Abstract
Severe Acute Respiratory Syndrome – Coronavirus 2 (SARS-CoV-2) emerged in Wuhan, China and spread to more than 114 countries resulting in a pandemic, which was declared by the WHO in March 2020. Tracking the spread of the virus raised a main concern in every country. Many researches proved the presence of SARS-CoV-2 in stool samples of patients, where the genes of this virus gave a positive signal several days prior to the occurrence of symptoms. The fact of viral shedding in stools provides an advantage in utilizing wastewater systems as a tool to monitor the viral prevalence. We tested more than 2900 municipal wastewater samples coming from 49 distinctive area in Dubai, where 28.6% showed positive results. We also looked into the wastewater samples from 198 commercial aircrafts arriving at Dubai Airport, giving a positive result percentage of 13.6%. The presence of SARS-CoV-2 genes was confirmed using TaqPath™ Covid-19 RT-PCR kit, which targets ORF1ab, N gene and S gene. This project shows the significance of utilizing wastewater-based epidemiology (WBE) in monitoring the prevalence of various infectious diseases such as SARS-CoV-2, which can assist the decision makers to determine the level of precautionary measures according to the areas of the outbreak. With this in mind, pricewise, WBE is considered cost-effective when comparing to clinical nasal swabs.
Keywords: SARS-CoV-2, COVID-19, Wastewater-based epidemiology, Aircraft sewage, Arab country
Graphical abstract
1. Introduction
In December 2019, a virus outbreak initiated in Wuhan, China identified as Severe Acute Respiratory Syndrome – Coronavirus 2 (SARS-COV-2), which was classified as a new strain of coronavirus (Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV), 2020). Novel Corona virus was found to mimic respiratory infections ranging from common cold to more severe diseases such as Poliovirus, Severe Acute Respiratory Syndrome (SARS), Middle East Respiratory Syndrome (MERS), and Aichivirus A, with symptoms including dry cough, fever, weakness in the respiratory system, diarrhea and tiredness (Petersen et al., 2020). After noticeable spread of the virus in 114 countries, WHO officially declared a world-wide pandemic on March 11, 2020 (WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020, 2020). The biological droplets released by an infected person by coughing or sneezing were found to be as a main factor responsible for the spread of this disease among individuals (Gu et al., 2020).
Health facilities & laboratories began to follow testing criteria on suspected coronavirus disease (COVID-19) cases and implement molecular testing of RT-qPCR (Real Time polymerase chain reaction) for diagnosis of the respiratory system through nasal swabs as the approved testing methodology for detection of SARS-COV-2 (Péré et al., 2020). Never the less, the presence of SARS-COV-2 (RNA) was also detected in other human specimens such as blood and urine in pre-symptomatic, asymptomatic and symptomatic individuals (Petersen et al., 2020; Woelfel et al., 2020; Zhang et al., 2020). As for human faeces, viral quantities can be found in them as a result of many factors, which include: absorbing of respiratory secretions from the upper respiratory tract into the stomach, deposits of infected immune cells, and viral replication in the intestines (Xiao et al., 2020). It has been observed that gastrointestinal symptoms may be a mutual symptom of different corona virus diseases as small percentage of viral nucleic acid was still found in stool samples 30 days after infection (Jefferson et al., 2020).
