Abstract
When the COVID-19 pandemic began, formal frameworks to collect data about affected patients were lacking. The COVID-19 and Cancer Consortium (CCC19) was formed to collect granular data on patients with cancer and COVID-19 at scale and as rapidly as possible. CCC19 has grown from five initial institutions to 125 institutions with >400 collaborators. More than 5,000 cases with complete baseline data have been accrued. Future directions include increased electronic health record integration for direct data ingestion, expansion to additional domestic and international sites, more intentional patient involvement, and granular analyses of still-unanswered questions related to cancer subtypes and treatments.
When the COVID-19 pandemic began, formal frameworks to collect data about affected patients were lacking. The COVID-19 and Cancer Consortium (CCC19) was formed to collect granular data on patients with cancer and COVID-19 at scale and as rapidly as possible. CCC19 has grown from five initial institutions to 125 institutions with >400 collaborators. More than 5,000 cases with complete baseline data have been accrued. Future directions include increased electronic health record integration for direct data ingestion, expansion to additional domestic and international sites, more intentional patient involvement, and granular analyses of still-unanswered questions related to cancer subtypes and treatments.
Main Text
Introduction
Patients with cancer have a higher risk of infection and subsequent morbidity and mortality because of their generally compromised immune systems. This is also the case for the novel coronavirus, SARS-CoV-2. Patients with cancer are twice as likely to die from this infection compared to the general population (Bakouny et al., 2020). The pandemic has also led to significant disruption in cancer screening, diagnosis, and treatment of cancer, which is anticipated to lead to an indirect increase in morbidity and mortality in this vulnerable population (Bakouny et al., 2020; van de Haar et al., 2020; Schrag et al., 2020). Thus, studying SARS-CoV-2 and its resultant COVID-19 in patients with cancer is highly warranted.
The complex nature of infectious disease research studies in patients with cancer is exacerbated during pandemics, when healthcare personnel are severely challenged by time and energy to collect and enter data in electronic survey instruments. The population of patients with cancer is extremely heterogenous, with differences in types of cancer; numerous treatment regimens; differences in survivorship/time from diagnosis of cancer; and differences in baseline characteristics such as age, gender, race, comorbidities, and other sociodemographic factors. Electronic health records (EHRs) are also not specifically designed for answering cancer- or infection-related questions. They generally lack specific structured fields on infection-specific information (e.g., prior use of antimicrobial treatment or any ongoing secondary prophylaxis, exposure history of infection, onset of symptoms, etc.) and cancer-specific information (e.g., Eastern Cooperative Oncology Group [ECOG] performance status, cancer status, treatment intent, treatment context, etc.). Furthermore, ascertainment of causality is often extremely difficult, and attribution of death to infection or cancer is almost impossible without an autopsy. Serious consideration for epidemiological and statistical challenges such as multicollinearity, measured and unmeasured confounding, interaction between various risk factors, bidirectional effect between infection and cancer, and multiple competing risks for outcomes is essential to prevent false positive and false negative claims and avoid wastage of precious resources during a pandemic. Despite these challenges, carefully designed cross-sectional and longitudinal cohort studies can rapidly answer many questions that prospectively designed clinical trials cannot within a practicable and feasible time frame.
At the beginning of the COVID-19 pandemic, information on the risks posed to patients with cancer was extremely scant (Liang et al., 2020). At the same time, the relatively new technologies of social media platforms and EHRs offered the opportunity to quickly undertake a multi-institutional and international effort to better understand the prognosis of infected patients, with an immediate goal of improving patient care.
Formation of The COVID-19 and Cancer Consortium (CCC19)
Prompted by the need for rapid assessment of clinical impact of COVID-19 in patients with cancer, and to identify and share the best practices to facilitate care during this pandemic, an active conversation took place on Twitter and other social media platforms, using the hashtag #COVID19nCancer. A dynamic discussion ensued, and the COVID-19 and Cancer Consortium (CCC19) was convened on March 15, 2020, by five founding institutions (Desai et al., 2020; Rubinstein et al., 2020). The driving goal and mission statement of CCC19 is “to collect and disseminate prospective, granular, uniformly organized information on people with cancer who are diagnosed with COVID-19—at scale and as rapidly as possible.”
