Abstract
The gut microbiota is a complex ecological community and widely recognized in many aspects of research, but little is known on the relation between gut microbiota and embryonic development in chickens. The aim of this study was to explore the dynamic distribution of gut microbiota in chickens' embryos during stages of developments (chicken embryos that had incubated until day 3 [E3], day 12 [E12], and day 19 [E19]). Here, 16S rRNA gene sequencing was performed on the gut microbiota in chicken embryos across different developmental stages. Twenty-one phyla and 601 genera were present in chicken embryos, and 96 genera such as Ochrobactrum, Phyllobacterium, and Amycolatopsis were the core microbiota in the 3 stages of development. Second, 94 genera of microbes were found to change significantly between E3 and E12, and 143 genera significantly differed between E12 and E19 in chicken embryos (P < 0.05). Ochrobactrum and Amycolatopsis decreased with growth changes: E3 (30.4%), E12 (25.1%), and E19 (13.6%) and E3 (11.5%), E12 (7.4%), and E19 (5.6%), respectively. Contrarily, Phyllobacterium increased to 47.9% at E19, indicating the growing trend of microbial diversity among the embryos' development. Moreover, the principal component analysis showed a high level of similarities between E3 and E12 compared with E19, whereas the alpha analysis showed more diversity of gut microbiota at E19. Furthermore, the functional predictions showed that metabolic pathways such as energy metabolism and genetic information processing were significantly enriched on day 3 and day 12 in our study, suggesting their strong influence on growth, development, and immunity of chicken embryos. Our findings provide insights into the understanding of dynamic shifts of gut microbiota during chicken embryonic growth.
Key words: gut microbiota, embryo development, 16S rRNA gene sequencing, chicken
Introduction
The gut microbiota population is dynamic (Gong et al., 2002; Wei et al., 2013), with widely recognized functions ranging from resistance to pathogens, maintenance of homeostasis (Thursby and Juge, 2017), intestinal physiology and regulation (Rodríguez et al., 2015), carbon and nitrogen metabolism (He et al., 2013), digestive performance (Fontaine et al., 2018), and host development and nutrition (Dominguez-Bello et al., 2019). Available evidence suggests that gut microbiota could promote animal health (Fouhse et al., 2016), and this has been demonstrated in adult mammals, wherein specific functional bacteria enhance the development of preventive and therapeutic strategies for chronic inflammation (Wu et al., 2018) and immunity (Shreiner et al., 2016). Although several studies focused on the functions of gut microbiota in adults and neonates (Weng and Walker, 2013; Grond et al., 2018), gut microbiota has been understudied during embryonic development.
Embryonic development commences with the fertilization of the female egg by a single male spermatozoon forming a diploid zygote (Bellairs and Osmond, 2014). In vertebrate embryos, it is formerly assumed that the fetus develops and resides within a sterile environment, and microbial colonization begins on the embryo's surface after birth (Penders et al., 2006; Palmer et al., 2007; Funkhouser and Bordenstein, 2013). However, this traditional view has been challenged by modern sequencing technology studies. These studies have shown that neither the fetus and placenta nor the amniotic fluid are sterile, thereby confirming that the colonization and establishment of the human gastrointestinal tract begins in the uterus (Aagaard et al., 2014; Collado et al., 2016).
The fertilization of the avian embryo occurs in vivo, whereas the reproduction and development occur in a closed egg, which is formed from the external environment but not in utero. Cisek and Binek (2014) showed that bacterial colonization of the avian gut begins immediately after hatching under the influence of the environmental microbe. Moreover, the inheritance of chicken embryo microbiota has been suggested to originate from the maternal cloaca, oviduct, or both (Lee et al., 2019). In addition, our previous study showed that gut microbiota inheritance in the chicken embryo is dependent on maternal hen-derived factors, which are influenced by environmental and host genetic variation during development (Ding et al., 2017).
Several studies have been conducted focusing majorly on the dynamic distribution and microbial composition in chicks and chickens (Sergeant et al., 2014; Ballou et al., 2016; Clavijo and Flórez, 2018; Thomas et al., 2019). However, there are limited reports on gut microbiota in different developmental stages and breeds of chicken embryos. It has been previously studied in chicken embryos that developmental stages are classified to early, middle, and late phases. The early and the middle stages control the formation of organs and systems, whereas the growth of organs and system maturation occur at the late phase (Hamburger and Hamilton, 1992).
In remedy, we investigated the microbial composition in the chicken embryos of Beijing You (Y), Shiqiza (S), and specific pathogen-free (SPF [B]) (from the breed of Babcock) chickens and also examined the dynamic changes of their gut microbiota across the stages of development. The Beijing You and Shiqiza breeds are native Chinese breeds. Babcock is an imported breed for free-range and commercial purposes. The hypothesis outlined in this study is that the growth stages of the chicken embryo affect gut microbiota composition.
Materials and methods
Ethical Considerations
All procedures used in this study were standardized by the Ethics Committee for the Care and Use of Laboratory Animals in Shanghai Jiao Tong University, China, approval number, 201812015 (December 17, 2018).
Study Design and Sampling
Fifty-four chicken eggs from 3 different breeds (Y, S, and B) were incubated in an Ova-Easy Advance Series 11 Digital Cabinet Egg Incubator (Brinsea, Weston Super Mare, UK) in the laboratory; on the same day, they were subjected to the same condition within a sanitized room at 38°C and 55 to 65% humidity. The Y and S breeds have a similar environmental background and were from the Animal Husbandry and Veterinary Research Institute Unit in Shanghai Academy of Agricultural Sciences, whereas we got the B eggs from Sais Poultry Co. Ltd., Jinan, China. To explore chicken embryo gut microbiota, 18 whole embryos that had been incubated until day 3 and 18 embryonic guts that had been incubated until days 12 and 19 were collected from the 54 chicken eggs of the 3 breeds (Supplementary Table 1). The sample collection of embryos and guts were performed inside a sterilized cabinet in a small room. We successfully collected a total number of 54 embryonic samples during the process of sample collection. Besides, 3 embryo-free samples were used as control groups to validate if there was contamination throughout the experimental process.
DNA Extraction and 16S rRNA Sequencing
DNA extraction was carried out immediately from the collected chicken embryo and intestinal samples using the TIANGEN (TIANamp) stool DNA kit (cat#DP328; TIANGEN Biotech Co., Ltd., Beijing, China). This process was carried out following the order given by the company. The DNA samples were stored at −20°C for further analysis. In addition, to assess the quantity and quality of the DNA, it was quantified using an UV Nanodrop spectrophotometer (Thermo Fisher Scientific Co., Ltd., Waltham, MA). PCR was performed to amplify the V4 hypervariable region of the 16S rRNA gene, using forward 5′-AYTGGGYDTAAAGNG-3′ and reverse 5′-TACNVGGGTATCTAATCC-3′ as sample-specific sequence barcoded fusion primers. The PCR was performed under the following conditions: 94°C for 5 min; 94°C for 30 s, 50°C for 30 s, and 72°C (extension); repeated for 27 cycles; and a final extension at 72°C for 7 min (Zhao et al., 2013). The PCR amplification product was detected by 2% agarose gel electrophoresis, and the target fragment was subjected to gelation recovery; we used the gel recovery kit of Axygen (Union City, CA) for recovery. PCR products were purified using the QIAGEN Quick Gel Extraction Kit (cat#28730, Qiagen, Hilden, Germany)
The purified PCR products from 57 samples were used to construct a sequencing library using Illumina TruSeq following the manufacturer's instructions (Illumina, San Diego, CA). 16S rRNA sequencing was carried out at Shanghai Personal Biotechnology Limited Company, Shanghai, China, using Illumina MiSeq (Illumina) sequencing platform.
