Abstract
Interleukin-10(IL-10) is an immunosuppressive cytokine and plays an important role in inflammation and cancers. Numerous studies have explored the association between single nucleotide polymorphisms of IL-10 and leukemia, but their results were conflicting, so we performed this meta-analysis to elucidate the association between 3 common single nucleotide polymorphisms of IL-10 (rs1800896, rs1800871 and rs1800872) and risk of leukemia.
We conducted a comprehensive research in Pubmed, Chinese Biomedical Literature Database disc and Embase using related terms. After strict selection, 18 studies with 2264 cases and 3846 controls were included into this meta-analysis. Odds ratio and 95% confidence interval were used to evaluate the strength of the association.
We found that polymorphism of IL-10 -1082A/G was associated with decreased risk of leukemia both in overall analysis and in stratified analysis according to ethnicity and cancer type. A strong relationship was also uncovered between polymorphism of IL-10 -592C/A and increased risk of leukemia in non-Chinese.
GG genotype of IL-10 -1082A/G is associated with decreased risk of leukemia, especially chronic lymphocytic leukemia. CC genotype of -592C/A is associated with decreased risk of leukemia in non-Chinese.
Keywords: interleukin-10, leukemia, meta-analysis, polymorphism
1. Introduction
Leukemia is 1 of the commonest cancers across the world, with a top ten incidence and mortality in both sex and all ages.[1] The incidence and mortality increased to the fifth place among children, adolescents and young adults.[2] However, the mechanisms underlying leukemia are complex and not fully known.
More and more researches revealed the indispensable participation of immunity during the process of cancers. Immunosuppressive cytokines such as interleukin-10 (IL-10) and transforming growth factor-β play important roles in the immunity of tumor.[3,4] IL-10 belongs to Th2 pathway. It can counterbalance the immune system by regulating both adaptive and innate immune system to protect host from exacerbated inflammation.[3,5] Altered expression of IL-10 may lose the control of immune homeostasis, thereby increasing the risk of cancers.[6]
Recent genetic studies disclosed the important role of single nucleotide polymorphisms (SNPs) in many kinds of cancers.[7] SNPs in gene regulating and coding areas can alter the expression level or function of these gene or the products they encode. So SNPs are increasingly found to be critical in the mechanisms of cancers.[8,9]
SNPs in the transcriptional start area of IL-10 gene influence the production of IL-10 thus may increase or decrease the risk of cancers,[10–12] including leukemia, but the conclusions are inconsistent. For example, Ovsepyan, V. A. et al[13] found SNP in the IL-10 -1082A/G was associated with the risk of chronic lymphoid leukemia, while Lo, W. J. et al[14] found no association between IL-10 -1082A/G and leukemia. Conflicting results may be due to small sample size. So we performed this pooled analysis to elucidate the relations between leukemia and 3 common SNPs of IL-10(-1082A/G, -819 C/T, -592C/A).
2. Material and method
2.1. Patient involvement and ethical approval
There was no patient involvement in this study. Ethical approval is not necessary for a meta-analysis.
2.2. Literature search and identification
We conducted a comprehensive search in Pubmed, Chinese Biomedical Literature Database disc, EMBASE with following related terms: (“IL-10” OR “IL10” OR “interleukin10” OR “rs1800896” OR “rs1800871 ” OR “rs1800872 ”) AND (“polymorphism∗” OR “variant∗” OR “gene∗” OR “variation∗” OR “SNP∗”) AND (“leukemia” OR “leukaemia”). All publications were updated to April 22, 2019. We included studies with inclusion criteria as follows:
-
(1)
written in English or Chinese;
-
(2)
human case-control studies about the association between leukemia and any 1 of the 3 SNPs mentioned above;
-
(3)
available genotype data for estimation of an odds ratio (OR) and 95% confidence interval (CI).
The exclusion criteria were:
-
(1)
reviews, letters, editorial articles or case reports;
-
(2)
without controls or genotype data. The latest study was adopted when overlapping with previous studies.
2.3. Data extract
Title and abstract of all articles were screened by 2 reviewers independently and eligible studies were identified according to the inclusion and exclusion criteria. Two investigators extracted data independently from selected studies. From each study, the following information was obtained: the first author's name, publication year, country where study was conducted, source of control, genotyping method, cancer type, and genotypes of cases and controls.
2.4. Statistics analysis
For each study, Pearson χ2 and P value were calculated to test whether the genotype frequencies in controls were consistent with Hardy-Weinberg equilibrium. We estimated odds ratio and 95% CI to assess the linkage between risk of leukemia and 3 SNPs under 4 comparisons (homozygote comparison, heterozygote comparison, dominant and recessive genetic model comparison). For IL-10 -1082A/G (rs1800896), they were GG vs AA, GA vs AA, GG+GA vs AA, GG vs GA+AA respectively. For IL-10 -819C/T (rs1800871), they were TT vs CC, TC vs CC, TT+TC vs CC, TT vs TC+CC. For IL-10 -592C/A (rs1800872), they were AA vs CC, AC vs CC, AA+AC vs CC, AA vs AC+CC. Subgroup analysis was conducted by ethnicity (Chinese or non-Chinese), phase (acute or chronic) and cancer type (acute myelocytic leukemia, acute lymphoblastic leukemia, chronic myelocytic leukemia and chronic lymphocytic leukemia [CLL]). Heterogeneity among studies was calculated using Q-statistics and I2 statistics. We thought there was no significant heterogeneity when P > 0.1 or I2 < 50%, then fixed-effects models were adopted, otherwise random-effects models were adopted.[15] We also performed stratified analysis to figure out the cause of heterogeneity and to reduce it. Z-test was conducted to exam the significance of ORs and 2-sided P < .05 was considered significant. Sensitivity analysis was conducted by recalculating overall ORs after omitting each individual study in turn. We also used Begg funnel plot and Egger linear regression test to detect publication bias.[16] All analysis was performed using Review Manager 5.0.23 (Cochrane Library Software, Oxford) and STATA14.0 software (STATA Corporation, College Station, Texas).
