Abstract
Both pathogenic as well as nonpathogenic species of staphylococci have been reported in poultry, but these studies have not compared staphylococcal flora of both farmed and household broiler chickens. Staphylococci from farmed (n = 51) and household chicken intestines (n = 43) were isolated and tested for resistance to antimicrobials, presence of resistance genes, and inhibitory activity against other bacteria; correlation of resistance phenotype and genotype was also evaluated. At least 12 staphylococcal species were identified; Staphylococcus carnosus subspecies carnosus was the predominant species from both sources. Most farmed chicken staphylococci were resistant to tigecycline (38/51; 74.8%) while the highest level of resistance among the household chicken staphylococci was to clindamycin (31/43; 72.1%). The mecA gene was only detected in staphylococci from household chickens, whereas ermC and tetK or tetM were found in staphylococci from both groups of birds. Multidrug resistance (resistance ≥ 2 antimicrobial classes) was observed in 88% of resistant staphylococci ranging from 2 to 8 classes and up to 10 antimicrobials. Isolates produced inhibitory activity against 7 clinical bacterial strains primarily Enterococcus faecalis (25/88; 28.4%) and Escherichia coli (22/88; 25%). This study demonstrated that the staphylococcal population among farmed and household chickens varies by species and resistance to antimicrobials. These results may reflect the influence of the environment or habitat of each bird type on the intestinal microflora. As resistance in the staphylococci to antimicrobials used to treat human infections was detected, further study is warranted to determine strategies to prevent transfer of these resistant populations to humans via contamination of the poultry meat.
Key words: staphylococcus, broiler chicken, antibiotic resistance, normal flora
Introduction
The poultry intestinal microflora is diverse, consisting of large numbers of microbial types, predominantly anaerobic bacteria, but the exact types of microorganisms present in poultry intestines largely depend on their diet and the environment in which they live (Salanitro et al., 1978). These normal flora bacteria may help in digestion and may also serve to protect animals from pathogenic bacteria. However, the presence of the bacteria in the poultry intestines is a risk for contamination of the resulting product if the intestinal contents become disseminated (Pan and Yu, 2014). This risk may be elevated if the bacteria are also resistant to antimicrobials.
Previous studies have shown that staphylococci are a common inhabitant of the chicken intestinal tract (Salanitro et al., 1974; Devriese et al., 1975; Salanitro et al., 1978). However, their presence in the intestines may not be without consequence as staphylococci can have dual roles as commensals and pathogens (Rosenstein and Gotz, 2013). Although they can be found as natural inhabitants of the skin of humans and animals, they (i.e., Staphylococcus aureus) may also cause infections in those same tissues as well as other diseases. In poultry, studies have also demonstrated an association of both coagulase positive staphylococci as well as coagulase negative staphylococci with poultry diseases (Al-Rubaye et al., 2015; Nazia et al., 2015). The coagulase negative staphylococci, Staphylococcus simulans, has been associated with endocarditis in chickens (Stepien-Pysniak et al., 2017) while a study conducted by Cheville et al. (Cheville et al., 1988) reported fibrinopurulent blepharitis and conjunctivitis caused by Staphylococcus hyicus (coagulase variable) in poultry. Both S. simulans and S. hyicus have also been reported to cause diseases in humans (Casanova et al., 2011; Tous Romero et al., 2016). Furthermore, antimicrobial-resistant staphylococci such as methicillin-resistant S. aureus (MRSA), once considered a laboratory and health-care–associated pathogen, have now been found commonly isolated from poultry and livestock as well as food sources (Cuny et al., 2015). Both livestock-associated and human lineages of MRSA have been isolated from poultry (Nemati et al., 2008).
In our previous studies, staphylococci were isolated from eggs of farmed and household chickens and examined for species prevalence and antimicrobial resistance (Syed et al., 2018; Syed et al., 2019). Those eggs may have become contaminated from the environment or the feces of the birds. Presence of staphylococci in poultry has been reported previously (Devriese et al., 1975; Kawano et al., 1996), but no comparative study covering both farmed and household chickens has been carried out. In addition, both poultry types may cause zoonotic staphylococcal infections in humans through poultry meat or direct contact with animals and serve as a possible reservoir of antimicrobial-resistant strains that may also be spread to humans, other animals, or the environment (Kim et al., 2018). The present study aimed to compare species diversity, phenotypic and genotypic antimicrobial resistance, and inhibitory activity of staphylococci from intestines of farmed and household chickens.
Materials and methods
Sample Collection
One hundred poultry intestines (50 each from farmed chickens and household chickens) were collected from September to December 2017 from poultry meat shops in Haripur and the Abbottabad districts of Khyber Pakhtunkhwa province of Pakistan where they were slaughtered. Farmed chickens were reared on poultry farms, and the household chickens were kept by the local villagers. Approximately 5 cm of intestines were removed from each slaughtered bird; both ends of the small intestines were tied with a string before removal, and the surface was disinfected by dipping in 70% ethanol for 30 s. The intestinal samples were then sealed in sterile bags and transported to the microbiology laboratory of the University of Haripur in cold containers.
Processing of Intestinal Segments
Each intestinal segment was placed in a sterile Petri plate in a safety cabinet and opened using sterile forceps. Intestinal contents were collected using a sterile spatula or sterile cotton swabs in Amies Transport Medium (Oxoid, Basingstoke, UK). Swab samples were inoculated on selective medium (i.e., mannitol salt agar) and incubated at 37°C for 24 h. Colonies appearing after 24 h were restreaked to obtain a pure culture and then subjected to preliminary identification using microscopy, catalase, coagulase, and DNAse testing. At least one colony was selected from each sample depending on morphology on the mannitol salt agar plate. Bacterial isolates were identified to the genus and species level using the Vitek 2 system (bioMérieux, Durham, NC) and the Vitek 2 gram-positive identification cards according to the manufacturer's directions.
