Abstract
Some monoclonal antibodies undergo liquid–liquid phase separation owing to self-attractive associations involving electrostatic and other soft interactions, thereby rendering monoclonal antibodies unsuitable as therapeutics. To mitigate the phase separation, formulation optimization is often performed. However, this is sometimes unsuccessful because of the limited time for the development of therapeutic antibodies. Thus, protein mutations with appropriate design are required. In this report, we describe a case study involving the design of mutants of negatively charged surface residues to reduce liquid–liquid phase separation propensity. Physicochemical analysis of the resulting mutants demonstrated the mutual correlation between the sign of second virial coefficient B2, the Fab dipole moment, and the reduction of liquid–liquid phase separation propensity. Moreover, both the magnitude and direction of the dipole moment appeared to be essential for liquid–liquid phase separation propensity, where electrostatic interaction was the dominant mechanism. These findings could contribute to a better design of mutants with reduced liquid–liquid phase separation propensity and improved drug-like biophysical properties.
Introduction
The number and variety of monoclonal antibodies (mAbs) approved for clinical trials and commercial release have been increasing continuously in recent decades [1–3]. However, unlike small-molecule drugs, manufacturing mAbs is challenging because their substantial complexity and labile conformational stability induce disorder in their higher-order structures. MAbs can be exposed to various stresses during their manufacturing processes, formulation, storage, delivery, and administration, which can result in undesirable fragmentation, aggregation, denaturation, and chemical modifications of the mAbs [4–6]. These alterations can affect the manufacturability as well as the efficacy and safety of therapeutic mAbs.
Therefore, all mAb drug candidates should undergo characterization of their physicochemical properties in order to mitigate the risks of significant instability, and this should be performed at the front end of the development process or during a discovery phase to reduce drug development costs [7, 8]. One of the key risks is the loss of colloidal stability, specifically undesirable liquid–liquid phase separation (LLPS) and increased viscosity in highly concentrated solutions [9–12]. The LLPS of mAbs could result in a poor appearance and inconsistent vial-to-vial concentrations [9].
The effect of chemical additives on formulation is systematically organized and well understood [13]. In fact, several formulation studies have reported mitigation of the LLPS propensity of mAbs [9, 14–18]. For example, adding sucrose or hydroxypropyl beta cyclodextrin [15], adding arginine glutamate [17], or adding salt or increasing the temperature mainly by decreasing protein–protein self-attractive interactions [18] mitigates the LLPS propensity of mAbs. Formulation studies by Reiche et al. [9] also showed that the overall hydrophobicity and charge distribution of proteins play a role in LLPS of mAbs. However, in our experience, such formulation studies for protein therapeutic candidates do not always work well within the limited time typically given by pharmaceutical companies for drug development.
Instead, the optimization of mAb sequence through protein engineering technology is an attractive approach for mitigating the LLPS propensity of mAbs because the molecular optimization could dramatically change it. Engineering can also be implemented in the early phase of drug development using very small amounts of mAbs. The effective in silico design of mAbs has previously been reported to reduce the risk of viscosity escalation [19–23]. In terms of LLPS, Du et al [24] successfully used in silico homology modelling and charged patch analysis to identify seven amino acids that were potentially related to self-association. They confirmed that mutations of the amino acids resulted in a lower LLPS propensity. Chow et al. [25] indicated that the disruption of the charged surface patch of an immunoglobulin (Ig) G4 suppressed its LLPS propensity. In their study, the variable region (Fv) dipole moment and the diffusion interaction parameter kD were qualitatively correlated with the LLPS propensity of a wild type (WT) and its four mutant mAbs.
In this study, we performed protein engineering to reduce the LLPS propensity of a humanized IgG1, mAb1, and showing strong LLPS propensity. mAb1 mutants were designed with the goal of eliminating the negatively charged surface patch. The physicochemical properties of the resultant clones were characterized, and their structures were analysed using homology models. The findings revealed that both the magnitude and direction of the antigen-binding fragment (Fab) dipole moment were essential contributors to LLPS propensity.
Materials and methods
MAb production
Wild type (WT) mAb1 and its mutants M1–M9 were produced by HEK293 cells and purified by successive two-step chromatography [26] with MAbSelect SuRe affinity resin (GE Healthcare, Chicago, IL, USA) and ceramic hydroxyapatite type I resin (Bio-Rad Laboratories, Hercules, CA, USA). The mAb concentrations were determined by size exclusion-high performance liquid chromatography (SE-HPLC) with absorbance detection at 280 nm. Molar extinction coefficients were calculated using Sednterp software version 1.09 (National Institutes of Health, Bethesda, MD, USA). Sodium chloride was purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan), and sodium phosphate and sodium acetate were obtained from Kanto Chemical Co., Inc. (Tokyo, Japan).
Solution preparation
We dialyzed mAbs against buffer A (25 mM sodium phosphate/15 mM NaCl/pH 6.0) or buffer B (25 mM sodium phosphate/150 mM NaCl/pH 6.0). We used mAbs in buffer A for all measurements and used its pH and ionic strength as parameters for in silico analyses, as described in the Analysis of full IgG and Fab model structures subsection. We also used mAbs in buffer B for dynamic light scattering measurements.
Analysis of full IgG and Fab model structures
Model structures of the full-length and antigen-binding fragment (Fab) of the WT mAb1 and its mutants were generated using the MODELLER implementation in Discovery studio 2017R2 software [27] (Dassault Systemes, Vélizy-Villacoublay, France) using crystal structures of an antibody (PDB ID, 1HZH) or a Fab (PDB ID, 5I19) as the template, respectively. The electrostatic potential of the surfaces was determined according to the Poisson–Boltzmann equation at pH 6.0 at an ionic strength of 43 mM, which corresponds to the ionic strength of buffer A (25 mM sodium phosphate/15 mM NaCl/pH 6.0) used for in vitro experiments. For force field assignment, the charge and bondi radii were applied as parameters. The +1 and −1 kBT/e isovalue surfaces were mapped onto the model structures, where kB is the Boltzmann constant. The angle θ (degree) of the Fab dipole moment between the WT and each mutant (Mi) was calculated with the component (Dipole X, Y, or Z) of each Fab dipole moment (S1 Table) according to Eq (1):
| (1) |
Isoelectric focusing (IEF)
Samples were electrophoresed on PhastGel IEF gels (GE Healthcare Life Sciences) using the PhastSystem electrophoresis workstation (GE Healthcare Life Sciences). An Isoelectric Focusing Calibration Kit High Range pI (pH 5–10.5) (GE Healthcare Life Sciences) was used for marker proteins. The gels were stained with PhastGel Blue R (GE Healthcare Life Sciences). The experiment was performed in one replicate.