3. Results
3.1. Characteristics of studies
After searching on Pubmed, Chinese Biomedical Literature Database disc, and Embase using retrieval strategy mentioned above, 425 articles were recruited. 387 studies were excluded (107 duplicates, 280 studies unrelated), with 38 articles being left for full-text review. Next, 20 articles were removed for the following reasons: 5 articles were not case-control studies; 6 articles lack available data; 6 articles were not about the association between leukemia and SNPs of IL-10; 1 study studied other SNPs of IL-10; 1 article overlapped with another study; 1 article was written in Turkish. Finally, there were 18 studies with 2264 cases and 3846 controls included into this meta-analysis. The process of selecting included studies is shown in Figure 1.
Figure 1.
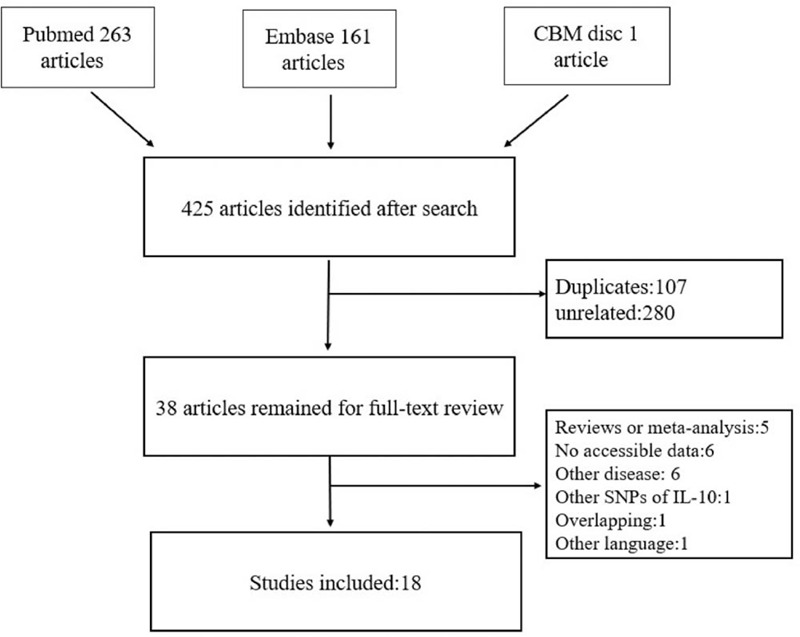
Process of selecting included studies in this meta-analysis.
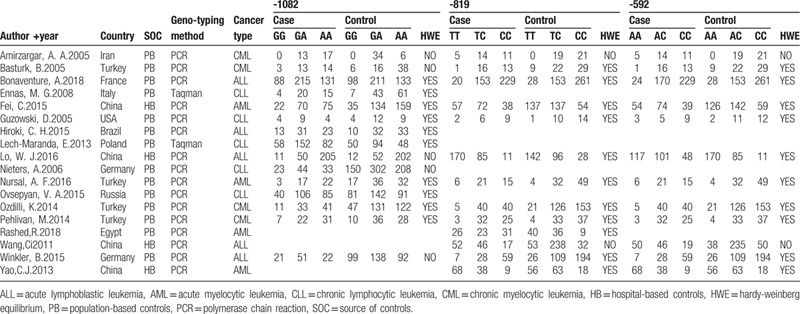
The details of eligible studies are presented in Table 1. Most studies were performed in non-Chinese ethnicity,[13,17–29] with 4 studies in Chinese population.[14,30–32] We found that controls in 6 studies were not consistent with Hardy-Weinberg equilibrium[14,17,18,24,29,30] maybe because of genotyping error or selection bias during controls’ recruitment.
Table 1.
Characteristics of published studies included in this meta-analysis.
3.2. Quantitative synthesis
The association between 3 SNPs of IL-10 and leukemia by overall analysis and subgroup analysis is shown in Table 2 and Table 3 . The overall analysis indicated that rs1800896 polymorphism was associated with risk of leukemia under recessive genetic model comparison (GG vs GA+AA: OR, 0.83; 95% CI, 0.83[0.71,0.98], P = .03, Fig. 2).
Table 2.
Overall analysis of association between 3 SNPs of IL-10 and risk of leukaemia.
