Abstract
International lineages, such as Salmonella Typhimurium sequence type (ST) 19, are most often associated with foodborne diseases and deaths in humans. In this study, we compared the whole-genome sequences of five S. Typhimurium strains belonging to ST19 recovered from clinical human stool samples in North Carolina, United States. Overall, S. Typhimurium strains displayed multidrug-resistant profile, being resistance to critically and highly important antimicrobials including ampicillin, ticarcillin/clavulanic acid, streptomycin and sulfisoxazole, chloramphenicol, tetracycline, respectively. Interestingly, all S. Typhimurium strains carried class 1 integron (intl1) and we were able to describe two genomic regions surrounding blaCARB-2 gene, size 4,062 bp and 4,422 bp for S. Typhimurium strains (HS5344, HS5437, and HS5478) and (HS5302 and HS5368), respectively. Genomic analysis for antimicrobial resistome confirmed the presence of clinically important genes, including blaCARB-2, aac(6’)-Iaa, aadA2b, sul1, tetG, floR, and biocide resistance genes (qacEΔ1). S. Typhimurium strains harbored IncFIB plasmid containing spvRABCD operon, as well as rck and pef virulence genes, which constitute an important apparatus for spreading the virulence plasmid. In addition, we identified several virulence genes, chromosomally located, while the phylogenetic analysis revealed clonal relatedness among these strains with S. enterica isolated from human and non-human sources obtained in European and Asian countries. Our results provide new insights into this unusual class 1 integron in virulent S. Typhimurium strains that harbors a pool of genes acting as potential hotspots for horizontal gene transfer providing readily adaptation to new surrounds, as well as being crucially required for virulence in vivo. Therefore, continuous genomic surveillance is an important tool for safeguarding human health.
Introduction
Non-typhoidal Salmonella (NTS) is one of the most important foodborne pathogens with unprecedented impact on global health [1]. Among NTS, Salmonella enterica subsp. enterica serovar Typhimurium represents a major threat, since its worldwide spread has been associated with a broad host range, which includes mostly humans and food-related sources [1, 2]. Besides that, the emergence of multidrug-resistant (MDR) S. enterica is another crucial aspect for food-related outbreaks globally, limiting our therapeutic options [3].
In addition to the high global burden of salmonellosis, extended-spectrum β-lactamase (ESBL)-producing S. enterica strains have been recognized as high-priority bacteria causing serious public health issue (https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed). Aside from this, the emergence of mobile genetic elements (MGEs), for instance, class 1 integrons play an essential role in the global spread of antimicrobial resistance [4, 5]. Another aspect to be considered is the wide range of virulence package that is typically associated with Salmonella Pathogenicity Islands (SPI), contributing to the infection process among diverse hosts [6–8]. In this context, while the surveys with genomic approach have helped in the development of mitigation strategies and clinical management, continuous active surveillance is urgently required.
Here, we describe the genomic characteristics of five MDR and virulent S. Typhimurium strains carrying the blaCARB-2 gene recovered from clinical human stool samples in North Carolina, United States.
Materials and methods
Ethics approval and consent to participate
The human patients from whom Salmonella were recovered were completely anonymous and even after all the analysis and tests, the human sample remained anonymous. As such, the NC State IRB (FWA: 00003429) indicated the study research did not need IRB approval because it does not meet the definition for human subjects research.
Bacterial strains and antimicrobial susceptibility testing
We conducted a genomic investigation on five clinical S. Typhimurium strains collected in 2014 in North Carolina, United States. The strains were subjected to phenotypic characterization using the microdilution panel susceptibility approach on Gram-negative Sensititre plates (CMV3AGNF and GNX2F, Trek Diagnostic Systems, OH, USA) following the interpretative criteria of Clinical and Laboratory Standards Institute [9, 10]. The MDR profile was defined as resistant to three or more classes of antimicrobials [11]. All S. Typhimurium strains underwent molecular screening for class 1 integron by PCR [12, 13] and were subsequently characterized by whole-genome sequencing (WGS) according to Pornsukarom et al. [14].
Whole-genome sequencing and phylogenetic analysis
Libraries were prepared using the Nextera XT DNA sample preparation kit (Illumina, San Diego, CA), which were multiplexed and sequenced on MiSeq platform (Illumina, San Diego, CA, USA) at a paired-end read (300 bp). Resulted raw sequence reads underwent a strict quality control, as well as we obtained the draft genomes by using default settings in CLC workbench 10.1.1 (Qiagen) as per Monte et al. [15]. The sequencing data were deposited in NCBI (PRJNA613764). For each strain, we uploaded the sequences into Center for Genomic Epidemiology (http://genomicepidemiology.org/) to detect multilocus sequence typing (MLST), resistome, plasmid incompatibility groups and Salmonella Pathogenicity Islands.
Virulome analyzes were performed by using default settings available in VFanalyzer [16]. Additionally, the genetic context of blaCARB-2 and presence of virulence genes were investigated using BLASTn analysis against the non-redundant (NR) database and manually curated using Geneious v. 11.1.5 (Biomatters Ltd., Auckland, New Zealand).
