Abstract
Toxoplasma gondii is a protozoan pathogen that widely affects the human population. The current antibiotics used for treating clinical toxoplasmosis are limited. In addition, they exhibit adverse side effects in certain groups of people. Therefore, discovery of novel therapeutics for clinical toxoplasmosis is imperative. The first step of novel antibiotic development is to identify chemical compounds showing high efficacy in inhibition of parasite growth using a high throughput screening strategy. As an obligate intracellular pathogen, Toxoplasma can only replicate within host cells, which prohibits the use of optical absorbance measurements as a quick indicator of growth. Presented here is a detailed protocol for a luciferase-based growth assay. As an example, this method is used to calculate the doubling time of wild-type Toxoplasma parasites and measure the efficacy of morpholinurea-leucyl-homophenyl-vinyl sulfone phenyl (LHVS, a cysteine protease-targeting compound) regarding inhibition of parasite intracellular growth. Also described, is a CRISPR-Cas9-based gene deletion protocol in Toxoplasma using 50 bp homologous regions for homology-dependent recombination (HDR). By quantifying the inhibition efficacies of LHVS in wild-type and TgCPL (Toxoplasma cathepsin L-like protease)-deficient parasites, it is shown that LHVS inhibits wild-type parasite growth more efficiently than Δcpl growth, suggesting that TgCPL is a target that LHVS binds to in Toxoplasma. The high sensitivity and easy operation of this luciferase-based growth assay make it suitable for monitoring Toxoplasma proliferation and evaluating drug efficacy in a high throughput manner.
Keywords: Immunology and Infection, Issue 158, Toxoplasma gondii, CRISPR-Cas9, gene manipulation, luciferase, growth assay, inhibitor efficacy, parasite
Introduction
Toxoplasma gondii is a highly successful obligate intracellular parasite that infects approximately one-third of the human population. Its high transmission rate is predominantly due to its diverse routes of transmission, including consumption of undercooked meat, exposure to mammalian reservoirs, and congenital transmission during birth. T. gondii mainly causes opportunistic infections that can lead to severe morbidity and mortality in immunocompromised individuals1,2,3,4,5,6. The antibiotics currently used for treating acute toxoplasmosis are particularly inefficient in treating congenital and latent infections and cause severe reactions in some individuals3,7,8. Thus, an urgent need to identify novel therapeutics exists. Understanding the differences in subcellular processes within Toxoplasma and its host will help to identify potential drug targets. Therefore, efficient and convenient genome manipulation techniques are required to study the roles of individual genes within Toxoplasma. Additionally, Toxoplasma belongs to the phylum Apicomplexa, which includes several other significant human pathogens, such as Plasmodium spp. and Cryptosporidium spp. Hence, Toxoplasma can be used as a model organism to help study basic biology in other apicomplexan parasites.
To identify novel antibiotics against microbial pathogens, high throughput screening of a library of chemical compounds is initially performed to determine their efficacy in the repression of microbial growth. So far, several microplate-based growth assays have been developed for measuring intracellular growth of T. gondii (i.e., radioactive 3H-uracil incorporation-based quantification9, quantitative ELISA-based parasite detection using T. gondii-specific antibodies10,11, reporter protein-based measurement using β-galactosidase or YFP-expressing Toxoplasma strains12,13, and a recently developed high-content imaging assay 14).
These individual strategies all have unique advantages; however, certain limitations also restrict their applications. For example, since Toxoplasma can only replicate within nucleated animal cells, autofluorescence and non-specific binding of anti-T. gondii antibodies to host cells cause interference in fluorescence-based measurements. Furthermore, usage of radioactive isotopes requires special safety compliance and potential safety issues. Some of these assays are more suitable for assessing growth at a single timepoint rather than continuous monitoring of growth.
Presented here is a luciferase-based protocol for the quantification of intracellular Toxoplasma growth. In a previous study, the NanoLuc luciferase gene was cloned under the Toxoplasma tubulin promoter, and this luciferase expression construct was transfected into wild-type (RHΔku80Δhxg strain) parasites to create an RHΔku80Δhxg∷NLuc strain (referred to as RHΔku80∷NLuc hereafter)15. This strain served as the parental strain for intracellular growth determination and gene deletion in this study. Using the RHΔku80∷NLuc strain, parasite growth in human foreskin fibroblasts (HFFs) was monitored over a 96 h period post-infection to calculate parasite doubling time.
In addition, the inhibition efficacy of LHVS against parasite growth can be determined by plotting Toxoplasma growth rates against serial LHVS concentrations to identify the IC50 value. Previous literature has reported that TgCPL is a major target of LHVS in parasites and that treatment with LHVS decreases the development of acute and chronic Toxoplasma infections16,17,18,19. Additionally, RHΔku80∷NLuc was used as the parental strain for genome modification to generate a TgCPL-deficient strain (RHΔku80Δcpl∷NLuc), and the inhibition of LHVS was measured against this mutant. By observing an upshift of IC50 values for LHVS in the TgCPL-deficient parasites compared to the WT strain, it was validated that TgCPL is targeted by LHVS in vivo.
In this protocol, RHΔku80∷NLuc is used as the parental strain, which lacks an efficient non-homologous end-joining pathway (NHEJ), thereby facilitating double crossover homology-dependent recombination (HDR)20,21. Additionally, 50 bp homologous regions are flanked at both ends of a drug resistance cassette by PCR. The PCR product serves as a repair template to remove the entire gene locus via HDR using CRISPR-Cas9-based genome editing tools. Such short homologous regions can be easily incorporated into primers, providing a convenient strategy for production of the repair template. This protocol can be modified to perform universal gene deletion and endogenous gene tagging.
