Highlights
-
•
We described an one-step end-point RT-PCR for molecular detection of the SARS-CoV-2 E gene.
-
•
The analytical sensitivity of the RT-PCR was about 7.15–9 copies of vRNA/μL.
-
•
The RT-PCR was able to detect SARS-CoV-2 infection in asymptomatic individuals.
-
•
Nonspecific amplifications were not observed in SARS-CoV-2 negative samples.
Keywords: SARS-CoV-2, COVID-19, Molecular diagnosis, RT-PCR, E gene
Abstract
Real-time reverse transcription-polymerase chain reaction (RT-qPCR) is considered the "gold standard" for the direct diagnosis of SARS-CoV-2 infections. However, routine diagnosis by RT-qPCR is a limitation for many laboratories, mainly due to the infrastructure and/or disproportionate relationship between demand and supply of inputs. In this context, and to increase the diagnostic coverage of SARS-CoV-2 infections, we describe an alternative, sensitive and specific one-step end-point RT-PCR for the detection of the SARS-CoV-2 E gene. The performance of the RT-PCR was evaluated in 43 clinical samples, of which 10 and 33 were previously identified as negative and positive, respectively, by RT-qPCR. Among the positive samples, 15 and 18 were from asymptomatic and symptomatic individuals, respectively. Here, 32/33 of the positive samples in the RT-qPCR, including from asymptomatic individuals, were found positive in the RT-PCR (Ct 15.94–34.92). The analytical sensitivity of the assay was about 7.15–9 copies of vRNA/μL, and nonspecific amplifications were not observed in SARS-CoV-2 negative samples. Importantly, the RT-PCR reactions were performed in a 10 μL final volume. Finally, considering specificity, analytical sensitivity and cost reduction, we believe that the RT-PCR platform described here may be a viable option for the diagnostic of SARS-CoV-2 infections in laboratories in which RT-qPCR is not available.
In November/December 2019, a new human coronavirus, Severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2), family Coronaviridae and genus Betacoronavirus, emerged in Hubei province, mainland China, and has been responsible for the coronavirus disease currently named COVID-19 (Zhu et al., 2020; Huang et al., 2020; ICTV, 2020). As of July 26, 2020, SARS-CoV-2 infections have been described in 235 countries/regions, totaling 39,442,444 cases and 1,106,181 deaths, numbers that increase daily (WHO, 2020a).
Among the diagnostic alternatives for COVID-19, gene amplification by real-time reverse transcription-polymerase chain reaction (RT-qPCR) remains the “gold standard” for direct diagnosis. Many in-house and commercial RT-qPCR, using several and/or combined viral genes, have been proposed for molecular detection of SARS-CoV-2 (WHO, 2020b). The Charité protocol, one of the first protocols established for COVID-19 diagnosis, employs the envelope (E) and RNA dependent RNA polymerase (RdRp) genes as targets for detection of SARS-CoV-2 in clinical samples (Corman et al., 2020). However, it is also possible/reliable to diagnose COVID-19 by detecting a single genetic target. Regarding the Charité protocol, as the detection of the E gene has demonstrated a slightly higher sensitivity, the Pan American Health Organization (PAHO) has recommend prioritizing the E gene as the selected target for molecular COVID-19 diagnosis (PAHO, 2020).
Unfortunately, routine diagnosis by RT-qPCR is still a limitation for many laboratories, especially in developing countries and regions. The current demand for inputs, material and equipment, associated with the few supplier companies, represents an additional (and real) obstacle for the timely diagnosis of SARS-CoV-2 infections. In this context, and in order to increase the diagnostic coverage of COVID-19, we describe a sensitive and specific protocol for the detection of the SARS-CoV-2 E gene through one-step end-point RT-PCR (conventional RT-PCR); a potential alternative for situations in which RT-qPCR is not possible and/or available.
