Abstract
Rare cardiac genetic diseases have generally been considered to be broadly Mendelian in nature, with clinical genetic testing for these conditions predicated on the detection of a primary causative rare pathogenic variant that will enable cascade genetic screening in families. However, substantial variability in penetrance and disease severity among carriers of pathogenic variants, as well as the inability to detect rare Mendelian variants in considerable proportions of patients, indicates that more complex aetiologies are likely to underlie these diseases. Recent findings have suggested genetic variants across a range of population frequencies and effect sizes may combine, along with non-genetic factors, to determine whether the threshold for expression of disease is reached and the severity of the phenotype. The availability of increasingly large genetically characterized cohorts of patients with rare cardiac diseases is enabling the discovery of common genetic variation that may underlie both variable penetrance in Mendelian diseases and the genetic aetiology of apparently non-Mendelian rare cardiac conditions. It is likely that the genetic architecture of rare cardiac diseases will vary considerably between different conditions as well as between patients with similar phenotypes, ranging from near-Mendelian disease to models more akin to common, complex disease. Uncovering the broad range of genetic factors that predispose patients to rare cardiac diseases offers the promise of improved risk prediction and more focused clinical management in patients and their families.
Keywords: Genetics, Rare cardiac disease, Genetic modifiers, Inherited cardiomyopathies, Ventricular arrhythmias, Genome-wide association studies
Introduction
Heritable cardiovascular diseases have generally been divided into two broad categories. The first category encompasses rare Mendelian genetic diseases such as inherited cardiomyopathies (e.g. hypertrophic cardiomyopathy, HCM) and ventricular arrhythmia syndromes (e.g. long QT syndrome, LQTS) that are usually caused by a rare genetic variant (a so-called ‘mutation’) that has a large risk-increasing effect size. In the second category are common complex diseases such as hypertension and coronary artery disease where environmental factors act in conjunction with a large number of common genetic variants, each with a small risk-increasing effect size. Discoveries into the genetic basis of cardiac diseases, and their application in the clinic, initially focused on the rarer Mendelian conditions. The familial inheritance observed in these diseases and the large effect sizes of the causative rare variants enabled the identification of the major disease genes through linkage studies in large family pedigrees.
Subsequent discoveries demonstrated the genetic heterogeneity of these disorders, with several causative genes identified through linkage and candidate gene studies, though the replication rate for the latter (as described below) has been poor, unless significant rare variant association has been demonstrated by case–control analysis.1 Sequencing of these genes has been recommended for a number of inherited cardiac conditions for several years and has become a standard aspect of clinical management in affected families. The primary benefit of this testing is to identify at risk carriers of the familial pathogenic variant (and non-carriers who are unlikely to develop disease), assuming a penetrant variant is identified that can be predicted with confidence to cause the disease. Clinical genetic testing for these conditions facilitates focused clinical screening of those relatives at risk of developing disease, has been shown to be cost-effective2 and can be considered as a success story in the application of genetics into clinical practice.
Limitations of Mendelian genetic approaches
The clinical impact of genetic testing for inherited cardiac conditions has been hampered however by two key factors—the substantial proportion of cases where a causative Mendelian variant is still not identified and, where a pathogenic variant is detected, the limited ability to predict clinical outcomes with this information. Diagnostic yields range from 75% to 80% in LQTS to only 20% in Brugada syndrome (BrS).3 Years of efforts to expand the genetic repertoire of inherited cardiac conditions through candidate gene studies have proved largely fruitless, with contemporary re-evaluation of these studies refuting many of these gene to disease associations. In particular, population genetics databases such as ExAC and gnomAD have shown that some implicated variants have population frequencies incompatible with causing rare diseases, as well as demonstrating that rare variation is collectively common for many genes. For example, all but one gene implicated in BrS (SCN5A) have now been repudiated by the ClinGen initiative,4 with similar findings observed for HCM5 , 6 (ClinGen curation for other diseases is ongoing). Similarly, 6.5–13.5% of variants associated with cardiomyopathies are now shown to have population frequencies incompatible with being penetrant variants for such diseases.7
Instead, several factors now suggest that the majority of cases where pathogenic variants are not identified (‘genotype-negative’) are likely to represent non-Mendelian forms of disease. Most large affected family pedigrees have now been genetically resolved (at least in Europe and North America), with genotype-negative patients more likely to present as sporadic cases and with a much lower family history of disease. In addition, genotype-negative cases can display substantially different phenotypic and clinical characteristics compared to genotype-positive cases—in HCM, this includes distinct left ventricular (LV) morphology and more benign outcomes8—suggesting a different genetic aetiology may underlie their disease.
Broad correlations can be observed between pathogenic variant classes (such as all variants in a particular gene) and phenotype for several inherited cardiac conditions. For example, differential arrhythmic risk profiles and response to drug therapy are observed amongst variants in the three main LQTS genes.9 Yet, despite the identification of disease genes and clinical risk factors, predicting disease severity and major complications such as heart failure and sudden cardiac death remains challenging. Even within family pedigrees carrying the same disease-causing variant, incomplete penetrance (variant carriers who do not develop disease) and variable expressivity (a wide range of severity amongst carriers) are common phenomena. Non-genetic factors are known to influence disease risk amongst pathogenic variant carriers in several cardiac diseases, ranging from generic factors such as age and sex to disease-specific modulators (e.g. obesity in HCM10 and QT prolonging drugs in LQTS11). It is also increasingly recognized that additional genetic factors may act to modulate the phenotype in individuals with a primary pathogenic variant and underlie a substantial proportion of the variability in penetrance and disease severity.
