Abstract
Aims
The recent outbreak of COVID-19 has become a global health concern. There are currently no effective treatment strategies and vaccines for the treatment or prevention of this fatal disease. The current study aims to determine promising treatment options for the COVID-19 through a computational drug repurposing approach.
Materials and methods
In this study, we focus on differentially expressed genes (DEGs), detected in SARS-CoV-2 infected cell lines including “the primary human lung epithelial cell line NHBE” and “the transformed lung alveolar cell line A549”. Next, the identified DEGs are used in the connectivity map (CMap) analysis to identify similarly acting therapeutic candidates. Furthermore, to interpret lists of DEGs, pathway enrichment and protein network analysis are performed. Genes are categorized into easily interpretable pathways based on their biological functions, and overrepresentation of each pathway is tested in comparison to what is expected randomly.
Key findings
The results suggest the effectiveness of lansoprazole, folic acid, sulfamonomethoxine, tolnaftate, diclofenamide, halcinonide, saquinavir, metronidazole, ebselen, lidocaine and benzocaine, histone deacetylase (HDAC) inhibitors, heat shock protein 90 (HSP90) inhibitors, and many other clinically approved drugs as potent drugs against COVID-19 outbreak.
Significance
Making new drugs remain a lengthy process, so the drug repurposing approach provides an insight into the therapeutics that might be helpful in this pandemic. In this study, pathway enrichment and protein network analysis are also performed, and the effectiveness of some drugs obtained from the CMap analysis has been investigated according to previous researches.
Keywords: COVID-19, SARS-CoV-2, Drug repurposing, Connectivity map, Transcriptomic profiling
Highlights
-
•
Computational genomics analysis can accelerate drug discovery for the COVID-19.
-
•
SARS-CoV-2 transcriptome showed a chemokine-dominant hyperinflammatory response.
-
•
Robust IFN response with significant expression of ISGs pathway is seen in COVID-19.
-
•
The drug repurposing can be used as an alternative to de novo drug development.
-
•
Many approved drugs and investigational compounds showed antiviral properties.
1. Introduction
Coronavirus disease 2019 (COVID-19) is an emerging global health concern. It initially appeared in December 2019 in China with cases of unknown origin pneumonia. Subsequently, a novel betacoronavirus was isolated from the throat sample of a patient, furtherly named 2019 novel coronavirus (2019-nCoV) and formally Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) with 79 and 52% sequence similarity with Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and the Middle East Respiratory Syndrome coronavirus (MERS-CoV), two other members of betacoronavirus family and leading causes of global outbreaks in past two decades, respectively (Wang et al., 2020; Ren et al., 2020; Paules et al., 2020).
As of 9 June 2020, there have been more than 7 million definite cases of COVID-19 with approximately 404,000 deaths worldwide (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/, 2020). Considering the urgency of identifying effective treatments for the COVID 19 outbreak, repositioning of available approved drugs for other indications, is an alternative to de novo drug development. Currently, many classes of United States Food and Drug Administration (FDA)-approved drugs have been repurposed to mitigate COVID-19 symptoms. Chloroquine, an antimalarial agent, was among the first proposed drugs. Several clinical trials have been done to assess its routine use for COVID-19 prophylaxis or treatment (Chowdhury et al., 2020). However, the results were contradictory and finally, the FDA emergency use authorization for hydroxychloroquine and chloroquine in the treatment of COVID-19 was revoked on 15 June 2020 (https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-chloroquine-and, 2020).
One of the repurposed drugs is remdesivir, an adenosine triphosphate (ATP) analog, inhibiting the RNA-dependent RNA polymerase (RdRp). It was initially described as an inhibitor of the Ebola virus and its antiviral properties against the CoVs family were furtherly demonstrated (Eastman et al., 2020). It was also proposed during the COVID-19 pandemic as a potential therapeutic agent, furtherly confirmed by in vitro experiments (Wang et al., 2020). Although some clinical trials have been conducted to evaluate its effectiveness for treating COVID-19 patients, the results were inconsistent (Wang and Guan, 2020). In a randomized, double-blind, placebo-controlled clinical trial conducted by Beigel et al. (2020), 1063 COVID-19 patients from 10 countries were enrolled to receive remdesivir or placebo infusion. Results showed remdesivir can significantly improve the clinical recovery of hospitalized patients (NCT04280705). However, another team (Wang et al., 2020) has conducted a double-blind, multicenter trial at 10 hospitals in China between Feb 6, 2020, and March 12, 2020. 237 confirmed COVID-19 patients have participated in this trial and were randomly grouped in a 2:1 ratio to cases receiving intravenous remdesivir and controls receiving placebo infusions for 10 days. Results revealed prescribing remdesivir for severe COVID-19 patients is not associated with significant clinical benefits (NCT04257656).
Lopinavir-ritonavir combination (Kaletra), two HIV protease inhibitors, has been under investigation for COVID-19 treatment since this pandemic. In a randomized open-label trial, 199 COVID-19 patients were evaluated for the effects of lopinavir-ritonavir treatment in comparison to standard supportive care. Results showed no benefit with lopinavir–ritonavir treatment beyond standard care (Chinese Clinical Trial Register number: ChiCTR2000029308) (Cao et al., 2020). In another exploratory randomized controlled trial, 86 COVID-19 patients were assigned to receive lopinavir-ritonavir or arbidol or no antiviral medication. Results revealed the little benefit of antiviral medications over supportive care (NCT04252885) (Li et al., 2020).
Besides, some other antiviral, anti-inflammatory, antineoplastic, and antiparasitic agents have been proposed and undergone in vitro and in vivo experiments; however, results were inconsistent and controversial (Heimfarth et al., 2020).
Different approaches are available for drug repurposing and one of the most commonly used strategies is signature matching. It is based on comparing the transcriptomic signature of a specific disease with the unique signature of a drug resulting in the creation of a connectivity map (CMap) (Pushpakom et al., 2019).
The CMap concept was initially presented by Lamb et al. (2006), in 2006. They generated a large-scale genomic signatures database to translate gene functions, diseases, and drug actions into the same language. It begins with identifying gene expression signature of a specific disease that contains a list of differentially expressed genes (DEGs), genes that are significantly up- or down-regulated in a specific disorder in comparison to its control sample. This signature is then compared with a collection of gene expression signatures of cell lines treated with various small molecules. The result of signature matching for each pair is shown with a connectivity score ranging from −1 to +1. A negative connectivity score reflects the potential effect of that small molecule to reverse the signature of a specific disorder. Therefore, it can be used as a therapeutic option.
Since the introduction of CMap, this method has been applied in pharmacological research to define drug-disease connections (Qu and Rajpal, 2012; Musa et al., 2018). In 2016, So et al. (2016) used the CMap concept to identify available approved drugs for seven common psychiatric disorders associated with promising results. Several other researchers have applied this approach to find potential novel therapies for different cancers (e.g. small cell lung carcinoma, gastric cancer, etc.) as well as rare disorders such as Hirschsprung disorder (HD) (Jahchan et al., 2013; Zhang et al., 2019; Xiao et al., 2018).
In this study, we set out to use a drug repurposing approach based on the CMap concept to identify possible treatments for COVID 19. Furthermore, to help interpret lists of DEGs, pathway enrichment and protein network analysis are performed. Genes are categorized into easily interpretable pathways based on their biological functions and overrepresentation of each pathway in the selected gene list is tested in comparison to what is expected randomly. The basic idea of this work was taken from Vidovik (2020). She conducted a CMap-based drug repurposing approach on available icSARS-CoV data on mouse models and suggested valproic acid as a therapeutic option. However, considering the genomic diversity of SARS-CoV-2 from icSARS-CoV and unique features of COVID-19 infection makes the study results less reliable and highlights the need for conducting further studies on SARS-CoV-2 data with Homo sapiens origin.
2. Materials and methods
2.1. Dataset selection and analysis
The NCBI (National Center for Biotechnology Information) Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) was used to identify microarray datasets (Barrett et al., 2007). The GEO database was manually searched using the terms “SARS-CoV-2” or “COVID-19”. The collected datasets were further selected if they met the following inclusion criteria: (a) whole-genome transcriptome profiling, and (b) species of origin were “Homo sapiens”. Finally, the GSE147507 (Blanco-Melo et al., 2020) dataset was selected.
