Abstract
Nisin, a food-grade antimicrobial peptide produced by lactic acid bacteria has been examined for its probable interaction with the human ACE2 (hACE2) receptor, the site where spike protein of SARS-CoV-2 binds. Among the eight nisin variants examined, nisin H, nisin Z, nisin U and nisin A showed a significant binding affinity towards hACE2, higher than that of the RBD (receptor binding domain) of the SARS-CoV-2 spike protein. The molecular interaction of nisin with hACE2 was investigated by homology modeling and docking studies. Further, binding efficiency of the most potent nisin H was evaluated through the interaction of hACE2:nisin H complex with RBD (receptor-binding domain) of SARS-CoV-2 and that of hACE2:RBD complex with nisin H. Here, nisin H acted as a potential competitor of RBD to access the hACE2 receptor. The study unravels for the first time that a globally used food preservative, nisin has the potential to bind to hACE2.
Keywords: Nisin, SARS-CoV-2, COVID-19, Human ACE2 receptor, Molecular docking, Therapeutics
1. Introduction
The ongoing global outbreak of COVID-19, a severe life-threatening infectious respiratory disease caused by a recently discovered severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has drastically affected human life with over eighteen millions of cases of infection globally (https://coronavirus.jhu.edu/map.html). Until now, no specific antiviral medication is available for COVID-19, but extensive efforts are underway worldwide. Although vaccines are thought to be the most powerful weapon to fight against virus invasion, it may take quite a long time to go from the lab to successful applications in humans. Considering the acute crisis of COVID-19 pandemic, there is an urgent need for developing effective antiviral therapeutics for the prevention and treatment of COVID-19. It is well accepted that the spike protein on the outer surface of SARS-CoV-2 is a crucial recognition factor for its attachment and entry to the host cells (Shang et al., 2020). The viral infection in humans is initiated by binding of RBD (receptor binding domain) of spike protein to human angiotensin-converting enzyme 2 (hACE2) receptor (Wang et al., 2020). Therefore, a therapeutic agent that blocks hACE2 might prevent the interaction of spike protein of SARS-CoV-2 and thereby could reduce the establishment of infection. Although small non-proteinaceous molecules are commonly preferred as therapeutics, they are not effective in blocking protein-protein interactions (PPIs) particularly, where a deep binding pocket may be missing at the interface (Arkin et al., 2014). On the contrary, peptides are more suitable for disrupting PPIs by specifically interacting with the interfaces. More importantly, small peptides have reduced immunogenicity (Sorolla et al., 2020). Hence, peptides are potentially the ideal candidates for application as novel therapeutics. The recently described peptides are all small, synthetic and costly, and have not produced promising results against SARS-CoV-2 (Du et al., 2005). The peptides recently designed computationally (Han and Král, 2020) against the SARS-CoV-2 has to be synthesized prior to practical application, hence such peptides are not natural and food-grade.
The present study attempts to investigate the ability of food-grade nisin A and its natural variants to block the interaction between hACE2 and the spike protein of SARS-CoV-2, a key step of COVID-19 disease initiation. Nisin, a pentacyclic antibacterial peptide with 34 residues, is produced by certain strains of food-grade Lactococcus lactis, widely used for cheese manufacturing (Fox and Wood, 1971; Lubelski et al., 2008; Juncioni et al., 2009). Nisin belongs to a group of cationic peptide antimicrobials collectively called Type A (I) lantibiotics (Smith and Hillman, 1016). It was first identified in fermented milk cultures and is now globally used as a natural and safe food preservative in a variety of food products around the world, such as processed cheese, dairy desserts, milk, fermented beverages, meat and canned foods (Hurst, 1981; Fons et al., 2009; Mitra et al., 2011). It has been approved by the European Union (E234), World Health Organization (WHO) as well as by the US Food and Drug Administration (FDA). Currently, nisin is licensed in over 50 countries (Shin et al., 2015). Because of the high safety profile over the past 40 years of usage and its strong antimicrobial action against a wide range of food spoilage and pathogenic bacteria, nisin has been extensively studied. It also has multiple applications in biomedicine