Summary
We present an integrated analysis of the clinical measurements, immune cells, and plasma multi-omics of 139 COVID-19 patients representing all levels of disease severity, from serial blood draws collected during the first week of infection following diagnosis. We identify a major shift between mild and moderate disease, at which point elevated inflammatory signaling is accompanied by the loss of specific classes of metabolites and metabolic processes. Within this stressed plasma environment at moderate disease, multiple unusual immune cell phenotypes emerge and amplify with increasing disease severity. We condensed over 120,000 immune features into a single axis to capture how different immune cell classes coordinate in response to SARS-CoV-2. This immune-response axis independently aligns with the major plasma composition changes, with clinical metrics of blood clotting, and with the sharp transition between mild and moderate disease. This study suggests that moderate disease may provide the most effective setting for therapeutic intervention.
Keywords: immune response, multi-omics, COVID-19, infection, proteomics, metabolomics, single-cell TCR-seq, single-cell RNA-seq, CITE-seq, single-cell secretome
Graphical Abstract

Highlights
-
•
Analysis of serial blood from 139 COVID-19 patients reveals immune coordination
-
•
A major immunological shift is seen between mild and moderate infection
-
•
Moderate and severe cases exhibit inflammation and a sharp drop in blood nutrients
-
•
Novel immune cell subsets emerge in moderate cases and increase with severity
Using serial blood draws from COVID-19 patients, Su et al. present an extensive multi-omics dataset of plasma and single PBMCs covering the first week of infection following clinical diagnosis, which includes information on plasma proteins, metabolites, and on PBMC transcriptomic and surface-protein data, immune receptor sequences, secreted proteins, and electronic health record data. Their integrated analysis identifies a major immunological shift between mild and moderate infection, which includes an increase in inflammation, drop in blood nutrients, and the emergence of novel immune cell subpopulations that intensify with disease severity.
Introduction
The novel coronavirus disease, COVID-19, has rapidly spread to become a global health challenge, with over 38 million cases and over 1 million associated fatalities as reported through mid-October 2020. 20%–31% of symptomatic patients require hospitalization, with intensive care unit (ICU) admission rates ranging from 4.9%–11.5%, and fatality rates ranging from 2%–10% (Iype and Gulati, 2020). Patients with more severe COVID-19 infections are distinguished by significant immune dysregulation, the nature of which is incompletely understood.
Most reports on immune dysfunction in COVID-19 patients have focused on severe disease. In particular, studies on peripheral blood mononuclear cells (PBMCs) using single-cell analytics, have revealed phenomena that tracked with disease severity, including robust HLA class II downregulation on monocytes (Wilk et al., 2020), lymphopenia (Cao, 2020), immune cell exhaustion (Zheng et al., 2020), and elevated levels of inflammatory cytokines (Del Valle et al., 2020). Larger patient population studies have revealed abnormal myeloid cell subsets in severe COVID-19 (Schulte-Schrepping et al., 2020; Silvin et al., 2020) and dysregulated mammalian target of rapamycin (mTOR) signaling in dendritic cells (Arunachalam et al., 2020). Mathew et al. (2020) reported on a large patient cohort characterized through clinical observations and flow cytometry single-cell proteomics to identify three patient immunotypes associated with clinical trajectories. Lucas et al. (2020) reported on maladapted cytokine profiles associated with severe COVID-19, including immune dysregulation arising from impaired type I interferon (IFN) response associated with elevated interleukin (IL)-6 (Hadjadj et al., 2020). Dysregulated plasma metabolites have been reported for serious infection (Shen et al., 2020). Of course, inflammation and immune activation are expected to accompany a viral infection, but understanding the coupling between elevated inflammation signals, plasma metabolite composition, and immune cell dysfunction requires a full characterization of both plasma and PBMCs in large cohorts of COVID-19 patients representing the spectrum of infection severities.
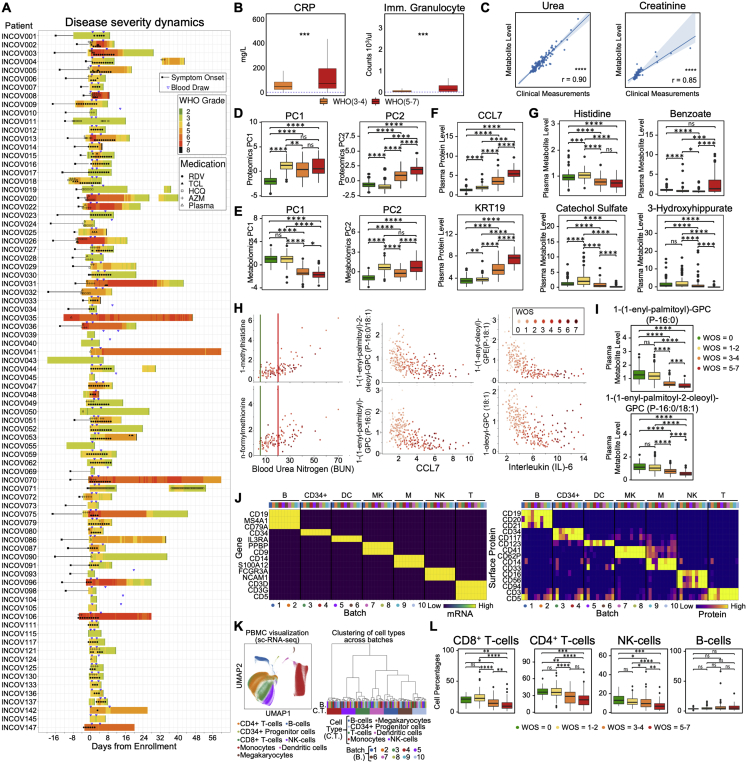
We characterized the circulating immune cell classes and the plasma multi-omic profiles from a cohort of 139 COVID-19 patients (265 samples from 2 longitudinal blood draws per patient) and 258 healthy donors. We interpret that data through the patients’ clinical features extracted from their electronic health records (EHR) (Figure 1A). One blood draw was collected shortly after the initial clinical diagnosis (time = T1), and the second was collected a few days later (T2) (Figure S1A). For each draw, the plasma levels of around 500 proteins and 1,000 metabolites were quantified along with single-cell multi-omic analyses of PBMCs in which whole transcriptome, 192 surface proteins, 32 secreted proteins, and T cell and B cell receptor gene sequences were measured (Figure 1A).
Figure 1.
Overview of the Multi-Omic Characterization of Immune Responses in COVID-19 Patients
(A) Overview of the ISB/Swedish INCOV study of COVID-19 patients. The bar graph represents the counts of patient samples across WHO ordinal score (WOS) of disease severity. The various analytic assays run on the plasma and isolated PBMCs are indicated.
(B) EHR clinical measurements for the COVID-19 patients. Correlation matrix of 34 clinical features from hospitalized patients. The square size corresponds to the absolute value of the Spearman rank correlation coefficient, with red (blue) color indicating a positive (negative) correlation. ∗FDR <0.05, ∗∗FDR <0.01, ∗∗∗FDR <0.001.
(C and D) Plasma protein (C) and metabolite (D) analysis. Left panel: PCA analysis of plasma proteins (metabolites). Each dot represents 1 plasma sample, color-coded for disease severity (see key). Right panel of (C): statistically significant (FDR <0.05) changed plasma protein from stepwise comparisons of disease severity from T1. The x axis indicates the numbers of significant protein changes, and the y axis indicates the magnitude of those changes (units = age, sex, BMI, and race/ethnicity adjusted NPX protein levels). The changes of four specific proteins are indicated by colored circles. Right panel of (D): statistically significant (FDR <0.05) changed plasma metabolites from stepwise comparisons of disease severity from T1. Changes are shown as upregulated and downregulated bar plot pairs, and each segment within a bar represents a superpathway (for color-code, see key) and is sized by the sum of the fold changes of the superpathway’s significantly changed metabolites.
(E) Circos plot visualizing WOS-dependent cross-omic interaction network. Proteins and clinical measures are only shown if they have degree ≥5. Complete results are provided in the Table S1.7. Positive main effects are represented as solid lines and negative as dashed lines. Significant associations with IL6, CCL7, and blood urea nitrogen are highlighted in black, green, and purple lines, respectively.
(F) Scatterplot depicting a specific connection from the circos plot. Each dot represents a patient sample color-coded for disease severity.
(G) Boxplot depicting percentages, from single cell transcriptome data, of major immune cell types among PBMCs from patients, grouped by WOS. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Figure S1.
Overview of the Multi-Omic Characterization of Immune Responses in COVID-19 Patients, Related to Figure 1
A. The swimmer plot depicting EHR-extracted WOS severity score dynamics for studied patients who were admitted to hospital. Symptom onset and pre-hospitalization (if available) are indicated by black rectangles and lines. WOS is calculated at 6-hour intervals during hospitalization. Blood draws are indicated by upside-down blue triangles and administered medications by symbols overlaid on the colored bands.
B. Boxplots of clinical data comparing moderate (orange) and severe (red) patient sample values. Ranges that specify normal limits are indicated by the dashed lines. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
C. Spearman Rank correlations between the measurement of the same analytes from EHR clinical labs and from metabolon metabolites. The regression line is shown in blue with 95% confidence area in shaded blue. Spearman Rank correlation coefficient and associated P value shown. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ****p < 0.0001).
D,E. Boxplot of principal component (PC) 1 and 2 from plasma proteomics data (D) or metabolomics data for donors, grouped by WOS (E). Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001)
F,G. Boxplots of select plasma protein (F) and metabolite(G) levels for donors, grouped by WOS. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
H. Scatterplots representing interactions drawn on the circos plot shown in Figure 1E. Each dot represents a patient sample with a color that corresponds to disease severity (see key).
I. Boxplots of select plasma lipid-related metabolites levels in donors grouped by different WOS. Metabolites were selected to illustrate (H) middle panel. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
J. Heatmap showing levels of well-known transcripts (left panel) and surface proteins (right panel) specific for each cell type and across different sequencing batches (see color-code). Cell types indicated by B: B cells, CD34+: CD34+ Progenitor cells, DC: Dendritic cells, MK: Megakaryocytes, M: Monocytes, NK: Natural killer cells, T: T cells.
K. UMAP visualization of single-cell RNA-seq data cell type annotation and analysis of batch effect. Left: 2D projection of single-cell RNA-seq data of all PBMCs from all samples using UMAP. Single cells are shown as dots, colored by their assigned cell type. Right: Hierarchical clustering of cell types across sequencing batches. Each column represents the expression profile of a cell type from a sequencing batch. Clustering was performed based on the expression of top 2000 most variable genes. Same cell types were clustered within the same hierarchical groups regardless of sequencing batch.
L. Boxplots depicting the percentages of major immune cell types within PBMCs for donors, grouped by WOS. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
Results
COVID-19 Patients Exhibit Clinical Profiles and Metabolic and Proteomic Plasma Compositions that Depend upon Disease Severity
The participating COVID-19 patients exhibited a range of clinical phenotypes, including many that exhibited statistically significant correlations with disease severity (quantified by the World Health Organization [WHO] ordinal scale [WOS]) (World Health Organization, 2020) (Figure 1B). Some correlations, including C-reactive protein (CRP) (Li et al., 2020), immature granulocyte count (Kuri-Cervantes et al., 2020), D-dimer (Mathew et al., 2020), and segmented neutrophils (Huang et al., 2020) have been previously reported (Figures 1B and S1B). Clinical metadata for these COVID-19 patients and healthy donors are presented in Tables S1.1–S1.2.
Analyzed plasma proteins and metabolites are listed in Tables S1.3–S1.4. Measurements of the same metabolites from Metabolon and clinical labs (EHR data) were highly correlated (Figure S1C). Principal component analysis (PCA) of both datasets, using both principal component (PC) 1 and PC2, resolved shifts in these plasma profiles in stepwise comparisons (healthy donors [WOS = 0] versus mild [WOS = 1–2]; mild versus moderate [WOS = 3–4], and moderate versus severe [WOS = 5–7]) (Figures 1C, 1D, S1D, and S1E). Proteomic PC1, which separated healthy donors and COVID-19 patients (Figure S1D), contains pro-inflammatory cytokines (e.g., CXCL6) and proteins associated with immune cell activation (e.g., CD244 and CD40) as high contributing factors (Table S1.5), implying immune activation in even mild COVID-19 cases. Proteomic PC2 distinguished mild from moderate patients (Figure S1D), which are differentiated by whether hospitalization is required (World Health Organization, 2020). The alterations in protein profiles can be appreciated by comparing the numbers of proteins significantly changed for each step in disease severity (Figure 1C, right panel). The highest number of significant protein changes were observed in the transition from healthy donors to mild infections (n = 307 proteins with only 27 downregulated, Table S1.6). A comparison of mild to moderate revealed relatively fewer changes (n = 132 total with 91 downregulated, Table S1.6), while few proteins changed from moderate to severe disease (n = 5, all upregulated, Table S1.6). Consistent with previous literature (Lucas et al., 2020), we identified the upregulation of CCL7, IL-10, and IL-6 (that barely misses significance between moderate and severe disease with p = 0.056) (Figures 1C, right panel, and S1F; Table S1.6). Keratin-19 (KRT19), which is involved in the organization of muscle fibers (Stone et al., 2007), is upregulated in all comparisons (Figures 1C, right panel, and S1F; Table S1.6) and may be a marker of tissue damage. This analysis suggests a striking similarity between moderate and severe COVID-19 cases.
Metabolomic PC1 resolved mild from moderate cases, while PC2 distinguished healthy and COVID-19 patients (Figure S1E). These changes were evaluated by the metabolic-change scores, which consider the sum of the fold changes observed in classes of metabolites (Figure 1D, right panel). From healthy to mild COVID-19, upregulated metabolites included molecules associated with amino acid (up = 58 versus down = 8), nucleotide (up = 14 versus down = 2), and carbohydrate (up = 11 versus down = 0) metabolism (Table S1.6), with lipids slightly more downregulated (up = 61 versus down = 79). Interestingly, for mild to moderate and moderate to severe transitions, there is a clear preference toward downregulation of lipids, which agrees with previous reports (Shen et al., 2020), and amino acid metabolites, which recalls reports of amino acid catabolism for severe COVID-19 (Thomas et al., 2020) (Figures 1D, right panel, and S1G; Table S1.6). Disruption of xenobiotic metabolism appears mostly associated with severe disease; the upstream xenobiotic benzoate is elevated while downstream products, such as catechol sulfate and 3-hydroxyhippurate, are repressed (Figure S1G; Table S1.6). Benzoate level may point to hepatic injury, because liver is the principal site for such metabolism (Alqahtani and Schattenberg, 2020). Therefore, the global metabolic profile alterations point to a major shift between mild and moderate cases with a disproportionate loss of several classes of circulating metabolites.
We explore the relationships between plasma analytes, clinical measures, and disease severity (WOS) in the circos interaction plot of Figure 1E. Linkages shown are analyte-analyte associations with significant (false discovery rate [FDR] <0.001) interaction effects with WOS (see STAR Methods). A representative severity-dependent linkage (IL-6 versus a lysoplasmalogen) is shown in Figure 1F. These interactions reveal many new insights. For example, blood urea nitrogen (BUN) (in hospitalized patients) has many connections with amino acid metabolism (Figures 1E and S1H, left panels), suggesting amino acid catabolism in advanced COVID-19 (Thomas et al., 2020). Several inflammation-associated proteins, including CCL7 and IL-6, are anti-correlated with many plasma lipids (Figures 1E and S1H, middle and right panels). For healthy donors or mildly infected patients, these lipid levels exhibit a range of values (Figure S1I). However, between mild and moderate infection, the lipid levels drop precipitously (Figure S1I). This general severity-dependent trend is seen for many interactions (Table S1.7). For example, out of the 32 significant connections of IL-6 in the circos plot, 29 are with lipids, and all negatively correlate with WOS (Table S1.7). The circos plot suggests that increased inflammatory signals are accompanied by a loss of metabolic resources and a drop in xenobiotic metabolism, seemingly reflecting a stressed environment that may influence the immune response in COVID-19 patients.
We further explored how COVID-19 severity is reflected in circulating immune cells. The PBMC single-cell transcriptome data were visualized as a two-dimensional (2D) projection via uniform manifold approximation and projection (UMAP) (Becht et al., 2018) (Figure S1K, left panel). No major batch effects were revealed (Figures S1J and S1K, right panel).
Relative percentages of different cell types were assessed relative to increasing WOS (Figures 1G and S1L). Consistent with previous reports (Mathew et al., 2020; Song et al., 2020), we observed a drop in the relative percentage of lymphocytes, including CD4+ T cells, natural killer (NK) cells, and especially CD8+ T cells (Figures 1G and S1L). By contrast, monocyte percentages were elevated (Figure 1G). B cell percentages did not significantly vary with WOS (Figure S1L), consistent with previous reports (Zhang et al., 2020). Healthy and mild participants exhibit similar immune cell compositions (Figure S1L), although mild and moderate disease exhibited the most significant changes (Figure 1G, p < 0.0001 for both). This observation aligns with the major shift we observed in plasma multi-omics between mild and moderate disease and prompted us to further investigate major immune cell types.