Initially, we evaluated the analytical sensitivity of the end-point RT-PCR for SARS-CoV-2 detection. The Brazilian isolate (SARS-CoV-2/SP02/human/2020/BRA, GenBank MT126808.1), previously inactivated, containing about 270,000 copies of viral RNA (vRNA)/μL, was kindly provided by Dr. Edison Durigon (Universidade de São Paulo, USP) and used as positive control throughout the study. The vRNA was extracted in the IndiMag48 automatic system (Indical Bioscience, Sachsen, Germany) and eluted in 100 μL of AVE buffer (Indical Bioscience, Sachsen, Germany). The eluate was then diluted [10-1 to 10-6, at the 0.1 dilution factor, i.e. 10-1.1, 10-1.2 (…) 10-6] and all dilutions were submitted to RT-PCR targeting the SARS-CoV-2 E gene.
The RT-PCR was performed with the SuperScript™ III One-Step RT-PCR System with Platinum™ Taq High Fidelity DNA Polymerase (Invitrogen, Carlsbad, California, USA) in a final volume of 10 μL: 5 μL of 2X reaction mix, 0.4 μL of SuperScript® III RT/Platinum® Taq High Fidelity Enzyme mix, 0.2 μL of primer E_Sarbeco_F1 (5′-ACAGGTACGTTAATAGTTAATAGCGT-3′) (10 μM), 0.2 μL of primer E_Sarbeco_R2 (5′-ATATTGCAGCAGTACGCACACA-3′) (10 μM) (Corman et al., 2020) and 4.2 μL of template. The amplification conditions were: 30 min at 55 °C and 2 min at 94 °C for cDNA synthesis and pre-denaturation, respectively, followed by PCR amplification in 40 cycles of 94 °C for 15 s for denaturation, 52 °C for 30 s for annealing, 68 °C for 10 s for extension, followed by a final extension at 68 °C for 5 min. The total volume of the PCR reaction (10 μL) was stained with GelRed (Biotium, Fremont, California, USA) and visualized under UV light on a 2% agarose gel. The analytical sensitivity was defined as the lowest concentration (highest dilution) in which it was possible to observe specific amplification.
The performance of the RT-PCR described above was evaluated in a combination of two nasopharyngeal and one oropharyngeal swab samples, according to recommendations of the Ministry of Health of Brazil (BRASIL, 2020), with genetic material extracted in the IndiMag48 automatic system (Indical Bioscience, Sachsen, Germany). We analyzed a total of 43 samples, of which 10 and 33 were previously identified as negative and positive, respectively, by the Hospital Universitário de Santa Maria (HUSM) (Rio Grande do Sul state, Brazil) by RT-qPCR targeting the SARS-CoV-2 E gene (Corman et al., 2020). Out of the 33 positive samples, 15 were from asymptomatic individuals and 18 from symptomatic patients (Table 1 ). Samples from asymptomatic individuals had cycle threshold (Ct) values between 18.93 and 34.92, and those from symptomatic patients showed Ct of 15.94–36.92 (Table 1). The Cts for all samples are available as Supplementary Data. The samples amplified by end-point RT-PCR were purified with the PureLink PCR Purification Kit (Invitrogen, Carlsbad, California, USA) and their identity was evaluated by gene sequencing, using BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fischer, Massachusetts, USA).
Table 1.
Positive samples by RT-qPCR and RT-PCR tests.
| RT-qPCR |
RT-PCR |
|||
|---|---|---|---|---|
| Asymptomatic (n = 15) | Symptomatic (n = 18) | Asymptomatic (n = 15) | Symptomatic (n = 17) | |
| Ct (range) | 18.93−34.92 | 15.94−36.92 | 18.93−34.92 | 15.94−34.91 |
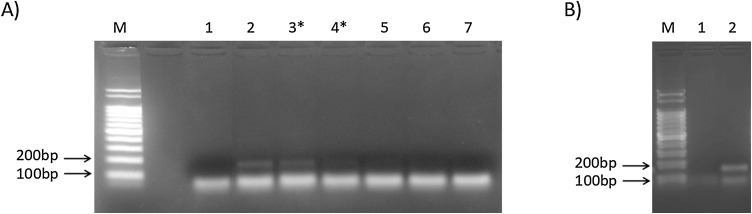
Here, the RT-PCR was able to specifically amplify the target region of the SARS-CoV-2 E gene. The 113bp amplicon, corresponding to the annealing region of the primers E_Sarbeco_F1 and E_Sarbeco_R2, precisely the 26,141−26,253 nt, was clearly observed in the positive control until 7.15–9 copies of vRNA/μL (i.e. 71.5–90 copies of vRNA) (Fig. 1 A). Although this sensitivity is lower than that of the described RT-qPCR (Corman et al., 2020; Peihua et al., 2020; WHO, 2020b), 32/33 of the positive samples in the RT-qPCR were found positive in the RT-PCR (Ct 15.94–34.92). The identity of the amplicons was confirmed by genetic sequencing.
