Abstract
The coronavirus disease-19 (COVID-19) outbreak that is caused by a highly contagious severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has become a zoonotic pandemic, with approximately 24.5 million positive cases and 8.3 lakhs deaths globally. The lack of effective drugs or vaccine provoked the research for drug candidates that can disrupt the spread and progression of the virus. The identification of drug molecules through experimental studies is time-consuming and expensive, so there is a need for developing alternative strategies like in silico approaches which can yield better outcomes in less time. Herein, we selected transmembrane protease serine 2 (TMPRSS2) enzyme, a potential pharmacological target against SARS-CoV-2, involved in the spread and pathogenesis of the virus. Since 3D structure is not available for this protein, the present study aims at homology modelling and validation of TMPRSS2 using Swiss-model server. Validation of the modelled TMPRSS2 using various online tools confirmed model accuracy, topology and stereochemical plausibility. The catalytic triad consisting of Serine-441, Histidine-296 and Aspartic acid-345 was identified as active binding site of TMPRSS2 using existing ligands. Molecular docking studies of various drugs and phytochemicals against the modelled TMPRSS2 were performed using camostat as a standard drug. The results revealed eight potential drug candidates, namely nafamostat, meloxicam, ganodermanontriol, columbin, myricetin, proanthocyanidin A2, jatrorrhizine and baicalein, which were further studied for ADME/T properties. In conclusion, the study unravelled eight high affinity binding compounds, which may serve as potent antagonists against TMPRSS2 to impact COVID-19 drug therapy.
Keywords: SARS-CoV-2, COVID-19, TMPRSS2, Homology modelling, Molecular docking, Drug repurposing
Highlights
-
•
3D structure of TMPRSS2 was constructed using Homology modeling and was validated.
-
•
50 K Ligand was utilized to reveal the active site of modelled TMPRSS2.
-
•
A total of twenty-five natural and marketed compounds were studied.
-
•
Nafamostatat, Meloxicam, Ganodermanontriol, Columbin, Myricetin, Proanthocyanidin A2, Jatrorrhizine and Baicalein shown good binding energies.
-
•
These potent TMPRSS2 inhibitors can prevent spread of SARS-CoV-2, aid in treatment of COVID-19.
1. Introduction
Coronavirus disease-19 (COVID-19) caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has turned out to be a life-threatening pandemic across the globe. The lack of effective treatments and a high mortality rate grabbed researcher's attention to develop effective therapeutic strategies and vaccines. Statistically, COVID-19 cases are increasing at alarming rate every day with approximately 24.5 million positive cases and 8.3 lakhs deaths worldwide (World Health Organization, 2020). The first step in SARS-CoV-2 infection is its entry and invasion into host lung epithelial cells through angiotensin converting enzyme-2 receptor by a transmembrane spike (S) glycoprotein. This entry mechanism also depends upon host cellular proteases which include TMPRSS2, cathepsins and human airway trypsin-like protease that cleaves SARS-CoV-2 spike protein at the site of Subunit-S1/S2 which is an arginine rich multi-basic site (Hoffmann et al., 2020). TMPRSS2 is an eccentric, androgen regulated, human airway and alveolar epithelial serine protease (Mikkonen et al., 2010), that is critical to promote influenza-A, SARS-CoV and Middle East respiratory syndrome (MERS) virus spread (Kleine-Weber et al., 2018). It plays a pivotal role in priming and activation of the viral spike protein for cell-cell fusion and entry into host cells. TMPRSS2 also weakens virus recognition by host's neutralizing antibodies, thus promoting viral pathogenesis (Glowacka et al., 2011). The redundant nature (Shen et al., 2017) along with its critical importance in SARS-CoV-2 pathogenesis, thus advocates TMPRSS2 as a potential target against COVID-19. Further, it has been reported that, TMPRSS2 inhibitors like camostat are useful in blocking viral host cell entry (McKee et al., 2020).
Recent advances in drug discovery process assumed greater use of synthetic molecules and phytochemicals for their antiviral activities which seemed to have promising future in treating the pandemic. So far, no studies yielded an approved drug for therapeutic use in COVID-19 patients for its complex mechanism. The identification of targets, drugs, vaccines through experimental studies is time-consuming, expensive and often yield few drug molecules to combat the disease. Consequently, we relied upon computational approaches which facilitate reliable results in less time. With the plethora of literature knowledge, we selected a variety of compounds of natural and synthetic origin with antiviral significance for the present study. The study aimed to construct and validate a model of TMPRSS2 using molecular modelling techniques. We further aimed to screen a list of compounds using docking studies against the modelled TMPRSS2, which might serve as potential antagonists in treating SARS-CoV-2.
2. Material and methods
2.1. Hardware and software
The complete work was performed on a state of art personal computer pre-installed with various online and offline bioinformatics tools like Protein Data Bank (PDB), UCSF Chimera, Procheck, ProSA, ProQ, Protter, Polyview, AutoDock, HADDOCK and ClusPro server for accomplishing the study.
2.2. Selection of active site
The 364-amino acid sequence of human TMPRSS2 was chosen from a c-DNA library (GenBank: O15393). The post-translational sequence of TMPRSS2 was searched at the Signal peptide website (http://www.signalpeptide.de). The Uniprot search (https://www.uniprot.org/) for selection of active domains and PSI-BLAST search of the obtained primary sequence to check 3D structure availability were performed.
2.3. Homology modelling and validation
The 3D structure of TMPRSS2 was built using Homology modeling due to the unavailability of complete structure in the protein data bank (PDB). The target sequence of TMPRSS2 was retrieved from UniProt, Genbank and utilized in search of similar structures or templates. The same was further confirmed with Pharos database (https://pharos.nih.gov/). PSI-BLAST search against the RCSB PDB was performed and templates with >40% identity and >90% query coverage were marked (Supplementary Table S1). The top model with best QMEAN score and high percentage similarity was selected as the base template. A search for extracellular domain and transmembrane domain were performed using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/), TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), SoSUI (http://harrier.nagahama-i-bio.ac.jp/sosui/), Protter (http://wlab.ethz.ch/protter), Polyview (http://polyview.cchmc.org/). The modelled TMPRSS2 was prospectively validated and assessed using protein structure and model assessment tools at the Swiss-Model server (http://swissmodel.expasy.org) using different estimation parameters (Schwede et al., 2003). The model with good range of >90% allowed and favoured region positions was validated through Procheck, ProSA and ProQ quality assessment tools (Laskowski et al., 1996; Khobragade et al., 2011). The stereochemical plausibility of the model for orientation of dihedral angles of phi (ϕ) and psi (ψ) was further confirmed by verify 3D, PROVE and ERRAT (Elfiky, 2020; Visegrády et al., 2001), (https://servicesn.mbi.ucla.edu/SAVES/). The acceptable ranges set for validation of the model were, Z-scores >3.00, LG score and Max Sub scores with >1.00 and > 2.00 respectively. Physico-chemical characterization studies were performed using EXPASY's ProtParam server to know the helix, sheets, turns and loops present in TMPRSS2 (https://web.expasy.org/), (Gasteiger et al., 2005). Based on the amino acid sequence, ligands and their structures were obtained from Pharos and Pubchem databases respectively, and were docked against TMPRSS2. Protoss hydrogen prediction of Protein plus (https://proteins.plus/) was utilized to add missing hydrogen atoms to the modelled TMPRSS2 (PDB-format). Sequence alignment was determined by using UCSF Chimera platform and the sequence of original TMPRSS2 obtained from UniProt was aligned with that of the developed homology protein structure.
2.4. Binding pocket and protein-protein network determination
The modelled TMPRSS2 was docked with the S1 domain of spike protein using HADDOCK and CLUSPRO to learn their possible interactive sites which aid for determination of active binding sites. The interactive pocket of the TMPRSS2 enzyme was accessed with UCSF Chimera and literature sources. Pockets with active catalytic triad were of principal significance for further study (Antalis et al., 2010). Dog site server was utilized to further determine and validate the different active pockets in the modelled 3D structure (Volkamer et al., 2012).
