.Molitch ME. Diagnosis and Treatment of Pituitary Adenomas: A Review. JAMA. 2017;317(5):516–524. doi: 10.1001/jama.2016.19699. [DOI] [PubMed] [Google Scholar]
- 3.Fernandez A, Karavitaki N, Wass JA. Prevalence of pituitary adenomas: a community-based, cross-sectional study in Banbury (Oxfordshire, UK) Clin Endocrinol (Oxf) 2010;72(3):377–382. doi: 10.1111/j.1365-2265.2009.03667.x. [DOI] [PubMed] [Google Scholar]
- 4.Day PF, Loto MG, Glerean M, Picasso MF, Lovazzano S, Giunta DH. Incidence and prevalence of clinically relevant pituitary adenomas: retrospective cohort study in a Health Management Organization in Buenos Aires, Argentina. Arch Endocrinol Metab. 2016;60(6):554–561. doi: 10.1590/2359-3997000000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daly AF, Rixhon M, Adam C, Dempegioti A, Tichomirowa MA, Beckers A. High prevalence of pituitary adenomas: a cross-sectional study in the province of Liege, Belgium. J Clin Endocrinol Metab. 2006;91(12):4769–4775. doi: 10.1210/jc.2006-1668. [DOI] [PubMed] [Google Scholar]
- 6.Kamilaris CDC, Faucz FR, Voutetakis A, Stratakis CA. Carney complex. Exp Clin Endocrinol Diabetes. 2019;127(2-03):156–164. doi: 10.1055/a-0753-4943. [DOI] [PubMed] [Google Scholar]
- 7.Vierimaa O, et al. Pituitary adenoma predisposition caused by germline mutations in the AIP gene. Science. 2006;312(5777):1228–1230. doi: 10.1126/science.1126100. [DOI] [PubMed] [Google Scholar]
- 8.Trivellin G, et al. Gigantism and acromegaly due to Xq26 microduplications and GPR101 mutation. N Engl J Med. 2014;371(25):2363–2374. doi: 10.1056/NEJMoa1408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinstein LS, Shenker A, Gejman PV, Merino MJ, Friedman E, Spiegel AM. Activating mutations of the stimulatory G protein in the McCune-Albright syndrome. N Engl J Med. 1991;325(24):1688–1695. doi: 10.1056/NEJM199112123252403. [DOI] [PubMed] [Google Scholar]
- 10.Schernthaner-Reiter MH, Trivellin G, Stratakis CA. Interaction of AIP with protein kinase A (cAMP-dependent protein kinase) Hum Mol Genet. 2018;27(15):2604–2613. doi: 10.1093/hmg/ddy166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bizzi MF, et al. Reduced protein expression of the phosphodiesterases PDE4A4 and PDE4A8 in AIP mutation positive somatotroph adenomas. Mol Cell Endocrinol. 2018;476:103–109. doi: 10.1016/j.mce.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Landis CA, Masters SB, Spada A, Pace AM, Bourne HR, Vallar L. GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature. 1989;340(6236):692–696. doi: 10.1038/340692a0. [DOI] [PubMed] [Google Scholar]
- 13.Lee EJ, et al. Absence of constitutively activating mutations in the GHRH receptor in GH-producing pituitary tumors. J Clin Endocrinol Metab. 2001;86(8):3989–3995. doi: 10.1210/jcem.86.8.7732. [DOI] [PubMed] [Google Scholar]
- 14.Chesnokova V, et al. p21(Cip1) restrains pituitary tumor growth. Proc Natl Acad Sci USA. 2008;105(45):17498–17503. doi: 10.1073/pnas.0804810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, et al. Pituitary tumor transforming gene (PTTG) expression in pituitary adenomas. J Clin Endocrinol Metab. 1999;84(2):761–767. doi: 10.1210/jcem.84.2.5432. [DOI] [PubMed] [Google Scholar]
- 16.Yu R, Lu W, Chen J, McCabe CJ, Melmed S. Overexpressed pituitary tumor-transforming gene causes aneuploidy in live human cells. Endocrinology. 2003;144(11):4991–4998. doi: 10.1210/en.2003-0305. [DOI] [PubMed] [Google Scholar]
- 17.Abbud RA, et al. Early multipotential pituitary focal hyperplasia in the alpha-subunit of glycoprotein hormone-driven pituitary tumor-transforming gene transgenic mice. Mol Endocrinol. 2005;19(5):1383–1391. doi: 10.1210/me.2004-0403. [DOI] [PubMed] [Google Scholar]
- 18.Boggild MD, et al. Molecular genetic studies of sporadic pituitary tumors. J Clin Endocrinol Metab. 1994;78(2):387–392. doi: 10.1210/jcem.78.2.8106627. [DOI] [PubMed] [Google Scholar]
