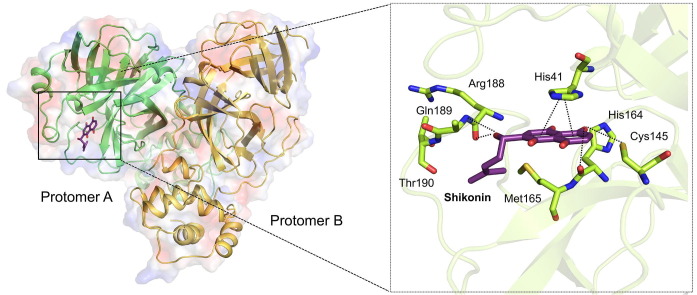
Graphical abstract
Almost everyone is susceptible to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), an RNA virus, which can cause many symptoms and even death among high-risk individuals [1], [2]. The main protease (Mpro, also known as 3CLpro) is a cysteine protease essential for producing infectious virions and thus, an attractive target for drug development. Up to now, many studies using either in silico ligand docking or drug discovery based on available structures have been performed to discover new Mpro-inhibiting agents [3], [4]. However, most studies have used either peptidomimetics or covalent inhibitors for Mpro, which may introduce non-specific reactions with host proteins. Here, we presented the structure of shikonin in a non-covalent binding configuration with Mpro and compared it with covalent bonding structures in pursuit of novel scaffolds capable of inhibiting the main protease.
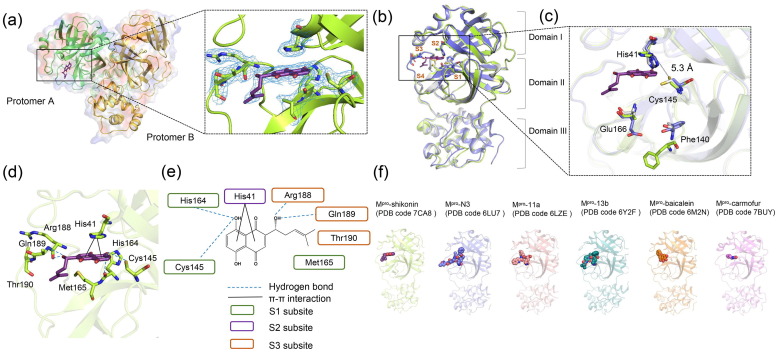
As shown in Fig. 1 , the crystal structure of Mpro in complex with shikonin (ShiMpro) is resolved at 2.45 Å (Fig. 1a and Table S1 online), and shikonin binds to only one of the protomers (i.e., protomer A) despite their overall structural similarity (Fig. S1 online, Supplementary materials and methods online). ShiMpro shows the same overall fold as for the apo structure of Mpro at pH 7.5 (apoMpro) [5]. The root mean square (RMS) difference of equivalent Cα positions between apo and ShiMpro is ~ 0.3 Å (Fig. 1b).
Fig. 1.
Crystal structure of SARS-CoV-2 main protease (Mpro) in complex with natural product inhibitor shikonin and comparison of SARS-CoV-2 Mpro structures. (a) Structure of the Mpro dimer. One protomer of the dimer with inhibitor shikonin is shown in green, the other is shown in yellow. A zoomed view of the shikonin binding pocket showing all residues within 4 Å, along with the 2mFo-DFc electron density (blue mesh) contoured at 1σ level. Shikonin is shown as sticks with purple carbons. (b) Structure of ShiMpro is shown in green. Structure of Mpro with N3 is shown in blue. Structure of apoMpro is shown in grey. Carbon atoms of shikonin are magenta, and oxygen atoms are red. Hydrogen bonds and π-π interactions are indicated by dashed black lines. Brown symbols S1, S2, S3, and S4 indicate the substrate binding pockets. (c) Conformational difference in catalytic site His41-Cys145. Residues of Mpro structure with shikonin are shown in green. (d) Strucuture of shikonin binding pocket. (e) Schematic interaction between shikonin and Mpro. Hydrogen bonds and π-π stacking interactions are shown as blue dashed lines and black solid lines, respectively. The green circle indicates conserved residues in S1 subsite. The purple circle indicates conserved residues in S2 subsite. The orange circle indicates conserved residues in S3 subsite. (f) Crystal structures of Mpro-inhibitor complexes from previously reported structures presenting diverse inhibitor-binding sites. Mpro structures are shown in cartoon representation and the inhibitors are shown as sphere models with transparent surfaces. The representative structures of Mpro along with covalent inhibitors, N3 (PDB code 6LU7), 11a (PDB code 6LZE), and 13b (PDB code 6Y2F) are shown. Similarly, structures for Mpro bound to natural products shikonin (PDB code 7CA8) and baicalein (PDB code 6M2N), and antineoplastic drug carmofur (PDB code 7BUY) are shown.
An overlay of the ShiMpro structure with the previously solved inhibitor-bound structures shows high spatial conservation (Fig. 1b and Fig. S2 online). The inhibitor binding pocket is surrounded by S1–S4 subsites, and shikonin forms multiple interactions with them (Fig. 1b). First, shikonin forms a hydrogen bond network with the protease polar triad Cys145 and His164 located on the S1 subsite. Second, the aromatic head groups of shikonin form a π-π interaction with His41 on the S2 subsite. Third, the hydroxy and methyl group of the isohexenyl side chain of shikonin tail form H-bonding with Arg188 and Gln189 on the S3 subsite, respectively.