2.5. Molecular docking simulations
The structure files (canonical smiles format) of seven marketed drugs, ulinastatin, 1-(3-(3-(amino imino methyl) phenyl)-2-(((4-methyl phenyl) sulfonyl) amino)-1-oxopropyl) piperidine, di-isopropyl flouorophosphate, meloxicam, 4-(2-Aminoethyl) benzene sulfonyl fluoride, P-amino benzamidine and nafamostat, and eighteen natural compounds, tetratriacontane (leaves of Azadiracta indica, Plectranthus amboinicus, Calligonum polygonoides, Crateva adansonii), baicalein (Scutellaria baicalensis), hesperetin and naringenin (fruits of citrus sps.), fisetin (fruits of Fragaria ananassa), myricetin (leaves of Isatis indigotica, Torreya nucifera), 3,4-dichloroisocoumarin (bark of Cinnamomum verum), 4-Amidinophenylmethanesulfonyl fluoride hydrochloride, aesculitannin B and proanthocynidine A2 (seeds of Litchi chinensis), ganoderiol D and ganodermanontriol (Ganoderma lucidum mushroom), columbin, ecdysterone, magnoflorine and jatrorrhizine (stem of Tinospora cardifolia), ciscapsaicin and dihydrocapsaicin (fruits of Capsicum baccatum) were collected from PubChem (Wishart et al., 2008), (Supplementary Tables S2 and S3). These structures were used for performing docking studies against the modelled TMPRSS2 using Autodock VINA. Camostat, a known TMPRSS2 inhibitor was used as standard drug for the study. The pdbqt files for protein and ligands were prepared and grid box creation was completed using AutoDock Tools (ADT), (Morris et al., 2009). Polar hydrogen was alone added to the protein with the later addition of Kollman and Gastegier charges. The grid spacing was configured at a default value of 0.375 Å. The grid box was set to 126 Å, 102 Å and 92 Å (x, y and z axis respectively) to include all the domains of protein (Dhanavade and Sonawane 2014). Blind docking studies were employed to decipher the potent inhibitors of TMPRSS2. All other values were set to default with maximum exhaustiveness and energy range 4. Two-dimensional data was analyzed using BIOVIA Discovery Studio (Temml et al., 2014). The protein and ligands were kept rigid during the procedure and the complex with root mean square deviation (RMSD) < 1.0 was clustered as the most favourable binding energy (Azizian et al., 2010). The docked complex with lowest binding energy was selected along with the receptor structure and was saved as pdb file for further studies. The entropy effects that play a predominant role in the interactions of receptor-ligand were given by their binding energies.
2.6. Protein dynamics and residual frustration analysis
Protein movements, frustration energetics in the structural topology, relative stability and dynamic motions of the modelled structure were calculated using Cartesian trajectory coordinates by DynaMut, DynaOmics and Frustratometer servers (Amir et al., 2019; Ahamad et al., 2019).
2.7. Physiochemical and ADME/T properties
Absorption, Distribution, Metabolism, Excretion/Toxicity (ADME/T) attributes of the selected compounds were evaluated using Swiss-ADME tool (Kaur et al., 2020). These played an important role in analyzing drug filtration and predict other pharmacodynamic parameters of complex consequentially aiding towards determination of potent TMPRSS2 inhibitors.
3. Results and discussion
3.1. TMPRSS2 – active site
The gene sequence of entire TMPRSS2 protein was retrieved from databanks namely Genbank (Id: 7113) and UniProt (Id: O15393) and sequence of Chromosome 21 - NC_000021.9 from Homo sapiens was downloaded in FASTA format (Supplementary Table S4). An attempt made by Signal peptide website search for post translational sequence of TMPRSS2 revealed no appropriate sequence. The Uniprot search provided some important information regarding active domains and the catalytic triad (Serine-441, Histidine-296 and Aspartic acid-345) of this protein. The three active domains revealed were 1. Topological (1–84): Cytoplasmic domain, 2. Membrane (85–105): Transmembrane domain and 3. Topological (106–492): Extracellular domain. Since the catalytic triad is in the topological extracellular domain, it is perceived as the active binding site of S1 domain of the spike protein, and the same domain sequence was considered as the active site of TMPRSS2.
The PSI-BLAST search of the entire amino acid sequence showed no existing 3D structures for TMPRSS2 in Protein data bank (Supplementary Fig. S1). Further, search for 3D model was also conducted on RCSB database and resulted in no deposited 3D structure for the host enzyme. Since the TMPRSS2 protein lacks any experimental structure, homology modelling was attempted using the SWISS-MODEL server. The search for extracellular domain and transmembrane domain using PSIPRED, TMHMM, SoSUI revealed that the domain is extracellular with no significant transmembrane helices (Supplementary Fig. S2). The results from ‘SoSUI server revealed the protein as extracellular, soluble protein and also gave information about alpha helix, beta strands and coils (Supplementary Table S5). PSIPRED, Polyview gave information about secondary structure, topological prediction and helix information of TMPRSS2. A recurring and periodic pattern of β-strand elements can be seen in the extracellular domain beginning at residue 158 (leucine) running through to residue 162, other residues seen were at 182, 209–212; 255–257; 269–274; 279–281; 284–285; 290–297; 307–315; 324–333; 346–350; 362–365; 378–382; 399–404; 424–427; 445–447; 452–460 and 473–476. These β-strand domains indicate a highly regular structure in the extracellular region. The C-terminal domain started from residue 109 (methionine) and other alpha helix residues were observed at 106 (trypsine) to 107; 183–184; 395–398; 406–410 and 477–490 as confirmed from Protter server (Supplementary Fig. S2). The amino acid sequence consisting of catalytic triad (106–492) in FASTA sequence format was provided to SWISS-MODEL to obtain the 3D structure of TMPRSS2. SWISS-MODEL resulted in 3 templates, of which, one template (5CE1 - Serine protease hepsin) was found to be most suitable with 89.6% identity with the amino acid sequence provided. The template 5CE1 was also available with published XRD data hence it was found to be most suitable. Four important ligands were identified from Pharos database with Pub med ID: L1 - 46899577, L2 - 56663319, L3 - 56677007 and L4 (50 K) - 121232387.
3.2. Validation of model
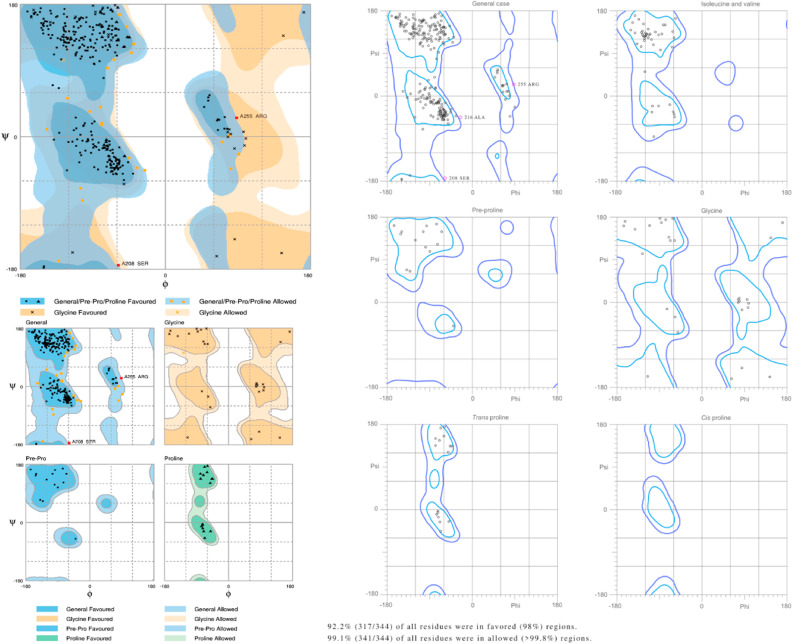
The modelled protein structure was further validated to know the model accuracy. The generated reliable model of TMPRSS2 was subjected to Procheck, ProSA and ProQ analysis. The stereochemical quality of a model is said to be good, when more than 90% residues are in the most favoured region of the Ramachandran plot (Ramachandran, 1963). Several online validation tools were used to gauge the validity and authenticity of the modelled structure. The stereochemical analysis was performed using Procheck by considering the overall structural geometry of the respective protein (Laskowski et al., 1996). The results clearly indicated that 98% of the residues were in favourable and allowed regions of the Ramachandran plot. Analysis by ProSA-web validation tool showed a Z-score value of −6.29. A Z-score value greater than −4 indicates that the structure is properly refined and can be used for further studies (Wiederstein and Sippl, 2007). Thus, our modelled structure was in compliance with the range of native conformations of crystal structures. The degree of closeness of the modelled structure to the native structure was analysed by Procheck and ProSA, while ProQ was used for the analysis of nearness to the most correct models as shown in Fig. 1 . The threshold value for a good model of a protein structure in ProQ is -log of the p value score (LG) which should be > 4. The modelled TMPRSS2 protein was found to have LG score of 5.615, which suggested that the model is of extremely good quality. The stereochemical plausibility of the model was further confirmed by verify 3D, PROVE and ERRAT (Supplementary Fig. S3). Physicochemical characterization, molecular weight, total number of positive and negative residues, extinction co-efficient, theoretical isoelectric point, instability index, aliphatic index and grand energy with hydropathy were computed using Expasy's ProtParam server (Supplementary Table S6).
