Abstract
Background
With the persistent COVID-19 pandemic, there is an urgent need to use rapid and reliable diagnostic tools for highly urgent cases. Antigen tests are disappointing with their lack of sensitivity. Among molecular tools allowing a diagnosis in less than an hour, only one, the Cepheid Xpert Xpress SARS-CoV-2 assay, has exhibited a good sensitivity. However, we are also facing a global shortage of reagents and kits. Thus, it is imperative to evaluate other point-of-care molecular tests.
Methods
We evaluated the VitaPCR™ RT-PCR assay, whose sample analysis time is of approximately 20 min, in nasopharyngeal secretions from 534 patients presenting to our Institute, for the diagnosis of COVID-19, and compared it to our routine RT-PCR assay. We also compared the two assays with tenfold dilutions of a SARS-CoV-2 strain.
Results
Compared to our routine RT-PCR and the previous diagnosis of COVID-19, the sensitivity, specificity, positive and negative predictive values of VitaPCR™ can be evaluated to be 99.3 % (155/156), 94.7 % (358/378), 88.6 % (155/175) and 99.7 % (358/359), respectively. Tenfold dilutions of a SARS-CoV-2 strain show that the VitaPCR™ was more sensitive that our routine RT-PCR assay.
Conclusion
The VitaPCR™ SARS-CoV-2 is an accurate rapid test, suitable for clinical practice that can be performed as part of a point-of-care testing, for the rapid diagnosis of COVID-19.
Keywords: SARS-CoV-2, COVID-19, Diagnosis, Rapid diagnostic test, Point-of-care
1. Introduction
The onset of the coronavirus disease 2019 (COVID-19) pandemic in March 2020 initiated a race to develop rapid detection tools for the causative virus, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in order to optimize the management and triage of patients [[1], [2], [3], [4]]. The congestion of emergency departments also required that we could offer an accurate point-of-service test that could be performed directly there. Antigen tests are easy to perform and can provide rapid diagnosis, but lack sensitivity, which makes them unreliable for the diagnosis of COVID-19 [1]. Therefore, RT-PCR assays remain the gold standard for the diagnosis of COVID-19. Several molecular tools allowing a diagnosis in less than an hour have been evaluated. Of these, the fastest two (the Abbott ID NOW and the Mesa Accula) with less than 30 min of delay between sampling and answer accumulated evidence of poorer diagnostic performance with a lack of sensitivity [5,6]. Only the Cepheid Xpert Xpress SARS-CoV-2 assay has shown to be a valuable tool with a run-time of 45−50 min with hands on time limited to 2−3 min [7]. However, due to the pandemic, we are also facing a global shortage of reagents and kits and uncertainty over the availability of Xpert Xpress cartridges [5]. It is therefore imperative to evaluate other point-of-care molecular tests for emergency diagnosis.
In this context, we evaluated the VitaPCR™ RT-PCR assay (Credo Diagnostics Biomedical, Singapore), whose sample analysis time is of approximately 20 min.
2. Material and methods
From September 28th to October 1st, 2020, 534 patients presenting to the Mediterranee Infection Institute (Marseille, France), for the diagnosis of COVID-19, were included in the study. Each patient benefited from two naso-pharyngeal swab samplings, one per nostril. VitaPCR™ SARS-CoV-2 was systematically compared to our routine in-house real-time RT-PCR as reference method [8,9].
The VitaPCR™ assay includes three detection systems: (1) one targeting the human β-globin gene, to check the quality of DNA extracts; (2) a second targeting a specific sequence on the nucleocapside N-encoding gene; (3) a third targeting a conserved sequence common to SARS-CoV-2, SARS-CoV, and SARS-like bat coronavirus, also located on the N-encoding gene. We strictly followed the manufacturer’s instructions for VitaPCR™ SARS-CoV-2 assay (Credo Diagnostics Biomedical, Singapore). For virus lysis and inactivation, the swab was discharged in the kit-provided collection buffer by stirring it 15 times. We allowed the lysis buffer to act for 5 min. Thirty μL of lysate were transferred to the tube containing the lyophilized PCR reagents. They were mixed well by pipetting. We avoided bubbles during all the process. The tube was then introduced into the apparatus in order to perform the analysis by RT-PCR, and then the latter returned the results in 20 min.
