Abstract
Clostridium perfringens is an important zoonotic microorganism. The present study was undertaken to investigate prevalence, serotype distribution, antibiotic resistance, and genetic diversity of C. perfringens isolates from 4 duck farms in Shandong, China. In total, 424 samples of cloacal swabs and environment were collected from 3 commercial meat-type duck farms in Tai'an, Liaocheng, and Weifang and one breeder duck farm in Liaocheng between December 2018 and June 2019, of which, 207 (48.82%) samples were determined to be positive for C. perfringens; a total of 402 isolates of C. perfringens were recovered, all of which were identified as type A; 30.85% of the isolates were positive for cpb2 gene; and cpe gene was found in 0.5% of the isolates. Antimicrobial susceptibility testing revealed that some of the isolates exhibited high antibiotic resistance, and 39.14% of the isolates were resistant to at least 5 classes of commonly used antibiotics. Multilocus sequence typing analysis showed that 85 representative isolates encompassed 54 different sequences types (STs), clustered in 5 clonal complexes (CCs) and 40 singletons. ST3, the most common ST in 54 STs, constituting 15.29% of all isolates, was also the most prevalent ST of isolates from the Liaocheng breeder duck farm (farm 3). CC1, the most prolific CC, containing 15.29% of the analyzed isolates, was the popular subtype of isolates from Liaocheng meat duck farm (farm 2). Although all the isolates belong to type A, the genetic diversity varied greatly in different regions; the Simpson's Diversity Index of STs for Liaocheng, Tai'an, and Weifang were 0.5941, 0.9198, and 0.9627, respectively. Some of cloacal isolates and environmental isolates were distributed in the same ST or CC, indicating close genetic relationship between cloacal isolates and environmental isolates. A portion of the strains from humans and ducks was found to be phylogenetically close. The close relationship between strains from humans and ducks, the high antibiotic resistance of C. perfringens, and the cpe-positive isolates indicated potential public health risks.
Key words: Clostridium perfringens, duck, antimicrobial resistance, multilocus sequence typing, phylogenetics
Introduction
Clostridium perfringens (C. perfringens) is an anaerobic, spore forming, gram-positive pathogen (Abakabir Mahamat et al., 2019), which is widely found in the digestive tract of humans and animals, as well as in soil, sewage, sediment, and feces, and can be transmitted horizontally through the environment (Hibberd et al., 2011; Abakabir Mahamat et al., 2019). C. perfringens causes a variety of diseases in animals and humans, such as necrotic enteritis (NE) in poultry and gas gangrene and food poisoning in humans (Guran and Oksuztepe, 2013). The pathogenicity of C. perfringens is largely attributable to its ability to produce a wide variety of exotoxins and enzymes, among which, toxins alpha (α), beta (β), epsilon (ε), and iota (ι) are the major lethal toxins, which are encoded by cpa, cpb, etx, and iap genes, respectively. According to these 4 toxins, C. perfringens strains are classified into 5 pathotypes (Miyamoto et al., 2004; Fohler et al., 2016), including type A (α), B (α, β, ε), C (ɑ, β), D (α, ε), and E (ɑ, ι).
C. perfringens can not only cause NE, increasing mortality, and growth retardation in poultry but also lead to contaminating along the slaughtering and processing chain, increasing the risk of food poisoning or digestive tract disease in humans. There have been previous reports of C. perfringens isolated from chicken, but few publications had evaluated prevalence and molecular characteristics of C. perfringens isolated from duck farms. China is one of the top duck-consuming countries. According to the data provided by the Food and Agriculture Organization, there were about 5.0 billion meat ducks in the world in 2013, and China accounted for more than 40% of the global total, making China the country with the largest meat duck production in the world. According to the statistics and calculations of China Animal Agriculture Association, the production of ducks in Shandong province accounts for more than 30% of the national production of ducks, and Shandong province is the largest meat duck–breeding base in China.
Serotyping is a classical method for classification of C. perfringens; the most reported serotype in poultry is type A, but this method cannot further classify these strains into subtypes. Multilocus sequence typing (MLST) is a genotyping method based on the nucleotide sequences of several pairs of housekeeping genes which are amplified by PCR; after sequencing, the obtained alleles were uniformed with reference to standard sequence (Urwin and Maiden, 2003). As an effective method to solve bacterial population genetics, MLST is highly reproducible and portable (Cao and Wei, 2012); sequence data can be held through a central database and queried through a Web server (Chan et al., 2001). Hence, this approach is of great value for genotyping and tracking of pathogens.
The detection of virulence genes is an important supplement to the evaluation of population phylogenetic characteristics (Nakano et al., 2017). C. perfringens enterotoxins and beta-2 toxins (β2), encoded by cpe and cpb2 genes, respectively, are considered to be significantly associated with human and animal intestinal diseases such as antibiotic-associated diarrhea, NE, and food poisoning (Lindström et al., 2011; Hu et al., 2018; Zhang et al., 2018). In addition, netB toxin produced by C. perfringens can cause NE in poultry (Keyburn et al., 2008; Shojadoost et al., 2012). Another toxin, TpeL, is also a potential virulence factor of NE. In a recent study, inoculation of broilers with both TpeL and netB positive strains was more likely to induce intestinal lesions typical of NE (Coursodon et al., 2012; Bailey et al., 2013).
Antibiotics can prevent disease and promote the growth of livestock and poultry (Wang et al., 2019). With the frequent usage of antibiotics, animal intestinal flora generates high antimicrobial resistance, which not only causes great difficulties in clinical treatment but also seriously threatens public health (Osman and Elhariri, 2013). The use of antibiotics varies greatly in different countries and regions, and limited information is available on the antibiotic resistance of C. perfringens from duck farms in China, so it is of great significance to investigate the antibiotic resistance of C. perfringens in different regions for effective control of diseases caused by C. perfringens and also provide data for public health.
This study was undertaken to investigate the prevalence, serotype distribution, virulence gene, antibiotic resistance, and genetic diversity of C. perfringens isolated from some duck farms in Shandong province and analyze genetic relationship of isolates from duck, environment, and humans. Hence, this epidemiological investigation of C. perfringens from duck farms was not only undertaken to provide reference for controlling duck diseases associated with this microorganism but also to provide data for public food safety and public health assessment.
Materials and methods
Sample Collection
In total, 317 cloacal swab samples of Cherry Valley ducks and 107 environmental samples from 3 commercial meat-type duck farms of Tai'an (farm 1), Liaocheng (farm 2), and Weifang (farm 4) and one breeder duck farm of Liaocheng (farm 3) were collected between December 2018 and June 2019 (Table 1). Environmental samples included water samples aseptically collected from nipple drinking fountains, feed samples, soil samples around the duck houses, duck feces samples in the environment, and other environmental samples (the PBS swabs of nets and troughs). Cherry Valley ducks were raised in all 4 farms, among which, commercial meat-type ducks (4–6 wk old) were raised by farm 1, 2, and 4; breeder ducks were raised (9 wk old) by farm 3. The feeding methods of 4 farms were different: In farm 1 and farm 2, ducks were raised on the plastic mesh (Plastic mesh was installed 100 cm above the floor.), while farm 3 and farm 4 were rearing on floor (The feeding ground was covered with soil and litter.). Antibiotics were used differently between the 4 farms: Ampicillin and lincomycin had been used as growth promoters by farm 1, and no antibiotics had been used by farm 2; farm 3 had a history of using antibiotics (cefepime, enoxacin, and tetracycline) to prevent and treat diarrhea in the previous 4 wk; ampicillin, florfenicol, lincomycin, neomycin, and amoxicillin had been used as growth promoters by farm 4. On each farm and sampling occasion, samples were taken randomly. Fresh cloacal swab samples and environmental samples were placed in fluid thioglycollate medium (FTG) broth immediately after collection. Samples were transported to the laboratory within 4 h in a freezer box.
Table 1.
The number of samples and positive rate of Clostridium perfringens from different samples.
| Source | No. (%) of cloacal swab samples | No. (%) of environmental samples |
No. (%) of all samples | No. of isolates1 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Feed | Water | Soil | Duck feces | Others | ||||
| TA (Farm 1) | 112 (49.12)a | 30 (23.33)b | 4 (50.00) | 8 (12.50) | 4 (25.00) | 4 (75.00) | 10 (0.00) | 142 (43.66)b | 120 |
| LC (Farm 2) | 40 (72.50)b | 11 (81.82)a | 2 (100.00) | 4 (50.00) | — | 2 (100.00) | 3 (100.00) | 51 (74.51)a | 96 |
| LC (Farm 3) | 55 (50.91)a | 11 (36.36)b | 2 (100.00) | 4 (50.00) | — | 2 (0.00) | 3 (0.00) | 66 (48.48)b | 70 |
| WF (Farm 4) | 110 (41.82)a | 55 (52.73)b | 17 (70.59) | 15 (6.67) | 3 (100.00) | 7 (100.00) | 13 (46.15) | 165 (45.45)b | 116 |
| In total | 317 (49.84)a | 107 (45.79)b | 25 (72.00) | 31 (19.35) | 7 (57.14) | 15 (80.00) | 29 (31.03) | 424 (48.82)b | 402 |
a,bMeans in the same column with different lowercase letters are significantly different (P < 0.05).