Antimicrobial Susceptibility Testing
Minimum inhibitory concentrations (MICs, μg/mL) for the isolated staphylococci were determined by broth microdilution using the Sensititre semi-automated antimicrobial susceptibility system (Trek Diagnostic Systems, Inc., Cleveland, OH) and the Sensititre Gram-Positive Plate GPN3F according to the manufacturer's directions. Antimicrobials and breakpoints were cefoxitin (≥8 μg/mL), clindamycin (≥4 μg/mL), daptomycin (>1 μg/mL), erythromycin (≥8 μg/mL), gentamicin (≥16 μg/mL), levofloxacin (≥4 μg/mL), linezolid (≥8 μg/mL), moxifloxacin (≥2 g/mL), mupirocin (256 μg/mL), nitrofurantoin (≥128 μg/mL), oxacillin (≥4 μg/mL), penicillin G (≥0.25 μg/mL), Synercid (quinupristin/dalfopristin) (≥4 μg/mL), tetracycline (≥16 μg/mL), tigecycline (>0.25 μg/mL), trimethoprim/sulfamethoxazole (≥4/76 μg/mL), and vancomycin (≥16 μg/mL). MIC values were manually recorded by using the Sensitouch system (Trek Diagnostic Systems, Inc., Cleveland, OH). Clinical and Laboratory Standards Institute standards were used to determine resistance (CLSI. 2018). Only susceptible breakpoints for daptomycin (≤1 μg/mL) and tigecycline (≤0.25 μg/mL) have been established by Clinical and Laboratory Standards Institute; resistance for these 2 drugs were defined as MICs greater than those values. S. aureus ATCC 29213 was used as a quality control strain.
Antimicrobial Resistance Genes
Staphylococcal isolates were tested for the presence of resistance genes to macrolides [erm(A), erm(C)], aminoglycosides (aacA-aphD), oxacillin [mecA, mecC/C1], tetracycline [(tet(K), tet(M)], and streptogramins [vat(A), vat(B), vat(C)] using multiplex PCR as previously described (Strommenger et al., 2003; Harrison et al., 2014). Positive controls were as follows: aacA-aphD-Enterococcus faecalis ATCC 49532, erm(A)-S. aureus RN1389, erm(C)-S. aureus RN4220, mecA-S. aureus ATCC 33591, mecC/C1-S. aureus NCTC 13552 (generously supplied by Gavin Paterson), tet(K)-S. aureus RB 36-1 (Jackson et al., 2013), tet(M)-En. faecalis OG1-SSp, vat(A)-S. aureus CIP 107907, vat(B)-S. aureus CIP 108540, and vat(C)-S. aureus CIP 107908.
Bacteriocin Activity
Staphylococcal isolates (n = 88) were screened for inhibitory activity against clinical bacteria including Escherichia coli, En. faecalis, Streptococcus pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, Salmonella enterica serotype Typhi, MRSA, and Klebsiella pneumoniae using the cross-streaking method as previously described (De Vliegher et al., 2004). Briefly, staphylococcal isolates were plated onto sheep blood agar (SBA; Becton Dickinson, Sparks, MD) and incubated overnight at 37°C. Bacterial colonies were diluted in phosphate buffered saline (PBS) until a density of McFarland 0.5 standard was obtained. The suspension was then used to inoculate a 5-mm-wide center streak on an SBA plate, which was then incubated at 37°C for 24 h. After incubation, the inoculated agar was loosened and flipped upside down onto the lid of the plate. Test bacteria were then applied by diluting an overnight culture grown on SBA to a 10−3 dilution in PBS of McFarland 0.5 standard and streaking 100 μL of the suspension to the surface of the inverted agar. After incubation at 37°C for 24 h, plates were examined for bacterial growth and inhibition of growth of the test bacteria. PBS was used as the negative control for the center streak; each staphylococcal isolate was also self-cross-streaked to exclude nutrient availability for inhibition of growth.
Statistical Analysis
The results of resistance phenotypes and genotypes were converted into binary values for correlation analysis, where antimicrobial resistance and the presence of resistance gene scored “1” and the sensitivity to antimicrobial and absence of resistance gene were represented as “0”. The correlation of binary values (0/1) of resistance phenotypes and genotypes was calculated using the “cor” function from the software “R” (R version 3.6.1; https://www.r-project.org/); significance was determined using the function “cor.test”. The significant correlations were visualized using the “corrplot” function from the “corrplot” package in the software “R”.
Results
Bacterial Isolation and Identification
Staphylococci were isolated from the intestines of 70% (35/50) farmed and 64% (32/50) of household chickens. Of those, a total of 51 staphylococcal isolates were recovered from farmed chickens and 43 from household chicken intestines (Table 1). Twelve staphylococcal species and presumptive species were identified using the Vitek 2 system from both chicken types. The most prevalent Staphylococcus species was Staphylococcus carnosus subspecies carnosus for both farmed (16/51; 31.4%) and household (15/43; 34.9%) chickens followed by Staphylococcus xylosus (11/51; 21.6%) and Staphylococcus cohnii subspecies urealyticus (8/51; 15.7%) for farmed chickens and S. cohnii subspecies urealyticus (9/43; 20.9%) and Staphylococcus lentus (6/43; 14%) for household chickens.
Table 1.
Staphylococcal species isolated from farmed and household chicken.
| Species | Farmed (n = 51) | Household (n = 43) |
|---|---|---|
| Staphylococcus caprae/Staphylococcus cohnii ssp cohnii | 2 (3.9) | 0 (0) |
| Staphylococcus carnosus ssp carnosus | 16 (31.4) | 15 (34.9) |
| Staphylococcus chromogenes | 0 (0) | 2 (4.7) |
| Staphylococcus cohnii ssp urealyticus | 8 (15.7) | 9 (20.9) |
| Staphylococcus gallinarum | 4 (7.8) | 3 (7.0) |
| Staphylococcus kloosii | 3 (5.9) | 1 (2.3) |
| Staphylococcus lentus | 0 (0) | 6 (14) |
| Staphylococcus saprophyticus | 0 (0) | 1 (2.3) |
| Staphylococcus simulans | 2 (3.9) | 1 (2.3) |
| Staphylococcus simulans/Staphylococcus saprophyticus | 2 (3.9) | 0 (0) |
| Staphylococcus species | 3 (5.9) | 0 (0) |
| Staphylococcus warneri | 0 (0) | 1 (2.3) |
| Staphylococcus xylosus | 11 (21.6) | 4 (9.3) |
Antimicrobial Phenotype and Genotype of Staphylococcal Isolates
Isolates were tested for susceptibility against a panel of 17 antimicrobials; farmed chicken staphylococci were resistant to 12 antimicrobials while staphylococci from household chicken intestines were resistant to 10 of the 17 tested ones (Table 2). Resistance was observed for a number of antimicrobials used in human clinical medicine. More than 50% of the isolates were resistant to 4 antimicrobials including erythromycin (66.7 and 69.8%), clindamycin (68.6 and 72.1%), tetracycline (66.7 and 65.1%), and tigecycline (74.5 and 58.1%) for farmed and household chickens, respectively.