For our routine assay, automated nucleic acid extraction was performed using a KingFisher™ Flex system (Thermo Fisher Scientific), following the manufacturer's instructions. Our routine SARS-CoV-2 RT-PCR assay, that targets the envelope protein E-encoding gene, was performed as previously reported [8]. Besides, PCR targeting the human β-actin gene was performed to check the quality of DNA extracts [9]. In routine, the cycle threshold (Ct) to conclude that an analysis is positive using our RT-PCR is less than or equal to 35 Ct. In parallel, we also assessed the impact of delayed testing on Ct values using VitaPCR™ assay for twelve positive samples tested directly and 3 h later.
Besides, we also determined the level of detection of the two molecular assays by analyzing tenfold dilutions of a suspension of Vero E6 cell-cultured SARS-CoV-2 IHUMI-3 strain [10]. This strain was obtained from a nasopharyngeal swab of an RT-PCR positive patient, as previously reported [10].
3. Results
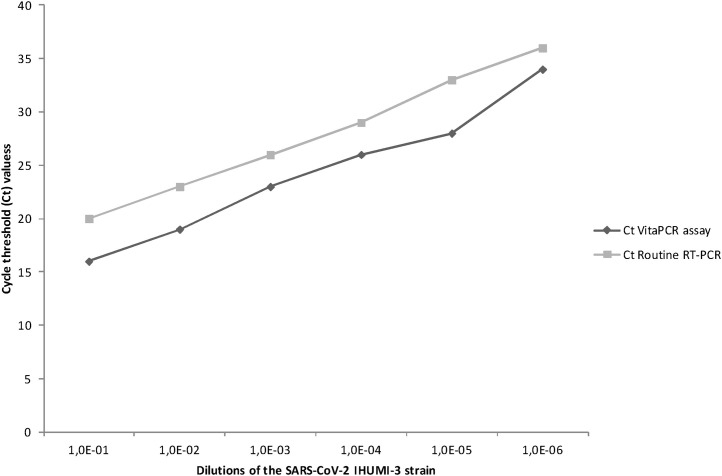
By analyzing tenfold dilutions of IHUMI-3 strain, from 780 × 106 copies/mL at a dilution of 10−1 to 1484 copies/mL at a dilution of 10−6, the Ct values were 16 and 34 for the highest (10−1) and lowest (10−6) using the VitaPCR™ and 20 and 36, respectively, using our routine PCR assay (Fig. 1 ).
Fig. 1.
Evaluation of the in vitro sensitivity of VitaPCR™ SARS-CoV-2 assay by comparison with our routine RT-PCR using tenfold dilutions of a suspension of Vero E6 cell-cultured SARS-CoV-2, IHUMI-3 strain.
Among the 534 analyzed samples, 119 were positive and 358 negative using both assays (Supplementary Figure). One from recent diagnosis of COVID-19 was positive only with our routine RT-PCR. Fifty-six were positive only with the VitaPCR™. Among them, nine were negative for β-actin PCR showing thus the poor quality of the DNA extracts and the impossibility of interpreting the SARS-CoV-2 results obtained by routine RT-PCR; in contrast, β-globin was correctly detected from the naso-pharyngeal swabs from these patients interpreted using VitaPCR™. Eighteen exhibited a cycle threshold (Ct) value from 35 to 38 using our routine RT-PCR (including 6 from patients with a recent diagnosis of COVID-19). In our laboratory, a threshold of Ct 35 was selected in order to prioritize diagnoses of putatively contaminant patients. Nine were also from recent diagnosis of COVID-19, including 6 with a Ct value greater than 31 with VitaPCR™. Overall, these data support false negative results from our routine RT-PCR due to a biased threshold or at least a lower sensitivity. Finally, among the other twenty patients, two were asymptomatic whereas eighteen exhibited clinical and biological data highly evocative of COVID-19, such as fever, cough, anosmia, ageusia, and eosinopenia (Table 1 ) [11]. Compared to our routine RT-PCR with a Ct less than or equal to 38 and the previous diagnosis of COVID-19, the sensitivity, specificity, positive, and negative predictive values of VitaPCR™ can be evaluated to be 99.3 % (155/156), 94.7 % (358/378), 88.6 % (155/175) and 99.7 % (358/359), respectively.
Table 1.