Fig. 1.
End-point RT PCR for SARS-CoV-2 E gene. A) M: Ladder 100bp marker (Ludwig Biotec, Rio Grande do Sul, Brazil); 1: negative control (water); 2-7 positive control at 10−4 (11.34 vRNA/μL), 10-4.1 (9 vRNA/μL), 10-4.2 (7.15 vRNA/μL), 10-4.3 (5.68 vRNA/μL), 10-5 (1.13 vRNA/μL) and 10-6 (0.11 vRNA/μL), respectively. B) M: Ladder 100bp marker (Ludwig Biotec, Rio Grande do Sul, Brazil); 1: negative sample; 2: positive sample (Ct 34.92). *Interval considered to define the end-point RT-PCR analytical sensitivity.
In detail, end-point RT-PCR detected 15/15 of samples from asymptomatic individuals (Ct 18.93–34.92) and 17/18 from symptomatic patients (Ct 15.94–34.91) (Table 1). The RT-qPCR positive sample not detected in RT-PCR came from a symptomatic patient and showed Ct of 36.92. Nonspecific amplifications were not observed in SARS-CoV-2 negative samples (Fig. 1B).
False-negative results have been an important concern in the RT-qPCR for SARS-CoV-2, so that several targets/primers and sample types have been evaluated to increase the sensitivity (Corman et al., 2020; Peihua et al., 2020; Tahamtan and Ardebili, 2020; Wang et al., 2020; WHO, 2020b; Woloshin et al., 2020). However, considering the sensitivity of these assays, it is likely that false-negative results are more related to other factors than the characteristics of the assays per se. For example, previous studies have reported that the SARS-CoV-2 viral load (or the so-called RNAemia) may vary over time and thus strongly influence the diagnosis (Zhang et al., 2020; Zheng et al., 2020; Zou et al., 2020). Failures related to the pre-analytical phase, such as sample collection, transport and/or storage, as well as aspects of the host and the theoretical possibility of viral mutation, may also negatively influence the diagnosis of COVID-19 (Tahamtan and Ardebili, 2020; Xiao et al., 2020).
It is possible that the factors described above may also influence the end-point amplification, which still presented lower sensitivity than the RT-qPCR. However, we do not believe that the sensitivity represents a real limitation for the implementation of a RT-PCR platform for detection of SARS-CoV-2 infection. Individuals with COVID-19 often have a high viral load, which is why the European Centre for Disease Prevention and Control (ECDC) recommends that samples with Ct >35 be retested so that the possibility of contamination is ruled out (ECDC, 2020). A minimum concentration of about 10 copies of vRNA/μL has also been reported in respiratory samples from individuals with mild or severe COVID-19 (Pan et al., 2020; Zheng et al., 2020). In this context, our RT-PCR showed a considerable and useful sensitivity-specificity, detecting samples with Ct 34.92, including samples from asymptomatic individuals, and not amplifying nonspecific sequences, which could require additional steps of analysis, such as gene purification and sequencing (Fig. 1, Table 1). Importantly, the RT-PCR was performed in a 10 μL final volume. This, in addition to not using specific inputs for RT-qPCR, can significantly reduce the cost of the assay.
Finally, when considering specificity, analytical sensitivity and cost, we believe that the RT-PCR platform described here may be a viable option for molecular detection of SARS-CoV-2 in laboratories in which RT-qPCR is not available and/or not possible for any reason.
