Abstract
Various diagnostic tests utilizing different principles are currently under development for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, these tests can occasionally produce discrepant results, causing confusion in their interpretation. Here, we evaluated the performance and features of three diagnostic assays: quantitative reverse transcription polymerase chain reaction (RT-qPCR), FilmArray Respiratory Panel (RP) v2.1, and the LUMIPULSE antigen test. Twenty-seven serial nasopharyngeal swabs were collected from a prolonged viral shedding patient who had been hospitalized for 51 days. We examined the SARS-CoV-2 detection rates of the three tests. The overall agreement rate was 81% between RT-qPCR and FilmArray RP v2.1, 63% between the antigen test and FilmArray RP v2.1, and 59% between the antigen test and RT-qPCR. We obtained concordant results in samples with high viral loads (low threshold cycle values) by all three tests. RT-qPCR and FilmArray RP v2.1 accurately detected SARS-CoV-2 at the early to intermediate phases of infection, but the results varied at the late phase. The antigen test also produced a positive result at the early phase but varied at the intermediate phase and consistently produced negative results at late phase of infection. These results demonstrated FilmArray RP v2.1 could detect SARS-CoV-2 with accuracy comparable to RT-qPCR. Further, there were discrepant results using different types of diagnostic tests during the clinical course of prolonged viral shedding patient. We provided insights into how to utilize different types of kits to assess and manage SARS-CoV-2 infections.
Keywords: COVID-19, SARS-CoV-2, RT-qPCR, FilmArray, Antigen
The ongoing outbreak of the emergent severe acute respiratory coronavirus 2 (SARS-CoV-2) started in December 2019 in Wuhan, China. The total number of infected patients currently stands at 40 million, with 1100,000 fatalities resulting from coronavirus disease 2019 (COVID-19) [1]. Diagnostic test kits used in clinical settings for SARS-CoV-2 typically use either nucleic acid amplification to detect viral RNA or antigen assays to detect protein targets (i.e. spike and nucleocapsid).
A number of easy-to-use in vitro diagnostic tests for SARS-CoV-2 are currently under development. FilmArray Respiratory Panel (RP) is a multiplex polymerase chain reaction (PCR) kit that detects 20 pathogens (17 viruses and three bacteria) related to respiratory disease in a single test. Recently, BioMérieux launched a new version of FilmArray RP (version 2.1; also known as BioFire RP 2.1) [2] which also targets SARS-CoV-2. This assay has been approved for emergency use by the Food and Drug Administration in the US. Because FilmArray RP v2.1 can detect the other respiratory pathogens including influenza virus, simultaneous detection of viruses will have more benefits in winter season.
While RT-qPCR remains the gold standard method and is widely conducted for detecting SARS-CoV-2, many laboratories are using diagnostic tests utilizing different principles. As these tests have unique nucleic acid sequence and antigen/epitope targets, they also differ in sensitivity and specificity. Such variability between diagnostic tests could cause confusion amongst clinicians and healthcare workers as to the appropriate test to select and the interpretation of results.
A 86-year-old woman with a medical history of chronic atrial fibrillation was transferred to the other hospital due to subdural hematoma. She had fever (37.6 °C), cough and poor oxygenation (SpO2: 90%). SARS-CoV-2 was detected in nasopharyngeal swabs by RT-qPCR and the diagnosis of COVID-19 infection was made. Computed tomography of the chest showed pneumonia with diffuse glass shadow in the bilateral lung fields. The infected patient with moderate symptom was transferred to our hospital and treated with camostat mesilate. Duration of hospitalization was 51 days.
We collected 27 serial nasopharyngeal swabs from the patient with persistent viral shedding during hospitalization. We analyzed these swab samples using quantitative reverse transcription PCR (RT-qPCR), FilmArray RP v2.1 (BioMérieux, Marcy-l'Etoile, France), and the LUMIPULSE antigen test (Fujirebio, Tokyo, Japan) to evaluate the performance of these assays at detecting SARS-CoV-2.
Total nucleic acid was automatically isolated from viral transport media [3,4], and subjected to RT-qPCR with the primers and probe targeting the SARS-CoV-2 nucleocapsid gene [5,6]. Threshold cycle (Ct) values were assigned to each PCR reaction, and the amplification curve was visually assessed. We also conducted multiplex PCR targeting using FilmArray RP v2.1 [7]. FilmArray RP v2.1 includes two primer sets targeting SARS-CoV-2 membrane and spike genes. The antigen level was determined with LUMIPULSE SARS-CoV-2 Ag kit on LUMIPULSE G600II [8].