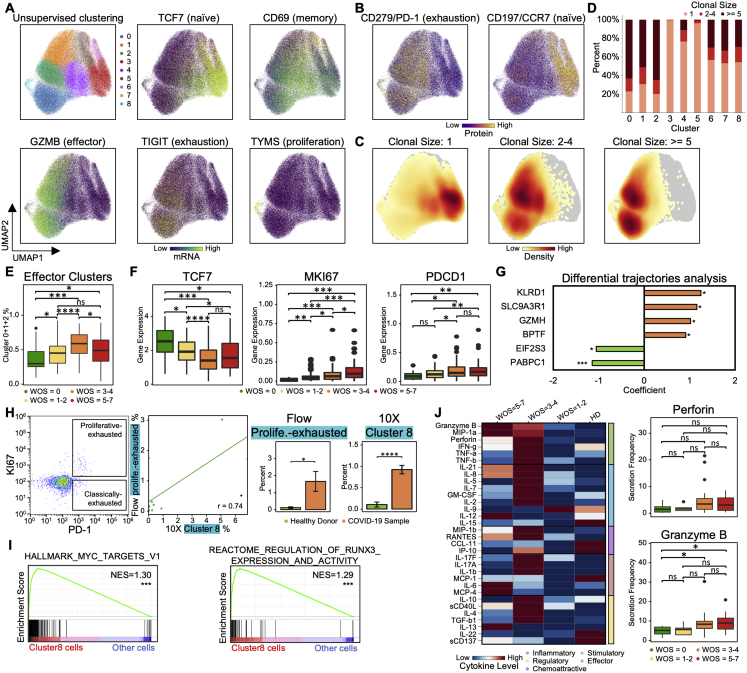
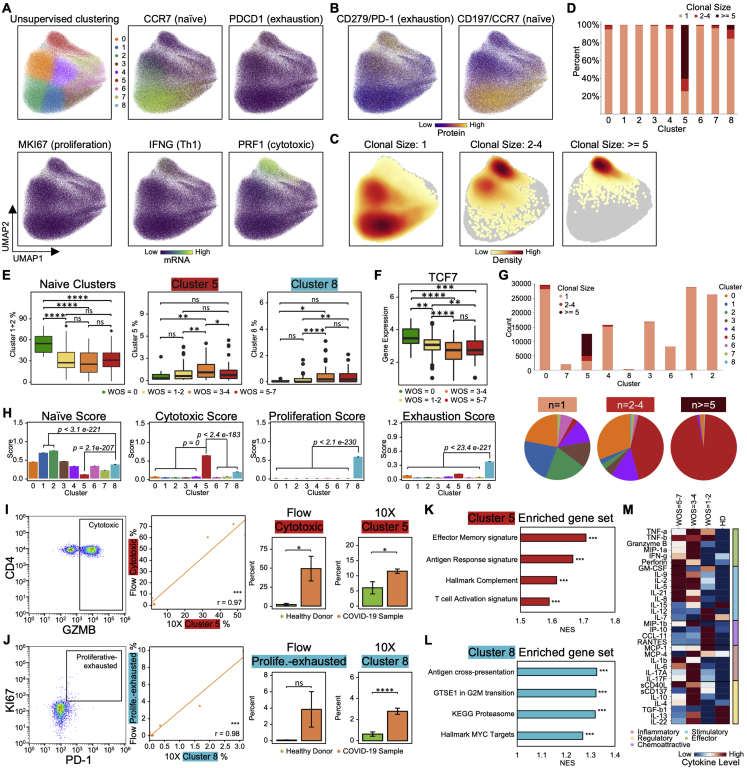
CD8+ T Cell Phenotypic Composition Evolves with COVID-19 Severity with a Non-monotonic Change of Activation Status
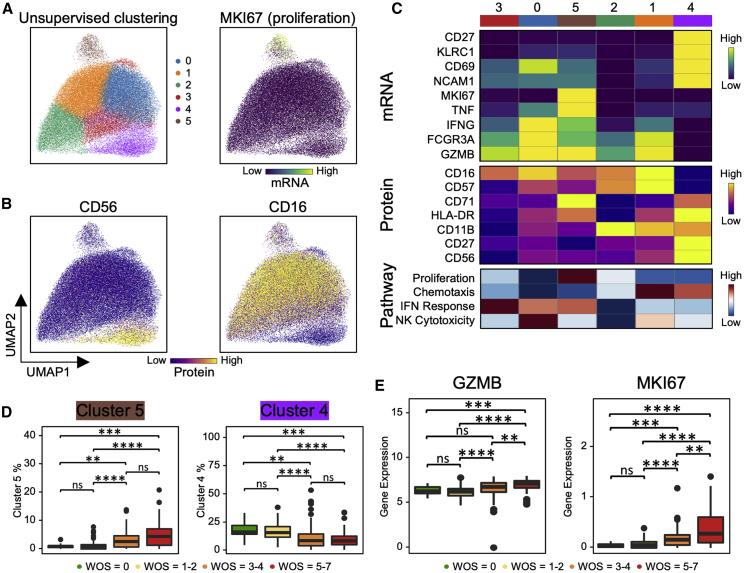
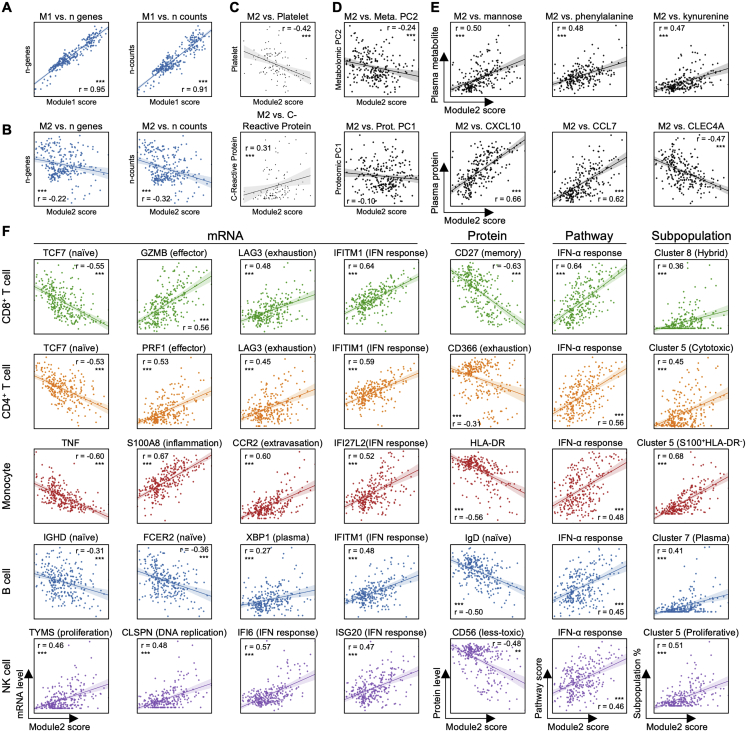
We projected the CD8+ T cell single-cell transcriptomic data onto a 2D UMAP and resolved nine subpopulations characterized by both protein and transcriptional signatures (Figures 2A, 2B, S2A, and S2B). We identified naive, memory, effector, exhausted-like, and proliferative phenotypes. For instance, the naive-related mRNA and protein markers LEF1, TCF7, and CD197 were all upregulated in the red-colored cluster 3, while effector markers, such as GZMB and PRF1, were elevated in clusters 0, 1, and 2 (Figures 2A, 2C, S2A, and S2B). The protein levels yielded a consistent picture. For example, the naive-like cluster 1 displayed a high CD45RA/CD45RO ratio and effector-like clusters 0, 1, and 2 displayed intermediate levels (Figure 2B). Exhausted CD8+ T cells have been documented in COVID-19 patients (Diao et al., 2020) and, indeed, exhaustion markers LAG3 and TIGIT were upregulated in both effector clusters (0 and 2) (Figures 2A, 2C, and S2A). Such inhibitory markers are also increased after T cell activation (Wherry, 2011; Wherry and Kurachi, 2015) and may not indicate dysfunction.
Figure 2.
CD8+ T Cell Heterogeneity in COVID-19 Patients and Its Association with Disease Severity
(A and B) UMAP embedding of all CD8+ T cells colored by unsupervised clustering (top left) and by selected mRNA transcript levels (other panels in A) or (B) the CD45RA/CD45RO surface protein ratio.
(C) Heatmaps showing the normalized levels of selected mRNA (top panel) and proteins (bottom panel) across each cell cluster.
(D) UMAP embedding of CD8+ T cells shaded by clonal expansion level.
(E) Boxplots showing the WOS-dependence of percentages of CD8+ T cell clusters. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(F) UMAP embedding density of CD8+ T cells for different blood draw samples, grouped by WOS. Selected clusters are encircled in the colors of the (A) clusters.
(G) Scatterplots showing the naive (x axis) and cytotoxic (y axis) signature scores of individual CD8+ T cells from all PBMC samples. Cluster 8 is encircled. Each point represents one cell. Cells are color coded with cluster-specific colors (left) or signature scores (middle and right).
(H) Pearson correlation between MKI67 and PDCD1 gene expression for cluster 8 cells. Correlation coefficient and p value shown. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(I) Clonal expansion score for CD8+ T cells from patients with different WOS. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(J) TCR clustering analysis. Left panel: hierarchical clustering of TCRs (columns) based on TCR sharing patterns across clusters (rows). The two distinct groups of TCRs identified are shaded with orange (group1) and green (group2). Middle panel: UMAP visualization of the embedding density of cells containing TCRs from group1 and group2 from the left panel. Right panel: boxplots represent ratio of cells containing TCRs from group1 over cells containing TCRs from group2 for samples of different WOS. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(K) Single cell polyfunctional strength index (PSI) of CD8+ T cells according to sample WOS. Data are represented as mean ± SEM. Pairwise statistical comparisons are shown in Table S2.3.
Figure S2.
CD8+ T Cell Heterogeneity in COVID-19 Patients and Its Association with Severity, Related to Figure 2
A,B. UMAP embedding of all CD8+ T cells colored by unsupervised clustering (top left of A) and by selected mRNA transcript levels (other panels in A) or (B) two selected surface proteins.
C. UMAP embedding of all CD8+ T cells colored by the density of cells characterized by different clonal expansion sizes (n = 1, n = 2-4, and n > = 5).
D. Clonal expansion sizes of each CD8+ T cell subset from unsupervised clustering. Bar plot shows the normalized clonal composition.
E. Boxplots represent percentages of effector CD8+ T cells (cluster 0, 1 and 2) over all CD8+ T cells for PBMCs in donors for different WOS.
F. Boxplots showing the mRNA expression levels of 3 transcripts in healthy donors (green), mild (yellow), moderate (orange) and severe (red) blood draws of COVID-19 patients.
G. Differential expression analysis of genes that uniquely change in patients who improve (T2 versus T1, WOS decreased) in comparison with patients who did not. Each row represents a gene. The x axis differential expression score with positive being upregulated and negative being downregulated. FDR-corrected P values was shown.
H. Flow cytometry validation of the proliferative-exhausted cluster 8 CD8+ T cell. First panel (starting from the left): Representative flow cytometry plots of the gating strategy for proliferative-exhausted CD8+ T cells (top box) and non-proliferative exhausted CD8+ T cells (bottom box). Gating was performed on all CD8+ T cells. Second panel: quantitative comparison of the % of gated proliferative-exhausted CD8+ T cells from flow cytometry (y axis) and % of cluster 8 CD8+ T cells quantified by 10X (x axis) of the same sample. Each dot represents a PBMC sample. The regression line is drawn in green. Pearson correlation coefficient and associated P value shown. Right two panels: Bar plots represent % of proliferative-exhausted CD8+ T cells quantified by flow and cluster 8 cells quantified by sc-RNA-seq, comparing samples from healthy donors and COVID-19 patients. Data are represented as mean ± SEM.
I. GSEA of top pathways enriched for cluster 8 cells. Normalized enrichment score (NES) and P values are shown. Full results provided in Table S2.2.
J. Functional characterization of CD8+ T cells using single-cell secretome analysis. Left panel: heatmap visualization of average cytokine secretion frequency for cells from healthy donors (HD), mild (WOS = 1-2), moderate (WOS = 3-4) and severe (WOS = 5-7) patients. Right panel: Boxplots indicate the percentage of CD8+ T cells secreting granzyme B, and perforin from samples grouped by different WOS. P values are shown.
Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
Certain CD8+ subpopulations correlated with disease severity. As expected, naive-like markers and cell cluster 3 exhibited the highest density in healthy donors and decreased in COVID-19 patients (Figures 2E, 2F, and S2F). Effector-like cells (clusters 0, 1, and 2) were enriched in COVID-19 samples (Figures 2F and S2E), also as expected (Mathew et al., 2020). Similarly, CD8+ T cells in patients who improved (defined as a decrease of WOS from T1 to T2) displayed higher levels of effector-associated transcripts such as GZMH, KLRD1, SLC9A3R1, etc. (Figure S2G; Table S2.1). Interestingly, the severe patients showed an increase of naive clusters and a decrease in activated effector T cells (Figures 2F and S2E) relative to moderate disease. Such non-monotonic change of T cell activation status with disease severity may resolve conflicting reports of both positive and negative correlations of T cell activation with disease severity (Lucas et al., 2020; Song et al., 2020) and underscores the importance of analyzing large patient cohorts in distinct disease stages to accurately sample the heterogeneity of COVID-19 disease manifestation.
A Proliferative Exhausted CD8+ T Cell Subpopulation Emerges in Moderate COVID-19 Patients
Cluster 8 displayed intermediate levels of effector markers, upregulated exhaustion markers (LAG3, TIGIT, and CD279), and, counterintuitively, exclusively expresses proliferation markers (MKI67 and TYMS) (Figures 2A, 2C, S2A, and S2B). In fact, transcript levels of exhaustion marker PDCD1 showed a positive correlation with MKI67 for cluster 8 cells (Figure 2H). Cluster 8 also displays high cytotoxic signatures and has not fully lost its naive signature (Figure 2G). This subpopulation emerges at the stage of moderate disease (Figure 2E). The relative percentages of this hybrid proliferative-exhausted phenotype were confirmed by flow cytometry (Figure S2H). This may reflect the proliferative hierarchy that maintains the exhausted T cells, which has been observed in chronic infections and cancer (Bengsch et al., 2018; Huang et al., 2017; Paley et al., 2012).
Pathway analysis of cluster 8 cells revealed that genes regulated by the MYC and the regulation of RUNX3 activity were both enriched (Figure S2I; Table S2.2). This may imply that cluster 8 is comprised of early-stage activated T cells, because MYC is rapidly but transiently induced during the early stage of activation (Nie et al., 2012).
Integrating Single-Cell Omics with TCR Datasets Resolved Two Distinct Groups of Phenotypic-Specific TCRs in CD8+ T Cells that Associate with Disease Severity
CD8+ T cell clonal expansion was enhanced in mild and moderate COVID-19 patients relative to healthy donors (Figures 2D and 2I). The effector-like clusters 0 and 2 showed the most clonal expansion, while T cells with a clonal expansion index of 1 were mainly within the naive-like cluster 3 (Figures S2C and S2D), suggesting the activated effector CD8+ T cells detected in the peripheral blood are clonally expanding, likely due to virus-antigen encounters.
Integrating the TCR and single-cell transcriptome datasets using hierarchical clustering revealed one set of TCRs (group 1) within the cytotoxic effector phenotype and a group 2 within the memory-like phenotype (Figure 2J). Group1 TCRs are mainly from patients with moderate or severe COVID-19 and the group2 TCRs are mostly from mild patients (Figure 2J). Thus, the expansion of CD8+ clones emphasizes the sharp transition between mild and moderate disease.
COVID-19 Severity Is Associated with Non-monotonic Changes of Polyfunctionality in CD8+ T Cells
Polyfunctional T cells produce multiple different cytokines, can release a substantially higher amount of cytokines relative to other T cells, and can dominate an immune response (Abel et al., 2010; Lu et al., 2015; Ma et al., 2013; Zhou et al., 2017). We measured 32 secreted cytokines from individual live CD8+ T cells. Polyfunctionality, as quantified by the polyfunctional strength index (PSI = the numbers of different proteins secreted × copy numbers secreted), is similar for healthy and mild cases, but upregulated at moderate severity (Figure 2K), with increased frequency of cells secreting granzyme B and perforin (Figure S2J). Polyfunctionality is reduced in severe patients (Figure 2K), consistent with the peak in CD8+ effector cluster percentage for moderate cases (Figure S2E). Thus, peripheral CD8+ T cells in COVID-19 patients with moderate illness reflect the highest polyfunctionality and the highest percentages of effector phenotypes.
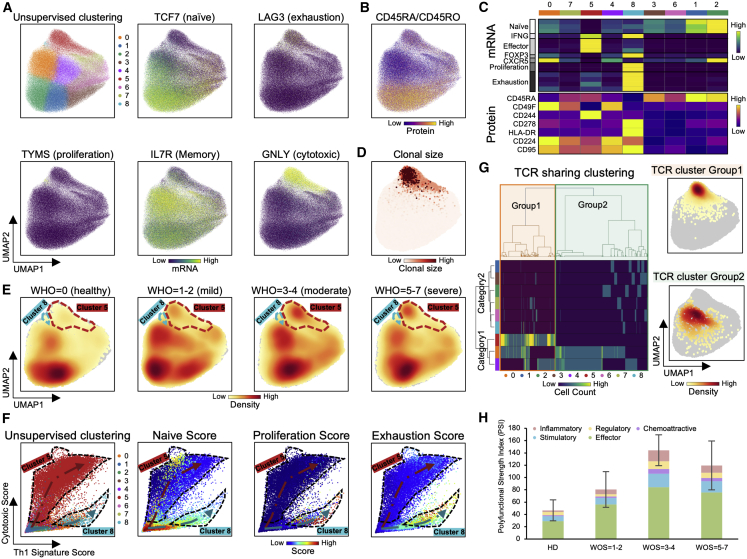
Two Distinct CD4+ T Cell Subpopulations Are Associated with COVID-19 Severity
UMAP representations of single CD4+ T cell transcriptomic data are provided in Figures 3A, 3B, S3A, and S3B. Projecting data for COVID-19 patients grouped by disease severity revealed how CD4+ T cell phenotype is influenced by infection (Figure 3E). For example, naive-like cells (clusters 1 [blue] and 2 [green]), which are characterized by elevated naive-related transcripts TCF7, CCR7, and CD197 surface protein, were reduced in COVID-19 patients (Figures 3E, S3E, and S3F), implying increased activation of CD4+ T cells with COVID-19 infection, as reported (Mathew et al., 2020).
Figure 3.
Two Distinct CD4+ T Cell Subpopulations Are Associated with COVID-19 Severity
(A and B) UMAP embedding of all CD4+ T cells colored by unsupervised clustering (top left of A) and by selected mRNA transcript levels (other panels in A) or (B) the CD45RA/CD45RO surface protein ratio.
(C) Heatmap showing the normalized expression of selected transcripts (top panel) and proteins (bottom panel) across each cell cluster.
(D) UMAP embedding of CD4+ T cells shaded by clonal expansion level.
(E) UMAP embedding density of CD4+ T cells for different samples, grouped by WOS. Selected clusters are encircled in the colors of the (A) clusters.
(F) Scatterplots showing the Th1 (x axis) and cytotoxic (y axis) gene signature scores of individual CD4+ T cells from all PBMC samples. Clusters 5 and 8 are encircled. Each point represents one cell. Plots are color coded for their cluster and functional signature as specified on top of each plot.
(G) TCR clustering analysis. Hierarchical clustering of TCRs (columns) based on TCR sharing patterns across clusters (rows). Left: the two distinct groups of TCRs identified are shaded with orange (group1) and green (group2). Right: UMAP visualization of the embedding density of cells containing TCRs from group1 and group2 defined by left panel.