Briefly, the GSE147507 dataset included independent biological triplicates of “primary human lung epithelial cell line NHBE”, “transformed lung alveolar cell line A549” and “transformed lung-derived Calu-3 cells” which were mock-treated or infected with SARS-CoV-2 for 24 h in addition to 4 months old ferrets infected with SARS-CoV-2 and lung biopsies of COVID-19 patients (supplementary file 1). For the generation of this dataset, total RNA from infected and mock-treated cells were extracted. Subsequently, mRNA enriched libraries were prepared from total RNA using TruSeq Stranded mRNA LP. cDNA libraries were sequenced using an Illumina NextSeq 500 platform. Raw sequencing reads were then aligned to the human genome (hg19) using the RNA-Seq Alignment App on Basespace (Illumina, CA, USA) (Blanco-Melo et al., 2020). For the differential expression analysis of the GSE147507 dataset, DESeq2 (Love et al., 2014) was used. Resulting p-values were adjusted for multiple testing using Benjamini and Hochberg's method of False Discovery Rate (FDR) (Benjamini and Hochberg, 1995). Studies have shown that statistical cut-offs for the selection of DEGs can significantly modulate the results of microarray analysis. That means applying more stringent cut-offs can select only the genes which vary significantly among the others and genes with subtle changes are not considered (Dalman et al., 2012). Therefore, we aimed to conduct a microarray study with different criteria. First, genes with an adjusted p value < 0.05 irrespective of the fold-change were identified as DEGs and selected for further analysis. Then, more stringent criteria for selection of the DEGs were applied (i.e., adj. p value < 0.05 and ǀfold-changeǀ > 2).
2.2. CMap analysis
The CMap library is a collection of gene-expression profiles of drug-treated human cancer cells, which has been widely used for investigation of polypharmacology and drug repurposing (Lamb et al., 2006; Lamb, 2007). In this study, PharmacoGx (Smirnov et al., 2016), an R package for analysis of large pharmacogenomic datasets, was used. The pre-processed CMap data (build 02), available in the PharmacoGX package, was used for further analysis. This current version (build 02; https://portals.broadinstitute.org/cmap/) of CMap collects more than 7000 gene-expression profiles representing 1309 compounds. Because most CMap compounds are the FDA-approved drugs, this database has become a powerful tool for drug repurposing (Montero-Melendez and Perretti, 2014). Gene expression signatures of SARS-CoV-2 infected cell lines, lists of significantly up- and down-regulated genes, resulting from the GSE147507 dataset were used as the input data and compared with gene expression signatures of available CMap drugs by using a pattern-matching algorithm based on the non-parametric rank-ordered Kolmogorov–Smirnov statistics (Hollander et al., 2013) to determine connectivity scores (Cheng et al., 2014). Connectivity scores were analysed for each drug-disease pair to investigate the correlation between the drug and the COVID-19 signature. Drugs with significant negative connectivity scores can potentially reverse the gene expression profile of SARS-CoV-2 infection and can be used as therapeutic options.
2.3. Enrichment analysis
Pathway enrichment analysis was performed with the Metascape web tool (http://metascape.org) to determine whether the identified DEGs are enriched for specific biological functions. Metascape is a web-based portal integrating over 40 bioinformatic knowledgebases within one portal, implementing an automated analysis workflow including conversion, gene annotation, membership, and enrichment analysis (Zhou et al., 2019). Pathway enrichment analysis was carried out for up-regulated and down-regulated genes of NHBE and A549 cell lines separately using Metascape with the following ontology sources: KEGG Pathway, GO Biological Processes, Reactome Gene Sets, Canonical Pathways, and CORUM. Terms with a p-value < 0.01, a minimum count of 3, and an enrichment factor > 1.5 were grouped into clusters based on their membership similarities. Kappa scores (Cohen, 1960) are used as the similarity metric and sub-trees with a similarity of >0.3 are considered a cluster. Each cluster was represented by the most statistically significant term. Enrichment factor (EF) showed how many folds more given pathway members are included in our gene list in comparison to what would have been expected by chance (http://metascape.org/blog/?p=122, 2019). The p-value is defined as the probability of obtaining the observed or more pathway members in our gene list and is calculated based on the cumulative hypergeometric distribution (Zar, 1999).
2.4. Protein-protein interaction
The protein-protein interaction (PPI) network was constructed, visualized, and analysed with the Metascape tool. Analysing DEGs in the context of protein interaction networks provides a framework for a better understanding of transcriptome and functional mechanisms underlying diseases. For each gene list, the PPI network was constructed using Metascape with the following protein interaction databases: BioGrid, InWeb_IM, and OmniPath. After constructing the network, the Molecular Complex Detection (MCODE) method was applied to identify closely connected network components (Bader and Hogue, 2003). Furthermore, pathway enrichment analysis was applied to each MCODE components independently. Terms were then ranked based on the p-value and the three best-scoring terms by p-value were shown as the functional description of the corresponding components.
3. Results
3.1. Drug prediction analysis using CMap
We aimed to study the genes differentially expressed upon SARS-CoV-2 infection, in NHBE cells and A549 cells separately. First, all genes with adjusted p-value<0.05, irrespective of the fold-change were considered as DEGs. For NHBE, a total of 548 genes were found to be modulated by SARS-CoV-2 (378 up-regulated and 170 down-regulated genes) and for A549, a total of 119 genes were found to be modulated by SARS-CoV-2(100 up-regulated and 19 down-regulated genes) (supplementary file 2). Then, a more stringent selection of the DEGs was applied (i.e., adj. p-value < 0.05 and ǀfold-changeǀ > 2). Applying these two criteria, a set of 72 genes (64 genes were up-regulated and 8 genes were down-regulated) from NHBE cells, and 46 genes (44 genes were up-regulated and 2 genes were down-regulated) from A549 cells were identified that showed evidence for differential expression. (supplementary file 3). These identified lists of DEGs were further analysed through CMap study separately.
For the CMap analysis, PharmacoGx was used. Lists of DEGs identified in the previous step were applied as the input data and compared with CMap compounds library. Compounds with p-value < 0.05 and negative connectivity scores were selected and analysed. First, based on genes with adjusted p-value < 0.05, 57 and 25 compounds were shown to significantly reverse genomic signatures of SARS-CoV-2 infected NHBE and A549 cells, respectively. After applying a more stringent selection of DEGs, results were limited to 8 and 6 effective compounds against infected NHBE and A549 cells, respectively. Table 1 summarizes the results of the CMap analysis.
Table 1.
Top-scoring CMap study results. Highlighted compounds are resulted from applying the more stringent criteria (i.e. DEGs with adjusted p-value < 0.05 and fold change ≥ 2). Compounds are ranked based on connectivity score, ranging from −1 (indicating the exactly reverse correlation of signatures between the compound and the disease) to +1. Resulting investigational molecules have been omitted from this table.
3.2. Enrichment analysis
To analyse the biological process of identified DEGs in SARS-CoV-2 infected cells, the Metascape tool was used. Pathway enrichment analysis was carried out for each gene list separately. Fig. 1, Fig. 2 show top clusters with their representative enriched terms for each gene list.
Fig. 1.
Bar graph of top clusters with their representative enriched terms (one per cluster) across input gene lists, colored by p-values. “Log10(P)” is the p-value in log base 10. (A) top enriched functional pathways based on upregulated genes in SARS-CoV-2 infected NHBE cells. (B) same as (A) for downregulated genes in SARS-CoV-2 infected NHBE cells.
Fig. 2.
Bar graph of top clusters with their representative enriched terms (one per cluster) across input gene lists, colored by p-values. “Log10(P)” is the p-value in log base 10. (A) top enriched functional pathways based on upregulated genes in SARS-CoV-2 infected A549 cells. (B) same as (A) for downregulated genes in SARS-CoV-2 infected A549 cells.
Interestingly, up-regulated genes in SARS-CoV-2 infected NHBE cells were mainly enriched in “cytokine-mediate signaling pathway”, “response to virus” and “regulation of cytokine production”. Furthermore, the most enriched pathways in up-regulated genes of infected A549 cells were “defense response to virus”, “response to interferon-gamma” and “antiviral mechanisms by IFN-stimulated genes” as well as “response to interferon-beta” and “response to interferon-alpha”.
However, down-regulated genes of infected NHBE cells were enriched in “kidney development”, “response to growth factor” and “inner ear development”. Down-regulated genes of A549 cells were mainly enriched in the “macroautophagy” pathway.
3.3. Protein-protein interaction
PPI enrichment network was constructed for each gene list with Metascape online tool. Fig. 3, Fig. 4 and Table 2, Table 3 show the PPI network and detailed proteins involved in each MCODE subcluster with their corresponding biological pathways based on up-regulated genes of infected NHBE and A549 cell lines separately. Down-regulated genes did not show a significant PPI network.