including bacterial infections, cancer, oral diseases and other veterinary and research field (Shin et al., 2015). Since the discovery of nisin A, eight natural variants of nisin have been discovered which include nisin A, Z, F, Q, H, U, U2 and P (Garcia-gutierrez et al., 2020). Nisin Z producing organisms are very common in nature (Mitra et al., 2011; Vos et al., 1993). The structures of eight variants of nisin were analyzed in the present study. All nisin peptides were aligned to show their identity and modeled on SWISS-Model web server. hACE2 and RBD domain of 2019-CoV-2 were also modeled on the same platform to increase the acceptability of the structures. All the peptides and RBD were docked with hACE2 using HADDOCK server. The binding affinity of the peptides was examined by docking analysis based on Z-score, binding affinity and buried surface area. Structurally, nisin is a unique molecule containing unusual amino acids including dehydroalanine and dehydrobutyrine, formed by dehydration of serine and threonine residues, respectively. These two residues are stereo-and regio-specifically coupled to the thiol group of the cysteines to form lanthionine and β-methyl lanthionine introduced enzymatically at post-translational level (Cotter et al., 2012). Nisin is thus a thioether-bridged pentacyclic peptide. The crystal structure of nisin has not been developed. The peptide molecule adopts different conformations depending on the environment. The structure of nisin cannot be described in terms of regular secondary-structure elements, due to the presence of the ring systems in which 65% of the residues are incorporated. However, the NMR structure is available in PDB database, which was used in this study as template to generate the model structures of the nisin variants. The NMR structure of nisin has determined two structured domains: an N-terminal domain (residues 3–19) containing three lanthionine rings, A, B and C; and a C-terminal domain (residues 22–28) containing two intertwined lanthionine rings numbered D and E (Hilbers, 1996). These domains are flanked by regions showing structural flexibility. The four-residue rings B, D and E of nisin all show a β-turn structure, which is closed by the thioether linkage. The backbones of the rings B and D form type I1 β-turns. The C-terminal domain consists of three consecutive β-turns. The NMR data will help us to locate residues in nisin interacting with hACE2.
The present study attempts to evaluate the potential of nisin variants to interact with hACE2 by predicting nisin binding site using nisin-hACE2 docking computation with the NMR structure of nisin in the PDB database. This is the first report on the potential of widely used food-grade antibacterial peptide nisin to bind with hACE2 and predicting the possibility of nisin as therapeutic against COVID-19. The work is significant in finding a solution to prevent the infection by novel coronavirus SARS-CoV-2.
2. Materials and methods
2.1. Data mining and alignment
Amino acids sequences of eight nisin variants: nisin Z (accession No: ABV64387.1), nisin A (accession No: AAA26948.1), nisin F (accession No: ABU45463.1), nisin Q (accession No: ADB43136.1), nisin H (accession No: AKB95119.1), nisin U (accession No: Q2QBT0.1), nisin U2 (accession No: ABO32538.1), nisin P (accession No: WP_105156946.1) were retrieved from Genbank database (https://www.ncbi.nlm.nih.gov/protein/). Full length amino acid sequence of ACE-2 of Homo sapiens (accession No: NP_001358344.1) and spike protein of SARS-CoV-2 (accession No:YP_009724390.1) were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/protein/ ). Multiple sequence alignment (MSA) of the nisin variants was performed using the ClustalW of Clustal Omega web server of the European Bioinformatics Institute (EMBL-EBI) (Park et al., 2019). Esprit 3 software (Robert and Gouet, 2014) was used to represent the MSA using BLOSUM 62 algorithm.
2.2. Homology modeling
Homology models of all nisin variants were done using the SWISS-MODEL web server (Waterhouse et al., 2018) using nisin Z (SMTL ID:1wco.1) as template. The steriochemical property of each of the models was evaluated by Ramachandran plot using Volume, Area, Dihedral Angle Reporter (VADAR) server (Willard et al., 2003) (Fig. S1). Similarly, the RBD (receptorbinding domain) of spike protein of SARS-CoV-2 and hACE2 receptor was modeled using SMTL ID: 6lzg.1 and SMTL ID: 6m18.1, respectively. All the models of nisin variants were superimposed together to determine their structural differences using read scoring matrix in PyMOL software. (The Pyolecular Graph).
