Abstract
In this study, specific primers and fluorescent probes were designed to target the thymidine kinase (TK) gene sequence of avian infectious laryngotracheitis virus (ILTV). Through specificity and sensitivity tests, a real-time fluorescence-based recombinase-aided amplification (RF-RAA) method for detecting ILTV was established. The results showed that the method was specific and could be used to accurately detect ILTV, and there was no cross-reaction with Newcastle disease virus (NDV), avian influenza virus (AIV), or infectious bronchitis virus (IBV). Real-time fluorescence-based recombinase-aided amplification had high sensitivity, and the lowest detectable limit (LDL) for ILTV could reach 10 copies/μL, 1,000 times more sensitive than conventional PCR (104 copies/μL), to rival that of real-time fluorescence-based quantitative PCR (RFQ-PCR) (10 copies/μL). This method and RFQ-PCR were used to detect 96 samples of chicken throat swabs with ILT initially diagnosed in clinic from the north of China, and the coincidence rate of the 2 methods was 100%. The RF-RAA reaction required only 20-30 minutes to completing, and its sensitivity was much higher than that of conventional PCR. Real-time fluorescence-based recombinase-aided amplification is similar to RFQ-PCR and has the advantages of specificity, sensitivity, and high efficiency, so it is suitable for early clinical detection and epidemiological investigation of ILTV.
Key words: infectious laryngotracheitis virus, real-time fluorescence-based recombinase-aided amplification, TK gene
Introduction
Infectious laryngotracheitis (ILT) is an acute respiratory infectious disease caused by infectious laryngotracheitis virus (ILTV) in Gallid herpesvirus 1 (Ali et al., 2019). Infectious laryngotracheitis often breaks out and spreads in autumn and winter, and all day-old chickens are susceptible to this disease. It first occurred in the United States in 1925 and then spread all over the world, with an infection rate as high as 90% and a mortality rate of about 20%. It is one of the major infectious diseases harmful to chickens and has caused huge economic losses to the poultry industry. The main clinical signs of ILT are dyspnea, cough, and exudates containing blood. In addition, swelling, bleeding, and erosion of laryngeal, tracheal mucous membrane can be seen during autopsy (Jorge et al., 2019). The clinical signs and pathological characteristics of ILT are similar to those of low pathogenic avian influenza (AI), Newcastle disease (ND), infectious bronchitis (IB), mucous fowlpox (MF), and so on. Therefore, cases of ILT are prone to misdiagnosis if relying solely on clinical (Giovanni et al., 2017), which may cause great economic losses or spread of disease. For these reasons rapid, sensitive and specific detection methods are very important for the prevention and control of ILTV. At present, a variety of ILTV detection methods have been established, but some of them were limited. Isolation and identification of ILVT is time consuming and isolation rate is low; there was a problem of nonspecific reaction in the detection of serum antibody level of ILTV by enzyme-linked immunosorbent assay (ELISA). PCR–agarose gel electrophoresis (AGE) requires toxic dyes; although loop-mediated isothermal amplification (LAMP) does not need special instruments, it places great demands on primer design and has some problems such as non-specific reaction (Zhang et al., 2018). In real-time fluorescence-based quantitative PCR (RFQ-PCR), fluorescence-labeled probes or corresponding fluorescent dyes are added on the basis of conventional PCR, and the changes of PCR products are monitored by collecting fluorescence signals. Real-time fluorescence-based quantitative PCR technology has been widely used in the detection of pathogens owing to its advantages in terms of high sensitivity and strong specificity.
Recombinase-aided amplification (RAA) is a technique for nucleic acid amplification under isothermal conditions by using recombinant enzyme, single-strand DNA-binding protein (SSB), and DNA polymerase. The principle is that the recombinant enzyme combines with the primer DNA to form a polymer, and when the primer finds a perfectly matched complementary sequence on the template DNA, it opens the double-stranded structure of template DNA with the help of SSB and forms a new DNA complementary strand under the action of DNA polymerase, and the amplification product grows exponentially (Daher et al., 2016). RAA has the advantages of more simplistic operation, faster reaction, simultaneous detection of multiple targets, and lower requirements on equipment. This technology has been widely used in human diseases, veterinary drugs, food industry, agriculture, and other fields. Real-time fluorescence-based RAA (RF-RAA) adds a fluorescence-labeled probe into RAA, which contains both fluorophores and quenchers joined by an internal tetrahydrofuran (THF) site. In the process of amplification, the probe is bound to the amplifier, and the exonuclease recognizes and cleaves the THF site so that the fluorophores are no longer bound by the quenchers and can be detected. With continued amplification, enhanced fluorescence signal is positively correlated with the accumulation of amplification products, so that the amplification products can be monitored in real time. Compared with conventional PCR, RF-RAA has short reaction time and clear results and can be used for qualitative and quantitative detection (Piepenburg et al., 2006).