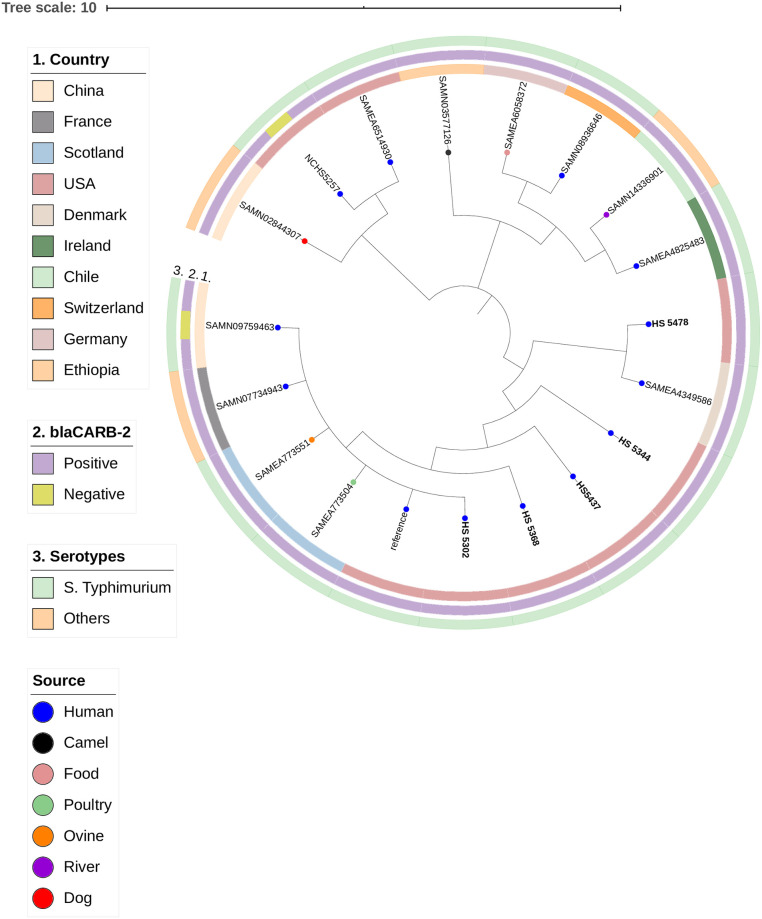
For phylogenetic purpose, we reconstructed a maximum likelihood phylogenetic tree based on single nucleotide polymorphism (SNP) using default settings of CSI Phylogeny version 1.4 [17]. SNP tree was reconstructed with five genomes of S. Typhimurium from this study in addition to thirteen genomes retrieved from GenBank database. Additional genomes of S. enterica strains were chosen from different sources (human, camel, food, poultry, ovine, river, and dog) and countries, including USA (SAMN10863500 and SAMEA6514930), France (SAMN07734943), Scotland (SAMEA773504 and SAMEA773551), Denmark (SAMEA4349586), Ireland (SAMEA4825483), Switzerland (SAMN08936646), Germany (SAMEA6058372), Chile (SAMN14336901), China (SAMN09759463 and SAMN02844307), and Ethiopia (SAMN03577126).
Results
Antimicrobial susceptibility testing and class 1 integron detection
All the five strains were classified as MDR, displaying resistance to critically important antimicrobials including ampicillin (100%), ticarcillin/clavulanic acid (100%), and streptomycin (60%), as well as to highly important antimicrobials comprising sulfisoxazole (100%), chloramphenicol (100%), and tetracycline (60%) (Table 1). Moreover, intermediate resistance to doxycycline was detected in three strains (HS5344, HS5437 and HS5478), and in a single strain (HS5437) to ceftazidime. In addition, we confirmed the presence of class 1 integron in all S. Typhimurium strains.
Table 1. Phenotypic and genomic features of Salmonella Typhimurium ST19 strains isolated from clinical human samples in United States.
| Strain ID | Serotype | Source | R-type (MIC)* | Resistance genotype | Plasmids | ST | Accession number |
|---|---|---|---|---|---|---|---|
| HS5302 | Typhimurium (O5-) | Stool | FIS-AMP-TIM2 | blaCARB-2, aac(6’)-Iaa, sul1 | IncFIB(S), IncFII(S) | 19 | JAATJP000000000 |
| HS5344 | Typhimurium (O5-) | Stool | CHL-TET-FIS-AMP-STR-TIM2 | blaCARB-2, aac(6’)-Iaa, aadA2b, sul1, tet(G), floR | IncFIB(S), IncFII(S) | 19 | JAATGY000000000 |
| HS5368 | Typhimurium (O5-) | Stool | FIS-AMP-TIM2 | blaCARB-2, aac(6’)-Iaa, sul1 | IncFIB(S), IncFII(S) | 19 | JAATJO000000000 |
| HS5437 | Typhimurium (O5-) | Stool | CHL-TET-FIS-AMP-STR-TIM2 | blaCARB-2, aac(6’)-Iaa, aadA2b, sul1, tet(G), floR | IncFIB(S), IncFII(S) | 19 | JAATGZ000000000 |
| HS5478 | Typhimurium | Stool | CHL-TET-FIS-AMP-STR-TIM2 | blaCARB-2, aac(6’)-Iaa, aadA2b, aph(3’)-Ia, sul1, tet(G), floR | IncFIB(S), IncFII(S) | 19 | JAATHA000000000 |
*FIS, sulfisoxazole; AMP, ampicillin; TIM2, ticarcillin/clavulanic acid constant 2; CHL, chloramphenicol; TET, tetracycline; STR, streptomycin.
