Abstract
The coronavirus disease 2019 (COVID-19) pandemic is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This pathogen has spread rapidly across the world, causing high numbers of deaths and significant social and economic impacts. SARS-CoV-2 is a novel coronavirus with a suggested zoonotic origin with the potential for cross-species transmission among animals. Antarctica can be considered the only continent free of SARS-CoV-2. Therefore, concerns have been expressed regarding the potential human introduction of this virus to the continent through the activities of research or tourism to minimise the effects on human health, and the potential for virus transmission to Antarctic wildlife. We assess the reverse-zoonotic transmission risk to Antarctic wildlife by considering the available information on host susceptibility, dynamics of the infection in humans, and contact interactions between humans and Antarctic wildlife. The environmental conditions in Antarctica seem to be favourable for the virus stability. Indoor spaces such as those at research stations, research vessels or tourist cruise ships could allow for more transmission among humans and depending on their movements between different locations the virus could be spread across the continent. Among Antarctic wildlife previous in silico analyses suggested that cetaceans are at greater risk of infection whereas seals and birds appear to be at a low infection risk. However, caution needed until further research is carried out and consequently, the precautionary principle should be applied. Field researchers handling animals are identified as the human group posing the highest risk of transmission to animals while tourists and other personnel pose a significant risk only when in close proximity (< 5 m) to Antarctic fauna. We highlight measures to reduce the risk as well as identify of knowledge gaps related to this issue.
Keywords: Antarctica, Coronavirus, COVID-19, Mitigation measures, Reverse zoonoses, Transmission
Graphical abstract
Highlights
-
•
Conditions in Antarctica could be favourable for SARS-CoV-2 stability.
-
•
In silico analyses revealed that Cetaceans have a higher susceptibility to the virus.
-
•
Seals and birds seem to have a lower risk of infection.
-
•
Wildlife researchers have the highest risk of transmission to Antarctic fauna.
-
•
Tourists can be potential vectors for SARS-CoV-2 transmission to Antarctic fauna.
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in the Hubei Province of China in late 2019. This virus has spread rapidly to most parts of the world, having caused over one million confirmed deaths as of October 2020, posing grave concerns to all aspects of human life, with significant social and economic impacts (World Health Organization, 2020). SARS-CoV-2 is a novel coronavirus closely related to coronaviruses found in bats and pangolins (Lu et al., 2020b; Zhang et al., 2020), suggesting a zoonotic origin (Andersen et al., 2020). Coronaviruses have a long history of cross-species transmission, whereby all members of the seven identified coronaviruses that infect humans are suspected to have zoonotic origins (Corman et al., 2018). In addition to coronavirus spread from animals to humans, there are numerous records of cross-species transmission of coronaviruses among non-human animals (Hemida et al., 2017; Zhou et al., 2018). In humans, transmission of SARS-CoV-2 is thought to occur predominantly via direct contact with airborne droplets projected when coughing, sneezing or talking, as well as indirectly through aerosols or contact with fomites (Allen and Marr, 2020; Cai et al., 2020; Liu et al., 2020; Ong et al., 2020). Additionally, concerns have arisen that humans can act as a transmission source to wild or domestic animal species (reverse zoonosis) and non-human hosts could serve as a source of infection for humans (zoonosis) (Franklin and Bevins, 2020).
Prior to mid-March 2020, Antarctica was the only continent presumed to be free of SARS-CoV-2. However, at the end of the 2019–2020 tourist season in March, at least one SARS-CoV-2 positive tourist visited several sites along the Antarctic Peninsula (Ing et al., 2020). This highly mobile event highlights the concerns regarding the potential human introduction of this virus to the continent through research activities or tourism. The potential effects of SARS-CoV-2 in Antarctica include those related primarily to human health and the risks of potential transmission of the virus to Antarctic wildlife. In this paper, we assess the potential risk of SARS-CoV-2 reverse-zoonotic transmission (i.e. from humans to other animals) to Antarctic vertebrates. We considered information available on host susceptibility and infection dynamics in humans, and risks of the possible contact interactions between humans and Antarctic wildlife as these elements are necessary (Plowright et al., 2017) for the emergence of SARS-CoV-2 in the Antarctic ecosystem. We identify important knowledge gaps and, recommend that the precautionary principle be applied to limit the transmission of SARS-CoV-2 to Antarctic fauna and propose measures to reduce such risk.
2. SARS-CoV-2 host range and evidence of cross-species transmission
Coronaviruses are enveloped single-stranded RNA viruses. The family Coronaviridae is composed of two subfamilies, Letovirinae with one genus (Alphaletovirus) and Orthocoronavirinae with four genera (Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus). Alphacoronaviruses and betacoronaviuses are common in mammals, including humans, domestic animals and bats (Chan et al., 2013). These viruses cause a range of diseases including upper and lower respiratory tract disease, gastroenteritis, and central nervous system infections. Zoonotic transmission, or spill-over from a wild or domestic animal host to humans, is a frequent occurrence (Morse et al., 2012) including SARS-CoV-1 from bats and palm civets (Li et al., 2005), MERS-CoV from camels (with an ultimate origin in bats) (Lau et al., 2013) and HCoV-OC43, which is believed to have its origin in cattle (Vijgen et al., 2005). Birds are the main reservoirs for gammacoronaviruses and deltacoronaviruses, with most of these viruses causing no disease in their wild bird hosts. Avian coronavirus, a Gammacoronavirus, was the first described coronavirus (ca. 1931) and the infections it causes in poultry have a large socioeconomic impact. Gammacoronaviruses and deltacoronaviruses have mammalian hosts with Beluga whale coronavirus SW1 (Gammacoronavirus) infecting cetaceans (Mihindukulasuriya et al., 2008) and Coronavirus HKU15 (Deltacoronavirus) infecting swine and mammals in wet markets (Woo et al., 2012). There are no known avian hosts for alphacoronaviruses or betacoronaviruses.
SARS-CoV-2 is a betacoronavirus closely related to coronaviruses found in bats and pangolins (Lu et al., 2020a, Lu et al., 2020b; Zhang et al., 2020), and it appears to have a wide vertebrate host range. The virus has been detected in companion animals such as cats and dogs living with infected owners (American Veterinary Medical Association, 2020) and has caused large outbreaks with lethal disease in farmed minks (Oreshkova et al., 2020). Experimental studies have shown that the virus can infect and replicate efficiently in cats, captive ferrets and hamsters, with transmission to conspecifics (Kim et al., 2020; Shi et al., 2020; Chan et al., 2020a, Chan et al., 2020b). In contrast, the virus replicates poorly in dogs, and experimental infection attempts in pigs, chickens and ducks have been unsuccessful, suggesting that these species may not be susceptible (Shi et al., 2020).