2.3. Molecular Docking
Molecular Docking was performed to test the binding affinity of all nisin variants towards hACE2. In order to understand the comparative binding strength, multi-body docking were done between hACE2:nisin H complex with the RBD of SARS-CoV-2 and hACE2:RBD complex with nisin H. The solvated docking software, HADDOCK (Melquiond et al., 2016) was used without defying any restraints for such study. Most reliable model was selected by lowest HADDOCK score value. The score is calculated as
| HADDOCK score = 1.0 * Evdw +0.2 * Eelec +1.0 * Edesol +0.1*Eair, |
where Evdw is the intermolecular van der waals energy, Eelec the intermolecular electrostatic energy, Edesol represents an empirical desolvation energy. Active site residues of hACE2 (K31, E35, D38, M82) responsible of RBD spike binding were selected for docking. The residues surrounding the active loci were considered as passive. The interacting residues were visualized using Discovery studio. (SYSTÈMES, 2017) Prodigy@Bonvin lab web server (Xue et al., 2016) was used to calculate ΔG to predict the affinity of nisin H for hACE2 at 25 °C with other parameters remained under default condition. Grand average of hydropathy score of hACE2 was calculated with ExapassyProtparam webserver. (Gasteiger et al.Walker, 2005)
3. Results and discussion
3.1. Sequence and structural alignment
In multiple sequence alignment (Fig. 1 ) of amino acid residues of eight nisin variants (nisin A, Z, Q, H, P, U, U2 and F), nisin Z shared 82.35% amino acid sequence similarity with nisin H, whereas nisin P, U, U2, Q and F shared only 70.97%, 67.74%,67.74%, 76.47% and 79.41%, respectively with nisin H (Table S2). Nisin A was found to be closely related to nisin Z (97.06% identity) with only a single amino acid difference (His27Asn). In contrast, nisin H differs from nisin A by five different amino acids at positions 1, 6, 18, 21 and 31 with 85.29% identity. Nisin P is shorter than nisin H (34 residues) by three residues from the C-terminus. Nisin H differs from nisin F by 7 residues, F1I, M6L, T18G, Y21 M, H27 N, I30V and K31H. Nisin Q is different from nisin H due to the presence of isoleucine, leucine, valine, glycine, leucine, asparagine, valine and histidine at positions 1, 6, 15, 18, 21, 27, 30 and 31, respectively. Nisin U and U2 differed from nisin H by ten amino acids. The residual surface accessibility is present at the bottom of the alignment (Fig. 1).
Fig. 1.
Multiple sequence alignment of nisin variants (nisin P, nisin U, nisin U2, nisin H, nisin Q, nisin F, nisin Z, nisin A). The red highlighted residues are conserved among the eight nisin variants. Surface accessible regions (dark blue) and buried regions (light blue) are shown schematically at the bottom.
3.2. Homology modeling
The model structures of all nisin variants, hACE2, RBD of spike protein built on using SWISS-MODEL Web Server were validated for steriochemical properties using Ramachandran plot (Fig. S2). We considered the number of amino acids in the disallowed regions except for glycine and proline because of their chirality and imino group, respectively. Homology model of nisin P and U2 had no disallowed amino acids. Nisin H and U had only one residue in disallowed region, whereas two residues were found in the disallowed region for nisinA, F, Q and Z. The RMSD (C-alpha) from all the superimposed variants of nisin was found 0.191. These signify that all the nisin models were structurally similar to one another. The binding efficiency of nisins with hACE2 was further evaluated from docking studies.
3.3. Molecular docking
Best HADDOCK model of nisin variants in complex with hACE2 was analyzed for three parameters viz. Z-score, Buried surface area, and binding affinity. The Z-score indicates how many standard deviations from the average of the cluster is located in terms of score (the more negative the better). Z-score of hACE2-SARS-CoV-2 RBD, hACE2-nisin A, hACE2-nisin Z, hACE2-nisin H, hACE2-nisin Q, hACE2-nisin U, hACE2-nisin U2, hACE2-nisin F, and hACE2-nisin P was predicted as −1.5,-1.6,-1.9,-2.1,-1.4,- 1.7,-0.8,-1.4, and −1.5. Hence, both nisin H and nisin Z were lowest than rest of the nisin variants as well as RBD of spike protein. Burried surface area of nisin Z and nisin H with hACE2 were found higher, 2332.4 Å2 and 2395.1 Å2, respectively in contrast to 2092 Å2 for the RBD. This suggests that nisin H and nisin Z had better binding efficiency for hACE2.