For instance, in our most recent publication, three protease genes, TgCPL, TgCPB (Toxoplasma cathepsin B-like protease), and TgSUB1 (Toxoplasma subtilisin-like protease 1), were genetically ablated in TgCRT (Toxoplasma chloroquine-resistance transporter)-deficient parasites using this method15. Additionally, TgAMN (a putative aminopeptidase N [TgAMN, TGGT1_221310) was endogenously tagged15. The Lourido lab also reported using short homologous regions in the range of 40-43 bp for the introduction of site-directed gene mutation and endogenous gene tagging in the Toxoplasma genome using a similar method22. These successful genome modifications suggest that a 40-50 bp homologous region is sufficient for efficient DNA recombination in the TgKU80-deficient strain, which greatly simplifies genome manipulation in Toxoplasma gondii.
Protocol
Toxoplasma gondii is categorized in Risk Group 2 and must be handled at a Biosafety Level 2 (BSL-2). The protocol has been reviewed and approved by the Institutional Biosafety Committee at Clemson University.
1. Luciferase-based Toxoplasma growth assay
-
Seed human foreskin fibroblasts (HFFs) 1 week before parasite inoculation to ensure that host cells are fully confluent. Perform a mock assay in a transparent plate to ensure that parasites remain intracellular throughout the evaluation period.
NOTE: Here, the assay is conducted in 96 well microplates. According to experimental needs, it can be scaled up to 384 or 1536 well microplates.
Pass Toxoplasma parasites into confluent HFFs 2 days prior to use by transferring ~0.3-0.4 mL of fully lysed parasites into a T25 flask. Incubate infected host cells at 37 °C with 5% CO2 for 2 days.
Syringe 5 mL of freshly lysed parasites through a 21 G safety needle 5x to liberate intracellular parasites, then pass through a 3 μm filter to remove host cell debris. Rinse residual parasites out of the flask using 7 mL of phenol red-free D10 medium, then pass through the filter again.
Centrifuge parasites at 1000 x g for 10 min at room temperature (RT). Pour off the supernatant and resuspend the pellet in 10 mL of phenol red-free D10 media.
Count parasites using a hemocytometer to determine the concentration.
Dilute parasites to 1 x 104 parasites/mL for the wild-type (WT) strain. For growth-deficient parasite strains, increase the concentration accordingly to observe a significant increase in luciferase signals.
Aspirate media carefully from 96 well microplates pre-seeded with HFFs and inoculate 150 μL of parasite resuspension into wells in a format of three columns and five rows, which represents three technical replicates and five timepoints.
Incubate the microplate at 37 °C and 5% CO2 for 4 h.
Aspirate media carefully from the wells to remove non-invaded parasites, then fill the wells with RT phenol red-free media in each row (except for the first row).
Mix equal volumes of PBS and 2x luciferase assay buffer and dilute the luciferase substrate to 12.5 μM.
Add 100 μL of dilute luciferase substrate into each well of the top row. Incubate the microplates at RT for 10 min to allow the cells to fully lyse.
Measure the luciferase activity using a microplate reader. The plate reader settings are listed in Table 1. Each reading represents the initial number of invaded parasites at 4 h post-infection.
Repeat steps 1.9-1.12 for each row every 24 h for 4 days without changing the medium. These readings reflect the total number of replicated parasites at 24 h, 48 h, 72 h, and 96 h post-infection.
Calculate the average readings at each timepoint and divide them by the average readings at 4 h to determine the fold changes in parasite growth over time.
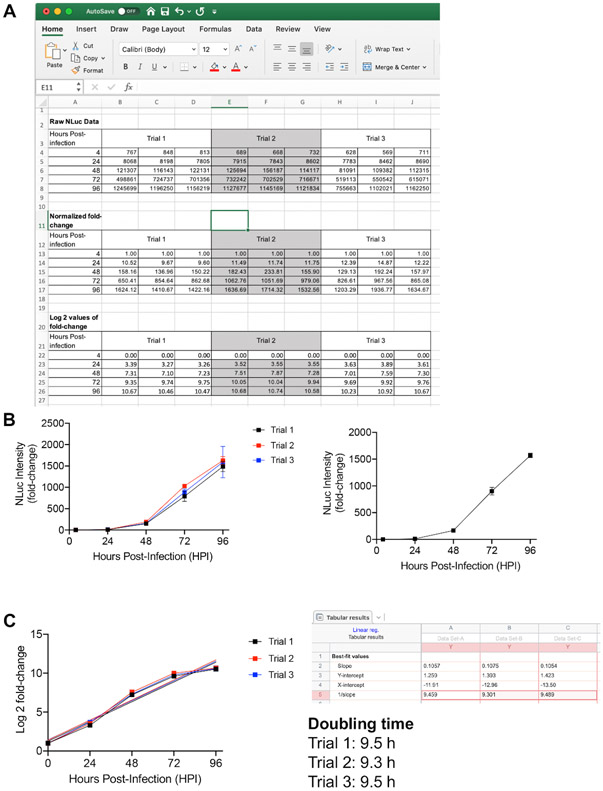
Plot the data using graphing software. A representative growth reading table and plots of RHΔku80∷NLuc parasites are shown in Figure 1A,B.
To calculate doubling time, plot the log2 values of fold changes at the individual timepoints over the incubation time. Use a linear regression function to calculate slope, which represents the doubling time of each strain (Figure 1A,C).
Table 1:
Microplate reader settings for luciferase activity measurement during luciferase-based Toxoplasma growth assay.
| Luciferase: | Endpoint |
| Integration time: | 1 s |
| Filter Set - Emission: | Full light |
| Optics: | Top |
| Gain: | 135 |
| Read speed: | Normal |
| Delay: | 100 ms |
| Read height: | 4.5 mm |
Figure 1: Intracellular growth quantification for Toxoplasma parasites using a luciferase-based method.