Identification of genetic risk variants in rare cardiac disease
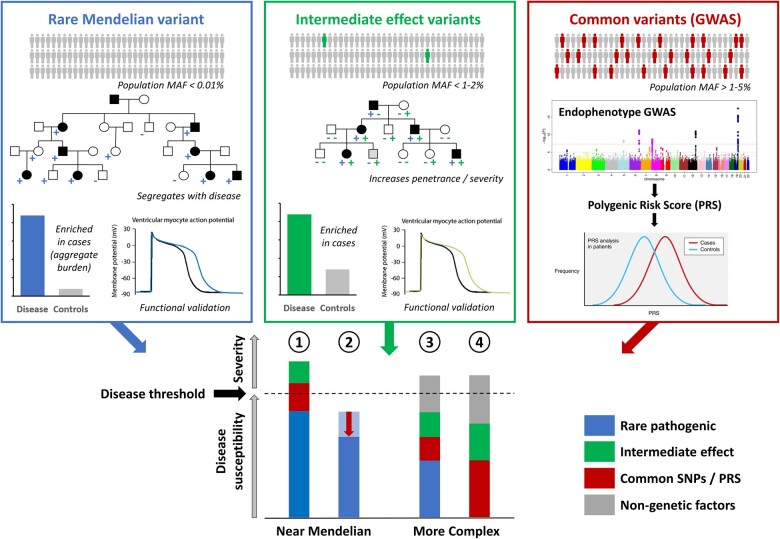
While all disease-associated genetic variants will lie on a spectrum of phenotype effect size and population frequency, it is useful to distinguish between Mendelian and non-Mendelian variants. The former are primary drivers of disease in affected family pedigrees and can be used for cascade genetic screening to identify at-risk individuals while the latter contribute to disease risk with smaller effect sizes and cannot be used in isolation to define risk. We can consider there to be broadly two types of non-Mendelian genetic risk variants that are detectable with current approaches and study sizes and could potentially contribute to disease risk in rare cardiac diseases. Common variants [usually defined as having a minor allele frequency (MAF) of >5%] identified through genome-wide association studies (GWAS) generally have individually small effect sizes but collectively have been shown to be associated with disease risk across a broad range of disease phenotypes. Intermediate effect variants (MAF < 1–5%) have effect sizes and frequencies between common and Mendelian variants. Preliminary research into genetic risk variants in cardiac disease (described in detail below) suggests that these factors may, to varying extents, influence penetrance in individuals with Mendelian genetic defects by pushing the genetic burden towards the threshold of disease12 (Figure 1). These variants are also likely to contribute to disease risk in individuals without primary Mendelian variants, with a larger burden of disease risk variants expected to be found in such genotype-negative individuals. With some exceptions, the contribution of such variants in determining disease severity remains largely unexplored.
Figure 1.
The increasingly complex aetiology of rare cardiac genetic diseases. Mendelian variants (blue) are ultra-rare in the population and have large effect sizes, though often not sufficient in isolation to yield a disease phenotype. Mendelian genes and variants can be identified through analysis of family pedigrees or burden analysis in case–control studies and further validated with functional assays. Common variants (red) with individually small effect sizes may collectively contribute to disease burden or modulate the effects of Mendelian variants. Intermediate effect variants (green) are emerging variant classes that usually have population frequencies and effect sizes between rare Mendelian and common variants and may act to increase severity and penetrance. Such variants can be identified by demonstrating enrichment in case cohorts and deleterious effects in established functional assays. These different variant classes can combine to reach the threshold of disease in patients with rare cardiac diseases and contribute to the variable expressivity/severity observed in patients (concept adapted from ref.12) Diseases such as HCM and LQTS are often near-Mendelian, where Mendelian variants of large effect sizes can combine with other variant classes to causes disease (1) or act as protective modifiers (e.g. regulatory variants affecting the expression ratio of the mutant vs. non-mutant alleles) (2). In contrast, diseases such as BrS and DCM may exhibit a more complex aetiology where substantial non-Mendelian genetic and non-genetic factors are required to reach disease threshold in the presence of a low penetrance rare variant (3) or in a non-Mendelian disease model.
Common variation and genome-wide association studies
Genome-wide association studies have had an enormous impact on elucidating the genetic basis of common complex diseases over the last 12 years, with thousands of robust associations identified and sample sizes in some studies now approaching 1 million.13 With increasing recognition of the genetic complexity of rare Mendelian disease, GWAS are now also starting to be applied to these conditions to identify variation that may underlie both variable penetrance and genotype-negative cases. Two approaches can be used for GWAS in rare genetic disease. The first employs a standard case–control study design including unrelated patients with the genetic disease and population-matched controls, which allows for direct detection of disease-associated variants as well as further stratified analyses based on factors such as pathogenic variant status and disease severity. Rare disease GWAS are limited however by the availability of disease samples, with multi-centre collaborations and meta-analysis required to achieve even moderately powered studies. To complement these efforts, quantitative studies on disease-relevant endophenotypes may be performed using population cohorts such as the UK Biobank. These are powered to detect a larger number of associations and can produce a polygenic risk score (PRS, a weighted aggregate of associated loci) underlying the endophenotype that can then be tested for association in patients with the rare disease.