Superimposing ShiMpro with other inhibitor-bound structures reveals a striking difference in the arrangement of the catalytic dyad His41-Cys145 and smaller, but substantial, differences in Phe140 and Glu166. First, in covalent-bonding structures, the inhibitor binds to the Sγ atom of Cys145, but in the current structure, the side chain of Cys145 adopts a different configuration to form a hydrogen bond with shikonin (Fig. 1c and d). Second, shikonin forms H-bonds with Arg188 and Gln189 in the S3 pocket (Fig. 1d and e). Third, the imidazole group of His41 points toward the binding pocket in covalent-bonding structures, but it flips outward in the current structure, opening a way for the entry of shikonin. Fourth, the distance between His41 Nε2 and Cys145 Sγ is 5.3 Å in ShiMpro structure, significantly longer than those observed in other Mpro structures (Fig. 1c) [6], [7], [8], [9]. Fifth, the phenyl ring of Phe140 in ShiMpro moves outward to the solvent and no longer has π-π interaction with His163. Lastly, the side chain of Glu166 is flexible in ShiMpro structures but is well ordered in covalent inhibitor binding structures (Fig. 1c). Glu166 is strictly conserved among all Mpro and is critical for forming a hydrogen bond with peptidomimetic inhibitors and N terminal residues from the other protomer [9]. The conformational change of Glu166 in the current ShiMpro structure may explain how the non-covalent binding of shikonin can inhibit protease activity.
Additionally, the apoMpro structure has two water molecules in the substrate-binding site (Fig. S3a online). Water 1 forms a hydrogen bond network involving Phe140, His163, and Glu166 located in the S1 pocket, stabilizing the oxyanion hole in the apo state structure [5]. Water 2 hydrogen-bonded with His41 and Cys145. However, these two water molecules are not observed in the ShiMpro structure, and the space for water 2 in the apo structure is now occupied by shikonin (Fig. S3b online), suggesting water molecule displacement may be part of the inhibitor mechanism.
Unlike ShiMpro, covalent and peptidomimetic inhibitors bind to the S1/S2/S4 site, carmofur binds to the S2 subsite, and baicalein binds the S1/S2 pocket (Fig. 1f) [6], [7], [8], [9], [10]. Therefore, the ShiMpro structure highlights a new mode of binding, and may serve as an invaluable resource to improve the design of novel antiviral drugs.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
Acknowledgments
This work was supported by the Thousand Young Talents Program of China, the National Natural Science Foundation of China (31770795, 81974514, 21961003, and 31971043), the Jiangxi Provincial Natural Science Foundation (20181ACB20014 and 20192BAB205114), the Open Project of Key Laboratory of Prevention and Treatment of Cardiovascular and Cerebrovascular Diseases, Ministry of Education (XN201904), Gannan Medical University (QD201910), Jiangxi "Double Thousand Plan", the Foreign Talent Project of Jiangxi Province, Talent Project of Jiangxi Province, Shenzhen Fundamental Research Program, and Ganzhou COVID-19 Emergency Research Project. We thank the team of Shanghai Synchrotron Radiation Facility (SSRF) BL17U1 beamline for data collection and processing.
Author contributions
Jian Li, Xuelan Zhou, Yan Zhang, Fanglin Zhong, and Cheng Lin made constructs for expression and determined the conditions used to enhance protein stability. Huan Zhou and Qisheng Wang carried out X-ray experiments, including data acquisition and processing. Jian Li and Jin Zhang built the atomic model. Jin Zhang, Jian Li, Yan Zhang, Yang Fu, Jun Luo, Feng Jiang, Peter J. McCormick, and Jingjing Duan drafted the manuscript. Jin Zhang and Jian Li supervised the research.
Biographies

Jian Li graduated from Shanghai Institute of Applied Physics, Chinese Academy of Sciences (CAS) with a Ph.D. degree and worked as a postdoctoral fellow at the Shanghai Institute of Materia Medica, CAS, and University of Kentucky. He is a professor at the College of Pharmaceutical Sciences, Gannan Medical University. His research focuses on structure-based drug discovery.

Jin Zhang is a professor at the School of Basic Medical Sciences, Nanchang University. He has a broad interest in structural biology and functional characterization of G protein-coupled receptors and ion pump/channels. He mainly focuses on drug discovery and development for chronic kidney disease and infectious diseases.
Footnotes
Supplementary materials to this article can be found online at https://doi.org/10.1016/j.scib.2020.10.018.
Appendix A. Supplementary materials
The following are the Supplementary data to this article:
References
- 1.Li Q., Guan X., Wu P. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N.a., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L., Lin D., Kusov Y. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J Med Chem. 2020;63:4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 4.Chang K.O., Kim Y., Lovell S. Antiviral drug discovery: norovirus proteases and development of inhibitors. Viruses. 2019;11:197. doi: 10.3390/v11020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou X., Zhong F., Lin C. Structure of SARS-CoV-2 main protease in the apo state. Sci China Life Sci. 2020 doi: 10.1007/s11427-020-1791-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai W., Zhang B., Jiang X.M. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin Z., Du X., Xu Y. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 8.Jin Z., Zhao Y., Sun Y. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat Struct Mol Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L., Lin D., Sun X. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su H., Yao S., Zhao W. Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro. bioRxiv. 2020 doi: 10.1101/2020.04.13.038687. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.