Fig. 3.
PPI network and MCODE components analysis identified for upregulated genes of infected NHBE cells. (A) The resultant protein interaction network for the given gene list containing the subset of proteins that form physical interactions with at least one other member in the list. (B) three most densely connected network components based on MCODE clustering.
Fig. 4.
PPI network and MCODE components analysis identified for upregulated genes of infected A549 cells. (A) The resultant protein interaction network for the given gene list containing the subset of proteins that form physical interactions with at least one other member in the list. (B) densely connected network components based on MCODE clustering.
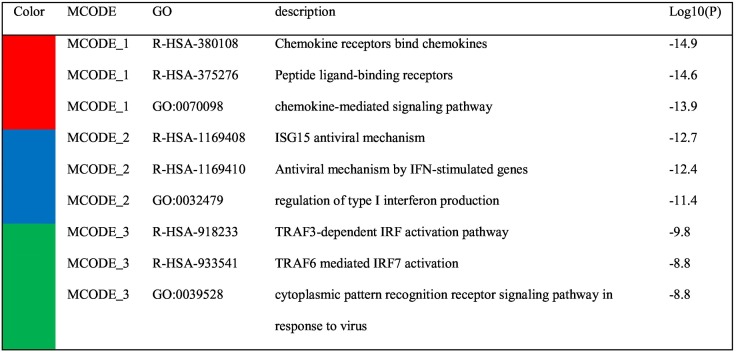
Table 2.
Pathway enrichment analysis applied to MCODE 1–3 components represented in Fig. 3 independently, and the three most significant terms by p-value represent the functional description of the corresponding components.
Table 3.
Pathway enrichment analysis applied to MCODE 1–3 components presented in Fig. 4 independently, and the three most significant terms by p-value represent the functional description of the corresponding components.
The results of PPI were compatible with the results of the pathway enrichment analysis mentioned above. In the PPI network constructed based on up-regulated genes of infected NHBE cells, 24 densely connected proteins were grouped as MCODE1 mainly associated with the “chemokine receptors bind chemokines” pathway. PPI network of A549 up-regulated genes also showed similar results; seven proteins including CXCL1, CXCL2, CXCL3, CXCL5, CXCL8, CCL20, and C3 were grouped as MCODE1. Pathway and process analysis showed that these seven proteins mainly belong to chemokine related pathways. Other MCODE subclusters were mainly associated with the regulation of interferon (IFN) production, IFN signaling pathways, and antiviral mechanisms by IFN-stimulated genes (ISG).
4. Discussion
The newly emerged COVID-19 has become a major global public health concern. However, complete life cycle details of SARS-CoV-2 and host response are not properly known yet. As long as our knowledge of this virus is limited, designing effective novel treatments are almost impossible. Transcriptome analysis of SARS-CoV-2 infected cells can aid our understanding of this virus and accelerate the discovery of novel therapies.
4.1. Drug repurposing
In this study, we conducted a drug repurposing strategy based on CMap to identify therapeutic candidates for COVID-19. Transcriptional profiling of NHBE and A549 cell lines infected with SARS-CoV-2 were prepared. Drugs tending to reverse transcriptional response to SARS-CoV-2 infection were considered as therapeutic options. Interestingly, many of the top-scoring drugs are justified by the available evidence and can be proposed as available therapeutic candidates for COVID-19. Since the publication of transcriptomic profiling of SARS-CoV-2, some researchers have analysed this data to better understand SARS-CoV-2 pathogenesis, host responses, and evaluate the efficacy of possible treatments. Fagone et al. (2020) have analysed and compared this data with the transcriptome profile of SARS-CoV from the 2003 pandemic as well as predicting potential drugs. They proposed Mitogen-activated protein kinase (MEK), serine-threonine kinase (AKT), mammalian target of rapamycin (mTOR), and I kappa B Kinase (IKK) inhibitors as candidate drugs.
Taguchi and Turki (2020) have applied the tensor decomposition (TD)-based unsupervised feature extraction (FE) method to gene expression profiles of COVID-19 infected cells summarized in this data to identify important genes and furtherly identify drug candidates that altered the expression of the genes selected by TD-based unsupervised FE.
However, our work differs from these studies. First, considering the urgency of the COVID-19 pandemic, repurposing already approved drugs with known pharmacokinetics/pharmacodynamics aspects would be more beneficial than investigational compounds. Therefore, the current version of the CMap dataset (build 02) was selected since available approved drugs account for a higher percentage of this dataset than investigational molecules, compatible with our results. Most of the connectivity map-based drug repurposing studies for COVID-19 have been done based on another dataset, the LINCS dataset, and some investigational compounds have been resulted as potential therapeutic options. However, these compounds need further preclinical and clinical experiments to assess their safety, appropriate timing and dosage, limiting their efficacy in controlling COVID-19 pandemic.
Second, as the previous studies showed, the consistency between prioritization results from the CMap dataset and other connectivity map libraries such as the LINCS library is low (Lim and Pavlidis, 2019). Therefore, LINCS does not reproduce CMap dataset drug prioritization. Since most of the connectivity map drug repurposing studies for COVID-19 have been done based on another dataset, the LINCS dataset, it would be reasonable to conduct a study based on the CMap dataset (build 02).
Besides, we have used a different tool, PharmacoGX, for our CMap analysis. Since the introduction of the CMap concept, perturbation datasets such as CMap and LINCS L1000 were created. However, lacking standard frameworks for annotation, storage, access, and analysis challenged the use of these datasets. Therefore, unifying integrative platforms could remove biases of different sources, profiling platforms, and cell-specific differences. PharmacoGx is one of the few or even only integrative platforms developed for this reason.
Some of the top-scoring compounds resulting from the CMap analysis have been discussed:
4.1.1. Lansoprazole
Lansoprazole, a proton pump inhibitor (PPI), is another candidate. Watanabe et al. (2020) have published an article in 2020, confirming potential antiviral properties of prazoles against different viruses such as HIV, Ebola virus (EBOV), and Epstein–Barr virus (EBV). Bojkova et al. (2020) have conducted in vitro experiments to identify novel inhibitors of SARS-CoV-2 replication and they showed the potential of omeprazole, another PPI, to increase the antiviral activity of remdesivir. It is known that inhibiting ATPase proton pumps with medications such as PPI may interfere with the acidification of endosomal pathways that seems an essential step for coronavirus infectivity. Chloroquine and hydroxychloroquine also are shown to have the same effects as prazoles to increase lysosomal PH (Touret et al., 2020). Besides this, other mechanisms should also be considered.
4.1.2. Folic acid
Folic acid is another option. Some previous studies have suggested folic acid as an adjunctive therapy (Sharma, 2020). Besides immune-boosting properties, folic acid can be used as an inhibitor of furin activity, an enzyme responsible for SARS-CoV-2 spike protein cleavage (Sheybani et al., 2020). Therefore, it can be considered as a prophylactic or therapeutic candidate for COVID-19, especially in initial phases. In silico studies have also proposed folic acid as a potential inhibitor of SARS-CoV-2 main protease (Serseg et al., 2020).
4.1.3. Sulfamonomethoxine
Another top-scoring compound is sulfamonomethoxine, a long-acting sulfonamide antibiotic (https://pubchem.ncbi.nlm.nih.gov/compound/Sulfamonomethoxine, 2019). Sulfonamides possess a wide spectrum of biological activities such as anti-bacterial, anti-inflammatory, anti-neoplastic, and antioxidant properties. A large number of sulfonamide derivatives have shown significant antiviral activities against HIV, HCV, HSV, influenza virus, coxsackie virus, etc. based on in vitro and in vivo studies (Khan et al., 2018). Some of the known antiviral compounds contain sulfonamide moieties in their structures (Supuran et al., 2003). As summarized in a study conducted by Supuran et al. (2004), sulfonamide derivatives have been synthesized and used as HIV protease inhibitors (amprenavir), non-nucleoside HIV reverse transcriptase inhibitors, HIV integrase inhibitors, and HIV entry inhibitors. Besides, they can inhibit the replication of retroviruses by targeting the ejection of zinc ions from viral zinc finger proteins.