Fig. 1.
Stereochemical properties of TMPRSS2 modelled structure generated using Ramachandran plot analysis (PROCHECK), ProSA and ProQ quality servers.
The ligands L1, L2, L3 and L4 were docked with the homology model and the docking score was relatively not significant. Protoss hydrogen prediction of Protein plus detected tautomers, reasonable protonation states and hydrogen coordinates of both protein and ligand molecules. Additionally, alternative conformations or overlapping entries which may be annotated in the original protein structure were eliminated, as they could disturb the analysis of molecular interactions. The resulting protein was considered and ligands L1, L2, L3 and L4 were docked again, revealing binding energies of −6.8, −6.7, −6.9 and −8.0 kcal/mol respectively (Supplementary Fig. S4, S5 and S6). The results showed that the ligand L4 (50 K) produced good binding energy and it was considered as active ligand. We found good agreement when comparing modelled sequence with original sequence of TMPRSS2 through sequence alignment (Supplementary Fig. S7). So, the resultant homology model was considered for further study.
3.3. Binding site detection
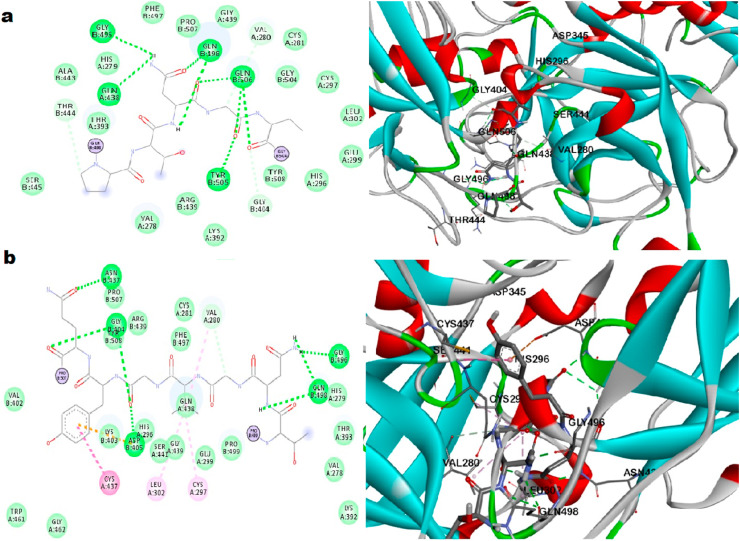
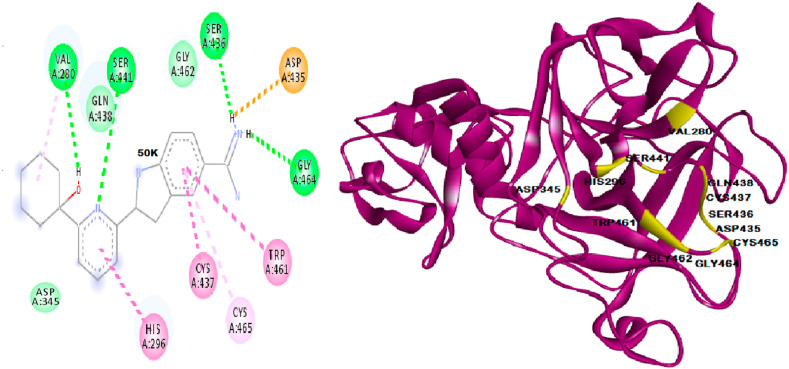
The interactive site of TMPRSS2 and S1 of spike protein from HADDOCK and CLUSPRO also confirmed the main binding site consisting of catalytic triad as shown in Fig. 2 . The important amino acid residues of TMPRSS2 involved in bond formation with S1 domain of spike protein are His 296, Ser 441, Asp 345, Gly 496, Gln 438, Gln 498, Gln 506, Tyr 505 and His 296, Ser 441, Asp 345, Asn 437 Gly 404, Asp 405, Gly 496, Gln 498 as obtained from HADDOCK and CLUSPRO respectively. Since the catalytic triad is found in the interactive site of TMPRSS2 and spike protein, it was confirmed as active binding site of TMPRSS2. Dog site study resulted in 16 active pockets with 14 subpockets, out of which, three pockets were targeted which consisted of the active catalytic triad of this enzyme TMPRSS2 that were very druggable (Supplementary Table S7 and Fig. S8 a, b, c, d). The docking of ligands to the developed 3D structure underline the importance of these pockets and active sites of this protein. The amino acid residues of catalytic triad; Ser 441 was found in the binding pocket 2 and 4, His 296 found in pocket 4 and Asp 345 found in pocket 9 with predicted pocket druggability of 70, 71 and 44% respectively. The literature mining marked the importance of several other amino acid residues at the binding pocket of TMPRSS2 (Antalis et al., 2010). The active ligand L4, 2-[6-(1-hydroxycyclohexyl) pyridin-2-yl]-1H-indole-5-carboximidamide (50 K) complexed with TMPRSS2 was analysed for the binding pocket elucidation and the same was utilized as active ligand in the study as depicted in Fig. 3 . The residues Ser 441, Val 280, Ser 436, Gly 464 were actively involved in the formation of hydrogen bonds and other residues His 296, Asp 345, Gly 462, Gln 438, Asp 435, Trp 461, Cys 437 and Cys 465 were also involved, thus confirming the active site of TMPRSS2 for further study.
Fig. 2.
The interactive site of TMPRSS2 and S1 of spike protein from a. HADDOCK and b. CLUSPRO.
Fig. 3.
50 K complexed with TMPRSS2 depicting active site, amino acid residues showing active site were given in stick model with violet colour. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. Docking analysis
Docking analysis is a computational approach to analyse the structural complexes between the target and the ligand and to comprehend the structural basis of the protein target specificity. A variety of compounds of natural and synthetic origin with different pharmacological profiles, that have been found to inhibit vital components in the processes of viral entry and replication were selected systematically and screened. The docking analysis revealed that the selected twenty-five compounds showed binding towards the interactive site of TMPRSS2. The results were further analysed by comparing the docking scores and binding orientation which led to short-list of eight active compounds namely nafamostat, meloxicam, ganodermanontriol, columbin, myricetin, proanthocyanidin A2, baicalein and jatrorrhizine for their good binding affinities towards TMPRSS2 as shown in Table 1 . The interaction of other seventeen compounds including five marketed and twelve natural compounds also exhibited binding towards active site of TMPRSS2 but with less affinity as shown in Table 1 (Supplementary Figs. S9a, b, c, d).
Table 1.