Abbreviations: LC, Liaocheng; TA, Tai'an; WF, Weifang.
At least 1 C. perfringens was collected from each positive sample.
Isolation and Identification of C. perfringens
The FTG broth containing the samples was incubated in anaerobic condition (90% N2, 10% CO2) for 8 h at 42°C with shaking at 180 rpm. Subsequently, the strains were obtained by plating the preenriched FTG broth on Tryptose Sulfite Cycloserine agar base (TSC) (Qingdao Hope Bio-Technology Co., Ltd., Qingdao, China) and then purified on a 5% defibrinated sheep blood agar (Qingdao Hope Bio-Technology Co., Ltd., Qingdao, China) and incubated anaerobically at 37°C for 24 h. C. perfringens was identified by colony morphology, Gram staining, and hemolytic characteristics (black colonies on TSC agar, gram-positive bacterium under a microscope, dual hemolysis on sheep blood agar). Two to 5 colonies suspected to be C. perfringens on TSC agar were identified and then purified on the sheep blood agar. All obtained strains were used for pathotyping and toxin genes detection; at least one strain from each positive sample was randomly selected for antimicrobial susceptibility tests, and strains for MLST were selected according to the antibiotic resistance profiles and origins (Mwangi et al., 2018).
DNA Extraction
The boiling technique was used to extract DNA from all the isolates. In total, 1 mL of bacterial suspension was transferred into a 1.5-mL eppendorf tube, centrifuged at 12,000 × g for 5 min. After discarding the supernatant, 100 μL of sterile double-distilled water was added to the eppendorf tube, boiled at 100°C for 10 min, and centrifuged at 12,000 × g for 5 to 8 min. Then, the supernatant was recovered.
Antimicrobial Susceptibility Test
Antibiotic susceptibility of C. perfringens was carried out using the Kirby-Bauer disk diffusion method (Xing et al., 2015), in accordance with the guidelines of British Society for Antimicrobial Chemotherapy (BSAC, 2015). A panel of 12 antibiotic discs (Hangzhou Microbial Reagent Co., Ltd., Hangzhou, China) were used in this study: penicillin (1 UI), cefotaxime (30 μg), cefepime (30 μg), imipenem (10 μg), florfenicol (30 μg), fosfomycin (200 μg), norfloxacin (5 μg), lincomycin (30 μg), erythromycin (15 μg), tetracycline (30 μg), bacitracin (10 μg), and gentamycin (10 μg). C. perfringens reference strain ATCC13124 was used as a quality control strain for antimicrobial susceptibility test.
Toxin Gene Detection
The isolates were characterized for the presence of cpa, cpb, etx, and iap genes by using a previously published multiplex PCR assay (Yoo et al., 1997), and isolates were also detected for the presence of cpb2, cpe, TpeL, and netB genes (Bailey et al., 2013; Hu et al., 2018). In this study, reference strains, C. perfringens NCTC 528 (cpa), NCTC 3180 (cpb), NCTC 4989 (cpb, cpb2), NCTC 8346 (etx), and NCTC 8084 (iap, cpe), were used as positive controls in the multiplex PCR.
Sequencing of Housekeeping Genes
The primers of 8 housekeeping genes ddla, dut, glpk, gmk, plc, sod, recA, and tpiA were synthesized by using the MLST scheme developed by Jost et al. (2006). PCR conditions for the 8 housekeeping genes were as described in previous studies (Liu et al., 2020). The PCR products were submitted to the sequencing company (Tsingke Biological Technology Company, Co., Ltd., Beijing, China) for sample purification and automated nucleotide sequencing in both directions.
Multilocus Sequence Typing and Evolutionary Relationship Analysis
Genetic relationship of 85 strains of C. perfringens was analyzed using MLST. Among these 85 strains, 57 strains were cloacal sources, and 28 strains were environmental sources. Eight housekeeping genes successfully sequenced by bidirectional sequencing were assembled by the DNASTAR 8.0 software package (available at http://www.dnastar.com), and ambiguities were resolved during assembling, after which, all examined genes were aligned and trimmed to an equal length by using the BioEdit software (available at http://bioedit.software.informer.com) according to the reference sequence of each allele. After assembling, data of all examined genes (fasta files) were imported into the BioNumerics software (Bionumerics, version 7.6 (3); Applied Maths, Inc., Austin, TX) to create an allele database. Sequence types (STs) were arbitrarily assigned on the basis of unique allelic profiles. Clonal complexes (CCs) were defined as groups of independent isolates that shared identical alleles at 7 or more of the 8 loci (in this study), and each CC was arbitrarily assigned a number. Both STs and CCs were considered to be C. perfringens MLST subtypes (Hibberd et al., 2011).
Besides, the START2 software package (http://pubmlst.org/software/analysis/start2/) was used to assemble and analyze concatenated sequence information for each ST. Based on a representative of each ST, the Maynard-Smith index of association (IA) was calculated to evaluate the recombination, and the ratio of synonymous to nonsynonymous mutations (dN/dS) was computed by the Nei-Gojobori method as a measure of selection (Nei and Gojobori, 1986). Concatenated sequence data for a representative of each distinct ST were imported into the MEGA 7.0 software package (http://www.megasoftware.net/) to examine the strain and ST relatedness at the sequence-level resolution. After complete deletion of alignment gaps, a total of 2,449 bp positions were used in each concatenated sequence as a data set for phylogeny calculations. An evolutionary phylogeny was constructed in MEGA 7.0 using the neighbor-joining method and maximum composite likelihood to estimate evolutionary distances (1,500 replicates), and the topology was validated by bootstrapping (Saitou and Nei, 1978; Tamura et al., 2004). To display antibiotic resistance profiles of examined isolates, each evolutionary cluster was attached to the corresponding resistance profile (heat map), which was constructed by using an online software (https://evolgenius.info/evolview-v2/). For comparison, 10 strains of C. perfringens sequences from broilers with NE previously analyzed by Hibberd et al. (2011) and Nakano et al. (2017) (ST21, ST27, and ST29 of Nakano's study; ST31, ST32, ST34, and ST39 of Hibberd's study) were also used for MLST in the minimum spanning tree. In addition, 7 strains of C. perfringens isolated from healthy humans reported (ST6, ST36 to ST41) by Liu et al. (2020) were also included.
Statistical Analysis and Simpson's Diversity Index
The positive rate and pairwise correlation between antibiotic resistance of C. perfringens samples collected from different duck farms were compared using a chi-square test. All analyses were performed by means of IBM SPSS Statistics 19 (SPSS Inc., Chicago, IL). P < 0.05 was considered as statistically significant. The genetic diversity of isolates from different regions was compared with the Simpson's Diversity Index (D) (Hunter and Gaston, 1988; Snelling et al. 1996).
Results
Occurrence of C. perfringens
The prevalence of C. perfringens from different sources in some duck farms in the Shandong province of China is shown in Table 1. Among the 424 samples, 207 samples (48.82%) were confirmed to be positive of C. perfringens. In total, 158 of 317 (49.84%) samples were positive in cloacal samples, and 49 of 72 (45.79%) samples were positive in environmental samples. Among the environmental samples, C. perfringens was isolated from 18 feed samples (72.00%) and 6 drinking water samples (19.35%); the percentage of samples that tested positive from soil, duck feces, and other environmental samples reached 57.14% (4/7), 80.00% (12/15), and 31.03% (9/29), respectively.
Among the 4 farms, farm 2 had the highest positive rate of 75.51% (38/51), whereas farm 1 showed the lowest positive rate of 43.66% (62/142). In terms of cloacal samples, the positive rates of farm 2, 3, 1, and 4 were 72.5% (29/40), 50.91% (28/55), 49.12% (55/112), and 41.82% (46/110), respectively. In addition, farm 2 had the highest positive rate of C. perfringens occurrence (81.82%) among the environmental samples. The results of the statistical analysis showed that the positive rate of isolates (including cloacal positive rate, environmental positive rate, and total positive rate) in farm 2 was significantly different from that in other farms (P < 0.01). At least one and at most 4°C. perfringens isolates from each positive sample were identified. A total of 402 isolates were obtained in all positive samples (Table 1)
Toxin Gene Screening
All isolates (n = 402) of different origins were identified as C. perfringens type A, which means that cpb, etx, and iap genes were not detected in all isolates. The cpb2 prevalence in all C. perfringens isolates was 30.85% (124/402). The detection rate of the cpb2 gene in farm 1 was the highest (40.83%; 49/120), followed by farm 3 (35.71%; 25/70), farm 4 (27.59%; 32/116), and farm 2 (18.75%; 18/96). In our study, the cpe gene was detected in 1°C. perfringens strain isolated from cloacal samples of farm 1 and one strain isolated from environmental samples of farm 4, respectively. The cpe prevalence in all C. perfringens isolates was 0.5% (2/402). The netB and TpeL toxin genes were not detected in all strains (Table 2, only the strains used for MLST are displayed).
Table 2.
