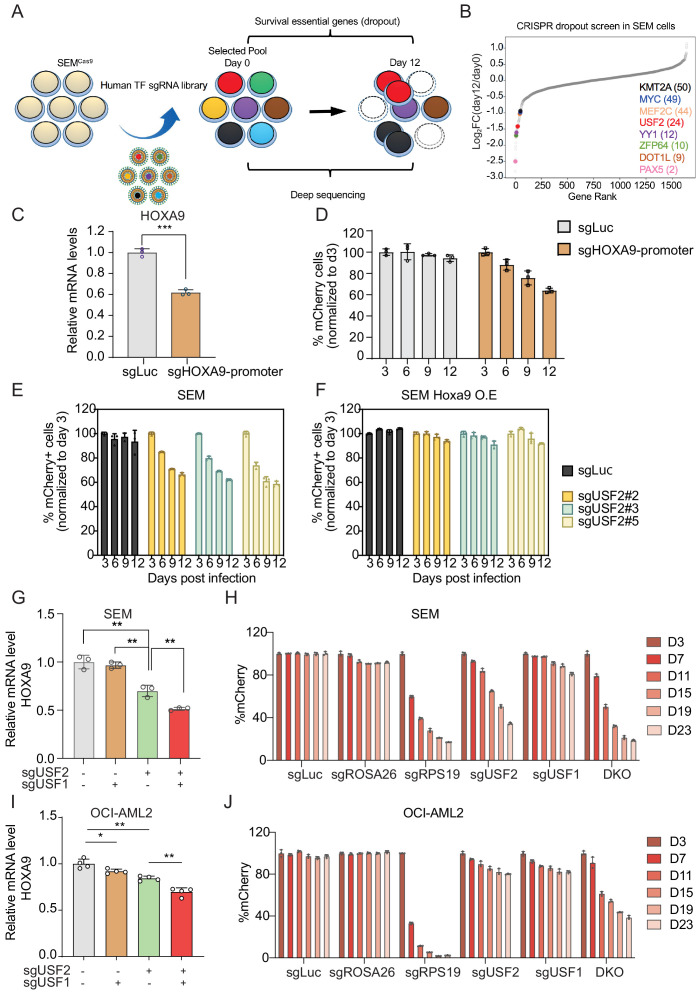
Figure 5. USF1 and USF2 synergistically regulate HOXA9 expression in MLLr leukemia.
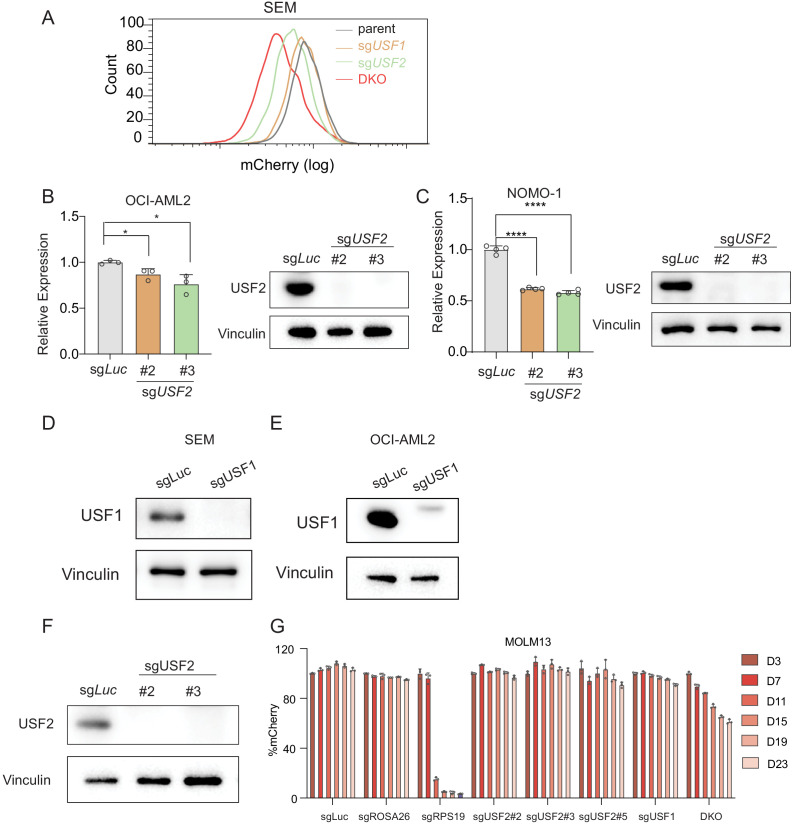
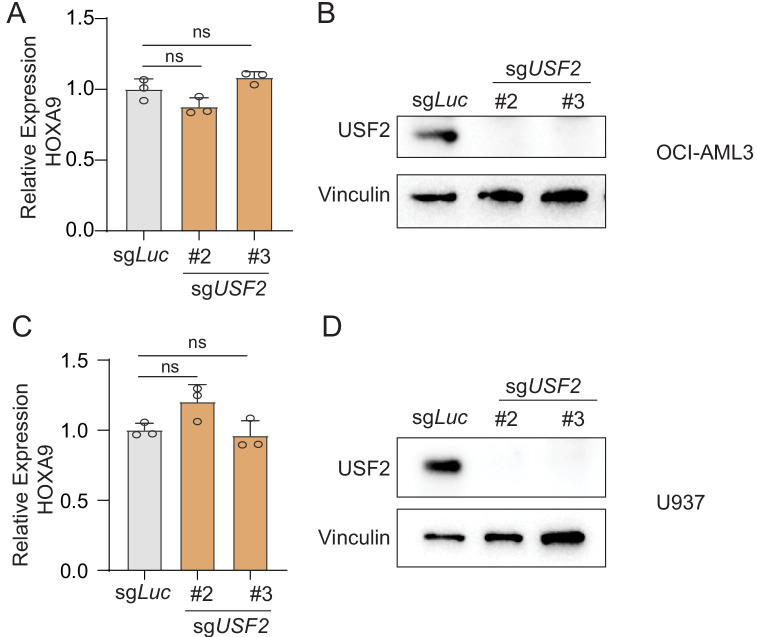
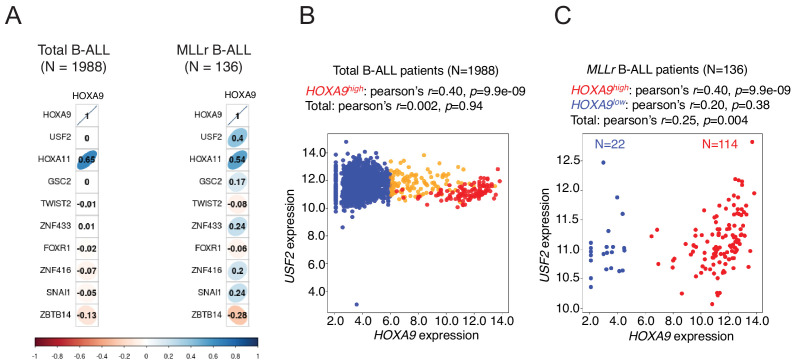
(A) Flow diagram of dropout CRISPR screening procedure. (B) Gene ranking of all transcription factors from dropout screening was illustrated. The enrichment score of seven sgRNAs against each transcription factor was combined by the MAGeCK algorithm. (C) Q-PCR was conducted to monitor HOXA9 expression upon CRISPR targeting on its promoter. (D) Competitive proliferation assay was conducted by infecting SEMCas9 cells with Lentiviral-mCherry-sgRNAs against HOXA9 promoter at about 50% efficiency. The mCherry% was quantified every three days by flow cytometry to evaluate the growth disadvantage. (E) Competitive proliferation assay was conducted by infecting SEMCas9 cells with Lentiviral-mCherry-sgRNAs against luciferase (sgLuc) and USF2 (sgUSF2#2, 2#3 and 2#5) at about 50% efficiency. The mCherry% was quantified every 3 days by flow cytometry to evaluate the growth disadvantage. (F) Rescued competitive proliferation assay was conducted by infecting SEM cells overexpressing ectopic Hoxa9 with Lentiviral-mCherry-sgRNAs against luciferase (sgLuc) and USF2 (sgUSF2#2, 2#3 and 2#5) at about 50% efficiency. The mCherry% was quantified every 3 days by flow cytometry to evaluate the growth disadvantage. (G) Q-PCR analysis