Host receptor binding to the angiotensin-converting enzyme 2 (ACE2) cell surface receptor is a critical determinant of the host range of some coronaviruses. In these viruses the receptor binding is mediated by the spike protein which is a glycoprotein expressed on their surface. Host susceptibility to the virus based on in silico simulations of the binding affinity of the viral spike protein to ACE2 receptors suggests that mammals including primates, cattle, hamsters, cetaceans, cats, dogs, bats, pigs, ferrets, civets and pangolins could present high susceptibility to SARS-CoV-2 (Damas et al., 2020; Luan et al., 2020; Piplani et al., 2020). In contrast, the in silico analysis of ACE2 receptors of a range of fish, amphibian, reptile and bird species predicted a very low risk for SARS-CoV-2 infection (Damas et al., 2020). Interestingly, in silico analysis also predicted a very low risk for SARS-CoV-2 infection of several bat and pangolin species, and contradictory results have been reported (Qiu et al., 2020), highlighting the limitations of in silico predictions. Additional factors that can contribute to the host range of coronaviruses, such as interactions between host proteases and the virus spike protein, remain poorly studied in non-model species (Millet and Whittaker, 2015). These additional factors could potentially explain the discrepancy between in silico analyses and experimental infection results. Overall, the host range of SARS-CoV-2 is yet to be determined, but the risk of infection of wild species, in particular mammals, appears to be potentially significant.
3. Risk assessment of emergence of SARS-CoV-2 in Antarctic fauna
3.1. Human-to-human transmission and risk of introduction
As of October 2020, no SARS-CoV-2 infection has been reported at any Antarctic research station. Considering that approximately 5000 research and support personnel and a growing number of tourists from around the world (close to 55,000 tourists on land in 2018/2019; iaato.org 2020) visit Antarctica annually (Hughes et al., 2019), the risk of humans introducing SARS-CoV-2 to Antarctica is non-negligible. It has been described several ways of transmission of SARS-CoV-2 through respiratory droplets, aerosols or through fomites (Allen and Marr, 2020; Cai et al., 2020; Liu et al., 2020; Ong et al., 2020). Epidemiological data suggest that indoor conditions are particularly favourable to transmission (Qian et al., 2020). Scientific and tourism activities in the Antarctic often involve close indoor confinement (vessels and research stations) for extended periods of time. Therefore, upon introduction, the risk of human-to-human transmission of the virus could be high under these circumstances, as illustrated by high transmission on cruise ships in the early phase of the pandemic (Xu et al., 2020).
Detection of SARS-CoV-2 is the first crucial step for effective infection control, however, its relatively long incubation period, up to two weeks (Lauer et al., 2020), and variability in symptoms are major obstacles for mitigating viral transmission (Li et al., 2020). Viral shedding by humans infected with SARS-CoV-2 starts prior to the appearance of clinical symptoms and is estimated to occur for several weeks after infection (Liu et al., 2020; Lu et al., 2020a, Lu et al., 2020b). Viral RNA has been detected in respiratory samples for more than three weeks after the onset of symptoms (He et al., 2020) and for six weeks in faecal samples (Wu et al., 2020). However, it is unclear whether these persistent viral particles are infectious or remnant. Considering these infection dynamics and the frequent movements of tourists, scientists and support staff within Antarctica, the spread of the virus to different sites is a plausible scenario.
3.2. Virus stability and infectivity in the Antarctic environment
SARS-CoV-2 is relatively stable in aerosols, with aerosolized particles remaining potentially viable for several hours (van Doremalen et al., 2020). Moreover, SARS-CoV-2 can also remain viable on a variety of materials, with viable virus being detectable for up to 72 h post-exposure on plastics and steel surfaces in classical indoor conditions (van Doremalen et al., 2020; Matson et al., 2020). Temperature and humidity are crucial for the viability, infectivity and transmission of enveloped viruses (Yang and Marr, 2012). SARS-CoV-2 stability is increased in cold conditions (Chin et al., 2020; Matson et al., 2020) and experiments conducted on other coronaviruses have demonstrated the higher stability of coronaviruses at extreme (low or high) levels of humidity (Ijaz et al., 1985; Casanova et al., 2010). The potential effect of climatic conditions on transmission dynamics from predictions based on modelling of SARS-CoV-2 epidemiological data and endemic human coronavirus data suggest that climatic conditions could play an even more important role during the post-epidemic phase (Baker et al., 2020). Together, these results suggest that the Antarctic continent could provide suitable environmental conditions for the presence and transmission of the virus.
Areas where research activities and tourist operations are concentrated, such as the South Shetland Islands, northern Antarctic Peninsula and Victoria Land (Convey and Peck, 2019), likely present the highest risk for viral introduction and transmission. While environmental conditions could have a limited impact on human-to-human transmission in the current pandemic phase (Baker et al., 2020), the unique climatic conditions in Antarctica could increase transmission risk, notably from humans to animals as most contacts occur outdoors. Cold conditions can potentially facilitate fomite transmission, especially via scientific equipment used by several people or in contact with both humans and other animals. However, aerosol transmission would be less likely outdoors due to the common occurrence of strong winds.
3.3. Transmission to Antarctic wildlife
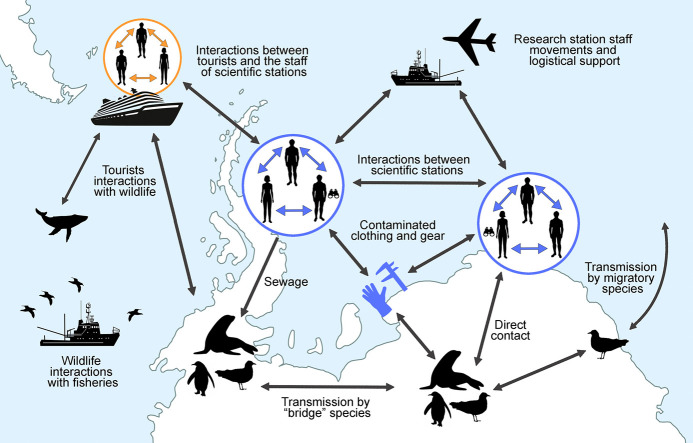
Contact between humans and wildlife in Antarctica occurs during research activities or tourist visitation and, to a lesser extent, during unexpected encounters associated with operational and logistical activities or fishing. As wildlife research often involves handling animals to collect biological samples (e.g. blood or swabs), attaching devices (e.g. satellite transmitters or behavioural loggers) or recording biometry data (e.g. body weight), there are various opportunities for Antarctic wildlife species to be exposed to respiratory droplets or other secretions from SARS-CoV-2 infected people. Resolution 3 on General Guidelines for Visitors to the Antarctic at the XXXIV ATCM (Antarctic Treaty Consultative Meeting) recommends a minimum distance of 5 m from animals; however, there are occasions when animals approach humans spontaneously. In some other instances, unintentional interaction with wildlife is possible while conducting other activities; species that purposely approach humans while defending territories (e.g. skuas protecting their chicks) or in an attempt to scavenge or steal food (e.g. skuas and sheathbills on land, albatrosses at sea) could be at a greater risk of exposure. Therefore, wildlife researchers, research support staff and tourists present potential sources of infection for Antarctic wildlife.