Bioinformatics Analysis
The sequence was identified using QIIME software (Quantitative Insights into Microbial Ecology, version 1.8.0, http://qiime.org/) (Caporaso et al., 2010). We first used sliding window method to screen the double-end sequence of FASTQ format. The window size is 10 bp, step 1 from the 5'-end of the first base position began to move, for window average quality acuity bases in Q20 sequencing or greater accuracy by an average of 99% (base), from the first value is lower than the average quality Q20 window of truncated sequence, and requirements after the truncation of 150 bp sequence length or greater, and do not allow the existing fuzzy bases (Ambiguous base). Then, we used FLASH software tool (version 1.2.7, http://ccb.jhu.edu/software/FLASH/) (Magoč and Salzberg, 2011), the quality by the sieve double ended at the beginning of the sequence according to the overlap base matching connection: Read 1 and Read 2 2 sequences overlapping base length of 10 or higher bp, and base mismatch number less than 10% of overlapping base length. Finally, based on the index information corresponding to each sample (i.e., barcode sequence, a small base sequence used to identify the sample at the beginning of the sequence), the connected sequence recognition is assigned to the corresponding sample (index sequence is required to match exactly) so as to obtain the valid sequence of each sample. The trimmed sequences were only uploaded. We eliminated the sequence with the mismatched base number of the primer at the 5'-end of >1; 2) and sequences with the same base number of > and 8 successively. Then, through the QIIME software (version 1.8.0, http://qiime.org/) call USEARCH (version 5.2.236, http://www.drive5.com/usearch/), the chimeric sequences were checked and removed (Haas et al., 2011). A total of 2,090,612 sequences that passed the quality screening from the V4 region of the 16S rRNA sequence from 57 samples, including the chicken embryo guts and control, were used for the study (Supplementary Table 2).
Microbial operational taxonomic unit (OTU) were drawn from the approved sequence of the PCR amplicon for the V4 hypervariable region of the 16S rRNA gene and were also compared with the Greengenes (DeSantis et al., 2006). For the representative sequence of each OTU, the default parameters are used in QIIME software to obtain taxonomic information corresponding to each OTU by comparing the OTU representative sequence with the template sequence of the corresponding database. For sequences of different categories, specific databases were used as template sequences for OTU classification status identification: a) 16S rRNA gene database for bacteria and archaea. Greengenes Database (Release 13.8, http://greengenes.secondgenome.com/) (DeSantis et al., 2006) and the RDP (Ribosomal Database Project) Database (Release 11.1, http://rdp.cme.msu.edu/) databases utilizing the uclust and blast functions in QIIME (Caporaso et al., 2010).
A total of 57,618 OTU were annotated from the 2,090612 amplicons and further divided at a specific taxonomical level known as phylum, class, order, family, genus, and species levels (Supplementary Table 3). The OTU whose abundance value was lower than 0.001% (1/100,000) of the total sequencing volume of all samples was removed (Bokulich et al., 2013), and the abundance matrix of the rare OTU was removed for a subsequent series of analysis. The OTU of each sample was arranged according to its abundance from large to small, and the abundance value is converted by Log2 logarithmic transformation, and R software (R Core Team, Vienna, Austria) was used to draw the abundance grade curve of each sample. GraPhlAn (Graphical Phylogenetic Analysis), a software tool used in producing quality phylogenetic trees, showcased abundant microbes at different taxa (Asnicar et al., 2015). Microbiota compositions in the chicken embryo at the third, 12th, and 19th day of developments were further analyzed using the linear discriminant analysis effect size method (Segata et al., 2011). Core microbiota has been defined as a wide collection of shared microbial genes among the sampled individuals (Turnbaugh et al., 2009). Alpha diversity indices that include Shannon, ACE, Chao1, and Simpson's index were calculated using QIIME software to measure the richness and diversity of the microbial community (Shannon, 1948; Simpson, 1949; Chao, 1984; Chao and Shen, 2004). Beta diversity analysis was conducted to investigate the similarity of the community structure between different samples. For beta diversity metrics, the nonmetric multidimensional scaling analysis for unweighted and weighted UniFrac distance matrices was also conducted using R software. The principal component analysis (PCA) was conducted using R software. Analysis of similarities was also carried out to validate the differences in the microbial community structure (Clarke, 1993; Warton et al., 2012) among the 3 developmental stages (third, 12th, and 19th day) and breeds (Y, S, and B) using mothur software. PICRUSt was used to predict the microbial functional profile (Langille et al., 2013). At 97% similarity, the QIIME's command (a general bioinformatics pipeline) was used to map the OTU to the Greengenes 13.5 reference database (DeSantis et al., 2006). The OTU abundance was regularized by 16S rRNA gene copy numbers from known bacterial genomes in the Integrated Microbial Genomes system. Kyoto Encyclopedia of Genes and Genomes helped in the alignment of the predicted genes and their functions (Kanehisa et al., 2014). The group differences and similarities were compared using STAMP software (Parks and Beiko, 2010). The Venn diagrams were obtained using Venny software (Oliveros, 2007). In the group analysis, the 2-sided Welch's t-test (Welch, 1947) and Benjamini–Hochberg false discovery rate correction were used.
Results
Composition of Microbiota in the Chicken Embryo
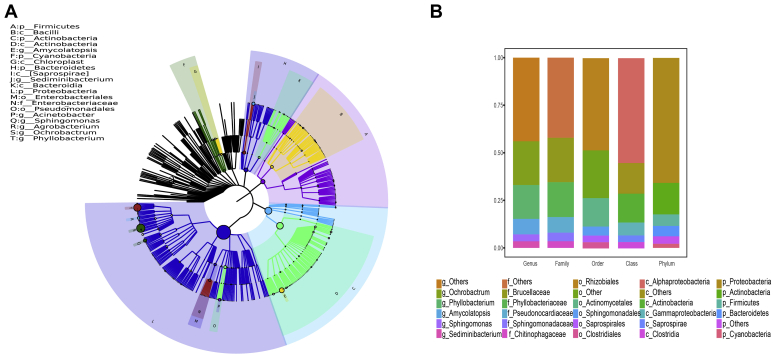
Fifty-four samples, collected from all breeds, included whole embryos that were incubated for 3 D and guts from chicken embryos that were incubated for 12 and 19 D. The operational taxonomical unit was clustered into 4,801 units in the chicken embryo. We used GraPhlAn software to construct a phylogenetic tree that identifies the dominant microbial population from the complex community data. The top abundant microbes the GraPhlAn software detected at each classification level were Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Cyanobacteria (phylum); Amycolatopsis, Sphingomonas, Agrobacterium, Acinetobacter, Sediminibacterium, Ochrobactrum, and Phyllobacterium (genus); Actinobacteria, Chloroplast, Bacteroidetes, Saprospirae, and Bacilli (class); Enterobacteriales and Pseudomonadales (order); and Enterobacteriaceae (family) (Figure 1A).