Oversight and Governance Structure
The consortium is governed by a steering committee comprised of members with a diverse clinical and research background in oncology, hematology, viral epidemiology, clinical informatics, and biostatistics. In addition to the steering committee, operational subcommittees include publications (to establish authorship guidelines for projects utilizing CCC19 data and/or resources), funding (to identify sources of funding for the consortium, disseminate this information to consortium members, and assist in the writing and critical revision of grants), epidemiology and biostatistics (to establish guidelines and provide support to investigators in designing and executing studies with the highest rigor, reproducibility, and impact), informatics (to develop and maintain the survey instrument[s] and oversee standardization of the data model, integration of data directly from EHRs, and visualization of data), and patient advocacy (to engage with cancer patient communities and advocacy networks and to coordinate with parties reaching out to CCC19).
Protocol and Website Development
The CCC19 survey was developed to create a de-identified centralized registry housed at Vanderbilt University Medical Center (VUMC), with participation limited to health care professionals or their proxies. No protected health information (PHI), as defined by the Health Insurance Portability and Accountability Act of 1996 (HIPAA), is collected by this centralized registry, which is IRB exempt (VUMC #200467). Participants voluntarily report details about patients with cancer under their direct care or at their institution who have been diagnosed with COVID-19. The survey respondents are anonymous and are not compensated by the consortium. Initially, the survey was open to any anonymous reporter in authorized countries; however, given data quality concerns and an inability to obtain follow-up from fully anonymous participants, eligibility was subsequently restricted to participating sites. Notably, while the site PI is identified to the consortium, the actual survey respondents remain anonymous. The participating sites may or may not implement their own separate IRB approval. Any site participating in CCC19 must execute a data transfer agreement following the standardized Federal Demonstration Partnership (FDP) template (https://thefdp.org/default/committees/research-compliance/data-stewardship/). This study is registered with ClinicalTrials.gov, NCT04354701, and is ongoing. A centralized website (https://ccc19.org/) has been created to direct potential participants to the survey, collect feedback, and disseminate results.
Eligibility criteria have been kept simple and non-restrictive to ensure capturing a wide range of patients with cancer and COVID-19. Inclusion criteria for entering a case include suspected (presumptive positive based on clinical presentation) COVID-19 or laboratory-confirmed SARS-CoV-2 and a current or past medical history of invasive malignancy (any type). No restriction is placed on how long ago a cancer diagnosis might have occurred, since patients may have received potentially lung-toxic therapy even many decades prior (e.g., bleomycin for Hodgkin lymphoma or testicular cancer). Exclusion criteria include participants located within countries not explicitly approved for participation by the VUMC legal counsel or reports from non-healthcare providers (or their proxies). Prior entry into another COVID-19 registry is allowed, although all respondents are asked to report on such reporting, when it does occur, to address concerns of duplicated data (Bauchner et al., 2020).
Leveraging Existing Health Informatics and Technology Platforms
A centralized database was developed using the REDCap web browser-based platform (Harris et al., 2009). REDCap also permits prospective and longitudinal data collection to enhance data capture of new variables or long-term outcomes as we gather more knowledge about COVID-19, and accommodates structured and free-text data entry. Given recent advances in natural language processing techniques, we provide free text entry for concepts difficult to capture with structured questionnaires (e.g., cancer-specific treatment, unanticipated COVID-19-related complications) (Savova et al., 2019). Whenever feasible, structured variables were mapped to existing terminologies to support future data harmonization efforts, e.g., SNOMED-CT for comorbidities and ATC for medication exposures. The HemOnc ontology was used to describe cancer-specific concepts such as context of systemic anti-cancer therapy (Warner et al., 2019). We deliberately chose vocabularies identified as being standard terminologies by the Observational Health Data Sciences and Informatics (OHDSI) consortium (Hripcsak et al., 2015). All variables contain an “Unknown” option.
Given the flexibility and extensibility of REDCap, some sites prefer to build and maintain local instances for direct data entry instead of reporting into the central VUMC instance. This also allows for the addition of site-specific variables for local needs.