Elucidation of docking scores and H-bonds of studied compounds by docking studies.
| S. No. | Compound | Docking Score (kcal/mol) | H-bond residues (Bond length Ao) | π-interactions | Amino acid residues involved in other interactions |
|---|---|---|---|---|---|
| 1 | Camostat | −8.4 | His 296 (1.89), Cys 281 (2.71), Glu 389 (2.8), Lys 390 (2.78), Gln 438 (2.72) |
Val 280, Ala 386 | Ser 441, His 279, Gly 439, Gly 391, Thr 393, Cys 297, Leu 302, Gly 391, Lys 392, Gly 385, Ser 460, Trp 461, Gly 462 |
| 2 | Columbin | −8.2 | His 296 (2.0). | Ser 460, Ser 436, Cys 437, Cys 465 | Gly 464, Gly 462, Trp 461, Gln 438, Gly 439, Ser 411, Thr 459 |
| 3 | Meloxicam | −8.3 | Gly 462 (1.91), Ser 441 (2.61), Ser 460 (1.81). |
Cys 437, Val 280, Trp 461, Lys 342 | Thr 459, Ser 436, Gly 439, Cys 465, Gln 438, His 279, Val 278, Leu 302, Ser 463, His 296 |
| 4 | Myricetin | −8.3 | Ser 460 (1.96), Glu 389 (2.88), His 296 (2.55). |
Cys 437, Trp 461, Glu 389 | Asp 435, Gly 472, Cys 465, Gly 464, Ser 436, Thr 459, Ser 441 |
| 5 | Ganodermanontriol | −8.1 | Gly 462 (2.77), Ser 441 (2.18). |
Trp 461 | Cys 437, Cys 465, Glu 389, Gln 438, Leu 419, Thr 341, Lys 340, Lys 342, Ser 460, His 296 |
| 6 | Nafamostat | −7.8 | Ser 436 (2.74), Asp 435 (2.35), Gly 461 (4.75). |
Asp 435, Trp 461, His 296, Cys 437, Cys 465 | Ala 466, Ser 441, Lys 342, Val 473, Gly 472, Thr 459, Ser 460, Gly 464, Gln 438, Leu 419. |
| 7 | Proanthocyanidin A2 | −7.9 | Gly 462 (2.83), Cys 465 (2.90), Ser 436 (2.71), Ser 441 (2.84), His 296 (2.62) |
Trp 461. | Ser 463, Lys 342, Asp 435, Gly 464, Cys 437, Gly 472, Val 473, Thr 459, Ser 460, Gln 438, Gly 439. |
| 8 | Baicalein | −7.7 | His 296 (2.46), Ser 460 (2.21), Ser 436 (2.15). |
Trp 461, Cys 437, Cys 465. | Gly 464, Asp 435, Gly 462, Gly 472, Val 473, Thr 459, Ser 441, Gln 438, Glu 389. |
| 9 | Jatrorrhizine | −7.5 | His 296 (2.66), Gly 439 (2.17). |
His 296, Leu 302, Pro 301, Val 275, Val 280 | His 279, Gln 438, Ser 441 |
| 10 | 3-TAPAP | −6.7 | Asn 192 (2.04), Arg 182 (2.55), Glu 289 (2.38) | Ile 242, Pro 288, Met 488 | Thr 287, Lys 191, Trp 290, Arg 489, Phe 194, Pro 354 |
| 11 | Di-isopropyl flouorophosphate | −4.2 | Gln 438 (2.48), Gly 462 (2.81) | – | His 296, Ser 441, Ser 460, Thr 459, Cys 465, Trp 461, Cys 437, Gly 464, Ser 436, Asp 435, Gly 472 |
| 12 | AEBSF | −5.5 | His 296 (2.13), Ser 436 (2.29) | Cys 465, Trp 461, Asp 435 | Ser 441, Ala 466, Gly 464, Cys 437, Gln 438, Gly 439, Thr 459, Ser 460, Gly 462, Gly 472, Val 473 |
| 13 | P-Amino benzamidine | −6.0 | Asp 440 (2.02), Gly 385 (2.91), Asp 435 (2.31) | Ala 400 | Thr 387, Ala 386, Trp 384, Gly 383, Ser 394, Asn 398, Ile 381, Gly 259, Val 434, Asn 433 |
| 14 | Ulinastatin | −5.2 | Ser 441 (2.94), His 296 (2.04) | Val 280 | Gln 438, Trp 461, Ser 460, Cys 281, Gly 439 |
| 15 | Aesculitannin B | −7.7 | Ser 441 (2.30) | Leu 419, Lys 342, Cys 465 | His 296, Ser 339, Lys 340, Gly 462, Gly 464, Ser 460, Gln 438, Thr 459, Ser 436, Cys 437, Tyr 416, Arg 470, Ser 463, Trp 461, Met 424, Thr 341 |
| 16 | Ganoderiol D | −7.4 | Ser 441 (2.26), Gly 462 (2.17) | Trp 461 | His 296, Gly 439, Gln 438, Ser 436, Thr 459, Cys 437, Ser 460, Cys 465, Lys 342, Leu 419, Thr 341, Lys 340 |
| 17 | Ecdysterone | −7.3 | Asn 192 (2.74), Glu 289 (2.36) | Phe 194 | Leu 355, Phe 357, Pro 288, Ile 242, Thr 287, Arg 182, Trp 290, Thr 356, Pro 354 |
| 18 | Magnoflorine | −6.9 | Gly 462 (2.34), Gly 464 (2.83) | His 296, Trp 461 | Ser 441, Leu 419, Ser 463, Lys 342, Ser 460 |
| 19 | Tetratriacontane | −6.1 | – | Pro 363, Ile 242, Ala 243, Pro 288, Pro 354, Phe 194, Val 246, Lys 362, Leu 248, Phe 357 | Arg 240, Cys 244, Glu 289, Asn 247 |
| 20 | Hesperetin | −7.6 | Ser 441 (2.92), Ser 436 (2.22) | Cys 437, Trp 461 | His 296, Val 280, Cys 281, Gly 439, Gln 438, Ser 460, Thr 459, Gly 472, Gly 462, Asp 435, Cys 465 |
| 21 | Fisetin | −6.9 | Arg 150 (2.70), Tyr 190 (2.76), Cys 241 (2.51) | Ile 452, Leu 151, Ala 243 | Val 246, Gly 153, Phe 156, Arg 240, Ile 242, Cys 244, Arg 147 |
| 22 | Naringenin | −6.8 | Arg 486 (2.09), Arg 489 (2.77), Arg 186 (2.05), Val 219 (2.26) | Ala 490, Arg 486, Arg 489 | Asn 218, Asp 220, Ala 216, Gly 217, Tyr 485, Ala 490 |
| 23 | Ciscapsaicin | −5.5 | Gly 462 (2.54) | Met 424, Lys 342, Trp 461, Leu 419, Cys 465 | His 296, Ser 441, Gly 439, Gln 438, Cys 437, Gly 464, Thr 341 |
| 24 | Dihydrocapsaicin | −5.8 | His 296 (2.72), Ser 460 (2.24), Gly 464 (2.13) | His 296, Leu 419, Trp 461, Lys 342 | Ser 441, Thr 341, Ser 463, Gly 462, Thr 459, Gln 438 |
| 25 | 3,4-Dichloro isocoumarin | −6.3 | Cys 148 (1.95), Val 149 (2.18) | Pro 369, Lys 223, Ile 221 | Asp 187, Arg 147, Asn 368, Gly 370 |
| 26 | APMSF | −6.2 | Val 331 (2.11), Val 298 (2.49), Asn 303 (2.33) | Val 331, Val 298 | Glu 329, Pro 305, Val 328, Lys 330, Ser 333 |
3.4.1. TMPRSS2-Camostat interaction
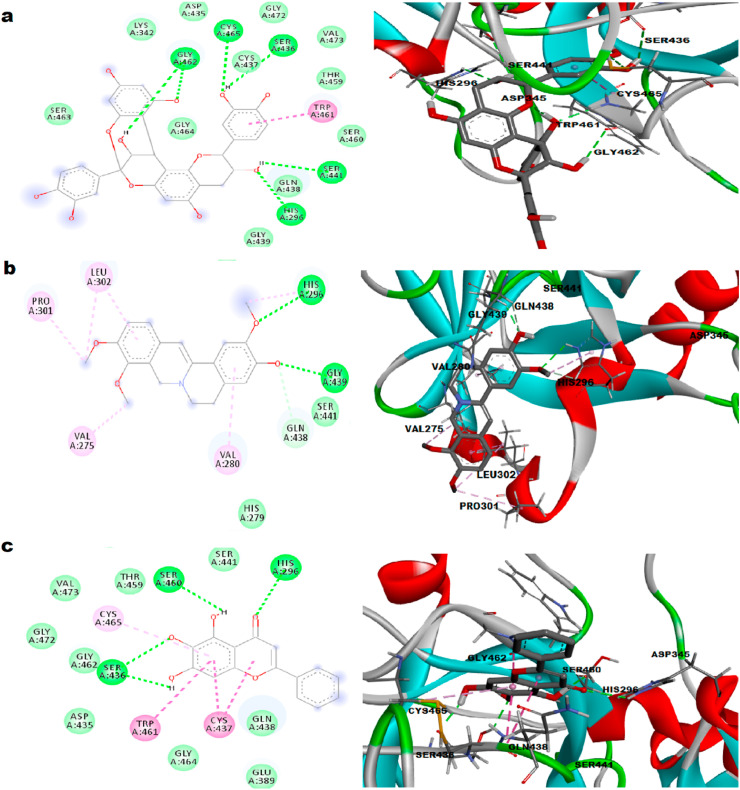
Camostat is a clinically established, well-tolerated commercial serine protease inhibitor that has been reported to block SARS-CoV-2 infection in a mouse model (McKee et al., 2020). So, it was used as reference compound for the study. The docking complex of TMPRSS2-Camostat was defined with highest binding energy of −8.4 kcal/mol. The molecular docking of camostat towards the TMPRSS2 active pocket was stabilized with the formation of five hydrogen bonds with His 296, Cys 281, Glu 389, Lys 390 and Gln 438 as represented in Fig. 4 a. Camostat's high affinity was also associated with the presence of van der Waal's forces with the amino acids Trp 461, Gly 462, Ser 460, Gly 385, His 279, Thr 393, Lys 392 and Gly 391 which created a strong cohesive environment, thereby stabilizing the complex formed. Other important interactions such as π-alkyl with Ala 386, Val 380 and Carbon–Hydrogen interaction with Ser 441, Gly 439, cys 297, Leu 302 were also observed. Two important amino acid residues of TMPRSS2 catalytic triad, Ser 441 and His 296 were involved in Carbon–Hydrogen interaction and Hydrogen bond formation respectively.