Differential scanning calorimetry
Differential scanning calorimetry (DSC) studies were conducted using a VP capillary DSC system (Malvern Panalytical, Malvern, UK). Samples (0.5 mg/mL in buffer A) were heated from 20°C to 100°C at a scanning rate of 200°C/h. For each sample, a relevant buffer blank was subtracted using Origin 7.0 software (OriginLab Corporation, Northampton, MA, USA). The apparent Tm was determined from the exothermic peak that provided the highest heat capacity (CP.) The experiment was performed in one replicate.
Phase separation experiments
The WT and mutants M1–M9 were concentrated to approximately 80 mg/mL and dialyzed against buffer A using an Xpress Micro Dialyzer MD 100 12–14 kDa cartridge (Scienova GmbH, Jena, Germany). The concentration of each dialysate was adjusted to 50 mg/mL by adding buffer A. The resulting solutions were transferred to 200-μL polymerase chain reaction (PCR) tubes (MicroAmp 8-cap strip; Thermo Fisher Scientific, Waltham, MA, USA) and stored at 4°C for 1–4 days to allow LLPS to occur. The samples in PCR tubes were mixed by gentle inversion, the temperature was raised from 4°C to 10°C, and the samples were stored overnight at 10°C. We repeated the 4°C LLPS experiments for M8 and M9 at a larger scale of 600 μL in 1.5-mL clear tubes, with WT as a reference. The tubes were incubated overnight at 4°C and then centrifuged for 48 h at 21,600 ×g at 4°C. Because of limited sample availability, the experiment was performed in one replicate.
Viscosity measurement
The mAbs in buffer A were concentrated to 130–140 mg/mL using a Vivapore 5 Static Concentrator (Sartorius Stedim Biotech GmbH, Göttingen, Germany), and the concentration was adjusted to 120 mg/mL with buffer A. The viscosity of the mAb solution was measured at 25°C using an m-VROC micro-viscometer (RheoSense Inc., San Ramon, CA, USA). Because of limited sample availability, all measurements were performed in one replicate.
Analytical ultracentrifugation-sedimentation equilibrium
Other than the following, all procedures were performed exactly as described in previous studies [28, 29]. For analytical ultracentrifugation-sedimentation equilibrium experiments, 97 μL of each mAb solution (1, 5, and 10 mg/mL) in buffer A was centrifuged in a Proteome XL-I instrument (Beckman Coulter, Inc., Brea, CA, USA). The partial specific volumes were calculated as 0.7291 (M1), 0.7291 (M2), 0.7291 (M3), 0.7293 (M4), 0.7293 (M5), 0.7293 (M6), 0.7295 (M7), 0.7292 (M8), 0.7292 (M9), and 0.7289 (WT), cm3/g using Sednterp software. We determined apparent molecular weights by nonlinear least-squares fitting using Origin software (OriginLab Corporation, Northampton, MA, USA). B2 was obtained from the slope of the plot between the inverse of the apparent molecular weight and protein concentration, as shown by Saito et al. [29, 30]. Because of limited sample availability, the experiment was performed in one replicate.
Dynamic light scattering
The mAb samples at concentrations of 2.5, 5.0, 7.5, and 10.0 mg/mL (after filtration through a 0.22-μm filter) in buffer A or buffer B (25 mM sodium phosphate/150 mM NaCl/pH 6.0) were plated into wells of 384-well optically transparent-bottom microtiter plates. The diffusion coefficient Ds was measured using a DynaPro Plate Reader version II (Wyatt Technology Corp., Santa Barbara, CA, USA) at 20°C. The diffusion interaction parameter kD was calculated using the protocol described in a previous paper [29]. The experiment was performed in duplicate.
PEG-mediated relative solubility
Polyethylene glycol (PEG)-6000 solutions (0.5%–18.5% w/v) at 2% increments in buffer A were prepared from 50% concentrated PEG-6000 stock solutions. The mAb samples were diluted to 1 mg/mL in buffer A, and 12 μL of the mAb samples and 28 μL of PEG solution were mixed, followed by filtration 20 min later. The absorbance at 280 nm was then measured. The PEGm, i.e., the weight% of PEG in solution required to reduce the protein concentration by 50%, was calculated as the relative solubility [31]. The experiment was performed in duplicate.
Results
Physicochemical analysis of WT mAb1
The WT mAb1 underwent LLPS at 50 mg/mL at 4°C in buffer A and a relatively high viscosity of 15.43 mPa·s at a concentration of 120 mg/mL (Table 1). The B2 was −4.08 × 10−5 mL·mol/g2 and the kD was −33 mL/g for the WT. The sign for B2 suggests the existence of attractive protein–protein intermolecular interactions [11, 20, 21, 26, 29, 30]. The addition of 150 mM NaCl increased the kD value from −33 mL/g to −12 mL/g, indicating that the protein–protein self–attractive interactions in low–ionic–strength buffer A could essentially be mediated by electrostatic interactions.
Table 1. Biophysical assessments using in silico and in vitro analysis of wild type (WT) and mutant (M) mAbs.