Figure 1.
Composition of gut microbiota in the chicken embryo. (A) Phylogenetic tree constructed from the taxa. Each classification unit is distinguished with different colors. Colored blocks in the outermost circle indicate phyla and in the inner circle indicate genera. (B) The abundant microbes at each level (genus, family, order, class, and phylum).
Further analysis was carried out to recognize the composition of gut microbiota in specific taxon groups. Twenty-one phyla, 95 classes, 184 orders, 330 families, and 601 genera existed in the chicken embryo (Supplementary Table 4). According to the phylum assignment result, Proteobacteria (65.6%) was the predominant phylum in all the samples, and Actinobacteria (16.6%) emerged as the second phylum, followed by Firmicutes (6.1%), Bacteroidetes (5.4%), and Cyanobacteria (2.2%). The relative abundance of the phylum Proteobacteria was seen in the chicken embryo considering the age and breeds (Figure 4D, Supplementary Figure 2). Apart from phylum distribution, we analyzed the abundance of microbiota at other taxonomic levels. At the class level, Alphaproteobacteria appeared as the microbiota with the highest proportion, followed by Actinobacteria. Moreover, the top abundant families were Phyllobacteriaceae, Brucellaceae, Pseudonocardiaceae, Sphingomonadaceae, Chitinophagaceae, Moraxellaceae, Rhizobiaceae, Micrococcaceae, Lactobacillaceae, Enterobacteriaceae, and Methylobacteriaceae. Interestingly, Ochrobactrum (23.0%) and Phyllobacterium (17.8%) that emerged as the most dominant at the genus level belong to the phylum of Proteobacteria (Figure 1B). We also observed a high proportion of Amycolatopsis, Sphingomonas, Sediminibacterium, Acinetobacter, and Agrobacterium in all the samples. These results affirmed the presence of microbiota in the chicken embryo, although at different proportions.
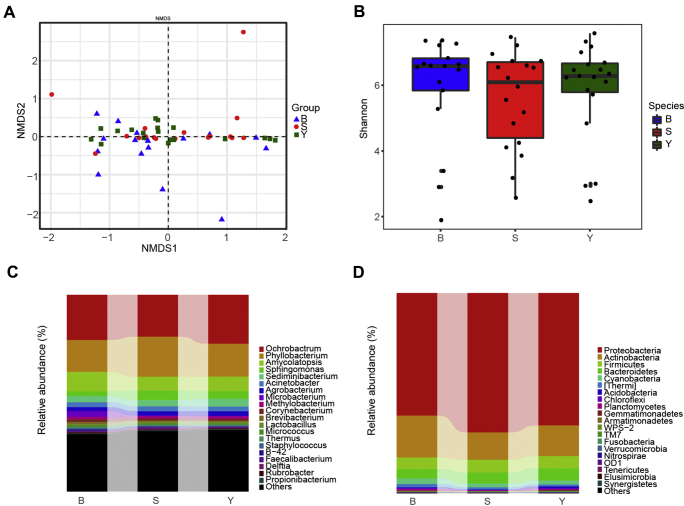
Figure 4.
Embryonic gut microbiota composition across different breeds: Beijing You (Y), Shiqiza (S), and SPF (Babcock) (B). (A) Beta analysis using NMDS across different breeds. Blue, red, and green represent SPF (Babcock) (B), Shiqiza (S), and Beijing You (Y) breeds, respectively. (B) The Shannon diversity index was not significant among the 3 groups (P > 0.05). The box shows the quartiles above and below the median, with a dark line at the center of the box denoting the median and black dots outside the box showing the outlier. (C) Relative abundance of bacteria among the 3 breeds at the genus level. (d) Relative abundance of bacteria among the 3 breeds at the phylum level. Abbreviations: NMDS, nonmetric multidimensional scaling; SPF, specific pathogen-free.
Chicken Embryonic Gut Microbial Differences in Three Developmental Stages
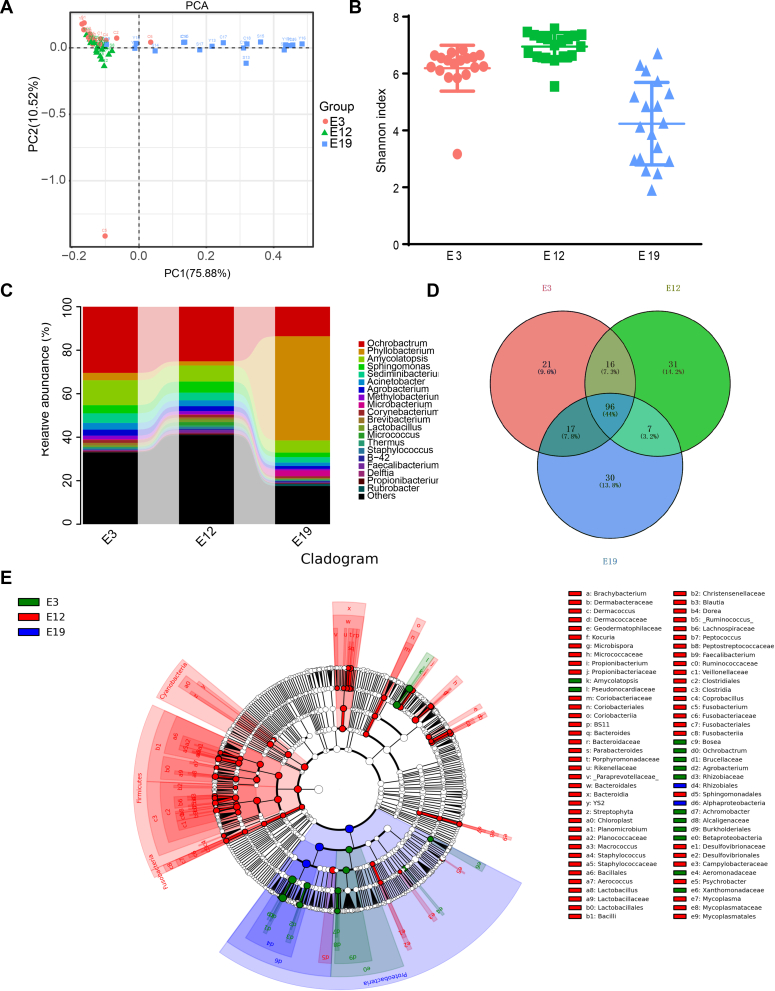
To measure microbial composition in the chicken embryo at different stages of development, chicken embryos that had been incubated until day 3 (E3), day 12 (E12), and day 19 (E19), we performed PCA. The PCA showed a high level of similarities between E3 and E12 compared with E19. In addition, the microbial composition at the late phase of development displayed a low degree of dissimilarity compared with the other early stages. It was cleared that the microbial composition was so clustered together at the early phase of the chicken embryo development compared with the mature stage of development (Figure 2A). Analysis of similarities displayed a significant difference in the microbial community structure among the 3 developmental stages (p-value < 0.05 [0.001]), and a higher p-value of more than 0.5 (r = 0.6059) specified that separation among different ages was significant (Supplementary Table 5A). Moreover, Figure 2B shows a diversity index estimating microbial richness and diversity. The alpha analysis showed the diversity of gut microbiota across different growth stages, which was verified by the Shannon index (P < 0.0001). We later compared the abundance of gut microbiota in the chicken embryo at different days. The most abundant phyla in all the 3 stages were Proteobacteria, Actinobacteria, and Firmicutes, followed by Bacteroidetes and Cyanobacteria. As the chicken embryo developed with time, we observed that the proportion of the phyla also changed. At E3, the percentage of Proteobacteria was 65.5%, which decreased to 54.8% at E12 and increased exponentially at E19 (76.7%) (Supplementary Figure 1, Supplementary Table 5B–D [P < 0.05]). The proportion of Actinobacteria increased from 17.8% (E3) to 18.4% (E12), but the