Clinical and biological data for the twenty patients positive only with the VitaPCR™and without previous diagnosis of COVID-19.
| Patients | Sex, age | Date of sample | Date of symptoms onset | Clinical and biological data |
|---|---|---|---|---|
| 1 | F, 20 y | 24 sept | None | Asymptomatic (another sample collected on 09/28 was negative by both techniques) |
| 2 | M, 83 y | 25 sept | None | Asymptomatic (another sample collected on 09/30 was negative by both techniques) |
| 3 | M, 31 y | 25 sept | 23 sept | Cough, aches, asthenia, leucopenia (3.6 Giga/l), eosinopenia (0.03 Giga/l), lymphopenia (0.87 Giga/l) |
| 4 | F, 47 y | 29 sept | 20 sept | Fever, cough, anosmia, ageusia, thoracic pain, rhinitis, diarrhea, eosinopenia (0 Giga/l), elevated CRP (42.1 mg/l), elevated ferritin (660 μg/l), elevated γGT (41 UI/l), elevated transaminases (ALT [45 UI/l] and AST [43 UI/l]), elevated LDH (272 UI/l), elevated fibrinogen (5.4 g/l) |
| 5 | F, 40 y | 29 sept | 28 sept | Cough, headache, leucopenia (3.8 Giga/l), lymphopenia (0.63 Giga/l) |
| 6 | M, 40 y | 28 sept | 19 sept | Fever, cough, anosmia, ageusia, diarrhea, headache, eosinopenia (0.02 Giga/l), elevated ferritin (943 μg/l), elevated CRP (21.8 mg/l), elevated transaminases (ALT [72 UI/l] and AST [63 UI/l]), elevated LDH (301 UI/l) |
| 7 | F, 39 y | 29 sept | 25 sept | Fever, anosmia, ageusia, headache, leucopenia (3.6 Giga/l), eosinopenia (0.08 Giga/l), thrombocytopenia (134 Giga/l), elevated fibrinogen (4.15 g/l), elevated d-dimers (3 μg/mL) |
| 8 | M, 24 y | 25 sept | 18 sept | Anosmia, ageusia, leucopenia (3.9 Giga/l), neutropenia (1.9 Giga/l), eosinopenia (0.02 Giga/l), elevated CRP (22.2 mg/l), elevated LDH (223 UI/l), elevated ferritin (10.8 μg/l) |
| 9 | M, 62 y | 01 oct | 26 sept | Fever, diarrhea, aches, abdominal pain; leucopenia (2,9 Giga/L), neutropenia (1.7 Giga/l), eosinopenia (0.03 Giga/l), lymphopenia (0.79 Giga/l), thrombocytopenia (129 giga/l), elevated CRP (16.3 mg/l), elevated ferritin (1400 μg/l), elevated fibrinogen (5.15 g/l), elevated LDH (242 UI/l) |
| 10 | M, 43 y | 28 sept | 14 sept | Fever, cough, headache, leucocytosis (23 Giga/l), neutrophilic leucocytosis (20 Giga/l), eosinopenia (0.04 Giga/l), elevated CRP (33 mg/l), elevated γGT (90 UI/l) |
| 11 | M, 35 y | 29 sept | 26 sept | Ageusia, headache, asthenia, aches, eosinopenia (0.02 Giga/l), elevated CRP (13.5 mg/l), elevated fibrinogen (4.6 g/l) |
| 12 | M, 19 y | 28 sept | 21 sept | Cough, ageusia, headache, asthenia, no biological abnormalities reported |
| 13 | M, 23 y | 28 sept | 21 sept | Rhinorrhea, headache, asthenia, eosinopenia (0.05 Giga/l) |
| 14 | F, 43 y | 28 sept | 22 sept | Cough, anosmia, rhinitis, aches, diarrhea, eosinopenia (0.01 Giga/l), elevated ferritin (212 μg/l) |
| 15 | F, 41 y | 29 sept | 22 sept | Fever, anosmia, ageusia, headache, diarrhea, rhinitis, chest pain, aches, eosinopenia (0.02 Giga/l), elevated LDH (222 UI/l) |
| 16 | M, 43 y | 30 sept | 20 sept | Anosmia, ageusia, eosinopenia (0.08 Giga/L), elevated ALAT (54 UI/l), elevated γGT (173 UI/l) |
| 17 | F, 16 y | 25 sept | 18 sept | Cough, anosmia, ageusia, headache, rhinitis, thoracic pain, aches, elevated transaminases (ALT [59 UI/l] and AST [46 UI/l]) |
| 18 | F, 67 y | 28 sept | 20 sept | Fever, diarrhea, breathlessness, aches, asthenia, eosinopenia (0.00 Giga/l), elevated ferritin (264 μg/l), elevated transaminases (ALT [53 UI/l] and AST [43 UI/l]), elevated LDH (276 UI/l), elevated fibrinogen (5.5 g/l), elevated d-dimers (0.67 μg/mL) |
| 19 | F, 34 y | 29 sept | 15 sept | Cough, anosmia, ageusia, eosinopenia (0.08 Giga/l) |
| 20 | F, 73 y | 29 sept | 14 sept | Cough, leucocytosis (11 Giga/l), neutrophilic leucocytosis (7.9 Giga/l), eosinopenia (0.08 Giga/L) |
CRP (C-reactive protein); γGT (Gamma glutamyl transpeptidase); ALT (alanine transaminase); AST (aspartate transaminase); LDH (lactate dehydrogenase).