Reverse zoonotic transmission of SARS-CoV-2 could also potentially occur via indirect contact, notably via faecal transmission (Franklin and Bevins, 2020). This transmission route has already been suggested for other infectious agents such as enteric bacteria infecting Antarctic scavengers (Cerdà-Cuéllar et al., 2019). Detection of high levels of SARS-CoV-2 RNA in wastewater in urban areas suggests high faecal shedding by infected individuals (Ahmed et al., 2020), and viable viral particles have previously been isolated from faecal samples (Xiao et al., 2020), although these studies were unable to detect infectious viral particles in faeces. While faecal-oral and faecal-respiratory transmission played a critical role in SARS-CoV-1 epidemiology (McKinney et al., 2006), the role played by these routes of transmission for SARS-CoV-2 remains unclear (Yeo et al., 2020). Faecal transmission of SARS-CoV-2 in many countries is probably partially limited by stringent wastewater treatment protocols (Foladori et al., 2020; Lesimple et al., 2020). However, this may not be the case for wastewater treatment facilities on Antarctic stations. Under the Environmental Protocol to the Antarctic Treaty (the Madrid Protocol, 1991) Annex III, the provisions for wastewater management are very general (Smith and Riddle, 2009). Thus a variety of wastewater treatments are in use, ranging from a lack of any form of treatment facility at stations with an average of fewer than 30 persons during the summer (37% of the permanent stations and 69% of the summer stations; Gröndahl et al., 2009) to advanced treatment systems generating final effluent quality that far exceeds secondary treatment standards (Law et al., 2006). Consequences of such differential treatment of wastewater is reflected by the introduction of enteric bacteria and antimicrobial resistant bacteria to the environment adjacent to research stations (Hernandez and González-Acuña, 2016).
Wildlife migration presents another potential route of transmission of SARS-CoV-2 with risks of migratory species encountering the virus in regions on their migration paths. This includes species that (a) breed in the Antarctic region and visit the coast or the coastal waters of South America, Africa, Australia or New Zealand and/or inhabited Subantarctic islands such as Antarctic terns (Sterna vittata; Tree and Klages, 2004), giant petrels (Macronectes spp.; de Souza Petersen et al., 2017), southern elephant seals (Mirounga leonina; Hindell et al., 2016) and humpback whales (Megaptera novaeangliae; Zerbini et al., 2006), (b) breed in Antarctica and overwinter in the northern hemisphere, such as South Polar skuas (Stercorarius maccormicki; Weimerskirch et al., 2015), or (c) breed in other regions and overwinter in Antarctica, such as Arctic terns (Sterna paradisaea; Fijn et al., 2013).
An additional potential risk for transmission is the chain of human-to-animal-to-animal, where a susceptible species that acquires the virus from humans could then serve as a source of infection for other species of Antarctic fauna. Through this transmission chain species that would not normally interact with humans, such as cetaceans, could be indirectly exposed to the virus via a species such as pinnipeds that could have occasional interaction with infected researchers or tourists. However, this last route of transmission seems highly implausible, as it would require at least two successive cross-species transmission events.
3.4. Coronaviruses of birds and marine mammals
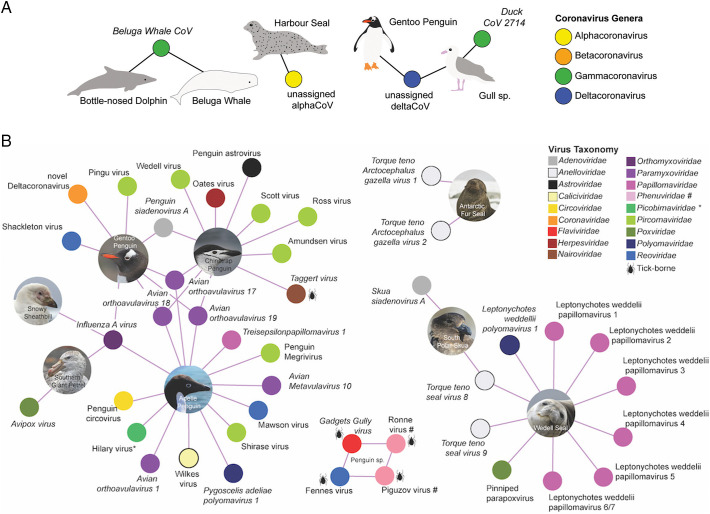
Birds are the main host for gammacoronaviruses and deltacoronaviruses, with five and seven described species, respectively (Fig. 1 ). There are no known alphacoronaviruses or betacoronaviruses for which birds are the confirmed hosts. In the gammacoronaviruses, the species Avian coronavirus, which comprises infectious bronchitis virus, Turkey coronavirus and Canada Goose coronavirus, are the only viral species associated with disease (Papineau et al., 2019). There are no reports of disease outcomes in wild birds caused by Duck coronavirus 2714, which is the species of gammacoronavirus most commonly detected in wild birds (e.g. Muradrasoli et al., 2010). There are no reports of disease associated with infections by deltacoronanviruses in avian species, with the exception of a disease event caused by Quail coronavirus (Domanska-Blicharz et al., 2019), a member of the Coronavirus HKU15 species. Interestingly, a putative species of deltacoronavirus has been detected in healthy Antarctic penguins (Wille et al., 2020). This viral species does not cause disease, has a broad geographic distribution and has been detected in a wide range of avian hosts, including falcons, bustards, pigeons, gulls and shorebirds (Wille et al., 2019b).
Fig. 1.