proportion decreased at E19 (13.4%). The proportion of Firmicutes increased from 5.3% (E3) to 10.6% (E12) and reduced to 2.3% at E19. The rate at which the phyla increased from E3 to E12 and then decreased at E19 corresponded to the proportion of Bacteroidetes (E3 [5.8%], E12 [6.7%], E19 [3.9%]) and Cyanobacteria (E3 [2.2%], E12 [3.4%], E19 [1.0%]) (Supplementary Figure 2). Likewise, time altered the microbial populations of the genera. Populations of Ochrobactrum (E3 [30.4%], E12 [25.1%], and E19 [13.6%]) and Amycolatopsis (E3 [11.5%], E12 [7.4%]. and E19 [5.6%]) reduced with growth changes (Figure 2C). In contrast, we observed an increase in the proportion of Phyllobacterium at the late phase (E19 [47.9%]), whereas less proportion was present in the early and middle stages (3.4% at E3 and 1.9% at E12) (Figure 2C; Supplementary Figure 3). Furthermore, we identified 96 genera as core microbes, which were present at E3, E12, and E19 (Figure 2D, Table 1), such as Ochrobactrum, Phyllobacterium, Amycolatopsis, Prevotella, Lactobacillus, Pseudomonas, and Sediminibacterium. Besides, 94 genera of microbes were significantly different between E3 and E12, 129 genera significantly changed between E3 and E19, and 143 genera significantly different between E12 and E19 (P < 0.05) (Supplementary Table 6A–C). Some of the genera that changed significantly across all stages were Sphingomonas, Sedimibacterium, Acinetobacter, Enterobacteriaceae, Agrobacterium, Methylobacterium, Microbacterium, Halomonas, and Aquicella. Furthermore, the linear discriminant analysis effect size method showed significant different microbial profiles at the 3 stages of the embryonic development (Figure 2E). In general, our results showed a significant and dynamic change in the gut microbiota of chicken embryos as they developed with time (Supplementary Figure 4).
Figure 2.
Embryonic gut microbial difference at different developmental stages. (A) PCA shows a high level of similarities between chicken embryos that had incubated until day 3 and 12 compared with those incubated until day 19. (B) Alpha analysis using the Shannon index showing the diversity of gut microbiota across different growth stages. (C) The relative abundance of gut microbiota in the chicken embryo at different days. (D) The Venn diagram shows the core microbes shared at different stages of development. (E) Taxonomic cladogram generated from LEfSe showing significant difference in the microbiota profile of 3 stages of development. Green, red, and blue represent the enriched taxa in chicken embryos that had been incubated until day 3, 12, and 19, respectively. Abbreviations: LEfSe, linear discriminant analysis effect size; PCA, principal component analysis.
Table 1.
Fifty core microbes that were common in all the 3 stages of chicken embryo development (E3, E12, and E19).
| Phylum | Genus | Phylum | Genus |
|---|---|---|---|
| [Thermi] | Thermus | Proteobacteria | Phyllobacterium |
| [Thermi] | B-42 | Proteobacteria | Sphingomonas |
| [Thermi] | Deinococcus | Proteobacteria | Acinetobacter |
| Actinobacteria | Amycolatopsis | Proteobacteria | Agrobacterium |
| Actinobacteria | Microbacterium | Proteobacteria | Methylobacterium |
| Actinobacteria | Corynebacterium | Proteobacteria | Delftia |
| Actinobacteria | Brevibacterium | Proteobacteria | Aminobacter |
| Actinobacteria | Micrococcus | Proteobacteria | Pseudochrobactrum |
| Actinobacteria | Propionibacterium | Proteobacteria | Achromobacter |
| Actinobacteria | Rubrobacter | Proteobacteria | Pseudomonas |
| Actinobacteria | Brachybacterium | Proteobacteria | Rhodoplanes |
| Actinobacteria | Actinomyces | Proteobacteria | Bosea |
| Actinobacteria | Bifidobacterium | Proteobacteria | Devosia |
| Actinobacteria | Arthrobacter | Proteobacteria | Comamonas |
| Armatimonadetes | Fimbriimonas | Proteobacteria | Shewanella |
| Bacteroidetes | Sediminibacterium | Proteobacteria | Bdellovibrio |
| Bacteroidetes | Prevotella | Proteobacteria | Brevundimonas |
| Bacteroidetes | Bacteroides | Proteobacteria | Enhydrobacter |
| Firmicutes | Lactobacillus | Proteobacteria | Novosphingobium |
| Firmicutes | Staphylococcus | Proteobacteria | Nelumbo |
| Firmicutes | Faecalibacterium | Proteobacteria | Serratia |
| Firmicutes | [Ruminococcus] | Proteobacteria | Sphingobium |
| Firmicutes | Streptococcus | Proteobacteria | Kaistobacter |
| Firmicutes | Bacillus | ||
| Firmicutes | Geobacillus | ||
| Fusobacteria | Fusobacterium | ||
| Proteobacteria | Ochrobactrum |
The unclassified microbes are removed.