Fig. 4.
Representation of 2D and 3D plots a. Camostat, b. Meloxicam, c. Nafamostat with TMPRSS2 and interactive site residues along with hydrogen bonds and other interactions.
3.4.2. TMPRSS2-Meloxicam interaction
Meloxicam, widely used as non-steroidal anti-inflammatory drug is a lysine analogue serine protease inhibitor which can be repurposed for TMPRSS2 inhibition. The docking complex of TMPRSS2-Meloxicam was defined with binding energy of −8.3 kcal/mol as shown in Fig. 4b. The complex revealed a stable conformation with the formation of three hydrogen bonds with Gly 462, Ser 441, and Ser 460. High affinity binding was associated with presence of van der Waal's forces with residues Gly 464, His 296, Thr 459, Gln 438, Cys 465, Ser 436, His 279 and Val 278. Other interactions such as π-π stacked, Amide-π Stacked and π-alkyl were also observed. Two amino acid residues of TMPRSS2 catalytic triad, Ser 441 and His 296 were involved in hydrogen bond and van der Waal's interactions respectively.
3.4.3. TMPRSS2-Nafamostat interaction
Nafamostat is an approved TMPRSS2 inhibitor by FDA to treat acute pancreatitis. Both camostat and nafamostat were reported to block SARS-CoV-2 infection in the human cells expressing TMPRSS2 (Guy et al., 2020). Therefore, these drugs are justified for repurposing in the treatment of COVID-19. The suitability of camostat and nafamostat for COVID-19 therapy is currently being examined in clinical trials. The docking interaction of nafamostat towards the TMPRSS2 active site was stabilized with the formation of three hydrogen bonds with Ser 436, Asp 435, Gly 461 having binding energy of −7.8 kcal/mol as presented in Fig. 4c. Interactions such as π-cation, π-π stacked, π-π T-shaped, amide-π stacked and π-alkyl, which largely involve charge transfer, help in intercalating the drug to the binding site of the receptor. Two amino acid residues of catalytic triad, Ser 441 and His 296 were involved in van der Waal's and π-interactions respectively. The present results justify the possible antiviral mechanism of camostat and nafamostat as TMPRSS2 inhibitors.
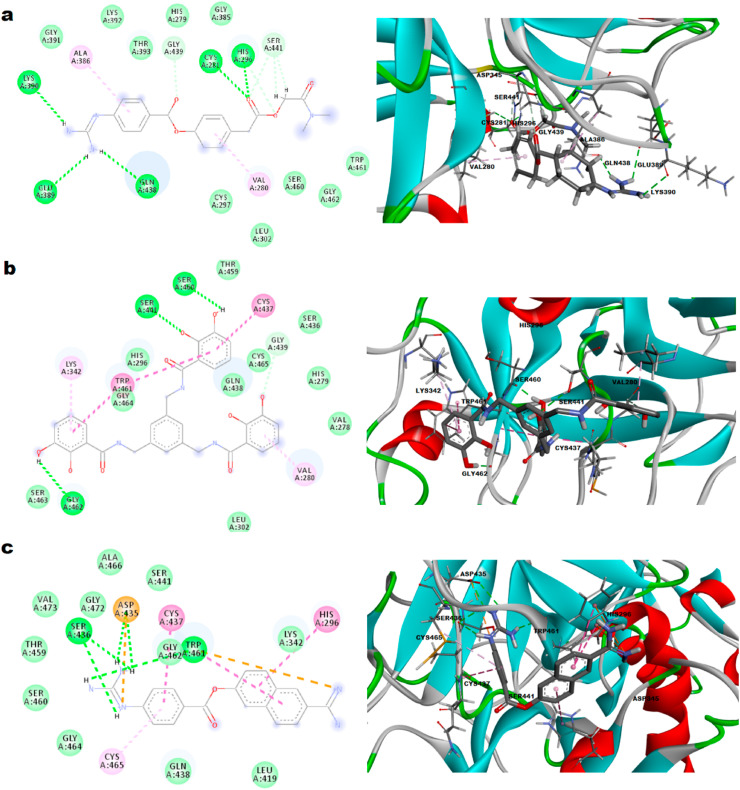
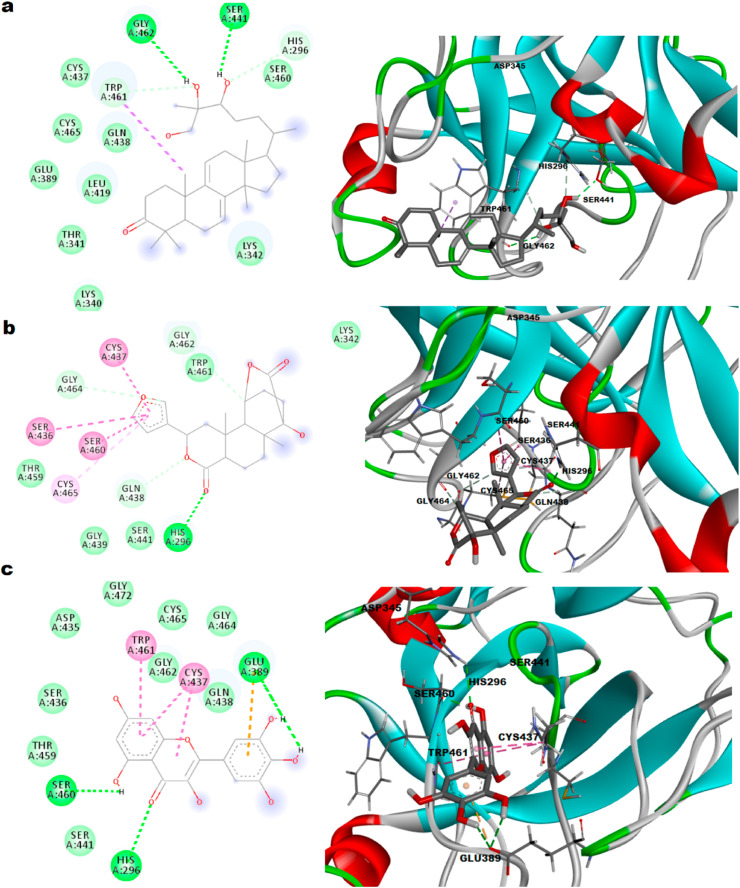
3.4.4. TMPRSS2-Ganodermanontriol interaction
Several pre-clinical and clinical studies on Ganoderma lucidum and its isolated compounds showed fruitful results in various medical disorders. Ganodermanontriol isolated from spores of G. lucidum, has been reported to possess potent anti-HIV-1 activity (protease inhibitory) in different in vivo and in vitro experiments (Boh et al., 2007). The TMPRSS2-Ganodermanontriol docked complex was identified with binding energy of - 8.1 kcal/mol. The complex formed two hydrogen bonds with Gly 462, Ser 441 as in Fig. 5 a. Other interactions such as van der Waal's with Lys 340, Thr 341, Leu 419, Glu 389, Gln 438, Cys 465, Cys 437, Lys 342, Ser 460 and Carbon–Hydrogen with His 296, Trp 461 attributed to high binding. Two amino acid residues of the catalytic triad, Ser 441 and His 296 were involved in hydrogen bond and Carbon–Hydrogen interactions respectively. The present results suggest the possible mechanism and usefulness of this compound in inhibiting TMPRSS2 and thus in treating COVID-19.