Since the COVID-19 pandemic, some studies have shown sulfonamide derivatives as potential therapeutic candidates for COVID-19 infection. Batra et al. (2020) have applied machine learning-based models and docking simulations to automatically screen potential therapeutic molecules for the COVID-19 pandemic. Two of the resulting top candidates, sulfaperine and Succinylsulfathiazole, were among sulfonamide derivatives. In another study conducted by Cavasotto and Di Filippo (2020), indisulam, another sulfonamide derivative with anti-neoplastic functions, was found as a potential inhibitor of SARS-CoV-2 papain-like protease (PLpro). This evidence highlights the need for further in vitro and in vivo experiments.
4.1.4. Tolnaftate
Another option is tolnaftate, a synthetic over-the-counter antifungal agent that inhibits ergosterol biosynthesis (https://go.drugbank.com/drugs/DB00525, 2018). Sultan et al. (2020) have conducted a drug repurposing study based on the genomic signature of the SARS family of coronaviruses (SARS-FCoVs)-infected patients, convalescent patients, and healthy controls. Tolnaftate was obtained as one of the potential effective agents. In another study, Mukundan Satyanarayanan (2020) used LIGANN, a drug design tool, to generate new ligands for SARS-CoV-2 main protease (Mpro) and the new ligands were evaluated with docking score, free binding energy, and binding probability and the top ligands were matched with already known drugs with structural similarity assessments. Tolnaftate was resulting in sharing a similar structure with one of the resulting new ligands.
Besides, some studies have introduced already known antifungal agents as antiviral drugs for COVID-19 treatment. Al-Khikani (2020) suggested amphotericin B, an antifungal drug, as a potential antiviral agent against enveloped viruses. Terbinafine, another antifungal agent with the same mechanism of action with tolnaftate (https://go.drugbank.com/drugs/DB00857, 2020), was also reported as a COVID-19 therapeutic option in some available literature. Blaess et al. (2020) believed that compounds with lysosomotropic properties such as terbinafine can be used as promising candidates for COVID-19 prevention and treatment. Lysosomotropic molecules can affect viral entry into host cells and inflammatory cytokines expression. Arya et al. (2020) also showed a significant binding affinity of terbinafine to SARS-CoV-2 PLpro.
Besides, SARS-CoV-2 infected patients are at increased risk of fungal co-infections such as pulmonary aspergillosis or candidiasis (Verweij et al., 2020; Al-Hatmi et al., 2020). Repurposing a drug with both antifungal and antiviral properties would be a more promising option.
4.1.5. Diclofenamide
Another top-scoring compound is diclofenamide with connectivity-score = −0.59, a carbonic anhydrase inhibitor used in the treatment of glaucoma (https://go.drugbank.com/drugs/DB01144, 2018). Some recent studies have suggested carbonic anhydrase inhibitors such as acetazolamide as adjunctive treatments in the management of COVID-19. Solaimanzadeh (2020) has reviewed features of COVID-19 pneumonia in parallel to high altitude pulmonary edema (HAPE) and revealed substantial similarities between these two conditions leading to ARDS. Therefore, it was hypothesized that medications commonly used in HAPE including acetazolamide, nifedipine, etc., can be effective for COVID-19 symptomatic therapy. In another study, Geier and Geier (2020), have also provided some evidence consistent with the previously mentioned study to confirm the potential effectiveness of carbonic anhydrase inhibitors for COVID-19 treatment. However, further investigations on the effectiveness of these compounds are needed.
4.1.6. Halcinonide
One of the other top-scoring compounds is halcinonide (https://go.drugbank.com/drugs/DB06786, 2018), a topical corticosteroid with 70% structural similarity with budesonide, another resulting compound in our analysis. Including two classes of corticosteroids, a topical steroid with one inhaled corticosteroid (ICS), highlights the need for further investigations on the effectiveness of these compounds for COVID-19 treatment. Although still some paradoxes about their effects on viral infections and in particular COVID-19 exist, some studies have revealed promising results.
In a retrospective cohort study conducted at Wuhan Jinyintan Hospital between December 25, 2019, and January 26, 2020, 201 confirmed COVID-19 patients were evaluated for risk factors associated with disease severity. They found that patients treated with methylprednisolone, another corticosteroid, had reduced risk of death (HR, 0.38; 95%CI, 0.20–0.72) (Wu et al., 2020).
In another study, Fadel et al. (2020) performed a quasi-experiment in Michigan from March 12 to March 27, 2020, on moderate to severe COVID-19 confirmed patients to evaluate the effect of methylprednisolone on the course of the disease. They showed a significant reduction of the need for ICU admission or mechanical ventilation and also the incidence of developing ARDS in patients receiving methylprednisolone.
In the Randomized Evaluation of COVID-19 therapy (RECOVERY) trial (The RECOVERY Collaborative Group, 2020), aimed to evaluate the effects of some possible treatments in comparison to usual care in COVID-19 patients, treatment with dexamethasone, another steroid, showed a significant reduction of death rates in COVID-19 patients with some degree of respiratory distress that need supplemental oxygen therapy or mechanical ventilation. Studies conducted to evaluate the effects of corticosteroids on COVID-19 patients have been reviewed by Singh et al. (2020).
Besides, previous in vitro models have shown significant inhibitory effects of ICS including budesonide on different members of the coronavirus family (Halpin et al., 2020; Yamaya et al., 2020; Matsuyama et al., 2020). Considering COVID-19 patient's comorbidities such as asthma or COPD, this evidence suggests continuing ICS in course of coronavirus infection.
4.1.7. Saquinavir
An approved drug with one of the highest connectivity scores is saquinavir, an HIV protease inhibitor (https://go.drugbank.com/drugs/DB01232, 2018). Virtual screening of clinically approved drugs has shown saquinavir as a potential inhibitor of SARS-CoV-2 main protease, RNA-dep RNA polymerase, and other non-structural proteins (nsp) (Hall and Ji, 2020; Ruan et al., 2020). Saquinavir can also suppress SARS-CoV-2 replication based on in vitro models in less than 10 μM concentration (Yamamoto et al., 2020). Other drugs of this group such as lopinavir and ritonavir are currently used as COVID-19 treatment.
4.1.8. Metronidazole
Another important drug is metronidazole, a commonly used antibiotic. Some recent studies have proposed metronidazole as an addition to the COVID-19 treatment regimen due to its immunomodulatory effects. Metronidazole can decrease the level of inflammatory cytokines which are increased during COVID-19 infection (Gharebaghi et al., 2020). Chakraborty and Das (2020) have proposed adding anaerobe-specific antibiotics such as metronidazole to COVID-19 treatment due to evidence of overexpression of anaerobic bacteria in COVID-19 patients.
4.1.9. Ebselen
Another top-scoring drug is ebselen. Ebselen is a small molecule with antioxidant, anti-inflammatory, and cytoprotective features (https://go.drugbank.com/drugs/DB12610, 2018). Previous computer-aided studies have revealed ebselen as a strong inhibitor of SARS-CoV-2 main protease, furtherly was approved through cell-based studies (Jin et al., 2020). It is also known that ebselen can covalently bind to the Zn-bound/catalytic cysteines in SARS-CoV-2 papain-like protease (PLpro) and nsp10, two critical proteins for viral replication, to inhibit them (Yang and Lim, 2020). In the last two decades, different analogs of ebselen were synthesized as antiviral agents (Wójtowicz et al., 2004). These data strongly emphasize the clinical potential of ebselen for treating COVID-19 patients.
4.1.10. Lidocaine and benzocaine
Two resulting compounds are lidocaine and benzocaine, local anesthetics with the same mechanisms of action (https://go.drugbank.com/drugs/DB12610, 2018; https://go.drugbank.com/drugs/DB01086, 2018). In a review article published by Cassuto et al. (2006), local anesthetics have been shown to inhibit different steps of the inflammatory cascade including leukocyte adhesion, migration, activation, and phagocytosis besides inhibitory effects on the release of inflammatory cytokines. Since COVID-19 is associated with a hyperinflammatory state known as “cytokine storm”, these compounds could be suggested as candidates for further in vitro and in vivo experiments.
In a recently published article, Ali and El-Mallakh (2020) have reviewed available evidence suggesting anti-inflammatory properties of local anesthetics and proposed nebulized lidocaine as a beneficial adjunctive treatment for COVID-19 associated hyperinflammatory state.
In another article, Finnerty and Buggy (2020) have discussed the role of neutrophil extracellular traps (NETs), web-like structures of DNA and proteins produced by neutrophils as an alternative way to kill circulating pathogens, in COVID-19 pathogenesis. Excessive production of NETs in COVID-19 patients would be associated with a hypercoagulable state, myocardial infarction, sepsis, respiratory distress, and contributing to cytokine storm. Since previous evidence has shown that local anesthetics can inhibit the production of NETs in patients undergoing surgery, they can be proposed as novel candidates to reduce COVID-19 severity and further complications. More studies are needed to confirm this hypothesis.