Functional Predictions of Chicken Embryonic Gut Microbiota
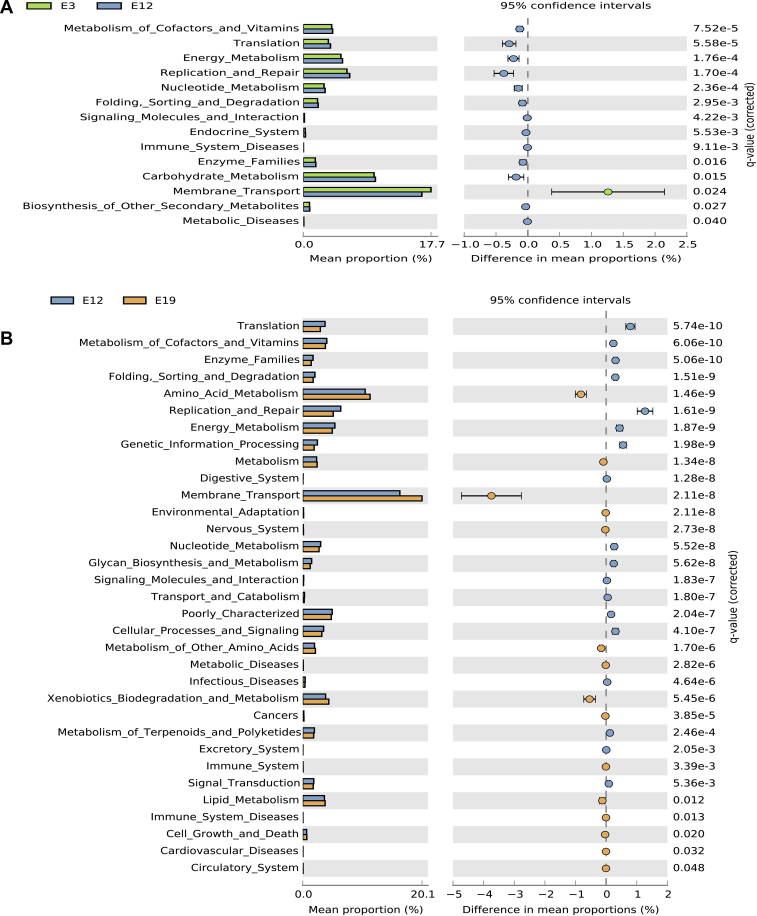
For further understanding of the metabolic profile of the microbial community, we analyzed the metagenomic functions of the bacteria. Comparing predicted microbial function between E3 and E12, we detected significant enrichment in the metabolism of cofactors, vitamins, energy, carbohydrates, nucleotides, membrane transport, translation, replication, and repair (Figure 3A). Pathways assigned for genetic information processing, amino acid metabolism, replication, translation, xenobiotic biodegradation, and metabolism were significantly enriched between E12 and E19 of chicken embryo development (Figure 3B). The abundance of functional capabilities in the developmental stages of embryo is presumed to be related to the growth of chicken embryos. The prevalence of xenobiotic degradation was impressive because this metabolic pathway links to several gut health benefits, and it involves both aerobic and anaerobic bacteria. The host's metabolism changes to promote excretion of many xenobiotics from the body and to support growth through production of energy and nutrients (Fetzner, 2002; Koppel et al., 2017).
Figure 3.
Functional profiles of the microbial community at different developmental stages (A) Functional pathways that changed significantly (P < 0.05) between E3 and E12. (B) Functional pathways that changed significantly (P < 0.05) between E12 and E19.
Other important functions that we detected in the chicken embryo belong to categories such as lipid metabolism, signal transduction, metabolism of terpenoids and polyketides, enzymes families, folding, sorting, and degradation. Notably, a significant increase in the membrane transport pathway that falls under environmental information processing metabolism dominated in the chicken embryo in all stages of development (Figures 3A, 3B). In addition, pathways related to cellular processing and signaling were detected in the chicken embryo. The functions of cellular processing and signaling pathways help in the activation of macrophages and dendritic cells in the presentation of antigens to T cells, which are known to fight against endemic diseases (Cella et al., 1997) that lead to embryonic death. Interestingly, the functional analysis also showed that the pathway allocated for genetic information processing increased in functional capabilities as the embryo grew from one stage to the other but decreased toward hatching. We also observed microbial functional pathways that are linked with diseases such as cardiovascular diseases, metabolic diseases, infectious diseases, and immune system diseases.
Embryonic Gut Microbial Composition Across Different Breeds
The microbial community diversity was also analyzed in all 3 major breeds that we used for this experiment. Both alpha and beta indices were examined. Surprisingly, the beta analysis using nonmetric multidimensional scaling–weighted analysis revealed that the samples clustered at the same location, which denoted that there were no differences between the microbiota across different breeds (Figure 4A). Likewise, the weighted and unweighted UniFrac distance from the analysis of similarities test showed no significant difference among the 3 breeds, with P = 0.156, r = 0.0212, and P = 0.275, r = 0.0106, respectively, affirming the similarities of the relative taxon abundance in the chicken embryo from different breeds (Supplementary Table 7).
Correspondingly, the Chao1 richness index, Simpson's index, and ACE index revealed no difference among the groups (Supplementary Table 8). The Shannon index showed a high level of similarities among the 3 groups (Y, S, and B) despite the difference in the genetic backgrounds (Figure 4B). In addition, we studied the composition and abundance of microbiota in the 3 breeds to deeply understand the microbial community of each breed. The results showed similar microbial populations at different proportions in all the selected breeds. At the genus level, Ochrobactrum was the most dominant in Beijing You, at the proportion of 24.8%, followed by SPF (Babcock) at 22.9% and Shiqiza at 21.2%. Phyllobacterium was the most dominant in Shiqiza, whereas it was almost of the same proportion in Beijing You and SPF (Babcock) breeds. Amycolatopsis was most dominant in the SPF (Babcock) breed, followed by Beijing You, whereas the proportion was the lowest in the Shiqiza breed (Figure 4C). According to the general phylum assignment result, phyla Proteobacteria and Firmicutes were most dominant in the Shiqiza breed, at the rate of 69.5 and 6.3%, respectively, whereas phyla Actinobacteria and Cyanobacteria were seen as most abundant in the SPF (Babcock) breed, at the rate of 20.9 and 2.8%, respectively. Bacteroidetes were observed to be dominant in Beijing You, whereas its proportion was lowest in the SPF (Babcock) breed (Figure 4D). These findings indicated similar microbial colonization in the chicken embryo among the 3 breeds.
Discussion
Lots of researchers have emphasized developmental stages from infancy to adulthood (Contreras et al., 2012; Ballou et al., 2016; Tanaka and Nakayama, 2017). In mammals, evidence is accumulating that gut microbiota change with age and stability could only be feasible in adults (Wall et al., 2009; Yatsunenko et al., 2012; Abrahamsson et al., 2014; Koleva et al., 2015; Kashtanova et al., 2016). Pan and Yu (2014) perceived similar phenomena in chickens, also stating that as chicken age increases, the gut microbiome becomes very diverse until it achieves stability, whereas few reports have focused on how the gut microbiota in chicken embryos developed and changed with time. Our study bridges this gap by examining the dynamic distribution of gut microbiota in the chicken embryo from 3 breeds at different developmental stages.