CRediT authorship contribution statement
José Valter Joaquim Silva Júnior: Conceptualization, Methodology, Investigation, Writing - original draft. Ingryd Merchioratto: Investigation, Writing - review & editing. Pablo Sebastian Britto de Oliveira: Investigation, Writing - review & editing. Thaísa Regina Rocha Lopes: Investigation, Writing - review & editing. Patrícia Chaves Brites: Resources, Writing - review & editing. Elehu Moura de Oliveira: Resources. Rudi Weiblen: Resources, Writing - review & editing. Eduardo Furtado Flores: Conceptualization, Supervision, Writing - review & editing, Resources, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Graduate students from the Programa de Pós-graduação em Medicina Veterinária, UFSM (IM and PSBO). PSBO and IM thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (Brazil) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) (Brazil), respectively, for their scholarships. TRRL thanks the Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS) (Brazil) for her technological and industrial development grant (process 20/2551-0000252-7). Eduardo Furtado Flores (process 301414/2010-6) and Rudi Weiblen (process 304153/2014-1) were supported by CNPq research fellowships. This study was supported by FAPERGS (Brazil), process 20/2551-0000252-7.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2020.114007.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Brasil . 2020. Guia de Vigilância Epidemiológica Emergência de Saúde Pública de Importância Nacional pela Doença pelo Coronavírus 2019.https://www.saude.gov.br/images/pdf/2020/April/06/GuiaDeVigiEp-final.pdf (accessed July 7, 2020) [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control . 2020. Questions and Answers Regarding Laboratory Topics on SARS-CoV-2.https://www.ecdc.europa.eu/en/all-topics-z/coronavirus/threats-and-outbreaks/covid-19/laboratory-support/questions (accessed July 7, 2020) [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Committee on Taxonomy of Viruses . 2020. Virus Taxonomy: 2019 Release.https://talk.ictvonline.org/taxonomy (accessed July 7, 2020) [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan American Health Organization . 2020. Laboratory Guidelines for the Detection and Diagnosis of COVID-19 Virus Infection.https://iris.paho.org/bitstream/handle/10665.2/52140/PAHOPHEIMSCOVID-19200003_eng.pdf?sequence=1&isAllowed= (accessed July 7, 2020) [Google Scholar]
- Peihua Niu., Roujian Lu., Li Zhao., Huijuan Wang., Baoying Huang., Fei Ye., Wang W., Tan W. Three novel real-time RT-PCR assays for detection of COVID-19 virus. China CDC Weekly. 2020;2:453–457. doi: 10.46234/ccdcw2020.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020:323. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection - challenges and implications. N. Engl. J. Med. 2020 doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- World Health Organization . 2020. Coronavirus Disease (COVID-19) Pandemic.https://www.who.int/emergencies/diseases/novel-coronavirus-2019?gclid=Cj0KCQjwo6D4BRDgARIsAA6uN18fJRvKcpt6tiLBwLBOVIsuqXny2LjmCLiptICCr4KiWZq2ytax-q0aAsysEALw_wcB (accessed October 17, 2020) [Google Scholar]
- World Health Organization . 2020. Molecular Assays to Diagnose COVID-19: Summary Table of Available Protocols.https://www.who.int/publications/m/item/molecular-assays-to-diagnose-covid-19-summary-table-of-available-protocols (accessed July 7, 2020) [Google Scholar]
- Xiao A.T., Tong Y.X., Zhang S. False negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J. Med. Virol. 2020 doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.F., Yan K., Ye H.H., Lin J., Zheng J.J., Cai T. SARS-CoV-2 turned positive in a discharged patient with COVID-19 arouses concern regarding the present standards for discharge. Int. J. Infect. Dis. 2020;97:212–214. doi: 10.1016/j.ijid.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X., Chen W., Wang Q., Zhang D., Liu Y., Gong R., Ma Z., Lu S., Xiao Y., Gu Y., Zhang J., Yao H., Xu K., Lu X., Wei G., Zhou J., Fang Q., Cai H., Qiu Y., Sheng J., Chen Y., Liang T. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369 doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., Team, C.N.C.I.a.R A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.