was conducted on the sgUSF2, sgUSF1 and sgUSF1/2-targeted SEM cells to monitor the reduction of HOXA9. Data shown are means ± SEM from three independent experiments. **p<0.01, two-tailed Student’s t test. (H) Competitive proliferation assay was conducted by infecting SEMCas9 cells with Lentiviral-mCherry-sgLuc, sgUSF1, sgUSF2, and sgUSF1/2 (DKO) at about 50% efficiency. The mCherry% was quantified at days 3, 7, 11, 15, 19, and 23 by flow cytometry to evaluate the growth disadvantage. A guide RNA targeting the survival essential gene RPS19 was included as a positive control for Cas9 activity. Guide RNAs targeting Luciferase gene (sgLuc) and the human ROSA26 gene (sgROSA26) were included as a negative control. (I) Q-PCR analysis was conducted on the sgUSF2, sgUSF1 and sgUSF1/2 targeted OCI-AML2 cells to monitor the reduction of HOXA9. Data shown are means ± SEM from three independent experiments. *p<0.05, **p<0.01, two-tailed Student’s t test. (J) Competitive proliferation assay was conducted by infecting OCI-AML2Cas9 cells with Lentiviral-mCherry-sgLuc, sgUSF1, sgUSF2, and sgUSF1/2 (DKO) at about 50% efficiency. The mCherry% was quantified at days 3, 7, 11, 15, 19, and 23 by flow cytometry to evaluate the growth disadvantage. A guide RNA targeting the survival essential gene RPS19 was included as a positive control for Cas9 activity. Guide RNAs targeting Luciferase gene (sgLuc) and the human ROSA26 gene (sgROSA26) were included as negative controls.