Data Collection Forms Development and Revisions
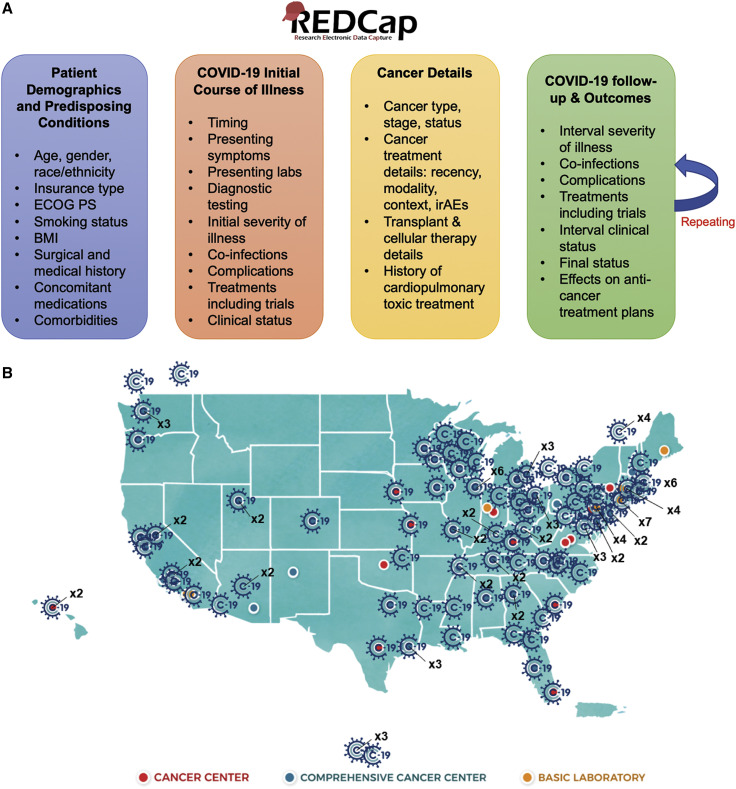
We designed customized forms for cancer and COVID-19 as per the limited literature on COVID-19 risk factors and outcomes available at the time of starting the registry and have continuously included emerging variables of clinical significance for more focused and precise analyses (Figure 1 A). Most questions in the survey are optional, with a subset being mandatory; evolution of the knowledge of prognostic factors has led to conversion of some variables from optional to mandatory (e.g., cancer status, which is strongly associated with 30-day all-cause mortality) (Kuderer et al., 2020a).
Figure 1.
CCC19 Data Collection Schema and Participating Institutions
(A) The general schema for data collection.
(B) Participating institutions as of October 3, 2020. Current participants include 10 NCI-Designated Cancer Centers; 43 NCI-Designated Comprehensive Cancer Centers; 25 NCI Community Oncology Research Program (NCORP) community sites, of which 10 are designated as Minority/Underserved; and 10 international sites (Canada and Mexico). Image source: NCI (public domain).
ECOG PS, Eastern Cooperative Group performance status; BMI, body mass index; irAEs, immune-related adverse events.
The survey is designed such that incompletely filled-out forms can be completed at a later time, using a unique link that is generated within the REDCap system and available only to the respondent. Data entry personnel can voluntarily return and add outcome data and/or complete the forms using this unique link provided to them as the patient’s clinical course evolves.
The survey includes the following five data collection forms: (1) Patient Demographics, (2) COVID-19 Details, (3) Cancer Details, (4) Respondent Details, and (5) Follow-up. Further details about the content of the forms can be found at https://ccc19.org/faqs.
Quality Assurance and Quality Improvement Processes
Due to the challenges of temporality as well as the need to define composite outcomes and risk factors, a large number of derived variables have been developed. These have evolved in parallel with the main survey instruments. Given that many of the variables are optional and that there is an “Unknown” option for each variable, missingness and excessive unknown responses are concerns for both raw and derived variables. In order to mitigate these concerns, we developed a quality score that is used to evaluate case reports for targeted improvement. Data problems are classified as minor (1 point), moderate (3 points), and major (5 points) (Table 1 ). Quality score metrics are periodically returned to sites, and the overall change in the cumulative distribution of quality scores is shared with the consortium on a regular basis.
Table 1.
Quality Score Metrics
| Major Problems (5 Points) | Moderate Problems (3 Points) | Minor Problems (1 Point) |
|---|---|---|
| High levels of baseline missingness | Cancer status missing | Cancer status unknown |
| Large number of unknowns | ECOG performance status missing | ECOG performance status unknown |
| 30-day f/u is 60+ days overdue | 30-day f/u is 30–59 days overdue | |
| Death status missing or unknown | Metastatic status missing or unknown | |
| Baseline COVID-19 severity missing or unknown | ICU status missing or unknown | |
| Hospitalization status missing or unknown | ||
| Intubation status missing or unknown | ||
| O2 need missing or unknown | ||
| Days to death missing or unknown | ||
| ADT missing or unknown (prostate cancer only) | ||
| Biomarkers missing or unknown (breast cancer only) | ||
| BCG exposure missing or unknown (bladder cancer only) |
ADT, androgen deprivation therapy; BCG, Bacillus Calmette-Guérin; ECOG, Eastern Cooperative Oncology Group.