| Name | Mutations | Magnitude of Fab dipole moment (debye) | Angle of Fab dipole moment (°) | LLPS at (4°C) | B2 (×10−5 mL·mol/g2) | kD (mL/g) in buffer A | kD (mL/g) in buffer B | PEGm (%) | Viscosity (mPa·s) | Observed pI |
|---|---|---|---|---|---|---|---|---|---|---|
| WT | 933.0 | - | Detected | −4.08 | −33 | −12 | 5.62 | 15.43 | 8.17 | |
| M1 | LC: E27A | 939.7 | 1.84 | Detected | −2.23 | −22 | −12 | 7.62 | 13.17 | 8.38 |
| M2 | LC: D28A | 848.2 | 3.03 | Detected | −2.62 | −25 | −14 | 7.49 | 11.93 | 8.44 |
| M3 | LC: D93A | 886.9 | 0.12 | Detected | −1.22 | −20 | −11 | 7.67 | 9.08 | 8.39 |
| M4 | LC: E27A, D28A | 776.9 | 9.50 | Not detected | 0.84 | −19 | −11 | 8.25 | 10.57 | 8.50 |
| M5 | LC: E27A, D93A | 760.1 | 9.30 | Not detected | 2.04 | −14 | −14 | 8.44 | 8.19 | 8.50 |
| M6 | LC: D28A, D93A | 715.1 | 5.65 | Not detected | 1.88 | −15 | −12 | 8.63 | 8.77 | 8.56 |
| M7 | LC: E27A. D28A. D93A | 674.1 | 10.07 | Not detected | 3.60 | −12 | −12 | 9.13 | 9.49 | 8.68 |
| M8 | HC: D31A | 940.6 | 16.05 | Opaque | −3.70 | −32 | −19 | 6.97 | 13.6 | 8.40 |
| M9 | LC: D56A | 969.8 | 7.08 | Opaque | −2.60 | −23 | −12 | 7.34 | 9.6 | 8.39 |
Mutant design
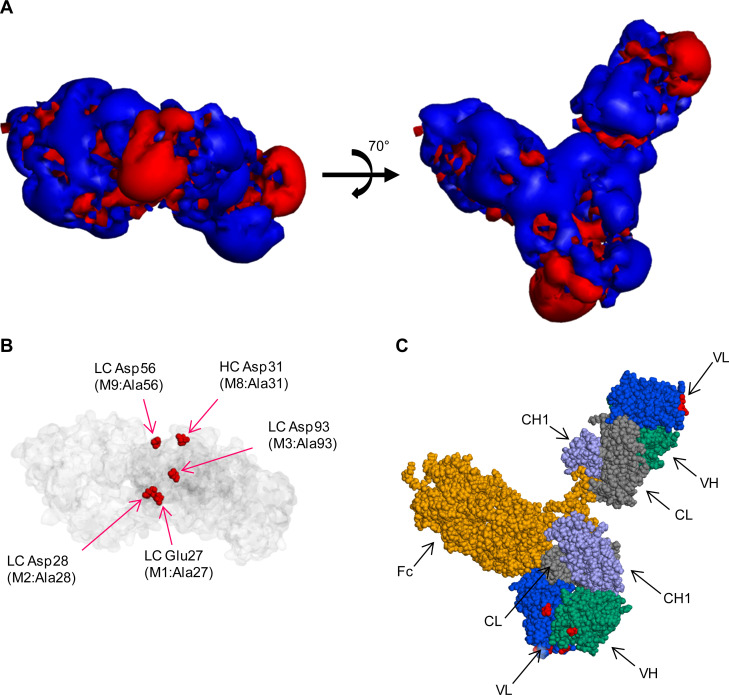
The relationship between the surface charge distribution and the location of negatively charged surface residues was demonstrated on a full-length IgG model of WT mAb1 (Fig 1). The negative charges on the surface of the WT mAb1 were unevenly distributed in the surface electrostatic potential maps (Fig 1A), which was previously believed to induce protein self-interactions and high viscosity [19, 20]. As shown in Fig 1C, mAb has two Fabs and an Fc; the Fab is composed of a constant domain (CL and CH1) and an Fv domain (VL and VH). Major negative surface charges were located in both Fv domains of the full-length model. They primarily arose from five acidic amino acid residues: light chain (LC) Glu27, Asp28, Asp56, Asp93, and heavy chain (HC) Asp31. These five negatively charged amino acids were highlighted on the Fv domain facing forward in Fig 1B, while not highlighted on another Fv domain located on the right side. Their positions corresponded to positions of the major negative charge patches in panel A, left. Of these, LC Glu27, Asp28, and Asp93 formed a major continuous patch of negative charges on the molecular surface.
Fig 1. Charge distribution on the surface of WT mAb1.
Three representations of full-length IgG were used. (A) Rotational view of mAb1 isovalue surface. The red and blue contours indicate the −1 kBT/e (negative) and +1 kBT/e (positive) isosurface potentials. (B). Connolly surface (transparent gray color) with Corey–Pauling–Koltun (CPK) representation (red color) of five negatively charged amino acids: light chain (LC) Glu27, LC Asp28, LC Asp93, LC Asp56, and heavy chain (HC) Asp31. (C) CPK representation of the WT, with yellow = Fc, blue = VL, green = VH, light purple = CH1, gray = CL, and red = mutated residues on Fv regions.
We designed combinations of Ala-substituted mutants at the LC amino acids Glu27, Asp28, and Asp93 to disrupt the continuous negative charge patch. Specific designs were single-charge deletion mutants (M1, M2, and M3), double-charge deletion mutants (M4, M5, and M6), and a triple-charge deletion mutant (M7). In addition to these mutants, Ala-substituted mutants for HC Asp31 (M8) and LC Asp56 (M9) were designed.
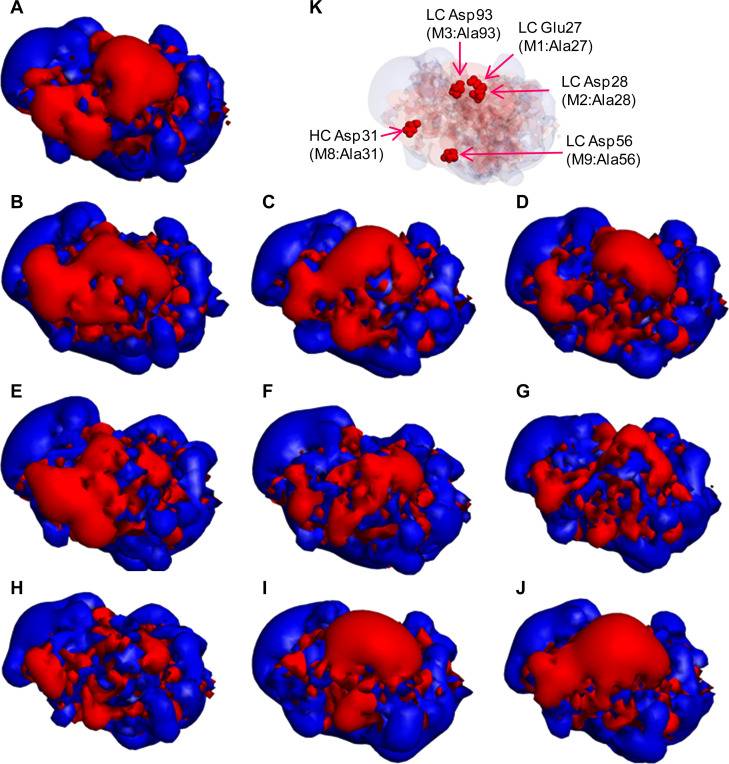
The surface electrostatic potential of the designed Fabs are presented in Fig 2. The apparent sizes of the negatively charged patches were reduced in M1–M7 in proportion to the number of depleted negative charges. In M8 and M9, the apparent sizes of rather small negative charge patches were reduced, while the continuous patch was not reduced, as designed.