In this study, specific primers and probes were designed to target the thymidine kinase (TK) gene of ILTV, and the reaction temperature and time were screened based on the criterion that strong fluorescence signals could be detected in the shortest reaction time. Finally, a rapid detection method for ILTV with RF-RAA was established and its specificity and sensitivity were verified. The throat swabs of 96 diseased chickens clinically diagnosed as ILT were detected by RF-RAA. This study provides an effective method for veterinary clinical diagnosis of ILT.
Materials and methods
Extraction of Viral DNA/RNA
According to the instructions of DNA/RNA extraction kit (Tiangen Biotech Co., Ltd., Beijing, China), the DNA of ILTV (AV195) was extracted; the RNA of IBV (AV1511), AIV, and NDV (The 2 strains from clinical cases identified by PCR) was extracted and reversely transcribed into cDNA, respectively. The viral DNA was extracted from 96 suspected ILTV-infected chicken throat swabs from northern China for follow-up experiments.
Primer and Probe Design for RF-RAA
According to the conserved and stable TK sequence in ILTV genome, one pairs of specific primers and one probe were designed according to the principle of RAA primer design (Chen, 2018). After its specificity was determined by BLAST comparison, it was synthesized by Sangon Biotech Co., Ltd. Primer sequence was ILTV-RAA-F: CAGTATCTGGCATCGCCTCATTTCTTTCTA, ILTV-RAA-R: CTCATCACTATCCTCCTCAACCTCCTCCTC; Probe sequence was: 5′-AGTTCCCCCGGCCGGAACTCCTCCACGACCC/i6FAMdT//idSp//iBHQ1dT/AGACGTTACTACAAGG-3′(C3 Spacer).
RF-RAA Reaction System
The reaction system was prepared according to the kit (Qitian Biotechnology Co., Ltd. Wuxi, China). VI buffer, 25 μL; purified water, 16.7 μL; upstream and downstream primers (10 μmol), 2.1 μL each; probe 0.6 μL; template, 1 μL. After mixing, the liquid was transferred into the reaction tubes, and 2.5 μL of magnesium acetate was added before reaction. It was put into RFQ-PCR (Roche Diagnostics Co., Ltd. Shanghai, China) instrument to react for 30 min. Carboxyfluorescein (FAM) signals were collected during the reaction.
Screening of RF-RAA Reaction Temperature and Time
In the experiment, the reaction was carried out at 37, 38, 39, 40, and 41°C, respectively, then the optimum reaction temperature was selected. The reaction time was set to 30 min, and the optimum reaction time was determined according to the amplification efficiency (fluorescence value). When a strong fluorescence signal could be detected in the shortest reaction time, it was the best reaction condition.
Specificity Verification of RF-RAA
In the aforementioned reaction system, the templates were DNA of ILTV and cDNA of NDV, AIV, and IBV (using purified water instead of template for negative reaction), which verified the specificity of RF-RAA method.
Sensitivity Verification of RF-RAA
Construction of Standard Plasmids
Using ILTV genome as template DNA, the length of ILTV-RAA-F/ILTV-RAA-R primers was modified (according to the requirements of conventional PCR reaction), and the sequence was F: 5′-CAGTATCTGGCATCGCCTCAT-3′, R: 5′-CTCATCACTATCCTCCTCAAC-3′. The reaction conditions were as follows: predenaturation at 94°C for 5 min; denaturation at 94°C for 45 s; annealing at 56°C for 45 s; extension at 72°C for 60 s, 35 cycles of amplification; extension at 72°C for 10 min, preservation at 4°C. The product was purified by gel extraction kit (Takara Biomedical Technology Co., Ltd. Dalian, China), and the target fragment was ligated to pMD20 plasmid. After the standard plasmid was selected by Blue-White Screening, the copy number of DNA per unit volume of plasmid was calculated according to Moore's law.