Table 2.
Antimicrobial resistance of staphylococcal species isolated from farmed and household chicken.
| Antimicrobial1 | No. resistant (%) |
No. (%) resistant isolates with targeted gene |
||||
|---|---|---|---|---|---|---|
| Breakpoint (μg/mL) | Farmed (n = 51) | Household (n = 43) | Targeted gene2 | Farmed | Household | |
| Oxacillin | ≥ 0.5 | 17 (33.3) | 19 (44.2) | mec A | 0 (0) | 3 (15.8) |
| Cefoxitin | ≥ 8 | 0 (0) | 3 (7) | mecA | 0 (0) | 0 (0) |
| Penicillin | ≥ 0.25 | 8 (15.7) | 19 (44.2) | mecA | 0 (0) | 3 (15.8) |
| Erythromycin | ≥ 8 | 34 (66.7) | 30 (69.8) | erm C | 23 (67.6) | 25 (83.3) |
| Clindamycin | ≥ 4 | 35 (68.6) | 31 (72.1) | erm C | 23 (65.7) | 25 (80.6) |
| Synercid (quinupristin/dalfopristin) | ≥ 4 | 15 (29.4) | 16 (37.2) | vat A, B, C | 0 (0) | 0 (0) |
| Daptomycin | ≥ 1 | 1 (2) | 0 (0) | n/t | – | – |
| Levofloxacin | ≥ 4 | 8 (15.7) | 14 (32.6) | n/t | – | – |
| Moxifloxacin | ≥ 2 | 4 (7.8) | 11 (25.6) | n/t | – | – |
| Mupirocin | 256 | 1 (2) | 0 (0) | n/t | – | – |
| Tetracycline | ≥ 16 | 34 (66.7) | 28 (65.1) | tet K/tet M | 20 (58.8) | 9 (32.1) |
| Tigecycline | ≥ 0.25 | 38 (74.5) | 25 (58.1) | n/t | – | – |
| Trimethoprim/sulfamethoxazole | ≥ 4/76 | 1 (2) | 0 (0) | n/t | – | – |
Abbreviation: n/t, not tested.
No isolates were resistant to gentamicin, linezolid, nitrofurantoin, or vancomycin.
Seven farmed isolates and 7 household isolates from chickens were positive for aac-aph, but susceptible to gentamicin.
Resistance to the macrolide antibiotics, erythromycin, and clindamycin was due in part to the presence of ermC (Table 2). Approximately 67.6 and 83.3% of erythromycin-resistant staphylococci and 65.7 and 80.6% of clindamycin-resistant staphylococci from farmed and household chickens, respectively, harbored ermC. Although isolates were also tested for the presence of ermA, none of the isolates were positive for that gene. Likewise, although 29.4% (15/51) and 37.2% (16/43) of isolates from farmed and household chickens were resistant to the streptogramin antibiotic, Synercid (quinupristin/dalfopristin), none of the resistant isolates harbored vatA, vatB, or vatC. Of the 2 tetracycline resistance genes tested, tetK and tetM, 58.8% of farmed chicken staphylococci and 32.1% of household chicken staphylococci contained one or both of those genes. Surprisingly, although resistance to the β-lactam antimicrobials (oxacillin, cefoxitin, and penicillin) ranged from 0 to 33.3% in staphylococci from farmed chickens and 7 to 44.2% in staphylococci from household birds, none of the β-lactam–resistant isolates from farmed chicken intestines and only 15.8% from the household birds (3/19 isolates resistant to both oxacillin and penicillin) contained mecA (Table 2). None of the isolates were resistant to linezolid, nitrofurantoin, or vancomycin; however, although all isolates were also susceptible to the aminoglycoside, gentamicin, 7 isolates, each from farmed and household chickens, were positive for the aminoglycoside resistance gene, aacA-aphD.
Multidrug resistance (MDR; resistance ≥ 2 antimicrobial classes) was observed for 88% of resistant staphylococci from both farmed and household chickens (Table 3). Resistance ranging from 2 to 8 classes and up to 10 antimicrobials was noted in one isolate of S. cohnii ssp urealyticus from farmed chickens. The most common MDR pattern was CliEryTetTig which included 27 isolates from 6 staphylococcal species, predominantly MDR S. carnosus found equally in farmed and household chickens (n = 11 each). This represents one of only 8 staphylococcal MDR patterns shared between the 2 sources; the vast majority of MDR patterns were observed for one or the other source, but not both. For example, MDR pattern OxaPenTetTig was found in one Staphylococcus kloosii from farmed chickens and one Staphylococcus saprophyticus from household chickens (Table 3). In contrast, MDR pattern CliEryLevMoxOxaPenSynTetTig, consisting of 9 resistances in 7 antimicrobial classes, was found in 4 staphylococcal species (6 isolates total) from household chickens only. In addition to being predominant species isolated from the intestinal samples from both sources, all S. carnosus (n = 31) and S. cohnii ssp urealyticus (n = 17) were MDR.
Table 3.