Strain number, source, clonal complex (CC), sequence type (ST), and toxin genes.
| CC | ST | Strains | Source | Farm, region | Toxin genes |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cpa | cpb | etx | iap | cpb2 | cpe | netB | TpeL | |||||
| CC1 | ST36 | G2 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − |
| CC1 | ST36 | B8 | Cloaca | Farm 1, Tai'an | + | − | − | − | − | − | − | − |
| CC1 | ST36 | C24 | Cloaca | Farm 1, Tai'an | + | − | − | − | + | − | − | − |
| CC1 | ST36 | BS3 | Water | Farm 1, Tai'an | + | − | − | − | − | − | − | − |
| CC1 | ST49 | D10 | Cloaca | Farm 2, Liaocheng | + | − | − | − | − | − | − | − |
| CC1 | ST49 | D11 | Cloaca | Farm 2, Liaocheng | + | − | − | − | − | − | − | − |
| CC1 | ST49 | D14 | Cloaca | Farm 2, Liaocheng | + | − | − | − | − | − | − | − |
| CC1 | ST49 | D19 | Cloaca | Farm 2, Liaocheng | + | − | − | − | − | − | − | − |
| CC1 | ST49 | D21 | Cloaca | Farm 2, Liaocheng | + | − | − | − | − | − | − | − |
| CC1 | ST49 | D28 | Cloaca | Farm 2, Liaocheng | + | − | − | − | − | − | − | − |
| CC1 | ST49 | DS1 | Water | Farm 2, Liaocheng | + | − | − | − | − | − | − | − |
| CC1 | ST49 | DS2 | Water | Farm 2, Liaocheng | + | − | − | − | − | − | − | − |
| CC1 | ST49 | ES1 | Water | Farm 3, Liaocheng | + | − | − | − | − | − | − | − |
| CC2 | ST64 | A20 | Cloaca | Farm 1, Tai'an | + | − | − | − | − | − | − | − |
| CC2 | ST65 | A25 | Cloaca | Farm 1, Tai'an | + | − | − | − | − | − | − | − |
| CC2 | ST65 | BSL1 | Feed | Farm 1, Tai'an | + | − | − | − | − | − | − | − |
| CC2 | ST68 | B15 | Cloaca | Farm 1, Tai'an | + | − | − | − | + | − | − | − |
| CC3 | ST39 | G47 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − |
| CC3 | ST20 | 2G2 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − |
| CC3 | ST30 | 3G18 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − |
| CC4 | ST38 | G32 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − |
| CC4 | ST34 | 3GSL1 | Feed | Farm 4, Weifang | + | − | − | − | + | − | − | − |
| CC4 | ST53 | DDF1 | Duck feces | Farm 2, Liaocheng | + | − | − | − | − | − | − | − |
| CC5 | ST47 | D2 | Cloaca | Farm 2, Liaocheng | + | − | − | − | + | − | − | − |
| CC5 | ST21 | 2G5 | Cloaca | Farm 4, Weifang | + | − | − | − | + | − | − | − |
| CC5 | ST66 | ATR1 | Soil | Farm 1, Tai'an | + | − | − | − | − | − | − | − |
| Singletons1 | ST3 | E1 | Cloaca | Farm 3, Liaocheng | + | − | − | − | − | − | − | − |
| ST3 | E3 | Cloaca | Farm 3, Liaocheng | + | − | − | − | − | − | − | − | |
| ST3 | E4 | Cloaca | Farm 3, Liaocheng | + | − | − | − | − | − | − | − | |
| ST3 | E5 | Cloaca | Farm 3, Liaocheng | + | − | − | − | − | − | − | − | |
| ST3 | E8 | Cloaca | Farm 3, Liaocheng | + | − | − | − | + | − | − | − | |
| ST3 | E16 | Cloaca | Farm 3, Liaocheng | + | − | − | − | − | − | − | − | |
| ST3 | E22 | Cloaca | Farm 3, Liaocheng | + | − | − | − | + | − | − | − | |
| ST3 | E30 | Cloaca | Farm 3, Liaocheng | + | − | − | − | − | − | − | − | |
| ST3 | E35 | Cloaca | Farm 3, Liaocheng | + | − | − | − | − | − | − | − | |
| ST3 | ES2 | Water | Farm 3, Liaocheng | + | − | − | − | − | − | − | − | |
| ST3 | 3G19 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST3 | 2GDL1 | Duck feces | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST3 | GTR3 | Soil | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST8 | 3G17 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST8 | 3G28 | Cloaca | Farm 4, Weifang | + | − | − | − | + | − | − | − | |
| ST8 | B18 | Cloaca | Farm 1, Tai'an | + | − | − | − | − | − | − | − | |
| ST31 | 3G26 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST31 | A22 | Cloaca | Farm 1, Tai'an | + | − | − | − | + | + | − | − | |
| ST19 | 3G11 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST19 | 3GDF2 | Duck feces | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST24 | 2G25 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST24 | 2GWF3 | Net | Farm 4, Weifang | + | − | − | − | + | − | − | − | |
| ST37 | G4 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST37 | G11 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST54 | DSL1 | Feed | Farm 2, Liaocheng | + | − | − | − | + | − | − | − | |
| ST54 | ESL1 | Feed | Farm 3, Liaocheng | + | − | − | − | − | − | − | − | |
| ST59 | C5 | Cloaca | Farm 1, Tai'an | + | − | − | − | − | − | − | − | |
| ST60 | C36 | Cloaca | Farm 1, Tai'an | + | − | − | − | + | − | − | − | |
| ST61 | C38 | Cloaca | Farm 1, Tai'an | + | − | − | − | + | − | − | − | |
| ST62 | A5 | Cloaca | Farm 1, Tai'an | + | − | − | − | − | − | − | − | |
| ST67 | B12 | Cloaca | Farm 1, Tai'an | + | − | − | − | + | − | − | − | |
| ST69 | C3 | Cloaca | Farm 1, Tai'an | + | − | − | − | − | − | − | − | |
| ST70 | C8 | Cloaca | Farm 1, Tai'an | + | − | − | − | + | − | − | − | |
| ST72 | CSL2 | Feed | Farm 1, Tai'an | + | − | − | − | − | − | − | − | |
| ST18 | 3G27 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST22 | 2G13 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST23 | 2G15 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST25 | 2G26 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST28 | 3G8 | Cloaca | Farm 4, Weifang | + | − | − | − | + | − | − | − | |
| ST29 | 3G9 | Cloaca | Farm 4, Weifang | + | − | − | − | + | − | − | − | |
| ST43 | 3H1 | Cloaca | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST44 | 3H24 | Cloaca | Farm 4, Weifang | + | − | − | − | + | − | − | − | |
| ST26 | 2GSL5 | Feed | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST27 | 2GSL7 | Feed | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST35 | 3GSL7 | Feed | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST41 | GSL1 | Feed | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST42 | GSL3 | Feed | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST45 | 3HSL2 | Feed | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST46 | 3HSL7 | Feed | Farm 4, Weifang | + | − | − | − | + | − | − | − | |
| ST32 | 3GDF3 | Duck feces | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST40 | GLC2 | Trough | Farm 4, Weifang | + | − | − | − | − | − | − | − | |
| ST33 | 3 GLC2 | Trough | Farm 4, Weifang | + | − | − | − | + | + | − | − | |
| ST48 | D8 | Cloaca | Farm 2, Liaocheng | + | − | − | − | + | − | − | − | |
| ST50 | D16 | Cloaca | Farm 2, Liaocheng | + | − | − | − | + | − | − | − | |
| ST51 | D18 | Cloaca | Farm 2, Liaocheng | + | − | − | − | − | − | − | − | |
| ST52 | D22 | Cloaca | Farm 2, Liaocheng | + | − | − | − | − | − | − | − | |
| ST55 | DSL2 | Feed | Farm 2, Liaocheng | + | − | − | − | − | − | − | − | |
| ST56 | E15 | Cloaca | Farm 3, Liaocheng | + | − | − | − | − | − | − | − | |
| ST57 | ESL2 | Feed | Farm 3, Liaocheng | + | − | − | − | − | − | − | − | |
Abbreviations: CC, clonal complex; ST, sequence type.
Independent STs not involved in forming any CC.
Antibiotic Resistance Profiles
Antimicrobial susceptibility testing showed that resistance against gentamicin was the most prevalent (95.72%), followed by bacitracin (71.05%), lincomycin (65.79%), and tetracycline (55.26%). Resistances against erythrocin, norfloxacin, and cefepime were 37.50, 32.89, and 19.08%, respectively. Resistance against florfenicol, penicillin, fosfomycin, cefotaxime, and imipenem was less than 10%. The resistance of isolates from each farm to different antibiotics is shown in Table 3.
Table 3.