Wastewater studies were used as a complimentary tool for investigating the circulation of the virus in human population (Lodder and de Roda Husman, 2020; Ahmed et al., 2020a, Ahmed et al., 2020b; Medema et al., 2020). Several researches revealed the presence of SARS-CoV-2 in stool specimens, which allowed the implementation of wastewater testing to track this novel disease (F. Wu et al., 2020; Xiao et al., 2020). In France, a test was performed to prove the assumption that the quantification of SARS-CoV-2 genomes in wastewaters should correlate with the number of symptomatic or non-symptomatic carriers during the lock down period. It was performed by collecting raw wastewater samples from several major wastewater treatment plants and performing a time-course quantitative analysis of SARS-CoV-2 by RT-qPCR. The results of the analysis confirmed that the increase of genome units in raw wastewaters followed the increase of human COVID-19 cases observed at the regional level accurately, which proved that the spread and exponential growth of the epidemic can be detected beforehand (Wurtzer et al., 2020). The objective of this research, which is still ongoing, is to utilize sewage wastewater in domestic areas in Dubai and Dubai International Airport Aircrafts for monitoring the spread of the virus and use this epidemiology as an early warning tool in case of upcoming waves. It is of vital importance to note that this paper looked into more than 3000 municipal's and aircrafts' wastewater, which is considered the highest number of samples among the published papers of this testing field. Moreover, it provides the first proof of detection of SARS-CoV-2 genes in wastewater samples from an Arab country.
2. Materials & methods
2.1. Sampling
In this project, the sample collection strategy was divided into two main stages. In the first stage, untreated wastewater samples were collected from 9 main pumping stations in three consecutive times totalling in 27 samples. The dates of collection were 22nd April, 28th April, and 4th May, 2020. The aim of this stage was to establish a wide screening procedure of the city of Dubai. In the second stage, collection intended to branch out and look closely within the distinctive areas of the city. Dubai consists of two main regions; Bur Dubai and Bur Deira. These two main regions comprise a total of 49 distinctive areas, noting that 25 areas belong to Bur Dubai and the other 24 belong to Bur Deira. Sampling was carried out by collecting 10 samples from each area once a fortnight for 6 consecutive times, from 7th of May to 7th of July 2020. In addition, this study has looked into the treated wastewater samples by collecting 3 samples from all three wastewater treated pumps (WWTPs) of Dubai.
2.2. Sample concentrating/extraction
Wastewater samples were collected in 1000 ml LDPE bottles (BNH1000BULK; Azlon ®, Staffordshire, UK) and stored at room temperature waiting for processing. The collection of samples followed grab sampling approach. A dedicated team from Dubai Airports managed to collect samples from arriving aircrafts directly from the excretory valve beneath the airplane, using a big bucket. The wastewater then transferred into the above mentioned 1000 ml bottles. Viral RNA was concentrated following a modified method, with an initial step of pipetting 10 ml of the wastewater sample through 11-μm-pore-size, 125-mm-diameter cellulose filter (Z240095; Whatman®), followed by centrifuging 1.5 ml of the filtered sample at 4750g for 30 min. Without disturbing the pellet, 400 μl of the supernatant was later centrifuged at 3500g for 15 min through MB Spin Column from the DNeasy PowerSoil Kit ® (12888-100; Qiagen, Germany), the eluted sample was collected for extraction. Viral RNA was extracted using MagMax Viral/Pathogen Kit (A42352; ThermoFisher Scientific™, Massachusetts, US) following the manufacturer manual using KingFisher™ Flex Purification System (5400610, ThermoFisher Scientific™, Massachusetts, US).
2.3. Real-Time PCR
The presence of SARS-CoV-2 RNA was tested using Real time PCR by the detection of three different genes specific to this virus; ORF1ab, N gene and S gene. The adopted qPCR methodology followed TaqPath™ Covid-19 RT-PCR Kit (A48067; ThermoFisher Scientific™, Massachusetts, US), using the manufacturer's protocol. MS2 is used as an internal standard, and nuclease free water as a negative control. Samples were prepared and set accordingly with the total volume of 25 μl. The reactions were carried out using QuantStudio™ 5 Real-Time PCR System (A34322; ThermoFisher Scientific™, Massachusetts, US). The results were analysed as instructed in the manual. Note that this kit is used as a qualitative method for the detection of SARS-CoV-2 in samples.