(A) Raw luciferase activity readings in a spreadsheet software. The readings at 24 h, 48 h, 72 h, and 96 h post-infection were normalized against the initial readings at 4 h post-infection for calculating the fold changes in parasite growth. (B) The normalized data were averaged and plotted. (C) The log2 values of the fold changes were also plotted and subjected to linear regression for determination of the parasite's doubling time.
2. Evaluation of chemical compound inhibition efficacy against Toxoplasma growth
NOTE: Here, evaluation of the inhibition of LHVS in Toxoplasma growth is presented as an example. Eight different concentrations of LHVS are tested, and three technical replicates are performed for each of the three biological replicates for both RHΔku80∷NLuc and RHΔku80Δcpl∷NLuc strains.
Prior to the parasite infection, seed HFFs to 96 well microplates in the format of three rows and nine columns for one biological replicate per compound per strain. Host cells will be allowed to grow for at least 7 days before use.
Pass RHΔku80∷NLuc and RHΔku80Δcpl∷NLuc parasites for 2 days prior to use. Follow steps 1.2-1.6 for parasite purification and quantification. Resuspend parasites in phenol red-free media at 1 x 104 parasites/mL.
Aspirate media from a plate containing confluent HFFs and inoculate each well with 150 μL of parasite resuspension. Incubate the microplate at 37 °C and 5% CO2 for 4 h.
-
Prepare LHVS at eight different concentrations in a 12 well reservoir by serial dilution. Generally, the concentrations are decreased by threefold in a serial dilution manner.
NOTE: The lowest concentration is reduced by 6,561-fold relative to the highest concentration. The fold change of the dilution can be adjusted accordingly based on different properties of individual compounds.
At 4 h post-infection, aspirate media to remove non-invaded parasites and fill each well from columns 2-9 with 150 μL of media supplemented with LHVS at different concentrations. Leave the first column filled with regular medium to serve as a nontreated control.
Incubate the microplate at 37 °C and 5% CO2 for an additional 96 h.
Perform steps 1.9-1.11 and measure luciferase activity of individual wells.
Average the luciferase activities of three technical replicates from wells of each individual LHVS concentration.
Divide the average luciferase activity for each LHVS concentration by the average luciferase activity derived from nontreated parasites to calculate the normalized luciferase activity as a percentage.
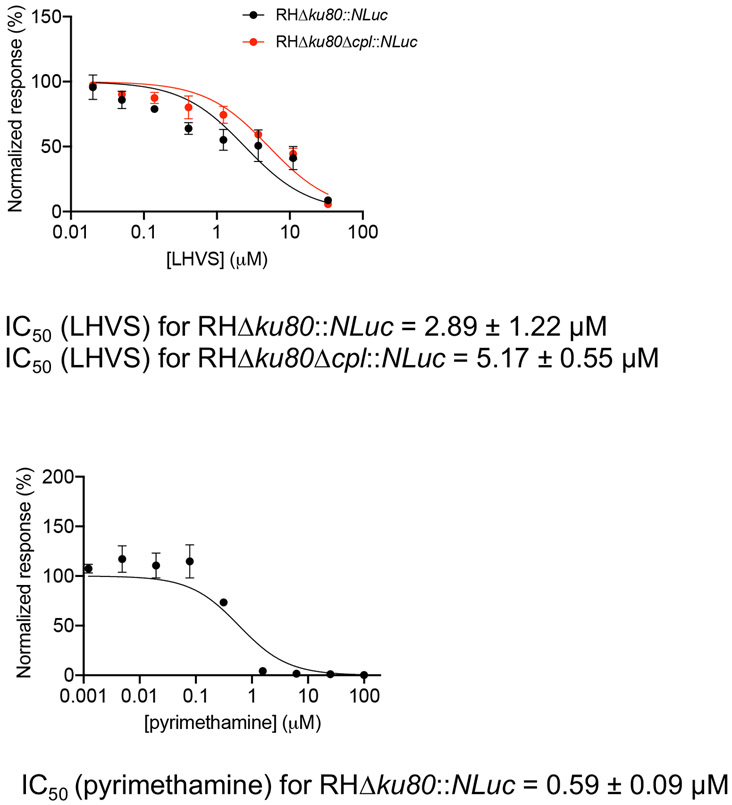
Plot the normalized luciferase activities against the individual LHVS concentrations using graphing software (Figure 2). Inhibition of pyrimethamine against parasite growth is also measured as a control. Pyrimethamine is a clinical antibiotic used to treat acute toxoplasmosis by inhibiting folic acid metabolism in Toxoplasma.
-
Calculate the IC50 values for individual compounds using the embedded method in the graphing software, normalized response vs. [inhibitor], under the "dose-response-inhibition" regression program. The IC50 is calculated using the following formula:
Where: Y represents normalized luciferase activities of infected cells under different concentrations of inhibitor, and X represents individual concentrations of inhibitor.
Figure 2: Inhibition efficacy assessment of LHVS and pyrimethamine using the luciferase-based growth assay.
Parasites were inoculated into a 96 well microplate for 4 h to allow for invasion of host cells. Non-invaded parasites were washed away, and the plate was filled with media containing different concentrations of LHVS or pyrimethamine and incubated for an additional 96 h before determination of luciferase activity. The measured luciferase readings for parasites treated with individual inhibitor concentrations were normalized against the signal detected from untreated parasites. The data were plotted in a graphing program, and a regression analysis for IC50 determination was performed. The assay was repeated in three biological replicates with three technical replicates each. Data represent mean ± SEM, n = 3 biological replicates.