Detection rates of SARS-CoV-2 were 70% (19/27), 67% (18/27), and 30% (8/27) by RT-qPCR, FilmArray RP v2.1, and the antigen test, respectively. These results suggested that nucleic acid amplification has higher sensitivity than antigen detection. FilmArray RP v2.1 did not detect other viruses or bacteria in the samples, indicating that the patient was not co-infected with other pathogens.
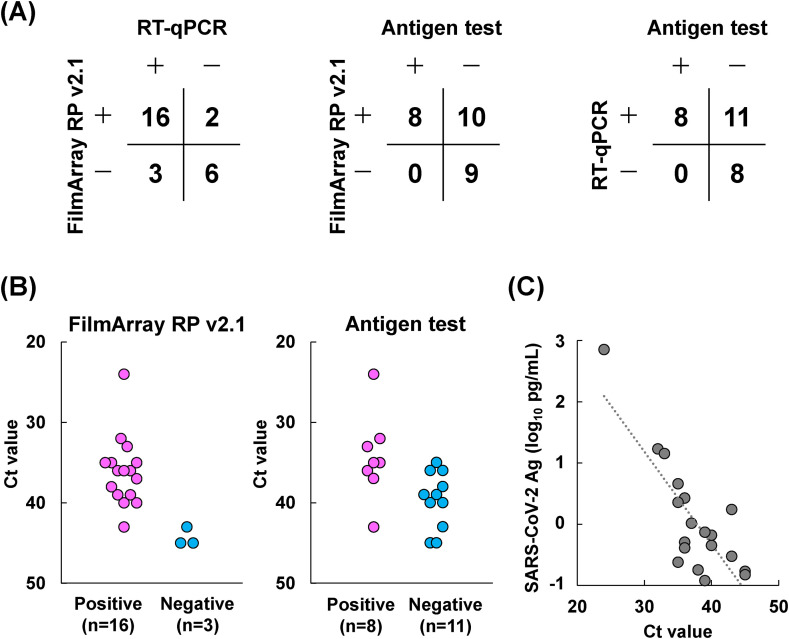
Overall agreement rates of SARS-CoV-2 detection were 81% (22/27) between RT-qPCR and FilmArray RP v2.1, 63% (17/27) between the antigen test and FilmArray RP v2.1, and 59% (16/27) between the antigen test and RT-qPCR (Fig. 1 A); the most concordant results were observed between RT-qPCR and FilmArray RP v2.1. All positive samples (n = 8) by antigen detection were also positive by both FilmArray RP v2.1 and RT-qPCR.
Fig. 1.
Comparison of the results of RT-qPCR, FilmArray RP v2.1 and the LUMIPULSE antigen test. (A) Overall agreement of three assays. Twenty-seven serial nasopharyngeal swabs were collected from a prolonged viral shedding inpatient. Positive (+) and negative (-) results were compared among three assays. (B) Threshold cycle (Ct) values of RT-qPCR in positive and negative results determined by FilmArray RP v2.1 and the LUMIPULSE antigen test. Nineteen samples were determined as positive by RT-qPCR. The relationship between Ct values of the 19 samples and results from (left panel) FilmArray RP v2.1 and (right panel) the LUMIPULSE antigen test were evaluated. (C) Correlation between Ct values and SARS-CoV-2 antigen (Ag) levels. A correlation (R2 = 0.663) was observed between Ct values and Ag level (log10 pg/mL).
We further examined the relationship between viral load and detection sensitivity. To this end, we analyzed the Ct values of RT-qPCR-positive samples (n = 19), with lower Ct values indicating higher viral loads (and vice versa) in the nasopharyngeal samples.
Average Ct values were 36.1 (range: 24–43) and 44.3 (range: 43–45) for positive (n = 16) and negative (n = 3) samples determined by FilmArray RP v2.1, respectively (Fig. 1B). These results indicated that FilmArray RP v2.1 did not detect SARS-CoV-2 in samples with extremely low viral RNA. A similar trend was observed for the antigen test, where average Ct values of 34.4 (range: 24–43) and 39.6 (range: 35–45) for positive (n = 8) and negative (n = 11) samples, respectively, were measured (Fig. 1B). The Ct value was correlated with the antigen level (R2 = 0.663) (Fig. 1C).