3. Results and discussion
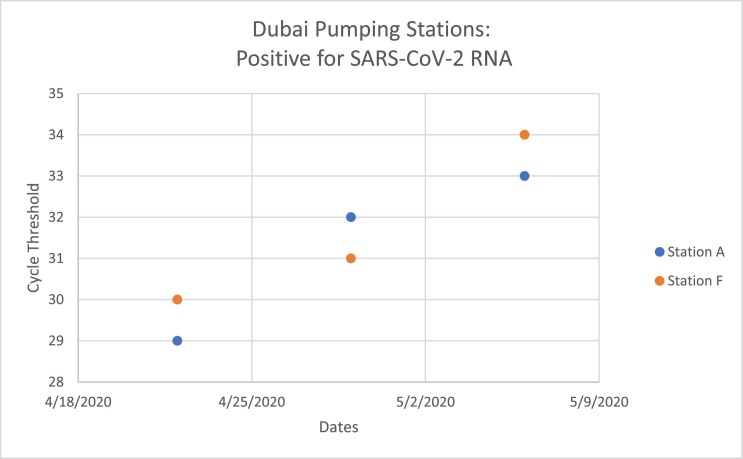
In the first stage of this project, nine main pumping stations, which pump 100% of the wastewater of Dubai sewage system were sampled. Among the 27 samples tested from the pumping stations on three different intervals, six samples from two specific stations (Stations A and F) turned out to be positive for SARS-CoV-2 RNA following the assays of ORF1ab, S protein and N protein as shown in Fig. 1 . The Ct values of the positive samples ranged between 32 and 34 cycles.
Fig. 1.
In the first stage of this project, out of 27 samples tested, 6 samples showed a positive signal to SARS-CoV-2 genes, all coming from the same two stations (stations A and F). The cycle threshold showed to increase in value, implying a decrease in the concentrations of the viral load within the samples throughout the short period of collection.
This stage provided a full scanning method to locate areas of high viral loads in Dubai and to consider the areas of concern in order to monitor the control of the precautionary measures. One of the first conducted studies on sewage samples to detect SARS-CoV-2 was carried out by Medema et al. (2020). They tested samples from 7 different cities of Netherlands, as well as, a sample from the airport (Medema et al., 2020). Moreover, in Australia, Ahmed et al., 2020a, Ahmed et al., 2020b tested two wastewater treatment plants (WWTP) and a pumping station. Both of these studies came into conclusion that testing samples from the sewage system provides a powerful tool in monitoring any increase or decrease in infection among certain population (Ahmed et al., 2020a). The rich information provided by such methodology to monitor SARS-CoV-2 RNA in sewage systems was used by many countries to expect any incoming waves and manage hospital admissions. A study carried out by Nemudryi et al. (2020) over a 17-day period showed a decreasing trend in the viral load which correlated with hospital admissions in Bozeman, United States (Nemudryi et al., 2020). Peccia et al. (2020) implied that SARS-CoV-2 RNA concentrations in sewage systems were a one week leading indicator upfront of submitting a COVID-19 test and local hospital admissions (Peccia et al., 2020).
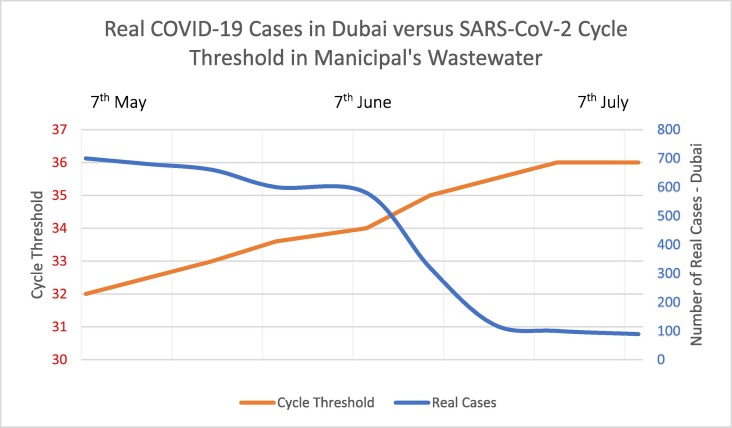
The second stage of this research project looked into untreated municipal wastewater samples that were collected from 49 different areas in Dubai. The research team of this study agreed to proceed with this stage, despite the large number of samples and the expended effort, in which each area was tested narrowly, having at least 10 municipal wastewater samples, which adds up the total number of tested samples to approximately 2940 over 3 months-period. Fig. 2 represents real COVID-19 Cases in Dubai versus SARS-CoV-2 cycle threshold detected from municipal's wastewater. The advantage of having tests done once a fortnight provided us a prevalence of any incoming waves, as well as, the opportunity to create a heat map of the whole city over 3 months' period, as seen in Fig. 3 .