Whole-genome sequencing and phylogenetic analysis
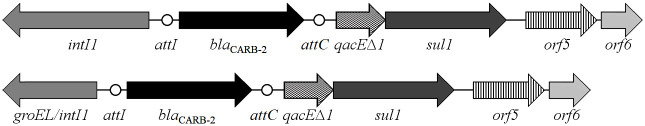
Genomic analysis revealed that all five S. Typhimurium strains belonged to the international sequence type (ST) ST19, while antimicrobial resistome confirmed the presence of critically important genes, such as carbenicillinase [blaCARB-2], aminoglycosides [aac(6’)-Iaa and aadA2b], sulfonamide [sul1], tetracycline [tetG], and florfenicol [floR]. The IncFIB(S) and IncFII(S) plasmid incompatibility groups were detected in all strains. We were also able to describe two schematic representations of the genetic context surrounding blaCARB-2 gene. First, three S. Typhimurium strains (HS5344, HS5437, and HS5478) analyzed in this study, shared a genomic environment with 4,062 bp in size composed by groEL/intI1-blaCARB-2-qacEΔ1-sul1-orf5 (acetyltransferase)-orf6 (hypothetical protein). Second, the remaining S. Typhimurium strains (HS5302 and HS5368) presented a genomic content slightly different with a 4,422 bp region composed by intI1-blaCARB-2-qacEΔ1-sul1-orf5 (acetyltransferase)-orf6 (hypothetical protein) (Fig 1). Additionally, the sul1, blaCARB-2, tetG, floR, and aadA2b resistance genes were harbored by a partial sequence of a complex class 1 integron (In104) from HS5344, HS5437, and HS5478. This sequence included duplications of parts of the integron conserved segments (CS), specifically, part of the intI1 gene from the 5’-CS and part of the 3’-CS (qacEΔ1 and partial sul1 genes). Consequently, the structure had two attI1 sites, into which the aadA2b gene cassette was inserted in one and the blaCARB-2 cassette in the other. The floR and tetG genes were identified between the two integron-derived regions. In HS5302 and HS5368, only the region containing the intI1-blaCARB-2-qacEΔ1-sul1-orf5-orf6 array was detected. Furthermore, while aac(6’)-Iaa was found at a site distant from the other resistance genes on the chromosome of all S. Typhimurium strains in this study, aph(3’)-Ia was identified in a partial transposon sequence from HS5478.
Fig 1. Schematic representation of the genetic context surrounding blaCARB-2 genes in Salmonella Typhimurium ST19 strains isolated from clinical human samples in United States.
Virulome analysis revealed presence of several Salmonella Pathogenicity Island (SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, and Centisome 63 Pathogenicity Island) as shown in Table 2. Upon encountering these SPI, we also identified important virulence genes involved in fimbrial adherence (fimA, C, D, F, H, I, W, Y, Z), non-fimbrial adherence (misL), invasion (InvA, B, C, E, F, G, H, I, J), secretion system (ssa, ssc, sse, and ssr), magnesium uptake (mgtB and mgtC), regulation (phoP, phoQ, and pipB), and translocated effector (sopB/sigD and sopE2) (Table 2).
Table 2. Genomic features of virulence factors of Salmonella Typhimurium ST19 strains isolated from clinical human samples in United States.