Our results showed significant changes in the gut microbiota population of the chicken embryo across developmental stages. We observed an increase in the gut microbial population as chicken embryos developed from E3 to E12. Simultaneously, the increase in the gut microbial population from E3 to E12 in the chicken embryo could be associated with different organ and system formation, which would have undergone in the chicken embryo at the early stage (Hamburger and Hamilton, 1992), which is associated with the endocrine, immune, excretory system, and genetic information pathway found in our study. Consistent with our previous study, we witnessed drastic reduction in the microbial population at E19, whereas some microbes such as those of the genus Thermogemmatisporaceae, Coprococcus, Planomicrobium, Aerococcus, Modestobacter, and Dactylosporangium were eliminated. This result supports the observation of Ding et al. (2017), who showed that substantial decline in the microbial population with the advancement in the growth stage is as a result of the genetic and environmental factors, which shortens the life span of these microbes, through continuous disappearance as chicken embryos attain hatching. More so, research has proven that on the 19th day of embryonic development, there is increase in the metabolic rate of the chicken embryo (Vleck and Vleck, 1987), which may contribute to the low abundance of microbiota species observed at this period. Besides, as organisms attain higher development gradually, the immune system becomes more complex and aggressive, whereby keeping the microbiota in an extracellular manner, making it out of reach (Rosenberg and Zilber-Rosenberg, 2013). Further evidence held the view that facultative aerobes colonized the chicken gut and later were interchanged by anaerobes (Awad et al., 2016). Possibly, the dominated gut microbiota at E3 and E12 are mainly facultative aerobes, which was substituted at the late phase, and this could probably be one of the striking reasons why most of the microbes faded away in the late phase of chicken embryonic development. In our study, it is noteworthy to mention that the 3rd and 12th day of chicken embryo development is characterized by a high proportion of gut microbiota. We envisaged that this stage in the chicken embryo is essential for microbiota development as this early colonization may contribute to the founding and maintenance of nonpathogenic gut microbiota, which is likely to play a prominent role in the growth and health of the chicken embryo, reducing embryonic diseases and death (Wall et al., 2009). However, future work is highly encouraged to validate this. The paradigm shift in the abundance of gut microbiota at different developmental stages in our study corresponds to the hologenome concept, supporting that variability in microbiota should be counted as a norm.
Across the different phases of development, the functional profiles of the microbiota changed. Meanwhile, metabolism such as energy, metabolism of vitamins, and carbohydrate metabolism significantly increased at E3 and E12 in the chicken embryo. Interestingly, we found the proportion of the genus Fluviicola, an aerobic microbe that is associated with using carbohydrates for growth (O'Sullivan et al., 2005), increased significantly at E3 and E12, which suggests that Fluviicola could be involved in the carbohydrate and energy metabolism pathway, which is significantly abundant at the early phase of chicken embryo development in our study and may be an indicator that Fluviicola helps in the growth and development of the chicken embryo. Moreover, Prevotella, a member of the core microbiota, which belongs to the phylum Bacteroidetes, has been previously studied and is known for its cellulolytic activity (Ley, 2016) and positive contribution to energy metabolism by increasing the stored glycogen (Kovatcheva-Datchary et al., 2015). Probably, the metabolic role played by Prevotella is related to the availability of glucose and amino acid, which successively promote growth in chicken embryos. Pseudomonas, another minute member of the core microbes, are reported to be involved in cellulolytic pathways mainly useful for breaking down cellulose (Huang et al., 2012). In addition, various species of Lactobacillus had been studied for its unique ability to induce differential cytokine expression in T cells of chickens in the regulation and maintenance of homeostasis in the gut (Brisbin et al., 2012; Parker et al., 2018), which may also be involved in the cellular process pathway found in our study. Further evidence revealed the importance of Lactobacillus in longevity of the life span (Ikeda et al., 2007).
In contrast to our findings, several reports showed that in human embryos, the vitamin metabolism pathway is significantly enriched in adult microbiomes compared with the microbiomes of babies; it was also reported that serum cobalamin decreased in the early stage of the human embryo, whereas it increased in the latter stage (Monsen et al., 2003; Yatsunenko et al., 2012). We found out that the vitamin metabolism pathway at E3, E12, and E19 was quite similar in its enrichment in the chicken embryo. Together, the findings suggest that most of the crucial functions are performed by the core microbiota regardless of the minute proportion of each. The richness of genetic information processing (translation) pathway at this early stage of development supports previous studies that pointed out that assurance of early development in an animal is on reliance on translation and utilization of the stored mRNA for later development (Curtis et al., 1995). The abundance of the core microbes in our study specified that chicken gut health is highly correlated to the homeostasis of its gut microbial association (Yao et al., 2018). We supposed that the 3 distinct stages of the chicken embryonic growth and development might be a major factor for the dominance of specific genera within the chicken core microbiota. Ochrobactrum, the most dominant genus in our study, has been reported to be ubiquitous and has a distinct ability to persist in the intestines under stressful conditions (Dharne et al., 2008; Dirksen et al., 2016; Kulkarni et al., 2017). A recent study emphasized the salient role of Ochrobactrum, revealing its influence on Caenorhabditis elegans energy metabolism, metabolism of specific amino acids and fatty acids, and also folate biosynthesis (Yang et al., 2019). We assumed that the richness of Ochrobactrum in the chicken embryo is as a result of its involvement in the active pathways such as energy metabolism, metabolism of vitamins and amino acids. Importantly, these functioning pathways are critical nutritional requirements for the chicken embryo; absence of these pathways might impede growth and development.
Contrary to expectations, the microbial diversity among the 3 breeds Beijing You, Shiqiza, and SPF (Babcock) used in this study was similar, although the proportion of some gut microbiota changed over time. Our most promising finding is the diverse gut microbiota harbored in SPF (Babcock) eggs, which are similar to those found in conventional eggs. Specifically, we found out that Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria dominated the chicken embryo, which is in accordance with previous studies (Ding et al., 2017; Grond et al.,2018). Rosenberg and Zilber-Rosenberge (2013) noted that this core microbiota could be supplemented with microorganisms from the environment throughout their life span. A study has justified that nearly all avian embryos are influenced by external environmental conditions and regulated by incubation conditions (Reed and Clark, 2011). Notably, the eggs from the 3 chicken breeds (Beijing you, Shiqiza, and SPF [Babcock]) have different genetic backgrounds but have similar microbial colonization. Regardless of the host genetics and environmental variations, most of the gut microbes harbored in the chick embryo are core microbes, although in different proportions.
In conclusion, our results noted time as a strong shaping factor in gut microbiota composition, and in general, similar intestinal microbiota were noticed among the 3 breeds with a significant change in the gut microbiota population as they developed with time. Our findings indicate the growing trend of microbial diversity in the chicken embryo's development and also establish a background for understanding the distribution of gut microbiota in the chicken embryo, across different developmental stages.
Acknowledgments
The authors are grateful to the National Key Research and Development Program of China (grant no. 2017YFD0500506) and the National Science Foundation of China (grant no. 31572384). The funding bodies provided funds for the purchase of samples, materials for the study and sequencing.
Conflict of Interest Statement: The authors state that the research was carried out in the absence of any commercial or financial issues that could be interpreted as a potential conflict of interest.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.06.016.