- 19.Szymas J, Schluens K, Liebert W, Petersen I. Genomic instability in pituitary adenomas. Pituitary. 2002;5(4):211–219. doi: 10.1023/A:1025313214951. [DOI] [PubMed] [Google Scholar]
- 20.Simpson DJ, Bicknell EJ, Buch HN, Cutty SJ, Clayton RN, Farrell WE. Genome-wide amplification and allelotyping of sporadic pituitary adenomas identify novel regions of genetic loss. Genes Chromosomes Cancer. 2003;37(3):225–236. doi: 10.1002/gcc.10216. [DOI] [PubMed] [Google Scholar]
- 21.Song ZJ, et al. The genome-wide mutational landscape of pituitary adenomas. Cell Res. 2016;26(11):1255–1259. doi: 10.1038/cr.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bi WL, et al. Landscape of genomic alterations in pituitary adenomas. Clin Cancer Res. 2017;23(7):1841–1851. doi: 10.1158/1078-0432.CCR-16-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colnaghi R, Carpenter G, Volker M, O’Driscoll M. The consequences of structural genomic alterations in humans: genomic disorders, genomic instability and cancer. Semin Cell Dev Biol. 2011;22(8):875–885. doi: 10.1016/j.semcdb.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Lauer S, Gresham D. An evolving view of copy number variants. Curr Genet. 2019;65(6):1287–1295. doi: 10.1007/s00294-019-00980-0. [DOI] [PubMed] [Google Scholar]
- 25.Frohman LA, Kineman RD. Growth hormone-releasing hormone and pituitary development, hyperplasia and tumorigenesis. Trends Endocrinol Metab. 2002;13(7):299–303. doi: 10.1016/S1043-2760(02)00613-6. [DOI] [PubMed] [Google Scholar]
- 26.Vitali E, et al. Cyclic adenosine 3’-5’-monophosphate (cAMP) exerts proliferative and anti-proliferative effects in pituitary cells of different types by activating both cAMP-dependent protein kinase A (PKA) and exchange proteins directly activated by cAMP (Epac) Mol Cell Endocrinol. 2014;383(1-2):193–202. doi: 10.1016/j.mce.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Cooper O. Silent corticotroph adenomas. Pituitary. 2015;18(2):225–231. doi: 10.1007/s11102-014-0624-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fountas A, Lavrentaki A, Subramanian A, Toulis KA, Nirantharakumar K, Karavitaki N. Recurrence in silent corticotroph adenomas after primary treatment: a systematic review and meta-analysis. J Clin Endocrinol Metab. doi: 10.1210/jc.2018-01956. https://doi.org/10.1210/jc.2018-01956. [DOI] [PubMed] [Google Scholar]
- 29.Pozhitkov AE, et al. Tracing the dynamics of gene transcripts after organismal death. Open Biol. 2017;7(1):160267. doi: 10.1098/rsob.160267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumari M, Cover PO, Poyser RH, Buckingham JC. Stimulation of the hypothalamo-pituitary-adrenal axis in the rat by three selective type-4 phosphodiesterase inhibitors: in vitro and in vivo studies. Br J Pharmacol. 1997;121(3):459–468. doi: 10.1038/sj.bjp.0701158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kopp B, Khoury L, Audebert M. Validation of the γH2AX biomarker for genotoxicity assessment: a review. Arch Toxicol. 2019;93(8):2103–2114. doi: 10.1007/s00204-019-02511-9. [DOI] [PubMed] [Google Scholar]
- 32.Collins A, et al. The comet assay as a tool for human biomonitoring studies: the ComNet project. Mutat Res Rev Mutat Res. 2014;759:27–39. doi: 10.1016/j.mrrev.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Møller P. The comet assay: ready for 30 more years. Mutagenesis. 2018;33(1):1–7. doi: 10.1093/mutage/gex046. [DOI] [PubMed] [Google Scholar]
- 34.Zhou C, et al. DNA damage evaluated by gammaH2AX foci formation by a selective group of chemical/physical stressors. Mutat Res. 2006;604(1-2):8–18. doi: 10.1016/j.mrgentox.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamberts SW, Krenning EP, Reubi JC. The role of somatostatin and its analogs in the diagnosis and treatment of tumors. Endocr Rev. 1991;12(4):450–482. doi: 10.1210/edrv-12-4-450. [DOI] [PubMed] [Google Scholar]