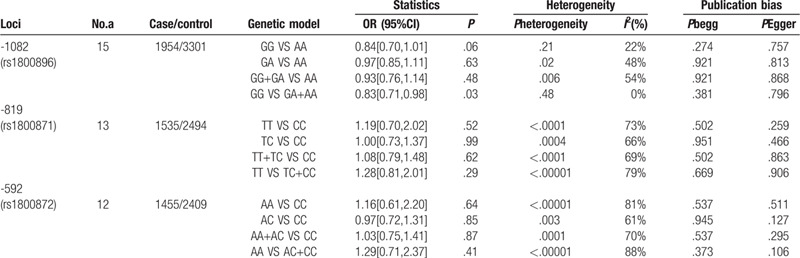
Table 3.
Stratified analysis of association between 3 SNPs of IL-10 and risk of leukemia.
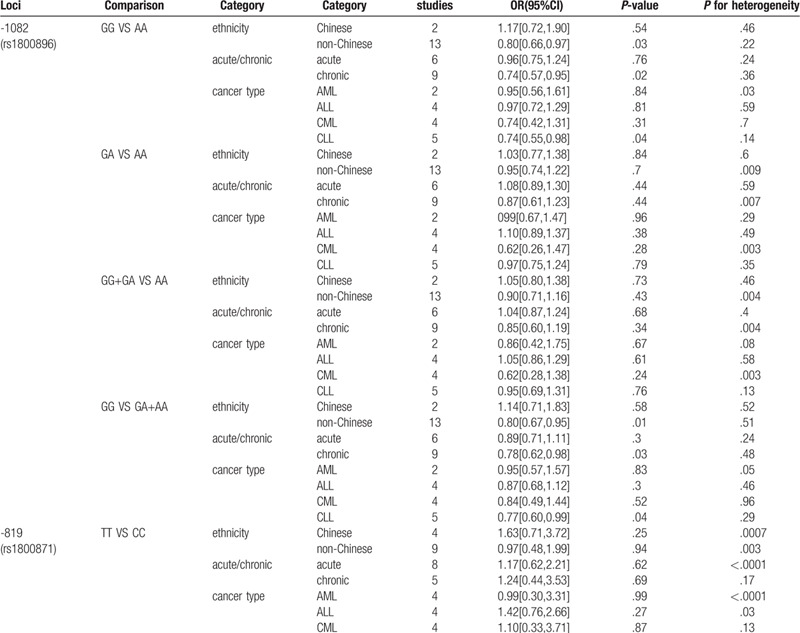
Figure 2.
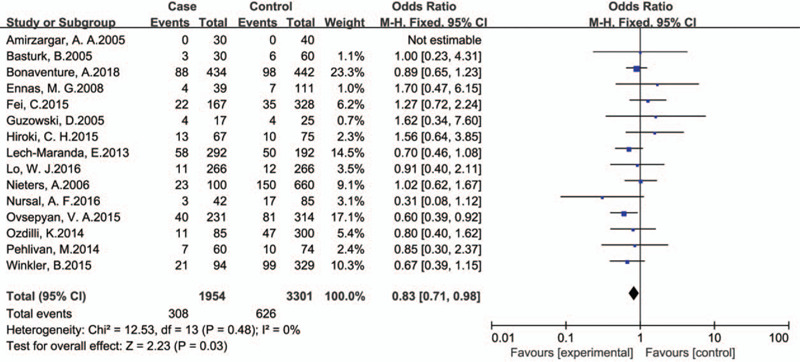
Overall analysis for the association between IL-10 rs1800896 polymorphism and leukemia risk (GG vs GA+AA).
Table 3 (Continued).
Stratified analysis of association between 3 SNPs of IL-10 and risk of leukemia.
Stratified analysis by ethnicity indicated that polymorphism of IL10 -1082 A/G(rs1800896) was associated with leukemia in non-Chinese under homozygote comparison and recessive genetic model comparison (GG vs AA: OR, 0.80; 95% CI, 0.80[0.66,0.97], P = .03; GG vs GA+AA: OR, 0.80; 95% CI, 0.80[0.67,0.95], P = .01). Besides, subgroup analysis of IL10 -1082 A/G by phase of leukemia indicated that GG genotype was associated with decreased risk of chronic leukemia (GG vs AA: OR, 0.74; 95% CI, 0.74[0.57,0.95], P = .02; GG vs GA+AA: OR, 0.78; 95% CI, 0.78[0.62,0.98], P = .03.), especially CLL, for stratified analysis by cancer type showed a strong link between this polymorphism and risk of CLL under homozygote comparison and recessive genetic model comparison (GG vs AA: OR, 0.74; 95% CI, 0.74[0.55,0.98], P = .04; GG vs GA+AA: OR, 0.77; 95% CI, 0.77[0.60,0.99], P = .04 Fig. 3).
Figure 3.