Prevalence (%) of antibiotic resistance in 304 strains of Clostridium perfringens.
| Antibiotics | No. (%) of antimicrobial-resistant isolates |
||||
|---|---|---|---|---|---|
| Farm 1 (n = 63) | Farm 2 (n = 96) | Farm 3 (n = 70) | Farm 4 (n = 75) | Total (%) (n = 304) | |
| Penicillin | 8 (12.70)a | 1 (1.04)b | 4 (5.71)a,b | 9 (12.00)a | 22 (7.24) |
| Cefotaxime | 1 (1.59)a | 2 (2.08)a | 5 (7.14)a | 1 (1.33)a | 9 (2.96) |
| Cefepime | 2 (3.17)b | 8 (8.33)b | 34 (48.57)a | 14 (18.67)c | 58 (19.08) |
| Imipenem | 0 (0.00)a | 0 (0.00)a | 0 (0.00)a | 0 (0.00)a | 0 (0.00) |
| Florfenicol | 2 (3.17)b | 17 (17.71)a | 3 (4.29)b | 8 (10.67)a,b | 30 (9.87) |
| Fosfomycin | 0 (0.00)b,c | 0 (0.00)b | 4 (5.71)a,c | 11 (14.67)a | 15 (4.93) |
| Norfloxacin | 13 (20.63)b | 26 (27.08)b | 48 (68.57)a | 13 (17.33)b | 100 (32.89) |
| Lincomycin | 57 (90.48)a | 68 (70.83)b | 16 (22.86)c | 59 (78.67)a,b | 200 (65.79) |
| Erythrocin | 34 (53.97)a | 30 (31.25)b | 14 (20.00)b | 36 (48.00)a | 114 (37.50) |
| Tetracycline | 33 (52.38)c | 29 (30.21)b | 62 (88.57)a | 44 (58.67)c | 168 (55.26) |
| Bacitracin | 52 (82.54)a | 58 (60.42)b | 60 (85.71)a | 46 (61.33)b | 216 (71.05) |
| Gentamicin | 62 (98.41)a,b | 94 (97.92)a | 67 (95.71)a,b | 68 (90.67)b | 291 (95.72) |
| No. (%) of multidrug-resistant isolates1 | 62 (98.41)a | 62 (64.58)c | 61 (87.14)b | 63 (84.00)b | 248 (81.58) |
a-cMeans in the same row with different lowercase letters are significantly different (P < 0.05).
The number and prevalence of multidrug-resistant isolates from 4 farms.
Resistance of the isolates from different farms against the same antibiotics varied. The drug resistance of the isolates from farm 3 to cefepime (48.57%), norfloxacin (68.57%), and tetracycline (88.57%) was significantly higher than that of isolates from the other 3 farms (P < 0.01), while resistance to lincomycin (22.86%) was the lowest among the 4 farms (P < 0.01). Compared with the other 3 farms, the resistance rate of isolates from farm 2 against tetracycline (30.21%) was the lowest (P < 0.01).
Strains resistant to 3 or more classes of antibiotics were defined as multidrug resistant; the proportion of multidrug-resistant isolates was 81.58% (248/304). The multidrug resistance rate of strains from farm 1 (98.41%) was the highest (P < 0.05), whereas resistance rate of strains from farm 2 (64.58%) was the lowest (P < 0.01) and significantly different from that of other farms. Strains that showed resistance to at least 5 classes of antibiotics accounted for 39.14% (119/304) of all strains. The proportion of strains which showed resistance to at least 5 classes of antibiotics in farm 1 to farm 4 was 36.51% (23/63), 26.04% (25/96), 58.57% (41/70), and 40% (30/75), respectively. The proportion of strains from farm 3 which showed resistance to at least 5 classes of antibiotics was the highest with a significant difference (P < 0.05) compared with the other 3 farms, while farm 2 was the lowest.
Allelic Analysis
The diversity of the MLST loci in 85 strains of C. perfringens is shown in Table 4. Polymorphism of the gmk gene was the lowest with only 7 different alleles, and the highest polymorphism was observed in the glpk gene with 25 alleles, followed by the plc gene and sod gene with 23 alleles. The average number of alleles for all loci was 16.88. The polymorphism index was determined by the percentage of polymorphic sites. The percentage of polymorphism for the sod gene was the highest. The mutation site accounted for 10.95% of all sites, whereas the percentage of polymorphism for the glpk and tpiA genes was the lowest, with 4.10 and 4.48% of all sites, respectively. All allelic sequences examined in this study were coding sequences; thus, the ratio of nonsynonymous to synonymous mutations was used as a measure of selective pressure on each allele. Based on this analysis, all genes possessed a dN/dS ratio of less than 1, indicating purifying selection. The tpiA gene possessed the minimum dN/dS value of 0. A significant linkage disequilibrium was detected between the genes examined, as determined by classical Maynard-Smith IA value of 0.0478 (P = 0.000, based on one representative of each ST).
Table 4.
Diversity at the Clostridium perfringens multilocus sequence typing (MLST) loci.
| Genes | Sequences (bp) | No. of alleles | %Of alleles1 | No. (%) of polymorphic loci2 | dN/dS3 |
|---|---|---|---|---|---|
| ddla | 265 | 20 | 14.81 | 19 (7.17) | 0.0570 |
| dut | 259 | 14 | 10.37 | 26 (10.04) | 0.0908 |
| glpk | 446 | 25 | 18.52 | 20 (4.48) | 0.0753 |
| gmk | 321 | 7 | 0.74 | 16 (4.98) | 0.0715 |
| plc | 327 | 23 | 17.04 | 24 (7.34) | 0.0791 |
| recA | 298 | 14 | 10.37 | 17 (5.70) | 0.0086 |
| sod | 265 | 23 | 17.04 | 29 (10.95) | 0.0051 |
| tpiA | 268 | 9 | 6.67 | 11 (4.10) | 0 |
Percentage of alleles to all isolated strains (n = 85).
Percentage of polymorphic loci to all alleles.
Calculated in the START2 software package (http://pubmlst.org/software/analysis/start2/) by the method of Nei-Gojobori.
STs and Minimum Spanning Tree Analysis
Eighty-five strains of C. perfringens from 4 farms were successfully divided into 54 STs. Among the 54 STs, 44 unique STs were identified; the most prolific ST was ST3 (15.29%; 13/85), followed by ST49 (10.59%; 9/85), ST36 (4.71%; 4/85), and ST8 (3.52%; 3/85). Strains in ST19, ST24, ST31, ST37, ST54, and ST65 accounted for 2.35% (2/85) of all examined strains. ST3 contained 13 strains from 2 farms (cloaca [n = 9], water [n = 1] of farm 3 and cloaca [n = 1], soil [n = 1], duck feces [n = 1] of farm 4), ST49 contained 9 strains from 2 farms (cloaca [n = 6], water [n = 2] of farm 2 and water [n = 1] of farm 3), ST36 contained 4 strains from 2 farms (cloaca [n = 2], water [n = 1] of farm 1 and water [n = 1] of farm 4), ST8 contained 3 strains from cloaca of farm 1 (n = 1) and farm 4 (n = 2), ST65 contained 2 strains from farm 1 (cloaca [n = 1] and feed [n = 1]) (Table 2).
In total, 57 strains of cloacal origin were divided into 37 STs, and 31 unique STs were identified, the most prolific ST was ST3 (17.54%; 10/57), followed by ST49 (10.53%; 6/57), ST36 (5.26%; 3/57), ST8 (5.26%; 3/57), ST31 (3.51%; 2/57), and ST37 (3.51%; 2/57); 28 strains of environment origin were divided into 23 STs, containing 20 unique STs, with the most common ST being ST3 (10.71%; 3/28) and ST49 (10.71%; 3/28), followed by ST54 (7.14%; 2/28). Among the STs of 4 farms, the most prolific ST of farm 1 was ST36 (16.67%; 3/18), followed by ST65 (11.11%; 2/18); the most common STs of farm 2 and farm 3 were ST49 (50%; 8/16) and ST3 (71.43%; 10/14), respectively; ST3 was the dominant ST in examined strains in farm 4, followed by ST8, ST19, ST24, and ST37 which accounted for 5.41% (2/37), respectively. The dominant genotypes in Liaocheng (farm 2 and farm 3) accounted for a high proportion of detected strains.
Diversity of the STs was calculated with the Simpson's Diversity Index (D), and the index of STs in our study was 0.9556. The genetic diversity of isolates in the Weifang farm (farm 4) was the most abundant (37 strains were divided into 31 STs, D = 0.9627), followed by the Tai'an farm (farm 1) (18 strains were divided into15 STs, D = 0.9198), Liaocheng meat-type duck farm (farm 2) (16 strains were divided into 9 STs, D = 0.7188), and Liaocheng breeder farm (farm 3) (14 isolates were divided into 5 STs, D = 0.4694).
