Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in December 2019 and quickly spread around the world, forcing global health authorities to develop protocols for its diagnosis. Here we report dimer formation in the N2 primers–probe set (CDC 2019-nCoV Real-Time RT-PCR) used in the diagnostic routine, and propose alternatives to reduce dimerization events. Late unspecific amplifications were visualized in 56.4% of negative samples and 57.1% of no-template control, but not in positive samples or positive control. In silico analysis and gel electrophoresis confirmed the dimer formation. The RT-qPCR parameters were optimized and the late unspecific amplifications decreased to 11.5% in negative samples and no-template control. The adjustment of PCR parameters was essential to reduce the risk of false-positives results and to avoid inclusive results requiring repeat testing, which increases the costs and generates delays in results or even unnecessary requests for new samples.
Keywords: Coronavirus, Diagnostic techniques and procedures, COVID-19, RT-qPCR
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in December 2019 in Wuhan, China. The virus spread rapidly to several countries, prompting the World Health Organization (WHO) to declare the disease a pandemic. Health authorities then had to quickly develop and implement diagnostic tools to fill the growing demand for tests. The reverse transcriptase quantitative PCR (RT-qPCR) is widely deployed in virology laboratory diagnostics. Among the currently available SARS-CoV-2 RT-qPCR protocols, the Charité protocol (Corman et al., 2020) was the World’s first publicly available test (Charité Universitätsmedizin Berlin, 2020). Later on, the Centers for Disease Control and Prevention (CDC) in the USA (CDC, 2020) proposed an alternative diagnostic protocol and it soon became popular among laboratories worldwide (Pan American Health Organization, 2020)
According to the CDC protocol (CDC, 2020), the single amplification of any of the specific virus targets (N1/N2) results in an ‘inconclusive result’, and the recommendation is to repeat the analysis. As observed in routine assays, late N2 amplifications frequently appear in negative samples, turning them ‘inconclusive’. Thus, it was decided to study the late unspecific amplifications in negative samples when using the CDC 2019-nCoV RT-qPCR protocol and to propose alternatives to reduce these events.
RNA was extracted from samples collected from the nasopharynx and oropharynx using TRIzol Reagent (Invitrogen). The RNA concentration and purity were assessed using a NanoDrop 2000c spectrophotometer (Thermo Scientific) by calculating the ratio of optical densities at wavelengths of 260/280. Samples presenting a 260/280 ratio in the range of 1.7–2.2 were subjected to RT-qPCR assays. The reactions were performed in a total volume of 25 μl using the One-Step RT-qPCR Low Rox Kit (LGC Biotecnologia) in a 7500 Fast Real-Time PCR System (Applied Biosystems). The CDC primers–probe panel (2019-nCoV RUO Kit, Integrated DNA Technologies (IDT)) (CDC, 2020) was used, which includes regions of the virus nucleocapsid gene (N1 and N2 targets) and an additional primers–probe set to detect the human RNase P gene (RP) (endogenous control). Initial RT-qPCR conditions followed the manufacturer’s recommended procedures and the CDC protocol for all targets: 12.5 μl Reaction Mix 2× (LGC Biotecnologia), 3 mM of MgSO4, 400 nM of each primer (forward and reverse), and 100 nM of probes. Thermal cycling was performed at 55 °C for 30 min for reverse transcription, followed by 95 °C for 15 min and then 40 cycles of 95 °C for 15 s, 60 °C for 1 min. All assays included a positive control (2019-nCoV_N Positive Control, IDT) and a no-template control (NTC).
The analysis of hairpin loops and self- and hetero-dimer formation was performed using OligoAnalyzer v.3.1 (IDT; https://www.idtdna.com/calc/analyzer) and NetPrimer (Premier Biosoft; http://premierbiosoft.com/) software. PCR products were electrophoresed in a 2.5% agarose gel plate and visualized with ethidium bromide staining.
This study was approved by the local research ethics committee (Comitê de Ética em Pesquisa com Seres Humanos do Hospital Universitário UFJF–CAAE: 32077320.0.0000.5133). This was a retrospective case series study. Written informed consent was obtained from participants.