The detection of common risk variants in inherited cardiac disease has initially focused on LQTS, given the directly applicable endophenotype (QT interval) readily available in large population studies. The latest QT-interval GWAS, conducted on 76 000 individuals of European descent, identified 35 loci with individually small effects but collectively explaining approximately 10% of QT interval variation in the general population.14 Studies with LQTS patients have shown that some of these variants can modulate QT interval and risk of arrhythmias and cardiac events—for variants at the NOS1AP locus,15 KCNQ1 locus,16 and with a PRS derived from 22 QT interval single polymorphisms (SNPs) in LQTS Type 2 patients (those with a KCNH2 pathogenic variant).17 These findings highlight the potential role for common variation in explaining phenotypic variability in LQTS patients, though clinical utility remains to be demonstrated.
In contrast, investigations into common variation in cardiomyopathies has so far been largely restricted to case–control studies with moderate sample sizes.18–20 For HCM and dilated cardiomyopathy (DCM) relevant endophenotypes related to LV dimensions and function are expensive to measure by cardiac magnetic resonance (CMR), difficult to accurately quantify by echocardiography and are less clearly correlated with disease phenotype. Such data are now becoming available through studies such as the EchoGen consortium21 and a CMR-scanned subset of the UK Biobank, with LV traits GWAS yielding association signals and PRS that may subsequently be assessed for roles in HCM/DCM disease susceptibility and severity.22 Indeed, two of the loci identified in the UK Biobank study were previously associated with DCM in a case–control GWAS19—BAG3 (associated with LV ejection fraction and end-systolic/diastolic volumes) and the CLCNKA/HSPB7 locus (associated with LV ejection fraction).22 Larger multicentre cardiomyopathy GWAS studies are needed to more fully elucidate the role of common variation in these conditions and assess the utility of these LV endophenotype PRS (Figure 2).
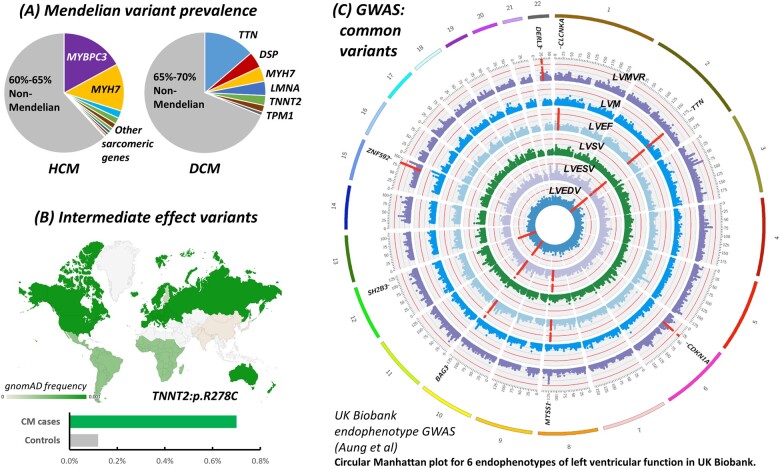
Figure 2.
Increasing genetic complexity of cardiomyopathies. (A) Mendelian variants are identified in less than half of HCM/DCM patients.7 (B) Additional genetic risk variants may include intermediate effect variants like TNNT2:p.R278C, enriched in European HCM cases31 , 32 and often occurring as a secondary sarcomeric variant (https://www.ncbi.nlm.nih.gov/clinvar/variation/12411/). (C) Common susceptibility variants may be identified using direct case–control GWAS. An alternative approach is to identify variants associated with relevant endophenotypes in population cohorts like UK Biobank22 and then test these for association with disease in patient datasets. The figure shows a Circular Manhattan plot (reproduced with permission from ref.22) highlighting significant loci associated with six traits of left ventricular function, relevant intermediate phenotypes for cardiomyopathies. Chromosomes are coloured in the outer band, with Manhattan plots for the six phenotypes in concentric circles. Significant loci are highlighted in red, with the closest gene indicated. Loci associated with multiple traits include those harbouring TTN and BAG3. Rare, truncating variants in both TTN and BAG3 are prominent Mendelian causes of DCM, while BAG3 was also significantly associated with DCM in a case-control GWAS.19
In 2013, a multicentre case–control GWAS in BrS (with 312 cases and 1115 controls) identified three independent association signals at the SCN5A, SCN10A, and HEY2 genes, confirming the central role for the sodium channel in this disorder.23 In aggregate, these three SNPs alone confer a high relative risk for disease (with an odds ratio >20 for carriers of >4 risk alleles compared to <2), accounting for 7% of the variance in disease susceptibility, though absolute risk is low given the rarity of the disease. The strong association signals and high cumulative effects on relative risk observed in this study, despite its limited sample size, highlight the distinctive genetic architecture of BrS. Rare variants in SCN5A are observed in only 20% of cases and, even in genotype-positive families, they have been shown to be neither necessary nor sufficient to cause disease (with reports of both incomplete penetrance in mutation carriers and non-segregation of putatively pathogenic variants, i.e. non-carriers with a BrS phenotype24). These findings point towards a highly polygenic nature of BrS, in contrast to other rare cardiac conditions. More recently, it has been shown that a BrS PRS could potentially be of clinical utility in the diagnostic strategy of BrS.25
As well as highlighting the increasing genetic complexity of diseases previously interpreted as principally Mendelian, GWAS and their derived PRS can also now be used to identify individuals in the general population with disease risks equivalent to those conferred by monogenic Mendelian variants. Khera et al. 26 found that 8% of the UK Biobank samples had a PRS that conferred ≥three-fold risk for coronary artery disease (similar to the risk for carriers of familial hypercholesterolaemia pathogenic variants), with similar risks observed for other common diseases (though at lower percentages). While familial inheritance will be different in such cases, this data could be used to identify individuals for intensive screening and/or preventive intervention. It also illustrates that genetic architecture and risk for many phenotypes should be viewed as a spectrum rather than a simple dichotomy of Mendelian and complex as previously assumed.