The binding affinity of docked structures of all eight variants of nisin in complex with hACE2 was calculated as ΔG derived from analysis with Prodigy for each complex in comparison with the RBD of spike protein of SARS-CoV-2. ΔG of hACE2-SARS-CoV-2, hACE2-nisin A, hACE2-nisin Z, hACE2-nisin H, hACE2-nisin Q, hACE2-nisin U, hACE2-nisin U2, hACE2-nisin F, and hACE2-nisin P was −11 Kcal/mol, −10.6 Kcal/mol, −10.8 Kcal/mol, −11.3 Kcal/mol, −10.5 Kcal/mol, −10.5 Kcal/mol, −12.3 Kcal/mol, −12.5 Kcal/mol, and −11.4 Kcal/mol, respectively. Thus ΔG of hACE2-nisin Z and hACE2-nisin H are much higher conferring strong binding affinity than that of hACE2-RBD.GRAVY score of nisin A, Z, H, Q, U, U2, F, P and RBD-SARS-CoV-2 was calculated as 0.415, 0.406, 0.185, 0.524, 0.542, 0.439, 0.171, 0.185, −0.258, respectively (Table 1 ). From the GRAVY score of all nisin variants, nisin H turned out to be more hydrophilic than nisin A and nisin Z and will thus more potent to interact with the hydrophobic groove of hACE2 than others variants of nisin.
Table 1.
Comparative affinity of interaction between nisin-variants and human ACE2.
| Molecule interacts ACEII | Binding affinity (ΔG Kcal/mol) | GRAVY | Z score | Burried surface area(Å2) |
|---|---|---|---|---|
| RBD SARS-CoV-2 | −11.0 | −0.258 | −1.5 | 2092.0 |
| Nisin H | −11.3 | 0.185 | −2.1 | 2395.1 |
| Nisin Z | −10.8 | 0.406 | −1.9 | 2332.4 |
| Nisin A | −10.6 | 0.415 | −1.6 | 2311.8 |
| Nisin U | −12.3 | 0.542 | −1.7 | 2347.5 |
| Nisin U2 | −12.5 | 0.439 | −0.8 | 2192.8 |
| Nisin F | −11.4 | 0.171 | −1.4 | 2377.8 |
| Nisin Q | −10.5 | 0.524 | −1.4 | 2297.7 |
| Nisin P | −12.6 | 0.185 | −1.5 | 2190.3 |
From the docking analysis it is evident that nisin Z and nisin H interacts to hACE2 more efficiently. The interacting residues and atoms are given in (Table S1). The hydrogen bonds (K31:C19, K31:T13, K31:K12, E35:K12, E35:C19, E35:N20, D38:K22, D38:C27, M82:C7, K353:N27) and hydrophobic bonds (M82:I4, M82:C7, K31:C19, Y83:C7, K353:C28) are the major binding force for hACE2-nisinZ interaction. Interacting residues of nisin Z was predicted as I4, C7, K12, T13, C19, N20, K22, and C27. All interacting residues of nisin Z were hydrophilic in nature. The residues in nisin H interacting with the hACE2 include hydrogen bond of T13:K31, C19:K31, K12:K31, T8:K31, P9:K31, K12:E35, K22:D38, N20:E35, C26:D38, H27:D38, C28:K353, T23:K353 (Fig. 2 ) and hydrophobic bond of C19:K31 and Y21:K31, C7:M82, A24:K353, C26:K353, C28:K353. Among all these interacting residues, T8, P9, C11, K12, T13, C19, K22, C26 were highly conserved among all the nisin variants. Like RBD, surface accessible hydrophilic residues, T8, P9, C11, K12, T13, K22, and C26 were found to be involved in binding to hydrophobic groove of hACE2. It was found that nisin Z and nisin H recognized five common residues (K31, E35, D38, M82, K353) in hACE2 that were also recognized by RBD of spike.
Fig. 2.
Docked structure of human-ACE2 and nisin H; binding interface with interacting residues is indicated in the box region (A) and further zoomed in (B) to show the interacting residues. Nisin H and human-ACE2 are highlighted with red and yellow color, respectively.