Fig 2. Electrostatic potential surfaces of Fabs of WT and mutants.
(A) WT, (B) M1, (C) M2, (D) M3, (E) M4, (F) M5, (G) M6, (H) M7, (I) M8, and (J) M9: The red and blue contours indicate the −1 and +1 kBT/e isosurface potentials. (K) Connolly surface (transparent gray color) with a CPK model (red color) of five negatively charged amino acids: light chain (LC) Glu27, LC Asp28, LC Asp93, LC Asp56, and heavy chain (HC) Asp31.
S1 Fig shows Fab dipole moments. The magnitude of Fab dipole moment was 933 debye for WT, 940–969 debye for single-charge deletion mutants, 715–777 for double-charge deletion mutants, and 674 debye for the triple-charge deletion mutant (Table 1). The angle between the WT and mutant dipole moments for M8 and M9 was larger compared with that for other single-charge deletion mutants: M1, M2, and M3.
Physicochemical analysis of mutants
IEF was performed to confirm the charge deletions of the mutants (Table 1). The isoelectric point (pI) of each mAb was distributed from 8.17 for the WT to 8.68 for M7. The pI values correlated well with the number of charge deletions as designed.
In addition to an alterlation in charges, mutations can often cause drastic changes in the protein structure [32], which might affect LLPS propensity, viscosity, and protein solubility. The structural identities were verified using DSC. Overall, DSC thermograms of the WT and mutants appeared almost identical and had a main peak Tm of ~80°C (S2 Fig), indicating that mutations do not likely affect the conformational stability of proteins.
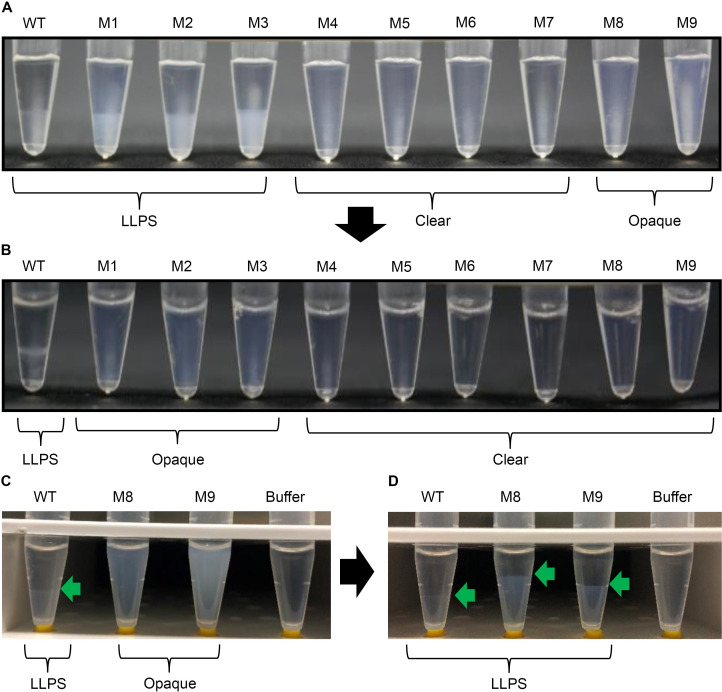
The LLPS propensity of all mAbs at 50 mg/mL in low-ionic-strength buffer A is shown in Fig 3A. Single-charge deletion mutants (M1, M2, and M3) still underwent LLPS at 4°C however, they required more time to undergo LLPS than the WT. The WT underwent LLPS within 3 h of incubation at 4°C, whereas the appearances of mutants M1, M2, and M3 remained cloudy and homogeneous for several hours. These mutants ultimately underwent LLPS following overnight incubation at 4°C. LLPS was clearly mitigated in double- and triple-charge deletion mutants (M4, M5, M6, and M7), even after overnight incubation. M8 and M9 did not undergo clear phase separation at 4°C; however, their solutions were opaque, unlike those of M4–M7. In general, the opaque appearance could be due to both LLPS and fine precipitation.
Fig 3. LLPS of WT and mutants at 4°C.
(A) We added 100 μL of WT and mutant antibodies in 200-μL PCR tubes and stored them overnight at 4°C. (B) The samples were then mixed by gentle inversion at 4°C and stored overnight at 10°C. (C) We added 600 μL of WT, M8, M9, and buffer A in 1.5-mL clear tubes and stored them overnight at 4°C. (D) The samples were centrifuged at 21,600 ×g for 48 h at 4°C. The sample tubes in panels C and D were placed on yellow ferrules (from the 1/8" o.d. tubing connectors of GE Healthcare for AKTA systems) to allow them to stand on the desk.
To explain the opacity of M8 and M9 solutions, we conducted a larger-scale LLPS experiment with centrifugation. M8 and M9 solutions showed opacity after 24 h incubation at 4°C (Fig 3C). Subsequent centrifugation for 48 h at 4°C resulted in clear phase separation for M8 and M9, indicating that the opaque appearances of M8 and M9 solutions are due to LLPS (Fig 3D).
With temperature elevation from 4°C to 10°C, LLPS was maintained in the WT and mitigated in M1, M2, M3, M8, and M9. At 10°C, only M1, M2, and M3 solutions were opaque (Fig 3B), whereas M8 and M9 solutions were clear. Therefore, the LLPS propensity rank order was WT > M1 = M2 = M3 > M9 = M8 > M4 = M5 = M6 = M7. With regard to the lower phase, M8 and M9 solutions in the 1.5-mL tubes (Fig 3D) were clearer than M1, M2, and M3 solutions in PCR tubes (Fig 3A), probably because only M8 and M9 were subject to strong centrifugation for a long time.