Viral species relevant to this risk assessment. (A) Coronavirus species detected in marine mammals and seabirds, globally. A filled circle refers to a virus, and is coloured according to the genera. Hosts are indicated by an image and connected by lines to the viruses from which they have been detected. A virus (filled circle) is connected to more than one library indicates that the same (putative) virus species has been found in both hosts. Ratified viral species are presented in italics, putative viral species are presented in regular text. Silhouettes generated by M. Wille. (B) Viral species detected in Antarctic birds and mammals. We have not included viral species detected by serology. Only viral species detected in Antarctica have been included; we have excluded viral species recorded on sub-antarctic islands such as South Georgia Island and Macquarie Island. A filled circle refers to a virus. Hosts are indicated by an image and connected by lines to the viruses from which they have been detected. Ratified viral species are presented in italics, putative viral species are presented in regular text. Viruses transmitted by ticks are indicated by a tick silhouette. Picobirnaviridae, indicated by an asterisk have previously been associated with vertebrate hosts. Other than Taggert Virus, in which viral reads were found in Chinstrap Penguins, the Antarctic hosts of tick viruses are not confirmed. Gadgets Gully virus has been detected in ticks in Antarctica and King Penguins in Macquarie Island. Ronne Virus and Piguzov virus, members of the Phenuvidiae are indicated with a # and their avian hosts have not been confirmed despite being detected in ticks adjacent to penguin colonies in two independent studies. All images were taken by M. Wille.
In marine mammals, two related gammacoronaviruses have been found in cetaceans: Beluga whale coronavirus and the closely related Bottlenose dolphin coronavirus (Schütze, 2016). Despite being gammacoronaviruses, these cetacean viruses differ from the gammacoronaviruses found in birds and are grouped in a different subgenus, consequently it seems unlikely to represent a recent cross-species transmission event. Also, it is unclear whether these cetacean viruses cause disease in their hosts. On the other hand, the record of an alphacoronavirus in harbor seals (Phoca vitulina) could be considered more concerning because it suggests that seals might be susceptible to mammalian coronaviruses. The Harbor seal coronavirus falls in a clade comprising Ferret coronavirus, Feline coronavirus, Canine coronavirus and Transmissible gastroenteritis virus (Nollens et al., 2010). Whether this is the detection of a virus species specific to seals, or the detection of cross-species spill-over, remains to be determined.
3.5. Virus presence in Antarctic wildlife
Information about the presence of viruses in Antarctic wildlife is scarce and fragmented (Barbosa and Palacios, 2009; Barbosa et al., 2015; Grimaldi et al., 2015; Grimaldi et al., 2018; Smeele et al., 2018). A major constraint contributing to this dearth of knowledge is that many of the early studies were limited to investigating only known viruses, most of which were associated with humans or domestic animals. However, the development of novel diagnostic tools has allowed the discovery of several new viruses in Antarctic wildlife over the last decade (Tables S1 and S2; Fig. 1). Despite coronaviruses being known to circulate in wildlife populations there has been no dedicated investigation of coronaviruses in Antarctic fauna. This is limiting because without dedicated studies it is not possible to determine which viral species circulate; which animals are potential reservoirs; what is the prevalence or burden of these viruses in animal hosts; and what is the genetic diversity of these viral species. This baseline information is essential to improve our understanding of the consequences of the potential introduction of SARS-CoV-2 to Antarctica and to assess the risk of recombination between local coronaviruses and SARS-CoV-2 (Su et al., 2016). In this context, new research to investigate the presence of coronavirus in Antarctic wildlife is urgently needed.
3.6. Susceptibility of Antarctic wildlife to infection by SARS-CoV-2
The lack of information about susceptibility to SARS-CoV-2 is a key knowledge gap for a risk assessment of the potential impacts of this virus for Antarctic wildlife. In the absence of this information, the in silico modelling results from Damas et al. (2020) combined with the current understanding of coronavirus host range can provide some early insight. Based on a comparative and structural analysis of the ACE2 receptor of 72 avian species, the Adélie penguin (Pygoscelis adeliae) and the emperor penguin (Aptenodytes forsteri) which inhabit Antarctic regions, showed “very low” binding affinity of the ACE2 receptor to the SARS-CoV-2 spike protein, as did all other studied bird species (Damas et al., 2020). This suggests that other Antarctic birds are likely to have low susceptibility to SARS-CoV-2 infection, which is further supported by the absence of infection in chickens and ducks experimentally inoculated with SARS-CoV-2 (Shi et al., 2020). Specifically, viral RNA was not detected in any swabs collected from these virus-inoculated animals or from naïve contact animals, and all animals were seronegative for SARS-CoV-2 when tested by ELISA at 14 days post-infection (Shi et al., 2020). This is perhaps not surprising given that birds are not reservoirs for betacoronaviruses. Based on this indirect information, transmission of SARS-CoV-2 to Antarctic birds seems unlikely. Nevertheless, further research on Antarctic birds is warranted to confirm this.
While none of the six pinniped species in the modelling study by Damas et al. (2020) occur in the Antarctic region, the pinnipeds evaluated had “very low” ACE2 binding affinity, suggesting that Antarctic pinnipeds could also have low susceptibility to SARS-CoV-2 infection. In contrast, 12 of the 14 cetacean species in that study received a “high” score for the binding affinity of their ACE2 receptor, including the Antarctic minke whale (Balaenoptera bonaerensis) and killer whale (Orcinus orca) (Damas et al., 2020). A third cetacean species occurring in Antarctic waters, the sperm whale (Physeter macrocephalus), showed “medium” ACE2 binding affinity (Damas et al., 2020). Based on these findings, cetaceans appear to have the highest risk of SARS-CoV-2 infection among Antarctic wildlife. However, the results from Damas et al. (2020) should be interpreted with caution, because some vertebrate species such as bats, which are known hosts of coronaviruses closely related to SARS-CoV-2, or ferrets, which have demonstrated susceptibility to experimental infection, also scored in the “low” or “very low” categories. This suggests that ACE2 receptor binding affinity might not be the only factor determining species susceptibility to SARS-CoV-2 and that alternative receptors could also be involved (Damas et al., 2020; Sigrist et al., 2020). It is therefore clear that further studies are necessary to evaluate the susceptibility of Antarctic mammals to coronaviruses, including SARS-CoV-2.