Data Sharing
In order to increase and maintain the transparency of the CCC19 project, the data dictionary is made freely available through a public GitHub repository: https://github.com/covidncancer/CCC19_dictionary. The code to create all derived variables is also maintained here. A static version of the data dictionary and a list of derived variables can be downloaded through the CCC19 website at https://ccc19.org/faqs. New variable requests are collected through a crowdsourced process at https://redcap.link/CCC19-variable-request.
Participating sites have a right to obtain their own data on demand. A fully de-identified aggregated extract of the CCC19 registry is made freely available to non-commercial and academic researchers after an embargo period of approximately 6 months from prespecified data submission deadlines, during which members of the consortium have the opportunity to conduct scientific inquiries and publish the results.
Outcome
Since its founding, the consortium has expanded to 120+ institutions and 400+ individual members (Figure 1B). As of October 14, 2020, there are 5,991 records in the central database, of which 4,959 (83%) have complete baseline data. Among participating sites, 81 have reported one or more cases, 68 have reported 10+ cases, and 17 have reported 100+ cases. Three analyses have been published to date: (1) an analysis of risk factors associated with 30-day all-cause mortality in N = 928 patients (Kuderer et al., 2020a), (2) an analysis of patterns of anti-COVID-19 medication treatments and their effect on mortality in N = 2,186 patients (Rivera et al., 2020), and (3) an update to the mortality rates and examination of causes of death in the initial cohort of N = 928 patients (Kuderer et al., 2020b).
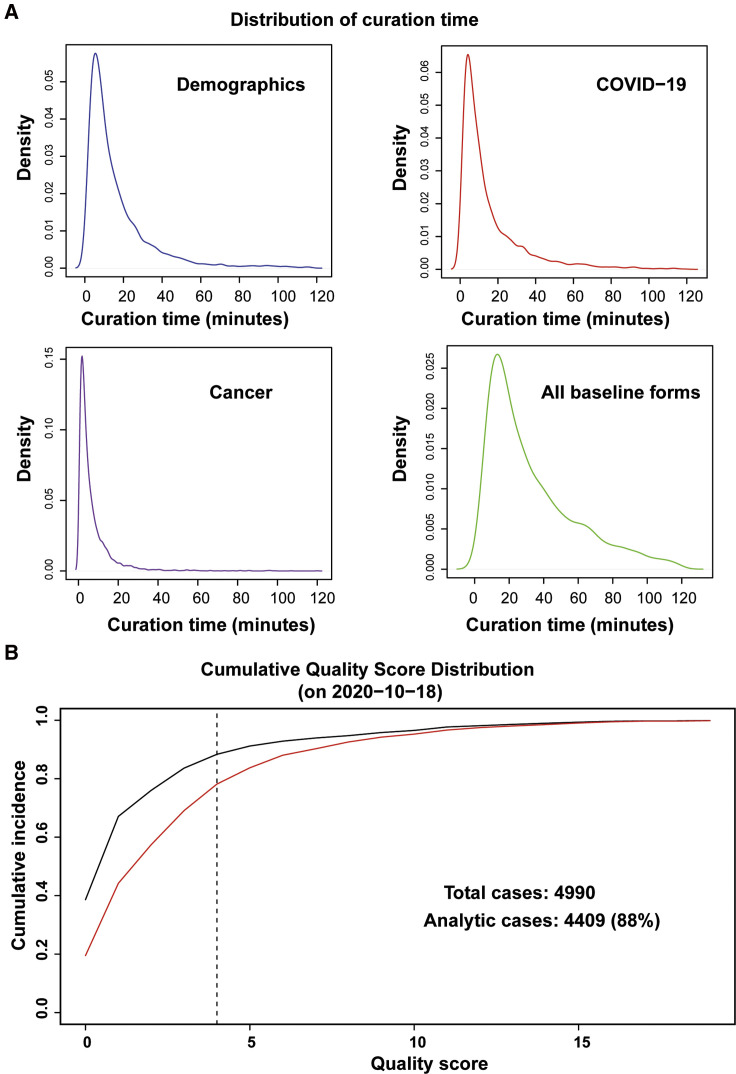
Initial beta testing indicated that the baseline information in the survey would take ∼5–15 min to complete. The practical experience after the first ∼4,000 cases have been entered indicates that the process follows a Poisson distribution after removal of outliers, with median of 25 min (10th percentile, 9 min; 90th percentile, 71 min) per case (Figure 2 A).