(H) Single-cell polyfunctional strength grouped according to patient WOS. Data are represented as mean ± SEM. Pairwise statistical comparison are shown in Table S3.3.
Figure S3.
Two Distinct CD4+ T Cell Subpopulations Are Associated with COVID-19 Severity, Related to Figure 3
A,B. UMAP embedding of all CD4+ T cells colored by unsupervised clustering (top left of A) and by selected mRNA transcript levels (other panels in A) or two selected surface proteins (B).
C. UMAP embedding of all CD4+ T cells colored by the density of cells characterized by different clonal expansion sizes (n = 1, n = 2-4, and n > = 5).
D. Clonal expansion sizes of each CD4+ T cell subset from unsupervised clustering. Bar plot shows the normalized clonal composition.
E. Boxplots showing percentages of naive CD4+ T cells (cluster 1 and 2), cluser5 and 8 over all CD4+ T cells in samples grouped by WOS.
F. Boxplots showing the mRNA expression levels in samples grouped by WOS.
G. Clonal expansion status, presented as bar plots, for each CD4+ T cell cluster, color-coded by clonal expansion sizes (n = 1, n = 2-4, n > = 5). The pie charts show the CD4+ T cell cluster composition for each clonal expansion sizes present in the bar plots
H. Bar plots representing four functional signature scores across different subclusters of CD4+ T cells. P values are shown.
I,J. Flow cytometry validation of cytotoxic CD4+ T cells (I) and proliferative-exhausted cluster 8 CD4+ T cells (J). Left panel: Representative flow cytometry plots of the gating strategy for cytotoxic CD4+ T cells (I), and proliferative-exhausted CD4+ T cells (J). Gating was performed on all CD4+ T cells. Middle panel: quantitative comparison of the % of gated cytotoxic (proliferative-exhausted) CD4+ T cells (y axis) and cluster 5 (cluster 8) CD4+ T cell % (x axis); each dot represents a PBMC sample. The regression line is drawn in orange. Pearson correlation coefficient and associated P value shown. Right panels: Boxplots represent % of cytotoxic (proliferative-exhausted) CD4+ T cells quantified by flow cytometry and % of cluster 5 (cluster 8) cells quantified by 10X single cell RNA-seq. Comparison was between samples from healthy donor and COVID-19 patients. Data are represented as mean ± SEM.
K, L. Bar plots represent GSEA of top pathways enriched for cluster 5 (K) and cluster 8 (L) cells. Normalized enrichment score (NES) and P values are shown. Full results provided in Tables S3.1 and S3.2.
M. Heatmap visualization of average cytokine secretion frequencies for cells from healthy donor (HD), mild (WOS = 1-2), moderate (WOS = 3-4) and severe (WOS = 5-7) patient samples.
Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
We identified two unusual CD4+ T cell subpopulations (clusters 5 and 8) that varied with WOS. Cluster 5 cells exhibited elevated cytotoxic transcripts PRF1 and GNLY (Figures 3A, 3C, and S3A) and increased in patients with moderate infections (Figure S3E), possibly suggesting repeated antigen exposure (Juno et al., 2017). In fact, more than 95% of the highly expanded CD4+ T cell clones (defined by n ≥ 5) were in cluster 5 (Figures 3D, S3C, S3D, and S3G), suggesting viral specificity for these cells. Cluster 8 expressed exhaustion markers, an elevated Th1 signature, the most elevated proliferation signature, but little clonal expansion (Figures S3D, S3G, and S3H). The single-cell RNA sequencing (scRNA-seq)-derived relative percentages of the hybrid and cytotoxic CD4+ T cell clusters were both quantitatively confirmed by flow cytometry (Figures S3I and S3J).
Both clusters 5 and 8 exhibited unique groups of enriched genes (Figures S3K and S3L; Tables S3.1 and S3.2). Cytotoxic cluster 5 cells showed elevated effector memory, antigen presentation signatures, and enrichment of the complement gene set (Figure S3K). Hybrid cluster 8 cells showed enrichment of antigen cross presentation and cell cycle G2M transition-related gene signatures (Figure S3L). Interestingly, cluster 8 CD4+ T cells were highly enriched for MYC targets, similar to the hybrid CD8+ T cell cluster, suggesting a transient intermediate cell-state during early-stage activation, potentially due to rapid activation.
The functional differences of all CD4+ T cell clusters were compared by projecting single cells onto a scatterplot where the x and y axes represent the Th1 and cytotoxic signature scores, respectively (Figure 3F). Cells with high naive scores reside at the bottom-left (Figure 3F). The two unique clusters extend out toward opposite corners of the map, while other clusters align along either of the branches (Figure 3F). For example, cluster 6 (pink) aligns along the upper branch with cluster 5, and cluster 0 (orange) aligns with cluster 8. No significant overlap is observed between the two branches (Figure 3F), implying that the branches may comprise two distinct cell state destinations for a given naive CD4+ T cell. These two clusters exhibit differences in other functional signatures. Cluster 8 displays uniquely upregulated proliferation and exhaustion signatures, intermediate levels of the naive markers, and low levels of the cytotoxic signature (Figure S3H). In contrast, cytotoxic cluster 5 shows opposite trends for these functional signatures. Importantly, these two clusters exhibit notable differences in their TCR sharing patterns (Figure 3G). Cells with group 1 TCRs were uniquely enriched in cytotoxic cluster 5 (Figure 3G). Thus, these two unique clusters may represent distinct cell state destinations for CD4+ T cells during infection.
Polyfunctionality of single, viable CD4+ T cells revealed that CD4+ T cells from COVID-19 patients exhibit polyfunctionality relative to those from healthy donors, peaking at patients with moderate illness (Figure 3H). Notably, the frequencies of CD4+ T cells that can secrete the Th1 cytokine IFN-γ, Th17 cytokines IL17-A, IL17-F, Th2 cytokine IL-4, and cytotoxic molecule granzyme B are increased in COVID-19 patient samples, again peaking for moderate patients (Figure S3M). This recalls trends seen in CD8+ T cells, but whether this is related to T cell exhaustion in the most severe patients is unclear.
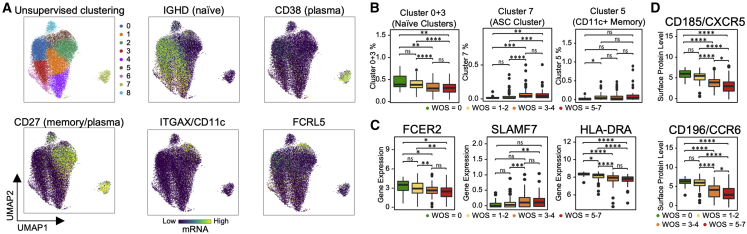
B Cell Heterogeneity in COVID-19 Patients and Its Association with Severity
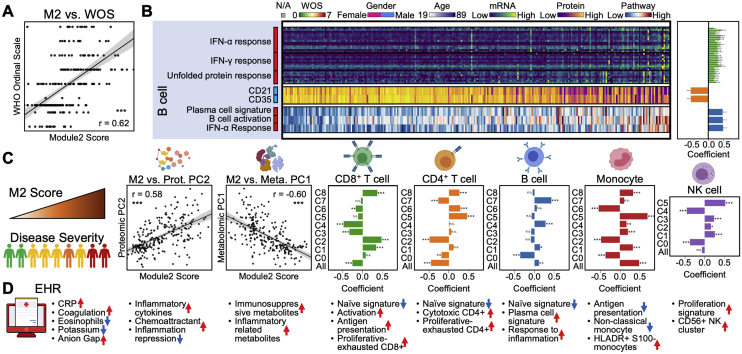
We observed significant activation in naive B cells (downregulation of FCER2 and upregulation of SLAMF7) and expansion of antibody-secreting cells (ASCs) in moderate and severe samples compared to healthy and mild samples (Figures 4A–4C). A distinct memory B cell population that has high expression in ITGAX and FCRL5 (Figure 4A), resembling a tissue-like memory B cell phenotype (Li et al., 2016), was found to be high in mild samples compared to healthy samples (Figure 4B). We found prominent downregulation of several HLA class II genes in moderate and severe COVID-19 patients (Figure 4C), suggesting dysregulation of immune cell crosstalk between the adaptive immune cell classes. Besides the loss of chemokine receptor CXCR5 in moderate and severe samples of COVID-19 patients (Figure 4D) that has been reported previously (Mathew et al., 2020), we also discovered a significant loss of chemokine receptor CCR6 in moderate and severe samples (Figure 4D). This could impair germinal center reactions (Reimer et al., 2017) and ultimately lead to the dysregulated humoral immunity responses found in early infection of COVID-19 (Kaneko et al., 2020).
Figure 4.
B Cell Heterogeneity in COVID-19 Patients and Its Association with Severity
(A) UMAP embedding of all B cells colored by unsupervised clustering (top left panel of A) and by selected mRNA transcript levels (other panels in A).
(B–D) Boxplots showing the WOS-dependence of specific B cell clusters (B), normalized levels of transcript (C), and proteins (D). p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
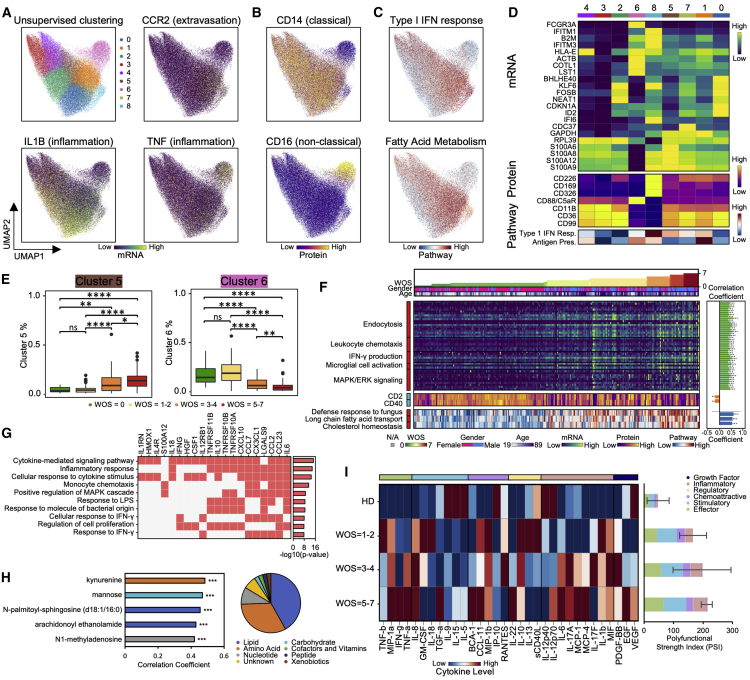
S100highHLA-DRlow Dysfunctional Monocyte Subpopulation Reflects Coordinated Changes with Both Plasma Multi-Omics Signals and COVID-19 Severity
UMAP visualization of all monocytes shows a clear separation of non-classical monocytes (CD14lowCD16high cluster 6), characterized by the upregulated transcripts FCGR3A, CX3CR1, and surface protein CD16 (Figures 5A–5D, S4A, and S4B). The non-classical monocyte fraction decreases in moderate and severe infections (Figures 5E and S4C), compared to a reported drop in severe patients (Schulte-Schrepping et al., 2020; Silvin et al., 2020; Wilk et al., 2020). Among the classical monocyte clusters (CD14highCD16low), the cluster 5 (brown) monocyte percentage increases for moderate and severe cases (Figures 5E and S4C). Cluster 5 exhibits expression of inflammation-related transcripts such as S100A4, S100A9, S100A12, and decreased HLA-DR surface protein level (Figures 5A–5D, S4A, and S4B). Interestingly, monocytes in patients who improved (defined as a decrease of WOS from T1 to T2) displayed higher levels of HLA-DRB5, S100A8, etc. (Figure S4D; Table S4.1). The increase of S100highHLA-DRlow cluster 5 subset with downregulated HLA class II is reminiscent of monocyte “immunoparalysis” in sepsis (Giamarellos-Bourboulis et al., 2020) and is further supported by the decrease in tumor necrosis factor alpha (TNF-α) transcripts (Figure S4E). Expression of HLA class II genes in monocytes were negatively correlated with IL-6 plasma level (Figure S4F), suggesting an influence of the hyper-inflammatory plasma environment on monocyte dysfunction, prompting us to more deeply explore this connection.
Figure 5.
S100highHLA-DRlow Dysfunctional Monocyte Subpopulation Reflects Coordinated Changes with Both Plasma Multi-Omics Signals and COVID-19 Severity
(A–C) UMAP embedding of all monocytes colored by unsupervised clustering (top left panel of A) and by selected mRNA transcript levels (other panels in A) or (B) surface proteins or (C) pathway enrichment scores.
(D) Heatmap displaying normalized expression of differentially expressed genes in each cluster (top), select proteins (middle) and pathway-enrichment scores (bottom) across each cell cluster. Full gene list of differential analysis is provided in Table S4.12.
(E) Boxplots showing the WOS-dependence of relative abundance of specific monocyte clusters from (A). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(F) Heatmap visualization of top genes, surface proteins, and pathways in monocytes that significantly correlated with disease severity (WOS). Each column represents a sample and each row corresponds to levels of mRNA, surface protein, or pathway enrichment score for the monocytes from that sample. Columns are ordered based on WOS in ascending order. The top three rows indicate the WOS, gender, and age. The heatmap keys are provided at the bottom. Sidebar on the left of each row represent correlation of that value with WOS, with red (blue) indicating positive (negative) correlations. Full list of the top genes, proteins, and pathways is provided in Tables S4.5–S4.11. Detailed correlation coefficient and FDR-corrected p values are indicated at the right panel bar plots. ∗FDR <0.05, ∗∗FDR <0.01, ∗∗∗FDR <0.001.
(G) GO enrichment analysis of the top50 plasma proteins that positively correlated with cluster 5 monocyte percentages. Each row represents one of the top10 enriched pathways, each column represents each plasma protein. The top10 most over-represented proteins are shown. Bar plot shows the -log10(p value) of the enriched pathways. Full enrichment results are provided Table S4.4.
(H) Analysis of the top50 plasma metabolites that positively correlated with cluster 5 monocyte percentage. Pie chart represents the super-pathway composition of the top50 positively correlated metabolites. Bar plot shows Pearson correlation coefficient of the top5 most significantly (p < 0.05) correlated metabolites (Table S4.3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(I) Functional characterization of monocytes. Left panel: heatmap visualization of average cytokine secretion frequencies for cells from samples grouped by WOS. Right panel: polyfunctional characterization of monocytes. Single cell polyfunctional strength is grouped according to sample WOS. Data are represented as mean ± SEM. Pairwise statistical comparison are shown in Table S4.13.
Figure S4.
S100highHLA-DRlow Dysfunctional Monocyte Subpopulation Reflects Coordinated Changes with Both Plasma Multi-Omics Signals and COVID-19 Severity, Related to Figure 5
A,B. UMAP embedding of all monocytes colored by unsupervised clustering (top left panel of A) and by selected mRNA transcript levels (other panels in A) or (B) surface proteins for non-classical (CD16) monocytes.
C. UMAP embedding density of monocytes for different blood draw samples, grouped by WOS. Selected clusters that display significant changes from WOS group to WOS group are encircled in the colors of the (A) clusters.
D. Differential expression analysis of monocyte genes that uniquely change in patients who improve (T2 versus T1, WOS decreased) in comparison with patients who did not. Each row represents a gene. The x axis represents differential expression score with positive being upregulated and negative being downregulated. FDR-corrected P values was shown. Significance is indicated by: (∗ FDR < 0.05, ∗∗ FDR < 0.01, ∗∗∗ FDR < 0.001).
E. Boxplots showing the mRNA expression levels of TNF transcripts from monocytes in samples grouped by WOS. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
F. Pearson correlation of HLA-DRA gene expression in monocytes with plasma IL-6 levels. Each dot represents a blood draw sample and is colored by disease severity (WOS, see key). Regression line indicated in black, with a 95% confidence interval shown in shaded gray. Pearson correlation coefficient and associated P value are shown. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
We hypothesized that monocyte responses to COVID-19 may correlate with the metabolic and proteomic plasma compositions that vary with WOS (Figure 1E). We first identified plasma proteins that correlated with cluster 5 monocyte percentages. Pathway analysis of the top50 proteins (Figure 5G; Table S4.2) suggests an association with cytokine signaling, inflammation response, monocyte chemotaxis, IFN-γ, and mitogen-activated protein kinase (MAPK) signaling (Figure 5G; Table S4.4). These plasma protein signatures, most of which are reflected in the cross-omic interaction network of Figure 1E, can be compared against monocyte gene expression signatures that increased with severity (Figure 5F). These include leukocyte chemotaxis, IFN-γ, and MAPK/extracellular signal-regulated kinase (ERK) signaling (Figure 5F). Metabolites that correlate with cluster 5 percentage are mainly within lipid and amino acid metabolisms (Figure 5H right panel), reminiscent of the many WOS-dependent cross-omic interaction connecting lipid and amino acid metabolites (Figure 1E). The top five positively correlated metabolites are N1-methyladenosine, two lipids, mannose, and kynurenine (Figure 5H; Table S4.3), of which kynurenine has been previously associated with COVID-19 severity (Shen et al., 2020). The associations between the cluster 5 subpopulation and the plasma analytes suggests that cluster 5 monocytes may play a potential role in the coordinated immune response for severe COVID-19 infections, and highly correlated plasma proteins, such as IL-6 and CCL23, could serve as potential biomarkers for rapidly assessing the dysfunctional state of monocytes in clinic.
We also characterized the polyfunctional strength index (PSI) of monocytes from COVID-19 patients. Unlike the case of CD8+ and CD4+ T cells (Figures 2K and 3H), the PSI monotonically increases with disease severity (Figure 5I), suggesting that monocytes contribute to the pro-inflammatory condition of moderate or severe COVID-19. One can speculate that the increased S100highHLA-DRlow monocyte percentage may be influenced by and contribute to the inflammatory proteomic and altered metabolomic plasma profile.