Knowledge of SARS-CoV-2 pathogenicity in non-human animals in general is limited. However, symptomatic infections, eventually resulting in death, have been recorded in experimentally infected cats, and in farmed mink in contact with infected humans (Oreshkova et al., 2020; Shi et al., 2020). For these reasons, the risk posed by SARS-CoV-2 infection to Antarctic wildlife health cannot be disregarded.
3.7. Risk assessment conclusions and perspectives
Globally wild animal and human interactions are increasing as natural areas are increasingly disrupted by anthropogenic activities (Johnson et al., 2020 and references therein). Increased interactions between wild animals and humans greatly facilitates the potential of pathogen spill-over and the emergence of zoonotic pathogens. Recognising this increased risk of cross-species transmission, there is a clear imperative for careful monitoring of human activities in Antarctica and for implementing mitigation measures. This can be achieved by the development and implementation of a set of clear and standardised biosecurity protocols to minimise potential pathogen transfer. These could include: quarantine periods for visitors to Antarctica, increased screening to identify pathogens, increased serological screening to detect the presence of antibodies against high risk pathogen families, and managing human movements to and within Antarctica.
With respect to the current pandemic of SARS-CoV-2 and based on the available knowledge, we can conclude that the environmental conditions in Antarctica are likely favourable for virus stability and thus infectivity. Transmission among humans could occur locally at research stations, onboard research vessels or tourist cruise ships as has already been documented (Ing et al., 2020; Xu et al., 2020), and at broader geographical scales depending on the movements of tourists and scientific research endeavours between different locations. Field researchers handling animals should be considered as posing the highest risk of transmission to animals due to direct and close contact with wildlife, while tourists and other personnel pose a significant risk only if in close proximity (< 5 m) to Antarctic fauna. Although Antarctic wildlife appears to be at low risk of infection when the predictions based on the structure of the virus receptors found in these animals is considered, these findings should be interpreted with caution until further data on transmission risk to Antarctic species can be obtained. Therefore, the precautionary principle should be applied, and measures should be taken to reduce the risk of introduction and transmission of the virus to Antarctic wildlife. In addition, it has been suggested that SARS-CoV-2 could become endemic to human populations (Kissler et al., 2020), implying that the risk of introduction of the virus to Antarctic ecosystems and its potential impact on the local fauna, could remain well after the epidemic phase. In the future, based on available knowledge and vaccine availability, mitigation measures would have to be revised.
4. Proposed mitigation measures
Based on the current knowledge on SARS-CoV-2, we propose a set of guidelines and additional measures to reduce the risk of SARS-CoV-2 and other infectious pathogens being introduced to Antarctic fauna. Considering the precautionary principle and this risk assessment, the following guidelines are recommended in addition to the biosecurity measures and regulations aimed at limiting animal disturbance already in place:
4.1. Antarctic research facilities staff (crew, scientific and technical personnel)
-
1.
It is recommended that all individuals participating in research facilities (crew, scientific and technical personnel) be tested for active virus infection by RT-qPCR or any validated rapid SARS-CoV-2 test prior to departure for Antarctica (at a minimum) in parallel to quarantine procedures for two weeks (preferable).
-
2.
As a general measure, any individual demonstrating any disease symptoms and specifically those compatible with COVID-19 should self-isolate, be tested for SARS-CoV-2 presence if possible, and not be permitted close contact with wildlife or humans.
-
3.
Individuals in the same station, field camp or vessel with a risk of local infection transmission due to close contact with a person showing symptoms compatible with COVID-19 should be excluded from close contact with wildlife.
-
4.
Due to the possibility of SARS-CoV-2 positive, asymptomatic individuals, extreme caution should be taken when handling animals to avoid the risk of transmission. Appropriate face masks (PFF-2 is recommended) must be worn whenever in close contact with animals. Additionally, eye protection, gloves (latex, nitrile or rubber) and specific clothing (i.e. overalls) should be used whenever possible (considering the use of cold weather clothing). Hand washing with soap or hydroalcoholic gel solution is highly recommended before and after animal handling. No animals should be handled in enclosed spaces.
-
5.
To avoid transmission through fomites, outer clothing should be disinfected using soap and warm water every day prior to and after work with animals and in between colonies (if researchers visit more than one colony per day).
-
6.
Field equipment disinfection procedures should be carried out prior to and after animal handling, especially when working in different areas or colonies. Wherever possible, it is recommended that field/sampling equipment not be shared between locations if appropriate disinfection cannot be achieved. The use of 70% ethanol, vaporized hydrogen peroxide, dry heat or UV lamps is recommended.
-
7.
To minimise animal exposure to equipment/potential fomite transmission, field equipment should not be left unattended and should be kept far from wildlife when not in use.
-
8.
Scientific equipment deployed in the field should be disinfected before deployment and after handling.
-
9.
Individuals should adhere to strict recommendations for personal hygiene at all times (frequent hand washing for the appropriate time and regular disinfection).
-
10.
Limit, except where considered essential for scientific and logistical purposes, movement of individuals between stations, field camps or research vessels, and restrict contact between tourists and all personnel at stations or research vessels.
For additional information for national program operators, which are independent of this recommendations, see SARS-CoV-2/COVID-19 information for Antarctic Operations; https://www.comnap.aq/members/covid-19-information/.
4.2. Tourists (including researchers/staff accompanying tourists)
-
1.
It is recommended that all individuals participating in a tourist expedition be tested for active virus infection by RT-qPCR or any validated rapid SARS-CoV-2 test at least prior to departure for Antarctica and maintain quarantine procedures for two weeks.
-
2.
Tourists showing symptoms compatible with COVID-19 disease must self-isolate, be tested for SARS-CoV-2 presence if possible and be excluded from close contact with wildlife or humans.
-
3.
Tourists in the same vessel with risk of infection due to the presence of an infected person (close contacts), should not be permitted to go anywhere near wildlife.
-
4.
Tourists and guides should always keep a minimum distance of 5 m from wildlife, in addition to strict adherence to IAATO guidelines given a greater distance may be required for different animal species and age cohorts. If an animal spontaneously approaches an individual or group, tourists and guides should retreat to ensure this minimum distance is rigorously adhered to.
-
5.
To minimise animal exposure to equipment/potential fomite transmission, field equipment should not be left unattended and should be kept far from wildlife.
-
6.
All tourists and staff should refrain from sitting on or lying on bare ground or rocks or leaving any equipment on bare ground or rocks close to animal activity (within 10 m of nests, haul out sites or pathways).
-
7.
Tourists and guides to adhere to strict recommendations for personal hygiene at all times (frequent hand washing for the appropriate time and regular disinfection).