Figure 2.
Distribution of Curation Times and Quantitative Improvement in Quality Score after the First Round of Feedback to Sites
(A) Time intervals are determined by taking the difference between timestamps as recorded by REDCap at the initiation of each of the baseline forms. Outliers (negative calculated time, or calculated time greater than 120 min) are removed.
(B) The red curve illustrates the state of the registry at the time of the fourth data lock (July 31, 2020); at that time, fewer than 80% of records met the quality threshold. Targeted feedback was provided to sites ∼2 weeks later, and after 2 months the Quality Score had improved such that 88% of cases met the threshold to qualify as analytic cases.
An analysis of data quality at the time of the fourth data lock (July 31, 2020) indicated that the overall quality of reports was acceptable but in need of improvement, with 78% of cases meeting the predetermined threshold (Figure 2B). After targeted queries to sites, the cumulative quality score improved significantly, with 88% of cases meeting the predetermined threshold.
Outlook
In a very short period of time, CCC19 formed to become one of the largest consortia focused on COVID-19 and its effects on patients with cancer. In addition to publications to date, a number of specific subprojects are underway to make the most of this large data resource.
Balancing Risks versus Benefits
The web-based survey asks for information that is collected during routine clinical care. There is an indirect risk to the patient that the respondent could inadvertently disclose PHI in a free text response field; all structured fields (such as age) are constructed such that PHI cannot be disclosed. Clear instructions are given within the survey that no PHI is to be recorded, and the survey respondent is also advised to speak with their Privacy Office if they have any concerns about sharing non-PHI clinical data. Breach of confidentiality poses a risk to institutions and to individuals. The benefits of critical information for understanding the burden of novel infection and methods to prevent and treat complications still outweigh the risks of data collection in such registries; thus local IRB offices are encouraged to approve the study with waiver of HIPAA Authorization.
A separate risk to scientific integrity is that of excessive missing or unknown data. In our early analyses, expediency was key, and a high level of missingness was deemed acceptable; standard statistical methods of multiple imputation were used to partially address this issue. Unknown responses are more problematic, although they may occasionally be medically appropriate, such as cancer status being unknown in the period between initiation of a treatment and the first assessment by imaging or physical exam. For example, in our initial analysis, we found that having an unknown number of comorbidities was associated with increased 30-day all-cause mortality, pAOR 6.77 (95% CI, 1.42–32.33) (Kuderer et al., 2020a)—this is likely a surrogate of an unmeasured confounder, e.g., skilled nursing facility residents may have fewer available medical records and thus more unknowns. With the introduction of the quality score described above, records not meeting a sufficient quality score (less than 5 points, i.e., no major problems and at most one moderate problem) will only be used for descriptive purposes in future CCC19 data reports; others are considered for full analyses, subject to standard additional project-specific exclusion criteria. This generally follows the model of “analytic cases” as used by the cancer registry community (Mallin et al., 2013).
Future Directions and Investment in Long-Term Infrastructure
It is imperative to conduct periodic critical evaluations to ensure established and ongoing objectives are being met. We will adapt over time to encompass emerging information on COVID-19 (e.g., antibody levels, SARS-CoV-2 genomic tests, etc.), as well as possibly additional patient data (e.g., patient-reported outcomes) to augment the existing clinical data. As a grassroots and member-driven organization, the scientific direction of CCC19 will be influenced by member interests and expertise. Given the wide range of EHR platforms across participating institutions and the regulatory restrictions, we were unable to utilize the integration of EHR for direct data capture; however, this research capacity is essential to quickly capture and disseminate findings via informatics platforms during novel pandemics in future. EHR integration will also allow better data harmonization efforts with other groups building complementary registries. Finally, this is a completely voluntary project, so identifying and securing funding is essential to maintain the sustainability of such endeavors.