| Strain ID | SPI-1 encode genes | SPI-2 encode genes* | SPI-3 encode genes | SPI-5 encode genes | Virulence plasmid* | Fimbrial adherence determinants | SPI* |
|---|---|---|---|---|---|---|---|
| HS5302 | inv (A, B, C, E, F, G, H, I, J); sopE2 | ssa (C, D, E, G, H, I, J, K, L, M, N, O, P, Q, R, T, U, V); ssc (A, B); sse (B, C, D, E); ssr (A, B) | mgtB, mgtC, misL | phoP, phoQ, pipB, sopB/sigD | spv (A, B, D, R) | fim (A, C, D, F, H, I, W, Y, Z) | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI |
| HS5344 | inv (A, B, C, E, F, G, H, I, J); sopE2 | ssa (C, D, E, G, H, J, K, L, M, N, O, P, Q, R, T, U, V); ssc (A, B); sse (B, C, D, E); ssr (A, B) | mgtB, mgtC, misL | phoP, phoQ, pipB, sopB/sigD | spv (A, B, D, R) | fim (A, C, D, F, H, I, W, Y, Z) | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI |
| HS5368 | inv (A, B, C, E, F, G, H, I, J); sopE2 | ssa (C, D, E, G, H, I, J, K, L, M, N, O, P, Q, R, T, U, V); ssc (A, B); sse (B, C, D, E); ssr (A, B) | mgtB, mgtC, misL | phoP, phoQ, pipB, sopB/sigD | spv (A, B, D, R) | fim (A, C, D, F, H, I, W, Y, Z) | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI |
| HS5437 | inv (A, B, C, E, F, G, H, I, J); sopE2 | ssa (C, D, E, G, H, I, J, K, L, M, N, O, P, Q, R, T, U, V); ssc (A, B); sse (B, C, D, E); ssr (A, B) | mgtB, mgtC, misL | phoP, phoQ, pipB, sopB/sigD | spv (A, B, D, R) | fim (A, C, D, F, H, I, W, Y, Z) | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI |
| HS5478 | inv (A, B, C, E, F, G, H, I, J); sopE2 | ssa (C, D, E, G, H, J, K, L, M, N, O, P, Q, R, T, U, V); ssc (A, B); sse (B, C, D, E); ssr (A, B) | mgtB, mgtC, misL | phoP, phoQ, pipB, sopB/sigD | spv (A, B, D, R) | fim (A, C, D, F, H, I, W, Y, Z) | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI |
*Letters highlighted in bold represents differences among strains.
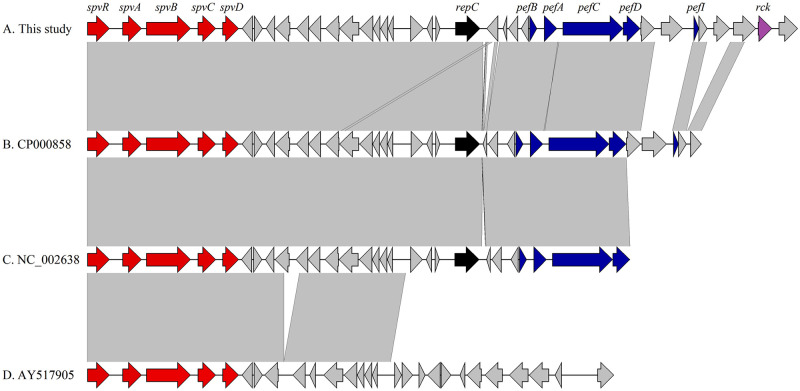
Interestingly, these strains possess a highly conserved spv operon composed by spvR, spvA, spvB, spvC, and spvD genes which are located upstream of the genes pefA (plasmid-encoded fimbriae) and rck (resistance to complement killing) in a virulence plasmid as shown in Fig 2. In addition, in silico analyses confirmed that these virulence genes were located on IncFIB plasmid.
Fig 2. Genomic comparison between genetic contexts of virulence plasmids carried by Salmonella Typhimurium strains from this study (A) and S. enterica strains B (CP000858), C (NC_002638), and D (AY517905) as out-group.
Genes and shotgun sequences were retrieved from the GenBank database. Arrows indicate the positions and directions of the genes; Regions with >99% identity are indicated with gray shading.
To achieve a better understanding of the clonal spread of these MDR strains, we reconstructed a phylogenetic tree based on SNPs. Indeed, these strains were found to be genetically related. The phylogenetic tree framed a major cluster composed by five S. Typhimurium strains from this study (HS5478, HS5344, HS5437, HS5302, and HS5368), which nested together with S. enterica strains from different sources (Human, poultry, ovine) and countries, including Denmark (SAMEA4349586), Scotland (SAMEA773504 and SAMEA773551), France (SAMN07734943), and China (SAMN09759463) as shown in Fig 3. Interestingly, S. Typhimurium strains within same cluster shared the same resistance phenotype and genotype profile.
Fig 3. SNP-based phylogenetic tree composed by five Salmonella Typhimurium and additional 14 Salmonella enterica strains.
This figure was generated with iTOL v.5.5 (https://itol.embl.de).
Discussion
The continuous dispersal of MDR S. enterica strains frequently deserves attention of the public health authorities, particularly the international lineages as S. Typhimurium ST19 that most often causes diseases and deaths [18, 19]. Owing to their importance, the ST19 members have been globally identified in a variety of sources, such as human clinical samples, animals, food products, and environmental samples [20–22]. Moreover, S. Typhimurium ST19 has shown broad resistance to a variety of critically important antimicrobials [23], including colistin (an antibiotic of last resort for some MDR infections) [24, 25]. Besides that, the occurrence of intermediate resistance reported here implies in possible treatment failure that should be noted by public health authorities.