Plasmid copy number (copy/L) = [plasmid concentration (g/μL) × 6.02 × 1,023]/[total fragment length (bp) × 660 g/mol]; total fragment length = vector length (bp) + fragment length (bp).
RF-RAA Reaction System
The constructed standard plasmid was diluted to 100-105 copies/μL. The reaction was based on the optimized RF-RAA reaction conditions, the negative control was set up, and the lowest detectable limit (LDL) was observed.
RFQ-PCR Reaction System
TB Green Premix DimerEraser (2X) (Takara Biomedical Technology Co., Ltd., Dalian, China), 12 μL; upstream primer (10 μmol), 0.75 μL; downstream primer (10 μmol), 0.75 μL; template, 2 μL (100∼106 copies/μL plasmid); the final volume was made up to 25 μL with water. The reaction system was as follows: predenaturation at 95°C for 30 s; denaturation at 95°C for 5 s; annealing at 55°C for 30 s; extension at 72°C for 30 s, 40 cycles.
Conventional PCR Reaction System
2 × Gflex PCR Buffer (Mg2+, dNTP plus), 12.5 μL; water, 10 μL; upstream primer (10 μmol), 0.5 μL; downstream primer (10 μmol), 0.5 μL; Tks Gflex DNA Polymerase (1.25 units/μL), 0.5 μL; template, 1 μL (100 ∼107copies/μL plasmid). The reaction system was as follows: predenaturation at 94°C for 1 min; denaturation at 98°C for 10 s; annealing at 55°C for 15 s; extension at 68°C for 30 s, 30 cycles; extension at 68°C for 5 min.
Detection of Clinical Samples of ILT by RF-RAA and RFQ-PCR
Throat swabs were collected from 96 chickens clinically diagnosed with ILT from 8 chicken farms in northern China,with the DNA was extracted and used as template, and the RF-RAA and RFQ-PCR were used to detect ILTV. The coincidence rate of the 2 detection results was calculated.
Results and discussion
Primer and Probe Design for RF-RAA
Recombinase-aided amplification reaction hinges in large measure on a good primer design. The length of RAA primers is required to be 30-35 bp, which is significantly longer than that of conventional PCR primers (15-30 bp), and the best length of amplification product is 100-200 bp (Zheng et al., 2019). The content of primer G and C is 30%-70%. It is best to have C and T at 5′, avoiding the repetition of G, and it is best to have G and C at 3′. Any base mismatch at the 3′ end will seriously affect the efficiency of RAA amplification. At that same time, primer dimers should be avoided (Chen, 2018). The primer designed in this experiment had low continuous complementarity and met the design requirements; a fluorescence probe was designed to label the primer, so that the RF-RAA method was constructed successfully.
Screening of RF-RAA Reaction Temperature and Time
In this experiment, the reaction temperature and reaction time of RF-RAA were screened, and when the strongest fluorescence signal could be detected in the shortest reaction time, it was the best reaction condition. Based on these parameters, optimum reaction conditions were determined to be 40°C for 22 minutes.
RAA is an isothermal amplification technique of nucleic acid, the whole process is very fast, and the detectable amplification products can be obtained within a short time. Based on RAA, RF-RAA designs exo probe between the upstream and downstream primers, and amplification products can be monitored in real time through RF-PCR instrument. Real-time fluorescence-based RAA can take the DNA extracted from infected tissue as a template, and there is no need for virus purification. The sample requirements are not strict, and it can be used to complete the testing work in a short period of time. Real-time fluorescence-based RAA has the advantages of high accuracy and efficiency and is expected to occupy the market of nucleic acid detection quickly and become the mainstream detection method.
Specificity Verification of RF-RAA
The results showed that there was no fluorescence curve for other viral nucleic acids and negative control except for ILTV, which indicated that the RF-RAA method could specifically detect ILTV and had no cross-reaction with viral nucleic acids of NDV, AIV, and IBV.