Acknowledgments
The consortium would like to acknowledge the writing committee who drafted this manuscript: Dimpy P. Shah, MD, PhD; Sanjay Mishra, MS, PhD; and Jeremy L. Warner, MD, MS. The consortium would also like to acknowledge the early contributions of Dr. Nicole M. Kuderer, MD, and Dr. Donna R. Rivera, PharmD, MsC. We thank all members of the CCC19 steering committee: Toni K. Choueiri, MD; Petros Grivas, MD, PhD; Gilberto de Lima Lopes, Jr., MD, MBA; Corrie A. Painter, PhD; Solange Peters, MD, PhD; Brian I. Rini, MD; Dimpy P. Shah, MD, PhD; Michael A. Thompson, MD, PhD; and Jeremy L. Warner, MD, MS, for their invaluable guidance of the CCC19 consortium. This study was partly supported by grants from the American Cancer Society and Hope Foundation for Cancer Research (MRSG-16-152-01-CCE; to D.P.S.) and the National Cancer Institute (P30 CA054174 to D.P.S.; P30 CA068485 to J.L.W. and S.M.; and U01 CA231840 to J.L.W.). REDCap is developed and supported by Vanderbilt Institute for Clinical and Translational Research grant support (UL1 TR000445 from NCATS/NIH). The funding sources had no role in the writing of the manuscript or the decision to submit it for publication.
Declaration of Interests
J.L.W. reports having received personal fees from Westat and IBM Watson Health and equity in HemOnc.org LLC outside the submitted work.
Contributor Information
The COVID-19 and Cancer Consortium:
Maheen Abidi, David M. Aboulafia, Melissa K. Accordino, Jared D. Acoba, Manmeet S. Ahluwalia, Syed A. Ahmad, Archana Ajmera, Saif I. Alimohamed, Jessica Altman, Anne H. Angevine, Ziad Bakouny, Michael H. Bar, Aditya Bardia, Jill S. Barnholtz-Sloan, Briana Barrow McCollough, Babar Bashir, Gerald Batist, Tanios S. Bekaii-Saab, Stephanie Berg, Eric H. Bernicker, Divaya Bhutani, Mehmet A. Bilen, Poorva Bindal, Rohit Bishnoi, Sibel Blau, Pamela Bohachek, Genevieve Boland, Mark Bonnen, Gabrielle Bouchard, Nathaniel Bouganim, Daniel W. Bowles, Fiona J. Busser, Omar Butt, Angelo Cabal, Wilhelmina D. Cabalona, Elwyn C. Cabebe, Paolo Caimi, Jian L. Campian, Theresa M. Carducci, James L. Chen, Alex Cheng, David D. Chism, Toni K. Choueiri, Melanie J. Clark, Jessica M. Clement, Jean M. Connors, Erin Cook, Catherine R. Curran, Ahmad Daher, Mark E. Dailey, Elizabeth J. Davis, Scott J. Dawsey, John F. Deeken, Salvatore A. Del Prete, George D. Demetri, Aakash Desai, Deborah B. Doroshow, Eric B. Durbin, Pamela C. Egan, Rawad Elias, Arielle Elkrief, Destry J. Elms, Amro Elshoury, Bryan Faller, Dimitrios Farmakiotis, Leslie A. Fecher, Lawrence E. Feldman, Cristiano Ferrario, Mark A. Fiala, Daniel B. Flora, Benjamin French, Christopher R. Friese, Julie C. Fu, Shirish M. Gadgeel, Justin Gainor, Matthew D. Galsky, Gerald Gantt, Jr., Jorge A. Garcia, Benjamin A. Gartrell, Margaret E. Gatti-Mays, David M. Gill, Erin A. Gillaspie, Antonio Giordano, (Mary) Grace Glace, Michael J. Glover, Sanjay Goel, Jerome J. Graber, Elizabeth A. Griffiths, Petros Grivas, Punita Grover, Anthony P. Gulati, Shuchi Gulati, Shilpa Gupta, Michael Gurley, Navid Hafez, Susan Halabi, Thorvardur R. Halfdanarson, Balazs Halmos, Daniel J. Hausrath, Jessica E. Hawley, Cassandra Hennessy, Roy S. Herbst, Dawn L. Hershman, Claire Hoppenot, Kent F. Hoskins, Irma Hoyo-Ulloa, Emily Hsu, Chih-Yuan Hsu, Clara Hwang, Jessica Yasmine Islam, Salma K. Jabbour, Chinmay Jani, Alokkumar Jha, Sachin