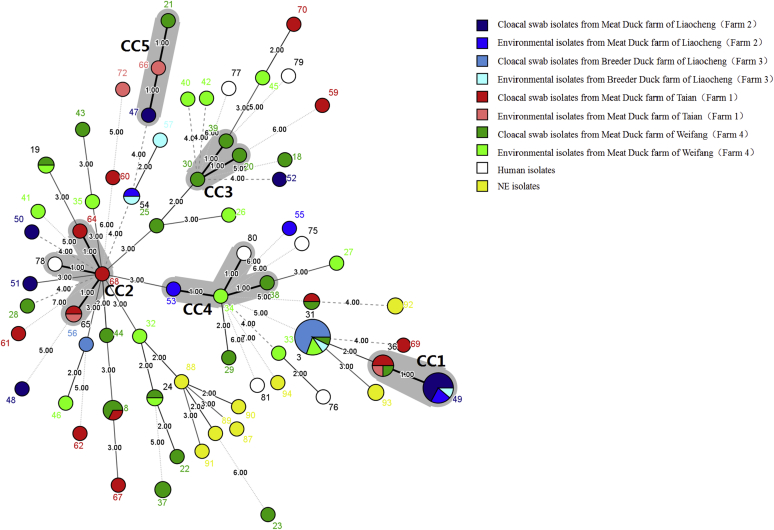
The minimum spanning tree of C. perfringens strains was drawn using the minimum spanning tree method in BioNumerics software based on alleles and STs (Figure 1). In total, 5 CC subtypes (CC1–CC5), containing 30.59% (26/85) of the 85 examined isolates from 4 farms, were identified. Forty STs were identified as singletons with no observed CC associations. CC1, the largest CC, contained cloacal isolates, drinking water isolates from 4 farms and 2 STs (ST36 and ST49), with a total of 13 strains which accounted for 15.30% (13/85) of all examined strains: CC2 grouped strains from cloaca and feed of farm 1 (ST64, ST65, and ST68); CC3 only contained strains from cloaca of farm 4 (ST20, ST30, and ST39); CC4 contained cloacal and feed source isolates from farm 4 (ST34 and ST38) and environmental isolates from farm 2 (ST53); CC5 contained cloacal isolates from farm 2 (ST47) and farm 4 (ST21), soil isolates from farm 1 (ST66).
Figure 1.
The minimum spanning tree of 102 Clostridium perfringens from different sources. The minimum spanning tree was constructed by using the Bionumerics software (Bionumerics, version 7.6 (3); Applied Maths, Inc., Austin, TX). Seven groups of human sequences (ST75 to ST81 in this study) from Liu et al., 5 groups sequences of necrotic enteritis (NE)-infected chicken (ST87 to ST91 in this study) from Nakano et al., and 5 groups sequences of NE-infected chicken (ST92 to ST94 in this study) from Hibberd et al. were also used for analysis. The shaded section represents 5 clonal complexes (CCs). The area of the circle represents the number of strains, different colors represent different sources, and the number on the branch represents the difference of alleles.
After adding 7 human origin isolates (ST75–ST81 in this study) and 10 NE isolates (ST87–ST94 in this study) donated by Liu et al. (2020), Nakano et al. (2017), and Hibberd et al. (2011), 2 CCs (CC2 and CC4) were expanded. One human source strain (ST78) enlarged CC2, one human source strain (ST80) enlarged CC4, and the NE isolates were not involved in the formation of any CC (Figure 1).
A portion of cloacal isolates and environmental (feed and drinking water) isolates were distributed in the same ST (e.g., ST3, ST36, ST49, and ST65) or CC (e.g., CC4). STs (including ST3, ST36, ST31, ST8, ST49, and ST54) and CCs (including CC1, CC4, and CC5) contained strains from different farms. Interestingly, we observed that human strains, environmental, and cloacal isolates were assigned to the same CC (e.g., CC2 and CC4), whereas NE isolates had relatively far genetic relationship to environmental and cloacal isolates.
Phylogenetic Analysis
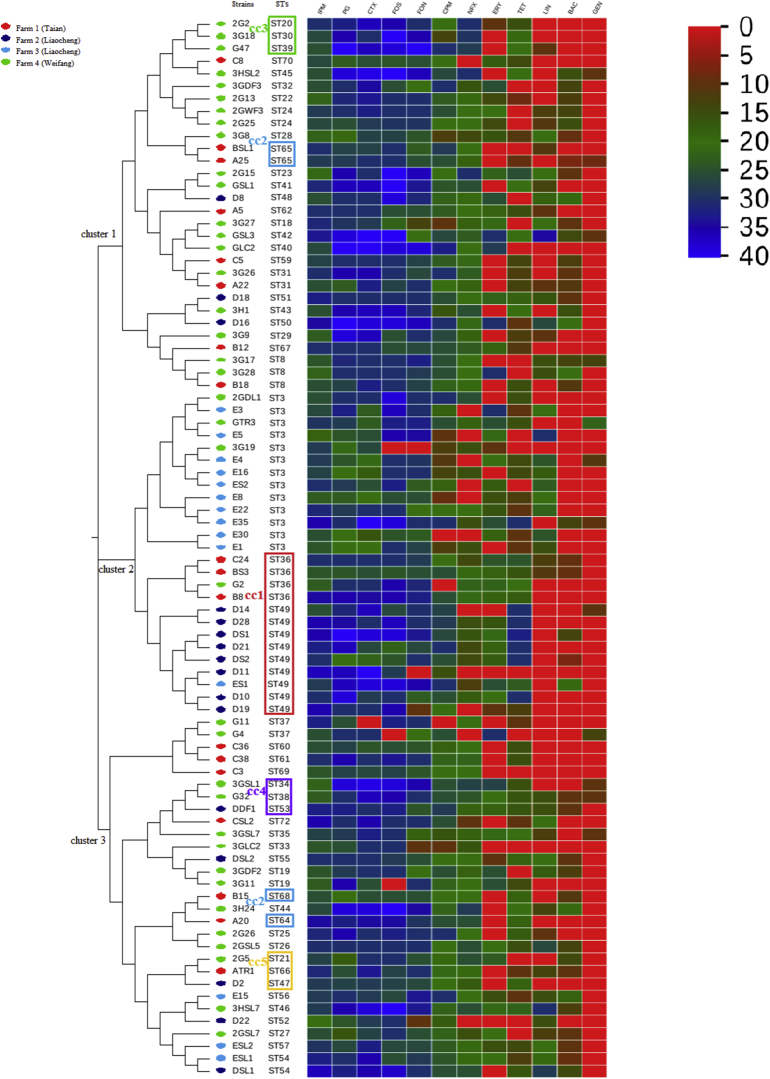
Viewing the whole phylogenetic trees, the dendrogram was found to be dominated by 3 large clusters, which contained all CCs, as well as a substantial number of closely related STs. ST3 and CC1 clustered together to form cluster 2, indicating that the 2 subtypes had closed evolutionary relationship. The isolates from 2 farms in Liaocheng were mainly concentrated in cluster 2, accounting for 63.33% (19/30) of the tested strains in this region; isolates in Tai'an and Weifang farms were concentrated in cluster 1 and cluster 3, which were more disperse than Liaocheng. Isolates from farm 3 (n = 14) had the highest concentration of STs, mainly concentrated in ST3 (71.43%; 10/14); the STs in farm 2 (n = 16) was mainly concentrated in the largest CC-CC1 (50%; 8/16); isolates from farm 1 (n = 18) were mainly concentrated in CC2 (22.22%; 4/18), followed by CC1 (16.67%; 3/18); STs of isolates from farm 4 was relatively fragmented, mainly concentrated in ST3 (8.11%; 3/37), CC3 (8.11%; 3/37), and followed by CC4 (5.41%; 2/37).
The results of phylogenetic trees are basically consistent with the minimum spanning tree, but not completely. Strains in the same CC were usually clustered together. Strains in CC1, CC3, CC4, and CC5 were clustered together, but there were exceptions. For example, 4 strains in CC2 were assigned to different clusters of the tree. Moreover, we also found that strains of feed origin and cloacal origin in Weifang were clustered together in the phylogenetic trees (ST42 and ST18; ST23 and ST41; ST25 and ST26), whereas the same phenomenon was not observed in the minimum spanning tree (Figure 1, Figure 2).
Figure 2.
Phylogenetic trees of sequence types (ST) of 85 C. perfringens from duck farms and heat map (antibiotic resistance profiles). The heat map was constructed by using an online software program (https://www.graphpad.com/register/confirmation/); 0 to 40 stands for inhibition zone. Abbreviations: BAC, Bacitracin; CPM, Cefepime; CTX, Cefotaxime; ERY, Erythromycin; FON, Florfenicol; FOS, Fosfomycin; GEN, Gentamicin; IPM, Imipenem; LIN, Lincomycin; NFX, Norfloxacin; PG, Penicillin; TET, Tetracycline.