Multidrug resistance patterns among staphylococci from farmed and household chickens.
| Pattern | No. resistances | No. resistance by class | Species | No. isolates |
|
|---|---|---|---|---|---|
| Farmed (n = 45) | Household (n = 38) | ||||
| CliDapEryLevMoxOxaPenSynTetTig | 10 | 8 | Staphylococcus cohnii ssp urealyticus (1) | 1 | 0 |
| CliEryLevMoxOxaPenSynTetTig | 9 | 7 | Staphylococcus xylosus (1) | 0 | 1 |
| Staphylococcus lentus (1) | 0 | 1 | |||
| Staphylococcus cohnii ssp urealyticus (2) | 0 | 2 | |||
| Staphylococcus lentus (2) | 0 | 2 | |||
| CliEryFoxLevMoxOxaPenSyn | 8 | 5 | Staphylococcus cohnii ssp urealyticus (1) | 0 | 1 |
| CliEryFoxLevOxaPenSynTet | 8 | 6 | Staphylococcus cohnii ssp urealyticus (1) | 0 | 1 |
| CliEryLevMoxPenSynTig | 7 | 6 | Staphylococcus cohnii ssp urealyticus (1) | 1 | 0 |
| CliEryLevMoxOxaPenTet | 7 | 5 | Staphylococcus chromogenes (1) | 0 | 1 |
| CliEryLevMoxPenTetTig | 7 | 6 | Staphylococcus chromogenes (1) | 0 | 1 |
| CliEryLevMoxSynTetTig | 7 | 6 | Staphylococcus cohnii ssp urealyticus (1) | 1 | 0 |
| CliEryOxaPenSynTetTig | 7 | 6 | Staphylococcus cohnii ssp urealyticus (1) | 0 | 1 |
| CliEryLevMoxSynTet | 6 | 5 | Staphylococcus cohnii ssp urealyticus (2) | 1 | 1 |
| CliEryLevOxaPenSyn | 6 | 5 | Staphylococcus cohnii ssp urealyticus (1) | 0 | 1 |
| CliEryOxaSynTetTig | 6 | 6 | Staphylococcus cohnii ssp urealyticus (1) | 1 | 0 |
| LevMoxOxaPenTetTig | 6 | 4 | Staphylococcus cohnii ssp urealyticus (1) | 0 | 1 |
| CliEryLevTetTig | 5 | 5 | Staphylococcus carnosus ssp carnosus (4) | 4 | 0 |
| CliEryLevSynTet | 5 | 5 | Staphylococcus kloosii (1) | 0 | 1 |
| CliEryPenSynTet | 5 | 5 | Staphylococcus cohnii ssp urealyticus (1) | 0 | 1 |
| CliEryOxaSynTet | 5 | 5 | Staphylococcus cohnii ssp urealyticus (1) | 1 | 0 |
| CliEryOxaSynTig | 5 | 5 | Staphylococcus caprae/Staphylococcus cohnii ssp cohnii (1) | 1 | 0 |
| CliErySynTetTig | 5 | 5 | Staphylococcus cohnii ssp urealyticus (1) | 1 | 0 |
| Staphylococcus kloosii (1) | 1 | 0 | |||
| CliEryTetTigTri | 5 | 5 | Staphylococcus carnosus ssp carnosus (1) | 1 | 0 |
| OxaPenSynTetTig | 5 | 4 | Staphylococcus xylosus (1) | 1 | 0 |
| CliEryMupTet | 4 | 4 | Staphylococcus simulans (1) | 1 | 0 |
| CliEryOxaSyn | 4 | 4 | Staphylococcus simulans/Staphylococcus saprophyticus (1) | 1 | 0 |
| CliEryTetTig | 4 | 4 | Staphylococcus caprae/Staphylococcus cohnii ssp cohnii (1) | 1 | 0 |
| Staphylococcus carnosus ssp carnosus (22) | 11 | 11 | |||
| Staphylococcus kloosii (1) | 1 | 0 | |||
| Staphylococcus simulans (1) | 1 | 0 | |||
| Staphylococcus simulans/Staphylococcus saprophyticus (1) | 1 | 0 | |||
| Staphylococcus spp (1) | 1 | 0 | |||
| OxaPenSynTig | 4 | 3 | Staphylococcus gallinarum (3) | 2 | 1 |
| OxaPenTetTig | 4 | 3 | Staphylococcus kloosii (1) | 1 | 0 |
| Staphylococcus saprophyticus (1) | 0 | 1 | |||
| CliEryTet | 3 | 3 | Staphylococcus xylosus (1) | 1 | 0 |
| OxaPenSyn | 3 | 2 | Staphylococcus gallinarum (4) | 2 | 2 |
| OxaTetTig | 3 | 3 | Staphylococcus warneri (1) | 0 | 1 |
| CliEry | 2 | 2 | Staphylococcus cohnii ssp urealyticus (1) | 1 | 0 |
| Staphylococcus carnosus spp carnosus (4) | 0 | 4 | |||
| OxaTig | 2 | 2 | Staphylococcus xylosus (4) | 3 | 1 |
| TetTig | 2 | 2 | Staphylococcus spp (1) | 1 | 0 |
| Staphylococcus xylosus (1) | 1 | 0 | |||
| Staphylococcus simulans (1) | 0 | 1 | |||
Abbreviations: Cli, clindamycin; Dap, daptomycin; Ery, erythromycin; Fox, cefoxitin; Lev, levofloxacin; Mox, moxifloxacin; Mup, mupirocin; Oxa, oxacillin; Pen, penicillin; Syn, Synercid (quinupristin/dalfopristin); Tet, tetracycline; Tig, tigecycline; Tri, trimethoprim/sulfamethoxazole.
Bacteriocin Activity
Although none of the staphylococcal species tested for potential bacteriocin activity in this study inhibited growth of the Salmonella Typhi isolate, almost half of the staphylococci tested (40/88; 45.5%) produced inhibitory activity to at least one of the remaining 7 clinical indicator strains (Table 4). Bacteriocin activity for 4 of the isolates was inconclusive and not included in the results. Most isolates that produced inhibitory activity did so against the En. faecalis strain (25/88 isolates; 28.4%), followed by inhibitory activity against the Es. coli strain (22/88; 25%). Fewer isolates were active against K. pneumoniae (9/88; 10.2%) as well as P. aeruginosa and A. baumannii (8/88; 9.1% for each). Slightly higher numbers inhibited growth of the MRSA isolate (12/88; 13.6%), whereas only 4 isolates (4.5%) demonstrated inhibitory activity against S. pneumoniae.
Table 4.