Supplementary data
References
- Aagaard K., Ma J., Antony K.M., Ganu R., Petrosino J., Versalovic J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamsson T.R., Jakobsson H.E., Andersson A.F., Björkstén B., Engstrand L., Jenmalm M.C. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy. 2014;44:842–850. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- Asnicar F., Weingart G., Tickle T.L., Huttenhower C., Segata N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ. 2015;3:e1029. doi: 10.7717/peerj.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad W.A., Mann E., Dzieciol M., Hess C., Schmitz-Esser S., Wagner M., Hess M. Age-related differences in the Luminal and mucosa-associated gut microbiome of broiler chickens and shifts associated with Campylobacter jejuni infection. Front. Cell. Infect. 2016;6:154. doi: 10.3389/fcimb.2016.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou A.L., Ali R.A., Mendoza M.A., Ellis J.C., Hassan H.M., Croom W.J., Koci M.D. Development of the chick microbiome: how early Exposure influences future microbial diversity. Front. Vet. Sci. 2016;3:1–12. doi: 10.3389/fvets.2016.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellairs R., Osmond M. Atlas of Chick Development. Academic Press; Cambridge, MA: 2014. The hen’s egg and its formation; pp. 1–6. [Google Scholar]
- Bokulich N.A., Subramanian S., Faith J.J., Gevers D., Gordon J.I., Knight R., Mills D.A., Caporaso J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisbin J.T., Parvizi P., Sharif S. Differential cytokine expression in T-cell subsets of chicken caecal tonsils co-cultured with three species of Lactobacillus. Benef. Microbes. 2012;3:205–210. doi: 10.3920/BM2012.0014. [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pẽa A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M., Sallusto F., Lanzavecchia A. Origin, maturation and antigen presenting function of dendritic cells. Curr. Opin. Immunol. 1997;9:10–16. doi: 10.1016/s0952-7915(97)80153-7. [DOI] [PubMed] [Google Scholar]
- Chao A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984;11:265–270. [Google Scholar]
- Chao A., Shen T.J. Nonparametric prediction in species sampling. J. Agric. Biol. Environ. Stat. 2004;9:253–269. [Google Scholar]
- Cisek A.A., Binek M. Chicken intestinal microbiota function with a special emphasis on the role of probiotic bacteria. Pol. J. Vet. Sci. 2014;17:385–394. doi: 10.2478/pjvs-2014-0057. [DOI] [PubMed] [Google Scholar]
- Clarke K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993;18:117–143. [Google Scholar]
- Clavijo V., Flórez M.J.V. The gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: a review. Poult. Sci. 2018;97:1006–1021. doi: 10.3382/ps/pex359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M.C., Rautava S., Aakko J., Isolauri E., Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016;6:23129. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras M., Magris M., Hidalgo G., Robert N., Anokhin A.P., Heath A.C., Warner B., Reeder J., Kuczynski J., Caporaso J.G., Lozupone C.A., Lauber C., Clemente J.C., Knights D., Knight R., Gordon J.I. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D., Lehmann R., Zamore P.D. Translational regulation in development. Cell. 1995;81:171–178. doi: 10.1016/0092-8674(95)90325-9. [DOI] [PubMed] [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharne M.S., Misra S.P., Misra V., Dwivedi M., Patole M.S., Shouche Y.S. Isolation of urease-positive Ochrobactrum intermedium in the stomach of a non-ulcer dyspeptic patient from north India. J. Microbiol. Immunol. Infect. 2008;41:183–186. [PubMed] [Google Scholar]
- Ding J., Dai R., Yang L., He C., Xu K., Liu S., Zhao W., Xiao L., Luo L., Zhang Y., Meng H. Inheritance and establishment of gut microbiota in chickens. Front. Microbiol. 2017;8:1–11. doi: 10.3389/fmicb.2017.01967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen P., Marsh S.A., Braker I., Heitland N., Wagner S., Nakad R., Mader S., Petersen C., Kowallik V., Rosenstiel P., Félix M.A., Schulenburg H. The native microbiome of the nematode Caenorhabditis elegans: Gateway to a new host-microbiome model. BMC Biol. 2016;14:38. doi: 10.1186/s12915-016-0258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello M.G., Godoy-Vitorino F., Knight R., Blaser M.J. Role of the microbiome in human development. Gut. 2019;68:1108–1114. doi: 10.1136/gutjnl-2018-317503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetzner S. Biodegradation of xenobiotics. In: Doelle H.W., Rokem J.S., Berovic M., editors. Biotechnology Volume 10, Encyclopedia of Life Support Systems (EOLSS) UNESCO, Eolss Publishers; Paris, France: 2002. pp. 1–8. [Google Scholar]
- Fontaine S.S., Novarro A.J., Kohl K.D. Environmental temperature alters the digestive performance and gut microbiota of a terrestrial amphibian. J. Exp. Biol. 2018;221:jeb187559. doi: 10.1242/jeb.187559. [DOI] [PubMed] [Google Scholar]
- Fouhse J.M., Zijlstra R.T., Willing B.P. The role of gut microbiota in the health and disease of pigs. Anim. Front. 2016;6:30–36. [Google Scholar]
- Funkhouser L.J., Bordenstein S.R. Mom Knows best: the Universality of maternal microbial Transmission. PLoS Biol. 2013;11:e1001631. doi: 10.1371/journal.pbio.1001631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Forster R.J., Yu H., Chambers J.R., Sabour P.M., Wheatcroft R., Chen S. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiol. Lett. 2002;208:1–7. doi: 10.1111/j.1574-6968.2002.tb11051.x. [DOI] [PubMed] [Google Scholar]
- Grond K., Sandercock B.K., Jumpponen A., Zeglin L.H. The avian gut microbiota: community, physiology and function in wild birds. J. Avian Biol. 2018;49:1–19. [Google Scholar]
- Haas B.J., Gevers D., Earl A.M., Feldgarden M., Ward D.V., Giannoukos G., Ciulla D., Tabbaa D., Highlander S.K., Sodergren E., Methé B., DeSantis T.Z., Petrosino J.F., Knight R., Birren B.W. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V., Hamilton H.L. A series of normal stages in the development of the chick embryo. Dev. Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- He S., Ivanova N., Kirton E., Allgaier M., Bergin C., Scheffrahn R.H., Kyrpides N.C., Warnecke F., Tringe S.G., Hugenholtz P. Comparative metagenomic and Metatranscriptomic analysis of Hindgut Paunch microbiota in Wood- and Dung-Feeding higher Termites. PLoS One. 2013;8:e61126. doi: 10.1371/journal.pone.0061126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Sheng P., Zhang H. Isolation and identification of cellulolytic bacteria from the gut of holotrichia parallela larvae (Coleoptera: Scarabaeidae) Int. J. Mol. Sci. 2012;13:2563–2577. doi: 10.3390/ijms13032563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T., Yasui C., Hoshino K., Arikawa K., Nishikawa Y. Influence of lactic acid bacteria on longevity of Caenorhabditis elegans and host defense against Salmonella enterica serovar Enteritidis. Appl. Environ. Microbiol. 2007;73:6404–6409. doi: 10.1128/AEM.00704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashtanova D.A., Popenko A.S., Tkacheva O.N., Tyakht A.B., Alexeev D.G., Boytsov S.A. Association between the gut microbiota and diet: fetal life, early childhood, and further life. Nutrition. 2016;32:620–627. doi: 10.1016/j.nut.2015.12.037. [DOI] [PubMed] [Google Scholar]
- Koleva P.T., Kim J.S., Scott J.A., Kozyrskyj A.L. Microbial programming of health and disease starts during fetal life. Birth Defects Res. Part C - Embryo Today Rev. 2015;105:265–277. doi: 10.1002/bdrc.21117. [DOI] [PubMed] [Google Scholar]
- Koppel N., Rekdal V.M., Balskus E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science. 2017;356:1246–1257. doi: 10.1126/science.aag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovatcheva-Datchary P., Nilsson A., Akrami R., Lee Y.S., De Vadder F., Arora T., Hallen A., Martens E., Björck I., Bäckhed F. Dietary Fiber-Induced Improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015;22:971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Kulkarni G., Gohil K., Misra V., Kakrani A.L., Misra S.P., Patole M., Shouche Y., Dharne M. Multilocus sequence typing of Ochrobactrum spp. isolated from gastric niche. J. Infect. Public Health. 2017;10:201–210. doi: 10.1016/j.jiph.2016.04.013. [DOI] [PubMed] [Google Scholar]
- Langille M.G.I., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., Clemente J.C., Burkepile D.E., Vega Thurber R.L., Knight R., Beiko R.G., Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., La T.M., Lee H.J., Choi I.S., Song C.S., Park S.Y., Lee J.B., Lee S.W. Characterization of microbial communities in the chicken oviduct and the origin of chicken embryo gut microbiota. Sci. Rep. 2019;9:6838. doi: 10.1038/s41598-019-43280-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E. Prevotella in the gut: choose carefully. Nat. Rev. Gastroenterol. Hepatol. 2016;13:69–70. doi: 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- Magoč T., Salzberg S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsen A.L.B., Refsum H., Markestad T., Ueland P.M. Cobalamin status and its Biochemical Markers Methylmalonic acid and Homocysteine in different age groups from 4 Days to 19 Years. Clin. Chem. 2003;49:2067–2075. doi: 10.1373/clinchem.2003.019869. [DOI] [PubMed] [Google Scholar]
- O’Sullivan L.A., Rinna J., Humphreys G., Weightman A.J., Fry J.C. Fluviicola taffensis gen. nov., sp. nov., a novel freshwater bacterium of the family Cryomorphaceae in the phylum “Bacteroidetes. Int. J. Syst. Evol. Microbiol. 2005;55:2189–2194. doi: 10.1099/ijs.0.63736-0. [DOI] [PubMed] [Google Scholar]
- Oliveros J.C. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007. http://bioinfogp.cnb.csic.es/tools/venny/index.html. bioinfogp.cnb.csic.es/tools/venny/index.html Accessed July 2020.