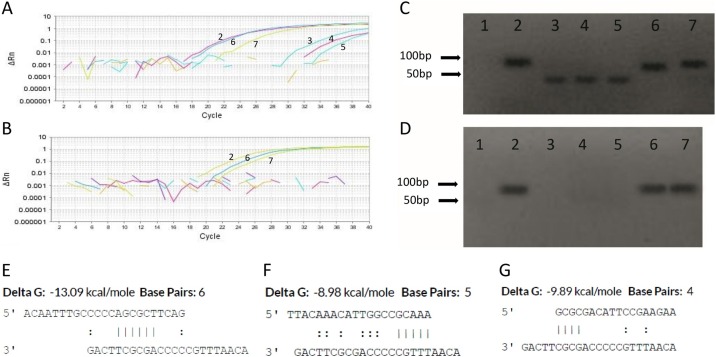
The singleplex RT-qPCR performed with the initial conditions demonstrated late unspecific amplifications in the N2 assay (Figure 1 A, curves 3, 4, and 5) in samples with results of ‘not detected’ for SARS-CoV-2. Of a total of 495 reactions, late amplifications were visualized in 56.4% of ‘not detected’ and 57.1% of NTC reactions, but not in positive samples or positive control. Neither N1 nor RP targets showed unspecific signals during the reactions. These results suggest the formation of dimers in N2 reactions of negative samples during RT-qPCR.
Figure 1.
Dimerization during RT-qPCR with the CDC N2 primers–probe set. Amplification plots of initial (A) and optimized (B) RT-qPCR conditions. Dimer formation can be visualized by the late signal produced in ‘not detected’ samples (curves 3, 4, and 5). Gel electrophoresis of initial (C) and optimized (D) RT-qPCR conditions. Dimers appear as diffuse bands (lanes 3, 4, 5) at the bottom of the gel (PCR products <50 bp). Partial sequence homologies between probe–probe (E), primer F–probe (F), and primer R–probe (G) estimated by OligoAnalyzer v.3.1. Key: 1 = no-template control (NTC); 2 = 2019-nCoV_N Positive Control (IDT); 3, 4, 5 = ‘not detected’ samples, 6, 7 = positive samples.
In order to confirm the dimer origin of the unspecific amplifications, in silico analysis of primers–probe sets and gel electrophoresis of the amplicons were performed. ΔG values of the N1, N2, and RP primers–probe sets showed stronger values for the N2 target (Supplementary Material File 1). Partial sequence homology between N2 probe–probe (self-dimer, Figure 1E), N2 forward primer–probe, and N2 reverse primer–probe (hetero-dimer, Figure 1F and G, respectively) of more than three base pairs was observed, with ΔG values of −13.09, −8.98, and −9.89 kcal/mol, respectively. Moreover, there was N2 forward primer–probe homology at the 3′ end of the sequence (Figure 1F). Hairpin loops in the N2 primers and probe showed ΔG values close to zero. Interestingly, a fragment size of less than 50 bp was observed in negative samples (Figure 1C, lanes 3, 4, and 5). As the expected fragment of 2019-nCoV_N Positive Control and positive samples is 72 bp (Figure 1C, lanes 2 and 6, 7, respectively), PCR products <50 bp confirm the dimerization of primers and/or probes.
In order to minimize the dimer formation in N2 reactions, different RT-qPCR conditions were tested, including the concentration of primers (133, 213, 266, 320, and 400 nM), probe (33, 54, 67.5, 81, and 100 nM), and MgSO4 (3, 4, 5, 6, and 6.5 mM), the annealing and extension temperature (60 °C, 63 °C, and 65 °C), and the reverse transcriptase time (12 min and 30 s, 15 min, and 30 min). Further details of the qPCR condition sets can be found in Supplementary Material File 2. The best performance was observed with the combination of the following conditions (set 7, Supplementary Material File 2): 213 nM of primers, 54 nM of probe, 6 mM of MgSO4, 63 °C of extension temperature, and 15 min of RT cycling. The most critical parameters were the concentration of MgSO4 and the annealing and extension temperature. The late unspecific amplifications decreased to 11.5% (n = 217 reactions) in ‘not detected’ samples and NTC samples; this absence can be noted in Figure 1B (amplification plot) and Figure 1D (gel electrophoresis).
However, fluorogenic probe-based reactions are not supposed to be influenced by dimerization in the N2 primers–probe and/or primer–primer from the CDC RT-qPCR recommended protocol used for SARS-CoV-2 diagnosis. Won et al. (2020) found unspecific amplifications when using the N2 and N3 primers–probe sets and then proposed an alternative primers–probe panel for the nucleocapsid target. Interestingly, Waggoner et al. (2020) corroborated the results of Won et al. and found late amplification in the N2 target (cycle threshold [Ct] = 44.8) in ‘not detected’ samples. The apparent occurrence of dimerization does not appear to be exclusive to nucleocapsid targets. Unspecific signals in the late cycles of the envelope protein gene (E target) assay using the Charité protocol (Konrad et al., 2020) and a mismatch of primer sequences (Pillonel et al., 2020) have been reported recently. The scientific community is discussing the technical limitations of the current SARS-CoV-2 RT-qPCR protocols (Marx, 2020) and their optimization is still underway.