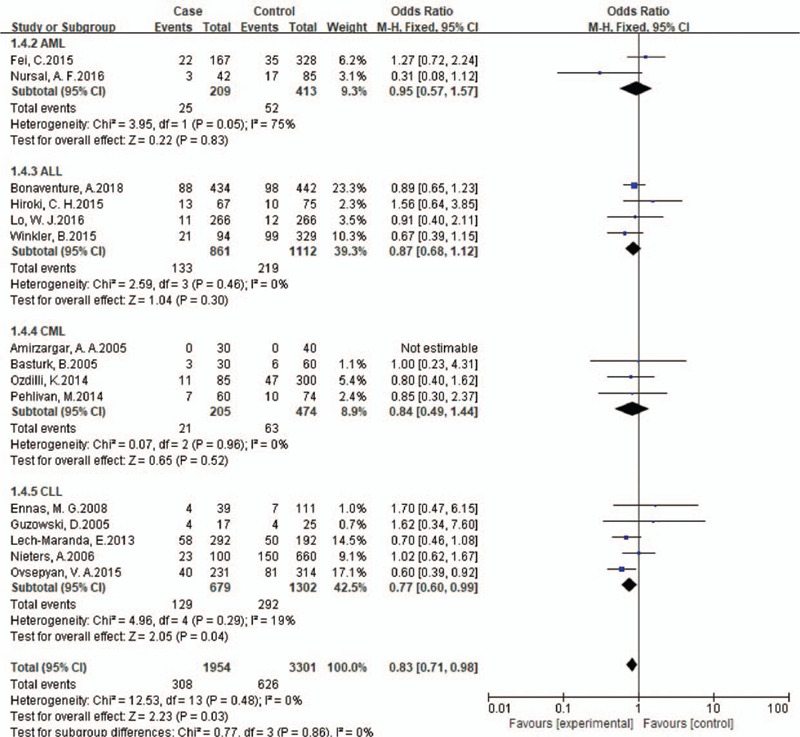
Subgroup analyses for the association between IL-10 rs1800896 polymorphism and leukemia risk according to cancer type (GG vs GA+AA).
We found no association between IL-10 -819 C/T (rs1800871) and leukemia in overall analysis and subgroup analysis.
We found no association between IL-10 -592C/A (rs1800872) and leukemia in overall analysis. But we found a strong association between rs1800872 and leukemia in non-Chinese under heterozygote comparison and dominant genetic model comparison. (AC vs CC: OR, 1.24; 95% CI, 1.24[1.02,1.50], P = .03, Fig. 4. AA+AC vs CC: OR,1.22; 95%CI,1.22[1.01,1.46], P = .04).
Figure 4.
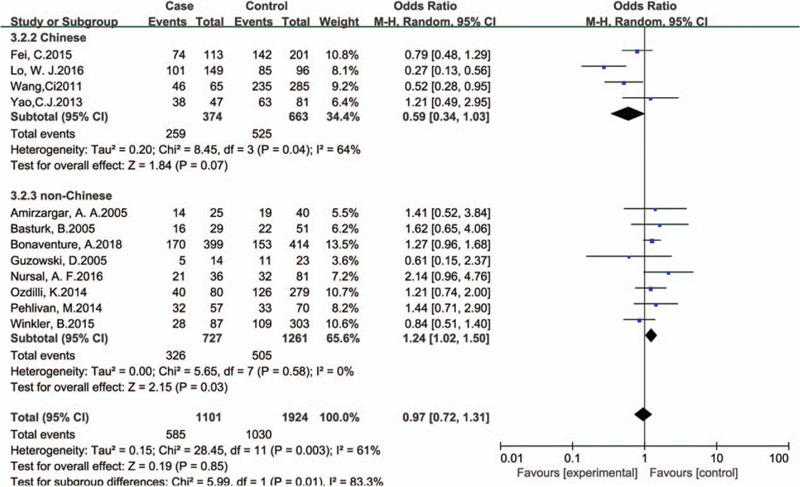
Subgroup analyses for the association between IL-10 rs1800872 polymorphism and leukemia risk according to ethnicity (AC vs CC).
3.3. Test of heterogeneity
For the overall test of heterogeneity, most comparisons had low or moderate heterogeneity. The heterogeneity of rs1800896 was low or moderate(GG vs AA: P = .21, I2 = 22%; GA vs AA:P = .02, I2 = 48%; GG+GA vs AA:P = .006, I2 = 54%; GG vs GA+AA: P = .48, I2 = 0%.), while the heterogeneity of rs1800871(TT vs CC: P < .0001, I2 = 73%; TC vs CC: P = .0004, I2 = 66%; TT+TC vs CC:P < .0001, I2 = 69%; TT vs TC+CC: P < .00001, I2 = 79%.) and rs1800872(AA vs CC: P < .00001, I2 = 81%; AC vs CC:P = .003, I2 = 61%; AA+AC vs CC:P = .0001, I2 = 70%; AA vs AC+CC: P < .00001, I2 = 88%.) was moderate or high. In the subgroup analysis according to ethnicity, the heterogeneity of rs1800872 was moderate or high in Chinese (AA vs CC: P < .00001, I2 = 93%; AC vs CC:P = .04, I2 = 64%; AA+AC vs CC:P = .0002, I2 = 84%; AA vs AC+CC: P < .00001, I2 = 96%.) while low or moderate in non-Chinese(AA vs CC: P = .15, I2 = 35%; AC vs CC:P = .58, I2 = 0%; AA+AC vs CC:P = .45, I2 = 0%; AA vs AC+CC: P = .17, I2 = 33%.).
3.4. Sensitivity analysis
Sensitivity analysis was conducted by omitting each study in turn. There was no big change when omitting each article, indicating the result was stable and trustworthy. Figure 5 shows the sensitivity analysis for the association between polymorphism of rs1800896 and risk of leukemia under recessive genetic model comparison (GG vs GA+AA).