Sensitivity Verification of RF-RAA
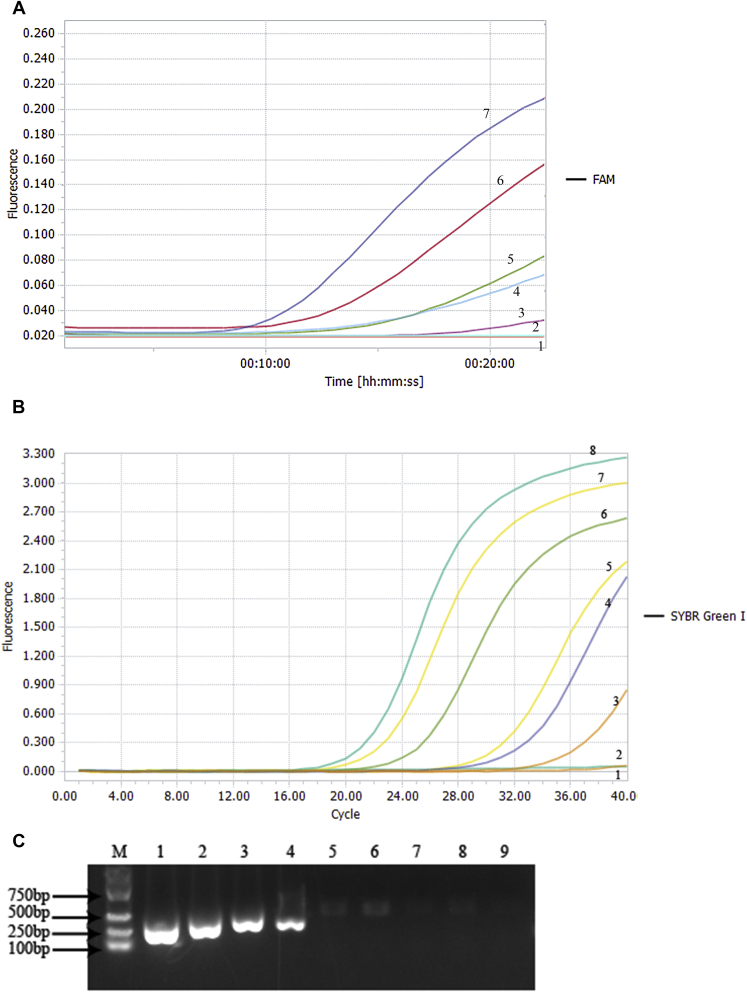
The results showed that LDL was 104 copies/μL for conventional PCR (Figure 1A). The RF-RAA detection method established in this study had very high sensitivity, its LDL was 10 copies/μL, had 1,000 times higher than that of conventional PCR. In terms of detection sensitivity, it could reach 10 copies/μL for both RF-RAA and RFQ-PCR (Figure 1B and 1C), but the time of RF-RAA detection was short and only 20-30 min was needed to complete detection while RFQ-PCR took more than 1 h. The laboratory can also be equipped with portable isothermal amplification instrument which is much cheaper than RFQ-PCR instrument, so it greatly reduces the investment in detection equipment. And it is easy to carry and use in clinic. The drawback is that RF-RAA needs to synthesize and label probes in the process of reaction, with the detection cost higher than that of conventional PCR.
Figure 1.
Sensitivity verification of 3 kinds of technology in ILTV detection. (A) Sensitivity verification of RF-RAA in ILTV detection.1: Negative control; 2-7: Template concentration was 100-105 copies/μL. The results showed that the lowest detectable limit of RF-RAA was 10 copies/μL. B. Sensitivity verification of RFQ-PCR in ILTV detection.1: Negative control; 2-8: Template concentration was 100-106 copies/μL. The results showed that the lowest detectable limit of RFQ-PCR was 10 copies/μL. C. Sensitivity verification of conventional PCR in ILTV detection. M: Marker; 1-8: Template concentration was 107-100 copies/μL; 9: Negative control. The results showed that the lowest detectable limit of conventional PCR was 104copies/μL. Abbreviations: RF-RAA, real-time fluorescence-based recombinase-aided amplification; RFQ-PCR, real-time fluorescence-based quantitative PCR.