The B2 values for these mAbs were determined using the AUC-SE method [28–30] (Table 1). A negative B2 value denotes attractive protein–protein interactions, whereas a positive value denotes mutual repulsion. The B2 values of all single-charge deletion mutants increased from that of the WT; however, their values were still negative. The double and triple-charge deletion mutants, which showed no LLPS, had positive B2 values. The triple-charge deletion mutant M7 had the highest positive B2 value of 3.6 among the tested mAbs.
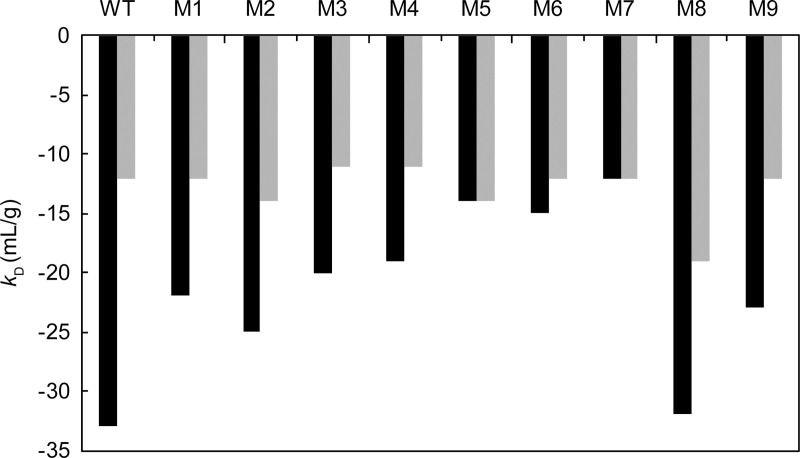
The kD value was determined by dynamic light scattering (Table 1) and was increased in proportion to the number of charge deletions. The kD values were −33 ml/g for WT, −20 to −25 mL/g for the other single-charge deletion mutants, −14 to −10 mL/g for the double-charge deletion mutants, and −12 mL/g for the triple-charge deletion mutant. Adding 150 mM salt increased the kD of the WT and mutants to a range of −11 to −19 mL/g (Fig 4). These results indicated that the deletion of negative charges effectively reduced attractive self-interactions and suggested that with regard to the WT, the electrostatic interaction could play a role as a a part of the soft interaction.
Fig 4. kD values in the presence of 15 mM NaCl and 150 mM NaCl.
Black bars show kD values in buffer A containing 15 mM NaCl. Gray bars show kD values in buffer B containing 150 mM NaCl.
The relative solubility of the mAbs was evaluated by PEG precipitation. The value of PEGm increased proportionally with the number of Ala-substituted acidic residues, which ranged from 5.62% to 9.13%. All mAbs showing LLPS propensity (WT, M1, M2, M3, M8, and M9) had PEGm values of less than 8%.
All the negative surface charge mutations showed reduced viscosity compared with the WT. M5 showed the lowest viscosity of 8.19 mPa·s (S3 Fig).
Discussion
Relationships between the dipole moment, B2, and LLPS
Our findings showed a strong correlation between B2 and the Fab dipole moment of single-, double-, and triple-charge deletion mutants (Fig 5A), indicating that the association can be described as being driven by a Fab dipole–dipole attraction. Kanai et al. [33] described the Fab–Fab interaction for protein–protein self-attractive interactions. Gentiluomo et al. [34] characterized the native reversible self-association of mAbs using Fab and Fc fragments and confirmed that self-associations such as hydrophobic and electrostatic interactions are driven by the Fab fragment. In addition, Du et al. [24] and Chow et al. [25] identified the amino acids responsible for the LLPS propensity of mAbs, located in the Fab region. In this study, we calculated Fab dipole moments to investigate the Fab–dipole-related association. In addition, limited numbers of entire IgG crystal structures are available, making the calculation of the entire IgG dipole moment difficult because the full-length model is only a single snapshot and cannot be interpreted as a representation of multiple hinge-angle forms. The dipole moment of the entire mAb differs in magnitude and direction from the Fab dipole moment. The computational approach we followed using a Fab model structure provided insight into a better design of LLPS mitigation mutants. However, further in vitro analysis, such as X-ray crystal structure analysis, is required to obtain evidence of Fab–Fab interaction mediating protein network formation of the mAb1 heavy phase.
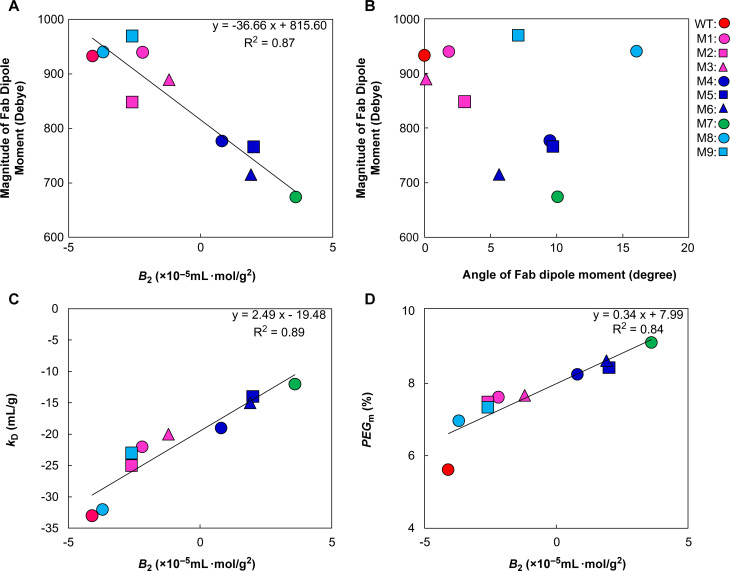
Fig 5. Correlations among various biophysical parameters of WT mAb1 and mutants in buffer A.
(A) The magnitude of the Fab dipole moments was plotted against B2 values; (B) The angle between the WT dipole moment and each mutant dipole moment was plotted against the magnitude of the Fab dipole moment; (C) The kD values were plotted against B2 values; (D) The PEGm values were plotted against B2 values. Least-squares linear regression lines, correlation equations, and correlational coefficients are shown in (A), (C), and (D).