Figure 5.
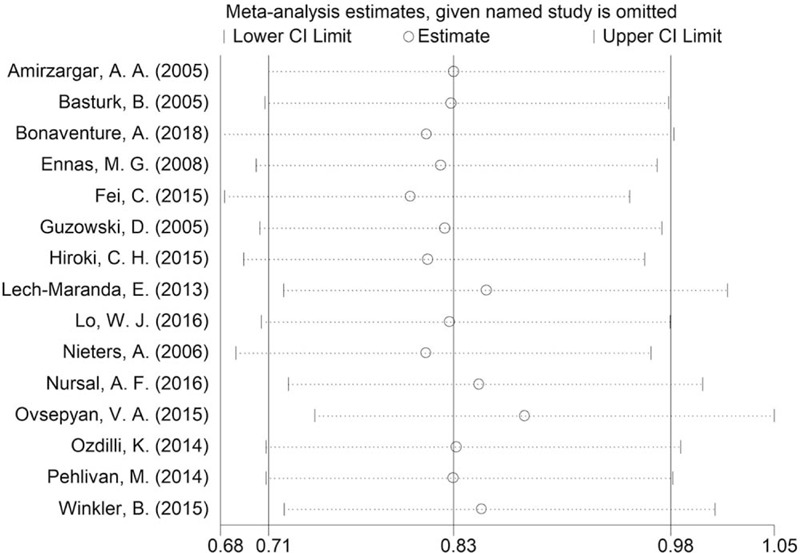
Sensitivity analysis for the association between polymorphism of rs1800896 and risk of leukemia under recessive genetic model comparison (GG vs GA+AA).
3.5. Publication bias
Figure 6 shows the Begg funnel plot for the association between polymorphism of rs1800871 and leukemia under heterozygote comparison (TC vs CC). There was no evidence of publication bias both in Begg funnel plots and Egger test. The symmetry of the Begg funnel plot was good and P values for Begg tests and Egger tests were >0.05. Begg funnel plots for the association between polymorphism of rs1800871 and leukemia under homozygote comparison (TT vs CC) in non-Chinese are shown in Figure 7. These data indicated that there was no significant publication bias in this meta-analysis.
Figure 6.
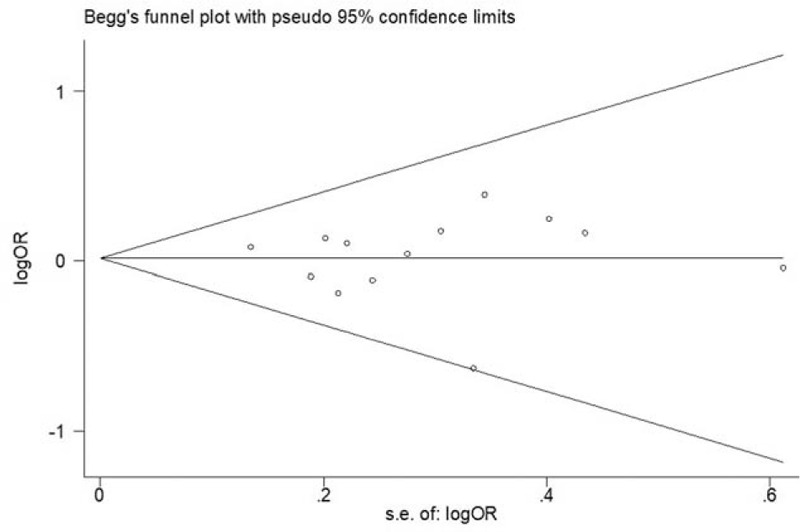
Begg funnel plot for publication bias of overall analysis for the association between polymorphism of rs1800871 and leukemia (TC vs CC; P for bias = .951).
Figure 7.
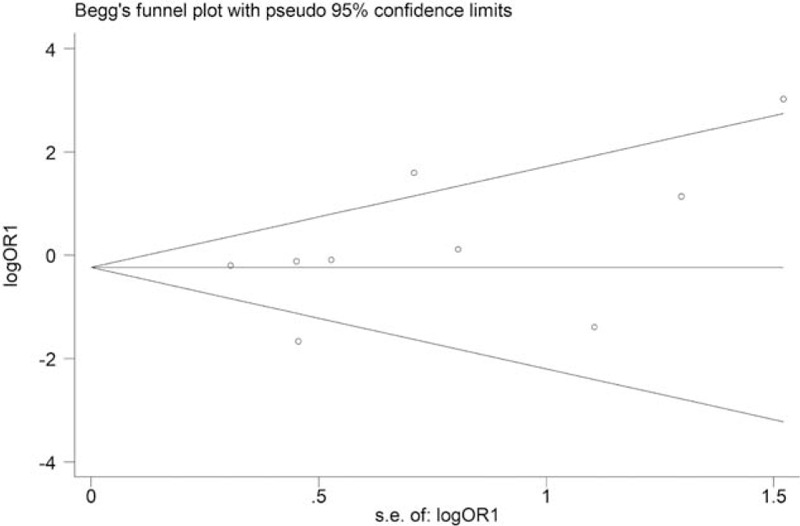
Begg funnel plot for publication bias of stratified analysis for the association between polymorphism of rs1800871 and leukemia in non-Chinese (TT vs CC; P for bias = .076).