3. CRISPR-Cas9-based gene deletion in Toxoplasma parasites
- Generation of a plasmid construct expressing guide RNA (sgRNA) and Cas9 for deleting a gene of interest
-
Go to www.ToxoDB.org and retrieve the entire gene coding sequence, including introns and exons, along with 1.5 kb 5'-UTRs and 3'-UTRs (untranslated regions).NOTE: Here, TgCPL (TGGT1_321530) is targeted as a representative example.
- Copy the retrieved TgCPL sequence into the sequence analysis software (refer to Table of Materials for the name and version) and label the 5'- and 3'-UTR regions.
- Select the Tools icon in the top menu bar, then select Cloning ∣ Find CRISPR Sites.
- Choose 3'(Cas9)' for the PAM site location and select the folder containing the Toxoplasma genome sequence in the specificity scoring section. Leave the rest of the settings as defaults.
- Choose a sgRNA with the following two criteria: 1) showing a high specificity score, generally >98%, and 2) lacking a G following the NGG, a protospacer adjacent motif (PAM) sequence. The selected sgRNA is usually located at sites close to the start and stop codons of the gene of interest.
-
Copy the sequence of the selected sgRNA and paste it into the following primer template.sgRNA.TgCPL.F: GTCGTCCTCGCCGTTGAGGAGTTTTAGAGCTAGAAATAGCsgRNA.R: AACTTGACATCCCCATTTACCAGAAGGCAAACACCThe portion in red represents the selected TgCPL sgRNA sequence. It can be replaced with different sgRNAs for various genes of interest.NOTE: If the selected sgRNA does not start with G, add G at the beginning of the sgRNA to help enhance its expression.
- Run the PCR product on an agarose gel to confirm successful amplification. A 10 kb PCR product is expected to be amplified (Figure 3B).
- Extract the PCR product using a DNA gel extraction kit and circularize it using a site-directed mutagenesis kit. Refer to Table 3 for the recipe. Incubate the reaction for 10-20 min at RT.
- Transform the circularized PCR product into E. coli and pick 10 clones for further verification of incorporation of designed sgRNA.
- Grow two clones and extract plasmids. Cut the purified plasmids with BamHI and EcoRV. The candidate plasmids will yield two bands at 2.4 kb and 7.2 kb (Figure 3C).
- Send the plasmids for Sanger sequencing using M13 reverse primers to confirm successful replacement of TgUPRT sgRNA with the designed sgRNA (Figure 3D).
-
- Generation of repair template for gene deletion via HDR mechanism
- According to the targeting sites of the selected sgRNA, locate 50 bp of 5'-UTRs or 3'-UTRs of the target gene for homology-dependent recombination (HDR, see discussion section). The selection of regions follows the criteria listed below, depending on the location the sgRNA targets.
- If the cleavage site by Cas9 is located upstream from the start codon, select the following: a 50 bp DNA sequence upstream from the cleavage site as the left HDR region, and a 50 bp DNA sequence downstream from the stop codon as the right HDR region.
- If the cleavage site by Cas9 is between the start and stop codons, select the following: a 50 bp DNA sequence upstream from the start codon as the left HDR region, and a 50 bp DNA sequence downstream from the stop codon as the right HDR region.
-
If the cleavage site by Cas9 is located downstream from the stop codon, select the following: a 50 bp DNA sequence upstream from the start codon as the left HDR region, and a 50 bp DNA sequence downstream from the cleavage site as the right HDR region.NOTE: For the TgCPL gene, the cleavage site is located between the start and stop codons. Thus, the following primers are designed for amplifying the repair template using pMDC64 as the template, which encodes a pyrimethamine resistance cassette. The sequences in black anneal to the pMDC64 plasmid for PCR amplification. The regions labeled in red are TgCPL-specific sequences for homologous recombination.TgCPLKO.DHFR.F:GAGTTTTGACCGTTTGTCCCTCTTACCCCGCGGCCTCGCTCTCGAACACCGTCCGCGTTCGTGAAATTCTCTgCPLKO.DHFR.R:CGTCTTCCTTCTCACTTCCTTCCACGAACTTCCCGAAAGCCACGGTCGCGCGCTCTAGAACTAGTGGATC
- Perform PCR using a PCR premix under the PCR conditions described in Table 4.
-
Run the PCR product on an agarose gel (Figure 3E), followed by gel extraction and standard nucleic acid quantification procedures.NOTE: If the expected band cannot be successfully amplified, optimize PCR conditions and/or switch primer pairs.
- Toxoplasma transfection
- Pass RHΔku80∷NLuc parasites for 2 days in a T25 flask containing confluent HFFs. A T25 flask of fully lysed parasites is sufficient for two to three transfections.
- Syringe and filter-purify parasites as described in step 1.2. Resuspend parasites in cytomix buffer and spin down at 1,000 x g for 10 min at RT.
- Wash pelleted parasites with 10 mL of cytomix buffer and spin down the parasites at 1,000 x g for 10 min at RT.
- Carefully pour off the supernatant and resuspend the parasites in the same buffer at a concentration of 1 x 108 parasites/mL.
- Mix 2 μg of repair template DNA with 20 μg of the sgRNA/Cas9 expression plasmids (mass ratio = 1:5, equivalent to a 1:3 molar ratio). If the amplification yield of repair template is low, reduce the input of both DNA pieces accordingly. A minimum of 0.5 μg of repair template can be used.
- Mix 400 μL of parasite resuspension, DNA, and 5 μL of 200 mM ATP/500 mM reduced glutathione (GSH) in a 1.5 mL centrifuge tube. Bring the total volume to 500 μL with cytomix buffer, if needed.
- Transfer the mixture of parasites and DNA to an electroporation cuvette (4 mm gap width) and perform electroporation (2 kV voltage, 50 Ω resistance) using an electroporation apparatus.
- Transfer electroporated parasites to a T25 flask containing confluent HFFs in fresh D10 medium. Apply appropriate antibiotic for drug selection after 24 h.