It is important to note that these strains can easily acquire such genes through mobile genetic elements such as plasmids, integrons, and genomic islands from other MDR clones, resulting in their rapid dissemination. The presence of class 1 integron in all S. Typhimurium ST19 strains constitutes a risk factor to the rapid spread of antimicrobial resistance (AMR) genes. Indeed, class 1 integron coding various resistance profiles has been widely reported in S. Typhimurium as well as in multiple serovars [5, 21, 26–30]. This genetic frame is crucial for the spread resistance markers, since they are able to capture AMR genes through chromosomal cassettes incorporating them by site-specific recombination [4, 12, 31]. Additionally, resistance genes located in class 1 integrons are often within Salmonella genomic islands (SGI), such as the conjugative Salmonella genomic island 1 (SGI1) (~43-kb) and its variants [32, 33].
The detection of quaternary ammonium compounds (QACs) raises a particular concern, since this qac-containing integrons typically harbors a pool of genes that are hotspots for horizontal gene transfer providing readily adaptation to new surrounds [34, 35]. The co-resistance of critically important antimicrobials and disinfectants QACs reinforces the evidence of the overuse of biocides in clinical settings [34], and their spread have been also described in Salmonella serotypes isolated from livestock [36].
The blaCARB-2 gene, earlier identified as blaPSE-1, is most often a part of the chromosomal cassette [37, 38]. To date, the occurrence of this carbenicillinase gene has been limited to a few reports in different bacteria species and countries, including Acinetobacter pittii and Salmonella serovars in Australia [38, 39], Salmonella Typhimurium from England and Wales [40], Salmonella Senftenberg in Mexico [41], S. Typhimurium in Canada [42], Pseudomonas aeruginosa in Netherlands [43], and Escherichia coli in Pakistan [44]. It is noteworthy that such genetic element has the ability to move among different lineages of S. enterica serovars on a global scale, contributing to AMR spread [28]. Indeed, the genetic contexts surrounding blaCARB-2 gene in this study are typically found in SGI1 and its variant SGI1-B.
Drug-resistant variants of SGI1 have been identified in numerous S. enterica serovars, and strains harboring them may be more virulent and have a tendency to rapidly disseminate [33, 39]. In fact, S. Typhimurium strains within this survey demonstrate to possess several virulence factors, which have been reported earlier [45–48]. Furthermore, we confirmed the presence of several plasmid-borne virulence genes (spvR, spvA, spvB, spvC, spvD, rck, and pefA) that denotes an important genomic apparatus for the spreading of this plasmid, and may provide fitness benefit as previously reported [28, 49, 50]. Increasing evidences have demonstrated that the spv operon affects the formation of autophagosomes, as well as highlight its association in killing of macrophages and neutrophils [6], being crucially required for virulence in vivo [8], including aggravated damage in zebrafish infection model [7]. Furthermore, the PhoP-regulated gene mig-14 that is required for virulence and resistance to antimicrobial peptides was detected in these strains. Yet, mig-14 contributes to Salmonella persistence in hosts, being also associated with resistance against polymyxin B and cathelin-related antimicrobial peptide (CRAMP) [51–54]. Thus, the clonal dissemination of MDR S. Typhimurium (mostly the invasive clones) constitutes an important issue to public health [55], especially S. Typhimurium ST19, which have been circulating worldwide (http://enterobase.warwick.ac.uk/) as demonstrated in this study, since our S. Typhimurium strains nested with international lineages from at least four countries (Fig 2).
In summary, we report the genomic features of virulent and MDR S. Typhimurium ST19 strains carrying the blaCARB-2 gene recovered from clinical human samples in United States. Our results provide new insights into this genetic environment that besides blaCARB-2, contains genes, coding resistance to quaternary ammonium compounds (qacEΔ1) and sulfonamides (sul1). Furthermore, our findings could aid in understanding the epidemiology of S. Typhimurium ST19, which are of great value to initiate preventive measures to safeguard human health. Given the high spread of this international lineage, especially among the young and the elderly or immunocompromised people, public health authorities and regulatory food agencies need to be aware of the potential impact in public health and in economy caused by such pandemic MDR S. Typhimurium ST19 lineage, with particular attention in high-burden areas.
Acknowledgments
We thank Lyndy Harden for generating the whole genome sequence profiles for the study.
Data Availability
All relevant data are within the paper file.
Funding Statement
The whole genome sequencing work is supported by the National Institutes of Health/Food and Drug Administration under award number 5U 18FD006194-02. NO.