The patient in this study had a long viral shedding duration. We examined the timing of positive and negative results determined by the three assays. In the early phase of infection (days 1–10 after admission), all five samples were determined as positive by the three tests (Table 1 ). During the intermediate phase (day 14–26), almost all samples (7/8 samples) were determined as positive by RT-qPCR and FilmArray RP v2.1, while the results fluctuated for the antigen test. In the late phase of infection (after 27 days), both RT-qPCR and FilmArray RP showed “positive to negative” or “negative to positive” fluctuations (Table 1), as we previously described [8]. The antigen test consistently returned negative results at the late phase of infection (Table 1).
Table 1.
Diagnostic results of 27 serial samples from a persistent viral shedding patient.
| Days from admission | RT-qPCR |
FilmArray RP v2.1 |
LUMIPULSE antigen testa |
||
|---|---|---|---|---|---|
| Judgement | Ct | Judgement | Judgement | pg/mL | |
| 1 | + | 24 | + | + | 714.23 |
| 3 | + | 32 | + | + | 17.01 |
| 5 | + | 33 | + | + | 14.30 |
| 7 | + | 43 | + | + | 1.74 |
| 10 | + | 35 | + | + | 4.59 |
| 14 | + | 36 | + | – | 0.51 |
| 16 | + | 40 | + | – | 0.45 |
| 18 | + | 36 | + | + | 2.68 |
| 21 | + | 40 | + | – | 0.66 |
| 22 | – | + | – | 0.55 | |
| 24 | + | 39 | + | – | 0.74 |
| 25 | + | 37 | + | + | 1.04 |
| 26 | + | 35 | + | + | 2.28 |
| 27 | + | 36 | + | – | 0.41 |
| 28 | – | – | – | 0.29 | |
| 29 | – | – | – | 0.38 | |
| 30 | + | 35 | + | – | 0.24 |
| 31 | – | – | – | 0.22 | |
| 32 | – | – | – | 0.18 | |
| 35 | + | 38 | + | – | 0.18 |
| 37 | – | – | – | 0.09 | |
| 38 | + | 43 | – | – | 0.30 |
| 39 | + | 45 | – | – | 0.17 |
| 42 | + | 45 | – | – | 0.15 |
| 43 | + | 39 | + | – | 0.12 |
| 44 | – | + | – | 0.06 | |
| 45 | – | – | – | 0.46 | |
+, positive; -, negative.
Combined with the results of the RT-qPCR test, antigen test determined as positive when antigen level ≥1.0 pg/mL.
RT-qPCR can quantitatively evaluate viral RNA levels with high sensitivity. Our results show that both RT-qPCR and FilmArray RP v2.1 can detect prolonged viral shedding [[9], [10], [11]]. Half of COVID-19 patients shed virus particles up to 25 days after the first positive PCR test [12]. Higher viral loads (lower Ct values) were observed in COVID-19 patients in the first week of the onset of symptoms and viable SARS-CoV-2 was cultured within 7–9 days of onset [9,11]. However, infectious SARS-CoV-2 are not always present in patients who have prolonged viral production and shedding [13,14]. As RT-qPCR and FilmArray RP v2.1 cannot distinguish between RNA of live viruses from that of non-infectious viruses, we should carefully interpret Ct values and viral load results, especially in the late phase of the infection period. We also consider that the contamination of PCR inhibitor reduces amplification efficiency, which may cause negative results in sample with low viral RNA.
In summary, the accuracy was comparable between RT-qPCR and FilmArray RP v2.1. Using different types of test kits, we encountered discrepancies between the results from a prolonged viral shedding patient depending on the viral load and the phase of infection. Although further studies are needed to clarify these results are observed in other patients, our results provide the strong and weak points of each test (Table 2 ). These features may help healthcare worker to properly interpret results. Combining diagnostic tests (i.e. PCR-based test and antigen test) could avoid false-positive and false-negative results, provide accurate results and be beneficial to better manage COVID-19 infected patients.
Table 2.
Features of RT-qPCR, FilmArray RP v2.1, and the LUMIPULSE antigen test.
| Assay | Principle | Strong points | Weak points |
|---|---|---|---|
| RT-qPCR | Detects SARS-CoV-2 RNA by PCR amplification |
|
|
| FilmArray RP v2.1 | Detects SARS-CoV-2 RNA by PCR amplification |
|
|
| LUMIPULSE antigen test | Detects SARS-CoV-2 N protein by chemiluminescent enzyme immunoassay |
|
|
Contributions
YH contributed to study design, data collection, data analysis and writing – review & editing. MM, MS, KA, YN, KH, and HS contributed to sample preparation and data collection, data analysis. MH and HM contributed to supervision. TT, YK and YM contributed to provide resources. MO contributed to supervision and writing – review & editing.