Discussion
Among the 424 samples collected in this study, the total positive rate of C. perfringens was 48.82% (207/424). Among which the positive rate of cloacal swab samples was 49.84% (158/317); this value was higher than those reported in chickens from central China and Taiwan (23.1 and 29.6%, respectively) (Fan et al., 2016; Zhang et al., 2018), 24.72 and 23.28% from 2 commercial farms in Canada, 43.23% in chickens of Jordan (Chalmers et al., 2008b; Gharaibeh et al., 2010) but lower than that reported in Egypt (57.9%) (Osman et al., 2012). In this study, the positive rate of samples differs in different farms, among which the positive rate (75.51%; 38/51) in farm 2 was the highest, and it was statistically higher than that of the other 3 farms (P < 0.01). The high positive rate of farm 2 might be related to its antibiotic-free farming model, antibiotic selection pressure reduced the C. perfringens–colonizing activity in the environment and duck intestines, and this explanation was consistent with a previous study (Osman et al., 2012). (Table 1)
Overall, our study shows a relatively high positive rate of C. perfringens in all collected samples. In the environmental samples, the contamination rate of feed was the highest with a value of 72.00%, indicating that the feed was seriously contaminated with C. perfringens. Sources of C. perfringens in feed ranged widely, including raw materials, processing, storage, sales, and transportation. This result suggested that we should pay attention to the contamination of C. perfringens in feed to evaluate the significance of feed contamination in the epidemiology of C. perfringens. The relatively low contamination rate of C. perfringens in drinking water (19.35%) may be due to the fact that the drinking water mainly came from relatively closed nipple drinking water fountains, which was not easily contaminated by the external environment. Multilocus sequence typing revealed that a portion of isolates from drinking water and cloacal swabs were phylogenetically close, indicating that drinking water pollution could also be an important source of C. perfringens in ducks. The collection of environmental samples can not only help us assess farm hygiene conditions but also help us analyze the possible source of C. perfringens in ducks.
Genotyping results showed that all isolates were identified as C. perfringens type A, which was consistent with previous reports in China and other countries (Fan et al., 2016; Zhang et al., 2018). Enterotoxin gene (cpe) was closely associated not only with antibiotic-associated diarrhea but also with outbreaks of food poisoning (Songer, 1996; Sarker et al., 2000; Osman et al., 2012; Gaucher et al., 2015). In this study, one isolate of type A carrying the cpe gene was detected from the cloacal swab samples of farm 1 and the other from farm 4, with a positive rate of 0.83 and 0.86%, respectively. The positive rate of cpe in all isolates was 0.5%, which was basically consistent with data reported in previous studies. For example, in Taiwan, Jordan, Sweden, and Canada, the positive rate of cpe in isolates from chicken farms was 0% (Engström et al., 2003; Chalmers et al., 2008b; Gharaibeh et al., 2010; Fan et al., 2016), and the cpe positive rate detected in chicken of central China was 3.08% (Zhang et al., 2018).
Beta-2 toxin encoded by the cpb2 gene, which can be produced by all types of C. perfringens, is associated with gastrointestinal disease in humans and animals (Jost et al., 2006; Hu et al., 2018). In this study, the cpb2 positive rate of strains from 4 farms was different, among which the highest positive rate was found in farm 1 (40.83%; 49/120), and the lowest was found in farm 2 (18.75%; 18/96). The positive rate of cpb2 in all isolates was 30.85% (124/402), which was higher than that of C. perfringens isolates from Sweden (9.52%) (Engström et al., 2003) and far lower than that from Egypt and the United States (62.6 and 95%) (Siragusa et al., 2006; Osman et al., 2012). The netB and TpeL toxin genes were not detected in all isolates (n = 402), and studies have shown that the netB and TpeL toxin genes were closely associated with NE-infected chickens (Chalmers et al., 2008a; Hibberd et al., 2011).
In recent years, the abuse of antibiotic feed additives has led to the increase of antimicrobial resistance of some intestinal flora, and a portion of zoonotic pathogens have developed multiple antimicrobial resistance (Wen and McClane, 2004). Previous studies have reported antibiotic resistance of C. perfrigens in chicken (Martel et al., 2004; Gharaibeh et al., 2010; Gaucher et al., 2015). In Egypt, C. perfrigens isolated from NE chickens were all resistant to gentamycin, lincomycin, erythromycin, and ciprofloxacin (Osman and Elhariri, 2013), whereas C. perfrigens isolates from Belgium chickens were sensitive to bacitracin, enrofloxacin, erythromycin, and florfenicol (Gholamiandehkordi et al., 2009). In this study, isolates from the 4 farms showed resistance against gentamicin (95.72%), bacitracin (71.05%), lincomycin (65.79%), and tetracycline (55.26%), completely sensitivity to imipenem and highly sensitive to cefotaxime. In general, isolates in this study showed a relatively high antibiotic resistance compared with those in Belgium chicken but lower resistance than those in Egypt; this phenomenon may be related to the ban of antibiotics in animal feed by the European Union since 2006 (Mwangi et al., 2018).
Different antimicrobial resistance of the isolates was observed in various farms. The antimicrobial resistance rate against cefotaxime, norfloxacin, and tetracycline in isolates of farm 3 was significantly higher than that in other 3 farms (P < 0.01). This might be related to the frequent usage of tetracycline, cephalosporins, quinolones antibiotics in Farm 3. The related study had shown that C. perfringens is extremely resistant against lincomycin (Osman and Elhariri, 2013), but in our study, the antimicrobial resistance rate against lincomycin in isolates from farm 3 (22.86%) was significantly lower than that in other 3 farms (P < 0.01) and other studies (Wen and McClane, 2004; Liu et al., 2020), indicating lincomycin can still be used as a choice for this farm to treat C. perfringens–related diseases. None of antibiotics were used in farm 2. Isolates showed low resistance to most tested antibiotics; however, the isolates of cloacal origin from this farm still had a certain degree of resistance against some antibiotics such as lincomycin (67.09%), erythrocin (31.65%), and tetracycline (29.11%). This phenomenon might be due to the antimicrobial resistance of strain from environment, such as soil, feed, and water. We found that isolates from drinking water were completely resistant against lincomycin (100.00%); strains from feed origin showed a high resistance against tetracycline (60.00%) and erythrocin (40.00%). Antibiotic resistance may not disappear completely because of local and short-term prohibitions.
In total, isolates from 4 duck farms showed high multiple-drug resistance (81.58%), and 39.14% of the isolates were resistant to at least 5 classes of commonly used antibiotics. As the proportion of strains which showed resistance to at least 5 classes of antibiotics, farm 3 was the highest with a significant difference (P < 0.05) compared with the other 3 farms, whereas farm 2 was the lowest. The multiple-drug resistance rate of isolates in farm 2 was significantly lower than that in other 3 farms (P < 0.01). This difference might be related to the antibiotic-free farming model in farm 2 and the use of a variety of antibiotics in farm 3. Differences in antibiotic resistance of isolates between farms were largely due to the history of antibiotic administration.
Even though the multidrug resistance rate of isolates in farm 2 (64.58%) was lower than that in other 3 farms, it was still higher than the multidrug resistance rate (53%) reported by Mwangi et al. (2018). Antimicrobial resistance poses significant challenges for current clinical care (Wang et al., 2019). Multidrug-resistant strains of C. perfringens have been extensively detected in this study, which means that in the event of a related disease outbreak, treatment will be difficult (Song et al., 2020). And if the antibiotic resistance is transmitted to humans through the food chain of related duck products, it will pose a serious threat to the public health. Therefore, antibiotics should be used more rationally in the future.
It was interesting that although the multiple resistance of C. perfringens in an antibiotic-free farm (farm 2) was significantly lower than that in other farms, the positive rate of C. perfringens was much higher than that in other farms. This result was consistent with a previous study (Gaucher et al., 2015). As the use of antibiotics as feed additives had been forbidden since 2020 in China, the high positive rate and low antibiotic resistance of C. perfringens might become more popular and apparent, which will have an impact on the occurrence and prevalence of C. perfringens; this phenomenon should be valued. The ban of antibiotics additive may also have an impact on other existing duck epidemics in China (Chen et al., 2020; Wang et al., 2020).
In this study, based on the MLST scheme previously published by Jost et al. (2006), C. perfringens strains of different origins were genotyped at multiple loci. Through the polymorphism of alleles, we realized that considerable genetic diversity existed in the core genome of isolates in this study. Multilocus sequence typing had been successfully applied to classify these isolates and compare the evolutionary relationships of C. perfringens from ducks, farm environment, and humans. All analyzed isolates (of cloacal origin and environmental origin) were divided into 54 STs, with 5 CCs accompanying, and alleles of the loci examined was 16.88 on average. In comparison, Jost et al. (2006) divided 132 strains of C. perfringens from different host species and toxinotypes into 80 STs and 3 CCs, with an average allele number of 24.4. Nakano et al. (2017) identified an average of 10.25 alleles, 30 STs, and 3 CCs among 40 strains from children and chicken. The average allele of our study was higher than that of Nakano's study but lower than that of JOST's study. A relatively abundant STs were found in the examined strains; Simpson's index which reveals genetic diversity was 0.9556, and this value indicated that our isolates had considerable genetic diversity. Although considerable polymorphism was observed in the loci analyzed, a significant linkage disequilibrium was observed among all genes examined, as determined by classical-Maynard-Smith IA (0.0478), indicating a low recombination rate in the genomes of the C. perfringens isolates examined. This hypothesis is substantiated by the observation that 26 of the 85 isolates (30.59%) were partitioned into 5 CCs.