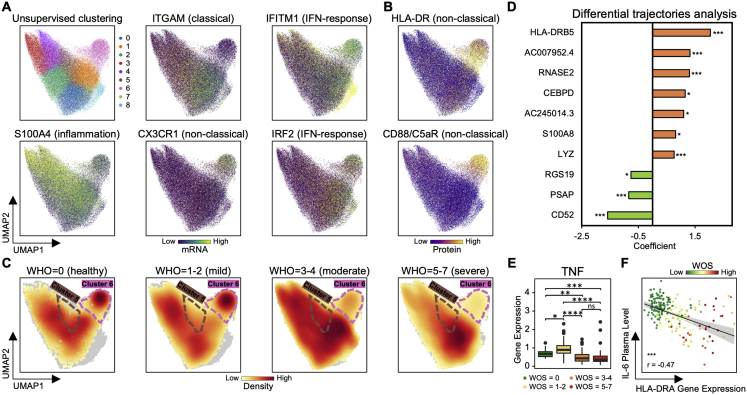
Proliferative NK Cell Subpopulation Is Associated with Increased COVID-19 Severity
Unsupervised clustering and UMAP visualization of NK cell transcriptomes resolved two well-known NK cell subsets: CD56bright (cluster 4) and CD56dimCD16bright (clusters 0–3 and 5) (Figures 6A–6C) (Miller and Lanier, 2019). The CD16bright subpopulations display high levels of cytotoxic transcripts (PRF1 and GZMB) and elevated levels of the terminal-differentiation marker CD57 and the exhaustion marker LAG3. The CD56bright subpopulation (cluster 4) expresses markers associated with less-differentiation, including IL7R, CD27, and CD62L (Figures 6A–6C, S5A, and S5B). Cluster 4 is significantly repressed in moderate and severe patients (Figures 6D and S5C), which may reflect the differentiation of NK cells toward more cytotoxic phenotypes and, again, points to the sharp transition between mild and moderate disease. This is consistent with the observed decrease of CD56 and CD127, and IL7R as well as the increase of PRF1, GZMB, and CD69 in moderate and severe patients (Figure S5E). The NK cell activation in COVID-19 PBMCs has been seen using flow cytometry (Maucourant et al., 2020).
Figure 6.
Proliferative NK Cell Subpopulation Is Associated with Increased COVID-19 Severity
(A and B) UMAP embedding of all NK cells colored by unsupervised clustering (top left panel of A) and by selected mRNA transcript levels (top right panel of A) or (B) selected surface proteins.
(C) Heatmap displaying normalized level of select mRNA (top), proteins (middle), and pathway-enrichment scores (bottom panel) in each cell cluster.
(D and E) Boxplots showing the WOS-dependence of relative abundance of (D) specific NK cell clusters from (A) and (E) normalized transcript levels. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Figure S5.
Proliferative NK Cell Subpopulation Is Associated with Increased COVID-19 Severity, Related to Figure 6
A,B. UMAP embedding of all NK cells colored by unsupervised clustering (top left of A) and by selected mRNA transcript levels (other panels in A) or (B) two selected surface proteins.
C. UMAP embedding density of NK cells for different blood draw samples, grouped by WOS. Selected clusters that display significant changes from WOS group to WOS group are encircled in the colors of the (A) clusters.
D. Differential expression analysis of NK cell genes that uniquely decrease in patients who improve (T2 versus T1, WOS decreased) in comparison with patients who did not. Each row represents a gene. The x axis represents differential expression score with positive being upregulated and negative being downregulated. FDR-corrected P values was shown. Significance is indicated by: (∗ FDR < 0.05, ∗∗ FDR < 0.01, ∗∗∗ FDR < 0.001). Full list provided in Table S5.2.
E. Boxplot showing the mRNA expression and protein levels of a few markers associated with NK cell functions in samples grouped by WOS. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
F. Scatterplots showing the Exhaustion gene signature score (x axis) and Undifferentiated gene signature score (y axis) of individual NK cells from all PBMC samples. Plots colored according to unsupervised clustering (from A), proliferation gene signature score, cytotoxic gene signature score, and KIR gene signature score are color-coded in each panel.
G. GSEA of top pathway enriched for cluster 5 cells. Normalized enrichment score (NES) and p values are shown. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). Full enrichment results are provided in Table S5.1.
Interestingly, among CD56dimCD16bright NK cells, one subpopulation (cluster 5) shows intermediate levels of CD16, CD57, and LAG3, and the highest level of the proliferation marker MKI67 (Figures 6A–6C, S5A, and S5B). The cluster 5 fraction increases from mild to moderate samples (Figures 6D and S5C). We further investigated cluster 5 by visualizing a few functional signature scores for each NK cell across different clusters (Figure S5F). As expected, CD56high cluster 4 cells reside mainly at the top-left region indicating a low level of exhaustion, while terminally differentiated cells with a high level of KIR transcripts are at the bottom-right branch (Figure S5F). Interestingly, the hybrid proliferative cluster 5 NK cells are at the intersection of these branches (Figure S5F), indicating that these cells may represent an intermediate transitional state during NK cell activation in response to virus. These cells also display upregulated signatures associated with fatty acid transport (Figure S5G; Table S5.1), potentially required to fulfill the proliferative capacity of this unique cluster.
We examined how NK cell transcriptomic signatures varied with disease trajectory and found an association between expression of the cytotoxic NK cell marker PRF1 and DNA repair marker DDIT4 and patient improvement (decrease of WOS between T1 and T2). For patients who did not improve, the nuclear factor κB (NF-κB)-inhibiting protein, COMMD6, increased (Figure S5D; Table S5.2). Thus, increased levels of cytotoxicity, DNA replication, and decreased inhibition of NFκB signaling in NK cells are associated with COVID-19 patient recovery.
Integrating Multi-Omic Profiles across Cell Types Resolves an Orchestrated Response Gene Module that Correlates with Clinical Features
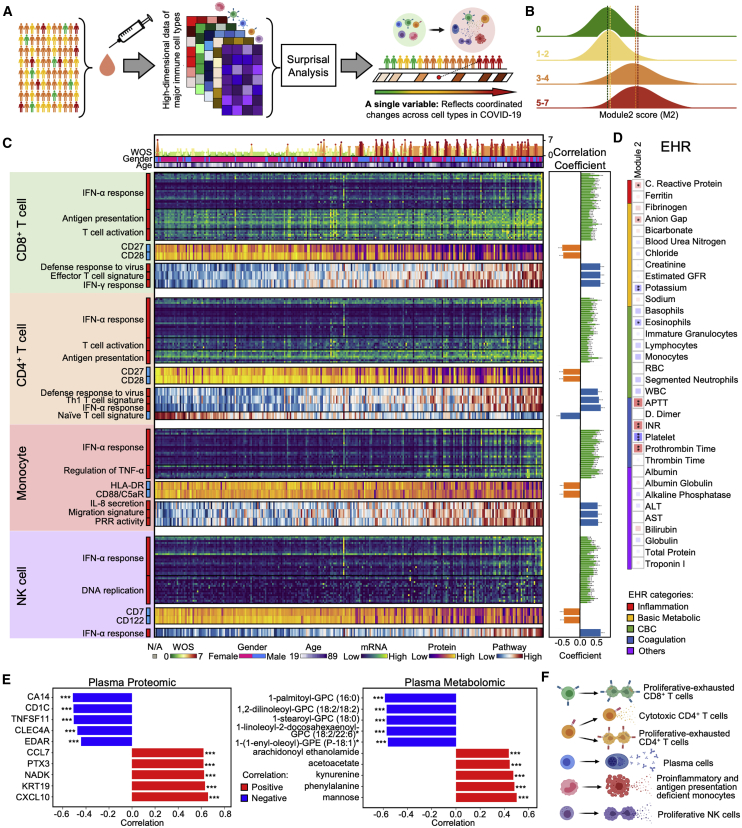
The successful control of SARS-CoV-2 infection requires immune system coordination. We explored for such coordination by analyzing how the transcriptomes of all cell types covary across patients with different levels of disease severity. The goal is to provide a unified view of the immune response in COVID-19 patients that can potentially provide important insights for patient treatment. For this purpose, we utilized surprisal analysis (Remacle et al., 2010), which has been applied to consolidate high-dimensional bulk and single-cell multi-omic data (Su et al., 2019b, 2019a, 2020). The basic hypothesis is that many genes co-vary across samples and thus can be viewed as functional gene modules. In this way, changes of thousands of correlated transcripts from different cell types are condensed into just one major gene module. In this view, each sample is reduced into a single dot along that gene module axis (Figure 7A). The module may then be mined to identify biological processes that co-vary across different cell types or correlate with clinical features and changes in plasma composition.
Figure 7.
Integrating Multi-Omic Profiles across Immune Cell Types Resolves a Coordinate Immune Response to SARS-CoV-2
(A) Cartoon illustration of the process of integrating data from different immune cell types from all samples, followed by reduction in single a dimensional representation (gene module). This characterizes the coordinated changes of cell types across COVID-19 patients.
(B) Distribution of individual PBMC datasets along gene module (M) 2 for healthy donors (WOS = 0, green), mild (WOS = 1–2, yellow), moderate (WOS = 3–4, orange), and severe (WOS = 5–7, red) patients.
(C) Heatmap visualization of selected top genes, surface proteins, and pathways for each cell type that significantly correlated with M2. Each column represents a sample and each row corresponds to levels of mRNA, surface protein, or pathway-enrichment score for the certain cell type of that sample. Columns are ordered based on M2 score in ascending order. The top three rows indicate the gender, age, and WOS. The heatmap keys are provided at the bottom. Sidebar on the left of each row represents the marker’s correlation with the M2 score, with red (blue) indicating positive (negative) correlation. Full list of the top genes, proteins, and pathways is provided in Table S6. Pearson correlation coefficients and FDR-corrected p values are indicated in the right panel bar plots. ∗FDR <0.05, ∗∗FDR <0.01, ∗∗∗FDR <0.001.
(D) Spearman rank correlations between M2 with clinical data. The square size corresponds to absolute value of the Spearman rank correlation coefficient. Blue indicates negative correlation and red indicates positive correlation. ∗FDR <0.05, ∗∗FDR <0.01, ∗∗∗FDR <0.001.
(E) Bar plot depicting Pearson correlation coefficient of the top 5 most significantly (p < 0.01) positively (red) and negatively (blue) correlated plasma proteins (left) or metabolites (right) with M2. Full list of correlated proteins (metabolites) is provided in Tables S6.36 and S6.37. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(F) Summary for coordinated immune response changes along M2 axis.
See also Figures S6 and S7 and Table S6.
The calculated most dominant module, module1 (M1), positively correlated (r = 0.95, p < 0.001, Figure S6A) with sequencing-depth-related information, reflecting technical details rather than biological information. The second most dominant gene module (M2), did not correlate with these technical parameters (Figure S6B), but correlated with many genes with biological functions (Figures 7C and S6B; Table S1, Table S2, Table S3, Table S4, Table S5, Table S6), and so we selected M2 for further analysis.
Figure S6.
Integrating Multi-Omic Profiles across Immune Cell Types Resolves Coordinate Immune Response to SARS-CoV2 Infection, Related to Figure 7
A. Pearson correlations of gene module (M) 1 score with two technical parameters: number of genes detected (left panel) and number of counts (right panel). The regression line is indicated in blue, with the 95% confidence area shown in shaded blue. Pearson correlation coefficient and associated P value are shown. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
B. Pearson correlations of gene module (M) 2 score with two technical parameters: number of genes detected (left panel) and number of counts (right panel). The regression line is indicated in blue, with the 95% confidence area shown in shaded blue. Pearson correlation coefficient and associated P value are shown. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
C. Spearman Rank correlations of gene module (M) 2 score with two clinical labs from EHR: Platelets (top panel) and CRP (bottom panel). The regression line is indicated in black, with the 95% confidence area shown in shaded gray. Spearman Rank correlation coefficient and associated P value shown. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
D. Pearson correlations of gene module (M) 2 score with principal component (PC) 1 values of plasma proteomic (bottom panel) and PC2 values of plasma metabolomic data (top panel). The regression line is indicated in black, with the 95% confidence area shown in shaded gray. Pearson correlation coefficient and associated p value shown. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
E. Pearson correlations of gene module (M) 2 score with top correlated plasma metabolites (top panel) and proteins (bottom panel). The regression line is indicated in black, with the 95% confidence area shown in shaded gray. Full list is provided in Tables S6.36 and S6.37. Pearson correlation coefficient and associated p value shown. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
F. Pearson correlations of the gene module (M) 2 score with levels of select top correlated mRNA, surface proteins, pathway enrichment scores, and subpopulation percentages from different immune cell types. Full list is provided in Table S6. The regression line is indicated in a cell-type specific color, with the 95% confidence area shown in shaded cell-type specific color. Pearson correlation coefficient and associated p value shown. Significance is indicated by: (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Although M2 was computed without considering clinical information, it clearly separates healthy and mild from moderate and severe patients (Figure 7B). This recalls the distinct shift from mild to moderate cases revealed in many analyses discussed above. Potentially because of this separation, the M2 score also correlates with disease severity (Figure S7A, Spearman correlation = 0.62, p < 0.001), as well as with several clinical observations including CRP, blood clotting metrics (APTT, INR, and prothrombin time), and negative correlations with eosinophils and platelets (Figures 7D and S6C).
Figure S7.
Integrating Multi-Omic Profiles across Immune Cell Types Resolves Coordinate Immune Response to SARS-CoV2 Infection, Related to Figure 7
A. Spearman Rank correlation of M2 with disease severity (WOS). Regression line is indicated in black, with the 95% confidence area in shaded gray. Spearman Rank correlation coefficient and associated P value shown. (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
B. Heatmap visualization of selected top genes, surface proteins and pathways for B cell that significantly correlated with M2. Each column represents a sample and each row corresponds to levels of mRNA, surface protein, or pathway enrichment score for B cells of that sample. Columns are ordered based on M2 score in ascending order. The heatmap keys are provided at the top. Sidebar on the left of each row represents the marker’s correlation with the M2 score, with red (blue) indicating positive (negative) correlation. Full list of the top genes, proteins and pathways is provided in Table S6. Pearson correlation coefficients and FDR-corrected P values are indicated in the right panel bar plots. (∗ FDR < 0.05, ∗∗ FDR < 0.01, ∗∗∗ FDR < 0.001).
C. Summary of the plasma proteomic, metabolomics and major immune subtypes correlation with M2. 1st panel: cartoon illustration of increase of severity along the M2 axis. 2nd −3rd panels, Pearson correlations of M2 with principal component (PC) 2 of the plasma proteomics data and PC1 of the plasma metabolomics data (PCA shown in Figures 1C and 1D). Regression lines are indicated in black, with 95% confidence area in shaded gray. Spearman Rank correlation coefficient and associated P value shown (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). Remaining panels: Bar plot depicting Pearson correlation coefficient of immune cell type percentages and subtype percentages with M2. (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
D. Summary for coordinated immune response changes along M2 axis.
We examined how properties of circulating immune cells tracked with increasing M2. Interestingly, the unusual phenotypes identified for each immune cell class (such as the hybrid CD8+ T cells or the cytotoxic CD4+ T cells, etc.) all exhibited significant positive correlations with M2 (Figures S6F and S7C; Table S6.38). This suggests a level of orchestration across these unique COVID-19 specific subpopulations that is not apparent by analyzing those phenotypes in isolation. Naive cell types were consistently anti-correlated with M2 (Figure S7C). In other words, for motion along the M2 axis, the adaptive immune cells become increasingly activated and differentiated, and various unique immune cell phenotypes emerge (Figure 7F).
Innate immune cells also orchestrated transcriptional alterations along the M2 axis. This includes an increasing IFN-α response for all cell classes, suggesting a role of type I IFN response to infection (Hadjadj et al., 2020). Different from the increased antigen presentation in both CD4+ and CD8+ T cells, monocytes showed decreased antigen presentation along M2 (Figure 7C). The unique S100highHLA-DRlow monocyte and the proliferative NK cell cluster also both correlate with M2 and emerge at the transition from mild to moderate disease (Figure 7C).
The M2 score also significantly correlated with the plasma proteomic principal component (PC) 2 and metabolomics PC1, both of which distinguish moderate and severe from mild disease (Figures S1D, S1E, S6D, and S7C). Many pro-inflammatory and chemo-attractive cytokines, including CCL7 and CXCL10, positively correlate with M2 while CLEC4A, a regulatory receptor for dendritic cells that impairs inflammation and T cell immunity (Uto et al., 2016), is negatively correlated (Figures 7E and S6E; Table S6.36). Metabolites that positively correlate with M2 include the endothelial relaxing factor kynurenine (Wang et al., 2010) that may be associated with hypotension seen in severe COVID-19 cases (Hanidziar and Bittner, 2020). A number of lipids negatively correlate with M2 (Figures 7E and S6E; Table S6.37), recalling the disproportionate loss of lipids in moderate and severe infections (Figure 1D, right panel). These results indicate that immune cell coordination, reflected in M2, is correlated with pro-inflammatory signals, loss of circulating lipids, and other biological processes associated with COVID-19.
In summary, we integrated the immune response, defined as whole transcriptomic changes, from all major cell types (with more than 120,000 variables) and condensed all of them into a single gene module (Figure 7A). This module M2 provides a detailed view for how different immune cell types coordinate across a highly heterogeneous cohort of COVID-19 patients (Figures 7F and S7D), and again suggests major physiological changes that distinguish mild from moderate and severe COVID-19 infections.
Discussion
A comprehensive understanding of immune responses in COVID-19 patients is fundamental to defining the effectiveness of treatments, predicting disease prognosis, and for understanding the reported heterogeneity of disease severities. We utilized computational methods to integrate clinical observations, single-cell characterizations, and plasma analytics, to develop a comprehensive and integrated view of COVID-19 during the week following initial diagnosis.
For a large cohort of patients representing the full spectrum of disease severities, we constructed a cross-omic interaction network (Figure 1E) that revealed, for example, COVID-19 severity-dependent connections between specific elevated cytokines and the downregulation of certain classes of metabolites and metabolic processes, suggesting an orchestration between increasing disease severity, elevated inflammation, and loss of key circulating nutrients. Further, the plasma multi-omic profiles captured a surprising similarity between moderate and severe COVID-19 and a sharp difference between mild and moderate infections. This major shift is marked by the preferential loss of lipids, amino acids, and xenobiotic metabolism (Figure 1D) and significant elevation of inflammatory cytokines (Figure 1C). The net implication is that of a stressed pro-inflammatory environment accompanied by decreased metabolic resources and signatures of possible hepatic dysfunction (Alqahtani and Schattenberg, 2020).