- Keep drug selective pressure until the growth of the transgenic parasites is stable.
- Purify genomic DNA from the knockout population and check for integration of the pyrimethamine resistance cassette into the TgCPL locus by PCR. After verified, proceed to section 3.4. If not, perform another round of parasite transfection and drug selection. Inability to detect the correct integration of the drug resistance cassette usually suggests that the target gene is essential or that the gene locus is not accessible.
- Cloning of knockout parasites
- Seed two 96 well microplates with HFF cells and incubate at 37 °C and 5% CO2 for 1 week prior to cloning parasites.
-
Pass ~0.3-0.4 mL of the population of transgenic parasites in a T25 flask containing confluent HFFs and grow them for 2 days. Consider passing more parasites if the mutant shows growth defects.NOTE: To achieve the best yield and viability, the host cells are heavily infected by the parasites, and most of the parasites are kept in the intracellular stage.
- Syringe infected host cells and filter-purify freshly lysed parasites as mentioned in step 1.3. Resuspend the parasites in D10 medium and spin them down at 1,000 x g for 10 min at RT.
- Resuspend the pelleted parasites in 10 mL of D10 medium.
- Count parasites using a hemocytometer to determine the parasite concentration.
- Conduct a two-step dilution to bring the concentration to 10 parasites/mL in D10 medium supplemented with the appropriate antibiotic. Usually, the initial parasite resuspension is diluted by 1,000-fold, followed by a second dilution to 10 parasites/mL.
- Aspirate media from 96 well microplates containing confluent HFFs and inoculate 150 μL of diluted parasites into each well.
- Incubate plates at 37 °C with 5% CO2 for 7 days without disturbance to allow plaque formation. The incubation period can be longer if transgenic parasites exhibit growth defects.
- Screen the plates using a phase-contrast microscope and mark only the wells containing a single plaque.
- Perform colony PCR to identify correct clones.
- Use pipette tips to scrape the bottom of each well to lift infected HFF monolayers.
- Pipet 75 μL of the cell resuspension from each marked well into 1.5 mL microcentrifuge tubes.
- Centrifuge tubes for 10 min at maximum speed at RT. Carefully aspirate the supernatant and resuspend the pellet in 10.25 μL of lysis buffer containing dilution buffer and DNA release additive provided in the kit (Table of Materials).
- Incubate the samples for 4 min at RT, then 2 min at 98 °C. Afterward, samples can be used for PCR or stored at −20 °C until use. Three sets of PCR reactions are used to test for the integration of the drug resistance cassette and loss of the gene of interest (Figure 4A). Refer to Table 5 for PCR reaction setup and Table 6 for thermocycler settings.
- Identify the correct clones and transfer four clones into T25 flasks containing confluent HFFs.
- After individual clones lyse host cells, purify genomic DNA for further PCR verification.
Materials
| Name | Company | Catalog Number | Comments |
|---|---|---|---|
| Agarose gel extraction kit | New England BioLabs | T1020L | |
| BamHI | New England BioLabs | R0316S | |
| Biotek Synergy H1 Hybrid Multi-Mode Microplate Reader | BioTek Instuments | ||
| BTX Gemini Twin Waveform Electroporation System | Harvard Apparatus | ||
| Chemically competent E. coli cells | New England BioLabs | C29871 | |
| CloneAmp HiFi PCR premix | Takara Bio | 639298 | |
| Coelenterazine h | Prolume | 301-10 hCTZ | |
| EcoRV | New England BioLabs | R3195S | |
| Phire Tissue Direct PCR Master Mix | Thermo Scientific | F170L | |
| Plasmid miniprep kit | Zymo Research | D4054 | |
| Q5 Site-Directed Mutagenesis kit | New England BioLabs | E0554S | |
| Software | |||
| Geneious software for sgRNA design (version: R11) | |||
| GraphPad Prism software (8th version) | |||
| SnapGene for molecular cloning (version: 4.2.11) |
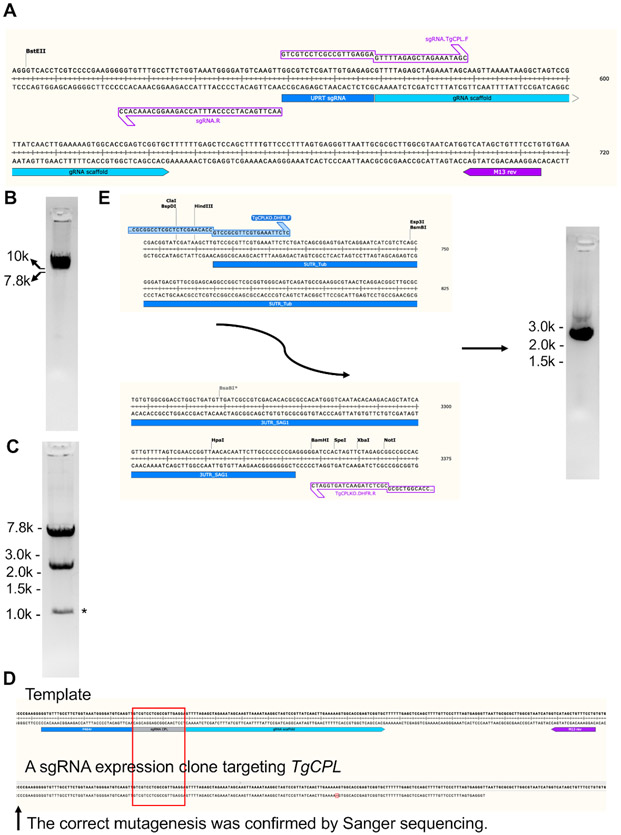
Figure 3: Generation of the plasmid construct expressing sgRNA targeting TgCPL and production of a repair template for TgCPL deletion.