Fig. 5.
Representation of 2D and 3D plots a. Ganodermanontriol, b. Columbin, c. Myricetin with TMPRSS2 and interactive site residues along with hydrogen bonds and other interactions.
3.4.5. TMPRSS2-Columbin interaction
Columbin, a major compound from Tinospora cordifolia has been reported to possess antiviral activity particularly anti-HIV activity due to its protease inhibitory potential (Lin et al., 2014). The TMPRSS2-columbin complex was defined with binding energy of −8.2 kcal/mol. The complex revealed stable conformation with the formation of one hydrogen bond with His 296 as shown in Fig. 5b. Interactions such as van der Waal's, Carbon–Hydrogen, Amide-π Stacked and π-alkyl, which helps in conformation of the drug within the receptor were also seen. Two catalytic triad's, amino acid residues like Ser 441 and His 296 were involved in van der Waal's and hydrogen bond respectively. The present results suggest the possible inhibitory action on TMPRSS2.
3.4.6. TMPRSS2-Myricetin interaction
Myricetin that is found in Isatis indigotica and Torreya nucifera has antiviral activity against HIV and influenza virus, also reported to inhibit helicase (nsp13), 3CL protease of SARS-CoV, reverse transcriptase and protease enzymes (da Silva Antonio et al., 2020). The molecular docking of myricetin towards the TMPRSS2 active pocket was stabilized with the formation of three hydrogen bonds at His 296, Ser 460, Glu 389 with bonding energy of −8.3 kcal/mol as presented in Fig. 5c. van der Waal's forces with residues Ser 441, Thr 459, Ser 436, Asp 435, Gly 472, Gly 462, Cys 465, Gly 464, Gln 438 and π-anion with Glu 389 contributed to stabilization of the drug-protein complex. Two amino acid residues of the catalytic triad, Ser 441 and His 296 were involved in van der Waal's and hydrogen bonds respectively. The present results suggest the possible intervention of TMPRSS2 inhibitory potential in the antiviral activity.
3.4.7. TMPRSS2-Proanthocyanidin A2 interaction
Proanthocyanidin A2 found in lychee seeds, Aesculus hippocastanum, Crataegus sinaica have been reported to exhibit in vitro as well as in vivo anti-viral activities against coxsackie B virus, herpes simplex virus type 1 (HSV-1), porcine reproductive and respiratory syndrome virus, canine distemper virus (Zhang et al., 2018). The molecular docking of Proanthocyanidin A2 towards TMPRSS2 active site showed a binding energy of −7.9 kcal/mol. The docking complex exhibited five hydrogen Bonds at Ser 441, His 296, Gly 462, Cys 465, Ser 436 as visualised in Fig. 6 a. Other interactions such as van der Waal's, amide-π Stacked were observed. Two amino acid residues of catalytic triad Ser 441 and His 296 of TMPRSS2 were involved in hydrogen bond. A similar compound aesculitannin B found in lychee seeds also showed a binding energy of −7.7 kcal/mol with a hydrogen bond and van der Waal's interactions (Table 1). These results indicate inhibition of TMPRSS2 as the possible pathway in the antiviral activity of proanthocyanidin A2 and aesculitannin B and thus consumption of lychees may be beneficial in treating viral infections.
Fig. 6.
Representation of 2D and 3D plots a. Proanthocyanidin A2, b. Jatrorrhizine, c. Baicalein with TMPRSS2 and interactive site residues along with hydrogen bonds and other interactions.
3.4.8. TMPRSS2-Jatrorrhizine interaction
Jatrorrhizine is an alkaloid obtained from Enantia chlorantha, Tenospora cardifolia, Mahonia aquifolium (Kuete V. 2014). It has been reported as an effective inhibitor of HIV Protease and H1N1 viral proteins. The present docking with TMPRSS2 showed a binding energy of −7.5 kcal/mol, with two hydrogen bonds formed in the complex at His 296 and Gly 439 as presented in Fig. 6b. Other interactions seen were π-interactions at Val 275, Val 280, Leu 302, Pro 301, Carbon–Hydrogen interactions at Gln 438 and van der Waal's at Ser 441, His 279. Two amino acid residues of the catalytic triad, His 296 and Ser 441 were involved in hydrogen bond and van der Waal's interaction respectively. The obtained results suggest protective effects of jatrorrhizine against SARS-CoV-2 possibly by inhibiting host TMPRSS2 enzyme.
3.4.9. TMPRSS2-Baicalein interaction
Baicalein is an isoflavanoidal compound commonly found in Scutellaria baicalensis which has been reported for in vitro virucidal properties against dengue virus, EV-A71, also possess inhibitory potential against H5N1 neuraminidase and HIV reverse transcriptases (Lalani et al., 2020; Moghaddam et al., 2014). The docking results from the present study revealed a binding energy of −7.7 kcal/mol. The complex of Baicalein with TMPRSS2 revealed stable conformation with the formation of three hydrogen bonds at His 296, Ser 460, Ser 436 and other interactions such as van der Waal's at Ser 441, Asp 435, Gln 438, Glu 389, Gly 464, Gly 462, Gly 472, Val 473, Thr 459, and π-interactions with Cys 437, Cys 465, Trp 461 as depicted in Fig. 6c. Two amino acid residues of the catalytic triad Ser 441 and His 296 were involved in van der Waal's and hydrogen bonds respectively. These results support the possible host TMPRSS2 inhibitory potential of baicalein and thus its usefulness in treating SARS-CoV-2 infection.