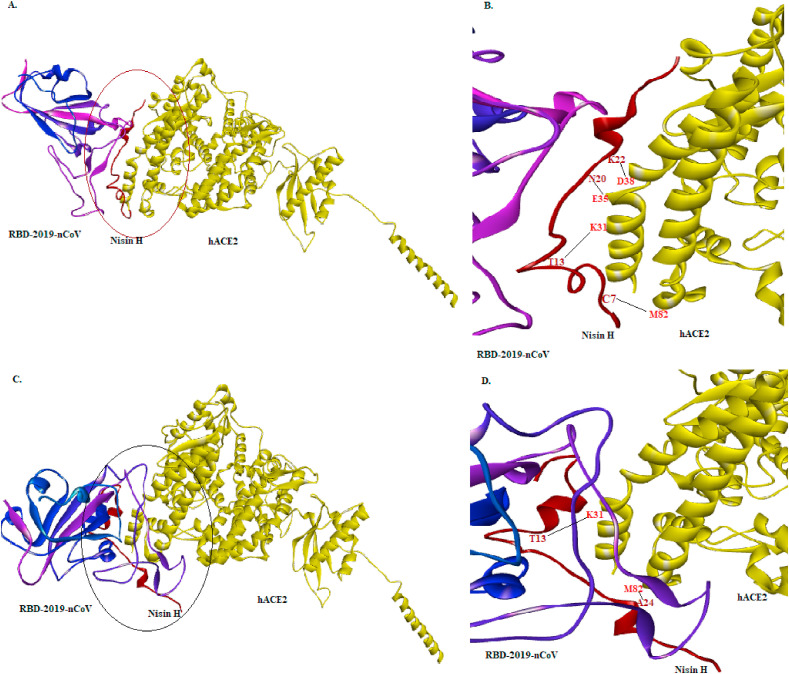
The binding efficiency of preformed hACE2:nisin H complex was performed by competitive tertiary docking with RBD of SARS-CoV-2 (Fig. 3 ). As, nisin H had already occupied the active site residues of hACE2 with strong hydrogen bond and hydrophobic interactions (Table S3), RBD of SARS-CoV-2 could not get access the active residues of hACE2 with reasonable efficiency by overcoming the binding strength of hACE2:nisin H interaction with ΔG of −11.3 kcal/mol (binding affinity of RBD:hACE2 complex is −11 kcal/mol). On the contrary when nisin H was allowed to interact with the hACE2:RBD complex, it was found that nisin H could be able to interact with active residues (K31 and M82) of hACE2 from the hACE2:RBD complex (Table S4). Nisin H, being more potent candidate could able to interfere in the interaction between RBD-hACE2. There is high possibility that nisin would be able to competitively displace bound SARS-CoV-2 because of its higher binding affinity towards the ACE2 receptor compared to that of the virus. Furthermore nisin being a non-synthetic molecule and smaller in size, will ensure high bioavailability. Based on such study, we hypothesize that nisin H, Z, A and U could be an eligible competitor of RBD of SARS-CoV-2 for having the same binding patch in hACE2. Recently, several peptides computationally designed to target the spike protein of SARS-CoV-2 have been reported (Han and Král, 2020; Baig et al., 2020) as a strategy to prevent their interaction with ACE 2 receptor for tackling COVID-19 infection. From an application perspective, it would be advantageous of using nisin as an effective treatment option over the reported designed peptides for several reasons, including its natural occurrence, food-grade status, extreme stability and ease of manufacturing through microbial fermentation, cost effectiveness, delivery at high concentration, etc. However, further experimental validation is required to confirm nisin binding to hACE2.
Fig. 3.
Competitive interaction of RBD of SARS-CoV-2, hACE2 and nisin H. (A) hACE2:nisin H complex was docked with RBD of SARS-CoV-2. Blue ribbons represent RBD of SARS-CoV-2, red ribbons represent Nisin H and yellow represents hACE2. (C) hACE2:RBD complex was docked with nisin H. Blue ribbons represent RBD of SARS-CoV-2, red ribbons represent Nisin H and yellow represents hACE2 (B) and (D) are magnified structures of (A) and (C), respectively to show the interacting residues.
4. Conclusions
Among all analyzed nisin variants, nisin Z, nisin A, nisin U and nisin H were most effective in interacting with human endothelial cell surface-receptor hACE2, the site where RBD of spike of SARS-CoV-2 binds to initiate infection. Compared to the RBD of viral spike protein, nisin binds with the hACE2 receptor with higher affinity. Nisin being a low molecular weight peptide and readily bioavailable in the system, its binding to hACE2 is expected to over-rule the interaction possibility of the RBD of spike of SARS-CoV-2 and could essentially exclude the virus entry to the host cell. Since nisin is a heat stable natural food grade peptide, can be produced cost effectively, even in large quantity through microbial fermentation, the present work will create greater interest among researchers to develop a new nisin-based treatment strategy for COVID-19, either through oral or nasal applications. However, further experimental validation is necessary to determine its doses and mechanistic application to check the competition of nisin and spike protein of SARS-CoV-2 for accessing the human.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgements
We most sincerely acknowledge the research grant (BT/PR16246/GET/119/73/2016) received from DBT, Department of Biotechnology, Govt. of India, New Delhi.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2020.10.002.