Detection of Clinical Samples
A total of 96 clinical samples were tested by RF-RAA and RFQ-PCR. The results of the 2 methods were the same, 46 samples were positive and 50 samples were negative. The coincidence rate of the 2 methods was 100%, and RF-RAA could be used for clinical detection of ILT. The technical comparison of RF-RAA, RFQ-PCR, and conventional PCR is shown in Table 1.
Table 1.
The comprehensive comparison of 3 techniques for ILTV detection.
| Method | Primer design | Enzyme components | Instrument price | Response time | Reaction temperature | Cross-reaction | LDL | Degree of detection |
|---|---|---|---|---|---|---|---|---|
| RF-RAA | 1 pair of primers, 1 probe | Recombinase, SSB, and polymerase | Cheap | 20–30 min | 37°C Constant | No | 1-10 copies/μL | Real time quantitative, qualitative |
| RFQ-PCR | 1 pair of primers | Taq polymerase | Expensive | 1–2 hour | High and variable | No | 10 copies/μL | Real time quantitative, qualitative |
| Conventional PCR | 1 pair of primers | Taq polymerase | Expensive | 2–3 hour | High and variable | No | 104 copies/μL | Qualitative |
Abbreviations: RF-RAA, real-time fluorescence-based recombinase-aided amplification; RFQ-PCR, real-time fluorescence-based quantitative PCR; SSB, single-strand DNA-binding protein.
The RF-RAA established in this experiment provides an effective method for the diagnosis of ILT and can be used for large-scale screening and rapid detection of chicken diseases, which is conducive to early intervention in diseases to reduce economic losses, and is of great significance to the poultry industry.
Acknowledgments
This work was supported by Scientific and Technological R&D Program of Shijiazhuang City (no. 201500412A), Key R&D Plan of Science and Technology Department of Hebei Province (no. 20326621D) and Open Fund of State Key Laboratory of Veterinary Etiological Biology (no. BYSWX2020KFKT02) from Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences. The funding agencies had no role in study design, in the collection, analysis, and interpretation of data, in the writing of the report, or in the decision to submit the article for publication.
Conflict of Interest Statement: The authors did not provide a conflict of interest statement.
References
- Ali S.A., Almofti Y.A., Abd-Elrahman K.A., Haspel N. Immunoinformatics approach for multiepitopes vaccine prediction against glycoprotein b of avian infectious laryngotracheitis virus. Adv. Bioinformatics. 2019;2019:1–23. doi: 10.1155/2019/1270485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Chinese Center for Disease Control and Prevention; Beijing: 2018. Establishment and Preliminary Evaluation of Recombinant Enzyme-Mediated Amplification Technique for Detection of Respiratory Syncytial Virus. [Google Scholar]
- Daher R.K., Stewart G., Boissinot M., Bergeron M.G. Recombinase polymerase amplification for diagnostic applications. Clin. Chem. 2016;62:947. doi: 10.1373/clinchem.2015.245829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanni F., Paola M., Maria T.C., Ilaria B., Giovanni T., Laura F., Massimo C., Antonio L., Mattia C., Ana M. Think globally, act locally: Phylodynamic reconstruction of infectious bronchitis virus (IBV) QX genotype (GI-19 lineage) reveals different population dynamics and spreading patterns when evaluated on different epidemiological scales. Plos One. 2017;12:e0184401. doi: 10.1371/journal.pone.0184401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorge B., Sandra M.R., Ricardo M., Ricardo C.G., Aldo R.N., Katherine P.A., Manolo F.D. Glycoprotein G (gG) production profile during infectious laryngotracheitis virus (ILTV) infection. PLoS One. 2019;14:e0219475. doi: 10.1371/journal.pone.0219475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLOS Biol. 2006;4:1115–1121. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Chen J., Wang C.G., Shi W.Y., Li D.G. The therapeutic effect of Yinhuangerchen mixture on Avian infectious laryngotracheitis. Poult. Sci. 2018;97:2690–2697. doi: 10.3382/ps/pey125. [DOI] [PubMed] [Google Scholar]
- Zheng X.C., Chen Y., Wen Z.Q., Ye W.T., Zhang J.X., Huang Q.J., Liang X.Q., Liu Q. Establishment of rapid fluorescent RAA detection method for carp edema virus. Chin. J. Prev. Vet. Med. 2019;41:721–725. [Google Scholar]