Detection of bacteriocin production by staphylococci from chickens.
| Clinical indicator strain1 | Bacteriocin-producing Staphylococcus spp (n = 88) (%) |
|---|---|
| Escherichia coli | 22 (25) |
| Enterococcus faecalis | 25 (28.4) |
| Streptococcus pneumoniae | 4(4.5) |
| Pseudomonas aeruginosa | 8 (9.1) |
| Acinetobacter baumannii | 8 (9.1) |
| Methicillin-resistant Staphylococcus aureus | 12 (13.6) |
| Klebsiella pneumoniae | 9 (10.2) |
| Overall bacteriocin production against any single indicator strain | 40 (45.5) |
No bacteriocin production was observed against Salmonella serotype Typhi.
Resistance Phenotype and Genotype Correlation
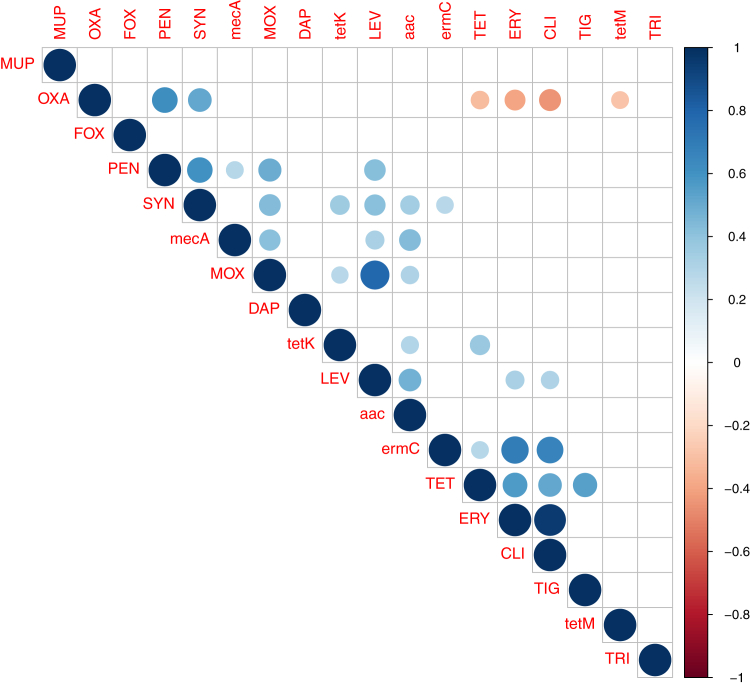
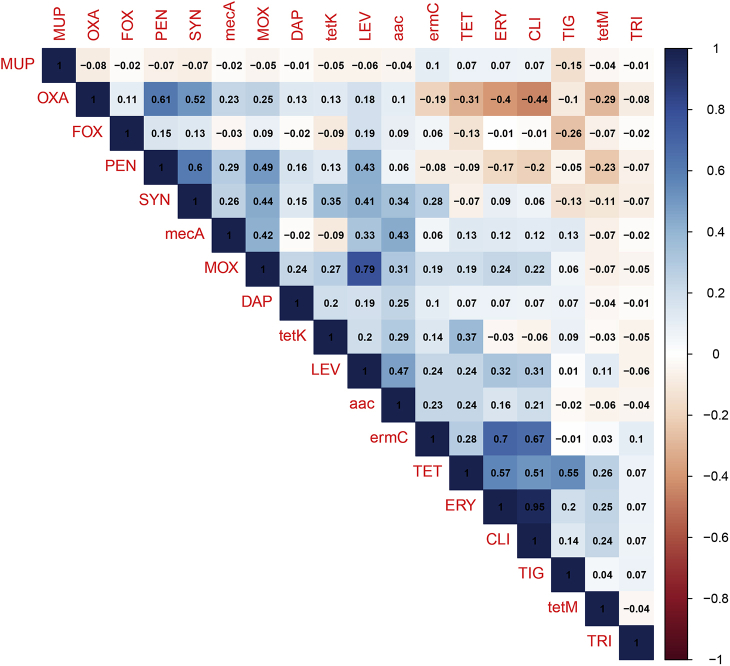
Correlation analyses of the resistance phenotypes and genotypes for the isolated staphylococcal species from both farmed and household chickens were determined (Figure 1 and Supplementary Figure 1). The phenotypic resistance analyses showed significant positive correlation between antimicrobials belonging to the same class as determined for the β-lactams (oxacillin and penicillin, r = 0.61) and fluoroquinolones (levofloxacin and moxifloxacin, r = 0.79). A significant positive correlation was also found between resistances to antimicrobials belonging to different classes. For instance, resistance to Synercid was significantly associated with penicillin (r = 0.6), oxacillin (r = 0.52), levofloxacin (r = 0.41), and moxifloxacin (r = 0.44). Tetracycline resistance was positively and significantly correlated to erythromycin (r = 0.57), clindamycin (r = 0.51), and tigecycline (r = 0.58). Furthermore, a significant positive correlation of levofloxacin resistance with penicillin (r = 0.43), erythromycin (r = 0.32), and clindamycin (r = 0.31) was observed. The correlation between erythromycin and clindamycin scored the highest value (r = 0.95). The only significant negative correlation was found between oxacillin resistance and resistance to tetracycline (r = −0.31), erythromycin (r = −0.4), and clindamycin (r = −0.44). Concerning the phenotypic/genotypic relationship, resistance genes were significantly correlated with their corresponding antimicrobials as determined between ermC and erythromycin (r = 0.7), tetK and tetracycline (r = 0.37), and mecA and penicillin (r = 0.29). Significant positive correlations were also found between resistance genes and antimicrobials other than the corresponding ones (e.g., mecA with fluoroquinolones; tetK, ermC, and aac with Synercid).
Figure 1.
Correlation matrix of resistance phenotypes and genotypes of Staphylococcus spp. isolated from farmed and household chickens using the “cor” and “cor.test” functions from the software “R”. Blue and red circles indicate positive and negative correlations, respectively. Blank boxes denote nonsignificant correlations.