4. Discussion
More and more researches concentrate on immunity when exploring the etiology of cancers. IL-10 plays a suppressive role on many types of cells in general,[33] but it has pleiotropic functions on tumor. IL-10 can impose both stimulatory and inhibitory impact on T cells.[34] There are studies revealing that IL-10 can fight against tumor by activating CD8+ T cells and cause the accumulation of tumor infiltrating lymphocytes (TILs),[33,35] while others indicate that IL-10 can promote the development and progression of tumors.[36–38] It has been reported that polymorphisms in the promoter region of IL-10 may influence the production of IL-10.[39] There are many studies and systematic analyses reporting the association between SNPs of IL-10 and other cancers, including gastric cancer40,[40] cervical cancer,[41] breast cancer,[42] and prostate cancer.[43]
Recently, numerous studies focus on the association of SNPs in the promoter of IL-10 (such as rs1800896, rs1800871 and rs1800872) and risk of leukemia, but their results are conflicting. There is 1 pooled analysis involving 4 studies found that there was no association between SNP of IL-10 and CLL,[44] but it only surveyed 1 SNP of IL-10, rs1800896. Besides, it only focused on CLL, ignoring the other 3 kinds of leukemia. In addition, it only took 4 studies into analysis. So we conducted this meta-analysis involving 18 articles with 2264 cases and 3846 controls to illuminate the link between 3 common SNPs of IL-10 and leukemia.
In the overall analysis, we found a strong association between polymorphism of rs1800896 and leukemia under recessive genetic model comparison (GG vs GA+AA) which means GG genotype of rs1800896 was associated with decreased risk of leukemia in overall population. However, rs1800871 and rs1800872 exerted no impact on the susceptibility to leukemia in overall analysis.
In the subgroup analysis according to ethnicity, a strong association was found between rs1800896 and leukemia in non-Chinese under homozygote comparison (GG vs AA) and recessive genetic model comparison (GG vs GA+AA) while the result was negative in Chinese under the same comparison. So in non-Chinese, individuals with GG genotype of rs1800896 may have lower risk of leukemia when compared with those carrying GA or AA genotype. When stratified by cancer types, we found GG genotype of rs1800896 was associated with decreased risk of chronic leukemia, especially CLL under homozygote comparison (GG vs AA) and recessive genetic model comparison (GG vs GA+AA). However, no association was found between rs1800871 and leukemia when stratified by ethnicity and cancer type. Stratified analysis by ethnicity has effectively minimized the heterogeneity. Heterogeneity was low or moderate in non-Chinese while moderate or high in Chinese, maybe because types of leukemia were different in these 4 Chinese studies (2 of them were acute myelocytic leukemia and the other 2 were acute lymphoblastic leukemia) or patients with different comorbidities were recruited or caused by other statistic or methodological reasons, so we adopted random-effects model. Under random-effects model, rs1800872 was associated with leukemia risk in non-Chinese under heterozygote comparison (AC vs CC) and dominant genetic model comparison (AA+AC vs CC), which means in non-Chinese, individuals with AA or AC genotype may have increased risk of leukemia. As for Chinese group, perhaps there was no significant association between rs1800872 and leukemia. No association was found when stratified by cancer type.
The fact that heterogeneities of some comparisons were high cannot be ignored. Although we have carried out a careful research when searching articles, selected eligible studies strictly following inclusion and exclusion criterion, paid attention to every process of this analysis, some comparisons still had high heterogeneity. Heterogeneity was effectively minimized after stratified analysis, but significant heterogeneity still existed in some comparisons, maybe because of different source of control (controls of some studies are hospital-based), different genotyping methods, sample error or other unidentified reasons. There was no evidence of publication bias both in Begg funnel plot and Egger test.
There are several limitations of this meta-analysis. Firstly, we did not perform a stratified analysis according to sex or age because of lacking relevant original data. Secondly, the number of studies about the association between rs1800871 or rs1800872 and CLL is too small, including only 1 original article. Thirdly, Chinese studies about the association between rs1800896 and leukemia are limited, including only 2 studies. More studies are needed. Lastly, we only took articles written in English or Chinese into analysis, which may produce unavoidable bias.
5. Conclusions
In conclusion, this meta-analysis indicated that GG genotype of rs1800896 was associated with decreased risk of leukemia when compared with GA + AA. Strong association was found between GG genotype of rs1800896 and decreased risk of leukemia in non-Chinese compared with AA or GA + AA. In addition, GG genotype of rs1800896 was associated with decreased risk of chronic leukemia, especially CLL compared with AA or GA + AA. Lastly, we found CC genotype of rs1800872 was associated with decreased risk of leukemia in non-Chinese compared with AA or AA + AC.
Acknowledgments
We are thankful to all participants of our study.
Author contributions
SG searched literature, selected eligible studies, extracted and analysed data and drafted manuscript. KT and JC participated in article selection and data extraction. JW supervised literature search, data extraction, data analysis and drafted manuscript.
Footnotes
Abbreviations: CI = confidence interval, CLL = chronic lymphocytic leukemia, IL-10 = interleukin-10, OR = odds ratio, SNPs = single nucleotide polymorphisms.