Ethical approval
The Institutional Review Board at Yamanashi Central Hospital, Japan, approved this study (Approval No. C2019-30 and C2020-9). Written informed consent was obtained from the patient to publish this case.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
This study was supported by a Grant-in-Aid for the Genome Research Project from Yamanashi Prefecture (to M.O. and Y.H.), the Japan Society for the Promotion of Science (JSPS) KAKENHI Early-Career Scientists JP18K16292 (to Y.H.), a Grant-in-Aid for Scientific Research (B) 20H03668 (to Y.H.), a Research Grant for Young Scholars (to Y.H.), the YASUDA Medical Foundation (to Y.H.), the Uehara Memorial Foundation (to Y.H.), and Medical Research Grants from the Takeda Science Foundation (to Y.H.).
We thank Shintaro Yagi, Satoshi Kojima, Hisashi Nojima, Masayasu Imaizumi, Yukie Ohtakagi, and Eri Inagaki (Fujirebio Inc.) for technical assistance and all of the medical and ancillary hospital staff and the patients for consenting to participate.
We thank Martin Cheung PhD, from Edanz Group (https://en-author-services.edanzgroup.com/ac) for editing a draft of this manuscript.
References
- 1.Organization W.H. Coronavirus disease (COVID-19) situation reports. https://wwwwhoint/emergencies/diseases/novel-coronavirus-2019/situation-reports
- 2.Creager H.M., Cabrera B., Schnaubelt A., Cox J.L., Cushman-Vokoun A.M., Shakir S.M., et al. Clinical evaluation of the BioFire® respiratory panel 2.1 and detection of SARS-CoV-2. J Clin Virol. 2020;129:104538. doi: 10.1016/j.jcv.2020.104538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K., et al. Pooling RT-qPCR testing for SARS-CoV-2 in 1000 individuals of healthy and infection-suspected patients. Sci Rep. 2020 doi: 10.1038/s41598-020-76043-z. [in press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirotsu Y., Maejima M., Nakajima M., Mochizuki H., Omata M. Environmental cleaning is effective for the eradication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in contaminated hospital rooms: a patient from the Diamond Princess cruise ship. Infect Control Hosp Epidemiol. 2020;41(9):1105–1106. doi: 10.1017/ice.2020.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirotsu Y., Mochizuki H., Omata M. Double-quencher probes improve detection sensitivity toward Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a reverse-transcription polymerase chain reaction (RT-PCR) assay. J Virol Methods. 2020;284:113926. doi: 10.1016/j.jviromet.2020.113926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., et al. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn J Infect Dis. 2020;73(4):304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- 7.Hirotsu Y., Maejima M., Shibusawa M., Amemiya K., Nagakubo Y., Hosaka K., et al. Analysis of Covid-19 and non-Covid-19 viruses, including influenza viruses, to determine the influence of intensive preventive measures in Japan. J Clin Virol. 2020;129:104543. doi: 10.1016/j.jcv.2020.104543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K., et al. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs including from 7 serially followed patients. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.08.029. S1201-9712(20):30658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 10.Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., et al. Positive RT-PCR test results in patients recovered from COVID-19. J Am Med Assoc. 2020;323(15):1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.COVID-19 Investigation Team Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med. 2020;26(6):861–868. doi: 10.1038/s41591-020-0877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ridgway J.P., Shah N.S., Robicsek A.A. Prolonged shedding of severe acute respiratory coronavirus virus 2 (SARS-CoV-2) RNA among patients with coronavirus disease 2019 (COVID-19) Infect Control Hosp Epidemiol. 2020;41(10):1235–1236. doi: 10.1017/ice.2020.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atkinson B., Petersen E. SARS-CoV-2 shedding and infectivity. Lancet. 2020;395(10233):1339–1340. doi: 10.1016/S0140-6736(20)30868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Widders A., Broom A., Broom J. SARS-CoV-2: the viral shedding vs infectivity dilemma. Infect Dis Health. 2020;25(3):210–215. doi: 10.1016/j.idh.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]