Discussion
Intestines of both healthy and diseased chickens serve as a reservoir for both pathogenic and nonpathogenic bacteria (Clavijo and Florez, 2018). However, little information is available on the staphylococcal normal flora of chickens in general and of household chickens in particular. When compared to the staphylococcal species from eggs from both farmed and household chickens (Syed et al., 2018; Syed et al., 2019), staphylococcal species from the intestines of both types of birds differed substantially. The only common staphylococcal species among farmed chicken eggs and intestines was S. simulans, while Staphylococcus gallinarum, S. lentus, and S. xylosus were shared species among eggs and intestines from household chickens indicating that the intestines of the birds were not major contributors to the staphylococcal populations of eggs from those bird types.
The poultry intestine microbiome has been found to be diverse (Pan and Yu, 2014; Clavijo and Florez, 2018). Similarly, the results of the present study also revealed that the intestinal staphylococcal flora is also diverse as 12 staphylococcal species and presumptive species from a small sample size of 100 chickens (50 of each chicken type) were identified. As both chicken types have differences in their diet, habitat, breed, environment, and life duration, differences in the staphylococcal normal flora were also likely. Household chickens were reared in the backyards or surroundings of farmers' houses in the villages, where chickens have wide space to move around on agricultural field. In contrast, farmed chickens share very little space in the commercial poultry house, where they are offered feed manufactured by commercial manufacturer. Hence, there is very little variation in the feed composition of a farm. These variations were reflected in the presence of some species in one group while absent in the other group. Nevertheless, the results of our study do not show major differences in the prevalence of staphylococcal species between both chicken types and instead revealed 6 species in common.
Surprisingly, no S. aureus isolates were identified from these samples. Previous studies on the microflora of chicken intestines have found staphylococci but have either not identified the species or selectively only targeted S. aureus as it is a dominant staphylococcal species and a known causative agent of disease in humans and animals (Salanitro et al., 1974; Devriese et al., 1975; Awan and Matsumoto, 1998). Although some of the staphylococcal species isolates from intestines are not known to cause diseases in humans and poultry, others are known pathogenic bacteria. Presence of these pathogenic species is of public health concern, as meat is not washed after slaughter in poultry shops in Pakistan, and there is a risk of human infections as well as outbreaks. For example, S. cohnii subspecies urealyticus has caused bacteremia in humans (Soldera et al., 2013) as well as joint infections in poultry (Tsai et al., 2015). S. simulans and Staphylococcus warneri have also been reported to cause serious skin infections, sepsis, and endocarditis in humans (Males et al., 1985; Kamath et al., 1992; Tous Romero et al., 2016).
The most prevalent species found in this study was S. carnosus subspecies carnosus followed by S. xylosus in farmed chickens and S. cohnii subspecies urealyticus in household chickens. The presence of all 3 species in sick birds has been reported previously (Awan and Matsumoto, 1998; Aarestrup et al., 2000), but other staphylococcal species found in the present study have not been previously reported from poultry intestines including Staphylococcus caprae, Staphylococcus chromogenes, S. saprophyticus, and S. warneri indicating differences between the staphylococcal population of sick and healthy birds. These distinctions in populations could be due to the various organs sampled (healthy intestines vs. diseased organs or blood) and methodology for identifying the staphylococcal species.
Contrary to the differences in species, antimicrobial resistance among staphylococci from farmed and household chicken intestines was very similar. Both groups exhibited the highest levels of resistance to erythromycin, clindamycin, tetracycline, and tigecycline. Resistance to the macrolides-lincosamides-streptogramins and tetracycline is common in staphylococcal isolates regardless of source, including poultry (Aarestrup et al., 2000; Frei et al., 2001; Bounar-Kechih et al., 2018), because of the widespread presence of resistance genes such as ermC and tetK/tetM, respectively (Levy et al., 1989; Aarestrup et al., 2000; Nawaz et al., 2000). However, of concern was resistance to antimicrobials used exclusively to treat human infections. For example, in addition to resistance to the newer antimicrobial tigecycline, resistance was also detected for daptomycin. Although comparison of resistance in poultry intestinal staphylococci to new antimicrobials was not possible as data are unavailable in the literature, resistance to these antimicrobials was observed in staphylococci from farmed and household chickens in our previous studies (Syed et al., 2018; Syed et al., 2019). Furthermore, most isolates were multidrug resistant with resistance up to 8 classes of antimicrobials, which is of concern for nonclinical isolates. Interestingly, no observable differences in antibiotic resistance patterns of staphylococci from both chicken types were detected indicating that antibiotic resistance is as common in farmed broiler chickens as in household chickens.
Irrespective of the isolate source, significant associations of resistance phenotypes were determined not only to the antimicrobials belonging to the same antimicrobial class but also to antimicrobials from different classes. Similarly, resistance genes were also positively correlated to their corresponding antimicrobials (e.g., mecA to β-lactams) as well as different noncorresponding antimicrobials (e.g., mecA with fluoroquinolones). It is not surprising that resistances, either at the phenotypic or genotypic level, to antimicrobials belonging to the same class yielded significant positive correlations (Oggioni et al., 2015). However, significant associations of resistance to different antimicrobials in the same isolate could be linked to co-localization of genes conferring resistances to different antimicrobials either on the chromosome itself or mobile genetic elements (Fatholahzadeh et al., 2008; Emaneini et al., 2013).
Bacteriocins, peptides produced by bacteria that are capable of inhibiting the growth of closely related bacteria (Cotter et al., 2005), were suspected in S. xylosus from our previous studies on household chicken eggs (Syed et al., 2019). Isolates from that study exhibited broad-spectrum inhibition of growth against the closely related gram-positive bacteria, MRSA and methicillin-susceptible S. aureus, as well as the gram-negative bacteria, Salmonella and Es. coli. Staphylococci from this study were also tested for their ability to inhibit growth of other bacteria. Similarly, staphylococci from both farmed and household chicken intestines were able to inhibit the growth of both gram-positive and gram-negative bacteria although none produced activity against S. Typhi. Bacteriocin activity in bacteria isolated from chicken ceca has been previously demonstrated (Line et al., 2008), but those were identified as enterococci, not staphylococci. Further study is needed to fully characterize these potential bacteriocins from chicken intestines.