According to the regional distribution, the genetic diversity varies in different farms, and the Simpson's index of STs for Liaocheng was lower than that for Tai'an and Weifang. In farm 2 (Liaocheng meat-type duck farm), ST49 was the prevalent ST, and the prevalent CC is CC1, which was mainly made up of ST49, which account for 61.54% (8/13) of all strains. In farm 3 (Liaocheng breeder duck farm), ST 3 was the prevalent ST (71.43%; 10/14) with no CC accompanied. According to the results (ratio of prevalent STs, Simpson's Diversity Index and the proportion of main ST in CC from 4 farms), we found that the strains from the 2 farms in the Liaocheng region (farm 2, farm 3) were more concentrated in genetic relationship than those in Tai'an and Weifang farms (farm 1, farm 4), which might be related to the region or company because the 2 farms belong to the same company. On the other hand, STs in Farm 3 was more concentrated than those in farm 2, which may be related to the fact that cephalosporins, tetracycline, and quinolones were frequently used in farm 3. Antibiotics were used as the selective pressure, the susceptible strains were eliminated as time went on, and antibiotic-resistant strains continuously gained advantages and became epidemic; the population diversity decreased in the end.
C. perfringens was considered to exist naturally in soil and sewage, which can be spread horizontally through the environment. In this study, a portion of cloacal isolates and environmental (feed and drinking water) isolates were found to be matched in the same CC or ST. For example, strains in ST36 (n = 3) were isolated from cloacal swabs (n = 2) and drinking water (n = 1) in farm 1, respectively; strains in ST49 (n = 8) isolated from cloacal swabs (n = 6) and drinking water (n = 2) in farm 2; and strains in ST3 (n = 10) isolated from cloacal swabs (n = 9) and drinking water (n = 1) in farm 3. It indicates that the water may be a source of C. perfringens in ducks. Moreover, strains in ST65 (n = 2) isolated from cloacal swab (n = 1) and feed (n = 1) in farm 1 and ST34 (including strains of feed origin in farm 4) and ST38 (including strains of cloacal swab origin in farm 4) were found to be matched in the same CC (CC4), indicating that the isolates of C. perfringens from ducks were closely related to isolates from feed, and the feed may be the source of infection in ducks. Therefore, in the process of duck breeding, measures should be taken to control the hygienic conditions of feed and water, so as to avoid the cross-contamination of C. perfringens.
According to the genetic relationship of the isolates, genetic relationship of the strains from the same region was relatively close; however, part of strains in different regions could also cluster together, existing in the same ST, which was consistent with the results of previous studies (Nakano et al., 2017; Zhang et al., 2018); for example, ST3 contains strains isolated from 2 regions and 3 farms, including cloacal swab samples and drinking water samples in farm 2 and farm 3 (Liaocheng), as well as cloacal swab samples, duck feces, and soil samples in farm 4 (Weifang). This phenomenon indicated that ST3 was a prevalent ST in investigated duck farms; the reason might be that ST3 was widespread in the environment or had elements suitable for epidemic. Antibiotic susceptibility testing showed that the antibiotic resistance of different ST3 strains was not exactly the same (Figure 2), indicating that the prevalence of ST3 might also be related to other characteristics of strains, which need further study. Similar phenomena also exist in other strains, ST36, ST31, and ST8 contained isolates from farm 1 and farm 4; CC1 (ST36 and ST49) and CC5 (ST47, ST21, and ST66) all contained isolates from all 4 farms; and CC4 (ST38, ST34, and ST53) contained isolates from farm 2, farm 3, and farm 4 (Figure 1).
Our study described the antibiotic resistance in the isolates from cloacal swabs and environment, and genetic relatedness was also observed in these isolates; antibiotic resistance profiles of the strains in the same ST or CC seem to be more similar than those of the strains in different CCs. Strains in the same CC were usually clustered together (not completely), and we also found that some strains had a relatively close genetic evolutionary relationship in phylogenetic trees, whereas these strains had a far relationship in the minimum spanning tree, and these phenomena were related to the number of point mutations in all alleles. ST3, the most prevalent ST of isolates from farm 3, had a relatively close evolutionary relationship with the most common genotype (CC1) of farm 2, and this phenomenon may be related to the fact that the 2 farms are owned by the same company in Liaocheng and that the feed materials used might be same. Therefore, the combination of minimum spanning tree and phylogenetic trees can help us to better analyze the genetic relationship between isolates (Figure 2).
Little has been reported on MLST of C. perfringens from duck farms; therefore, it is quite difficult to find a control on the website. To observe the evolutionary relationship of C. perfringens isolates between animals and humans, we added strains isolated from NE-infected chicken and healthy human by other researchers for control. It is interesting to note that the isolates from NE-infected chickens were distantly related with healthy ducks in evolution, whereas 2 human strains (ST78 and ST80) were assigned to CC2 and CC4, respectively. The fact indicated that strains of duck origin could pose a potential threat to humans through the food chain. This same phenomenon has been found in previous studies (Nakano et al., 2017; Liu et al., 2020).
This is the first report showing a MLST scheme of C. perfringens isolates from duck farms; the prevalence of C. perfringens from various duck farms in parts of the Shandong province was investigated. The results showed that the positive rate of C. perfringens was relatively high; the prevalent serotype was type A with 0.5% positive of cpe. The antibiotic resistance of isolates varied in different duck farms, and multidrug-resistant strains were widespread. Multilocus sequence typing showed that the genetic diversity of C. perfringens isolates from different duck farms was significantly different; ST3 and CC1 were the prevalent genotypes in some duck farms of the Shandong province. Some isolates from cloacal swab and environmental samples were contained in the same ST or CC, indicating that the duck cloacal isolates were possibly related to drinking water and feed; a portion of the strains from humans and ducks was found to be phylogenetically close, indicating potential public health risk. Therefore, measure should be taken to control the hygienic conditions of the duck farms, and reasonable usage of antibiotics is essential to avoid high antibiotic resistance of bacteria and to reduce the potential risk of public health.
Acknowledgments
The authors are grateful to Lixue Liu, Xiaojun Cong, and Wenping Xu for sample collection. This study was supported by the National Key Research and Development Program of China (grant number: 2018YFD0500500) and funds of the Shandong “Double Tops” Program (grant number: SYL2017YSTD11).
Conflict of Interest Statement: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- Abakabir Mahamat A., Radomski N., Delannoy S., Djellal S., LENEGRATE M., Hadjab K., Fach P., Hennekinne J.A., Mistou M.Y., Firmesse O. Large-scale genomic analyses and toxinotyping of Clostridium perfringens Implicated in Foodborne outbreaks in France. Front. Microbiol. 2019;10:777. doi: 10.3389/fmicb.2019.00777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M.A., Macklin K.S., Krehling J.T. Use of a multiplex PCR for the detection of toxin-Encoding genes netB and tpeL in strains of Clostridium perfringens. ISRN Vet. Sci. 2013;2013:865702. doi: 10.1155/2013/865702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- British Society for Antimicrobial Chemotherapy. BSAC; Birmingham, United Kingdom: 2015. Methods for Antimicrobial Susceptibility Testing, Version 14.0, May 2015. [Google Scholar]
- Cao Y., Wei D. Multi-locus sequence typing (MLST) and repetitive extragenic palindromic polymerase chain reaction (REP-PCR), characterization of Shigella spp. over two decades in Tianjin China. Int. J. Mol. Epidemiol. Genet. 2012;3:321–332. [PMC free article] [PubMed] [Google Scholar]
- Chalmers G., Bruce H.L., Hunter D.B., Parreira V.R., Kulkarni R.R., Jiang Y.F., Prescott J.F., Boerlin P. Multilocus sequence typing analysis of Clostridium perfringens isolates from necrotic enteritis outbreaks in broiler chicken populations. J. Clin. Microbiol. 2008;46:3957–3964. doi: 10.1128/JCM.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers G., Martin S.W., Hunter D.B., Prescott J.F., Weber L.J., Boerlin P. Genetic diversity of Clostridium perfringens isolated from healthy broiler chickens at a commercial farm. Vet. Microbiol. 2008;127:116–127. doi: 10.1016/j.vetmic.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Chan M.S., Maiden M.C., Spratt B.G. Database-driven multilocus sequence typing (MLST) of bacterial pathogens. Bioinformatics. 2001;17:1077–1083. doi: 10.1093/bioinformatics/17.11.1077. [DOI] [PubMed] [Google Scholar]