R. Jhawar, Douglas B. Johnson, Monika Joshi, Anup Kasi, Kaitlin Kelleher, Hagen F. Kennecke, Ali Raza Khaki, Hina Khan, Mahir Khan, Jordan Kharofa, Goetz Kloecker, Jeanna L. Knoble, Amit A. Kulkarni, Vaibhav Kumar, Philip E. Lammers, John C. Leighton, Jr., Christopher A. Lemmon, Mark A. Lewis, Ang Li, Xuanyi Li, Stephen V. Liu, K.M. Lo, Arturo Loaiza-Bonilla, Barbara B. Logan, Elizabeth T. Loggers, Gilberto de Lima Lopes, Jr., Jonathan M. Loree, Patricia LoRusso, Clarke A. Low, Maryam B. Lustberg, Gary H. Lyman, Ryan C. Lynch, Subha Madhavan, Daruka Mahadevan, Sana Z. Mahmood, Abdul-Hai Mansoor, Michelle Marcum, Merry-Jennifer Markham, Sandeep H. Mashru, Tyler Masters, Blanche H. Mavromatis, Rana R. McKay, Christopher McNair, Shannon McWeeney, Alvaro G. Menendez, Harry Menon, Ruben A. Mesa, Vasil Mico, Chaim Miller, Sanjay Mishra, Ryan S. Monahan, Alicia K. Morgans, Mary F. Mulcahy, Daniel Mundt, Sarah Mushtaq, Gayathri Nagaraj, Sarah Nagle, Elizabeth S. Nakasone, John M. Nakayama, Heather H. Nelson, Eneida R. Nemecek, Ryan H. Nguyen, Amanda Nizam, Anju Nohria, Pier Vitale Nuzzo, Nitin Ohri, Adam J. Olszewski, Susie Owenby, Corrie A. Painter, Joshua D. Palmer, Orestis A. Panagiotou, Cathleen Park, Mary M. Pasquinelli, Jaymin M. Patel, Kanishka G. Patel, Prakash Peddi, Nathan A. Pennell, Solange Peters, Christine Pilar, Clement Pillainayagam, Matthew Puc, Amelie G. Ramirez, Joerg Rathmann, Deepak Ravindranathan, Sonya A. Reid, Daniel Y. Reuben, Sanjay G. Revankar, Kerry L. Reynolds, Young Soo Rho, Terence D. Rhodes, Robert L. Rice, Jonathan Riess, Brian I. Rini, Cameron Rink, Lane R. Rosen, Lori J. Rosenstein, Rachel P. Rosovsky, Bertrand Routy, Marc A. Rovito, Samuel M. Rubinstein, M. Wasif Saif, Mary Salazar, Miriam Santos Dutra, Lidia Schapira, Andrew L. Schmidt, Brett A. Schroeder, Gary K. Schwartz, Candice Schwartz, Michael T. Schweizer, Oscar K. Serrano, Danielle A. Shafer, Pankil K. Shah, Dimpy Shah, Mansi R. Shah, Sumit A. Shah, Chintan Shah, Grace Shaw, Justin A. Shaya, Yu Shyr, David A. Slosky, Melissa Smits, Carmen C. Solorzano, Karen Stauffer, Keith E. Stockerl-Goldstein, Daniel G. Stover, Jamie Stratton, Catherine Stratton, Mitrianna Streckfuss, Suki Subbiah, Lisa Tachiki, Eyob Tadesse, Michael A. Thompson, Umit Topaloglu, Matthew D. Tucker, Eliezer M. Van Allen, Susan Van Loon, Karen Vega-Luna, Neeta K. Venepalli, Amit Verma, Praveen Vikas, Shaveta Vinayak, Donald C. Vinh, Michael J. Wagner, Sarah Wall, Lucy L. Wang, Jeremy L. Warner, Firas H. Wehbe, Paul L. Weinstein, Matthias Weiss, Lisa B. Weissmann, Tanya M. Wildes, Nicole Williams, Trisha M. Wise-Draper, William A. Wood, Julie Tsu-Yu Wu, Elizabeth M. Wulff-Burchfield, Zhuoer Xie, Wenxin Xu, Albert C. Yeh, Irene S. Yu, Peter Paul Yu, Rosemary Zacks, Qamar Ul Zaman, Howard Zaren, Tian Zhang, Alice Y. Zhou, Huili Zhu, Rebecca L. Zon, and Leyre Zubiri
References
- Bakouny Z., Hawley J.E., Choueiri T.K., Peters S., Rini B.I., Warner J.L., Painter C.A. COVID-19 and cancer: current challenges and perspectives. Cancer Cell. 2020;38 doi: 10.1016/j.ccell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauchner H., Golub R.M., Zylke J. Editorial concern—possible reporting of the same patients with COVID-19 in different reports. JAMA. 2020;323:1256. doi: 10.1001/jama.2020.3980. [DOI] [PubMed] [Google Scholar]