From the findings of the present study, it may be concluded that a number of staphylococcal species are present in intestines from farmed and household chickens, including reported human and animal pathogenic species. Nonetheless, there is no indication of significant differences in staphylococcal microbial flora of poultry intestines of both chicken types. Of concern is the level of resistance in isolated staphylococcal species against different tested antibiotics. Both poultry meat sold in the markets and close contact of humans with household chickens as egg sources and pets may be a source of transmission of antibiotic-resistant pathogenic strains of staphylococci to humans. Further studies are needed to ascertain the origin of antimicrobial resistance in staphylococci in farmed and household chickens and the risk of dissemination of those bacteria to consumers and owners.
Acknowledgements
This research was funded in part by the US Department of Agriculture, Agricultural Research Service, and the University of Haripur, Pakistan. M.A.S. is grateful to Maria Gul and Riaz Uddin Afridi for technical help.
Conflict of Interest Statement: The authors did not provide a conflict of interest statement.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.05.051.
Supplementary data
Supplementary Figure 1.
Correlation matrix of resistance phenotypes and genotypes of Staphylococcus spp. isolated from domestic and farmed chicken using the “cor” function from the software “R”.
References
- Aarestrup F.M., Agers L.Y., Ahrens P., JC J.L., Madsen M., Jensen L.B. Antimicrobial susceptibility and presence of resistance genes in staphylococci from poultry. Vet. Microbiol. 2000;74:353–364. doi: 10.1016/s0378-1135(00)00197-8. [DOI] [PubMed] [Google Scholar]
- Al-Rubaye A.A., Couger M.B., Ojha S., Pummill J.F., Koon J.A., 2nd, Wideman R.F., Jr., Rhoads D.D. Genome analysis of Staphylococcus agnetis, an agent of lameness in broiler chickens. PLoS One. 2015;10:e0143336. doi: 10.1371/journal.pone.0143336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awan M.A., Matsumoto M. Heterogeneity of staphylococci and other bacteria isolated from six-week-old broiler chickens. Poult. Sci. 1998;77:944–949. doi: 10.1093/ps/77.7.944. [DOI] [PubMed] [Google Scholar]
- Bounar-Kechih S., Taha Hamdi M., Aggad H., Meguenni N., Cantekin Z. Carriage methicillin-Resistant Staphylococcus aureus in poultry and cattle in Northern Algeria. Vet. Med. Int. 2018;2018:4636121. doi: 10.1155/2018/4636121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova C., Iselin L., von Steiger N., Droz S., Sendi P. Staphylococcus hyicus bacteremia in a farmer. J. Clin. Microbiol. 2011;49:4377–4378. doi: 10.1128/JCM.05645-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheville N.F., Tappe J., Ackermann M., Jensen A. Acute fibrinopurulent blepharitis and conjunctivitis associated with Staphylococcus hyicus, Escherichia coli, and Streptococcus sp. in chickens and turkeys. Vet. Pathol. 1988;25:369–375. doi: 10.1177/030098588802500506. [DOI] [PubMed] [Google Scholar]
- Clavijo V., Florez M.J.V. The gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: a review. Poult. Sci. 2018;97:1006–1021. doi: 10.3382/ps/pex359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI. Clinical and Laboratory Standards Institute; Wayne, PA: 2018. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. [Google Scholar]
- Cotter P.D., Hill C., Ross R.P. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 2005;3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- Cuny C., Wieler L.H., Witte W. Livestock-associated MRSA: the impact on humans. Antibiotics (Basel) 2015;4:521–543. doi: 10.3390/antibiotics4040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vliegher S., Opsomer G., Vanrolleghem A., Devriese L.A., Sampimon O.C., Sol J., Barkema H.W., Haesebrouck F., de Kruif A. In vitro growth inhibition of major mastitis pathogens by Staphylococcus chromogenes originating from teat apices of dairy heifers. Vet. Microbiol. 2004;101:215–221. doi: 10.1016/j.vetmic.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Devriese L.A., Devos A.H., van Damme L.R. Quantitative aspects of the Staphylococcus aureus flora of poultry. Poult. Sci. 1975;54:95–101. doi: 10.3382/ps.0540095. [DOI] [PubMed] [Google Scholar]
- Emaneini M., Bigverdi R., Kalantar D., Soroush S., Jabalameli F., Noorazar Khoshgnab B., Asadollahi P., Taherikalani M. Distribution of genes encoding tetracycline resistance and aminoglycoside modifying enzymes in Staphylococcus aureus strains isolated from a burn center. Ann. Burns Fire Disasters. 2013;26:76–80. [PMC free article] [PubMed] [Google Scholar]
- Fatholahzadeh B., Emaneini M., Gilbert G., Udo E., Aligholi M., Modarressi M.H., Nouri K., Sedaghat H., Feizabadi M.M. Staphylococcal cassette chromosome mec (SCCmec) analysis and antimicrobial susceptibility patterns of methicillin-resistant Staphylococcus aureus (MRSA) isolates in Tehran, Iran. Microb. Drug Resist. 2008;14:217–220. doi: 10.1089/mdr.2008.0822. [DOI] [PubMed] [Google Scholar]
- Frei A., Goldenberger D., Teuber M. Antimicrobial susceptibility of intestinal bacteria from Swiss poultry flocks before the ban of antimicrobial growth promoters. Syst. Appl. Microbiol. 2001;24:116–121. doi: 10.1078/0723-2020-00004. [DOI] [PubMed] [Google Scholar]