4.1.11. Aminocaproic acid
Another drug is aminocaproic acid, a plasminogen activator inhibitor (PAI) (https://go.drugbank.com/drugs/DB00513, 2018). It is known that PAIs can inhibit other membrane-anchored serine proteases such as transmembrane serine protease 2 (TMPRSS2), a critical factor for cell entry of SARS-CoV-2 (Jankun, 2020). Therefore, they can be used as a therapeutic or even prophylactic agent for COVID-19. Shen et al. (2013) have shown the potential effect of aminocaproic acid for inhibiting the influenza virus through suppressing the cleavage of hemagglutinin (HA) precursor, a critical glycoprotein for viral binding and entry. Besides, our results would suggest other unknown antiviral mechanisms for aminocaproic acid.
4.1.12. Molindone
One of the resulting compounds is molindone, an indole derivative acting as dopamine D2 receptors antagonist, serotonin 5-HT1A, 5-HT2, and 5-HT2A receptors antagonist that has been used as an antipsychotic agent (https://go.drugbank.com/drugs/DB01618, 2018). Some literature hypothesized the role of dopamine in COVID-19 infection. Sen (2020) hypothesized that using dopamine antagonists can boost innate and adaptive immune systems by increasing the serum level of prolactin to combat COVID-19 infection. Prolactin, the lactogenic hormone with other biological activities, is produced from the anterior pituitary gland as well as other sites such as immune cells. Interestingly, prolactin plays a significant role in immune hemostasis. Hypoprolactinemia can cause lymphocytic depletion leading to nosocomial infections. Using dopamine antagonists to enhance serum prolactin levels has been shown effective for HIV control by increasing lymphocyte proliferation. It can also boost the innate immune response by stimulating NK cells, neutrophils, and macrophages. Chlorpromazine, another dopamine antagonist, has also been suggested as a therapeutic candidate for COVID-19 with unknown antiviral mechanisms (Liu et al., 2020).
4.1.13. Benzethonium chloride
Two other top-scoring compounds are benzethonium chloride (https://go.drugbank.com/drugs/DB11125, 2018) with surfactant, antiseptic, and broad-spectrum antimicrobial properties and ambroxol (https://go.drugbank.com/drugs/DB06742, 2018), a secretolytic agent used in respiratory diseases and stimulant of surfactant synthesis. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 receptor, is expressed in type 2 alveolar pneumocytes, cells producing surfactant, and infection of these cells with SARS-CoV-2 leads to down-regulation of surfactant synthesis and release, contributing to acute respiratory distress syndrome (ARDS) (Bombardini and Picano, 2020). Therefore, these medications and other drugs with the same mechanisms would be potent adjunctive therapies for COVID-19.
4.1.14. Monorden
Monorden or radicicol is a heat shock protein 90 (HSP90) inhibitor (https://go.drugbank.com/drugs/DB03758, 2018). Connor et al. (2007) showed that HSP90 is essential for viral replication and its inhibitors such as radicicol or geldanamycin could block the replication of RNA viruses. A recent study conducted by Wyler et al. (2020) has examined the effect of HSP90 inhibitors on SARS-CoV-2 based on in vitro models and proposed this group as potent antiviral and anti-inflammatory agents.
4.1.15. Sodium phenylbutyrate
Sodium phenylbutyrate is a histone deacetylase (HDAC) inhibitor (https://go.drugbank.com/drugs/DB06819, 2018). They act as epigenetic modulators and regulate expression of genes involved in the cell cycle, proliferation, differentiation, and also inflammation (Seto and Yoshida, 2014). A number of these compounds have been suggested for SARS-CoV-2. In vitro experiments have shown an inhibitory effect of valproate, an HDAC inhibitor, on the NF-KB pathway, and subsequently suppressing TNF-α and IL 6 (Bhargava et al., 2020). Besides, they can be used as antifibrotic agents in pulmonary fibrosis (Lyu et al., 2019).
Some previous studies have also shown the effectiveness of some of the other resulting drugs (e.g. propylthiouracil, simvastatin, etc.) (Shrivastava-Ranjan et al., 2018; Reiner et al., 2020; Santos-Beneit et al., 2020). These mentioned drugs can be utilized for further in vitro and in vivo experiments to better examine their efficacy.
4.2. Enrichment analysis
Regarding the results of pathway enrichment analysis, analysing the transcriptome of SARS-CoV-2 infected cells has identified significant up-regulation of pathways involved in cytokine production and signaling pathways. Cytokines are small proteins, secreted by various cell lines and in particular macrophages and helper T cells as part of the immune response to infections with both pro-inflammatory and anti-inflammatory properties (Dinarello, 2000; Zhang and An, 2007). Although the immune response is the main line of defense against viral infections, the dysregulated immune response to infections and other stimuli can cause multi-organ damage leading to increased morbidity and mortality. Available evidence has suggested a maladaptive immune response in COVID-19 severely ill patients with a high level of pro-inflammatory cytokines such as IL-1B, IL-6, TNF, and chemokines (CCL-2, CCL-3, CCL-5, etc.) named as “cytokine storm” (Ye et al., 2020).
The results of our study have confirmed this evidence. Most of the up-regulated enriched pathways shown in Fig. 1, Fig. 2 belong to cytokine stimulation, production, and signaling mechanisms. These results can also guide further drug repurposing studies and propose therapeutic candidates to alleviate disease severity.
As a result, compounds with immunosuppressive properties would be beneficial in COVID-19 patients with signs of hyperinflammatory responses. Some have been already evaluated; however, further clinical trials should be conducted to confirm their effectiveness (Henderson et al., 2020; Bhaskar et al., 2020). Commonly used anti-inflammatory and immunosuppressive compounds with potential therapeutic effects are listed below:
-
1.
Corticosteroids: a class of steroids with anti-inflammatory and immunosuppressive features. Results of our CMap analysis have also confirmed their effectiveness. However, appropriate timing and dosage of corticosteroids are critical and should be limited to critically ill patients (Wu et al., 2020; Fadel et al., 2020; The RECOVERY Collaborative Group, 2020; Singh et al., 2020).
-
2.
Intravenous immunoglobulin (IVIG): an immunomodulating agent with various indications. Xie et al. (2020) have conducted a retrospective study on 58 severe COVID-19 confirmed cases admitted at Wuhan Third Hospital from January to February 2020 to evaluate the effect of IVIG utilization. They showed that prescribing IVIG within 48 h of admission can substantially reduce the need for mechanical ventilation, ICU admission and ultimately reducing 28-day mortality.
-
3.
IL-1 antagonists: In a retrospective study conducted at the San Raffaele Hospital in Milan, Italy on COVID-19 patients with signs of ARDS, effects of adjunctive therapy with anakinra, an IL-1 antagonist, were evaluated. Results confirmed clinical effectiveness and safety of high-dose anakinra administration (Cavalli et al., 2020). In another prospective cohort study conducted by Huet et al. (2020) (the Ana-COVID study) at Groupe Hospitalier Paris Saint-Joseph (Paris, France), treatment with subcutaneous anakinra reduced both mortality rates and the need for mechanical ventilation in severe COVID-19 cases without serious side-effects.
-
4.
IL-6 antagonists: Guaraldi et al. (2020) have conducted a retrospective, observational cohort study to evaluate the role of tocilizumab, an IL-6 antagonist, on COVID-19 severity and mortality. Results showed intravenous or subcutaneous tocilizumab can significantly reduce mortality rates and the risk of invasive mechanical ventilation. Some other studies have also confirmed the effectiveness of tocilizumab for COVID-19 disease (Luo et al., 2020; Xu et al., 2020).
-
5.
Other immunosuppressive agents such as TNF inhibitors, JAK inhibitors, etc. should also be considered.
4.3. Protein-protein interaction
PPI analysis revealed some densely connected proteins including CXCL1, CXCL2, CXCL3, CXCL5, CXCL8, CCL20, and C3, that are mainly involved in chemokine related pathways. Chemokines are a subset of cytokines, produced by various cell lines. Binding chemokines to their receptors, a class of G protein-coupled receptors mainly on leukocytes, leads to migration of immune cells to the site of infection known as chemo-attractant property. Besides chemotaxis, previous literature showed the role of two well-known chemokines, CXCL10 and CXCL8, in respiratory tract infections. CXCL10 and CXCL8 levels in serum or bronchoalveolar lavage fluid (BALF) correlate with the severity of respiratory symptoms. Some clinical studies have shown the relationship between chemokine dysregulation and COVID-19 severity (Coperchini et al., 2020). However, related evidence is scant and more experiments are needed. Considering this evidence, compounds targeting the chemokine/receptor-related immune response would be appropriate candidates to limit SARS-CoV-2 induced ARDS.