Fig. 2.
The results of the second stage of this project showed a direct correlation with the number of real COVID-19 cases recorded in the city of Dubai. During the 3-month period, the Ct values showed an increasing trend, implying a decrease in the concentration of SARS-CoV-2 genes in wastewater samples, which corresponds to the fact of the decrease in the record of real cases in the city.
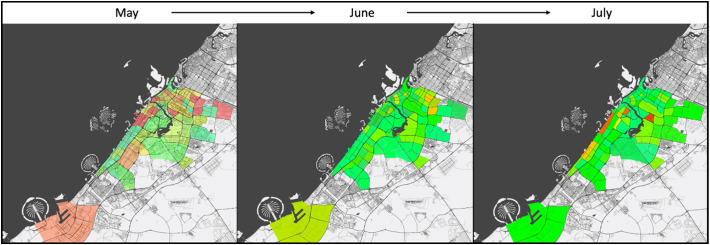
Fig. 3.
The results of second stage displayed as a heat map of the whole city over 3 months' period. The results showed decreasing of viral load concentration over the 3-months period. Heat maps provide a visualization of the overall prevalence throughout the city. Dark red color represents high concentration of viral load; yellow color represents moderate concentration of viral load; and green color represents low concentration of viral load. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Many experiments were done in detecting the viral nucleic acid of SARS-CoV-2 in faeces samples in order to measure its quantity and concentration. One of these studies tested 65 faecal samples of hospitalized infected people, where the authors have found that 31 of these samples showed positive results, including those with no any gastrointestinal symptoms (Lin et al., 2020). Another study carried out by Xiao et al. in 2020 tested for the presence of viral RNA in 73 hospitalized infected people, in which 53.4% of the samples showed a positive signal of SARS-CoV-2 RNA. Moreover, 20% of these patients were still showing positive results in their stool samples even when the respiratory samples resulted in negative signals (Xiao et al., 2020). In addition to the findings of this study, in 2020, Y. Wu and colleagues found out that over half of the 74 tested patients had stools which remained positive for SARS-CoV-2 RNA for an average of 11.2 days after the nasal swabs became negative. The authors have implied that the virus might be able to replicate in the gastrointestinal tract (Y. Wu et al., 2020). The findings might as well raise a main topic of utilizing such information from sewage systems to track any increase or decrease in the concentration of the viral load. In our case, for example, if any area showed to have an increase of viral nucleic acid concentration, a report would be sent to the authorities to act necessarily. Reports from such approach could also assist the decision makers in any country to apply or eliminate a lockdown. In addition to all of that, sewerage system testing is considered much cheaper in cost than that of clinical nasal swabs. To enumerate, if a screening test would be done for a neighborhood with a population of one thousand, the approximate difference of the operating cost between testing wastewater samples and clinical nasal swabs would be 97.76%. It is important to be aware that wastewater samples could represent hundreds or thousands of people, which makes this method cost-effective comparing to that of nasal swabs when measuring the outbreak of the disease.
Testing human faecal samples to trace COVID-19 cases are not limited on the municipal's sewerage systems or WWTPs. Ahmed et al. (2020b) have considered testing the wastewater of commercial aircrafts and cruise ships as a surveillance tool detect infections of SARS-CoV-2 among travelers. They have tested a total of 3 aircrafts and 2 samples of a cruise ship, where the results showed positive SARS-CoV-2 signals though concentrations were close to the limit of detection. The authors suggested prioritizing medical testing and interaction tracking among the travelers. They have also recommended using both strategies of wastewater and nasal clinical swabs testing to maximize the detection of SARS-CoV-2 infections among travelers (Ahmed et al., 2020b).