- Palmer C., Bik E.M., DiGiulio D.B., Relman D.A., Brown P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D., Yu Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes. 2014;5:108–119. doi: 10.4161/gmic.26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker A., Lawson M.A.E., Vaux L., Pin C. Host-microbe interaction in the gastrointestinal tract. Environ. Microbiol. 2018;20:2337–2353. doi: 10.1111/1462-2920.13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D.H., Beiko R.G. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26:715–721. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- Penders J., Thijs C., Vink C., Stelma F.F., Snijders B., Kummeling I., Van Den Brandt P.A., Stobberingh E.E. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- Reed W.L., Clark M.E. Beyond maternal effects in birds: Responses of the embryo to the environment. Integr. Comp. Biol. 2011;51:73-80. doi: 10.1093/icb/icr032. [DOI] [PubMed] [Google Scholar]
- Rodríguez J.M., Avershina E., Ross R.P., Murphy K., Stanton C., Rudi K., Kober O.I., Marchesi J.R., Narbad A., Juge N., Collado M.C., Jenmalm M.C. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Heal. Dis. 2015;26:26050. doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E., Zilber-Rosenberg I. Springer International Publishing; Switzerland: 2013. The Hologenome Concept: Human, Animal and Plant Microbiota. [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant M.J., Constantinidou C., Cogan T.A., Bedford M.R., Penn C.W., Pallen M.J. Extensive microbial and functional diversity within the chicken cecal microbiome. PLoS One. 2014;9:e91941. doi: 10.1371/journal.pone.0091941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:623–656. [Google Scholar]
- Shreiner A.B., Kao J.Y., B Y.V. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2016;31:69–75. doi: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson E.H. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- Tanaka M., Nakayama J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. 2017;66:515–522. doi: 10.1016/j.alit.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Thomas M., Wongkuna S., Ghimire S., Kumar R., Antony L., Doerner K.C., Singery A., Nelson E., Woyengo T., Chankhamhaengdecha S., Janvilisri T., Scaria J. Gut microbial dynamics during Conventionalization of Germfree chicken. mSphere. 2019;4:1–12. doi: 10.1128/mSphere.00035-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thursby E., Juge N. Introduction to the human gut microbiota. Biochem. J. 2017;474:1823–1836. doi: 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., Sogin M.L., Jones W.J., Roe B.A., Affourtit J.P., Egholm M., Henrissat B., Heath A.C., Knight R., Gordon J.I. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleck C.M., Vleck D. Metabolism and energetics of avian embryos. J. Exp. Zool. Suppl. 1987;1:111–125. [PubMed] [Google Scholar]
- Wall R., Ross R., Ryan C., Hussey S., Murphy B., Fitzgerald G., Stanton C. Role of gut microbiota in early infant development. Clin. Med. Pediatr. 2009;3:45–54. doi: 10.4137/cmped.s2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warton D.I., Wright S.T., Wang Y. Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol. Evol. 2012;3:89–101. [Google Scholar]
- Wei S., Morrison M., Yu Z. Bacterial census of poultry intestinal microbiome. Poult. Sci. 2013;92:671–683. doi: 10.3382/ps.2012-02822. [DOI] [PubMed] [Google Scholar]
- Welch B.L. The generalisation of student’s problems when several different population variances are involved. Biometrika. 1947;34:28–35. doi: 10.1093/biomet/34.1-2.28. [DOI] [PubMed] [Google Scholar]
- Weng M., Walker W.A. The role of gut microbiota in programming the immune phenotype. J. Dev. Orig. Health Dis. 2013;4:1–23. doi: 10.1017/S2040174412000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Wu Y., Li J., Bao Y., Guo Y., Yang W. The dynamic changes of gut microbiota in muc2 deficient mice. Int. J. Mol. Sci. 2018;19:2809. doi: 10.3390/ijms19092809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Petersen C., Pees B., Zimmermann J., Waschina S., Dirksen P., Rosenstiel P., Tholey A., Leippe M., Dierking K., Kaleta C., Schulenburg H. The inducible Response of the nematode Caenorhabditis elegans to members of its Natural microbiota across development and adult life. Front. Microbiol. 2019;10:1793. doi: 10.3389/fmicb.2019.01793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M., Magris M., Hidalgo G., Baldassano R.N., Anokhin A.P., Heath A.C., Warner B., Reeder J., Kuczynski J., Caporaso J.G., Lozupone C.A., Lauber C., Clemente J.C., Knights D., Night R., Gordon J.I. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Yang K., Huang L., Huang X., Qiuqian L., Wang K., Zhang D. Disease outbreak accompanies the dispersive structure of shrimp gut bacterial community with a simple core microbiota. AMB Expr. 2018;8:120. doi: 10.1186/s13568-018-0644-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Wang G., Siegel P., He C., Wang H., Zhao W., Zhai Z., Tian F., Zhao J., Zhang H., Sun Z., Chen W., Zhang Y., Meng H. Quantitative genetic background of the host influences gut microbiomes in chickens. Sci. Rep. 2013;3:1163. doi: 10.1038/srep01163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.