- 36.Ben-Shlomo A, Melmed S. Pituitary somatostatin receptor signaling. Trends Endocrinol Metab. 2010;21(3):123–133. doi: 10.1016/j.tem.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melmed S. Mechanisms for pituitary tumorigenesis: the plastic pituitary. J Clin Invest. 2003;112(11):1603–1618. doi: 10.1172/JCI20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newey PJ, et al. Whole-exome sequencing studies of nonfunctioning pituitary adenomas. J Clin Endocrinol Metab. 2013;98(4):E796–E800. doi: 10.1210/jc.2012-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Välimäki N, et al. Whole-genome sequencing of growth hormone (GH)-secreting pituitary adenomas. J Clin Endocrinol Metab. 2015;100(10):3918–3927. doi: 10.1210/jc.2015-3129. [DOI] [PubMed] [Google Scholar]
- 40.Sharma AK, Eils R, König R. Copy number alterations in enzyme-coding and cancer-causing genes reprogram tumor metabolism. Cancer Res. 2016;76(14):4058–4067. doi: 10.1158/0008-5472.CAN-15-2350. [DOI] [PubMed] [Google Scholar]
- 41.Fortini P, Ferretti C, Dogliotti E. The response to DNA damage during differentiation: pathways and consequences. Mutat Res. 2013;743-744:160–168. doi: 10.1016/j.mrfmmm.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Simonatto M, Latella L, Puri PL. DNA damage and cellular differentiation: more questions than responses. J Cell Physiol. 2007;213(3):642–648. doi: 10.1002/jcp.21275. [DOI] [PubMed] [Google Scholar]
- 43.Seuntjens E, Denef C. Progenitor cells in the embryonic anterior pituitary abruptly and concurrently depress mitotic rate before progressing to terminal differentiation. Mol Cell Endocrinol. 1999;150(1-2):57–63. doi: 10.1016/S0303-7207(99)00028-3. [DOI] [PubMed] [Google Scholar]
- 44.Kim GL, et al. Generation of immortal cell lines from the adult pituitary: role of cAMP on differentiation of SOX2-expressing progenitor cells to mature gonadotropes. PLoS ONE. 2011;6(11):e27799. doi: 10.1371/journal.pone.0027799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burton FH, Hasel KW, Bloom FE, Sutcliffe JG. Pituitary hyperplasia and gigantism in mice caused by a cholera toxin transgene. Nature. 1991;350(6313):74–77. doi: 10.1038/350074a0. [DOI] [PubMed] [Google Scholar]
- 46.Kineman RD, Teixeira LT, Amargo GV, Coschigano KT, Kopchick JJ, Frohman LA. The effect of GHRH on somatotrope hyperplasia and tumor formation in the presence and absence of GH signaling. Endocrinology. 2001;142(9):3764–3773. doi: 10.1210/endo.142.9.8382. [DOI] [PubMed] [Google Scholar]
- 47.Hernández-Ramírez LC, Trivellin G, Stratakis CA. Cyclic 3’,5’-adenosine monophosphate (cAMP) signaling in the anterior pituitary gland in health and disease. Mol Cell Endocrinol. 2018;463:72–86. doi: 10.1016/j.mce.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 48.Stergiopoulos SG, Stratakis CA. Human tumors associated with Carney complex and germline PRKAR1A mutations: a protein kinase A disease! FEBS Lett. 2003;546(1):59–64. doi: 10.1016/S0014-5793(03)00452-6. [DOI] [PubMed] [Google Scholar]
- 49.Bates B, et al. Characterization of Gpr101 expression and G-protein coupling selectivity. Brain Res. 2006;1087(1):1–14. doi: 10.1016/j.brainres.2006.02.123. [DOI] [PubMed] [Google Scholar]
- 50.Thorner MO, et al. Somatotroph hyperplasia. Successful treatment of acromegaly by removal of a pancreatic islet tumor secreting a growth hormone-releasing factor. J Clin Invest. 1982;70(5):965–977. doi: 10.1172/JCI110708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barkan AL, Shenker Y, Grekin RJ, Vale WW, Lloyd RV, Beals TF. Acromegaly due to ectopic growth hormone (GH)-releasing hormone (GHRH) production: dynamic studies of GH and ectopic GHRH secretion. J Clin Endocrinol Metab. 1986;63(5):1057–1064. doi: 10.1210/jcem-63-5-1057. [DOI] [PubMed] [Google Scholar]