In our recent study, we have tested a total of 198 commercial flights arriving in Dubai from 59 airport destinations from all six continents, where the percentage of positive signals showed to be 13.6%, having Ct values that ranged from 33 to 36. Such information can be accumulated to give a leading point to decision makers in any country in order to terminate any flights coming from a specific location. For example, out of 16 flights coming from Pakistan, 10 were found to be positive for SARS-CoV-2 RNA. This implies that such aircrafts coming from Pakistan could be terminated in order to help minimize the entry of infected passengers to the country.
The presence of SARS-CoV-2 traces within wastewater raised many questions regarding the fate of this virus in WWTPs. Normally, excreta from human travels through the sewerage systems into the wastewater treatment plants, which then directs the treated samples to many destinations such as rivers or lakes. Rivers and lakes are usually used as a water supply and for recreation purposes, which all direct back to the community. Furthermore, WWTP could be directed for irrigation purposes which can fire backwards again onto the members of community through crops and aerosols (Lesimple et al., 2020). Many studies have considered this issue and tested treated wastewater samples. Wurtzer et al. (2020) examined treated wastewater samples from three different Parisian WWTPs, where the results came to be positive (Wurtzer et al., 2020). Another experiment performed by Randazzo et al., 2020 looked into 18 secondary effluent samples from 6 WWTPs of the region of Murcia, Spain. The authors found that 2 of the 18 samples were positive (Randazzo et al., 2020). In our study, we have tested 3 samples of treated wastewater samples from the main three WWTP of Dubai. All of the samples showed no signal of SARS-CoV-2 RNA. The WWTPs in Dubai used to treat wastewater by applying a certain dose of chloride disinfectant, however, with the occurrence of this pandemic, we were informed that the chloride dosage was increased, however, we have no information on the concentration of the chloride used in local WWTPs.
4. Conclusion
Based on our experiment, we totally agree with Daughton (2020) in that WBE could be utilised in rapidly determining any increase or decrease of the spread of SARS-CoV-2 infections. This approach could also aid in regards to incoming viral waves or in the occasion of potential viral epidemics. Infection levels and trends could be observed by detecting the viral load in the sewage system. A way of communication should be established between scientists in this field and the decision-makers in every country to be aware of the disease prevalence and thus implement the required precautionary measures in the outbreak regions. Daughton also mentioned that it is of vital importance that coordination of approaches and sharing of data is linked between the scientists to elucidate if WBE can be applied successfully. For instance, what is the viral load detection limit that could raise a concern? How to translate scientific information to the decision-makers who normally have limited scientific background? Standardization of sampling methods and distributing analytical methods would help in building a strong foundation for estimating the reliability of the population served (Daughton, 2020; Foladori et al., 2020).
CRediT authorship contribution statement
Abdulla Albastaki: Writing, sampling, conceptualisation.
Mohammed Naji: Writing, sampling, conceptualisation.
Reem Lootah: Methodology.
Reem Almeheiri: Writing.
Hanan Almulla: Visualisation, Data Analysis.
Iman Almarri: Sampling, methodology.
Afra Alreyami: Methodology.
Ahmed Aden: Resources, coordination.
Rashed Alghafri: Supervision, Revision.
Acknowledgments and declaration of competing interest
The authors wish to confirm that there are no known conflicts of interest associated with this publication and would like to acknowledge and thank Dubai Municipality, Dubai Airports and other governmental entities in Dubai who played a key role in enabling this project.