Finally, a 3 -h delayed testing using VitaPCR™ assay has an impact on Ct values with an increase in these (Table 2 ).
Table 2.
Cycle threshold values obtained using VitaPCR™ assay when analyses were performed directly after the sampling and 3 h later.
| VitaPCRTM cycle threshold values |
||
|---|---|---|
| Patients | Directly performed | Performed 3 h later |
| 1 | 22 | 22 |
| 2 | 30 | 32 |
| 3 | 19 | 20 |
| 4 | 17 | 25 |
| 5 | 16 | 27 |
| 6 | 28 | 30 |
| 7 | 25 | 31 |
| 8 | 19 | 20 |
| 9 | 28 | 30 |
| 10 | 20 | 22 |
| 11 | 27 | 30 |
| 12 | 22 | 26 |
4. Discussion
Our study shows that the VitaPCR™ assay exhibits a high sensitivity for SARS-CoV-2 detection in nasopharyngeal samples. Moreover, the apparent lack of specificity must be heavily weighted with the patient data which suggests a potential lack of sensitivity of our routine RT-PCR. Of note, the manufacturer has reported no cross-reactivity with human coronavirus 229E, human adenovirus 1, influenza A virus (H1N1, H2N3), influenza B virus, and respiratory syncytial virus A. The assay is not only fast but also easy to handle. After the nasopharyngeal sampling, the swab is discharged into a specific lysis buffer and tested directly using ready-for-use reagents, stored at room temperature. The results are automatically interpreted, limiting human interpretation bias. The training required for operators is simple and does not last long. It required us an hour to train technicians, medical students and pharmacy students from collecting the sample to analyzing it. The device is not bulky and can therefore be installed in a delocalized laboratory, close to patients to be tested. Finally, the system is secure as the virus is inactivated by the kit-provided collection buffer.
Potential limits are that only one sample is processed by apparatus at a time (3 tests per hour) but several devices can be used concomitantly by a single person. Besides, extracted viral RNAs in lysis buffer is rapidly degraded, which prevents delayed testing.
Overall, the VitaPCR™ is a highly sensitive test that enables to deliver results in less than half an hour and to decentralize testing for SARS-CoV-2 in acute-care hospital or emergency departments where rapid triage decisions are required for the establishment of specific isolation for contagious patients, the use of adequate personal protective elements for the healthcare workers, and for the management of the patient. The VitaPCR™ can therefore be included in point-of-care tests.
Authors’ contributions
PEF and FF conceptualized and supervised the study. CZ, EP, LN, MM, HTD, and PC performed the investigations and experiments. PEF and FF wrote the initial draft of the manuscript. All authors agreed to the publication of this version of the manuscript.
Transparency declaration
This study was supported by the Méditerranée-Infection Foundation and the French Agence Nationale de la Recherche under the program “Investissements d’Avenir”, reference ANR-10-IAHU-03.
The patients gave an informed consent for this study.