It is known that the primer–probe sets are the pivotal component of a qPCR assay (Bustin and Huggett, 2017) and if dimerization occurs in a staggered manner, some extension can occur and become more abundant as cycling progresses. The in silico analysis showed that probe–probe self-dimer, for instance, had the potential to bind at the 5′ and 3′ ends, requiring a high amount of energy to fully break a secondary DNA structure (ΔG = −13.09 kcal/mol). Despite this, the optimization presented here drastically reduced the dimerization events.
The main concern with primer dimer formation is that they may cause false-positive results. In our experience, 56.4% of the ‘not detected’ reactions (negative samples) showed late unspecific amplification. The strict adjustment of the RT-qPCR conditions conducted in the present study was decisive for the optimization of the reaction and reduced the occurrence of late unspecific amplifications (11.5%). In addition, the single amplification of any of the specific virus targets (N1/N2)–even though it is an unspecific amplification–results in an ‘inconclusive result’ that requires repeat testing, increasing the costs and generating delays in results, or even unnecessary requests for new samples.
There was no decrease in the efficiency of the reactions in the positive control and positive samples, even though dimerization events with the N2 primers–probe set is suggested. This may be due to preferential annealing of the primers and probe to cDNA template of positive samples, which occurs in earlier cycles of PCR (cycles 10–30, depending on the amount of viral genetic material). Although the detection of SARS-CoV-2 in positive samples seems not to be affected by unspecific signals, these signals are of great importance in the assessment of negative samples, leading to inconclusive results.
Finally, we recommend that RT-qPCR users adjust primers–probe and magnesium concentrations, the duration of the reverse transcriptase step, and the thermal cycle temperature, independent of the master mix kit used, to minimize dimer formation and to avoid extensive test repetition and the waste of resources.
Funding statement
This work was supported by Universidade Federal de Juiz de Fora (UFJF, Brazil).
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgement
We thank M.I.N. Di Azevedo for editing the agarose gel images.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2020.10.079.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Bustin S., Huggett J. qPCR primer design revisited. Biomol Detect Quantif. 2017;14:19–28. doi: 10.1016/j.bdq.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . 2020. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel.https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcrpanel-primer-probes.html/ [Accessed 10 April 2020] [Google Scholar]
- Charité Universitätsmedizin Berlin . 2020. Researchers Develop First Diagnostic Test for Novel Coronavirus in China.https://www.charite.de/en/service/press_reports/artikel/detail/researchers_develop_first_diagnostic_test_for_novel_coronavirus_in_china/ [Accessed 8 June 2020] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad R., Eberle U., Dangel A., Treis B., Berger A., Bengs K. Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Euro Surveill. 2020;25(9) doi: 10.2807/1560-7917.ES.2020.25.9.2000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. Coronavirus jolts labs to warp speed. Nat Methods. 2020;17(5):465–468. doi: 10.1038/s41592-020-0827-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan American Health Organization . 2020. Laboratory Guidelines for Detection and Diagnosis of the Novel Coronavirus (2019-nCoV) Infection.https://www.paho.org/en/documents/laboratory-guidelines-detection-and-diagnosis-novel-coronavirus-2019-ncov-infection/ [Accessed 20 May 2020] [Google Scholar]
- Pillonel T., Scherz V., Jaton K., Greub G., Bertelli C. Letter to the editor: SARS-CoV-2 detection by real-time RT-PCR. Euro Surveill. 2020;25:2000880. doi: 10.2807/1560-7917.ES.2020.25.21.2000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner J.J., Stittleburg V., Pond R., Saklawi Y., Sahoo M.K., Babiker A. Triplex Real-Time RT-PCR for Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis. 2020;26(7) doi: 10.3201/eid2607.201285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won J., Lee S., Park M., Kim T.Y., Park M.G., Choi B.Y. Development of a laboratory-safe and low-cost detection protocol for SARS-CoV-2 of the coronavirus disease 2019 (COVID-19) Exp Neurobiol. 2020;29(2):107–119. doi: 10.5607/en20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.