References
- 1.Campos J, Mourão J, Peixe L, Antunes P. Non-typhoidal Salmonella in the Pig Production Chain: A Comprehensive Analysis of Its Impact on Human Health. Pathogens. 2019; 8(1):E19 10.3390/pathogens8010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida F, Seribelli AA, Medeiros MIC, Rodrigues DDP, de Mello Varani A, Luo Y, et al. Phylogenetic and antimicrobial resistance gene analysis of Salmonella Typhimurium strains isolated in Brazil by whole genome sequencing. PLoS One. 2018; 13(8):e0201882 10.1371/journal.pone.0201882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monte DF, Lincopan N, Fedorka-Cray P, Landgraf M. Current Insights on High Priority Antibiotic-Resistant Salmonella enterica in Food and Foodstuffs: A review. Curr Opin Food Sci. 2019; 26:35–46. 10.1016/j.cofs.2019.03.004. [DOI] [Google Scholar]
- 4.Ghaly TM, Chow L, Asher AJ, Waldron LS, Gillings MR. Evolution of class 1 integrons: Mobilization and dispersal via food-borne bacteria. PLoS One. 2017; 12(6):e0179169 10.1371/journal.pone.0179169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pornsukarom S, Thakur S. Horizontal Dissemination of Antimicrobial Resistance Determinants in Multiple Salmonella Serotypes following Isolation from the Commercial Swine Operation Environment after Manure Application. Appl Environ Microbiol. 2017; 83(20):e01503–17. 10.1128/AEM.01503-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu SY, Wang LD, Li JL, Xu GM, He ML, Li YY, et al. Salmonella spv locus suppresses host innate immune responses to bacterial infection. Fish Shellfish Immunol. 2016; 58:387–396. 10.1016/j.fsi.2016.09.042. [DOI] [PubMed] [Google Scholar]
- 7.Wu SY, Wang LD, Xu GM, Yang SD, Deng QF, Li YY, et al. spv locus aggravates Salmonella infection of zebrafish adult by inducing Th1/Th2 shift to Th2 polarization. Fish Shellfish Immunol. 2017; 67:684–691. 10.1016/j.fsi.2017.06.057. [DOI] [PubMed] [Google Scholar]
- 8.Passaris I, Cambré A, Govers SK, Aertsen A. Bimodal Expression of the Salmonella Typhimurium spv Operon. Genetics. 2018; 210(2):621–635. 10.1534/genetics.118.300822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2019. Performance standards for antimicrobial susceptibility testing. 29th ed. CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Clinical Laboratory Standards Institute. 2015. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. 3rd ed. CLSI Supplement VET01S. CLSI, Wayne, PA.
- 11.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012; 18(3):268–81. 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 12.Gebreyes WA, Thakur S, Davies PR, Funk JA, Altier C. Trends in antimicrobial resistance, phage types and integrons among Salmonella serotypes from pigs, 1997–2000. J Antimicrob Chemother. 2004; 53(6):997–1003. 10.1093/jac/dkh247. [DOI] [PubMed] [Google Scholar]
- 13.Rao S, Maddox CW, Hoien-Dalen P, Lanka S, Weigel RM. Diagnostic accuracy of class 1 integron PCR method in detection of antibiotic resistance in Salmonella isolates from swine production systems. J Clin Microbiol. 2008; 46(3):916–20. 10.1128/JCM.01597-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pornsukarom S, van Vliet AHM, Thakur S. Whole genome sequencing analysis of multiple Salmonella serovars provides insights into phylogenetic relatedness, antimicrobial resistance, and virulence markers across humans, food animals and agriculture environmental sources. BMC Genomics. 2018; 19(1):801 10.1186/s12864-018-5137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monte DF, Nelson V, Cerdeira L, Keelara S, Greene S, Griffin D, et al. Multidrug- and colistin-resistant Salmonella enterica 4,[5],12:i:- sequence type 34 carrying the mcr-3.1 gene on the IncHI2 plasmid recovered from a human. J Med Microbiol. 2019; 68(7):986–990. 10.1099/jmm.0.001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019; 47(D1):D687–D692. 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One. 2014; 9(8):e104984 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization. 2015. WHO estimates of the global burden of foodborne diseases, 2007–2015 WHO, Geneva, Switzerland.
- 19.Carroll LM, Wiedmann M, den Bakker H, Siler J, Warchocki S, Kent D, et al. Whole-Genome Sequencing of Drug-Resistant Salmonella enterica Isolates from Dairy Cattle and Humans in New York and Washington States Reveals Source and Geographic Associations. Appl Environ Microbiol. 2017; 83(12): e00140–17. 10.1128/AEM.00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gymoese P, Sørensen G, Litrup E, Olsen JE, Nielsen EM, Torpdahl M. Investigation of Outbreaks of Salmonella enterica Serovar Typhimurium and Its Monophasic Variants Using Whole-Genome Sequencing, Denmark. Emerg Infect Dis. 2017; 23(10):1631–1639. 10.3201/eid2310.161248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain P, Sudhanthirakodi S, Chowdhury G, Joshi S, Anandan S, Ray U, et al. Antimicrobial resistance, plasmid, virulence, multilocus sequence typing and pulsed-field gel electrophoresis profiles of Salmonella enterica serovar Typhimurium clinical and environmental isolates from India. PLoS One. 2018; 13(12):e0207954 10.1371/journal.pone.0207954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panzenhagen PHN, Paul NC, Conte CA Jr, Costa RG, Rodrigues DP, Shah DH. Genetically distinct lineages of Salmonella Typhimurium ST313 and ST19 are present in Brazil. Int J Med Microbiol. 2018; 308(2):306–316. 10.1016/j.ijmm.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Critically important antimicrobials for human medicine– 5th rev. Geneva: World Health Organization. 2017; License: CC BY-NC-SA 3.0 IGO.