(A) The original pSAG1-Cas9-sgRNA-UPRT plasmid23 was modified via a site-directed mutagenesis kit for replacement of the sgRNA targeting the TgUPRT gene to TgCPL. The sgRNA coding region is enlarged to show areas to which the primers anneal. After PCR, the mutated plasmid was linearized and loaded into a 1% agarose gel for verification of successful amplification, followed by gel extraction. (B) The gel image of the PCR-amplified linearized sgRNA expression construct. (C) After gel-extraction, the PCR product was circularized and subsequently transformed into E. coli. The clones containing the expected plasmids were screened by restriction endonuclease digestion and DNA sequencing. The band sizes after DNA digestion were 7.2 bp and 2.4 kb. The band generated by nonspecific cleavage from endonucleases is labeled by asterisk. (D) The M13 reverse primer labeled in the figure was used to sequence the mutated guide RNA region within the generated TgCPL-targeting sgRNA expression vector. The sequenced DNA region was aligned to the plasmid template to confirm successful mutagenesis. (E) In this study, 50 bp homologous regions matching to the 5'- and 3'-UTRs of TgCPL were engineered into the primers for amplification of the repair template and flanked at the 5'- and 3'-ends of the pyrimethamine resistance cassette by PCR, respectively. Agarose gel electrophoresis was used to verify the correct size of the PCR product before gel extraction. The expected size of the repair template is ~2.7 kb. Usually, 5-6 μg of repair template can be obtained from 200 μL of PCR reaction.
Table 2:
Thermocycler settings for generation of sgRNA expression vector.
| Initial denaturation: | 98 °C for 5 min |
| 25 cycles of | |
| Denaturing: | 98 °C for 5 s |
| Annealing: | 60 °C for 15 s |
| Extension: | 72 °C for 1 min |
| Final extension: | 72 °C for 10 min |
Table 3:
Reaction recipe for circularization of sgRNA expression vector.
| Sample | Volume (μl) |
|---|---|
| PCR product (10-50 ng) | 1 |
| 2X KLD (kinase, ligase, DpnI) Reaction Buffer | 5 |
| 10X KLD Enzyme Mix | 1 |
| Nuclease-free water | 3 |
| Total | 10 |
Table 4:
Thermocycler setting for generation of repair template.
| Initial denaturation: | 98 °C for 5 min |
| 35 cycles of | |
| Denaturing: | 98 °C for 15 s |
| Annealing: | 58 °C for 15 s |
| Extension: | 72 °C for 30 s per kb |
| Final extension: | 72 °C for 10 min |
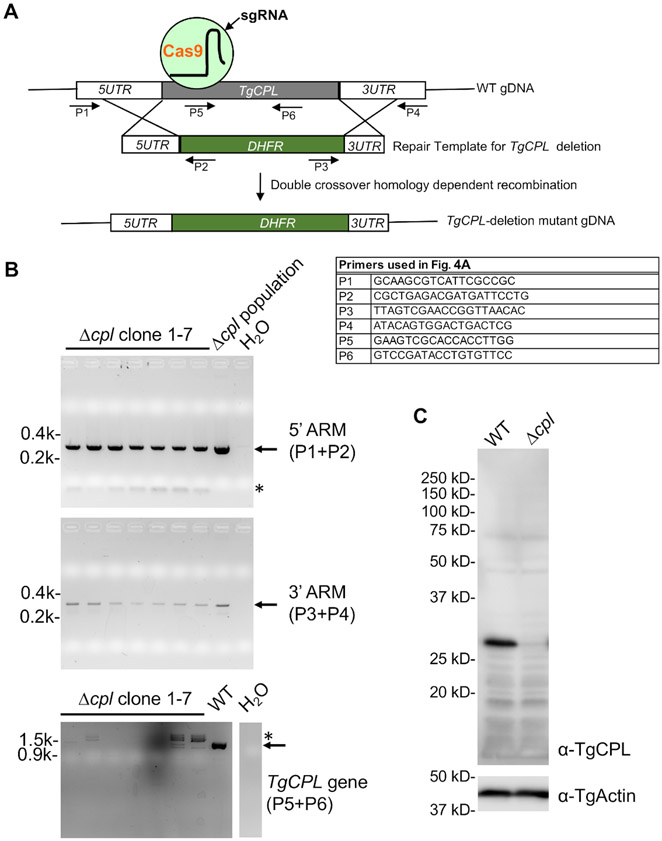
Figure 4: PCR and immunoblotting confirmation of TgCPL-deficient parasites.
(A) A schematic diagram depicting the general strategies of TgCPL-deletion in Toxoplasma and PCR-based screening of the correct TgCPL knockout clones. The primers used for the screening are labeled. (B) PCR and agarose gel electrophoresis were used to select clones containing the correct integration of the pyrimethamine resistance cassette into the TgCPL locus and loss of the TgCPL gene. The genomic DNA of the Δcpl population served as a positive control for 5'- and 3'-ARM detection, while the WT genomic DNA was used for the detection of the TgCPL gene as a positive control. Water was used instead of DNA template in the PCR reactions to serve as a negative control. The expected bands are denoted by arrows, whereas nonspecific PCR amplifications are labeled by asterisks. (C) Clone 1 identified by PCR screening was grown in tissue culture for cell lysate preparation and further immunoblotting analysis to confirm the loss of TgCPL expression in the knockout. TgActin was used as a loading control.