Some scientists are conducting studies to evaluate the effect of two C-C chemokine receptor type 5 (CCR5) antagonists, leronlimab and maraviroc, on COVID-19 infection. Another study has evaluated the effect of anti-CXCL8 agent BMS-986253 on cancer patients with COVID-19. Besides, inhibiting CXCR2, a cytokine receptor, has improved pulmonary inflammatory disease in an influenza-induced lung injury mouse model (Koenig et al., 2020). Therefore, more studies should be conducted to evaluate usage of CXCR1 and CXCR2 antagonists (e.g. navirixin, danixirin, etc.) for COVID-19 related hyperinflammatory response.
In addition, the inflammatory response is a double-edged sword. On the one hand, it is critical to limit viral replication. On the other hand, exaggerated responses can lead to multiorgan failure including ARDS.
Moreover, the results of SARS-CoV-2 infected cells confirm a robust IFN response with significant expression of ISGs pathway with known antiviral, inflammatory, and immune regulatory functions. However, previous analysis has shown impaired type 1 IFN production in severe COVID-19 cases and proposed different hypotheses whether IFN production is exhausted after the initial overexpression or SARS-CoV-2 can evade immune pathways (Li et al., 2020b; Hadjadj et al., 2020; Sallard et al., 2020). However, COVID-19 immunopathology is not properly known and more comprehensive studies are needed to clarify dynamic immune response to SARS-CoV-2.
It is promising that the CMap strategy has identified a number of therapeutic candidates that could be repurposed for COVID-19. Since SARS-CoV-2 infection can change host transcriptome in the way optimized for viral replication, transcriptome profiling and further CMap analysis can help better understand virus pathophysiology and may discover new unknown genes and pathways. However, data used in this study was obtained 24 h post-infection with SARS-CoV-2 and further studies should be conducted in variable time points to assess the dynamics of virus pathogenesis and host response. Furthermore, although CMap libraries have been improved since its introduction in 2006, all available small molecules are much more than current CMap libraries.
5. Conclusion
The newly emerged COVID-19 pandemic is the major global health crisis of our time affecting more than 217 countries and territories around the world. Several studies are ongoing to find effective therapies in particular through drug repurposing approaches. In the present study, we suggested potential antiviral candidates based on the CMap concept. Available gene expression profiles of SARS-CoV-2 infected NHBE and A549 cell lines were analysed to identify DEGs. Subsequently, these signatures were compared with genomic signatures of cell lines treated with variable compounds available in CMap databases. Among these compounds, lansoprazole, folic acid, sulfamonomethoxine, tolnaftate, diclofenamide, halcinonide, saquinavir, metronidazole, ebselen, lidocaine and benzocaine, histone deacetylase (HDAC) inhibitors, heat shock protein 90 (HSP90) inhibitors, and many other clinically approved drugs showed promising results to reverse gene expression profiles of SARS-CoV-2 infected cells. Pathway enrichment and PPI analysis also revealed that early immunomodulatory interventions might prevent severe cases and the need for invasive ventilation. Further in vitro and in vivo studies should be conducted to test potential antiviral effects of these suggested compounds against SARS-CoV-2.
Declaration of Competing Interest
The authors declare that there are no conflicts of interest. All authors deny any financial and personal relationships with other people or organizations that could inappropriately influence (bias) this work.
Acknowledgments
This research was partly funded through a research grant from Iran's National Elites Foundation called Ahmadi Rowshan grant. The study sponsor had no involvement in the study design, in the collection, analysis, and interpretation of data; in the writing of the manuscript; and in the decision to submit the manuscript for publication.
References
- Al-Hatmi A.M., Mohsin J., Al-Huraizi A., Khamis F. COVID-19 associated invasive candidiasis. J. Infect. 2020 doi: 10.1016/j.jinf.2020.08.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z.A., El-Mallakh R.S. Nebulized lidocaine in COVID-19, an hypothesis. Med. Hypothe. 2020;144 doi: 10.1016/j.mehy.2020.109947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khikani F.H.O. Amphotericin B as antiviral drug: possible efficacy against COVID-19. Ann. Thor. Med. 2020;15(3):118. doi: 10.4103/atm.ATM_147_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R., Das A., Prashar V., Kumar M. Potential inhibitors against papain-like protease of novel coronavirus (COVID-19) from FDA approved drugs. ChemRxiv. 2020 [Google Scholar]
- Bader G.D., Hogue C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003 Dec 1;4(1):2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Troup D.B., Wilhite S.E., Ledoux P., Rudnev D., Evangelista C. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucl. Acids Res. 2007;35(suppl_1):D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra R., Chan H., Kamath G., Ramprasad R., Cherukara M.J., Sankaranarayanan S.K. Screening of therapeutic agents for COVID-19 using machine learning and ensemble docking studies. J. Phys. Chem. Lett. 2020;11(17):7058–7065. doi: 10.1021/acs.jpclett.0c02278. [DOI] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C. Remdesivir for the treatment of Covid-19—preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007764. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57(1):289–300. [Google Scholar]
- Bhargava P., Panda P., Ostwal V., Ramaswamy A. Repurposing valproate to prevent acute respiratory distress syndrome/acute lung injury in COVID-19: a review of immunomodulatory action. Cancer Res. Stat. Treat. 2020;3(5):65. [Google Scholar]
- Bhaskar S., Sinha A., Banach M., Mittoo S., Weissert R., Kass J.S. Cytokine storm in COVID-19—immunopathological mechanisms, clinical considerations, and therapeutic approaches: The REPROGRAM Consortium position paper. Front. Immunol. 2020;11:1648. doi: 10.3389/fimmu.2020.01648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaess M., Kaiser L., Sauer M., Deigner H.-P. 2020. Lysosomotropic Active Compounds—Hidden Protection Against COVID-19/SARS-CoV-2 Infection? [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., McGreig J.E., McLaughlin K.-M., Masterson S.G., Widera M., Kraehling V. SARS-CoV-2 and SARS-CoV differ in their cell tropism and drug sensitivity profiles. bioRxiv. 2020 [Google Scholar]
- Bombardini T., Picano E. Angiotensin converting enzyme 2 as the molecular bridge between epidemiologic and clinical features of COVID-19. Can. J. Cardiol. 2020;36(5):784. e1–784. e2. doi: 10.1016/j.cjca.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassuto J., Sinclair R., Bonderovic M. Anti-inflammatory properties of local anesthetics and their present and potential clinical implications. Acta Anaesthesiol. Scand. 2006;50(3):265–282. doi: 10.1111/j.1399-6576.2006.00936.x. [DOI] [PubMed] [Google Scholar]
- Cavalli G., De Luca G., Campochiaro C., Della-Torre E., Ripa M., Canetti D. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2020;2:e325–e331. doi: 10.1016/S2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavasotto C.N., Di Filippo J.I. In silico drug repurposing for COVID-19: targeting SARS-CoV-2 proteins through docking and consensus ranking. Mol. Inform. 2020;39:1–9. doi: 10.1002/minf.202000115. 2000115. [DOI] [PubMed] [Google Scholar]
- Chakraborty S., Das G. Secondary infection by anaerobic bacteria possibly ensues a battle for oxygen in SARS-Cov2 infected patients: anaerobe-targeting antibiotics (like doxycycline/Metronidazole) to supplement Azithromycin in the treatment regimen of COVID19? OSF Preprints. 2020 [Google Scholar]
- Cheng J., Yang L., Kumar V., Agarwal P. Systematic evaluation of connectivity map for disease indications. Genome Med. 2014;6(12):95. doi: 10.1186/s13073-014-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury M.S., Rathod J., Gernsheimer J. A rapid systematic review of clinical trials utilizing chloroquine and hydroxychloroquine as a treatment for COVID-19. Acad. Emerg. Med. 2020;27(6):493–504. doi: 10.1111/acem.14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960;20:27–46. [Google Scholar]