- 24.Rau RB, de Lima-Morales D, Wink PL, Ribeiro AR, Martins AF, Barth AL. Emergence of mcr-1 Producing Salmonella enterica serovar Typhimurium from Retail Meat: First Detection in Brazil. Foodborne Pathog Dis. 2018; 15(1):58–59. 10.1089/fpd.2017.2346. [DOI] [PubMed] [Google Scholar]
- 25.Rau RB, de Lima-Morales D, Wink PL, Ribeiro AR, Barth AL. Salmonella enterica mcr-1 Positive from Food in Brazil: Detection and Characterization. Foodborne Pathog Dis. 2020; 17(3):202–208. 10.1089/fpd.2019.2700. [DOI] [PubMed] [Google Scholar]
- 26.Keelara S, Scott HM, Morrow WM, Hartley CS, Griffin DL, Gebreyes WA, et al. Comparative phenotypic and genotypic characterization of temporally related nontyphoidal Salmonella isolated from human clinical cases, pigs, and the environment in North Carolina. Foodborne Pathog Dis. 2014; 11(2):156–64. 10.1089/fpd.2013.1630. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed HA, El-Hofy FI, Shafik SM, Abdelrahman MA, Elsaid GA. Characterization of virulence-associated genes, antimicrobial resistance genes and Class 1 integrons in Salmonella enterica serovar Typhimurium isolates from chicken meat and humans in Egypt. Foodborne Pathog Dis. 2016; 13:281–288. 10.1089/fpd.2015.2097. [DOI] [PubMed] [Google Scholar]
- 28.Lopes GV, Michael GB, Cardoso M, Schwarz S. Antimicrobial resistance and class 1 integron-associated gene cassettes in Salmonella enterica serovar Typhimurium isolated from pigs at slaughter and abattoir environment. Vet Microbiol. 2016; 194:84–92. 10.1016/j.vetmic.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, Ju Z, Yang Y, Zhao X, Jiang Z, Sun S. Serotype, antimicrobial susceptibility and genotype profiles of Salmonella isolated from duck farms and a slaughterhouse in Shandong province, China. BMC Microbiol. 2019; 19(1):202 10.1186/s12866-019-1570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siriken B, Al G, Erol I. Prevalence and Antibiotic Resistance of Salmonella Enteritidis and Salmonella Typhimurium in Ground Beef and Meatball Samples in Samsun, Turkey. Microb Drug Resist. 2020; 26(2):136–144. 10.1089/mdr.2018.0481. [DOI] [PubMed] [Google Scholar]
- 31.Argüello H, Guerra B, Rodríguez I, Rubio P, Carvajal A. Characterization of Antimicrobial Resistance Determinants and Class 1 and Class 2 Integrons in Salmonella enterica spp., Multidrug-Resistant Isolates from Pigs. Genes (Basel). 2018; 9(5): E256 10.3390/genes9050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doublet B, Boyd D, Mulvey MR, Cloeckaert A. The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol. 2005; 55(6):1911–24. 10.1111/j.1365-2958.2005.04520.x. [DOI] [PubMed] [Google Scholar]
- 33.Mulvey MR, Boyd DA, Olson AB, Doublet B, Cloeckaert A. The genetics of Salmonella genomic island 1. Microbes Infect. 2006; 8(7):1915–22. 10.1016/j.micinf.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 34.Hegstad K, Langsrud S, Lunestad BT, Scheie AA, Sunde M, Yazdankhah SP. Does the wide use of quaternary ammonium compounds enhance the selection and spread of antimicrobial resistance and thus threaten our health? Microb Drug Resist. 2010; 16(2):91–104. 10.1089/mdr.2009.0120. [DOI] [PubMed] [Google Scholar]
- 35.Rajamohan G, Srinivasan VB, Gebreyes WA. Biocide-tolerant multidrug-resistant Acinetobacter baumannii clinical strains are associated with higher biofilm formation. J Hosp Infect. 2009; 73(3):287–289. 10.1016/j.jhin.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 36.de Quadros CL, Manto L, Mistura E, Webber B, Ritterbusch GA, Borges KA, et al. Antimicrobial and Disinfectant Susceptibility of Salmonella Serotypes Isolated from Swine Slaughterhouses. Curr Microbiol. 2020; 77(6):1035–1042. 10.1007/s00284-020-01904-9. [DOI] [PubMed] [Google Scholar]
- 37.Huovinen P, Jacoby GA. Sequence of the PSE-1 beta-lactamase gene. Antimicrob Agents Chemother. 1991; 35:2428–2430. 10.1128/aac.35.11.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamolvit W, Derrington P, Paterson DL, Sidjabat HE. A case of IMP-4-, OXA-421-, OXA-96-, and CARB-2-producing Acinetobacter pittii sequence type 119 in Australia. J Clin Microbiol. 2015; 53(2):727–30. 10.1128/JCM.02726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levings RS, Lightfoot D, Partridge SR, Hall RM, Djordjevic SP. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J Bacteriol. 2005; 187(13):4401–9. 10.1128/JB.187.13.4401-4409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker RA, Lindsay E, Woodward MJ, Ward LR, Threlfall EJ. Variation in clonality and antibiotic-resistance genes among multiresistant Salmonella enterica serotype Typhimurium phage-type U302 (MR U302) from humans, animals, and foods. Microb Drug Resist. 2001; 7(1):13–21. 10.1089/107662901750152701. [DOI] [PubMed] [Google Scholar]