Intermediate effect variants
Intermediate effect genetic variants lie on a wide spectrum between common variants investigated by GWAS and rare pathogenic Mendelian variants, as measured by both variant frequency in the population and phenotypic effect size in patients. The identification and characterization of such variants is an emerging topic for inherited cardiac conditions and faces substantial challenges in identifying candidate variants (requiring sequencing of large cohorts given their lower frequency) and distinguishing them from both pathogenic Mendelian and benign variants. Once identified, new clinical genetics guidelines will be required for classifying such variants (methods are currently designed for classic Mendelian genetics) and defining actionability when they are detected in patients and their families.
Population genetic resources such as gnomAD have highlighted that many variants previously associated with disease are in fact too common in the population to be penetrant pathogenic variants in rare genetic diseases. While many such variants are likely to be benign and had only been incidentally detected in patients, some may have an effect on disease susceptibility or phenotypic severity. A high burden of proof should be required to define a variant as a potential risk variant, which could include a significant enrichment in case cohorts compared to ethnically matched controls, a demonstrated effect on phenotypic severity within cases carrying a pathogenic variant and well-characterized functional assays.
The classic example of a genetic risk variant in inherited cardiac disease is the p.Asp85Asn missense variant in the KCNE1 gene. With a gnomAD MAF in non-Finnish Europeans of 0.012, the variant is at the lower end of detection frequencies for GWAS and has been associated with prolonged QT interval in the general population, with the largest effect size of any of the associated SNPs (7.42 ms per minor allele).14 KCNE1:p.Asp85Asn was also reported to modulate the QT interval by 26 ms in male LQTS patients with the Finnish founder mutation KCNQ1:p.Gly589Asp.27 It is enriched in LQTS patients from Japan28 and USA29 where, interestingly, the majority of carriers were genotype-negative for rare pathogenic LQTS variants, suggesting p.Asp85Asn may also contribute substantially to the genetic burden in non-Mendelian LQTS cases. Functional studies have confirmed the deleterious role of this variant by demonstrating significant reductions on repolarizing potassium channel-encoded currents.28
As intermediate effect variants are often population-specific, expanding genetic sequencing to non-European populations will be as important as increasing GWAS sample diversity. With a fixed number of variants in the major cardiac genes that are candidate intermediate effect variants, systematic approaches to evaluate their function and effects sizes, using functional assays or population biobanks, may be feasible in the near future.
Prospects for clinical genetics and precision medicine
It is increasingly evident that the genetic architecture of rare cardiac disease is more complex than accounted for by simple Mendelian models. A range of genetic variants of different frequencies and effect sizes may combine to produce an overall genetic burden that, in conjunction with non-genetics factors, may determine whether the threshold of disease is reached in each individual and the severity of disease in patients (Figure 1). While we are still very much in the early stages of discovering these modifying genetic factors and understanding how they affect disease risk, these findings have a number of implications for how we understand the aetiology of these diseases and how they will be applied in clinical practice.
The development of PRS from large GWAS to inform clinical risk prediction is currently an active area of research for common diseases such as coronary artery disease and atrial fibrillation. PRS that identify individuals with high risk of developing disease have the potential for improved prediction over and above current risk factors, but their clinical utility (particularly in non-European populations) remains to be established. For rarer genetic diseases, the integration of the different classes of variants contributing to disease, particularly accounting for the variable effects of primary pathogenic variants, will make clinical adoption even more challenging. Classification guidelines will need to be developed for intermediate effect variants that effectively assess both the likelihood of contribution to the disease phenotype and the estimated effect size.30 Finally, communicating this complex genetic profile to both clinicians and patients, who are accustomed to receiving deterministic findings from genetic testing, will present additional challenges.
However, more comprehensive assessment of genetic risk for rare cardiac diseases offers enormous potential for improved risk prediction and clinical management in patients and their families. Current Mendelian-based genetics is effective at broadly identifying at-risk individuals but is a blunt tool for individualized risk prediction. For unaffected carriers of cardiomyopathy pathogenic variants, near life-long periodic clinical screening is performed given the age-dependent penetrance. By more effectively stratifying pathogenic variant carriers according to their broader genetic risk profile, we will be able to identify those at most risk of developing disease and severe cardiac events, facilitating increased monitoring and interventional therapy where appropriate. At the same time, unaffected family members of cardiomyopathy patients, who currently undergo regular clinical screening to detect onset of disease (at considerable cost in healthcare provision), could be discharged if their overall risk was determined to be low.