There guidelines are proposed based on the knowledge summarized above and mitigation measure protocols available in the literature (Chin et al., 2020; Fischer et al., 2020).
4.3. Additional measures
-
1.
Surveillance of the presence of SARS-CoV-2 in wastewater from cruise ships, research stations and research vessels is recommended.
-
2.
As a reminder, human waste from camps or field parties is to be removed from the field and returned to research stations or vessels for wastewater management.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is an outcome of the Working Group of Wildlife Health Monitoring of the SCAR Expert Group of Birds and Marine Mammals.
AB is supported by the Spanish Research Agency project (CTM2015-64720). AG is supported by the Defence Advanced Research Projects Agency (DARPA PREEMPT Cooperative Agreement No. D18AC00031). The content of the information does not necessarily reflect the position or the policy of the US government, and no official endorsement should be inferred. AV is supported by National Science Foundation (USA) Polar program (award # 1947040). CRM is supported by Australia's Integrated Marine Observing System. IMOS is enabled by the National Collaborative Research Infrastructure Strategy (NCRIS). It is operated by a consortium of institutions as an unincorporated joint venture, with the University of Tasmania as Lead Agent.
JID is supported by Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Universidad Nacional de La Plata UNLP (N#859). VM is supported by National Science Foundation (USA) Polar program (PLR 1543459). TB is supported by CNRS, French Polar Institute Project ECOPATH (IPEV 1151), ZATA and OSU OREME. DGA is supported by INACH T-23-19 project.
Editor: Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2020.143352.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J., Marr L. 2020. Re-thinking the Potential for Airborne Transmission of SARS-CoV-2. [DOI] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AVMA (American Veterinary Medical Association) SARS-CoV-2 in animals. 2020. https://www.avma.org/resources-tools/animal-health-and-welfare/covid-19/sars-cov-2-animals-including-pets
- Baker R.E., Yang W., Vecchi G.A., Metcalf C.J.E., Grenfell B.T. Susceptible supply limits the role of climate in the early SARS-CoV-2 pandemic. Science. 2020 doi: 10.1126/science.abc2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa A., Palacios M.J. Health of Antarctic birds: a review of their parasites, pathogens and diseases. Polar Biol. 2009;32:1095–1115. doi: 10.1007/s00300-009-0640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa A., Schneider Costa E., Dewar M., González-Acuña D., Gray R., Power M., Vanstreels R.E.T. Antarctic Wildlife Diseases. 2015. https://www.environments.aq/information-summaries/antarctic-wildlife-diseases/ Antarctic Environments Portal.
- Cai J., Sun W., Huang J., Gamber M., Wu J., He G. Indirect virus transmission in cluster of COVID-19 cases, Wenzhou, China, 2020. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2606.200412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L.M., Jeon S., Rutala W.A., Weber D.J., Sobsey M.D. Effects of air temperature and relative humidity on coronavirus survival on surfaces. Appl. Environ. Microb. 2010;76:2712–2717. doi: 10.1128/AEM.02291-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdà-Cuéllar M., Moré E., Ayats T., Aguilera M., Muñoz-González S., Antilles N., Ryan P.G., González-Solís J. Do humans spread zoonotic enteric bacteria in Antarctica? Sci. Total Environ. 2019;654:190–196. doi: 10.1016/j.scitotenv.2018.10.272. [DOI] [PubMed] [Google Scholar]
- Chan R.W.Y., Chan M.C.W., Agnihothram S., Chan L.L.Y., Kuok D.I.T., Fong J.H.M., Guan Y., Poon L.L.M., Baric R.S., Nicholls J.M., Peiris J.S.M. Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J. Virol. 2013;87:6604–6614. doi: 10.1128/JVI.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., To K.K.W., Tse H., Jin D.Y., Yuen K.Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2020;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Yuan S., Zhang A.J., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y., Fan Z., Li C., Liang R., Cao J., Tang K., Luo C., Cheng V.C.-C., Cai J.-P., Chu H., Chan K.-H., To K.K.-W., Sridhar S., Yuen K.-Y. Surgical mask partition reduces the risk of non-contact transmission in a golden Syrian hamster model for coronavirus disease 2019 (COVID-19) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1:e1–e43. doi: 10.1016/S2666-5247/(20)3003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Convey P., Peck L.S. Antarctic environmental change and biological responses. Sci. Adv. 2019;5(11):eaaz0888. doi: 10.1126/sciadv.aaz0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Muth D., Niemeyer D., Drosten C. Chapter eight - hosts and sources of endemic human coronaviruses. In: Mettenleiter T.C., Roossinck M.J., editors. Kielian, M. Academic Press; Advances in Virus Research: 2018. pp. 163–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J., Hughes G.M., Keough K.C., Painter C.A., Persky N.S., Corbo M., Hiller M., Koepfli K.-P., Pfenning A.R., Zhao H., Genereux D.P., Swofford R., Pollard K.S., Ryder O.A., Nweeia M.T., Lindblad-Toh K., Teeling E.C., Karlsson E.K., Lewin H.A. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. U.S.A. 2020;8(117(36)):22311–22322. doi: 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza Petersen E., de Araujo J., Krüger L., Seixas M.M., Ometto T., Thomazelli L.M., Walker D., Durigon E.L., Petry M.V. First detection of avian influenza virus (H4N7) in Giant petrel monitored by geolocators in the Antarctic region. Mar. Biol. 2017;164:62. doi: 10.1007/s00227-017-3086-0. [DOI] [Google Scholar]
- Domanska-Blicharz K., Kuczkowski M., Sajewicz-Krukowska J. Whole genome characterisation of quail deltacoronavirus detected in Poland. Virus Genes. 2019;55:243–247. doi: 10.1007/s11262-019-01639-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijn R.C., Hiemstra D., Phillips R.A., van der Winden J. Arctic terns Sterna paradisaea from the Netherlands migrate record distances across three oceans to Wilkes Land, East Antarctica. Ardea. 2013;101:3–12. [Google Scholar]