Declaration of Competing Interest
The authors report no declarations of interest.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2020.104682.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Dinnes J., Deeks J.J., Adriano A., Berhane S., Davenport C., Dittrich S., Emperador D., Takwoingi Y., Cunningham J., Beese S., Dretzke J., Ferrante di Ruffano L., Harris H.M., Price M.J., Taylor-Phillips S., Hooft L., Leeflang M.M.G., Spijker R., Van den Bruel A. Cochrane COVID-19 diagnostic test accuracy group. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection (Review) Cochrane Database Syst. Rev. 2020 doi: 10.1002/14651858.CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pabbaraju K., Wong A.A., Douesnard M., Ma R., Gill K., Dieu P., Fonseca K., Zelyas N., Tipples G.A. A public health laboratory response to the COVID-19 pandemic. J. Clin. Microbiol. 2020;58(8):e01110–01120. doi: 10.1128/JCM.01110-20. https://jcm.asm.org/content/58/8/e01110-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ransom E.M., Potter R.F., Wallace M.A., Mitchell K.F., Yarbrough M.L., Burnham C.A.D., Anderson N.W., Parikh B.A. Comparison of extraction methods and thermocyclers for SARS-CoV-2 molecular detection using clinical specimens. J. Clin. Microbiol. 2020;58(10):e01622–20. doi: 10.1128/JCM.01622-20. https://jcm.asm.org/content/58/10/e01622-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hogan C.A., Garamani N., Lee A.S., Tung J.K., Sahoo M.K., Huang C., Stevens B., Zehnder J., Pinsky B.A. Comparison of the accula SARS-CoV-2 test with a laboratory-developed assay for detection of SARS-CoV-2 RNA in clinical nasopharyngeal specimens. J. Clin. Microbiol. 2020;58(8):e01072–20. doi: 10.1128/JCM.01072-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hogan C.A., Sahoo M.K., Huang C., Garamani N., Stevens B., Zehnder J., Pinsky B.A. Five-minute point-of-care testing for SARS-CoV-2: not there yet. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolters F., van de Bovenkamp J., van den Bosch B., van den Brink S., Broeders M., Chung N.H., Favié B., Goderski G., Kuijpers J., Overdevest I., Rahamat-Langedoen J., Wijsman L., Melchers W.J., Meijer A. Multi-center evaluation of cepheid xpert® xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amrane S., Tissot-Dupont H., Doudier B., Eldin C., Hocquart M., Mailhe M., Dudouet P., Ormières E., Ailhaud L., Parola P., Lagier J.C., Brouqui P., Zandotti C., Ninove L., Luciani L., Boschi C., La Scola B., Raoult D., Million M., Colson P., Gautret P. Rapid viral diagnosis and ambulatory management of suspected COVID-19 cases presenting at the infectious diseases referral hospital in Marseille, France, - January 31st to March 1st, 2020: a respiratory virus snapshot. Travel Med. Infect. Dis. 2020;36 doi: 10.1016/j.tmaid.2020.101632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mediannikov O., Fenollar F., Socolovschi C., Diatta G., Bassene H., Molez J.F., Sokhna C., Trape J.F., Raoult D. Coxiella burnetiiin humans and ticks in rural Senegal. PLoS Negl. Trop. Dis. 2010;4(2010):e654. doi: 10.1371/journal.pntd.0000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaafar R., Aherfi S., Wurtz N., Grimaldier C., Hoang Van Thuan, Colson P., Raoult D., La Scola B. Correlation between 3790 qPCR positive samples and positive cell cultures including 1941 SARS-CoV-2 isolates. In press Clin Infect Dis. 2020;(2020) doi: 10.1093/cid/ciaa1491. [DOI] [Google Scholar]
- 11.Lagier J.C., Million M., Gautret P., Colson P., Cortaredona S., Giraud-Gatineau A., Honoré S., Gaubert J.Y., Fournier P.E., Tissot-Dupont H., Chabrière E., Stein A., Deharo J.C., Fenollar F., Rolain J.M., Obadia Y., Jacquier A., La Scola B., Brouqui P., Drancourt M., Parola P., Raoult D. IHU COVID-19 Task force. Outcomes of 3,737 COVID-19 patients treated with hydroxychloroquine/azithromycin and other regimens in Marseille, France: a retrospective analysis. Travel Med. Infect. Dis. 2020;36(101791) doi: 10.1016/j.tmaid.2020.101791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.