These new insights into the genetics of rare cardiac diseases also compel us to re-evaluate the aetiology of genotype-negative disease and the clinical management of such cases. The majority of such cases, especially in families with no prior history of disease, are likely to be caused by a range of small to intermediate effect variants and non-genetic factors. Consequently, the risk to family members is likely substantially lower than in pedigrees with penetrant Mendelian variants, such that guidelines for cardiomyopathies might soon consider it feasible to release currently phenotype-negative relatives from ongoing clinical screening. The further development of disease-specific genome-wide risk scores could aid in this decision making by quantifying the risk to relatives through inexpensive genotyping assays. Questions also arise as to how these conditions are defined—are these cases in fact the extreme end of population polygenic risk whose phenotypes converge with Mendelian diseases? Finally, it has become evident that the language and dichotomous classifications that are pervasive in genetic disease (Mendelian/complex disease, pathogenic/benign variants) are increasingly inadequate for describing the genetic basis and risk of disease. Developments in understanding genetic cardiac disease over the next few years are likely to reveal ever increasing complexity but yield improvements in risk prediction for patients and their families.
Funding
R.W. received support from a post-doctoral grant from the Amsterdam Cardiovascular Sciences. R.T. received support from the Canadian Heart Rhythm Society’s George Mines Award, the European Society of Cardiology research award, and the Philippa and Marvin Carsley Cardiology Chair and is currently a clinical research scholar of the Fonds de Recherche du Québec-Santé. C.R.B. acknowledges the support from the Dutch Heart Foundation (CVON Predict 2 project, CVON Concor-genes project), the Netherlands Organization for Scientific Research (VICI fellowship, 016.150.610), and Fondation Leducq.
Conflict of interest: none declared.
Contributor Information
Roddy Walsh, Department of Clinical and Experimental Cardiology, Heart Centre, Amsterdam Cardiovascular Sciences, Amsterdam UMC, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, Netherlands.
Rafik Tadros, Department of Medicine, Cardiovascular Genetics Center, Montreal Heart Institute and Faculty of Medicine, Université de Montréal, 5000 Belanger, Montreal, QC H1T 1C8, Canada.
Connie R Bezzina, Department of Clinical and Experimental Cardiology, Heart Centre, Amsterdam Cardiovascular Sciences, Amsterdam UMC, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, Netherlands.
References
- 1. Herman DS, Lam L, Taylor MRG, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Di Lenarda A, Sinagra G, Bos JM, Ackerman MJ, Mitchell RN, Murry CE, Lakdawala NK, Ho CY, Barton PJR, Cook SA, Mestroni L, Seidman JG, Seidman CE. Truncations of titin causing dilated cardiomyopathy. N Engl J Med 2012;366:619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wordsworth S, Leal J, Blair E, Legood R, Thomson K, Seller A, Taylor J, Watkins H. DNA testing for hypertrophic cardiomyopathy: a cost-effectiveness model. Eur Heart J 2010;31:926–935. [DOI] [PubMed] [Google Scholar]
- 3. Bezzina CR, Lahrouchi N, Priori SG. Genetics of sudden cardiac death. Circ Res 2015;116:1919–1936. [DOI] [PubMed] [Google Scholar]
- 4. Hosseini SM, Kim R, Udupa S, Costain G, Jobling R, Liston E, Jamal SM, Szybowska M, Morel CF, Bowdin S, Garcia J, Care M, Sturm AC, Novelli V, Ackerman MJ, Ware JS, Hershberger RE, Wilde AAM, Gollob MH; National Institutes of Health Clinical Genome Resource Consortium. Reappraisal of reported genes for sudden arrhythmic death. Circulation 2018;138:1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walsh R, Buchan R, Wilk A, John S, Felkin LE, Thomson KL, Chiaw TH, Loong CCW, Pua CJ, Raphael C, Prasad S, Barton PJ, Funke B, Watkins H, Ware JS, Cook SA. Defining the genetic architecture of hypertrophic cardiomyopathy: re-evaluating the role of non-sarcomeric genes. Eur Heart J 2017;38:3461–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ingles J, Goldstein J, Thaxton C, Caleshu C, Corty EW, Crowley SB, Dougherty K, Harrison SM, McGlaughon J, Milko LV, Morales A, Seifert BA, Strande N, Thomson K, Peter van Tintelen J, Wallace K, Walsh R, Wells Q, Whiffin N, Witkowski L, Semsarian C, Ware JS, Hershberger RE, Funke B. Evaluating the clinical validity of hypertrophic cardiomyopathy genes. Circ Genomic Precis Med 2019;12:e002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Mazzarotto F, Blair E, Seller A, Taylor JC, Minikel EVExome Aggregation ConsortiumMacArthur DG, Farrall M, Cook SA, Watkins H. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med 2017;19:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ho CY, Day SM, Ashley EA, Michels M, Pereira AC, Jacoby D, Cirino AL, Fox JC, Lakdawala NK, Ware JS, Caleshu CA, Helms AS, Colan SD, Girolami F, Cecchi F, Seidman CE, Sajeev G, Signorovitch J, Green EM, Olivotto I. Genotype and lifetime burden of disease in hypertrophic cardiomyopathy: insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe). Circulation 2018;138:1387–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zareba W, Moss AJ, Locati EH, Lehmann MH, Peterson DR, Hall WJ, Schwartz PJ, Vincent GM, Priori SG, Benhorin J, Towbin JA, Robinson JL, Andrews ML, Napolitano C, Timothy K, Zhang L, Medina A; International Long QT Syndrome Registry. Modulating effects of age and gender on the clinical course of long QT syndrome by genotype. J Am Coll Cardiol 2003;42:103–109. [DOI] [PubMed] [Google Scholar]
- 10. Olivotto I, Maron BJ, Tomberli B, Appelbaum E, Salton C, Haas TS, Gibson CM, Nistri S, Servettini E, Chan RH, Udelson JE, Lesser JR, Cecchi F, Manning WJ, Maron MS. Obesity and its association to phenotype and clinical course in hypertrophic cardiomyopathy. J Am Coll Cardiol 2013;62:449–457. [DOI] [PubMed] [Google Scholar]
- 11. Weeke PE, Kellemann JS, Jespersen CB, Theilade J, Kanters JK, Hansen MS, Christiansen M, Marstrand P, Gislason GH, Torp-Pedersen C, Bundgaard H, Jensen HK, Tfelt-Hansen J. Long-term proarrhythmic pharmacotherapy among patients with congenital long QT syndrome and risk of arrhythmia and mortality. Eur Heart J 2019;40:3110–3117. [DOI] [PubMed] [Google Scholar]
- 12. Schwartz PJ, Crotti L, George AL. Modifier genes for sudden cardiac death. Eur Heart J 2018;39:3925–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet 2017;101:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arking DE, Pulit SL, Crotti L, van der Harst P, Munroe PB, Koopmann TT, Sotoodehnia N, Rossin EJ, Morley M, Wang X, Johnson AD, Lundby A, Gudbjartsson DF, Noseworthy PA, Eijgelsheim M, Bradford Y, Tarasov KV, Dörr M, Müller-Nurasyid M, Lahtinen AM, Nolte IM, Smith AV, Bis JC, Isaacs A, Newhouse SJ, Evans DS, Post WS, Waggott D, Lyytikäinen L-P, Hicks AA, Eisele L, Ellinghaus D, Hayward C, Navarro P, Ulivi S, Tanaka T, Tester DJ, Chatel S, Gustafsson S, Kumari M, Morris RW, Naluai ÅT, Padmanabhan S, Kluttig A, Strohmer B, Panayiotou AG, Torres M, Knoflach M, Hubacek JA, Slowikowski K, Raychaudhuri S, Kumar RD, Harris TB, Launer LJ, Shuldiner AR, Alonso A, Bader JS, Ehret G, Huang H, Kao WHL, Strait JB, Macfarlane PW, Brown M, Caulfield MJ, Samani NJ, Kronenberg F, Willeit JCARe Consortium; COGENT ConsortiumSmith JG, Greiser KH, Meyer zu Schwabedissen H, Werdan K, Carella M, Zelante L, Heckbert SR, Psaty BM, Rotter JI, Kolcic I, Polašek O, Wright AF, Griffin M, Daly MJ, Arnar DO, Hólm H, Thorsteinsdottir U, Denny JC, Roden DM, Zuvich RL, Emilsson V, Plump AS, Larson MG, O'Donnell CJ, Yin X, Bobbo M, D'Adamo AP, Iorio A, Sinagra G, Carracedo A, Cummings SR, Nalls MA, Jula A, Kontula KK, Marjamaa A, Oikarinen L, Perola M, Porthan K, Erbel R, Hoffmann P, Jöckel K-H, Kälsch H, Nöthen MM, den Hoed M, Loos RJF, Thelle DS, Gieger C, Meitinger T, Perz S, Peters A, Prucha H, Sinner MF, Waldenberger M, de Boer RA, Franke L, van der Vleuten PA, Beckmann BM, Martens E, Bardai A, Hofman N, Wilde AAM, Behr ER, Dalageorgou C, Giudicessi JR, Medeiros-Domingo A, Barc J, Kyndt F, Probst V, Ghidoni A, Insolia R, Hamilton RM, Scherer SW, Brandimarto J, Margulies K, Moravec CE, Greco M F. D, Fuchsberger C, O'Connell JR, Lee WK, Watt GCM, Campbell H, Wild SH, El Mokhtari NE, Frey N, Asselbergs FW, Leach IM, Navis G, van den Berg MP, van Veldhuisen DJ, Kellis M, Krijthe BP, Franco OH, Hofman A, Kors JA, Uitterlinden AG, Witteman JCM, Kedenko L, Lamina C, Oostra BA, Abecasis GR, Lakatta EG, Mulas A, Orrú M, Schlessinger D, Uda M, Markus MRP, Völker U, Snieder H, Spector TD, Ärnlöv J, Lind L, Sundström J, Syvänen A-C, Kivimaki M, Kähönen M, Mononen N, Raitakari OT, Viikari JS, Adamkova V, Kiechl S, Brion M, Nicolaides AN, Paulweber B, Haerting J, Dominiczak AF, Nyberg F, Whincup PH, Hingorani AD, Schott J-J, Bezzina CR, Ingelsson E, Ferrucci L, Gasparini P, Wilson JF, Rudan I, Franke A, Mühleisen TW, Pramstaller PP, Lehtimäki TJ, Paterson AD, Parsa A, Liu Y, van Duijn CM, Siscovick DS, Gudnason V, Jamshidi Y, Salomaa V, Felix SB, Sanna S, Ritchie MD, Stricker BH, Stefansson K, Boyer LA, Cappola TP, Olsen JV, Lage K, Schwartz PJ, Kääb S, Chakravarti A, Ackerman MJ, Pfeufer A, de Bakker PIW, Newton-Cheh C. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet 2014;46:826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crotti L, Monti MC, Insolia R, Peljto A, Goosen A, Brink PA, Greenberg DA, Schwartz PJ, George AL. NOS1AP is a genetic modifier of the long-QT syndrome. Circulation 2009;120:1657–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duchatelet S, Crotti L, Peat RA, Denjoy I, Itoh H, Berthet M, Ohno S, Fressart V, Monti MC, Crocamo C, Pedrazzini M, Dagradi F, Vicentini A, Klug D, Brink PA, Goosen A, Swan H, Toivonen L, Lahtinen AM, Kontula K, Shimizu W, Horie M, George AL, Trégouët D-A, Guicheney P, Schwartz PJ. Identification of a KCNQ1 polymorphism acting as a protective modifier against arrhythmic risk in long-QT syndrome. Circ Cardiovasc Genet 2013;6:354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kolder I, Tanck MWT, Postema PG, Barc J, Sinner MF, Zumhagen S, Husemann A, Stallmeyer B, Koopmann TT, Hofman N, Pfeufer A, Lichtner P, Meitinger T, Beckmann BM, Myerburg RJ, Bishopric NH, Roden DM, Kääb S, Wilde AAM, Schott J-J, Schulze-Bahr E, Bezzina CR. Analysis for genetic modifiers of disease severity in patients with long-QT syndrome type 2. Circ Cardiovasc Genet 2015;8:447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wooten EC, Hebl VB, Wolf MJ, Greytak SR, Orr NM, Draper I, Calvino JE, Kapur NK, Maron MS, Kullo IJ, Ommen SR, Bos JM, Ackerman MJ, Huggins GS. Formin homology 2 domain containing 3 variants associated with hypertrophic cardiomyopathy. Circ Cardiovasc Genet 2013;6:10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Villard E, Perret C, Gary F, Proust C, Dilanian G, Hengstenberg C, Ruppert V, Arbustini E, Wichter T, Germain M, Dubourg O, Tavazzi L, Aumont MC, DeGroote P, Fauchier L, Trochu JN, Gibelin P, Aupetit JF, Stark K, Erdmann J, Hetzer R, Roberts AM, Barton PJ, Regitz-Zagrosek VCardiogenics ConsortiumAslam U, Duboscq-Bidot L, Meyborg M, Maisch B, Madeira H, Waldenström A, Galve E, Cleland JG, Dorent R, Roizes G, Zeller T, Blankenberg S, Goodall AH, Cook S, Tregouet DA, Tiret L, Isnard R, Komajda M, Charron P, Cambien F. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J 2011;32:1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Esslinger U, Garnier S, Korniat A, Proust C, Kararigas G, Müller-Nurasyid M, Empana J-P, Morley MP, Perret C, Stark K, Bick AG, Prasad SK, Kriebel J, Li J, Tiret L, Strauch K, O'Regan DP, Marguiles KB, Seidman JG, Boutouyrie P, Lacolley P, Jouven X, Hengstenberg C, Komajda M, Hakonarson H, Isnard R, Arbustini E, Grallert H, Cook SA, Seidman CE, Regitz-Zagrosek V, Cappola TP, Charron P, Cambien F, Villard E. Exome-wide association study reveals novel susceptibility genes to sporadic dilated cardiomyopathy. PLoS One 2017;12:e0172995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wild PS, Felix JF, Schillert A, Teumer A, Chen M-H, Leening MJG, Völker U, Großmann V, Brody JA, Irvin MR, Shah SJ, Pramana S, Lieb W, Schmidt R, Stanton AV, Malzahn D, Smith AV, Sundström J, Minelli C, Ruggiero D, Lyytikäinen L-P, Tiller D, Smith JG, Monnereau C, Di Tullio MR, Musani SK, Morrison AC, Pers TH, Morley M, Kleber ME, Aragam J, Benjamin EJ, Bis JC, Bisping E, Broeckel U, Cheng S, Deckers JW, Del Greco M F, Edelmann F, Fornage M, Franke L, Friedrich N, Harris TB, Hofer E, Hofman A, Huang J, Hughes AD, Kähönen M, Investigators K, Kruppa J, Lackner KJ, Lannfelt L, Laskowski R, Launer LJ, Leosdottir M, Lin H, Lindgren CM, Loley C, MacRae CA, Mascalzoni D, Mayet J, Medenwald D, Morris AP, Müller C, Müller-Nurasyid M, Nappo S, Nilsson PM, Nuding S, Nutile T, Peters A, Pfeufer A, Pietzner D, Pramstaller PP, Raitakari OT, Rice KM, Rivadeneira F, Rotter JI, Ruohonen ST, Sacco RL, Samdarshi TE, Schmidt H, Sharp ASP, Shields DC, Sorice R, Sotoodehnia N, Stricker BH, Surendran P, Thom S, Töglhofer AM, Uitterlinden AG, Wachter R, Völzke H, Ziegler