- Desai A., Warner J., Kuderer N., Thompson M., Painter C., Lyman G., Lopes G. Crowdsourcing a crisis response for COVID-19 in oncology. Nat. Cancer. 2020 doi: 10.1038/s43018-020-0065-z. [DOI] [PubMed] [Google Scholar]
- Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hripcsak G., Duke J.D., Shah N.H., Reich C.G., Huser V., Schuemie M.J., Suchard M.A., Park R.W., Wong I.C.K., Rijnbeek P.R., et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud. Health Technol. Inform. 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- Kuderer N.M., Choueiri T.K., Shah D.P., Shyr Y., Rubinstein S.M., Rivera D.R., Shete S., Hsu C.-Y., Desai A., de Lima Lopes G., Jr., et al. COVID-19 and Cancer Consortium Clinical impact of COVID-19 on patients with cancer (CCC19): a cohort study. Lancet. 2020;395:1907–1918. doi: 10.1016/S0140-6736(20)31187-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuderer N.M., Wulff-Burchfield E., Rubinstein S.M., Grivas P., Warner J.L. Cancer and COVID-19—authors’ reply. Lancet. 2020;396:1067–1068. doi: 10.1016/S0140-6736(20)32065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Guan W., Chen R., Wang W., Li J., Xu K., Li C., Ai Q., Lu W., Liang H., et al. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21:335–337. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallin K., Palis B.E., Watroba N., Stewart A.K., Walczak D., Singer J., Barron J., Blumenthal W., Haydu G., Edge S.B. Completeness of American Cancer Registry Treatment Data: implications for quality of care research. J. Am. Coll. Surg. 2013;216:428–437. doi: 10.1016/j.jamcollsurg.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera D.R., Peters S., Panagiotou O.A., Shah D.P., Kuderer N.M., Hsu C.-Y., Rubinstein S.M., Lee B.J., Choueiri T.K., de Lima Lopes G., et al. Utilization of COVID-19 treatments and clinical outcomes among patients with cancer: a COVID-19 and Cancer Consortium (CCC19) cohort study. Cancer Discov. 2020 doi: 10.1158/2159-8290.CD-20-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein S.M., Steinharter J.A., Warner J., Rini B.I., Peters S., Choueiri T.K. The COVID-19 and cancer consortium: a collaborative effort to understand the effects of COVID-19 on patients with cancer. Cancer Cell. 2020;37:738–741. doi: 10.1016/j.ccell.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savova G.K., Danciu I., Alamudun F., Miller T., Lin C., Bitterman D.S., Tourassi G., Warner J.L. Use of natural language processing to extract clinical cancer phenotypes from electronic medical records. Cancer Res. 2019;79:5463–5470. doi: 10.1158/0008-5472.CAN-19-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrag D., Hershman D.L., Basch E. Oncology practice during the COVID-19 pandemic. JAMA. 2020;323:2005–2006. doi: 10.1001/jama.2020.6236. [DOI] [PubMed] [Google Scholar]
- van de Haar J., Hoes L.R., Coles C.E., Seamon K., Fröhling S., Jäger D., Valenza F., de Braud F., De Petris L., Bergh J., et al. Caring for patients with cancer in the COVID-19 era. Nat. Med. 2020;26:665–671. doi: 10.1038/s41591-020-0874-8. [DOI] [PubMed] [Google Scholar]
- Warner J.L., Dymshyts D., Reich C.G., Gurley M.J., Hochheiser H., Moldwin Z.H., Belenkaya R., Williams A.E., Yang P.C. HemOnc: A new standard vocabulary for chemotherapy regimen representation in the OMOP common data model. J. Biomed. Inform. 2019;96:103239. doi: 10.1016/j.jbi.2019.103239. [DOI] [PMC free article] [PubMed] [Google Scholar]