Table 5:
Colony PCR reaction recipe for screening single Toxoplasma clones.
| Sample | Volume (μl) |
|---|---|
| total Toxoplasma genomic DNA | 1 |
| Forward primer (25 μM) | 0.2 |
| Reverse primer (25 μM) | 0.2 |
| 2x PCR master premix | 5 |
| Nuclease-free water | 3.6 |
| Total | 10 |
Table 6:
Thermocycler setting for screening single Toxoplasma clones.
| Initial denaturation: | 98 °C for 5 min |
| 35 cycles of | |
| Denaturing: | 98 °C for 5 s |
| Annealing: | 55 - 62 °C for 5 s |
| Extension: | 72 °C for 20 s per kb |
| Final extension: | 72 °C for 1 min |
Representative Results
Figure 1 represents an example of a growth curve for the RHΔku80∷NLuc strain and the derived calculation for its doubling time. Generally, the assay is performed in three technical replicates for each of the three biological replicates to account for variations of luciferase activity readings. In order to calculate the normalized fold change of parasite growth, each reading at 24-96 h post-infection was divided by the initial reading at 4 h post-infection, which reflects the starting amount of live parasites in the assay (Figure 1A,B). In terms of determining parasite doubling time, the log2 values of the normalized fold changes of parasite growth were plotted against each timepoint. Next, the plot was subjected to a linear regression function to obtain the slope, which represents doubling time (Figure 1C).
The inhibition efficacies of LHVS in wild-type and Δcpl strains were determined by plotting luciferase activities against eight inhibitor concentrations in Figure 2. It is essential to include infected cells without inhibitor treatment for normalization of raw luciferase activities in the assay. In addition, a mock experiment performed in a clear microplate is required for the assay to ensure that parasites are still in the intracellular stage at the end of the assay period.
In Figure 3, the generation and validation of a sgRNA expression construct targeting TgCPL and the production of a repair template for TgCPL deletion are shown. The 20 bp sgRNA matching to the TgUPRT gene encoded in the original plasmid was mutated to the DNA sequence targeting the TgCPL gene via PCR-based site-directed mutagenesis. To achieve this, the DNA sequences coding for the sgRNAs that recognize different genes were engineered to the forward primer, while the reverse primer was kept unchanged to simplify primer design.
Figure 3A shows a zoomed-in region of the sgRNA DNA sequences targeting the TgUPRT gene in the original template plasmid as well as the primer set used for the generation of the linearized sgRNA expression vector. Figure 3B shows a representative gel picture of the linearized TgCPL-targeting sgRNA expression plasmid. Figure 3C shows the restriction endonuclease digestion of the circularized TgCPL-targeting sgRNA expression plasmid. A M13 reverse primer was used to sequence the incorporated guide RNA within the sgRNA expression vector generated for the specific gene. In Figure 3D, the sequenced DNA region was aligned to the plasmid template for the confirmation of successful mutagenesis. Figure 3E illustrates the start and end regions of the pyrimethamine resistance cassette, showing where the primers can anneal for production of the repair template for TgCPL gene deletion. The repair template was PCR-amplified and loaded into a 1% agarose gel for size verification and gel extraction.
The overall strategy for TgCPL knockout generation and screening is shown in Figure 4. Three sets of primers shown in Figure 4A were used to screen TgCPL-deletion parasites for the correct integration of 5'- and 3'-ARMs and deletion of the TgCPL-coding sequence. As shown in Figure 4B, generally, seven to eight clones are selected for screening initially. The screening usually starts with checking for deletion of the coding sequence for the gene of interest. This is followed by detection of 5' and 3'-ARMs, which helps minimize the total number of clones to be screened. Further verification by immunoblotting displayed in Figure 4C can be completed if an antibody recognizing the target protein is available.
Discussion
++This protocol describes a luciferase-based protocol to assess intracellular Toxoplasma growth and evaluate the inhibition efficacy of chemical compounds against parasite growth. Compared to the existing strategies available for measuring intracellular Toxoplasma growth, this method exhibits high sensitivity and specificity. While monitoring parasite growth, a mock assay in a clear 96 well microplate is recommended to confirm that the tested strain does not prematurely lyse host cells before the end of the evaluation period. Otherwise, the luminescence readings will not accurately reflect parasite growth, since Toxoplasma only replicates within host cells.
It has been observed that phenol red dye quickly quenches luciferase activity, which can result in significant differences in the luciferase readings among technical replicates due to a delay in individual well measurements by the plate reader. Therefore, it is optimal to prepare HFFs in phenol red-free medium prior to seeding in the 96 well microplates. Also, in the case of high luciferase activity, cross-well interference may lead to significant variation among neighboring wells exhibiting strong luciferase activity. Hence, it is recommended to place an empty column between each strain.
Specifically, for the RHΔku80∷NLuc strain, 1,500 parasites are inoculated into each well for the growth assay. Since the doubling time for WT Toxoplasma parasites is ~6-8 h24, it is expected to see an increase in luciferase activity by 8- to 16-fold at 24 h post-infection. However, certain strains with significant growth defects will only yield a slight increase in luciferase activity. Therefore, if the initial parasite inoculum is low, the inherent variation in luciferase activity will mask the observation of an increase in luminescence over the growth period. Hence, it is recommended to inoculate a higher number of parasites to achieve an accurate fold change for strains with growth deficiencies.
In the protocol, the guide RNA design follows the general rules used for CRISPR-Cas9-based genome modification in mammalian cells25. Currently, many types of software and online platforms provide services for guide RNA design in various organisms, such as CHOPCHOP26, E-CRISP27, and EuPaGDT28. Here, commercial software (Table of Materials) is used to design sgRNA. Compared to the previously mentioned online programs, this software provides a local environment for sgRNA design. It calculates activity and specificity scores for each candidate sgRNA using a previously published algorithm25,29.