Author's contributions
RB curated, analyzed and interpreted the data. AMG helped in docking studies. S Mitra, S Mandal and SRB supervised the work. All the authors write, review and edited the manuscript.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- Arkin M.R., Tang Y., Wells J.A. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem. Biol. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig M.S., Alagumuthu M., Rajpoot S., et al. Identification of a potential peptide inhibitor of SARS-CoV-2 targeting its entry into the host cells. Drugs R. 2020;20:161–169. doi: 10.1007/s40268-020-00312-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P.D., Ross R.P., Hill C. Bacteriocins — a viable alternative to antibiotics? Nat. Rev. Microbiol. 2012;11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- Du Q., Wang S., Wei D., Sirois S., Chou K.C. Molecular modeling and chemical modification for finding peptide inhibitor against severe acute respiratory syndrome coronavirus main proteinase. Anal. Biochem. 2005;337:262–270. doi: 10.1016/j.ab.2004.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fons M., Gomez A., Karjalainen T. Vol. 2235. 2009. (Microbial ecology in health and disease mechanisms of colonisation and colonisation resistance of the digestive tract part 2: bacteria/bacteria interactions mechanisms of colonisation and colonisation resistance of the digestive tract). [DOI] [Google Scholar]
- Fox, J. & Wood, S. (s, 3). 2919, 4634–4635 (1971).
- Garcia-gutierrez E., et al. First evidence of production of the lantibiotic nisin P. Sci. Rep. 2020:1–15. doi: 10.1038/s41598-020-60623-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., et al. In: The Proteomics Protocols Handbook. Walker J.M., editor. Springer Protocols Handbooks.Human Press; 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–608. [DOI] [Google Scholar]
- Han Y., Král P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbers C.W. Surface location and orientation of the lantibiotic nisin bound to membrane-mimicking micelles of dodecylphosphocholine and of sodium dodecylsulphate. Eur. J. Biochem. 1996;403:394–403. doi: 10.1111/j.1432-1033.1996.00394. [DOI] [PubMed] [Google Scholar]
- Hurst A. Vol. 27. 1981. (Introduction, I. A. hurst.). [DOI] [Google Scholar]
- Juncioni L., Arauz D., Faustino A., Vessoni T.C. Nisin biotechnological production and application: a review. Trends Food Sci. Technol. 2009;20:146–154. doi: 10.1016/j.tifs.2009.01.056. [DOI] [Google Scholar]
- Lubelski J., Rink R., Khusainov R., Moll G.N., Kuipers O.P. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell. Mol. Life Sci. 2008;65:455–476. doi: 10.1007/s00018-007-7171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E, Melquiond A.S.J., van Dijk M., de Vries S.J., Bonvin A.M.J.J. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428(4):720–725. doi: 10.1016/j.jmb.2015.09.014. 22. [DOI] [PubMed] [Google Scholar]
- Mitra S., Mukhopadhyay B.C., Biswas S.R. Potential application of the nisin Z preparation of Lactococcus lactis W8 in preservation of milk. Lett. Appl. Microbiol. 2011:98–105. doi: 10.1111/j.1472-765X.2011.03075.x. [DOI] [PubMed] [Google Scholar]
- Park Y., et al. The EMBL-EBI search and sequence analysis tools APIs in 2019 Fabio. Nucleic Acids Res. 2019;47:636–641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.M., et al. Biomedical applications of nisin. J. Appl. Microbiol. 2015:1–17. doi: 10.1111/jam.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, L. & Hillman, J. D. Therapeutic Potential of Type A ( I ) Lantibiotics , a Group of Cationic Peptide Antibiotics. DOI:10.1016/j.mib.2008.09.008. [DOI] [PMC free article] [PubMed]
- Sorolla A., et al. Precision medicine by designer interference peptides: applications in oncology and molecular therapeutics. Oncogene. 2020;39:1167–1184. doi: 10.1038/s41388-019-1056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Systèmes D. Release; 2017. Dassault Syst Mes BIOVIA, Discovery Studio Modeling Environment.http://accelrys.com/products/collaborative-science/biovia-discovery-studio/ 2016).at. [Google Scholar]
- The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
- Vos W.M.D.E., Mulders J.W.M., Siezen R.J., Hugenholtz J., Kuipers O.P. Properties of nisin Z and distribution of its gene , nisZ , in Lactococcus lactis. Appl. Environ. Microbiol. 1993;59:213–218. doi: 10.1128/AEM.59.1.213-218.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018:1–8. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard L., et al. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003;31:3316–3319. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L.C., Rodrigues J.P., Kastritis P.L., Mjj A. 2016. Structural Bioinformatics PRODIGY: a Web Server for Predicting the Binding Affinity of Protein-Protein Complexes; pp. 2014–2016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.