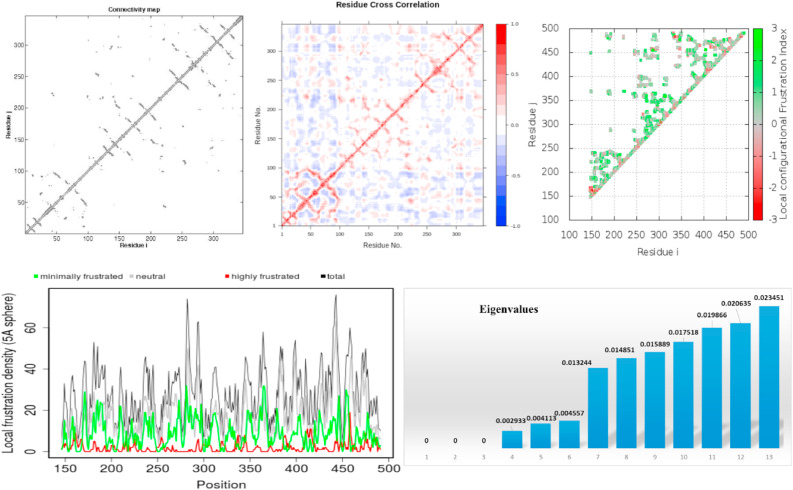
3.5. Essential dynamics
The eigenvectors calculations and eigenvalues that are identified by the matrix give the essential protein dynamics (Ahamad et al., 2019). The dynamic behaviour of the modelled structure of TMPRSS2 was found stable with the parameters mentioned in Table 2 . The projections that can be visualized manually define the cluster of stability between two states apparently forming the plots. The results revealed that the clusters are well defined in the TMPRSS2 model covering minimum region as shown in Fig. 7 . Overall, the protein structure was examined with a diagonalized covariance matrix to know the positional fluctuations on Cα-atoms. The reduction in the residual displacement in a large scale was observed on receptor protein, revealing the stable residues in the protein. Anisotropic Network Model (ANM) works on the covariance matrix which will be developed based on the trajectories after removal of the rotational and translational movements within the macromolecule (Amir et al., 2019). Magnitude of motion and the point direction was calculated using ANM by DynOmics web server for the TMPRSS2 modelled structure. The analysis was indicated with two colour representations, where small fluctuations were given blue and red colour denoted large fluctuations. The amplitude and the intensity of blue were magnified in the protein structure with stable residual frustration. The minimal frustrations principle and the free energy landscape of the given proteins are well characterized with the global energy minima occurrence which also includes local minima. The frustratometer follow the mentioned principle and the analysis revealed the location of frustration was stable in the TMPRSS2 modelled structure. The results showed stable frustration at positions 400–450 and 450–500 and a few residues around ~350–400 and around ~450–500. The overall analysis provided that the gain of functional properties of TMPRSS2 modelled structure showing large scale shift in the patterns. This could probably be responsible for the stability and functionality of protein.
Table 2.
ADME/T properties of the selected compounds using Swiss-ADME online server.
| ADME/T properties | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. No | Compound | Molecular weight (g/mol) | Rotatable bonds | LogP | Blood brain barrier | Lipinski rule of five | Absorption | LogS | H-bond accept | H-bond donor | TPSA (A2) | log Kp (cm/s) |
| 1 | Camostat | 374.4 | 6 | 0.31 | No | 0 | Low | −4.22 | 3 | 4 | 115.56 | −6.43 |
| 2 | Columbin | 392.5 | 4 | 3.37 | No | 0 | Low | −4.14 | 4 | 3 | 162.88 | −6.64 |
| 3 | Meloxicam | 443.41 | 6 | 2.68 | No | 0 | Low | −4.61 | 4 | 4 | 141.85 | −6.94 |
| 4 | Myricetin | 409.31 | 6 | −2.85 | No | 1 | Low | −4.31 | 3 | 5 | 123.7 | −6.7 |
| 5 | Ganodermanontriol | 380.79 | 5 | 2.8 | No | 0 | Low | −4.89 | 3 | 5 | 157.83 | −5.07 |
| 6 | Nafamostat | 419.68 | 4 | 2.8 | No | 1 | Low | −4.12 | 3 | 4 | 150.7 | −5.23 |
| 7 | Proanthocyanidin A2 | 576.50 | 2 | 1.61 | No | 3 | Low | −6.44 | 12 | 9 | 209.76 | −8.12 |
| 8 | Baicalein | 446.36 | 4 | 0.22 | No | 2 | Low | −4.63 | 11 | 6 | 187.12 | −8.23 |
| 9 | Jatrorrhizine | 338.38 | 3 | 2.31 | Yes | 0 | High | −4.19 | 4 | 1 | 51.80 | −5.94 |
| ADME/T properties | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S. No | Compound | Lead-likeness | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor | ESOL Solubility (mg/ml) | Bioavailability | Synthetic accessibility |
| 1 | Camostat | 1 | No | No | No | No | No | 2.35E-02 | 0.55 | 2.83 |
| 2 | Columbin | 1 | No | No | No | No | No | 2.87E-02 | 0.55 | 4.17 |
| 3 | Meloxicam | 1 | No | No | No | No | No | 1.09E-02 | 0.55 | 3.79 |
| 4 | Myricetin | 1 | No | No | No | No | No | 2.02E-02 | 0.55 | 3.44 |
| 5 | Ganodermanontriol | 1 | No | No | No | No | No | 2.86E-03 | 0.17 | 3.43 |
| 6 | Nafamostat | 1 | No | No | No | No | No | 3.92E-02 | 0.55 | 5.41 |
| 7 | Proanthocyanidin A2 | 1 | No | No | Yes | No | No | −5.21 | 0.17 | 5.85 |
| 8 | Baicalein | 1 | No | No | No | No | No | −3.41 | 0.11 | 5.09 |
| 9 | Jatrorrhizine | 1 | Yes | No | No | Yes | Yes | −4.37 | 0.55 | 3.06 |
Minimal Ranges: MW = Molecular Weight (130.0/725.0), Rotor = No. of Rotatable Bonds (0.0/15.0), logP o/w = log P for octanol/water (−2.0/6.5), logBB = log BB for brain/blood (Low), logS = log S for aqueous solubility (−6.5/0.5), % Human Oral Absorption in GI (High), Lipinski Rule of 5 Violations (maximum is 4), accPthB = Acceptor - Hydrogen Bonds (2.0/20.0), donorHB = Donor - Hydrogen Bonds (0.0/6.0), log Kp = log Kp for skin permeability (Kp in cm/s), TPSA = topological polar surface area (0.0/450.0), ESOL = Estimated SOLubility (−2.78/10.0), Bioavailability=(0.00/0.55), Synthetic accessibility=(2.0/3.0).
Fig. 7.
The dynamic behaviour of the TMPRSS2 modelled structure generated using Frustratometer and covariance matrix revealing the local frustration density, eigen values, connectivity map and the residue cross section.
3.6. Physiochemical and ADME/T properties
The selection of compounds depends on both pharmacological and ADME/T screening for a better probability of clinical success. The physicochemical properties of the selected molecules were analysed based on the Lipinski rule of five which includes molecular weight, rotatable bonds, hydrogen bond acceptor, log P octanol-water partition coefficient and hydrogen bond donor. The output of Swiss-ADME for blood-brain barrier penetration, log P range (−100 to 3), lead-likeliness (1–2), intestinal absorption (low), permeability (−8 to −13), bioavailability (0.11–0.55) and solubility of nafamostat, meloxicam, columbin, myricetin, ganodermanontriol, proanthocyanidin A2, baicalein and jatrorrhizine were well within the acceptable range as that of camostat. The compounds obeyed the rule of five suggesting good absorption in the biological systems. The drug-like properties were exhibited by effective values of molecular weight, hydrogen bonding capacity, rotatable bond count and surface polar atoms (Antalis et al., 2010). Nonetheless, jatrorrhizine showed few deviations like blood-brain barrier penetration and high intestinal absorption when compared to camostat as represented in Table 2.
Thus, the utilization of computational approaches like molecular modelling and molecular docking gave an insight to identify potent molecules against TMPRSS2. Targeting host cellular elements is a fairly novel antiviral approach that may lessen or avoid the occurrence of escape mutants. Out of twenty five compounds studied, eight compounds namely nafamostat, meloxicam, ganodermanontriol, columbin, myricetin, proanthocyanidin A2, jatrorrhizine and baicalein showed good binding energies in comparison to the standard TMPRSS2 inhibitor, camostat. The results also revealed important residues like His 296, Ser 441, Asp 345 (catalytic triad), and others like Ser 411, Ser 436, Gly 439, Cys 437, Gln 438, Thr 459, Trp 461, Gly 462, Ser 460, Gly 464 and Cys 465 which were actively involved in the hydrogen bond formation and other interactions to stabilize the receptor-drug complex. The compounds were predicted to show good absorption in the biological systems and drug likeliness as per Lipinski rule of five without any violation of nominal ranges. Therefore, the selected compounds, that are capable of binding to the TMPRSS2 enzyme might act as potential inhibitors and halt the entry of the virus into the host cell, thus preventing the spread of SARS-CoV-2. Thus, the active molecules identified can be further investigated through in vitro and in vivo studies for their clinical efficacy and can be used in the treatment and management of COVID-19.