Similarly, a sharp difference between mild and moderate cases is observed in peripheral immune cells. This is characterized by the significant elevation of activated adaptive immune cells, and the emergence of unusual phenotypes. An interesting example is that of CD4+ T cells, which exhibit both a proliferative exhausted phenotype and a clonally expanded CD4+ cytotoxic phenotype. In fact, these two CD4+ phenotypes exhibit distinct functional signatures, distinct TCR sharing patterns, and may represent two divergent destinations for naive CD4+ T cells. Whether these phenotypes are harmful or protective remains unclear, but their relative abundances increase with infection severity, starting at the transition between mild and moderate disease.
Similar to a previous report (Schulte-Schrepping et al., 2020), we find a relative decrease in non-classical monocytes and the emergence of S100highHLA-DRlow monocytes at the stage of moderate disease. We also find that this subset highly correlated with global changes within the plasma proteome and metabolome. In general, unusual or dysfunctional adaptive or innate immune cell phenotypes that are unique to severe cases are not seen. Rather such phenotypes are seen in moderately ill patients and are only relatively increased in severe patients, further emphasizing the similarity between severe and moderate disease and the sharp difference between mild and moderate cases.
Gene module M2 (Figure 7) further reflects the sharp transition between mild and moderate disease and yields a coordinated view of the immune response to infection. M2, which was calculated using the immune cell transcriptomic data, readily distinguishes healthy donors and mild patients from moderate and severe cases, positively correlates with all observed unique immune cell phenotypes, with pro-inflammatory signals in the plasma, with the loss of specific metabolite classes, and with multiple clinical metrics (e.g., CRP). These correlative findings suggest that routine clinical measures may provide surrogate biomarkers for the immune dysfunction that emerges post the stage of mild disease and may yield markers that can anticipate disease course or provide surrogate endpoints for COVID-19 trials. The recognition of a non-monotonic change occurring between mild and moderate COVID-19 is potentially of high value, because therapeutic interventions at the stage of moderate disease are likely to be most effective. The resources provided from this work could prove valuable in developing such interventions, such as anti-inflammatory therapies that preserve the IFN-α antiviral response, and target coagulation defects or metabolic resource starvation. This broad systems immunology approach should also be applicable toward understanding immune responses in a plethora of other infectious diseases.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3 | eBioscience | Cat# 16-0037-85; RRID: AB_468855 |
| CD28 | eBioscience | Cat# 16-0289-85; RRID: AB_468927 |
| CD4 | Biolegend | Cat# 317416; RRID: AB_571945 |
| CD8 | Biolegend | Cat# 344721; RRID: AB_2075390 |
| PD-1 | Biolegend | Cat# 329904; RRID: AB_940479 |
| KI67 | Biolegend | Cat# 350504; RRID: AB_10660752 |
| Granzyme B | Biolegend | Cat# 372206; RRID: AB_2687030 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RBC lysis buffer | Miltenyi Biotech | Cat# 130-094-183 |
| RPMI 1640 Medium | GIBCO | Cat# 11875-093 |
| TotalSeq-C0251 anti-human Hashtag 1 Antibody | Biolegend | Cat# 394661 |
| TotalSeq-C0252 anti-human Hashtag 2 Antibody | Biolegend | Cat# 394663 |
| TotalSeq-C0253 anti-human Hashtag 3 Antibody | Biolegend | Cat# 394665 |
| TotalSeq-C0254 anti-human Hashtag 4 Antibody | Biolegend | Cat# 394667 |
| TotalSeq-C0255 anti-human Hashtag 5 Antibody | Biolegend | Cat# 394669 |
| TotalSeq-C0256 anti-human Hashtag 6 Antibody | Biolegend | Cat# 394671 |
| TotalSeq-C0257 anti-human Hashtag 7 Antibody | Biolegend | Cat# 394673 |
| TotalSeq-C0258 anti-human Hashtag 8 Antibody | Biolegend | Cat# 394675 |
| TotalSeq-C0259 anti-human Hashtag 9 Antibody | Biolegend | Cat# 394677 |
| TotalSeq-C0260 anti-human Hashtag 10 Antibody | Biolegend | Cat# 394679 |
| TotalSeq-C Custom Human panel | Biolegend | Cat# 99814 |
| CD8 Microbeads | Miltenyi Biotech | Cat# 130-045-201 |
| CD4 Microbeads | Miltenyi Biotech | Cat# 130-045-101 |
| Pan Monocyte Isolation Kit | Miltenyi Biotech | Cat# 130-096-537 |
| Lipopolysaccharide | Sigma Aldrich | Cat# L2654 |
| Intracellular Staining Permeabilization Wash Buffer (10X) | Biolegend | Cat# 421002 |
| Ficol-Paque PLUS Media | GE Healthcare | Cat# 17-1440-03 |
| CryoStor CS-10 | Biolife Solutions Inc | Cat# 210102 |
| autoMACS Rinsing Solution | Miltenyi Biotec | Cat# 130-091-222 |
| PBS, 1X | Fisher Scientific | Cat# 21-040-CV |
| AO/PI Cell Viability Kit - dual stain for cell counter | Logos Bio | Cat# F23001 |
| Critical Commercial Assays | ||
| Chromium Next GEM Single Cell 5′ Library and Gel Bead Kit v1.1 | 10x Genomics | Cat# 1000165 |
| Chromium Next GEM Chip G Single Cell Kit | 10x Genomics | Cat# 1000120 |
| Single Cell Polyfunctional Strength Panel Human | Isoplexis | Cat# PANEL-1001 |
| Single Cell Polyfunctional Strength ISOCODE Chip | Isoplexis | Cat# ISOCODE-1000 |
| Single Cell Innate Myeloid Panel Human | Isoplexis | Cat# PANEL-3L02 |
| Single Cell Innate and Myeloid ISOCODE Chip | Isoplexis | Cat# ISOCODE-3000 |
| Cardiovascular II panel | Olink | Cat# 95500 |
| Inflammation panel | Olink | Cat# 95302 |
| Metabolism panel | Olink | Cat# 95340 |
| Immune Response panel | Olink | Cat# 95320 |
| Organ Damage panel | Olink | Cat# 95331 |
| Deposited Data | ||
| scRNA-seq raw data | This paper | phs002275.v1.p1 |
| Processed scRNA-seq data | This paper | ArrayExpress: E-MTAB-9357 |
| Tables S1, S2, S3, S4, S5, and S6 | This paper | Mendeley Data: http://dx.doi.org/10.17632/tzydswhhb5.5 |
| Software and Algorithms | ||
| CellRanger v3.1.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
| GSVA v3.11 (R package) | Hänzelmann et al., 2013 | https://github.com/rcastelo/GSVA |
| MAST (R package) | Finak et al., 2015 | https://github.com/RGLab/MAST |
| FlowJo | FlowJo | https://www.flowjo.com/ |
| Scrublet v0.2.1 (Python package) | Wolock et al., 2019 | https://github.com/AllonKleinLab/scrublet |
| Scanpy v1.6.0 (Python package) | Wolf et al., 2018 | https://github.com/theislab/scanpy |
| Leiden v0.8.0 (Python package) | Traag et al., 2018 | https://github.com/vtraag/leidenalg |
| bbKNN v1.3.12 (Python package) | Polański et al., 2020 | https://github.com/Teichlab/bbknn |
| pySCENIC v0.10.3 (Python package) | Van de Sande et al., 2020 | https://github.com/aertslab/pySCENIC |
| MAGIC v2.0.3 (Python package) | van Dijk et al., 2018 | https://github.com/KrishnaswamyLab/MAGIC |
| GSEA v4.1.0 | Subramanian et al., 2005 | https://software.broadinstitute.org/cancer/software/gsea/wiki/index.php/Main_Page |
| Scirpy v0.3 (Python package) | Sturm et al., 2020 | https://github.com/icbi-lab/scirpy |
| IsoSpeak Software | Isoplexis | Product Code: ISOSPEAK-1000-1 |
| Other | ||
| IsoLight System | Isoplexis | Product Code: ISOLIGHT-1000-1 |
| Data visualization website for this publication | This paper | https://atlas.fredhutch.org/isb/covid/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. James R. Heath (jim.heath@isbscience.org).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All blood scRNA-seq data used in this study can be accessed by Array Express under the accession number: E-MTAB-9357. Additional Supplemental Items, including the metabolomic and proteomic datasets, are available from Mendeley Data at http://dx.doi.org/10.17632/tzydswhhb5.5.
Experimental Model and Subject Details
COVID-19 patients and healthy control samples
The study sample consisted of N = 139 COVID-19 patients (60 males and 79 females) and 258 healthy controls (18 for single-cell analysis; 133 for metabolomics; and 124 for proteomics, 18 of which overlap with metabolomics controls). Enrolled COVID-19 patients have an age range from 18 to 89 (median = 58). For ethnicity, 16 patients were recorded as Hispanic or Latino, 118 were reported as not Hispanic or Latino, and 5 had no ethnicity recorded. For race, 82 patients were recorded as White, 17 patients as Asian, 13 as Black or African American, 4 as Native Hawaiian or other Pacific Islander, 2 as American Indian or Alaska Native, 1 as more than one race, and 20 had no race recorded. A large percentage of patients presented with chronic hypertension (42%), and among other comorbidities, obesity (BMI ≥ 30 kg/m2; 37%; BMI ranged from 13.9 to 56.0 with Mean = 29.7, SD = 7.8), type 2 diabetes mellitus (19%), coronary artery disease (5%), and pulmonary disease (i.e., asthma or COPD; 24 or 19%) were the most prevalent. Patients in this study received a number of medications, including tocilizumab (an IL-6 receptor inhibitor), remdesivir (an antiviral), hydroxychloroquine, and antibiotics. 38% of draws were collected from the patient’s home (mobile phlebotomy), 38% of draws were collected from non-ICU hospitalized patients, 21% of draws were from patients in the ICU, and 3% from the clinic. All participants in this study provided written informed consent, in accordance with 45 CFR 46. De-identified proteomic and metabolomic data from matched healthy controls were previously collected from individuals enrolled in a wellness program at baseline (Arivale, Seattle, WA) (Manor et al., 2018), and processed using the same proteomic and metabolomic protocol. Healthy control samples for single-cell analyses were obtained from Bloodworks Northwest (Seattle, WA). All healthy control participants provided informed consent and authorization and permission for their de-identified data to be used for scientific research. Control samples for proteomics and metabolomics were matched on age and sex with enrolled patients. Information on age, sex, medication, and co-morbidities is listed in Table S1. This study was designed to characterize the immunological responses in COVID-19 patients without the intention of the development of new treatments or new diagnostics, and therefore sample size estimation was not included in the original study design. Procedures for the current study were approved by the Institutional Review Board (IRB) at Providence St. Joseph Health with IRB Study Number [STUDY2020000175] and the Western Institutional Review Board (WIRB) with IRB Study Number 20170658.
Method Details
EHR extraction and illness status designation
WOS for time of blood draw were determined by manual expert review. WOS for Figure S1A were automatically generated from data extracted from the electronic health record for hospitalized patients, and plotted for 6-hour time intervals based on end-interval grade. Automated results were compared against manual expert review for 15% of study subjects.
The following data were collected from the subject’s electronic health record (EHR): complete blood count (CBC) with differential, comprehensive metabolic panel, APTT, D-dimer, fibrinogen, prothrombin time, thrombin time ,and troponin I. Lab data were extracted from the nearest time point to each blood draw, if available within a window ± two days. First blood draw (n = 76), second blood draw (n = 54). Blood draws were classified as WOS = 3-4 (n = 83) and WOS = 5-7 (n = 47). We used an unpaired Wilcoxon-test to determine the statistical difference between WOS = 3-4 and WOS = 5-7, and P values were FDR adjusted. Spearman correlation coefficient was calculated using R package ‘corrplot v0.84’ to observe the associations between EHR labs and WOS disease severity, and the correlation significance was reported as FDR adjusted P values.
Plasma and PBMC isolation
Plasma and PBMC isolation were conducted with standard protocols from Bloodworks Northwest (Seattle, WA). Patient blood was collected in BD Vacutainer (EDTA) tubes (Becton, Dickinson and Company, Franklin Lakes, NJ). Plasma fractions were collected after centrifuged at 800 x g at 4°C for 10 min, aliquoted, and stored until use at −80°C. The rest of the blood was diluted with PBS (pH7.2) to 2X of the original volume and layered over 15 mL Ficoll (GE Healthcare, Waukesha, WI) in SepMate-50 tubes (StemCell, Vancouver, BC). After centrifuged at 800 x g for 15 min at room temperature, the PBMC layer (did not include granulocytes (such as neutrophils)) was poured into a 50 mL conical tube. The cells were washed twice with autoMACS Rinsing Solution (Miltenyi Biotec, Auburn, CA) and centrifuge at 250 x g for 10 min, at RT. PBMC pellets were gently resuspended in 5 mL Rinsing Solution and a 5 μL aliquot was diluted 1:10 v/v for cell counting. Cells in 18 μL of diluted samples were first mixed with 2 μL of Acridine Orange / Propidium Iodide Stain (Logos Biosystems, Annandale, VA), 10 μL was then loaded to a PhotonSlide (Logos Biosystems) and counted in a LUNA FL Dual Fluorescence cell counter (Logos Biosystems). Cryopreservation freeze media CryoStor CS-10 (Biolife Solutions, Bothell, WA) was slowly added to make a concentration of 2.5 million PBMC/ml. Cells were aliquoted in 2.0 mL Cryotube vials (ThermoFisher, Waltham, MA) and frozen in CoolCell LX Cell Freezing Container (Corning, Corning, NY) at −80°C for at least 2 hours before stored in liquid nitrogen until use.
Plasma proteomics
Plasma concentrations of proteins were measured using the ProSeek Cardiovascular II, Inflammation, Metabolism, Immune Response, and Organ Damage panels (Olink Biosciences, Uppsala, Sweden). Health control plasma samples were processed at Olink facilities in Boston, MA. Plasma samples from COVID-19 participants were assayed at the Institute for Systems Biology. Proteins from patient plasma were measured using proximity extension assay (PEA) (Olink Proteomics, Uppsala, Sweden) which allows for the simultaneous analysis of 92 protein biomarkers on each panel. Five panels including Inflammation, Cardiovascular II, Organ Damage, Immune Response, and Metabolism were run using 328 patient plasma samples as well as 8 replicates of a pooled healthy control. One microliter of plasma was incubated overnight and allowed to bind with oligonucleotide-labeled antibody pairs to form specific DNA duplexes. This template was then extended and pre-amplified, and the individual protein markers were measured using high-throughput microfluidic real-time PCR (Fluidigm, South San Francisco, CA). The resulting Ct values were normalized against an extension control, an inter-plate control, and adjusted with a correction factor according to the manufacturer’s instructions to calculate a normalized protein expression value (NPX) in log2 scale. Samples were processed in batches with pooled quality control samples included in each batch. Specifically, 8 technical replicates from a pooled blood sample served as overlapping reference samples in each plate. Raw expression values are then batch corrected by normalizing to the overlapping reference samples within each plate.
Plasma metabolomics
Metabolon (Morrisville, NC, USA) conducted the metabolomics assays for all participant plasma samples used in this study. Data were generated with the Global Metabolomics platform via ultra-high-performance liquid chromatography/tandem accurate mass spectrometry. 100 μl of plasma was aliquoted and transported on dry ice to Metabolon Inc. for analysis. Sample handling and quality control were performed by Metabolon in their CLIA-certified laboratory. Mass spectrometry was performed using Metabolon’s ultra-high-performance liquid chromatography/tandem mass spectrometry (UHPLC/MS/MS) Global Platform, which consisting of four independent UPLC–MS/MS instruments, each with a Waters ACQUITY ultra-performance liquid chromatography (UPLC) and Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer at 35,000 mass resolution. Data extraction, along with biochemical identification, data curation, quantification, and data normalizations were performed by Metabolon’s hardware and software. Samples were processed in batches with pooled quality control samples included in each batch; pooled quality control samples were consistent across batches. Potential batch effects for each metabolite were adjusted by dividing by the corresponding average value identified in the pooled quality control samples from the same batch. For analysis, the raw metabolomics data were median scaled within each batch such that the median value for each metabolite was one.
Single-cell multiplex secretome assay
Cryopreserved PBMCs were thawed and incubated in the RPMI 1640 for overnight recovery at 37°C, 5% CO2. Recovered cell viability was measured at > 95% for all samples. After overnight recovery, CD4+ and CD8+ T cell populations and monocytes were magnetically isolated using the CD4+, CD8+ Microbeads and Pan Monocyte Isolation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) sequentially.
The enriched CD4+ and CD8+ T cells (100,000 cells/well in a 96 well-plate) were stimulated for 6 hours with plate-bound anti-CD3 antibodies (pre-coated at 10 μg/mL overnight at 4°C) and soluble anti-CD28 antibodies (5 μg/mL) in complete RPMI-1640 at 37°C, 5% CO2 . The enriched monocytes at 1 × 105/ml were stimulated with 10 ng/ml LPS for 12 hours.
After stimulation, the activated cells were collected, washed, and stained with membrane stain (included in the IsoPlexis kit). The stained cells were then loaded onto the chip consisting of the 12,000 chambers pre-coated with an array of 32 cytokine capture antibodies. The chip was inserted into the fully automated IsoLight for further incubation for 16 hours and the cytokines were detected by a cocktail of detection antibodies followed by the fluorescent labeling. The scanned fluorescent signal was analyzed by IsoSpeak software to calculate the numbers of cytokine-secreting cells, the intensity level of cytokines, and polyfunctional strength index (PSI).
See the below for the measured cytokines in each panel.
Single-Cell Adaptive Immune cytokine panel including the following subsets of cytokines. Effector: Granzyme B, IFN-γ, MIP-1α, Perforin, TNF-α, TNF-β; Stimulatory: GM-CSF, IL-2, IL-5, IL-7, IL-8, IL-9, IL-12, IL-15, IL-21; Chemoattractive: CCL11, IP-10, MIP-1β, RANTES; Regulatory: IL-4, IL-10, IL-13, IL-22, TGFβ1, sCD137, sCD40L; Inflammatory: IL-1β, IL-6, IL-17A, IL-17F, MCP-1, MCP-4.
Single-Cell Innate Immune cytokine panel including the following subsets of cytokines. Effector: IFN-γ, MIP-1α, TNF-α, TNF-β; Stimulatory: GM-CSF, IL-8, IL-9, IL-15, IL-18, TGF-α, IL-5; Chemoattractive: CCL11, IP-10, MIP-1β, RANTES, BCA-1; Regulatory: IL-10, IL-13, IL-22, sCD40L; Inflammatory: IL-1β, IL-6, IL-12-p40, IL-12, IL-17A, IL-17F, MCP-1, MCP-4, MIF; Growth Factors: EGF, PDGF-BB, VEGF.