- 52.Borson-Chazot F, et al. Acromegaly induced by ectopic secretion of GHRH: a review 30 years after GHRH discovery. Ann Endocrinol (Paris) 2012;73(6):497–502. doi: 10.1016/j.ando.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 53.Mayo KE, Hammer RE, Swanson LW, Brinster RL, Rosenfeld MG, Evans RM. Dramatic pituitary hyperplasia in transgenic mice expressing a human growth hormone-releasing factor gene. Mol Endocrinol. 1988;2(7):606–612. doi: 10.1210/mend-2-7-606. [DOI] [PubMed] [Google Scholar]
- 54.Ben-Shlomo A, Wawrowsky K, Melmed S. Constitutive activity of somatostatin receptor subtypes. Meth Enzymol. 2010;484:149–164. doi: 10.1016/B978-0-12-381298-8.00008-3. [DOI] [PubMed] [Google Scholar]
- 55.Nalepa G, Clapp DW. Fanconi anaemia and cancer: an intricate relationship. Nat Rev Cancer. 2018;18(3):168–185. doi: 10.1038/nrc.2017.116. [DOI] [PubMed] [Google Scholar]
- 56.Niraj J, Färkkilä A, D’Andrea AD. The Fanconi anemia pathway in cancer. Annu Rev Cancer Biol. 2019;3:457–478. doi: 10.1146/annurev-cancerbio-030617-050422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bi WL, et al. Clinical identification of oncogenic drivers and copy-number alterations in pituitary tumors. Endocrinology. 2017;158(7):2284–2291. doi: 10.1210/en.2016-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharpless NE, DePinho RA. p53: good cop/bad cop. Cell. 2002;110(1):9–12. doi: 10.1016/S0092-8674(02)00818-8. [DOI] [PubMed] [Google Scholar]
- 59.Toufektchan E, Toledo F. The guardian of the genome revisited: p53 downregulates genes required for telomere maintenance, DNA repair, and centromere structure. Cancers (Basel) 2018;10(5):E135. doi: 10.3390/cancers10050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu K, Jiang SW, Couch FJ. p53 mediates repression of the BRCA2 promoter and down-regulation of BRCA2 mRNA and protein levels in response to DNA damage. J Biol Chem. 2003;278(18):15652–15660. doi: 10.1074/jbc.M211297200. [DOI] [PubMed] [Google Scholar]
- 61.Jaber S, Toufektchan E, Lejour V, Bardot B, Toledo F. p53 downregulates the Fanconi anaemia DNA repair pathway. Nat Commun. 2016;7:11091. doi: 10.1038/ncomms11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chesnokova V, et al. Excess growth hormone suppresses DNA damage repair in epithelial cells. JCI Insight. 2019;4(3):125762. doi: 10.1172/jci.insight.125762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tatsi C, Stratakis CA. The genetics of pituitary adenomas. J Clin Med. 2019;9(1):E30. doi: 10.3390/jcm9010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bertherat J, Chanson P, Montminy M. The cyclic adenosine 3′,5′-monophosphate-responsive factor CREB is constitutively activated in human somatotroph adenomas. Mol Endocrinol. 1995;9(7):777–783. doi: 10.1210/mend.9.7.7476961. [DOI] [PubMed] [Google Scholar]
- 65.Mayo KE, Miller T, DeAlmeida V, Godfrey P, Zheng J, Cunha SR. Regulation of the pituitary somatotroph cell by GHRH and its receptor. Recent Prog Horm Res. 2000;55:237–266; discussion 266. [PubMed] [Google Scholar]
- 66.Pregi N, Belluscio LM, Berardino BG, Castillo DS, Cánepa ET. Oxidative stress-induced CREB upregulation promotes DNA damage repair prior to neuronal cell death protection. Mol Cell Biochem. 2017;425(1-2):9–24. doi: 10.1007/s11010-016-2858-z. [DOI] [PubMed] [Google Scholar]
- 67.Wolf Horrell EM, Jarrett SG, Carter KM, D’Orazio JA. Divergence of cAMP signalling pathways mediating augmented nucleotide excision repair and pigment induction in melanocytes. Exp Dermatol. 2017;26(7):577–584. doi: 10.1111/exd.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huston E, et al. EPAC and PKA allow cAMP dual control over DNA-PK nuclear translocation. Proc Natl Acad Sci USA. 2008;105(35):12791–12796. doi: 10.1073/pnas.0805167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cho EA, Juhnn YS. The cAMP signaling system inhibits the repair of γ-ray-induced DNA damage by promoting Epac1-mediated proteasomal degradation of XRCC1 protein in human lung cancer cells. Biochem Biophys Res Commun. 2012;422(2):256–262. doi: 10.1016/j.bbrc.2012.04.139. [DOI] [PubMed] [Google Scholar]