How to cite this article: Gao S, Tang K, Chen J, Wang J. The single nucleotide polymorphisms of interleukin-10 are associated with the risk of leukaemia: evidence from 18 case-control studies. Medicine. 2020;99:44(e23006).
Ethics approval and consent to participate was not applicable.
Consent for publication was not applicable.
All data and material are provided in this article.
The authors have no funding and conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [2].Fitzmaurice C, Allen C, Barber RM, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol 2017;3:524–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ouyang W, O’Garra A. IL-10 Family Cytokines IL-10 and IL-22: from Basic Science to Clinical Translation. Immunity 2019;50:871–91. [DOI] [PubMed] [Google Scholar]
- [4].Melief CJ. Cancer immunotherapy by dendritic cells. Immunity 2008;29:372–83. [DOI] [PubMed] [Google Scholar]
- [5].Chen Z, Bozec A, Ramming A, et al. Anti-inflammatory and immune-regulatory cytokines in rheumatoid arthritis. Nature Rev Rheumatol 2019;15:9–17. [DOI] [PubMed] [Google Scholar]
- [6].Rad R, Dossumbekova A, Neu B, et al. Cytokine gene polymorphisms influence mucosal cytokine expression, gastric inflammation, and host specific colonisation during Helicobacter pylori infection. Gut 2004;53:1082–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature 2009;458:719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Freedman ML, Monteiro AN, Gayther SA, et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet 2011;43:513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shah MY, Ferracin M, Pileczki V, et al. Cancer-associated rs6983267 SNP and its accompanying long noncoding RNA CCAT2 induce myeloid malignancies via unique SNP-specific RNA mutations. Genome Res 2018;28:432–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chen H, Tang JL, Shen N, et al. Interleukin 10 gene rs1800896 polymorphism is associated with the risk of prostate cancer. Oncotarget 2017;8:66204–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Men T, Yu C, Wang D, et al. The impact of interleukin-10 (IL-10) gene 4 polymorphisms on peripheral blood IL-10 variation and prostate cancer risk based on published studies. Oncotarget 2017;8:45994–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kong F, Liu J, Liu Y, et al. Association of interleukin-10 gene polymorphisms with breast cancer in a Chinese population. J Exp Clin Cancer Res 2010;29:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ovsepyan VA, Gabdulkhakova A, Shubenkiva AA, et al. Role of interleukin-10 gene promoter region polymorphism in the development of chronic lymphoid leukemia. Bull Exp Biol Med 2015;160:275–7. [DOI] [PubMed] [Google Scholar]
- [14].Lo WJ, Chang WS, Hsu HF, et al. Significant association of interleukin-10 polymorphisms with childhood leukemia susceptibility in Taiwan. In vivo (Athens, Greece) 2016;30:265–9. [PubMed] [Google Scholar]
- [15].Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539–58. [DOI] [PubMed] [Google Scholar]
- [16].Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Amirzargar AA, Bagheri M, Ghavamzadeh A, et al. Cytokine gene polymorphism in Iranian patients with chronic myelogenous leukaemia. Int J Immunogenet 2005;32:167–71. [DOI] [PubMed] [Google Scholar]
- [18].Basturk B, Evke E, Tunali A, et al. Interleukin-10 and interferon-gamma cytokine gene polymorphisms may be risk factors for chronic myelogenous leukemia. Turk J Haematol 2005;22:191–6. [PubMed] [Google Scholar]
- [19].Bonaventure A, Orsi L, Rudant J, et al. Genetic polymorphisms of Th2 interleukins, history of asthma or eczema and childhood acute lymphoid leukaemia: findings from the ESCALE study (SFCE). Cancer Epidemiol 2018;55:96–103. [DOI] [PubMed] [Google Scholar]
- [20].Ennas MG, Moore PS, Zucca M, et al. Interleukin-1B (IL1B) and interleukin-6 (IL6) gene polymorphisms are associated with risk of chronic lymphocytic leukaemia. Hematol Oncol 2008;26:98–103. [DOI] [PubMed] [Google Scholar]
- [21].Guzowski D, Chandrasekaran A, Gawel C, et al. Analysis of single nucleotide polymorphisms in the promoter region of interleukin-10 by denaturing high-performance liquid chromatography. J Biomol Tech 2005;16:154–66. [PMC free article] [PubMed] [Google Scholar]
- [22].Hiroki CH, Amarante MK, Petenuci DL, et al. IL-10 gene polymorphism and influence of chemotherapy on cytokine plasma levels in childhood acute lymphoblastic leukemia patients: IL-10 polymorphism and plasma levels in leukemia patients. Blood Cells Mol Dis 2015;55:168–72. [DOI] [PubMed] [Google Scholar]