A, Münzel T, März W, Cappola TP, Hirschhorn JN, Mitchell GF, Smith NL, Fox ER, Dueker ND, Jaddoe VWV, Melander O, Russ M, Lehtimäki T, Ciullo M, Hicks AA, Lind L, Gudnason V, Pieske B, Barron AJ, Zweiker R, Schunkert H, Ingelsson E, Liu K, Arnett DK, Psaty BM, Blankenberg S, Larson MG, Felix SB, Franco OH, Zeller T, Vasan RS, Dörr M. Large-scale genome-wide analysis identifies genetic variants associated with cardiac structure and function. J Clin Invest 2017;127:1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aung N, Vargas JD, Yang C, Cabrera CP, Warren HR, Fung K, Tzanis E, Barnes MR, Rotter JI, Taylor KD, Manichaikul AW, Lima JAC, Bluemke DA, Piechnik SK, Neubauer S, Munroe PB, Petersen SE. Genome-wide analysis of left ventricular image-derived phenotypes identifies fourteen loci associated with cardiac morphogenesis and heart failure development. Circulation 2019;140:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bezzina CR, Barc J, Mizusawa Y, Remme CA, Gourraud JB, Simonet F, Verkerk AO, Schwartz PJ, Crotti L, Dagradi F, Guicheney P, Fressart V, Leenhardt A, Antzelevitch C, Bartkowiak S, Borggrefe M, Schimpf R, Schulze-Bahr E, Zumhagen S, Behr ER, Bastiaenen R, Tfelt-Hansen J, Olesen MS, Kääb S, Beckmann BM, Weeke P, Watanabe H, Endo N, Minamino T, Horie M, Ohno S, Hasegawa K, Makita N, Nogami A, Shimizu W, Aiba T, Froguel P, Balkau B, Lantieri O, Torchio M, Wiese C, Weber D, Wolswinkel R, Coronel R, Boukens BJ, Bézieau S, Charpentier E, Chatel S, Despres A, Gros F, Kyndt F, Lecointe S, Lindenbaum P, Portero V, Violleau J, Gessler M, Tan HL, Roden DM, Christoffels VM, Le Marec H, Wilde AA, Probst V, Schott JJ, Dina C, Redon R. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet 2013;45:1044–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Probst V, Wilde AAM, Barc J, Sacher F, Babuty D, Mabo P, Mansourati J, Le Scouarnec S, Kyndt F, Le Caignec C, Guicheney P, Gouas L, Albuisson J, Meregalli PG, Le Marec H, Tan HL, Schott J-J. SCN5A mutations and the role of genetic background in the pathophysiology of Brugada syndrome. Circ Cardiovasc Genet 2009;2:552–557. [DOI] [PubMed] [Google Scholar]
- 25. Tadros R, Tan HLESCAPE-NET InvestigatorsEl Mathari S, Kors JA, Postema PG, Lahrouchi N, Beekman L, Radivojkov-Blagojevic M, Amin AS, Meitinger T, Tanck MW, Wilde AA, Bezzina CR. Predicting cardiac electrical response to sodium-channel blockade and Brugada syndrome using polygenic risk scores. Eur Heart J 2019;40:3097–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018;50:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lahtinen AM, Marjamaa A, Swan H, Kontula K. KCNE1 D85N polymorphism–a sex-specific modifier in type 1 long QT syndrome? BMC Med Genet 2011;12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishio Y, Makiyama T, Itoh H, Sakaguchi T, Ohno S, Gong Y-Z, Yamamoto S, Ozawa T, Ding W-G, Toyoda F, Kawamura M, Akao M, Matsuura H, Kimura T, Kita T, Horie M. D85N, a KCNE1 polymorphism, is a disease-causing gene variant in long QT syndrome. J Am Coll Cardiol 2009;54:812–819. [DOI] [PubMed] [Google Scholar]
- 29. Lane CM, Giudicessi JR, Ye D, Tester DJ, Rohatgi RK, Bos JM, Ackerman MJ. Long QT syndrome type 5-Lite: defining the clinical phenotype associated with the potentially proarrhythmic p. Asp85Asn-KCNE1 common genetic variant. Hear Rhythm 2018;15:1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Senol-Cosar O, Schmidt RJ, Qian E, Hoskinson D, Mason-Suares H, Funke B, Lebo MS. Considerations for clinical curation, classification, and reporting of low-penetrance and low effect size variants associated with disease risk. Genet Med 2019;21:2765–2773. [DOI] [PubMed] [Google Scholar]
- 31. Alfares AA, Kelly MA, McDermott G, Funke BH, Lebo MS, Baxter SB, Shen J, McLaughlin HM, Clark EH, Babb LJ, Cox SW, DePalma SR, Ho CY, Seidman JG, Seidman CE, Rehm HL. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med 2015;17:880–888. [DOI] [PubMed] [Google Scholar]
- 32. Mazzarotto F, Girolami F, Boschi B, Barlocco F, Tomberli A, Baldini K, Coppini R, Tanini I, Bardi S, Contini E, Cecchi F, Pelo E, Cook SA, Cerbai E, Poggesi C, Torricelli F, Walsh R, Olivotto I. Defining the diagnostic effectiveness of genes for inclusion in panels: the experience of two decades of genetic testing for hypertrophic cardiomyopathy at a single center. Genet Med 2019;21:284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]