4. Conclusion
Molecular modelling and molecular docking techniques have been employed for the search of possible inhibitors that are available in the established databases. The study used structural homology modelling, validation and docking studies against TMPRSS2. The developed model can be used as a binding site target for further in silico studies. It is exciting to uncover that the compounds nafamostat, meloxicam, ganodermanontriol, columbin, myricetin, proanthocyanidin A2, jatrorrhizine and baicalein have shown good affinity towards the active site of TMPRSS2. The selected compounds in the present study have potentiality to inhibit host cell entry of SARS-CoV-2 associated with TMPRSS2 enzyme. Therefore, the compounds serve as candidate molecules for the drug development by wet-lab evaluations in anti-viral treatment regimen.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
credit authorship contribution statement
Pooja M: Conceptualization, Methodology, Software, Validation, Investigation, Writing - original draft, Visualization. Gangavaram Jyothi Reddy: Conceptualization, Methodology, Investigation, Writing - review & editing. Kanipakam Hema: Methodology, Software, Validation, Writing - review & editing. Sujatha Dodoala: Conceptualization, Data curation. Bharathi Koganti: Supervision, Writing - review & editing.
Declaration of competing interest
Authors declare no conflicts over the work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejphar.2020.173688.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Ahamad S., Islam A., Ahmad F., Dwivedi N., Hassan M.I. 2/3D-QSAR, molecular docking and MD simulation studies of FtsZ protein targeting benzimidazoles derivatives. Comput. Biol. Chem. 2019;78:398–413. doi: 10.1016/j.compbiolchem.2018.12.017. [DOI] [PubMed] [Google Scholar]
- Amir M., Ahamad S., Mohammad T., Jairajpuri D.S., Hasan G.M., Dohare R., Islam A., Ahmad F., Hassan M.I. Investigation of conformational dynamics of tyr89cys mutation in protection of telomeres 1 gene associated with familial melanoma. J. Biomol. Struct. Dyn. 2019:1–10. doi: 10.1080/07391102.2019.1705186. [DOI] [PubMed] [Google Scholar]
- Antalis T.M., Buzza M.S., Hodge K.M., Hooper J.D., Netzel-Arnett S. The cutting edge: membrane-anchored serine protease activities in the pericellular microenvironment. Biochem. J. 2010;428(3):325–346. doi: 10.1042/BJ20100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizian H., Bahrami H., Pasalar P., Amanlou M. Molecular modeling of Helicobacter pylori arginase and the inhibitor coordination interactions. J. Mol. Graph. Model. 2010;28(7):626–635. doi: 10.1016/j.jmgm.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Boh B., Berovic M., Zhang J., Zhi-Bin L. Ganoderma lucidum and its pharmaceutically active compounds. Biotechnol. Annu. Rev. 2007;13:265–301. doi: 10.1016/S1387-2656(07)13010-6. [DOI] [PubMed] [Google Scholar]
- da Silva Antonio A., Wiedemann L.S.M., Veiga-Junior V.F. Natural products' role against COVID-19. RSC Adv. 2020;10(39):23379–23393. doi: 10.1039/D0RA03774E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanavade M.J., Sonawane K.D. Insights into the molecular interactions between aminopeptidase and amyloid beta peptide using molecular modeling techniques. Amino Acids. 2014;46(8):1853–1866. doi: 10.1007/s00726-014-1740-0. [DOI] [PubMed] [Google Scholar]
- Elfiky A.A. Life sciences; 2020. Anti-HCV, Nucleotide Inhibitors, Repurposing against COVID-19; p. 117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Wilkins M.R., Appel R.D., Bairoch A. The Proteomics Protocols Handbook. Humana press; 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [Google Scholar]
- Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy R.K., DiPaola R.S., Romanelli F., Dutch R.E. Rapid repurposing of drugs for COVID-19. Science. 2020;368(6493):829–830. doi: 10.1126/science.abb9332. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A., Garg S., Shiekh B.A., Singh N., Singh P., Bhatti R. In silico studies and in vivo MAOA inhibitory activity of coumarins isolated from Angelica archangelica extract: an approach toward Antidepressant activity. ACS Omega. 2020;5(25):15069–15076. doi: 10.1021/acsomega.0c00887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khobragade C.N., Beedkar S.D., Bodade R.G., Vinchurkar A.S. Comparative structural modeling and docking studies of oxalate oxidase: possible implication in enzyme supplementation therapy for urolithiasis. Int. J. Biol. Macromol. 2011;48(3):466–473. doi: 10.1016/j.ijbiomac.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Kleine-Weber H., Elzayat M.T., Hoffmann M., Pöhlmann S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci. Rep. 2018;8(1):1–11. doi: 10.1038/s41598-018-34859-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuete V. Toxicological Survey of African Medicinal Plants. Elsevier; 2014. Health effects of alkaloids from African medicinal plants; pp. 611–633. [DOI] [Google Scholar]
- Lalani S.S., Anasir M.I., Poh C.L. Antiviral activity of silymarin in comparison with baicalein against EV-A71. BMC complementary medicine and therapies. 2020;20(1):1–12. doi: 10.1186/s12906-020-2880-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmann J.A.C., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8(4):477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Lin L.T., Hsu W.C., Lin C.C. Antiviral natural products and herbal medicines. J. Trad. Comp. Med. 2014;4(1):24–35. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee D.L., Sternberg A., Stange U., Laufer S., Naujokat C. Pharmacological Research; 2020. Candidate Drugs against SARS-CoV-2 and COVID-19; p. 104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkonen L., Pihlajamaa P., Sahu B., Zhang F.P., Jänne O.A. Androgen receptor and androgen-dependent gene expression in lung. Mol. Cell. Endocrinol. 2010;317(1–2):14–24. doi: 10.1016/j.mce.2009.12.022. [DOI] [PubMed] [Google Scholar]
- Moghaddam E., Teoh B.T., Sam S.S., Lani R., Hassandarvish P., Chik Z., Yueh A., Abubakar S., Zandi K. Baicalin, a metabolite of baicalein with antiviral activity against dengue virus. Sci. Rep. 2014;4:5452. doi: 10.1038/srep05452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran G.N. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- Schwede T., Kopp J., Guex N., Peitsch M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L.W., Mao H.J., Wu Y.L., Tanaka Y., Zhang W. TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017;142:1–10. doi: 10.1016/j.biochi.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temml V., Kaserer T., Kutil Z., Landa P., Vanek T., Schuster D. Pharmacophore modeling for COX-1 and-2 inhibitors with LigandScout in comparison to discovery Studio. Future Med. Chem. 2014;6(17):1869–1881. doi: 10.4155/fmc.14.114. [DOI] [PubMed] [Google Scholar]
- Visegrády B., Than N.G., Kilár F., Sümegi B., Than G.N., Bohn H. Homology modelling and molecular dynamics studies of human placental tissue protein 13 (galectin-13) Protein Eng. 2001;14(11):875–880. doi: 10.1093/protein/14.11.875. [DOI] [PubMed] [Google Scholar]
- Volkamer A., Kuhn D., Rippmann F., Rarey M. DoGSiteScorer: a web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics. 2012;28(15):2074–2075. doi: 10.1093/bioinformatics/bts310. [DOI] [PubMed] [Google Scholar]
- Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(Suppl. l_2):W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Knox C., Guo A.C., Cheng D., Shrivastava S., Tzur D., Gautam B., Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Suppl. l_1):D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; Geneva, Switzerland: 2020. WHO Coronavirus Disease (COVID-19) Dashboard.https://covid19.who.int/ Accessed. [Google Scholar]
- Zhang M., Wu Q., Chen Y., Duan M., Tian G., Deng X., Sun Y., Zhou T., Zhang G., Chen W., Chen J. Inhibition of proanthocyanidin A2 on porcine reproductive and respiratory syndrome virus replication in vitro. PLoS One. 2018;13(2) doi: 10.1371/journal.pone.0193309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.