- [23].Lech-Maranda E, Mlynarski W, Grzybowska-Izydorczyk O, et al. Polymorphisms of TNF and IL-10 genes and clinical outcome of patients with chronic lymphocytic leukemia. Genes Chromosomes Cancer 2013;52:287–96. [DOI] [PubMed] [Google Scholar]
- [24].Nieters A, Beckmann L, Deeg E, et al. Gene polymorphisms in Toll-like receptors, interleukin-10, and interleukin-10 receptor alpha and lymphoma risk. Genes Immun 2006;7:615–24. [DOI] [PubMed] [Google Scholar]
- [25].Nursal AF, Pehlivan M, Sahin HH, et al. The Associations of IL-6, IFN-gamma, TNF-alpha, IL-10, and TGF-beta1 Functional Variants with Acute Myeloid Leukemia in Turkish Patients. Genet Test Mol Biomarkers 2016;20:544–51. [DOI] [PubMed] [Google Scholar]
- [26].Ozdilli K, Pehlivan S, Oret YD, et al. Cytokine gene polymorphisms in Turkish patients with chronic myeloid leukaemia and in healthy controls. Nobel Med 2014;10:74–8. [Google Scholar]
- [27].Pehlivan M, Sahin HH, Pehlivan S, et al. Prognostic importance of single-nucleotide polymorphisms in IL-6, IL-10, TGF-beta1, IFN-gamma, and TNF-alpha genes in chronic phase chronic myeloid leukemia. Genetic testing and molecular biomarkers 2014;18:403–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rashed R, Shafik RE, Shafik NF, et al. Associations of interleukin-10 gene polymorphisms with acute myeloid leukemia in human (Egypt). J Cancer Res Ther 2018;14:1083–6. [DOI] [PubMed] [Google Scholar]
- [29].Winkler B, Taschik J, Haubitz I, et al. TGFbeta and IL10 have an impact on risk group and prognosis in childhood ALL. Pediatric blood & cancer 2015;62:72–9. [DOI] [PubMed] [Google Scholar]
- [30].CNKI WCGX. Association of IL -1 0 gene single nucleotide polymorphisms with the acute susceptibility to acute lymphocyte leukemia. Ch in J Lab M ed 2011. [Google Scholar]
- [31].Fei C, Yao XM, Sun Y, et al. Interleukin-10 polymorphisms associated with susceptibility to acute myeloid leukemia. Genet Mol Res 2015;14:925–30. [DOI] [PubMed] [Google Scholar]
- [32].Yao CJ, Du W, Chen HB, et al. Associations of IL-10 gene polymorphisms with acute myeloid leukemia in Hunan, China. Asian Pac J Cancer Prev 2013;14:2439–42. [DOI] [PubMed] [Google Scholar]
- [33].Fujii S, Shimizu K, Shimizu T, et al. Interleukin-10 promotes the maintenance of antitumor CD8(+) T-cell effector function in situ. Blood 2001;98:2143–51. [DOI] [PubMed] [Google Scholar]
- [34].Ouyang W, Rutz S, Crellin NK, et al. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol 2011;29:71–109. [DOI] [PubMed] [Google Scholar]
- [35].Mumm JB, Emmerich J, Zhang X, et al. IL-10 elicits IFNgamma-dependent tumor immune surveillance. Cancer Cell 2011;20:781–96. [DOI] [PubMed] [Google Scholar]
- [36].Hattori E, Okumoto K, Adachi T, et al. Possible contribution of circulating interleukin-10 (IL-10) to anti-tumor immunity and prognosis in patients with unresectable hepatocellular carcinoma. Hepatol Res 2003;27:309–14. [DOI] [PubMed] [Google Scholar]
- [37].Stewart CA, Metheny H, Iida N, et al. Interferon-dependent IL-10 production by Tregs limits tumor Th17 inflammation. J Clin Investigation 2013;123:4859–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang Y, Sun SN, Liu Q, et al. Autocrine complement inhibits IL10-dependent T-cell-mediated antitumor immunity to promote tumor progression. Cancer Discov 2016;6:1022–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kingo K, Ratsep R, Koks S, et al. Influence of genetic polymorphisms on interleukin-10 mRNA expression and psoriasis susceptibility. J Dermatol Sci 2005;37:111–3. [DOI] [PubMed] [Google Scholar]
- [40].Pan F, Tian J, Pan YY, et al. Association of IL-10-1082 promoter polymorphism with susceptibility to gastric cancer: evidence from 22 case-control studies. Mol Biol Rep 2012;39:7143–54. [DOI] [PubMed] [Google Scholar]
- [41].Zhang S, Kong YL, Li YL, et al. Interleukin-10 gene -1082 G/A polymorphism in cervical cancer and cervical intraepithelial neoplasia: meta-analysis. J Int Med Res 2014;42:1193–201. [DOI] [PubMed] [Google Scholar]
- [42].Moghimi M, Ahrar H, Karimi-Zarchi M, et al. Association of IL-10 rs1800871 and rs1800872 polymorphisms with breast cancer risk: a systematic review and meta-analysis. Asian Pac J Cancer Prev 2018;19:3353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zou YF, Wang F, Feng XL, et al. Lack of association of IL-10 gene polymorphisms with prostate cancer: evidence from 11,581 subjects. Euro J cancer (Oxford, England: 1990) 2011;47:1072–9. [DOI] [PubMed] [Google Scholar]
- [44].Zintzaras E, Kitsios GD. Synopsis and synthesis of candidate-gene association studies in chronic lymphocytic leukemia: the CUMAGAS-CLL information system. Am J Epidemiol 2009;170:671–8. [DOI] [PubMed] [Google Scholar]