Flow cytometry analysis
For surface marker and intracellular cytokine staining, PBMCs were surface stained for 30 minutes at 4°C and washed twice. Cells were then fixed in 4% paraformaldehyde solution for 20 minutes and washed twice with 1x permeabilization solution. Intracellular cytokines were stained using an antibody cocktail and incubated for 30 min at 4°C. The washed cells were analyzed with Attune NxT and the data was analyzed with FlowJo software. Antibodies against human CD4, CD8, PD-1, Ki-67, and Granzyme B were purchased from Biolegend.
Single cell multi-omics assay
Chromium Single Cell Kits (10x Genomics) were utilized to analyze the transcriptomic, surface protein levels and, TCR sequences simultaneously from the same cell. Experiments were performed according to the manufacturer’s instructions.
Briefly, cryopreserved PBMCs were thawed and 1X red blood cell lysis solution (BioLegend) was used to lyse any remaining red blood cells in the PBMC samples. Cells were stained with a panel of TotalSeq-C human antibodies that includes hashtag multiplexing antibodies (BioLegend) detailed in Table S1.8 according to the manufacturer’s protocol. Stained cells were then loaded onto a Chromium Next GEM chip G. Cells were lysed for reverse transcription and complementary DNA (cDNA) amplification in the Chromium Controller (10X Genomics). The polyadenylated transcripts were reverse-transcribed inside each gel bead-in-emulsion afterward. Full-length cDNA along with cell barcode identifiers were PCR-amplified and sequencing libraries were prepared and normalized. The constructed library was sequenced on the Novaseq platform (Illumina).
Quantification and Statistical Analysis
Quantifying disease severity via WOS
Severity of COVID-19 was clinical status assessed throughout a patient’s encounter in accordance to the 9-point the World Health Organization (WHO) Ordinal Scale (WOS) for clinical improvement consisting of the following categories: 0) uninfected - no evidence of infection; 1) ambulatory - no limitation of activities; 2) ambulatory - limitation of activities; 3) hospitalized, mild - no oxygen therapy; 4) hospitalized, mild - oxygen by mask or nasal prongs; 5) hospitalized, severe - non-invasive ventilation or high-flow oxygen; 6) hospitalized, severe - intubation and mechanical ventilation; 7) hospitalized, severe - ventilation + additional organ support; and 8) dead – death (World Health Organization, 2020). For Figure S1A, WOS was calculated in 6-hour intervals, and the score was used that corresponded to the window in which the blood was drawn.
Correlation between EHR and other dataset
We conducted Spearman correlation analysis to evaluate the association between gene module2 (M2), obtained from surprisal analysis of sc-RNA-seq data, and disease severity (WOS) with clinical labs as well as CBCs. The correlation significance was reported as FDR adjusted P values.
Statistical Analysis of Plasma-omics
For quality control analysis, a threshold of less than 20% missing values was set for each protein and metabolite. Missing values for proteins were imputed to be the limits of detection while missing values for metabolites were imputed to the lowest measured value. A total of 463 proteins and 877 metabolites were included in further statistical analyses.
For association with COVID-19 status and severity, we assessed the differential abundance of protein and metabolite levels between healthy and COVID-19 samples. Specifically, we compared samples from uninfected participants with samples from mild (WHO Ordinal Scale 1-2), moderate (WHO Ordinal Scale (3-4), and severe (WHO Ordinal Scale 5-7) patients. For all analyses, only samples from the first blood draw were considered.
Prior to association analysis, levels of proteins and metabolites were first adjusted for age, sex, BMI, and race/ethnicity. We then performed Welch’s t tests between uninfected and mild; mild and moderate; and moderate and severe samples. Analyses were performed for proteins and metabolites separately. Multiple testing correction was subsequently applied to each comparison using the Benjamini-Hochberg (BH) procedure with false discovery rate (FDR) at 5%. Age, sex, BMI, and race/ethnicity adjusted protein and metabolite levels were tested separately and multiple testing correction was performed using BH FDR < 5%.
Further metabolites that significantly changed (FDR < 0.05) in the aforementioned stepwise comparisons were first grouped by their superpathway (e.g., lipid, xenobiotic, etc.). Then the fold changes of each metabolite within a superpathway group was summed and utilized for the stacked bar plots shown in Figure 1D, right panel. Similarly, proteins that significantly changed (FDR < 0.05) in the aforementioned stepwise comparisons were utilized for the plot shown in Figure 1C, right panel.
For association with gene module2 score of single-cell data, pearson correlation was also performed to test the association of adjusted metabolite and protein levels with the M2 score derived from surprisal analysis of single-cell data. Calculated Pearson correlation coefficients and associated P values were subsequently used.
Principal component analysis of plasma-omics
PCA was performed separately for metabolomic and proteomic data using metabolites and proteins that passed quality control, respectively. Values were centered and scaled from 0 to 1 prior to dimensional reduction. In the case of proteins, all proteins that could not be determined for any patient were removed from the dataset, and PCA was run on the remaining 352 proteins per patient using scikit-learn in python (Pedregosa et al., 2011). In the case of metabolites, several more values could not be determined and so deterministic PCA was not feasible. Instead, all metabolites that could not be determined for > 20% of patients were removed. This left 766 of 1053 metabolites remaining, and only 4.98% of the filtered dataset undetermined. Then, the dimensional reduction was applied to the filtered data via probabilistic PCA in MATLAB (Tipping and Bishop, 1999). PCA plots were colored by the severity of the patient based on the WOS at the time of the blood draw.
WOS-dependent cross-omic network analysis
Metabolite data (N = 123 patients and k = 243 observations) and protein data (N = 127 patients and k = 249 observations) collected from plasma were combined with clinical test data extracted from patent medical records (N = 79 patients and k = 137 observations) as well as patient baseline demographics. Clinical test data collected within two days of blood draws were used for this analysis. Multiple observations were available for most patients. Proteins and metabolites with greater than 50% missingness were dropped from the dataset.
First, we identified a set of significant pairwise Spearman correlations between inter-omic analytes, adjusting for potential confounding variables. This initial step was performed to lower the computational burden for downstream model fitting. Generalized estimating equations (GEE) were fit for each of N = 1,591 analytes using the model analyte ∼age_at_baseline + C(sex) + C(race) + C(ethnicity) + BMI. For skewed analytes (), a Gamma distribution with a log link function was used; otherwise a Gaussian distribution with an identity link function was used. The Exchangeable dependence structure was used to account for the non-independence of multiple observations within each patient. Residuals from each GEE were used to compute Spearman correlations between each pair of inter-omic analytes (e.g., metabolite-protein, metabolite-clinical, or clinical-protein). P values were adjusted for multiple hypotheses using the False Discovery Rate (FDR) method of Benjamini and Hochberg (1995).
Disease severity at each patient blood draw was calculated using the WHO ordinal scale. For each of 27,386 highly significant (FDR < 0.001) pairwise inter-omic Spearman correlations calculated in the previous step, we fit a GEE using the model analyte1 ∼age_at_baseline + C(sex) + C(race) + C(ethnicity) + BMI + who_ordinal_scale∗analyte2 on the original (non-residualized) data. For skewed analytes (), a Gamma distribution with a log link function was used; otherwise a Gaussian distribution with an identity link function was used. The Exchangeable dependence structure was used to account for the non-independence of multiple observations within each patient. Interaction p values were adjusted for multiple hypotheses.
Single cell RNA-seq data processing
Droplet-based sequencing data were aligned and quantified using the Cell Ranger Single-Cell Software Suite (version 3.0.0, 10x Genomics) against the GRCh38 human reference genome. Cells from each demultiplexed sample were first filtered for cells that expressed a minimum of 200 genes, then they were filtered based on three metrics: 1) the total number of unique molecular identifiers (UMI) counts per cell (library size) must be less than 10000; 2) the number of detected genes per cell must be less than 2500; and 3) the proportion of mitochondrial gene counts (UMIs from mitochondrial genes / total UMIs) must be less than 10%. Doublets were either simultaneously identified in sample demultiplexing or identified using scrublet (Wolock et al., 2019) and were removed prior to the aforementioned filtering. After QC metric filtering, a total of 559,583 cells were retained for downstream analysis. Scanpy (Wolf et al., 2018) was used to normalize cells via CPM normalization (UMI total count of each cell was set to 106) and log1p transformation (natural log of CPM plus one).
Single cell RNA-seq cell type identification
PCA was performed on the normalized, ln(CPM+1), gene expression matrix of all cells passing the previously mentioned QC metrics, “arpack” was set as the SVD solver. A neighborhood graph was built with n-neighbors set to 15 and all 50 calculated PCs as inputs. This neighborhood graph was utilized to calculate a UMAP (McInnes et al., 2018) and clusters were determined via the Leiden algorithm (Traag et al., 2018). Clusters identified in this first round of clustering were annotated based on the expression of canonical marker genes. Clusters that were not uniform in their expression of well-known marker genes were extracted and a second round of dimension reduction and clustering was performed on these subsets (recalculation of UMAP and Leiden, this was mainly utilized to separate T cells and NK cells, and rare cell type populations). Clusters that simultaneously expressed canonical markers from two or more major cell types were identified as potential doublets or low-quality cells and were removed from downstream analysis. 10,433 low-quality cells were removed resulting in 549,210 total cells for further analyses.
Identified T cells from the first and second round of clustering were extracted for CD4+ and CD8+ T cell identification. A CD4+ T cell score was obtained by taking the sum of the scaled values (scale the ln(CPM + 1) values to be from 0 to 1) of the CD4 transcript and CD4 surface protein. A CD8+ T cell score was obtained by taking the sum of the scaled values of the CD8A and CD8B transcripts, and the CD8 surface protein. Both scores were subsequently scaled to be from 0 to 1 and utilized as inputs for a scatterplot for manual gating of CD4+ T cells and CD8+ T cells. Other T cells were then defined as T cells that did not confidently categorize as CD4+ or CD8+.
Single cell RNA-seq batch information
All 50 computed PCs from the PCA performed on the normalized gene expression data were used as inputs to calculate a bbKNN graph (Polański et al., 2020) which was set to correct across batch-specific patient samples (this treats patient samples which were obtained from different sequencing batches/reactions as different). This bbKNN graph was then utilized to calculate batch corrected UMAP coordinates, which are projected in Figure S1K (left panel). Batch correction (based on bbKNN) was only used for UMAP visualization purposes in Figure S1K (left panel). No major batch effects were found as examined through heatmap visualization of batch-specific cell-types with cell-type markers in Figure S1J and hierarchical clustering of batch-specific cell-types in Figure S1K (right panel). Downstream statistical analysis was performed on the uncorrected data as recommended in (Amezquita et al., 2020).
Single cell RNA-seq gene regulatory networks
Major immune cell types CD4+ T cells, CD8+ T cells, monocytes, B cells, and NK cells were further analyzed via a construction of their single-cell regulatory networks by inputting their normalized gene expression matrices into pySCENIC (Van de Sande et al., 2020). Patient 4 B cells were removed from the B cell transcriptomic dataset inputted into pySCENIC as they had chronic lymphocytic leukemia and a resultant cancerous population of B cells. Due to the large number of identified monocytes (n = 175,635), it was not computationally feasible to perform single-cell regulatory network calculation on the entire monocyte dataset and thus monocytes were subsetted by taking one-third of the monocytes from each sample resulting in 58,542 monocytes being utilized to calculate transcription factor modules. The resultant single-cell transcription factor modules from each cell type were subject to PCA. Batch-specific PC dimensions, determined via plotting of sequencing batches and canonical subpopulation markers onto reduced dimensional space (PC or UMAP), were removed. This entailed the removal of PC7 from CD8+ T cells, PC9 from CD4+ T cells, and PC4 from NK cells. Further dimensional reduction was calculated via a neighborhood graph (n-neighbors was set to 15 and all PCs (49 if a PC was removed, 50 if no PCs were removed) were utilized) and subsequently a UMAP with the neighborhood graph as input. Clustering was calculated via the Leiden algorithm with resolution set to 0.8 for CD8+ T cells, 1.0 for CD4+ T cells, 0.95 for B cells, 0.95 for monocytes, and 0.7 for NK cells. All reduced dimensions (PCA, neighborhood graph, UMAP) and clusters (Leiden) for all of the aforementioned single cell RNA-seq data were calculated via Scanpy (Wolf et al., 2018).
Single cell RNA-seq signature score
Signatures scores were calculated for CD8+ T cells, CD4+ T cells, and NK cells for further characterization of cell type specific subpopulations. For all of the aforementioned cell types for which signature scores were computed, the single cell transcriptomic matrix was first subject to MAGIC imputation (van Dijk et al., 2018). Imputed gene expression values were then utilized to calculate the signature scores, unless otherwise specified the signature scores consist of the sum of the scaled values (genes were scaled to be from 0 to 1) of a gene set. For CD8+ T cells naive (TCF7, LEF1, SELL, CCR7), cytotoxic (NKG7, CCL4, CST7, PRF1, GZMA, GZMB, IFNG, CCL3), exhaustion (PDCD1, TIGIT, LAG3, HAVCR2, CTLA4), and proliferation (MKI67, TYMS) signature scores were calculated. For CD4+ T cells naive (TCF7, LEF1, SELL, CCR7), cytotoxic (GZMB, PRF1, GNLY), exhaustion (PDCD1, TIGIT, LAG3, HAVCR2, CTLA4), and proliferation (MKI67, TYMS) signature scores were calculated. The CD4+ T cell Th1 signature score was calculated via the average of a HLA class II signature score (HLA-DRA, HLA-DRB1, HLA-DRB5, HLA-DQA1, HLA-DQA2, HLA-DQB1, HLA-DMA, HLA-DMB, HLA-DPA1, HLA-DPB1) and the imputed expression of the IFNG transcript. Genes utilized in the HLA class II signature score were selected via filtering for transcripts with non-sparse expression as determined via plotting all HLA class II transcripts onto the single cell transcription factor module determined UMAP for CD4+ T cells. For NK cells undifferentiated (SELL, NCAM1), exhaustion (PDCD1, TIGIT, LAG3, HAVCR2, CTLA4), cytotoxic (NKG7, CCL4, CST7, PRF1, GZMA, GZMB, IFNG, CCL3), proliferation (MKI67, TYMS), and KIR (KIR2DL1, KIR2DL3, KIR3DL1, KIR3DL2) signature scores were calculated.
Gene set enrichment/variation analysis
To quantify the pathway enrichment score across samples or cells, GSVA was performed using the R package GSVA (v.3.11) (Hänzelmann et al., 2013) to identify the most changed pathways between samples/cells with the averaged log2 (CPM+1) data as input.
Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) was performed using GSEA v4.1.0 software with “weighted” enrichment statistics. Transcriptomic pseudo-bulk of Leiden clusters (per cell type) and transcriptomic pseudo-bulk of samples (patient blood draws and healthy donor samples, also per cell type) were subject to this analysis. Pseudo-bulk of Leiden clusters were calculated by taking the mean normalized (ln(CPM + 1)) gene expression values of each Leiden cluster, and pseudo-bulk of samples were similarly calculated by taking the mean normalized (ln(CPM + 1)) gene expression values of each sample. Each pseudo-bulk was then separately (only one pseudo-bulk was used at a time) used as the input with “diff_of_classes” methods used as the metric for ranking genes. Normalized enrichment score (NES) was assessed across the curated Molecular Signatures Database (MSigDB) Hallmark, C2 curated gene sets, C5 Gene ontology, and C7 Immunological signature.
Single cell RNA-seq marker selection
Figures S2F and S3F are showing well-reported naive-related gene TCF7 (Gullicksrud et al., 2017), proliferation gene MKI67 (Miller et al., 2018), and exhaustion gene PDCD1 (encodes PD-1) (Riley, 2009) in T cells.
Similarly, Figure 4C showed the FCER2 gene which is known to be highly expressed in naive B cell populations but low in class-switched B cells (Liu et al., 2016); the SLAMF7 gene that is upregulated in activated B cells (Kim et al., 2016); and the HLA-DRA gene, one of the HLA class II genes that play essential roles in T cell-dependent B cell activation (Adler et al., 2017; Katikaneni and Jin, 2019). In Figure 4D, we included two important chemokine receptors in B cells (CD185 and CD196) (Henneken et al., 2005; Reimer et al., 2017) that showed significant changes in COVID-19 patients compared to healthy donors. Figure S4E is investigating if TNF-a, another marker for monocyte immunoparalysis (Giamarellos-Bourboulis et al., 2020) is also downregulated with increasing disease severity, which we found to be such. Figures 6E and S5E show a few well-reported genes associated with NK cell states. Specifically, we included GZMB and PRF1 that are highly expressed in cytotoxic NK cells (Yoon et al., 2015), the proliferation gene MKI67 (Hudspeth et al., 2019), the CD56 surface marker for the classification of two canonical NK cell subtypes (Vivier et al., 2008), the activation marker CD69 (Borrego et al., 1999), the surface marker CD62L that is highly expressed on CD56bright NK cells and differentially expressed during various maturation stages of NK cells (Lima et al., 2017), and the IL7R gene (encodes CD127) that is expressed specifically on CD56bright cells and enhances the survival of this NK subset (Michaud et al., 2010).