The 10 mAbs were divided into two groups. The first group comprised four mAbs containing double and triple mutants that did not show LLPS propensity and had positive B2. The second group comprised the remaining six mAbs that showed LLPS propensity and had negative B2. Therefore, the sign of B2 could appear as an indicator of the LLPS propensity for mAb1. More precisely, B2 should be normalized to B2* while considering the excluded volume, as shown in Eq (2) [35]. Here, B2ex indicates the contribution of the excluded volume to the virial coefficient. Negative B2* denotes protein–protein self-attractive interactions, while positive B2* denotes mutual repulsion. We are unaware of the actual contribution of B2ex to the sign of B2*, but the sign of B2 appears to be well correlated with LLPS propensity in this study.
| (2) |
The LLPS propensity correlated with B2, and the B2 value had a linear relationship with the dipole moment. While LLPS propensity is a macroscopic phenomenon observed at high protein concentrations, B2 values are measured at relatively low protein concentrations. Accordingly, the dipole moment certainly contributes to B2, even at the high protein concentrations in this study. It is natural that not only the dipole moment but also other soft protein–protein interactions might play a role in determining B2. Saito et al. [36] also indicated a positive correlation between B2 values and viscosity in highly concentrated mAb solutions. They speculated that a similar force component, such as the dipole moment, could work not only at low concentration but also at high concentration. This supported the findings of our study.
Significance of the dipole moment as an index for LLPS mitigation
The LLPS propensity correlated well with the magnitude of the Fab dipole moment, except for the outside-patch mutants M8 and M9. Despite exhibiting a Fab dipole moment of almost the same magnitude as the WT, the outside-patch M8 and M9 mutants did not undergo clear LLPS without centrifugation (Fig 3). The M8 and M9 LLPS propensity rank order could not be interpreted from the magnitude of the Fab dipole moment.
We further investigated the reason why M8 and M9 had lower LLPS propensities by plotting the angle between the WT dipole moment and each mutant dipole moment against the magnitude of the Fab dipole moment (Fig 5B). M8 and M9 had larger angles of the dipole moments than the other three single-charge deletion mutants (M1, M2, and M3). Our study indicated that the dipole moment could play a role in LLPS propensity. It is expected that the angle of Fab dipole moment affects the alignment of the molecule and protein–protein network formation in a crowded heavy phase. Therefore, the weak LLPS propensity of M8 and M9 could be explained by the angle to the WT Fab dipole moment. The results revealed that both the magnitude and the direction of the dipole moment could be essential for the LLPS propensity of mAb1, and the dipole moment could be the referential index for risk mitigation of the LLPS propensity.
Chow et al. [25] used X-ray crystal structure analysis to determine the amino acid residues of mutations that would mitigate the LLPS propensity. In the present study, we designed mutants based on tertiary structural models alone, without laborious in vitro experiments such as crystallization. We also calculated the Fab dipole of 10 mAbs from their primary structure within half a day. This in silico analytical approach could work in conjunction with high-throughput LLPS assessment in the course of drug candidate screening. In addition, the approach provides a reliable protocol for designing a mutant that does not show LLPS propensity in cases where electrostatic interaction is the dominant mechanism for LLPS. It reduces the number of candidate molecules required for in vitro LLPS assessment, thereby greatly improving operational efficiency.
Relationships among various biophysical parameters other than the dipole moment
The kD values strongly correlated with the B2 value (Fig 5C; squared correlation coefficient = 0.89). A kD value of −20 mL/g was a threshold for LLPS where the B2 value indicated zero in this case study. Previous studies [28–30, 35–38] have also demonstrated a linear relationship between the kD and B2 values. The investigations by Connolly et al. [28] indicated this relationship among eight different mAbs and corroborated our results.
All the negative surface charge mutations clearly reduced the viscosity of the WT. Moderate correlation between viscosity and the B2 value was shown with a squared correlation coefficient of 0.58 (S3 Fig). Previous studies assessing viscosity [19, 20] also suggested that the elimination of the uneven distribution of surface charges reduced viscosity. Because both LLPS and viscosity are a result of protein–protein self-attractive interactions, the two phenomena are correlated. LLPS is qualitatively correlated with B2. The viscosity of the WT and mutants is moderately correlated with B2, probably because of the nonlinear dependence of viscosity on the protein concentration. The viscosity of mAbs could be proportional to the exponential function of concentration [39].
The squared correlation coefficient between B2 and PEGm was 0.84, indicating a strong correlation. The excluded volume theory predicts that PEG can trap H2O, thereby sterically excluding proteins from the solvent regions occupied by PEG and causing a phase transition of proteins [40]. Here, the electrostatic interaction could be the major interaction in a low-ionic-strength buffer such as buffer A. The electrostatic interaction also reflected interaction parameters such as B2 for our charge mutants in buffer A. Thus, the surface charge mutants showed correlations of B2 with PEGm. (Fig 5D).
Conclusions
We demonstrated the mitigation of LLPS by deletion of negative charge patches from the surface of the mAb1 Fv region. LLPS propensity was eliminated for the mutants with positive B2 values, with reducing theoretical Fab dipole moments. Therefore, the sign of B2 may be useful for predicting LLPS propensity. Protein–protein soft interactions are complex; however, both the magnitude and the direction of the dipole moment could be part of the essential contributors to the LLPS propensity where the dominant mechanism for LLPS is electrostatic interaction. Further studies are required to apply these findings to other mAbs. The insights obtained in this study could open up new avenues for the design and selection of well-behaved therapeutic mAbs.
Supporting information
(A) WT, (B) M1, (C) M2, (D) M3, (E) M4, (F) M5, (G) M6, (H) M7, (I) M8, and (J) M9. Red and blue contours indicate −1 and +1 kBT/e isosurface potentials. (K) Corresponding transparent isosurface potential depicted next to each model to highlight the dipole moment. The dipole moment is illustrated in both transparent and dense isosurface potential maps. The length does not show the dipole moment magnitude but indicates the scalar amount of the magnitude projected on a plane parallel to the paper.
(TIF)
(TIF)
The correlation equation was determined by least-squares linear regression analysis.
(TIF)
aDipole: X, Dipole: Y, and Dipole: Z are the x-, y-, and z-components of the Fab dipole moment. The angle between the WT dipole moment and each mutant dipole moment was calculated from the Dipole X, Dipole Y, Dipole Z values.