- Chen H., Zhang B., Yan M., Diao Y., Tang Y. First report of a novel goose astrovirus outbreak in Cherry Valley ducklings in China. Transbound. Emerg. Dis. 2020;67:1019–1024. doi: 10.1111/tbed.13418. [DOI] [PubMed] [Google Scholar]
- Coursodon C.F., Glock R.D., Moore K.L., Cooper K.K., Songer J.G. TpeL-producing strains of Clostridium perfringens type A are highly virulent for broiler chicks. Anaerobe. 2012;18:117–121. doi: 10.1016/j.anaerobe.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Engström B., Fermer C., Lindberg A., Saarinen E., Båverud V., Gunnarsson A. Molecular typing of isolates of Clostridium perfringens from healthy and diseased poultry. Vet. Microbiol. 2003;94:225–235. doi: 10.1016/s0378-1135(03)00106-8. [DOI] [PubMed] [Google Scholar]
- Fan Y.C., Wang C.L., Wang C., Chen T.C., Chou C.H., Tsai H.J. Incidence and Antimicrobial susceptibility to Clostridium perfringens in premarket broilers in Taiwan. Avian Dis. 2016;60:444–449. doi: 10.1637/11315-110915-Reg. [DOI] [PubMed] [Google Scholar]
- Fohler S., Klein G., Hoedemaker M., Scheu T., Seyboldt C., Campe A., Jensen K.C., Abdulmawjood A. Diversity of Clostridium perfringens toxin-genotypes from dairy farms. BMC Microbiol. 2016;16:199. doi: 10.1186/s12866-016-0812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher M.L., Quessy S., Letellier A., Arsenault J., Boulianne M. Impact of a drug-free program on broiler chicken growth performances, gut health, Clostridium perfringens and Campylobacter jejuni occurrences at the farm level. Poult. Sci. 2015;94:1791–1801. doi: 10.3382/ps/pev142. [DOI] [PubMed] [Google Scholar]
- Gharaibeh S., Al Rifai R., Al-Majali A. Molecular typing and antimicrobial susceptibility of Clostridium perfringens from broiler chickens. Anaerobe. 2010;16:586–589. doi: 10.1016/j.anaerobe.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Gholamiandehkordi A., Eeckhaut V., Lanckriet A., Timbermont L., Bjerrum L., Ducatelle R., Haesebrouck F., Van Immerseel F. Antimicrobial resistance in Clostridium perfringens isolates from broilers in Belgium. Vet. Res. Commun. 2009;33:1031–1037. doi: 10.1007/s11259-009-9306-4. [DOI] [PubMed] [Google Scholar]
- Guran H.S., Oksuztepe G. Detection and typing of Clostridium perfringenvs from retail chicken meat parts. Lett. Appl. Microbiol. 2013;57:77–82. doi: 10.1111/lam.12088. [DOI] [PubMed] [Google Scholar]
- Hibberd M.C., Neumann A.P., Rehberger T.G., Siragusa G.R. Multilocus sequence typing subtypes of poultry Clostridium perfringens isolates demonstrate disease niche partitioning. J. Clin. Microbiol. 2011;49:1556–1567. doi: 10.1128/JCM.01884-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.S., Kim H., Koo O.K. Molecular genotyping, biofilm formation and antibiotic resistance of enterotoxigenic Clostridium perfringens isolated from meat supplied to school cafeterias in South Korea. Anaerobe. 2018;52:115–121. doi: 10.1016/j.anaerobe.2018.06.011. [DOI] [PubMed] [Google Scholar]
- Hunter P.R., Gaston M.A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost B.H., Trinh H.T., Songer J.G. Clonal relationships among Clostridium perfringens of porcine origin as determined by multilocus sequence typing. Vet. Microbiol. 2006;116:158–165. doi: 10.1016/j.vetmic.2006.03.025. [DOI] [PubMed] [Google Scholar]
- Keyburn A.L., Boyce J.D., Vaz P., Bannam T.L., Ford M.E., Parker D., Di Rubbo A., Rood J.I., Moore R.J. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 2008;4:e26. doi: 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström M., Heikinheimo A., Lahti P., Korkeala H. Novel insights into the epidemiology of Clostridium perfringens type A food poisoning. Food Microbiol. 2011;28:192–198. doi: 10.1016/j.fm.2010.03.020. [DOI] [PubMed] [Google Scholar]
- Liu Y., Xiu L., Miao Z.M., Wang H.R. Occurrence and multilocus sequence typing of Clostridium perfringens isolated from retail duck products in Tai'an region. China. Anaerobe. 2020;62 doi: 10.1016/j.anaerobe.2019.102102. 102102. [DOI] [PubMed] [Google Scholar]
- Martel A., Devriese L.A., Cauwerts K., De Gussem K., Decostere A., Haesebrouck F. Susceptibility of Clostridium perfringens strains from broiler chickens to antibiotics and anticoccidials. Avian Pathol. 2004;33:3–7. doi: 10.1080/0307945031000163291. [DOI] [PubMed] [Google Scholar]
- Miyamoto K., Wen Q., McClane B.A. Multiplex PCR genotyping assay that distinguishes between isolates of Clostridium perfringens type A carrying a chromosomal enterotoxin gene (cpe) locus, a plasmid cpe locus with an IS1470-like sequence, or a plasmid cpe locus with an IS1151 sequence. J. Clin. Microbiol. 2004;42:1552–1558. doi: 10.1128/JCM.42.4.1552-1558.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwangi S., Timmons J., Fitz-Coy S., Parveen S. Characterization of Clostridium perfringens recovered from broiler chicken affected by necrotic enteritis. Poult. Sci. 2018;98:128–135. doi: 10.3382/ps/pey332. [DOI] [PubMed] [Google Scholar]
- Nakano V., Ignacio A., Llanco L., Bueris V., Sircili M.P., Ávila-Campos M.J. Multilocus sequence typing analyses of Clostridium perfringens type A strains harboring tpeL and netB genes. Anaerobe. 2017;44:99–105. doi: 10.1016/j.anaerobe.2017.02.017. [DOI] [PubMed] [Google Scholar]
- Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Osman K.M., Elhariri M. Antibiotic resistance of Clostridium perfringens isolates from broiler chickens in Egypt. Rev. Sci. Tech. 2013;32:841–850. doi: 10.20506/rst.32.2.2212. [DOI] [PubMed] [Google Scholar]
- Osman K.M., Soliman Y.A., Amin Z.M.S., Aly M.A.K. Prevalence of Clostridium perfringens type A isolates in commercial broiler chickens and parent broiler breeder hens in Egypt. Rev. Sci. Tech. Off. Int. Epiz. 2012;31:931–941. doi: 10.20506/rst.31.3.2169. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1978;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sarker M.R., Shivers R.P., Sparks S.G., Juneja V.K., McClane B.A. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid genes versus chromosomal enterotoxin genes. Appl. Environ. Microbiol. 2000;66:3234–3240. doi: 10.1128/aem.66.8.3234-3240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Siragusa G.R., D Danyluk M., L Hiett K., Wise M.G., Craven S.E. Molecular subtyping of poultry-associated type A Clostridium perfringens isolates by repetitive-element PCR. J. Clin. Microbiol. 2006;44:1065–1073. doi: 10.1128/JCM.44.3.1065-1073.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Yu L., Zhang Y., Dai Y., Wang P., Feng C., Liu M., Sun S., Xie Z., Wang F. Prevalence and characteristics of multidrug-resistant mcr-1-positive Escherichia coli isolates from broiler chickens in Tai’an, China. Poult. Sci. 2020;99:1117–1123. doi: 10.1016/j.psj.2019.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songer J.G. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 1996;9:216. doi: 10.1128/cmr.9.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojadoost B., Vince A.R., Prescott J.F. The successful experimental induction of necrotic enteritis in chickens by Clostridium perfringens: a critical review. Vet. Res. 2012;43:74. doi: 10.1186/1297-9716-43-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelling A.M., Gerner-Smidt P., Hawkey P.M., Heritage J., Parnell P., Porter C., Bodenham A.R., Inglis T. Validation of use of whole-cell repetitive extragenic palindromic sequence-based PCR (REP-PCR) for typing strains belonging to the Acinetobacter calcoaceticus-Acinetobacter baumannii complex and application of the method to the investigation of a hospital outbreak. J. Clin. Microbiol. 1996;34:1193–1202. doi: 10.1128/jcm.34.5.1193-1202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Nei M., Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwin R., Maiden M.C. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 2003;10:479–487. doi: 10.1016/j.tim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Wang H., Gao B., Liu X., Zhang S., Diao Y., Tang Y. Pathogenicity of a variant duck orthoreovirus strain in Cherry Valley Ducklings. Vet. Microbiol. 2020;242:108546. doi: 10.1016/j.vetmic.2019.108546. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang B., Du H., Zhang H., Li H., Wang F., Zhao X. Effects of Diutina rugosa SD-17 on growth performance, intestine morphology, and immune status of chickens. Poult. Sci. 2019;98:6311–6318. doi: 10.3382/ps/pez428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Q., McClane B.A. Detection of enterotoxigenic Clostridium perfringens type A isolates in American retail foods. Appl. Environ. Microbiol. 2004;70:2685–2691. doi: 10.1128/AEM.70.5.2685-2691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L., Yu H., Qi J., Jiang P., Sun B., Cui J., Ou C., Chang W., Hu Q. ErmF and ereD are responsible for erythromycin resistance in Riemerella anatipestifer. PLoS One. 2015;10:e0131078. doi: 10.1371/journal.pone.0131078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H.S., Lee S.U., Park K.Y., Park Y.H. Molecular typing and epidemiological survey of prevalence of Clostridium perfringens types by multiplex PCR. J. Clin. Microbiol. 1997;35:228–232. doi: 10.1128/jcm.35.1.228-232.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Zhang W., Ai D., Zhang R., Lu Q., Luo Q., Shao H. Prevalence and characterization of Clostridium perfringens in broiler chickens and retail chicken meat in central China. Anaerobe. 2018;54:100–103. doi: 10.1016/j.anaerobe.2018.08.007. [DOI] [PubMed] [Google Scholar]