The selection of transcripts and proteins for heatmaps in Figures 2C and 3C is based on knowledge of well-documented T cell phenotype genes and proteins. We show genes TCF7, CCR7, LEF1 that have high expression in naive T cells (Campbell et al., 2001; Willinger et al., 2006); memory-like/intermediate genes AQP3, CD69, GZMK (Szabo et al., 2019); effector-like cytotoxic genes GZMB, PRF1, GNLY (Hidalgo et al., 2008); proliferation genes MKI67 and TYMS (Miller et al., 2018; Shichijo et al., 2004); exhaustion-related genes PDCD1, LAG3, TIM3, TIGIT (Anderson et al., 2016) to help annotate T cell subpopulations. We also selected the IFNG gene that is highly expressed by T helper type 1 cells (Swanson et al., 2001), CXCR5 for T follicular helper cells (Crotty, 2014), and FOXP3 for regulatory T cells (Li et al., 2015). For proteins in Figure 2C, we show naive CD8+ T cell marker CD45RA, CD28 (Berard and Tough, 2002; Vallejo et al., 1999); long-lived memory CD8+ T cell marker CD127 (Huster et al., 2004); CD8+ effector T cell markers CD95, KLRG1, CD11b (Arens et al., 2005; Fiorentini et al., 2001; Herndler-Brandstetter et al., 2018); T cell activation marker CD71 (Caruso et al., 1997); terminally differentiated CD8+ T cell marker CD57 (Kared et al., 2016); and the immunoregulatory receptor CD244 (Agresta et al., 2018). For CD4+ T cell proteins in the heatmap in Figure 3C, we show naive T cell marker CD45RA (Berard and Tough, 2002); the activation marker CD278 for CD4+ T cells (Maawy and Ito, 2019); the memory T cell marker CD49F (Abitorabi et al., 1996); HLA class II protein HLA-DR that can be upregulated after CD4+ T cell activation (Mangalam et al., 2006); the dual function receptor CD95 that has both apoptotic and anti-apoptotic effects (Paulsen and Janssen, 2011); immunoregulatory receptor CD244 (Agresta et al., 2018) and the inhibitory receptor CD224 (Shuai et al., 2016). For monocytes, the selection of transcripts for Figure 5D are based on the combined list of the top5 most differentially expressed genes (determined via the Wilcoxon Rank-Sum test) for each monocyte cluster, only one copy of duplicate genes were kept in the final gene list and genes were hierarchically clustered via the “complete” linkage method (also known as the Farthest Point algorithm). The full list of top genes and associated statistics can be found in our supplementary Table S4.12.
The selection of mRNA, protein, and pathway for Figure 7C, is based on M2 score Pearson correlation coefficient and P value. The top300 most positively/negatively correlated genes with M2 from each cell type were separately enriched for gene ontology biological processes, then genes related to said biological processes were utilized for transcriptomic heatmaps. Proteins are selected from the top5 most positively/negatively correlated proteins, pathways are selected from top50 most positively/negatively correlated pathways. Full lists of the top genes, proteins, and pathways and their associated statistics are provided in Table S6 and the gene ontology analysis of the top300 genes are also provided in Table S6. P values for all correlations plotted are all less than 0.05.
The selection of transcripts, proteins, and pathways for Figure 5F is based on the WOS correlation coefficient and P value. The top300 most positively/negatively correlated genes with WOS from monocytes were separately enriched for gene ontology biological processes, then genes related to said biological processes were utilized for transcriptomic heatmaps. Proteins are selected from the top5 most positively/negatively correlated proteins, pathways are selected from top50 most positively/negatively correlated pathways. Full lists of the top genes, proteins, and pathways and their associated statistics are provided in Table S4 and the gene ontology analysis of the top300 genes are also provided in the Table S4. P values for all correlations plotted are all less than 0.05.
Surprisal analysis
The gene module score was calculated based on surprisal analysis, an information-theoretical analysis technique that integrates and applies principles of thermodynamics and maximal entropy (Agmon et al., 1979; Levine, 2005). It has already been well-used in a spectrum of disciplines including engineering, physics, chemistry, and biomedical engineering (Levine, 1978, 2005; Levine and Bernstein, 1974). Recently, it has been extended to characterize the state of living cells, specifically monitoring and characterizing biological processes using transcriptional data (Remacle et al., 2010; Zadran et al., 2014).
When applying surprisal analysis to our particular scenario, we have a dataset characterized by a cohort of patient samples with different immune profiles (defined by gene expression from all five cell types), Equation 1 from surprisal analysis can de-convolute the changes of thousands of genes into one unchanged gene expression baseline, , and a series of gene expression modules, . Each module contains a group of genes that are coordinately changing together across patient samples. In the equation, value represent the contribution of each gene from each cell type (with n going from 1 to (#cell types) x (#genes)), onto module j, and values represent the overall score of this gene module j across patient sample k.
The mathematical derivation and computational implementation details have been well documented in the previous publication (Remacle et al., 2010). Briefly, we are fitting the sum of terms, as shown on the right-hand side of Equation 1 to the logarithm of the measured expression level of cell-type transcript n (with n going from 1 to (#cell types) x (#genes)) at the given patient sample k. This is repeated for every sample in the pseudo-bulk which contains (#cell types) x (#genes) rows and (#patient samples) columns of the normalized (ln(CPM+1)) gene expression values. A suitable general method has been published (Agmon et al., 1979) and extensively applied mRNA microarrays monitor the expression levels of thousands of genes at a time. As the analysis of the data needs to be handled by numerical procedures that are compatible with such a large dataset, we have adapted singular value decomposition to handle the optimization problem as detailed in (Remacle et al., 2010).
| Equation 1 |
In this study, the most dominant constraint, λ1 was reflecting sequencing technical variations (Figure S6A), thus the 2nd most dominant constraint, λ2, which is associated with many biological processes (Figure 7C) was further investigated as the immune response module M2, scores for analyzed blood draw samples are in Table S6.39.
Single cell -omics disease severity analysis
The dataset collected here can be analyzed, at a high level, in two different ways. First, each blood draw corresponds to a time point at which that patient was characterized by a level of disease severity (WOS) (Table S1.1). Grouping each blood draw according to WOS, a contemporary measure of disease severity, can provide insight into how various immune cell populations, regulatory networks, and other biomarkers vary with disease severity level by sample. A second view is to consider each patient separately, so that the two blood draws are used to capture an individual patient’s disease trajectory. Here, patients that improve between T1 and T2 can be differentiated from patients advancing to more severe infection. We utilize both types of analyses here. Disease trajectories the time of the T1 and T2 blood draws, and WOS measures of disease severity, are provided in Figure S1A. These trajectories start at the self-reported day of symptom onset and give WOS during the time of hospital admission.
Statistical analysis of single cell -omics
Differential expression analysis was performed using the R package MAST (Finak et al., 2015). For each cell type (CD8+ T cells, CD4+ T cells, monocytes, B cells, and NK cells), we investigated the WHO-score delta by comparing patients getting better (having a WHO score that decreases (T2-T1)) to patients staying stable (having a WHO score that does not decrease/increase (T2-T1)). The normalized (but not batch corrected) gene-cell barcode was used as input, and we adjusted for all factors in our model as recommended in Amezquita et al. (2020). Our MAST models included as covariates the batch and patient ID, as well as the cellular detection rate (CDR) to correct for biological and technical nuisance factors. Multiple hypothesis testing correction was performed by controlling the false discovery rate (FDR). Genes were declared significantly differentially expressed at a FDR of 5% and a fold-change > 1.5.
Single cell TCR-seq data processing
Droplet-based sequencing data for T cell receptor sequences were aligned and quantified using the Cell Ranger Single-Cell Software Suite (version 3.1.0, 10x Genomics) against the GRCh38 human VDJ reference genome.
Single cell TCR & RNA-seq integration
Filtered annotated contigs for T cell receptor sequences were analyzed via scirpy (Sturm et al., 2020). The aforementioned contig files were read in as TCRs, filtered for either CD4+ or CD8+ T cells (as identified via single cell RNA-seq analysis) and then subject to clonotype definition and clonal expansion analysis utilizing nucleotide sequences. Samples were then concatenated together and merged with gene expression data for simultaneous single cell TCR and RNA data visualization.
Both the integrated CD4+ and CD8+ T cell datasets were subject to filtering for cells with complete TCR sequences, defined as a detectable TRA and TRB. This filtering resulted in 107,397 of the 169,019 CD4+ T cells, and 55,994 of the 91,810 CD8+ T cells for downstream TCR analysis. Each integrated T cell subset was grouped by Leiden clusters resulting in a TCR matrix consisting of Leiden clusters as rows and unique TCRs as columns with the number of cells with a given TCR in a certain Leiden cluster as values. The TCR matrix was then filtered for shared TCRs (TCR sequence is present in at least two clusters) and then transformed via log1p transformation (formula = ln(value + 1)) and values were clipped at 2 (any value greater than 2 was set to 2). Both TCRs and Leiden clusters were subject to hierarchal clustering (scanpy.tl.dendrogram) with the method set to “ward.” TCR clusters were determined utilizing the fcluster function from scipy (Virtanen et al., 2020) with the criterion set to “distance” and the distance set to 800 for CD8+ T cells and 175 for CD4+ T cells.
Additional Resources
All data has been integrated for further visualization and analysis in ISB COVID-19 data explorer at https://atlas.fredhutch.org/isb/covid/.
Acknowledgments
We are grateful to all participants in this study and to the medical teams at Swedish Medical Center for their support. We thank the Northwest Genomic Center for help with sequencing services, Allison Kudla for the graphical visualization, and Gretchen Krenn for the help with building the data visualization portal. We appreciate the insightful discussion from Prof. David Baltimore, Prof. Ilya Shmulevich and the ISB COVID-19 Study Group: Inyoul Lee, Kay Chinn, Scott Bloom, Guenther Kahlert, David Baxter, Zac Simon, Matt Idso, Rachel Calder, William Chour, Alphonsus Ng, Jimmi Hopkins, John Heath, Jingyi Xie, Jessica Yee, Kari Tetrault, Lee Rowen, Rachel Liu, Rongyu Zhang, Shannon Fallen, Simran Sidhu, Yue Lu, David Gibbs, Michael Strasser, and Gracie Baumgartner; the Swedish COVID-19 Research Steering Committee: John Pauk, Bill Berrington, Michael Bolton, Mary Micikas, Sonam Nyatsatsang, Cynthia Maree, Shane O’Mahony, Kelly Sweerus, Anne Lipke, George Pappas, Mark Sullivan, Karen Koo, Chris Dale, George Lopez, Naomi Diggs, Hank Kaplan, Krish Patel, Livia Hegerova, Vanessa Dunleavy, Phil Gold, John Pagel, Joshua Mark, Doug Kieper, Jim Scanlan, Evonne Lackey, Jodie Davila, Justin Rueda, Julie Wallick, Heather Algren, Jennifer Hansberry, and Elizabeth Wako; and the ISB-Swedish COVID-19 Biobanking Unit: Rick Edmark, John Heath, Simran Sidhu, Jessica Yee, Cara McCoy, Jeremy Johnson, Renee Duprel, Audri Hubbard, Theresa Davis, Julie Thatcher, Zuraya Aziz, Thea Swanson, Yong Zhou, Lesley Jones, Sarah Li, Paula Manner, Andrea Drouhard, Stephanie Johnson, Julia Karr, Clementine Chalal, Mattie Sader, Margo Badman, Allison Everett, Adel Islam, Jodie Davila, and Julie Wallick. We thank Amazon Web Services for their support through cloud computing credits provided by the AWS Diagnostic Development Initiative (DDI). We acknowledge funding support from the Wilke Family Foundation (J.R.H.), the Murdock Trust (J.R.H.), Gilead Sciences (J.R.H.), the Swedish Medical Center Foundation (J.D.G.), the Parker Institute for Cancer Immunotherapy (J.R.H., M.M.D., P.D.G., L.L.L., and J.A.B.), Merck, and the Biomedical Advanced Research and Development Authority (HHSO10201600031C to J.R.H.). K.W. was funded by DOD (W911NF-17-2-0086), NIH (R01 DA040395 and UG3TR002884). R.R. was funded by National Center For Advancing Translational Sciences of the NIH (OT2 TR003443). R.G. was funded by the NIH Human Immunology Project Consortium (U19AI128914) and the Vaccine and Immunology Statistical Center (Bill and Melinda Gates Foundation OPP1032317). Further funding by NIH (AI068129 to L.L.L. and R21 AI138258 to N.S.).
Author Contributions
Conceptualization, Y.S., J.D.G., and J.R.H.; Resources, J.D.G., and J.R.H.; Methodology, Y.S., D.C., J.S., and J.R.H.; Investigation, Y.S., D.C., D.Y., C.L., J.C., C.L.D., V.V., K.S., P.T., V.R.D., P.B., G.Q., B.S., S.A.K., C.R., J.J.H., R.Z., J.X., A.X., J.L., S.D., A.R., J.Z., K.M., R.E., S Hong., L.J., Y.Z., L.R., R.L., S. L., R.R., S.M., C.R.D., J.A.W., H.A.A., Z.A.M., A.T.M., W.W., N.D.P., S Huang., N.S., K.W., J.E.H., L.H., A.A., J.A.B., L.L.L., P.D.G., R.G., M.M.D., J.D.G., and J.R.H.; Formal Analysis, Y.S., D.C., D.Y., C.L., C.L.D., V.V., V.R.D., J.J.H., J.E., A.T.M., R.G., and J.R.H.; Writing – Original Draft, Y.S., D.C., D.Y., and J.R.H.; Writing – Review & Editing, Y.S., D.C., C.L., D.Y., J.C., C.L.D., V.V., K.S., P.T., V.R.D., P. B., G.Q., B.S., S.A.K., C.R., A.X., A.R., J.Z., A.T.M., W.W., N.D.P., S Huang, K.W., N.S., J.J.H., L.H., A.A., J.A.B., L.L.L., P.D.G., R.G., M.M.D., J.D.G., and J.R.H.
Declaration of Interests
J.R.H. is founder and board member of Isoplexis and PACT Pharma. M.M.D. is a member of the Scientific Advisory Board of PACT Pharma. J.A.B. is a member of the Scientific Advisory Boards of Arcus, Celsius, and VIR. J.A.B. is a member of the Board of Directors of Rheos and Provention. J.A.B. has recently joined Sonoma Biotherapeutics as President and CEO. Sonoma Biotherapeutics is involved in developing novel Treg-based cell therapies for the treatment of autoimmune diseases. R.G. has received consulting income from Juno Therapeutics, Takeda, Infotech Soft, Celgene, Merck and has received research support from Janssen Pharmaceuticals and Juno Therapeutics, and declares ownership in CellSpace Biosciences. P.D.G is on the Scientific Advisory Board of Celsius, Earli, Elpiscience, Immunoscape, Rapt, and Nextech, was a scientific founder of Juno Therapeutics, and receives research support from Lonza. J.D.G. declared contracted research with Gilead, Lilly, and Regeneron. The remaining authors declare no competing interests.
Published:October 28, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.10.037.
Contributor Information
Jason D. Goldman, Email: jason.goldman@swedish.org.
James R. Heath, Email: jim.heath@isbscience.org.