- Fischer R.J., Morris D.H., van Doremalen N., Sarchette S., Matson M.J., Bushmaker T., Yinda C.K., Seifert S.N., Gamble A., Williamson B.N., Judson S.D., de Wit E., Lloyd-Smith J.O., Munster V.J. Effectiveness of N95 respirator decontamination and reuse against SARS-CoV-2 virus. Emerg. Infect. Dis. 2020;26:2253–2255. doi: 10.3201/eid2609.201524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743:140444. doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin A.B., Bevins S.N. Spillover of SARS-CoV-2 into novel wild hosts in North America: a conceptual model for perpetuation of the pathogen. Sci. Total Environ. 2020;733:139358. doi: 10.1016/j.scitotenv.2020.139358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi W.W., Seddon P.J., Lyver P.O’.B., Nakagawa S., Tompkins D.M. Infectious diseases of Antarctic penguins: current status and future threats. Polar Biol. 2015;38:591–606. [Google Scholar]
- Grimaldi W., Ainley D.G., Massaro M. Multi-year serological evaluation of three viral agents in the Adélie Penguin (Pygoscelis adeliae) on Ross Island. Antarctica. Polar Biol. 2018;41:2023–2031. [Google Scholar]
- Gröndahl F., Sidenmark J., Thomsen A. Survey of waste water disposal practices at Antarctic research stations. Polar Res. 2009;28:298–306. doi: 10.1111/j.1751-8369.2008.00056.x. [DOI] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Hemida M.G., Chu D.K.W., Perera R.a.P.M., Ko R.L.W., So R.T.Y., Ng B.C.Y., Chan S.M.S., Chu S., Alnaeem A.A., Alhammadi M.A., Webby R.J., Poon L.L.M., Balasuriya U.B.R., Peiris M. Coronavirus infections in horses in Saudi Arabia and Oman. Transbound. Emerg. Dis. 2017;64:2093–2103. doi: 10.1111/tbed.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez J., González-Acuña D. Anthropogenic antibiotic resistant genes mobilization to the polar regions. Infect. Ecol. Epidemiology. 2016;6(1) doi: 10.3402/iee.v6.32112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindell M.A., McMahon C.R., Bester M.N., Boehme L., Costa D., Fedak M.A., Guinet C., Herraiz-Borreguero L., Harcourt R.G., Huckstadt L., Kovacs K.M., Lydersen C., McIntyre T., Muelbert M., Patterson C., Roquet F., Williams G., Charrassin J.-B. Circumpolar habitat use in the southern elephant seal: implications for foraging success and population trajectories. Ecosphere. 2016;7 [Google Scholar]
- Hughes K.A., Convey P., Pertierra L.R., Vega G.C., Aragón P., Olalla-Tárraga M.Á. Human-mediated dispersal of terrestrial species between Antarctic biogeographic regions: a preliminary risk assessment. J. Environ. Manag. 2019;232:73–89. doi: 10.1016/j.jenvman.2018.10.095. [DOI] [PubMed] [Google Scholar]
- Ijaz M.K., Brunner A.H., Sattar S.A., Nair R.C., Johnson-Lussenburg C.M. Survival characteristics of airborne human coronavirus 229E. J. Gen. Virol. 1985;66:2743–2748. doi: 10.1099/0022-1317-66-12-2743. [DOI] [PubMed] [Google Scholar]
- Ing A.J., Cocks C., Green J.P. COVID-19: in the footsteps of Ernest Shackleton. Thorax. 2020;75:693–694. doi: 10.1136/thoraxjnl-2020-215091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.K., Hitchens P.L., Pandit P.S., Rushmore J., Evans T.S., Young C.C.W., Doyle M.M. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. P. Roy. Soc. B. 2020;287:20192736. doi: 10.1098/rspb.2019.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-I., Kim S.-G., Kim S.-M., Kim E.-H., Park S.-J., Yu K.-M., Chang J.-H., Kim E.J., Lee S., Casel M.A.B., Um J., Song M.-S., Jeong H.W., Lai V.D., Kim Y., Chin B.S., Park J.-S., Chung K.-H., Foo S.-S., Poo H., Mo I.-P., Lee O.-J., Webby R.J., Jung J.U., Choi Y.K. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020;368:860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Li K.S.M., Tsang A.K.L., Lam C.S.F., Ahmed S., Chen H., Chan K.-H., Woo P.C.Y., Yuen K.-Y. Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of Pipistrellus bat coronavirus HKU5 in Japanese Pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S.A., Grantz H.K., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172(9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law K.P., Kulchawik R.J., Zenz D.R., Bouchard T.B. Overview of the design, construction, and operation of the McMurdo wastewater treatment facility in Antarctica. Water Environ. Fed. 2006;12:1439–1448. doi: 10.2175/193864706783749909. [DOI] [Google Scholar]
- Lesimple A., Jasim S.Y., Johnson D.J., Hilal N. The role of wastewater treatment plants as tools for SARS-CoV-2 early detection and removal. J. Water Process Eng. 2020;38:101544. doi: 10.1016/j.jwpe.2020.101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li R., Pei S., Chen B., Song Y., Zhang T., Yang W., Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ning Z., Chen Y., Guo M., Liu Y., Gali N.K., Sun L., Duan Y., Cai J., Westerdahl D., Liu X., Xu K., Ho K.-f. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582:557–560. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Lu J., Gu J., Li K., Xu C., Su W., Lai Z., Zhou D., Yu C., Xu B., Yang Z. COVID-19 outbreak associated with air conditioning in restaurant, Guangzhou, China, 2020. Emerg. Infect. Dis. 2020;26:1628–1631. doi: 10.3201/eid2607.200764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan J., Jin X., Lu Y., Zhang L. SARS-CoV-2 spike protein favors ACE2 from Bovidae and Cricetidae. J. Med. Virol. 2020;92:1649–1656. doi: 10.1002/jmv.25817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson M.J., Yinda C.K., Seifert S.N., Bushmaker T., Fischer R.J., van Doremalen N., Lloyd-Smith J.O., Munster V.J. Effect of environmental conditions on SARS-CoV-2 stability in human nasal mucus and sputum. Emerg. Infect. Dis. 2020;26:2276–2278. doi: 10.3201/eid2609.202267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney K.R., Gong Y.Y., Lewis T.G. Environmental transmission of SARS at Amoy gardens. J. Environ. Health. 2006;68(26):26–30. [PubMed] [Google Scholar]