- 41.Delgado-Suárez EJ, Ortíz-López R, Gebreyes WA, Allard MW, Barona-Gómez F, Rubio-Lozano MS. Genomic surveillance links livestock production with the emergence and spread of multi-drug resistant non-typhoidal Salmonella in Mexico. J Microbiol. 2019; 57(4):271–280. 10.1007/s12275-019-8421-3. [DOI] [PubMed] [Google Scholar]
- 42.Ng LK, Mulvey MR, Martin I, Peters GA, Johnson W. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob Agents Chemother. 1999; 43(12):3018–21. 10.1128/AAC.43.12.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Zee A, Kraak WB, Burggraaf A, Goessens WHF, Pirovano W, Ossewaarde JM, et al. Spread of Carbapenem Resistance by Transposition and Conjugation Among Pseudomonas aeruginosa. Front Microbiol. 2018; 9:2057 10.3389/fmicb.2018.02057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohsin M, Azam M, Ur Rahman S, Esposito F, Sellera FP, Monte DF, et al. Genomic background of a colistin-resistant and highly virulent MCR-1-positive Escherichia coli ST6395 from a broiler chicken in Pakistan. Pathog Dis. 2019; 77(7):ftz064 10.1093/femspd/ftz064. [DOI] [PubMed] [Google Scholar]
- 45.Marcus SL, Brumell JH, Pfeifer CG, Finlay BB. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2000; 2:145–56, 10.1016/S1286-4579(00)00273-2. [DOI] [PubMed] [Google Scholar]
- 46.Tang T, Cheng A, Wang M, Li X. Reviews in Salmonella Typhimurium PhoP/PhoQ two-component regulatory system. Rev Med Microbiol. 2013; 24:18–21, 10.1097/MRM.0b013e32835a9490. [DOI] [Google Scholar]
- 47.Abd El Ghany M, Shi X, Li Y, Ansari HR, Hill-Cawthorne GA, Ho YS, et al. Genomic and Phenotypic Analyses Reveal the Emergence of an Atypical Salmonella enterica Serovar Senftenberg Variant in China. J Clin Microbiol. 2016; 54(8):2014–22. 10.1128/JCM.00052-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monte DF, Lincopan N, Berman H, Cerdeira L, Keelara S, Thakur S, et al. Genomic Features of High-Priority Salmonella enterica Serovars Circulating in the Food Production Chain, Brazil, 2000–2016. Sci Rep. 2019; 9(1):11058 10.1038/s41598-019-45838-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guerra B, Soto S, Helmuth R, Mendoza MC. Characterization of a self-transferable plasmid from Salmonella enterica serotype Typhimurium clinical isolates carrying two integron-borne gene cassettes together with virulence and drug resistance genes. Antimicrob Agents Chemother. 2002; 46(9):2977–81. 10.1128/aac.46.9.2977-2981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009; 53(6):2227–38. 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Navarre WW, Halsey TA, Walthers D, Frye J, McClelland M, Potter JL, et al. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol Microbiol. 2005; 56(2):492–508. 10.1111/j.1365-2958.2005.04553.x. [DOI] [PubMed] [Google Scholar]
- 52.Brodsky IE, Ghori N, Falkow S, Monack D. Mig-14 is an inner membrane-associated protein that promotes Salmonella Typhimurium resistance to CRAMP, survival within activated macrophages and persistent infection. Mol Microbiol. 2005; 55(3):954–72. 10.1111/j.1365-2958.2004.04444.x. [DOI] [PubMed] [Google Scholar]
- 53.Brodsky IE, Ernst RK, Miller SI, Falkow S. mig-14 is a Salmonella gene that plays a role in bacterial resistance to antimicrobial peptides. J Bacteriol. 2002; 184(12):3203–13. 10.1128/jb.184.12.3203-3213.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheng X, Wang W, Chen L, et al. Mig-14 may contribute to Salmonella enterica serovar Typhi resistance to polymyxin B by decreasing the permeability of the outer-membrane and promoting the formation of biofilm. Int J Med Microbiol. 2019; 309(2):143–150. 10.1016/j.ijmm.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 55.GBD 2017 Non-Typhoidal Salmonella Invasive Disease Collaborators. The global burden of non-typhoidal Salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis. 2019; 19(12):1312–1324. 10.1016/S1473-3099(19)30418-9. [DOI] [PMC free article] [PubMed] [Google Scholar]