Supplemental Information
Human Subject Details, Plasma Proteomic and Metabolomic Datasets and Analysis, and CITE-Seq Antibodies, Related to Figures 1 and S1
CD8+ T Cell-Related Analysis, Related to Figures 2 and S2
CD4+ T Cell-Related Analysis, Related to Figures 3 and S3
Monocytes-Related Analysis, Related to Figures 5 and S4
NK Cell-Related Analysis, Related to Figures 6 and S5
Integrated Immune Response Gene Module Analysis, Related to Figures 7, S6, and S7
References
- Abel B., Tameris M., Mansoor N., Gelderbloem S., Hughes J., Abrahams D., Makhethe L., Erasmus M., de Kock M., van der Merwe L. The novel tuberculosis vaccine, AERAS-402, induces robust and polyfunctional CD4+ and CD8+ T cells in adults. Am. J. Respir. Crit. Care Med. 2010;181:1407–1417. doi: 10.1164/rccm.200910-1484OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abitorabi M.A., Mackay C.R., Jerome E.H., Osorio O., Butcher E.C., Erle D.J. Differential expression of homing molecules on recirculating lymphocytes from sheep gut, peripheral, and lung lymph. J. Immunol. 1996;156:3111–3117. [PubMed] [Google Scholar]
- Adler L.N., Jiang W., Bhamidipati K., Millican M., Macaubas C., Hung S.C., Mellins E.D. The Other Function: Class II-Restricted Antigen Presentation by B Cells. Front. Immunol. 2017;8:319. doi: 10.3389/fimmu.2017.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agmon N., Alhassid Y., Levine R.D. An algorithm for finding the distribution of maximal entropy. J. Comp. Physiol. 1979;30:250–258. [Google Scholar]
- Agresta L., Hoebe K.H.N., Janssen E.M. The Emerging Role of CD244 Signaling in Immune Cells of the Tumor Microenvironment. Front. Immunol. 2018;9:2809. doi: 10.3389/fimmu.2018.02809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqahtani S.A., Schattenberg J.M. Liver injury in COVID-19: The current evidence. United European Gastroenterol. J. 2020;8:509–519. doi: 10.1177/2050640620924157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amezquita R.A., Lun A.T.L., Becht E., Carey V.J., Carpp L.N., Geistlinger L., Marini F., Rue-Albrecht K., Risso D., Soneson C. Orchestrating single-cell analysis with Bioconductor. Nat. Methods. 2020;17:137–145. doi: 10.1038/s41592-019-0654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A.C., Joller N., Kuchroo V.K. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity. 2016;44:989–1004. doi: 10.1016/j.immuni.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arens R., Baars P.A., Jak M., Tesselaar K., van der Valk M., van Oers M.H.J., van Lier R.A.W. Cutting Edge: CD95 Maintains Effector T Cell Homeostasis in Chronic Immune Activation. J. Immunol. 2005;174:5915–5920. doi: 10.4049/jimmunol.174.10.5915. [DOI] [PubMed] [Google Scholar]
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- Bengsch B., Ohtani T., Khan O., Setty M., Manne S., O’Brien S., Gherardini P.F., Herati R.S., Huang A.C., Chang K.-M. Epigenomic-Guided Mass Cytometry Profiling Reveals Disease-Specific Features of Exhausted CD8 T Cells. Immunity. 2018;48:1029–1045. doi: 10.1016/j.immuni.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- Berard M., Tough D.F. Qualitative differences between naïve and memory T cells. Immunology. 2002;106:127–138. doi: 10.1046/j.1365-2567.2002.01447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrego F., Robertson M.J., Ritz J., Peña J., Solana R. CD69 is a stimulatory receptor for natural killer cell and its cytotoxic effect is blocked by CD94 inhibitory receptor. Immunology. 1999;97:159–165. doi: 10.1046/j.1365-2567.1999.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J.J., Murphy K.E., Kunkel E.J., Brightling C.E., Soler D., Shen Z., Boisvert J., Greenberg H.B., Vierra M.A., Goodman S.B. CCR7 Expression and Memory T Cell Diversity in Humans. J. Immunol. 2001;166:877–884. doi: 10.4049/jimmunol.166.2.877. [DOI] [PubMed] [Google Scholar]
- Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso A., Licenziati S., Corulli M., Canaris A.D., De Francesco M.A., Fiorentini S., Peroni L., Fallacara F., Dima F., Balsari A., Turano A. Flow cytometric analysis of activation markers on stimulated T cells and their correlation with cell proliferation. Cytometry. 1997;27:71–76. doi: 10.1002/(sici)1097-0320(19970101)27:1<71::aid-cyto9>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle D.M., Kim-Schulze S., Huang H.-H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentini S., Licenziati S., Alessandri G., Castelli F., Caligaris S., Bonafede M., Grassi M., Garrafa E., Balsari A., Turano A. CD11b Expression Identifies CD8+CD28+ T Lymphocytes with Phenotype and Function of Both Naive/Memory and Effector Cells. J. Immunol. 2001;166:900–907. doi: 10.4049/jimmunol.166.2.900. [DOI] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.-E., Katsaounou P. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020;27:992–1000. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullicksrud J.A., Li F., Xing S., Zeng Z., Peng W., Badovinac V.P., Harty J.T., Xue H.-H. Differential Requirements for Tcf1 Long Isoforms in CD8+ and CD4+ T Cell Responses to Acute Viral Infection. J. Immunol. 2017;199:911–919. doi: 10.4049/jimmunol.1700595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanidziar D., Bittner E.A. Hypotension, Systemic Inflammatory Response Syndrome, and COVID-19: A Clinical Conundrum. Anesth. Analg. 2020;131:e175–e176. doi: 10.1213/ANE.0000000000005062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneken M., Dörner T., Burmester G.-R., Berek C. Differential expression of chemokine receptors on peripheral blood B cells from patients with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Res. Ther. 2005;7:R1001–R1013. doi: 10.1186/ar1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndler-Brandstetter D., Ishigame H., Shinnakasu R., Plajer V., Stecher C., Zhao J., Lietzenmayer M., Kroehling L., Takumi A., Kometani K. KLRG1+ Effector CD8+ T Cells Lose KLRG1, Differentiate into All Memory T Cell Lineages, and Convey Enhanced Protective Immunity. Immunity. 2018;48:716–729. doi: 10.1016/j.immuni.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo L.G., Einecke G., Allanach K., Halloran P.F. The transcriptome of human cytotoxic T cells: similarities and disparities among allostimulated CD4(+) CTL, CD8(+) CTL and NK cells. Am. J. Transplant. 2008;8:627–636. doi: 10.1111/j.1600-6143.2007.02128.x. [DOI] [PubMed] [Google Scholar]
- Huang A.C., Postow M.A., Orlowski R.J., Mick R., Bengsch B., Manne S., Xu W., Harmon S., Giles J.R., Wenz B. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545:60–65. doi: 10.1038/nature22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudspeth K., Wang S., Wang J., Rahman S., Smith M.A., Casey K.A., Manna Z., Sanjuan M., Kolbeck R., Hasni S., Autoimmunity Molecular Team Natural killer cell expression of Ki67 is associated with elevated serum IL-15, disease activity and nephritis in systemic lupus erythematosus. Clin. Exp. Immunol. 2019;196:226–236. doi: 10.1111/cei.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huster K.M., Busch V., Schiemann M., Linkemann K., Kerksiek K.M., Wagner H., Busch D.H. Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc. Natl. Acad. Sci. USA. 2004;101:5610–5615. doi: 10.1073/pnas.0308054101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iype E., Gulati S. Understanding the asymmetric spread and case fatality rate (CFR) for COVID-19 among countries. medRxiv. 2020 doi: 10.1101/2020.04.21.20073791. [DOI] [Google Scholar]
- Juno J.A., van Bockel D., Kent S.J., Kelleher A.D., Zaunders J.J., Munier C.M.L. Cytotoxic CD4 T Cells-Friend or Foe during Viral Infection? Front. Immunol. 2017;8:19. doi: 10.3389/fimmu.2017.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko N., Kuo H.-H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J. Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell. 2020;183:143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kared H., Martelli S., Ng T.P., Pender S.L.F., Larbi A. CD57 in human natural killer cells and T-lymphocytes. Cancer Immunol. Immunother. 2016;65:441–452. doi: 10.1007/s00262-016-1803-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katikaneni D.S., Jin L. B cell MHC class II signaling: A story of life and death. Hum. Immunol. 2019;80:37–43. doi: 10.1016/j.humimm.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.R., Mathew S.O., Mathew P.A. Blimp-1/PRDM1 regulates the transcription of human CS1 (SLAMF7) gene in NK and B cells. Immunobiology. 2016;221:31–39. doi: 10.1016/j.imbio.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine R.D. Information Theory Approach to Molecular Reaction Dynamics. Annu. Rev. Phys. Chem. 1978;29:59–92. [Google Scholar]
- Levine R.D. Cambridge University Press; 2005. Molecular Reaction Dynamics. [Google Scholar]
- Levine R.D., Bernstein R.B. Energy disposal and energy consumption in elementary chemical reactions. Information theoretic approach. Acc. Chem. Res. 1974;7:393–400. [Google Scholar]
- Li Z., Li D., Tsun A., Li B. FOXP3+ regulatory T cells and their functional regulation. Cell. Mol. Immunol. 2015;12:558–565. doi: 10.1038/cmi.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Borrego F., Nagata S., Tolnay M. Fc Receptor-like 5 Expression Distinguishes Two Distinct Subsets of Human Circulating Tissue-like Memory B Cells. J. Immunol. 2016;196:4064–4074. doi: 10.4049/jimmunol.1501027. [DOI] [PubMed] [Google Scholar]
- Li Q., Ding X., Xia G., Chen H.-G., Chen F., Geng Z., Xu L., Lei S., Pan A., Wang L., Wang Z. Eosinopenia and elevated C-reactive protein facilitate triage of COVID-19 patients in fever clinic: A retrospective case-control study. EClinicalMedicine. 2020;23:100375. doi: 10.1016/j.eclinm.2020.100375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima J.F., Oliveira L.M.S., Pereira N.Z., Duarte A.J.S., Sato M.N. Polyfunctional natural killer cells with a low activation profile in response to Toll-like receptor 3 activation in HIV-1-exposed seronegative subjects. Sci. Rep. 2017;7:524. doi: 10.1038/s41598-017-00637-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Richard K., Wiggins M., Zhu X., Conrad D.H., Song W. CD23 can negatively regulate B-cell receptor signaling. Sci. Rep. 2016;6:25629. doi: 10.1038/srep25629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Xue Q., Eisele M.R., Sulistijo E.S., Brower K., Han L., Amir A.D., Pe’er D., Miller-Jensen K., Fan R. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc. Natl. Acad. Sci. USA. 2015;112:E607–E615. doi: 10.1073/pnas.1416756112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., Yale IMPACT Team Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Cheung A.F., Chodon T., Koya R.C., Wu Z., Ng C., Avramis E., Cochran A.J., Witte O.N., Baltimore D. Multifunctional T-cell analyses to study response and progression in adoptive cell transfer immunotherapy. Cancer Discov. 2013;3:418–429. doi: 10.1158/2159-8290.CD-12-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maawy A.A., Ito F. Future of immune checkpoint inhibitors. In: Ito F., Ernstoff M., editors. Immune Checkpoint Inhibitors in Cancer. Elsevier; 2019. pp. 227–243. [Google Scholar]
- Mangalam A., Rodriguez M., David C. Role of MHC class II expressing CD4+ T cells in proteolipid protein(91-110)-induced EAE in HLA-DR3 transgenic mice. Eur. J. Immunol. 2006;36:3356–3370. doi: 10.1002/eji.200636217. [DOI] [PubMed] [Google Scholar]
- Manor O., Zubair N., Conomos M.P., Xu X., Rohwer J.E., Krafft C.E., Lovejoy J.C., Magis A.T. A Multi-omic Association Study of Trimethylamine N-Oxide. Cell Rep. 2018;24:935–946. doi: 10.1016/j.celrep.2018.06.096. [DOI] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maucourant C., Filipovic I., Ponzetta A., Aleman S., Cornillet M., Hertwig L., Strunz B., Lentini A., Reinius B., Brownlie D. Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 2020;5:eabd6832. doi: 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L., Healy J., Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv. 2018 https://arxiv.org/abs/1802.03426 arXiv:1802.03426. [Google Scholar]
- Michaud A., Dardari R., Charrier E., Cordeiro P., Herblot S., Duval M. IL-7 enhances survival of human CD56bright NK cells. J. Immunother. 2010;33:382–390. doi: 10.1097/CJI.0b013e3181cd872d. [DOI] [PubMed] [Google Scholar]
- Miller J.S., Lanier L.L. Natural Killer Cells in Cancer Immunotherapy. Annu. Rev. Cancer Biol. 2019;3:77–103. [Google Scholar]
- Miller I., Min M., Yang C., Tian C., Gookin S., Carter D., Spencer S.L. Ki67 is a Graded Rather than a Binary Marker of Proliferation versus Quiescence. Cell Rep. 2018;24:1105–1112. doi: 10.1016/j.celrep.2018.06.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W., Wang R., Green D.R., Tessarollo L., Casellas R. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley M.A., Kroy D.C., Odorizzi P.M., Johnnidis J.B., Dolfi D.V., Barnett B.E., Bikoff E.K., Robertson E.J., Lauer G.M., Reiner S.L., Wherry E.J. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 2012;338:1220–1225. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen M., Janssen O. Pro- and anti-apoptotic CD95 signaling in T cells. Cell Commun. Signal. 2011;9:7. doi: 10.1186/1478-811X-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- Polański K., Young M.D., Miao Z., Meyer K.B., Teichmann S.A., Park J.-E. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics. 2020;36:964–965. doi: 10.1093/bioinformatics/btz625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer D., Lee A.Y.S., Bannan J., Fromm P., Kara E.E., Comerford I., McColl S., Wiede F., Mielenz D., Körner H. Early CCR6 expression on B cells modulates germinal centre kinetics and efficient antibody responses. Immunol. Cell Biol. 2017;95:33–41. doi: 10.1038/icb.2016.68. [DOI] [PubMed] [Google Scholar]
- Remacle F., Kravchenko-Balasha N., Levitzki A., Levine R.D. Information-theoretic analysis of phenotype changes in early stages of carcinogenesis. Proc. Natl. Acad. Sci. USA. 2010;107:10324–10329. doi: 10.1073/pnas.1005283107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley J.L. PD-1 signaling in primary T cells. Immunol. Rev. 2009;229:114–125. doi: 10.1111/j.1600-065X.2009.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L. Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell. 2020;182:1419–1440. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell. 2020;182:59–72. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichijo S., Azuma K., Komatsu N., Ito M., Maeda Y., Ishihara Y., Itoh K. Two proliferation-related proteins, TYMS and PGK1, could be new cytotoxic T lymphocyte-directed tumor-associated antigens of HLA-A2+ colon cancer. Clin. Cancer Res. 2004;10:5828–5836. doi: 10.1158/1078-0432.CCR-04-0350. [DOI] [PubMed] [Google Scholar]
- Shuai Z., Leung M.W.Y., He X., Zhang W., Yang G., Leung P.S.C., Eric Gershwin M. Adaptive immunity in the liver. Cell. Mol. Immunol. 2016;13:354–368. doi: 10.1038/cmi.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvin A., Chapuis N., Dunsmore G., Goubet A.-G., Dubuisson A., Derosa L., Almire C., Hénon C., Kosmider O., Droin N. Elevated Calprotectin and Abnormal Myeloid Cell Subsets Discriminate Severe from Mild COVID-19. Cell. 2020;182:1401–1418. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.-W., Zhang C., Fan X., Meng F.-P., Xu Z., Xia P., Cao W.-J., Yang T., Dai X.-P., Wang S.-Y. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 2020;11:3410. doi: 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone M.R., O’Neill A., Lovering R.M., Strong J., Resneck W.G., Reed P.W., Toivola D.M., Ursitti J.A., Omary M.B., Bloch R.J. Absence of keratin 19 in mice causes skeletal myopathy with mitochondrial and sarcolemmal reorganization. J. Cell Sci. 2007;120:3999–4008. doi: 10.1242/jcs.009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm G., Szabo T., Fotakis G., Haider M., Rieder D., Trajanoski Z., Finotello F. Scirpy: A Scanpy extension for analyzing single-cell T-cell receptor sequencing data. bioRxiv. 2020 doi: 10.1101/2020.04.10.035865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Bintz M., Yang Y., Robert L., Ng A.H.C., Liu V., Ribas A., Heath J.R., Wei W. Phenotypic heterogeneity and evolution of melanoma cells associated with targeted therapy resistance. PLoS Comput. Biol. 2019;15:e1007034. doi: 10.1371/journal.pcbi.1007034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Lu X., Li G., Liu C., Kong Y., Lee J.W., Ng R., Wong S., Robert L., Warden C. Kinetic Inference Resolves Epigenetic Mechanism of Drug Resistance in Melanoma. bioRxiv. 2019 doi: 10.1101/724740. [DOI] [Google Scholar]
- Su Y., Ko M.E., Cheng H., Zhu R., Xue M., Wang J., Lee J.W., Frankiw L., Xu A., Wong S. Multi-omic single-cell snapshots reveal multiple independent trajectories to drug tolerance in a melanoma cell line. Nat. Commun. 2020;11:2345. doi: 10.1038/s41467-020-15956-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M.A., Lee W.T., Sanders V.M. IFN-gamma production by Th1 cells generated from naive CD4+ T cells exposed to norepinephrine. J. Immunol. 2001;166:232–240. doi: 10.4049/jimmunol.166.1.232. [DOI] [PubMed] [Google Scholar]
- Szabo P.A., Levitin H.M., Miron M., Snyder M.E., Senda T., Yuan J., Cheng Y.L., Bush E.C., Dogra P., Thapa P. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun. 2019;10:4706. doi: 10.1038/s41467-019-12464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T., Stefanoni D., Reisz J.A., Nemkov T., Bertolone L., Francis R.O., Hudson K.E., Zimring J.C., Hansen K.C., Hod E.A. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5:e140327. doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipping M.E., Bishop C.M. Probabilistic Principal Component Analysis. J. R. Stat. Soc. Ser. B. Stat. Methodol. 1999;61:611–622. [Google Scholar]
- Traag V., Waltman L., van Eck N.J. From Louvain to Leiden: guaranteeing well-connected communities. arXiv. 2018 doi: 10.1038/s41598-019-41695-z. https://arxiv.org/abs/1810.08473 arXiv:1810.08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uto T., Fukaya T., Takagi H., Arimura K., Nakamura T., Kojima N., Malissen B., Sato K. Clec4A4 is a regulatory receptor for dendritic cells that impairs inflammation and T-cell immunity. Nat. Commun. 2016;7:11273. doi: 10.1038/ncomms11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo A.N., Brandes J.C., Weyand C.M., Goronzy J.J. Modulation of CD28 Expression: Distinct Regulatory Pathways During Activation and Replicative Senescence. J. Immunol. 1999;162:6572–6579. [PubMed] [Google Scholar]
- Van de Sande B., Flerin C., Davie K., De Waegeneer M., Hulselmans G., Aibar S., Seurinck R., Saelens W., Cannoodt R., Rouchon Q. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 2020;15:2247–2276. doi: 10.1038/s41596-020-0336-2. [DOI] [PubMed] [Google Scholar]
- van Dijk D., Sharma R., Nainys J., Yim K., Kathail P., Carr A.J., Burdziak C., Moon K.R., Chaffer C.L., Pattabiraman D. Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell. 2018;174:716–729. doi: 10.1016/j.cell.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., SciPy 1.0 Contributors SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E., Tomasello E., Baratin M., Walzer T., Ugolini S. Functions of natural killer cells. Nat. Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu H., McKenzie G., Witting P.K., Stasch J.-P., Hahn M., Changsirivathanathamrong D., Wu B.J., Ball H.J., Thomas S.R. Kynurenine is an endothelium-derived relaxing factor produced during inflammation. Nat. Med. 2010;16:279–285. doi: 10.1038/nm.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry E.J. T cell exhaustion. Nat. Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- Wherry E.J., Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willinger T., Freeman T., Herbert M., Hasegawa H., McMichael A.J., Callan M.F.C. Human naive CD8 T cells down-regulate expression of the WNT pathway transcription factors lymphoid enhancer binding factor 1 and transcription factor 7 (T cell factor-1) following antigen encounter in vitro and in vivo. J. Immunol. 2006;176:1439–1446. doi: 10.4049/jimmunol.176.3.1439. [DOI] [PubMed] [Google Scholar]
- Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock S.L., Lopez R., Klein A.M. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst. 2019;8:281–291. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; 2020. COVID-19 Therapeutic Trial Synopsis. [Google Scholar]
- Yoon S.R., Kim T.-D., Choi I. Understanding of molecular mechanisms in natural killer cell therapy. Exp. Mol. Med. 2015;47:e141. doi: 10.1038/emm.2014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadran S., Arumugam R., Herschman H., Phelps M.E., Levine R.D. Surprisal analysis characterizes the free energy time course of cancer cells undergoing epithelial-to-mesenchymal transition. Proc. Natl. Acad. Sci. USA. 2014;111:13235–13240. doi: 10.1073/pnas.1414714111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-Y., Wang X.-M., Xing X., Xu Z., Zhang C., Song J.-W., Fan X., Xia P., Fu J.-L., Wang S.-Y. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21:1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Kaiser A., Ng C., Karcher R., McConnell T., Paczkowski P., Fernandez C., Zhang M., Mackay S., Tsuji M. CD8+ T-cell mediated anti-malaria protection induced by malaria vaccines; assessment of hepatic CD8+ T cells by SCBC assay. Hum. Vaccin. Immunother. 2017;13:1625–1629. doi: 10.1080/21645515.2017.1304333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Human Subject Details, Plasma Proteomic and Metabolomic Datasets and Analysis, and CITE-Seq Antibodies, Related to Figures 1 and S1
CD8+ T Cell-Related Analysis, Related to Figures 2 and S2
CD4+ T Cell-Related Analysis, Related to Figures 3 and S3
Monocytes-Related Analysis, Related to Figures 5 and S4
NK Cell-Related Analysis, Related to Figures 6 and S5
Integrated Immune Response Gene Module Analysis, Related to Figures 7, S6, and S7
Data Availability Statement
All blood scRNA-seq data used in this study can be accessed by Array Express under the accession number: E-MTAB-9357. Additional Supplemental Items, including the metabolomic and proteomic datasets, are available from Mendeley Data at http://dx.doi.org/10.17632/tzydswhhb5.5.