- Mihindukulasuriya K.A., Wu G., St. Leger J., Nordhausen R.W., Wang D. Identification of a novel coronavirus from a Beluga whale by using a Panviral microarray. J. Virol. 2008;82:5084–5088. doi: 10.1128/JVI.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse S.S., Mazet J.A.K., Woolhouse M., Parrish C.R., Carroll D., Karesh W.B., Zambrana-Torrelio C., Lipkin W.I., Daszak P. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muradrasoli S., Balint A., Wahlgren J., Waldenström J., Belák S., Blomberg J., Olsen B. Prevalence and phylogeny of coronaviruses in wild birds from the Bering Strait area (Beringia) PLoS One. 2010;5 doi: 10.1371/journal.pone.0013640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollens H.H., Wellehan J.F., Archer L., Lowenstine L.J., Gulland F.M. Detection of a respiratory coronavirus from tissues archived during a pneumonia epizootic in free-ranging Pacific harbor seals Phoca vitulina richardsii. Dis. Aquat. Org. 2010;90:113–120. doi: 10.3354/dao02190. [DOI] [PubMed] [Google Scholar]
- Ong S.W.X., Tan Y.K., Chia P.Y., Lee T.H., Ng O.T., Wong M.S.Y., Marimuthu K. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. J. Amer. Med. Assoc. 2020;323:1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreshkova N., Molenaar R.-J., Vreman S., Harders F., Munnink B.B.O., Hakze R., Gerhards N., Tolsma P., Bouwstra R., Sikkema R., Tacken M., de Rooij M.M.T., Weesendorp E., Engelsma M., Bruschke C., Smit L.A.M., Koopmans M., van der Poel W.H.M., Stegeman A. SARS-CoV2 infection in farmed mink, Netherlands, April and May 2020. Euro Surveill. 2020;25(23):2001005. doi: 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papineau A., Berhane Y., Wylie T.N., Wylie K.M., Sharpe S., Lung O. Genome organization of Canada goose coronavirus, a novel species identified in a mass die-off of Canada geese. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-42355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piplani S., Singh P.K., Winkler D.A., Petrovsky N. 2020. In Silico Comparison of Spike Protein-ACE2 Binding Affinities Across Species; Significance for the Possible Origin of the SARS-CoV-2 Virus. arXiv:2005.06199. [Google Scholar]
- Plowright R.K., Parrish C.R., McCallum H., Hudson P.J., Ko A.I., Graham A.L., Lloyd-Smith J.O. Pathways to zoonotic spillover. Nat. Rev. Microbiol. 2017;15:502–510. doi: 10.1038/nrmicro.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H., Miao T., Liu L., Zheng X., Luo D., Li Y. 2020. Indoor Transmission of SARS-CoV-2. medRxiv 2020.04.04.20053058. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Zhao Y.-B., Wang Q., Li J.-Y., Zhou Z.-J., Liao C.-H., Ge X.-Y. Predicting the angiotensin converting enzyme 2 (ACE2) utilizing capability as the receptor of SARS-CoV-2. Microbes Infect. 2020;22:221–225. doi: 10.1016/j.micinf.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütze H. Coronaviruses in aquatic organisms. Aquaculture Virology. 2016;2016:327–335. doi: 10.1016/B978-0-12-801573-5.00020-6. [DOI] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C.J., Bridge A., Le Mercier P. A potential role for integrins in host cell entry by SARS-CoV-2. Antivir. Res. 2020;177:104759. doi: 10.1016/j.antiviral.2020.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeele Z.E., Ainley D.G., Varsani A. Viruses associated with Antarctic wildlife: from serology based detection to identification of genomes using high throughput sequencing. Virus Res. 2018;243:91–105. doi: 10.1016/j.virusres.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.J., Riddle M.J. Sewage disposal and wildlife health in Antarctica. In: Kerry K.R., Riddle M., editors. Health of Antarctic Wildlife: A Challenge for Science and Policy. Springer; Berlin, Heidelberg: 2009. pp. 271–315. [DOI] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree A.J., Klages N.T. Population size, distribution and origins of Antarctic terns Sterna vittata wintering in South Africa. Mar. Ornithol. 2004;15:55–61. [Google Scholar]
- Van Doremalen N., Morris D.H., Holbrook M.G., Gamble A., Williamson M.N., Tamin A., Lloyd-Smith J.O., de Wit E. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moës E., Thoelen I., Wollants E., Lemey P., Vandamme A.-M., Van Ranst M. Genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimerskirch H., Tarroux A., Chastel O., Delord K., Cherel Y., Descamps S. Population-specific wintering distributions of adult south polar skuas over three oceans. Mar. Ecol-Prog. Ser. 2015;538:229–237. doi: 10.3354/meps11465. [DOI] [Google Scholar]
- WHO (World Health Organization) https://covid19.who.int/
- Wille M., Shi M., Klaassen M., Hurt A.C., Holmes E.C. Virome heterogeneity and connectivity in waterfowl and shorebird communities. ISME J. 2019;13:2603–2616. doi: 10.1038/s41396-019-0458-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille M., Harvey E., Shi M., Gonzalez-Acuña D., Homes E.C., Hurt A.C. Sustained RNA virome diversity in Antarctic penguins and their ticks. ISME J. 2020;14:1768–1782. doi: 10.1038/s41396-020-0643-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao J., Huang J., Zhao J. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Qian H., Miao T., Yen H.-I., Tan H., Cowling B.J., Li Y.J. 2020. Transmission Routes of Covid-19 Virus in the Diamond Princess Cruise ship. medRxiv 2020.04.09.20059113. [DOI] [Google Scholar]
- Yang W., Marr L.C. Mechanisms by which ambient humidity may affect viruses in aerosols. Appl. Environ. Microbiol. 2012;78:6781–6788. doi: 10.1128/AEM.01658-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbini A.N., Andriolo A., Heide-Jørgensen M.P., Pizzorno J.L., Maia Y.G., VanBlaricom G.R., DeMaster D.P., Simões-Lopes P.C., Moreira S., Bethlem C. Satellite-monitored movements of humpback whales Megaptera novaeangliae in the Southwest Atlantic Ocean. Mar. Ecol-Prog. Ser. 2006;313:295–304. [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30:1346–1351.e2. doi: 10.1016/j.cub.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X.-L., Shi W.-F., Zhang W., Zhu Y., Zhang Y.-W., Xie Q.-M., Mani S., Zheng X.-S., Li B., Li J.-M., Guo H., Pei G.-Q., An X.-P., Chen J.-W., Zhou L., Mai K.-J., Wu Z.-X., Li D., Anderson D.E., Zhang L.-B., Li S.-Y., Mi Z.-Q., He T.-T., Cong F., Guo P.-J., Huang R., Luo Y., Liu X.-L., Chen J., Huang Y., Sun Q., Zhang X.-L.-L., Wang Y.-Y., Xing S.-Z., Chen Y.-S., Sun Y., Li J., Daszak P., Wang L.-F., Shi Z.-L., Tong Y.-G., Ma J.-Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material