(DOCX)
Acknowledgments
The authors express their sincere gratitude to Prof. Susumu Uchiyama for constructive discussions and contributions to the manuscript. The authors also thank Dr. Masato Amano for helpful discussions and comments on the manuscript. Finally, the authors thank the members of the Protein Production Research Group at Daiichi Sankyo RD Novare Co., Ltd. for the mAb production.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work. Daiichi Sankyo Co., Ltd provided support in the form of salaries for all authors, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Filtz IN, Karow M, Wasserman SM. Evolution and emergence of therapeutic monoclonal antibodies. What cardiologists need to know Circulation. 2013;127:2222–30. 10.1161/CIRCULATIONAHA.113.002033 . [DOI] [PubMed] [Google Scholar]
- 2.Elvin JG, Coulston RG, van der Walle CF. Therapeutic antibodies: Market considerations, disease targets and bioprocessing. Int J Pharm. 2013;440:83–98. 10.1016/j.ijpharm.2011.12.039 . [DOI] [PubMed] [Google Scholar]
- 3.Ecker DM, Jones SD, Levine HL. The therapeutic monoclonal antibody market. mAbs. 2015;7:9–14. 10.4161/19420862.2015.989042 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dengl S, Wehmer M, Hesse F, Lipsmeier F, Popp O, Lang K. Aggregation and chemical modification of monoclonal antibodies under upstream processing conditions. Pharm Res. 2013;30:1380–99. 10.1007/s11095-013-0977-8 . [DOI] [PubMed] [Google Scholar]
- 5.Shire SJ, Shahrokh Z, Liu J. Challenges in the development of high protein concentration formulations. J Pharm Sci. 2004;93:1390–1402. 10.1002/jps.20079 . [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Nguyen MDH, Andya JD, Shire SJ. Reversible self-association increases the viscosity of a concentrated monoclonal antibody in aqueous solution. J Pharm Sci. 2005;94:1928–40. 10.1002/jps.20347 [DOI] [PubMed] [Google Scholar]
- 7.Jarasch A, Koll H, Regula JT, Bader M, Papadimitriou A, Kettenberger H. Developability assessment during the selection of novel therapeutic antibodies. J Pharm Sci. 2015;104:1885–98. 10.1002/jps.24430 . [DOI] [PubMed] [Google Scholar]
- 8.Zurdo J, Arnell A, Obrezanova O, Smith N, de la Cuesta G, Gallagher TRA, et al. Early implementation of QbD in biopharmaceutical development: a practical example. Biomed Res Int. 2015;1–19. 10.1155/2015/605427 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiche K, Hartl J, Blume A, Garidel P. Liquid-liquid phase separation of a monoclonal antibody at low ionic strength: Influence of anion charge and concentration. Biophys Chem. 2017;220:7–19. 10.1016/j.bpc.2016.08.003 . [DOI] [PubMed] [Google Scholar]
- 10.Jiménez M, Rivas G, Minton AP. Quantitative characterization of weak self-association in concentrated solutions of immunoglobulin G via the measurement of sedimentation equilibrium and osmotic pressure. Biochemistry. 2007;46:8373–8378. 10.1021/bi7005515 . [DOI] [PubMed] [Google Scholar]
- 11.Chi EY, Krishnan S, Kendrick BS, Chang BS, Carpenter JF, Randolph TW. Roles of conformational stability and colloidal stability in the aggregation of recombinant human granulocyte colony-stimulating factor. Protein Sci. 2003;12:903–913. 10.1110/ps.0235703 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Liu Y. Recent progresses of understanding the viscosity of concentrated protein solutions. Curr Opin Chem Eng. 2017;16:48–55. 10.1016/j.coche.2017.04.001 .21538214 [DOI] [Google Scholar]
- 13.Timasheff SN. Control of protein stability and reactions by weakly interacting cosolvents: the simplicity of the complicated. Adv Protein Chem. 1998;51:355–432. 10.1016/s0065-3233(08)60656-7 [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Lomakin A, Latypov RF, Benedek GB. Phase separation in solutions of monoclonal antibodies and the effect of human serum albumin. Proc Natl Acad Sci U S A. 2011;108:16606–11. 10.1073/pnas.1112241108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raut AS, Kalonia DS. Effect of excipients on liquid-liquid phase separation and aggregation in dual variable domain immunoglobulin protein solutions. Mol Pharm. 2016;13:774–83. 10.1021/acs.molpharmaceut.5b00668 [DOI] [PubMed] [Google Scholar]
- 16.Casaz P, Boucher E, Wollacott R, Pierce BG, Rivera R, Sedic M. 2014. Resolving self-association of a therapeutic antibody by formulation optimization and molecular approaches. mAbs. 2014;6:1533–1539. 10.4161/19420862.2014.975658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kheddo P, Bramham JE, Dearman RJ, Uddin S, van der Walle CF, Golovanov AP. Investigating liquid-liquid phase separation of a monoclonal antibody using solution-state NMR spectroscopy: effect of Arg·Glu and Arg·HCl. Mol Pharm. 2017;14:2852–60. 10.1021/acs.molpharmaceut.7b00418 . [DOI] [PubMed] [Google Scholar]
- 18.Nishi H, Miyajima M, Nakagami H, Noda M, Uchiyama S. Fukui K. Phase separation of an IgG1 antibody solution under a low ionic strength condition. Pharm Res. 2010;27:1348–60. 10.1007/s11095-010-0125-7 . [DOI] [PubMed] [Google Scholar]
- 19.Yadav S, Laue TM, Kalonia DS, Singh SN, Shire SJ. The influence of charge distribution on self-association and viscosity behavior of monoclonal antibody solutions. Mol Pharm. 2012;9:791–802. 10.1021/mp200566k . [DOI] [PubMed] [Google Scholar]
- 20.Nichols P, Li L, Kumar S, Buck PM, Singh SK, Goswami S, et al. Rational design of viscosity reducing mutants of a monoclonal antibody: hydrophobic versus electrostatic inter-molecular interactions. mAbs. 2015;7:212–30. 10.4161/19420862.2014.985504 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geoghegan JC, Fleming R, Damschroder M, Bishop SM, Sathish HA, Esfandiary R. Mitigation of reversible self-association and viscosity in a human IgG1 monoclonal antibody by rational, structure-guided Fv engineering. mAbs. 2016;8:941–50. 10.1080/19420862.2016.1171444 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomar DS, Singh SK, Li L, Broulidakis MP, Kumar S. In silico prediction of diffusion interaction parameter (kD), a key indicator of antibody solution behaviors. Pharm Res. 2018;35:193 10.1007/s11095-018-2466-6 . [DOI] [PubMed] [Google Scholar]