- Connor J.H., McKenzie M.O., Parks G.D., Lyles D.S. Antiviral activity and RNA polymerase degradation following Hsp90 inhibition in a range of negative strand viruses. Virology. 2007;362(1):109–119. doi: 10.1016/j.virol.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalman M.R., Deeter A., Nimishakavi G., Duan Z.H. Fold change and p-value cutoffs significantly alter microarray interpretations. InBMC bioinformatics. BioMed Central. 2012 Dec;Vol. 13(No. S2) doi: 10.1186/1471-2105-13-S2-S11. p. S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello C.A. Proinflammatory cytokines. Chest. 2000;118(2):503–508. doi: 10.1378/chest.118.2.503. [DOI] [PubMed] [Google Scholar]
- Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., Hall M.D. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Central Science. 2020;6:672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadel R., Morrison A., Vahia A., Smith Z.R., Chaudhry Z., Bhargava P. Early short course corticosteroids in hospitalized patients with COVID-19. medRxiv. 2020 doi: 10.1093/cid/ciaa601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagone P., Ciurleo R., Lombardo S.D., Iacobello C., Palermo C.I., Shoenfeld Y. Transcriptional landscape of SARS-CoV-2 infection dismantles pathogenic pathways activated by the virus, proposes unique sex-specific differences and predicts tailored therapeutic strategies. Autoimmun. Rev. 2020;102571 doi: 10.1016/j.autrev.2020.102571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnerty D.T., Buggy D.J. A novel role for lidocaine in COVID-19 patients? Br. J. Anaesth. 2020;125(4):e391–e394. doi: 10.1016/j.bja.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier M.R., Geier D.A. Respiratory conditions in coronavirus disease 2019 (COVID-19): important considerations regarding novel treatment strategies to reduce mortality. Med. Hypotheses. 2020;140:1–5. doi: 10.1016/j.mehy.2020.109760. 109760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharebaghi R., Heidary F., Moradi M., Parvizi M. Metronidazole; a POTENTIAL NOVEL ADDITION to the COVID-19 treatment regimen. Archiv. Acad. Emerg. Med. 2020;8(1) [PMC free article] [PubMed] [Google Scholar]
- Guaraldi G., Meschiari M., Cozzi-Lepri A., Milic J., Tonelli R., Menozzi M. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. 2020;2(8):e474–e484. doi: 10.1016/S2665-9913(20)30173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Pere H. Impaired type I interferon activity and exacerbated inflammatory responses in severe Covid-19 patients. medRxiv. 2020 doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D.C., Jr., Ji H.-F. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med. Infect. Dis. 2020;35:1–3. doi: 10.1016/j.tmaid.2020.101646. 101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin D.M., Singh D., Hadfield R.M. Inhaled corticosteroids and COVID-19: a systematic review and clinical perspective. Eur. Respir. Soc. 2020;55(5):1–4. doi: 10.1183/13993003.01009-2020. 2001009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimfarth L., Serafini M.R., Martins-Filho P.R.S., Quintans J.S.S., Júnior L.J.Q. Drug repurposing and cytokine management in response to COVID-19: a review. Int. Immunopharmacol. 2020;106947 doi: 10.1016/j.intimp.2020.106947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson L.A., Canna S.W., Schulert G.S., Volpi S., Lee P.Y., Kernan K.F. On the alert for cytokine storm: immunopathology in COVID-19. Arthrit. Rheumatol. 2020;72(7):1059–1063. doi: 10.1002/art.41285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2019. http://metascape.org/blog/?p=122
- 2018. https://go.drugbank.com/drugs/DB00513
- 2018. https://go.drugbank.com/drugs/DB00525
- 2018. https://go.drugbank.com/drugs/DB01086
- 2018. https://go.drugbank.com/drugs/DB01144
- 2018. https://go.drugbank.com/drugs/DB01232
- 2018. https://go.drugbank.com/drugs/DB01618
- 2018. https://go.drugbank.com/drugs/DB03758
- 2018. https://go.drugbank.com/drugs/DB06742
- 2018. https://go.drugbank.com/drugs/DB06786
- 2018. https://go.drugbank.com/drugs/DB06819
- 2018. https://go.drugbank.com/drugs/DB11125
- 2018. https://go.drugbank.com/drugs/DB12610
- 2019. https://pubchem.ncbi.nlm.nih.gov/compound/Sulfamonomethoxine
- 2020. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-chloroquine-and
- 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/
- Hollander M., Wolfe D.A., Chicken E. John Wiley & Sons; 2013. Nonparametric Statistical Methods. [Google Scholar]
- Huet T., Beaussier H., Voisin O., Jouveshomme S., Dauriat G., Lazareth I. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2020;2(7):e393–e400. doi: 10.1016/S2665-9913(20)30164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahchan N.S., Dudley J.T., Mazur P.K., Flores N., Yang D., Palmerton A. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 2013;3(12):1364–1377. doi: 10.1158/2159-8290.CD-13-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankun J. COVID-19 pandemic; transmembrane protease serine 2 (TMPRSS2) inhibitors as potential drugs. Translation. 2020;7:1–5. [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. bioRxiv. 2020 [Google Scholar]
- Khan B., Saddique F., Kanwal A., Aslam S. Antiviral activity of organic molecules having sulfonamide moiety: an insight of recent research. Afinidad. 2018;75(582) [Google Scholar]
- Koenig L.M., Boehmer D.F., Metzger P., Schnurr M., Endres S., Rothenfusser S. Blocking inflammation on the way: rationale for CXCR2 antagonists for the treatment of COVID-19. J. Exp. Med. 2020;217(9) doi: 10.1084/jem.20201342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J. The Connectivity Map: a new tool for biomedical research. Nat. Rev. Cancer. 2007;7(1):54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P. Coronavirus infections and immune responses. J. Med. Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Xie Z., Lin W., Cai W., Wen C., Guan Y. Efficacy and safety of lopinavir/ritonavir or arbidol in adult patients with mild/moderate COVID-19: an exploratory randomized controlled trial. Medicine. 2020;1:1–9. doi: 10.1016/j.medj.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Nathaniel, Pavlidis Paul. Evaluation of Connectivity Map shows limited reproducibility in drug repositioning. bioRxiv. 2019 doi: 10.1038/s41598-021-97005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Chan W.K., Wang Z., Hur J., Xie J., Yu H. 2020. Ontological and Bioinformatic Analysis of Anti-coronavirus Drugs and Their Implication for Drug Repurposing Against COVID-19. [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P., Liu Y., Qiu L., Liu X., Liu D., Li J. Tocilizumab treatment in COVID-19: a single center experience. J. Med. Virol. 2020;92(7):814–818. doi: 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu X., Hu M., Peng J., Zhang X., Sanders Y.Y. HDAC inhibitors as antifibrotic drugs in cardiac and pulmonary fibrosis. Ther. Adv. Chron. Dis. 2019;10:1–19. doi: 10.1177/2040622319862697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Kawase M., Nao N., Shirato K., Ujike M., Kamitani W. The inhaled corticosteroid ciclesonide blocks coronavirus RNA replication by targeting viral NSP15. bioRxiv. 2020 doi: 10.1128/JVI.01648-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Melendez T., Perretti M. Connections in pharmacology: innovation serving translational medicine. Drug Discov. Today. 2014;19(7):820–823. doi: 10.1016/j.drudis.2013.11.022. [DOI] [PubMed] [Google Scholar]
- Mukundan Satyanarayanan D.V. Ligands Based Drug Design for Covid 19 - A MultiFaceted Approach using Ligand Design, Molecular Docking and Binding Probability Calculation. International Journal for Research in Applied Science & Engineering Technology (IJRASET) 2020;8:844–850. [Google Scholar]
- Musa A., Ghoraie L.S., Zhang S.-D., Glazko G., Yli-Harja O., Dehmer M. A review of connectivity map and computational approaches in pharmacogenomics. Brief. Bioinform. 2018;19(3):506–523. doi: 10.1093/bib/bbw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections—more than just the common cold. Jama. 2020;323(8):707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- Qu X.A., Rajpal D.K. Applications of Connectivity Map in drug discovery and development. Drug Discov. Today. 2012;17(23–24):1289–1298. doi: 10.1016/j.drudis.2012.07.017. [DOI] [PubMed] [Google Scholar]