Editor: Damia Barcelo
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J., Choi P., Kitajima M., Simpson S., Li J., Tscharke B., Verhagen R., Smith W., Zaugg J., Dierens L., Hugenholtz P., Thomas K., Mueller J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K., Hosegood I., Hugenholtz P., Jiang G., Kitajima M., Sichani H., Shi J., Shimko K., Simpson S., Smith W., Symonds E., Thomas K., Verhagen R., Zaugg J., Mueller J. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. Journal of Travel Medicine. 2020;27(5) doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C. The international imperative to rapidly and inexpensively monitor community-wide Covid-19 infection status and trends. Sci. Total Environ. 2020;726 doi: 10.1016/j.scitotenv.2020.138149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal–oral transmission. Gastroenterology. 2020;158(6):1518–1519. doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson T., Spencer E., Brassey J., Heneghan C. SARS-CoV-2 and the Role of Orofecal Transmission: Evidence Brief. Analysis of the Transmission Dynamics of COVID-19: An Open Evidence Review. 2020. https://www.cebm.net/wp-content/uploads/2020/07/SARS-CoV-2-and-the-Role-of-Orofecal-Transmission-Evidence-Brief-2.pdf [online] Available at.
- Lesimple A., Jasim S., Johnson D., Hilal N. The role of wastewater treatment plants as tools for SARS-CoV-2 early detection and removal. Journal of Water Process Engineering. 2020;38 doi: 10.1016/j.jwpe.2020.101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Jiang X., Zhang Z., Huang S., Zhang Z., Fang Z., Gu Z., Gao L., Shi H., Mai L., Liu Y., Lin X., Lai R., Yan Z., Li X., Shan H. 2020. Gastrointestinal Symptoms of 95 Cases With SARS-Cov-2 Infection. [DOI] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A. SARS-CoV-2 in wastewater: potential health risk, but also data source. The Lancet Gastroenterology & Hepatology. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. 2020. Presence of SARS-coronavirus-2 RNA in Sewage and Correlation With Reported COVID-19 Prevalence in the Early Stage of the Epidemic in the Netherlands. [DOI] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Reports Medicine. 2020;1(6):100098. doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D., Grubaugh N., Kaplan E., Casanovas-Massana A., Ko A., Malik A., Wang D., Wang M., Warren J., Weinberger D., Omer S. 2020. SARS-CoV-2 RNA Concentrations in Primary Municipal Sewage Sludge as a Leading Indicator of COVID-19 Outbreak Dynamics. [Google Scholar]
- Péré H., Podglajen I., Wack M., Flamarion E., Mirault T., Goudot G., Hauw-Berlemont C., Le L., Caudron E., Carrabin S., Rodary J., Ribeyre T., Bélec L., Veyer D. Nasal swab sampling for SARS-CoV-2: a convenient alternative in times of nasopharyngeal swab shortage. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00721-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen E., Koopmans M., Go U., Hamer D., Petrosillo N., Castelli F., Storgaard M., Al Khalili S., Simonsen L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020;20(9):e238–e244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Who.int Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-Ncov) 2020. https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov) [online] Available at.
- Woelfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Mueller M.A., Niemeyer D., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Bruenink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological Assessment of Hospitalized Cases of Coronavirus Disease. Vol. 2019. 2020. Clinical presentation and virological assessment of hospitalized cases of coronavirus disease 2019 in a travel-associated transmission cluster. [Google Scholar]
- World Health Organization WHO Director-General's Opening Remarks at the Media Briefing on COVID-19 - 11 March 2020. 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the- media-briefing-on-covid-19---11-march-2020 [online] Available at.
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. The Lancet Gastroenterology & Hepatology. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Moniz K., Erickson T., Chai P., Thompson J., Alm E. 2020. SARS-CoV-2 Titers in Wastewater Are Higher Than Expected From Clinically Confirmed Cases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J., Maday Y., Teyssou R., Richard E., Almayrac J., Moulin L. 2020. Evaluation of Lockdown Impact on SARS-CoV-2 Dynamics Through Viral Genome Quantification in Paris Wastewaters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen C., Zhu S., Shu C., Wang D., Song J., Song Y., Zhen W., Feng Z., Wu G., Xu J., Xu W. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) China CDC Weekly. 2020;2(8):123–124. [PMC free article] [PubMed] [Google Scholar]