- 70.Spada A, et al. Clinical, biochemical, and morphological correlates in patients bearing growth hormone-secreting pituitary tumors with or without constitutively active adenylyl cyclase. J Clin Endocrinol Metab. 1990;71(6):1421–1426. doi: 10.1210/jcem-71-6-1421. [DOI] [PubMed] [Google Scholar]
- 71.Larkin S, Reddy R, Karavitaki N, Cudlip S, Wass J, Ansorge O. Granulation pattern, but not GSP or GHR mutation, is associated with clinical characteristics in somatostatin-naive patients with somatotroph adenomas. Eur J Endocrinol. 2013;168(4):491–499. doi: 10.1530/EJE-12-0864. [DOI] [PubMed] [Google Scholar]
- 72.Buchfelder M, Fahlbusch R, Merz T, Symowski H, Adams EF. Clinical correlates in acromegalic patients with pituitary tumors expressing GSP oncogenes. Pituitary. 1999;1(3-4):181–185. doi: 10.1023/a:1009905131334. [DOI] [PubMed] [Google Scholar]
- 73.Cuevas-Ramos D, et al. A structural and functional acromegaly classification. J Clin Endocrinol Metab. 2015;100(1):122–131. doi: 10.1210/jc.2014-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Syro LV, Rotondo F, Serna CA, Ortiz LD, Kovacs K. Pathology of GH-producing pituitary adenomas and GH cell hyperplasia of the pituitary. Pituitary. 2017;20(1):84–92. doi: 10.1007/s11102-016-0748-8. [DOI] [PubMed] [Google Scholar]
- 75.Kiseljak-Vassiliades K, et al. Growth hormone tumor histological subtypes predict response to surgical and medical therapy. Endocrine. 2015;49(1):231–241. doi: 10.1007/s12020-014-0383-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams AB, Schumacher B. p53 in the DNA-damage-repair process. Cold Spring Harb Perspect Med. 2016;6(5):a026070. doi: 10.1101/cshperspect.a026070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stefana B, Ray DW, Melmed S. Leukemia inhibitory factor induces differentiation of pituitary corticotroph function: an immuno-neuroendocrine phenotypic switch. Proc Natl Acad Sci USA. 1996;93(22):12502–12506. doi: 10.1073/pnas.93.22.12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.International HapMap 3 Consortium. et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Forbes SA, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinformatics. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38(9):1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 92.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 93.Jetté L, et al. Human growth hormone-releasing factor (hGRF)1-29-albumin bioconjugates activate the GRF receptor on the anterior pituitary in rats: identification of CJC-1295 as a long-lasting GRF analog. Endocrinology. 2005;146(7):3052–3058. doi: 10.1210/en.2004-1286. [DOI] [PubMed] [Google Scholar]
- 94.Ionescu M, Frohman LA. Pulsatile secretion of growth hormone (GH) persists during continuous stimulation by CJC-1295, a long-acting GH-releasing hormone analog. J Clin Endocrinol Metab. 2006;91(12):4792–4797. doi: 10.1210/jc.2006-1702. [DOI] [PubMed] [Google Scholar]
- 95.Bennike TB, et al. Proteome stability analysis of snap frozen, RNAlater preserved, and formalin-fixed paraffin-embedded human colon mucosal biopsies. Data Brief. 2016;6:942–947. doi: 10.1016/j.dib.2016.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thacker JS, Yeung DH, Staines WR, Mielke JG. Total protein or high-abundance protein: Which offers the best loading control for Western blotting? Anal Biochem. 2016;496:76–78. doi: 10.1016/j.ab.2015.11.022. [DOI] [PubMed] [Google Scholar]
- 97.Rivero-Gutiérrez B, Anzola A, Martínez-Augustin O, de Medina FS. Stain-free detection as loading control alternative to Ponceau and housekeeping protein immunodetection in Western blotting. Anal Biochem. 2014;467:1–3. doi: 10.1016/j.ab.2014.08.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.