- Reiner Ž., Hatamipour M., Banach M., Pirro M., Al-Rasadi K., Jamialahmadi T. Statins and the COVID-19 main protease: in silico evidence on direct interaction. Arch. Med. Sci. 2020;16(3):490–496. doi: 10.5114/aoms.2020.94655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L.-L., Wang Y.-M., Wu Z.-Q., Xiang Z.-C., Guo L., Xu T. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin. Med. J. 2020;133(9):1015–1024. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Z., Liu C., Guo Y., He Z., Huang X., Jia X. 2020. Potential Inhibitors Targeting RNA-Dependent RNA Polymerase Activity (NSP12) of SARS-CoV-2. Preprints. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallard E., Lescure F.-X., Yazdanpanah Y., Mentre F., Peiffer-Smadja N., Florence A. Type 1 interferons as a potential treatment against COVID-19. Antivir. Res. 2020;178:1–4. doi: 10.1016/j.antiviral.2020.104791. 104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Beneit F., Raškevičius V., Skeberdis V.A., Bordel S. 2020. A Metabolic Modeling Approach Reveals Promising Therapeutic Targets and Antiviral Drugs to Combat COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A. Repurposing prolactin as a promising immunomodulator for the treatment of COVID-19: are common antiemetics the wonder drug to fight coronavirus? Med. Hypotheses. 2020;110208 doi: 10.1016/j.mehy.2020.110208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serseg T., Benarous K., Hispidin Yousfi M., Lepidine E. 2020. Two Natural Compounds and Folic Acid as Potential Inhibitors of 2019-novel Coronavirus Main Protease (2019-nCoVMpro), Molecular Docking and SAR Study. arXiv preprint arXiv:200408920. [DOI] [PubMed] [Google Scholar]
- Seto E., Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014;6(4) doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma L. Dietary management to build adaptive immunity against COVID-19. J. Peer Sci. 2020;2(2) [Google Scholar]
- Shen X., Zhang X., Liu S. Novel hemagglutinin-based influenza virus inhibitors. J. Thor. Dis. 2013;5(Suppl. 2):S149. doi: 10.3978/j.issn.2072-1439.2013.06.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheybani Z., Dokoohaki M.H., Negahdaripour M., Dehdashti M., Zolghadr H., Moghadami M. The role of folic acid in the management of respiratory disease caused by COVID-19. chemRxiv. 2020 [Google Scholar]
- Shrivastava-Ranjan P., Flint M., Bergeron É., McElroy A.K., Chatterjee P., Albariño C.G. Statins suppress Ebola virus infectivity by interfering with glycoprotein processing. MBio. 2018;9(3):e00660–e00718. doi: 10.1128/mBio.00660-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Majumdar S., Singh R., Misra A. Role of corticosteroid in the management of COVID-19: a systemic review and a clinician’s perspective. Diabet. Metabol. Syndr. 2020;14(5):971–978. doi: 10.1016/j.dsx.2020.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov P., Safikhani Z., El-Hachem N., Wang D., She A., Olsen C. PharmacoGx: an R package for analysis of large pharmacogenomic datasets. Bioinformatics. 2016;32(8):1244–1246. doi: 10.1093/bioinformatics/btv723. [DOI] [PubMed] [Google Scholar]
- So H.-C., Chau C.K., Chiu W.-T., Ho K.-S., Lo C.-P., Yim S.H.-Y. When GWAS meets the Connectivity Map: drug repositioning for seven psychiatric disorders. bioRxiv. 2016 [Google Scholar]
- Solaimanzadeh I. Acetazolamide, nifedipine and phosphodiesterase inhibitors: rationale for their utilization as adjunctive countermeasures in the treatment of coronavirus disease 2019 (COVID-19) Cureus. 2020;12(3) doi: 10.7759/cureus.7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan I., Howard S., Tbakhi A. 2020. Drug Repositioning Suggests a Role for the Heat Shock Protein 90 Inhibitor Geldanamycin in Treating COVID-19 Infection. [Google Scholar]
- Supuran C.T., Casini A., Scozzafava A. Protease inhibitors of the sulfonamide type: anticancer, antiinflammatory, and antiviral agents. Med. Res. Rev. 2003;23(5):535–558. doi: 10.1002/med.10047. [DOI] [PubMed] [Google Scholar]
- Supuran C.T., Innocenti A., Mastrolorenzo A., Scozzafava A. Antiviral sulfonamide derivatives. Mini Rev. Med. Chem. 2004;4(2):189–200. doi: 10.2174/1389557043487402. [DOI] [PubMed] [Google Scholar]
- Taguchi Y., Turki T. 2020. A New Advanced In Silico Drug Discovery Method for Novel Coronavirus (SARS-CoV-2) With Tensor Decomposition-based Unsupervised Feature Extraction. Preprints. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The RECOVERY Collaborative Group Dexamethasone in Hospitalized Patients with Covid-19 — Preliminary Report. N Engl J Med. 2020 doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touret F., Gilles M., Barral K., Nougairède A., Decroly E., de Lamballerie X. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. bioRxiv. 2020 doi: 10.1038/s41598-020-70143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij P.E., Gangneux J.-P., Bassetti M., Brüggemann R.J., Cornely O.A., Koehler P. Diagnosing COVID-19-associated pulmonary aspergillosis. Lancet Microb. 2020;1(2):e53–e55. doi: 10.1016/S2666-5247(20)30027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovik Tinka. Using Connectivity Map Data and PharmacoGx R Package to Find Drugs Against Coronaviruses. 2020. https://github.com/tinkavidovik/competition
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Guan Y. COVID-19 drug repurposing: a review of computational screening methods, clinical trials, and protein interaction assays. Med. Res. Rev. 2020 doi: 10.1002/med.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S.M., Ehrlich L.S., Strickland M., Li X., Soloveva V., Goff A.J. Selective targeting of virus replication by proton pump inhibitors. Sci. Rep. 2020;10(1):1–15. doi: 10.1038/s41598-020-60544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójtowicz H., Kloc K., Maliszewska I., Młochowski J., Piętka M., Piasecki E. Azaanalogues of ebselen as antimicrobial and antiviral agents: synthesis and properties. Il Farmaco. 2004;59(11):863–868. doi: 10.1016/j.farmac.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Wu C., Chen X., Cai Y., Zhou X., Xu S., Huang H. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyler E., Mösbauer K., Franke V., Diag A., Gottula L.T., Arsie R. Bulk and single-cell gene expression profiling of SARS-CoV-2 infected human cell lines identifies molecular targets for therapeutic intervention. bioRxiv. 2020 [Google Scholar]
- Xiao S.-j., Zhu X.-c., Deng H., Zhou W.-p., Yang W.-y., Yuan L.-k. Gene expression profiling coupled with Connectivity Map database mining reveals potential therapeutic drugs for Hirschsprung disease. J. Pediatr. Surg. 2018;53(9):1716–1721. doi: 10.1016/j.jpedsurg.2018.02.060. [DOI] [PubMed] [Google Scholar]
- Xie Y., Cao S., Li Q., Chen E., Dong H., Zhang W. Effect of regular intravenous immunoglobulin therapy on prognosis of severe pneumonia in patients with COVID-19. J. Infect. 2020;81(2):340–343. doi: 10.1016/j.jinf.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Han M., Li T., Sun W., Wang D., Fu B. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. 2020;117(20):10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Matsuyama S., Hoshino T., Yamamoto N. Nelfinavir inhibits replication of severe acute respiratory syndrome coronavirus 2 in vitro. bioRxiv. 2020 [Google Scholar]
- Yamaya M., Nishimura H., Deng X., Sugawara M., Watanabe O., Nomura K. Inhibitory effects of glycopyrronium, formoterol, and budesonide on coronavirus HCoV-229E replication and cytokine production by primary cultures of human nasal and tracheal epithelial cells. Respir. Investig. 2020;58(3):155–168. doi: 10.1016/j.resinv.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.S., Lim C. Multi-targeting of functional cysteines in multiple conserved SARS-CoV-2 domains by clinically safe Zn-ejectors. chemRxiv. 2020 doi: 10.1039/d0sc02646h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Wang B., Mao J. The pathogenesis and treatment of theCytokine Storm’in COVID-19. J. Infect. 2020;80(6):607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar J.H. 4th edn. NJ Prentice Hall; 1999. Biostatistical Analysis; p. 523. [Google Scholar]
- Zhang J.-M., An J. Cytokines, inflammation and pain. Int. Anesthesiol. Clin. 2007;45(2):27. doi: 10.1097/AIA.0b013e318034194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Kang W., Lu X., Ma S., Dong L., Zou B. Weighted gene co-expression network analysis and connectivity map identifies lovastatin as a treatment option of gastric cancer by inhibiting HDAC2. Gene. 2019;681:15–25. doi: 10.1016/j.gene.2018.09.040. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10(1):1–10. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]