- Harrison E.M., Paterson G.K., Holden M.T., Ba X., Rolo J., Morgan F.J., Pichon B., Kearns A., Zadoks R.N., Peacock S.J., Parkhill J., Holmes M.A. A novel hybrid SCCmec-mecC region in Staphylococcus sciuri. J. Antimicrob. Chemother. 2014;69:911–918. doi: 10.1093/jac/dkt452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C.R., Davis J.A., Barrett J.B. Prevalence and characterization of methicillin-resistant Staphylococcus aureus isolates from retail meat and humans in Georgia. J. Clin. Microbiol. 2013;51:1199–1207. doi: 10.1128/JCM.03166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath U., Singer C., Isenberg H.D. Clinical significance of Staphylococcus warneri bacteremia. J. Clin. Microbiol. 1992;30:261–264. doi: 10.1128/jcm.30.2.261-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano J., Shimizu A., Saitoh Y., Yagi M., Saito T., Okamoto R. Isolation of methicillin-resistant coagulase-negative staphylococci from chickens. J. Clin. Microbiol. 1996;34:2072–2077. doi: 10.1128/jcm.34.9.2072-2077.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.B., Seo K.W., Jeon H.Y., Lim S.K., Lee Y.J. Characteristics of the antimicrobial resistance of Staphylococcus aureus isolated from chicken meat produced by different integrated broiler operations in Korea. Poult. Sci. 2018;97:962–969. doi: 10.3382/ps/pex357. [DOI] [PubMed] [Google Scholar]
- Levy S.B., McMurry L.M., Burdett V., Courvalin P., Hillen W., Roberts M.C., Taylor D.E. Nomenclature for tetracycline resistance determinants. Antimicrob. Agents Chemother. 1989;33:1373–1374. doi: 10.1128/aac.33.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Line J.E., Svetoch E.A., Eruslanov B.V., Perelygin V.V., Mitsevich E.V., Mitsevich I.P., Levchuk V.P., Svetoch O.E., Seal B.S., Siragusa G.R., Stern N.J. Isolation and purification of enterocin E-760 with broad antimicrobial activity against gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 2008;52:1094–1100. doi: 10.1128/AAC.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Males B.M., Bartholomew W.R., Amsterdam D. Staphylococcus simulans septicemia in a patient with chronic osteomyelitis and pyarthrosis. J. Clin. Microbiol. 1985;21:255–257. doi: 10.1128/jcm.21.2.255-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz M.S., Khan S.A., Khan A.A., Khambaty F.M., Cerniglia C.E. Comparative molecular analysis of erythromycin-resistance determinants in staphylococcal isolates of poultry and human origin. Mol. Cell Probes. 2000;14:311–319. doi: 10.1006/mcpr.2000.0320. [DOI] [PubMed] [Google Scholar]
- Nazia K.K.M., Durrani N.U., Kamboh A.A., Lakho S.A., Rind R., Abro S.H., Soomro N.M. Prevalence of septic arthritis caused by Staphylococcus aureus in poultry birds at Tandojam, Pakistan. J. Anim. Health Prod. 2015;3:73–77. [Google Scholar]
- Nemati M., Hermans K., Lipinska U., Denis O., Deplano A., Struelens M., Devriese L.A., Pasmans F., Haesebrouck F. Antimicrobial resistance of old and recent Staphylococcus aureus isolates from poultry: first detection of livestock-associated methicillin-resistant strain ST398. Antimicrob. Agents Chemother. 2008;52:3817–3819. doi: 10.1128/AAC.00613-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oggioni M.R., Coelho J.R., Furi L., Knight D.R., Viti C., Orefici G., Martinez J.L., Freitas A.T., Coque T.M., Morrissey I., consortium B. Significant differences characterise the correlation coefficients between biocide and antibiotic susceptibility profiles in Staphylococcus aureus. Curr. Pharm. Des. 2015;21:2054–2057. doi: 10.2174/1381612821666150310103238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D., Yu Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes. 2014;5:108–119. doi: 10.4161/gmic.26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein R., Gotz F. What distinguishes highly pathogenic staphylococci from medium- and non-pathogenic? Curr. Top Microbiol. Immunol. 2013;358:33–89. doi: 10.1007/82_2012_286. [DOI] [PubMed] [Google Scholar]
- Salanitro J.P., Blake I.G., Muirhead P.A. Studies on the cecal microflora of commercial broiler chickens. Appl. Microbiol. 1974;28:439–447. doi: 10.1128/am.28.3.439-447.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanitro J.P., Blake I.G., Muirehead P.A., Maglio M., Goodman J.R. Bacteria isolated from the duodenum, ileum, and cecum of young chicks. Appl. Environ. Microbiol. 1978;35:782–790. doi: 10.1128/aem.35.4.782-790.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldera J., Nedel W.L., Cardoso P.R., d'Azevedo P.A. Bacteremia due to Staphylococcus cohnii ssp. urealyticus caused by infected pressure ulcer: case report and review of the literature. Sao Paulo Med. J. 2013;131:59–61. doi: 10.1590/S1516-31802013000100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien-Pysniak D., Wilczynski J., Marek A., Smiech A., Kosikowska U., Hauschild T. Staphylococcus simulans associated with endocarditis in broiler chickens. Avian Pathol. 2017;46:44–51. doi: 10.1080/03079457.2016.1203392. [DOI] [PubMed] [Google Scholar]
- Strommenger B., Kettlitz C., Werner G., Witte W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 2003;41:4089–4094. doi: 10.1128/JCM.41.9.4089-4094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed M.A., Jackson C.R., Ramadan H., Afridi R., Bano S., Bibi S., Fatima B., Tabassum S., Jamil B., Khan M.F., Barrett J.B., Woodley T.A. Detection and molecular characterization of staphylococci from eggs of household chickens. Foodborne Pathog. Dis. 2019;16:550–557. doi: 10.1089/fpd.2018.2585. [DOI] [PubMed] [Google Scholar]
- Syed M.A., Shah S.H.H., Sherafzal Y., Shafi-Ur-Rehman S., Khan M.A., Barrett J.B., Woodley T.A., Jamil B., Abbasi S.A., Jackson C.R. Detection and molecular characterization of methicillin-resistant Staphylococcus aureus from table eggs in Haripur, Pakistan. Foodborne Pathog. Dis. 2018;15:86–93. doi: 10.1089/fpd.2017.2336. [DOI] [PubMed] [Google Scholar]
- Tous Romero F., Gutierrez Garcia-Rodrigo C., Velasco Tamariz V., Llamas Martin R. Acute Infection by Staphylococcus simulans in the hand of a man. JAMA Dermatol. 2016;152:1060. doi: 10.1001/jamadermatol.2016.0959. [DOI] [PubMed] [Google Scholar]
- Tsai S., Chen L.J., Shih C.-Y., Chang T.-C., Chiou M.-T., Chuang K.P. Joint lesions in Taiwan native colored broiler chicken with natural and experimental Staphylococcus aureus or S. cohnii infection. Taiwan Vet. J. 2015;41:237–244. [Google Scholar]