- 23.Buck PM, Chaudhri A, Kumar S, Singh SK. Highly viscous antibody solutions are a consequence of network formation caused by domain-domain electrostatic complementarities: insights from coarse-grained simulations. Mol Pharm. 2015;12:127–139. 10.1021/mp500485w . [DOI] [PubMed] [Google Scholar]
- 24.Du Q, Damschroder M, Pabst TM, Hunter AK, Wang WT, Luo H. Process optimization and protein engineering mitigated manufacturing challenges of a monoclonal antibody with liquid-liquid phase separation issue by disrupting inter-molecule electrostatic interactions. mAbs. 2019;11:789–802. 10.1080/19420862.2019.1599634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chow CK, Allan BW, Chai Q, Atwell S, Lu J. Therapeutic antibody engineering to improve viscosity and phase separation guided by crystal structure. Mol Pharm. 2016;13:915–23. 10.1021/acs.molpharmaceut.5b00817 . [DOI] [PubMed] [Google Scholar]
- 26.Gagnon P. Monoclonal antibody purification with hydroxyapatite. N Biotechnol. 2009;25:287–93. 10.1016/j.nbt.2009.03.017 . [DOI] [PubMed] [Google Scholar]
- 27.Fasnacht M, Butenhorf K, Goupil-Lamy A, Hemandez-Guzman F, Huang H, Yan L. Automated antibody structure using Accelrys tools: results and best practice. Proteins. 2014:82:1583–98. 10.1002/prot.24604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connolly BD, Petry C, Yadav S, Demeule B, Ciaccio N, Moore JM, et al. Weak interactions govern the viscosity of concentrated antibody solutions: high-throughput analysis using the diffusion interaction parameter. Biophys J. 2012;103:69–78. 10.1016/j.bpj.2012.04.047 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito S, Hasegawa J, Kobayashi N, Kishi N, Uchiyama S, Fukui K. Behavior of monoclonal antibodies: relation between the second virial coefficient (B(2)) at low concentration and aggregation propensity and viscosity at high concentrations. Pharm Res. 2012;29:397–410. 10.1007/s11095-011-0563-x . [DOI] [PubMed] [Google Scholar]
- 30.Saito S, Uchiyama S. Biopharmaceutical evaluation of intermolecular interactions by AUC-SE Analytical Ultracentrifugation. 2016:419–40. 10.1007/978-4-431-55985-6_21 [DOI] [Google Scholar]
- 31.Gibson TJ, McCarty K, McFadyen IJ, Cash E, Dalmonte P, Hinds KD, et al. Application of a high-throughput screening procedure with PEG-induced precipitation to compare relative protein solubility during formulation development with IgG1 monoclonal antibodies. J Pharm Sci. 2011;100:1009–21. 10.1002/jps.22350 . [DOI] [PubMed] [Google Scholar]
- 32.Diskin R, Lebendiker M, Engelberg D, Livnah O. Structures of p38alpha active mutants reveal conformational changes in L16 loop that induce autophosphorylation and activation. J Mol Biol. 2007;365:66–76. 10.1016/j.jmb.2006.08.043 . [DOI] [PubMed] [Google Scholar]
- 33.Kanai S, Liu J, Patapoff TW, Shire SJ. Reversible self-association of a concentrated monoclonal antibody solution mediated by fab-fab interaction that impacts solution viscosity. J Pharm Sci. 2008;97:4219–27. 10.1002/jps.21322 . [DOI] [PubMed] [Google Scholar]
- 34.Gentilumo L, Roessner D, Streicher W, Mahapatra S, Harris P, Freiβ W, J PharmSci. 2020;109:443–51. 10.1016/j.xpha.2019.09.021 . [DOI] [PubMed] [Google Scholar]
- 35.Corbeit D, Bye JW, Curtis RA, Measuring nonspecific protein-protein interactions by dynamic light scattering. Methods Mol Biol. 2019;2039:3–21. 10.1007/978-1-4939-9678-0_1 . [DOI] [PubMed] [Google Scholar]
- 36.Saito S, Hasegawa J, Kobayashi N, Tomitsuka T, Uchiyama S, Fukui K. Effects of ionic strength and sugars on the aggregation propensity of monoclonal antibodies: influence of colloidal and conformational stabilities. Pharm Res. 2013;30:1263–80. 10.1007/s11095-012-0965-4 . [DOI] [PubMed] [Google Scholar]
- 37.Harding SE, Johanson P. The concentration-dependence of macromolecular parameters. Biochem. J. 1985;231:543–547. 10.1042/bj2310543 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lejermayr C, Mahler HC, Mader K, Fischer S. Assessment of net charge and protein-protein interactions of different monoclonal antibodies. J Phar Sci. 2011;100:2551–62. 10.1002/jps.22506 [DOI] [PubMed] [Google Scholar]
- 39.Ross PD, Minton AP. Hard quasispherical model for the viscosity of hemoglobin solutions. Biochem. Biophys. Res. Commun. 10.1016/0006-291x(77)90950-0 [DOI] [PubMed] [Google Scholar]
- 40.Großhans S, Wang G, Hubbuch J. Water on Hydrophobic Surfaces: Mechanistic Modeling of Polyethylene Glycol-Induced Protein Precipitation. Bioprocess Biosyst Eng. 2019;42:513–20. 10.1007/s00449-018-2054-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) WT, (B) M1, (C) M2, (D) M3, (E) M4, (F) M5, (G) M6, (H) M7, (I) M8, and (J) M9. Red and blue contours indicate −1 and +1 kBT/e isosurface potentials. (K) Corresponding transparent isosurface potential depicted next to each model to highlight the dipole moment. The dipole moment is illustrated in both transparent and dense isosurface potential maps. The length does not show the dipole moment magnitude but indicates the scalar amount of the magnitude projected on a plane parallel to the paper.
(TIF)
(TIF)
The correlation equation was determined by least-squares linear regression analysis.
(TIF)
aDipole: X, Dipole: Y, and Dipole: Z are the x-, y-, and z-components of the Fab dipole moment. The angle between the WT dipole moment and each mutant dipole moment was calculated from the Dipole X, Dipole Y, Dipole Z values.
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.