Technically, any sgRNA located within a gene of interest that contains a high specificity score can efficiently mediate the cleavage of genomic DNA for downstream homology-dependent recombination. In practice, sgRNA targeting a region close to the start or stop codon is preferred. Genes of interest can be endogenously epitope-tagged using the sgRNA, which generates a double-stranded gap at the end of the gene. Endogenously tagging a gene prior to its deletion will help confirm gene loss via immunoblotting detection in the case that an antibody against the protein of interest is not available. Epitope-tagging of a gene can also help determine the subcellular location of the protein of interest via immunofluorescence microscopy. Furthermore, if the target gene is essential, sgRNA recognizing the start region of the gene can be used for the replacement of its cognate promoter to a tetracycline-responsive promoter to generate a conditional knockout.
Additionally, the protocol describes a technique for gene deletion in Toxoplasma by replacing the TgCPL gene with a pyrimethamine resistance cassette. By using different plasmid templates encoding other drug resistance cassettes, investigators can modify primer sequences to incorporate other antibiotic resistance genes into the repair template via PCR. Furthermore, this protocol can be modified to perform other genome modifications, such as endogenous gene tagging, promoter replacement, and site-directed mutagenesis. It is noteworthy that the homologous regions used in this protocol are only 50 bp in length. A separate study successfully used 40-43 bp homologous DNA sequences to introduce single-nucleotide mutations and gene epitope tagging in Toxoplasma parasites22. Homologous DNA sequences in such short length can be easily incorporated into primers. Although we did not quantitatively evaluate the HDR efficiency for this particular length of homologous region, it seems that a 40-50 bp region is sufficient for efficient DNA recombination in the TgKu80-deficient Toxoplasma strain, as evidenced by the successful genetic manipulation of several genes recently achieved15,22.
During the efficacy determination of chemical compounds, if the prospective drug target gene is essential, a comparison of shifts in IC50 values between the wild-type and knockout strains is not practical. In this scenario, an assay directly measuring the ability of chemical compounds to inhibit recombinant protein activity is required to evaluate efficacy and specificity of the drugs. Recent literature reported the fitness scores of individual genes in Toxoplasma by performing a genome wide CRISPR screen30, which can serve as a guide to help assess the difficulty of generating a straight knockout mutant for the gene of interest.
Taken together, the protocol describes successful completion of a luciferase-based intracellular Toxoplasma growth assay and an evaluation strategy for chemical inhibitors against Toxoplasma growth. Also detailed is a CRISPR-Cas9-based genome editing protocol for gene deletion in Toxoplasma parasites, which has been widely used in the field. Individual labs can modify the described protocol according to experimental needs, such as endogenous gene tagging, switching drug selection markers, and altering the evaluation period for intracellular parasite growth.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. Sibley and Carruthers for sharing pSAG1-Cas9-sgRNA-TgUPRT plasmid and anti-TgCPL and TgActin antibodies. This work was supported by the Clemson Startup fund (to Z.D.), Knights Templar Eye Foundation Pediatric Ophthalmology Career-Starter Research Grant (to Z.D.), a pilot grant of an NIH COBRE grant P20GM109094 (to Z.D.), and NIH R01AI143707 (to Z.D.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Disclosures
The authors have nothing to disclose.
Video Link
The video component of this article can be found at https://www.jove.com/video/60985/
References
- 1.Blader IJ, Coleman BI, Chen C-T, Gubbels M-J Lytic Cycle of Toxoplasma gondii: 15 Years Later. Annual Review of Microbiology. 69 (1), 1–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones JL, Kruszon-Moran D, Rivera H, Price C, Wilkins PP Toxoplasma gondii Seroprevalence in the United States 2009-2010 and Comparison with the Past Two Decades. The American Journal of Tropical Medicine and Hygiene. 90 (6), (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kieffer F, Wallon M Congenital toxoplasmosis. Handbook of Clinical Neurology. 112, 1099–1101 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann S, Batz MB, Morris GJ Annual cost of illness and quality-adjusted life year losses in the United States due to 14 foodborne pathogens. Journal of Food Protection. 75 (7), 1292–1302 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Dubey J Toxoplasmosis. Journal of the American Veterinary Medical Association. 205 (11), 1593–1598 (1994). [PubMed] [Google Scholar]
- 6.Lindsay D, Dubey J Toxoplasma gondii: the changing paradigm of congenital toxoplasmosis. Parasitology. 138 (14), 1–3 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Deng Y, Wu T, Zhai S, Li C Recent progress on anti-Toxoplasma drugs discovery: Design, synthesis and screening. European Journal of Medicinal Chemistry. 183, 111711 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Butler NJ, Furtado JM, Wnthrop KL, Smith JR Ocular toxoplasmosis II: clinical features, pathology and management. Clinical & Experimental Ophthalmology. 41 (1), 95–108, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfefferko E, Pfefferko LC Specific Labeling of Intracellular Toxoplasma gondii with Uracil. Journal of Eukaryotic Microbiology. 24 (3), 449–453 (1977). [DOI] [PubMed] [Google Scholar]
- 10.Merli A, Canessa A, Melioli G Enzyme immunoassay for evaluation of Toxoplasma gondii growth in tissue culture. Journal of Clinical Microbiology. 21 (1), 88–91 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derouin F, Chastang C Enzyme immunoassay to assess effect of antimicrobial agents on Toxoplasma gondii in tissue culture. Antimicrobial Agents and Chemotherapy. 32 (3), 303–307 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McFadden D, Seeber F, Boothroyd J Use of Toxoplasma gondii expressing beta-galactosidase for colorimetric assessment of drug activity in vitro. Antimicrobial Agents and Chemotherapy. 41 (9), 1849–1853 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gubbels M-J, Li C, Striepen B High-Throughput Growth Assay for Toxoplasma gondii Using Yellow Fluorescent Protein. Antimicrobial Agents and Chemotherapy. 47 (1), 309–316 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Touquet B et al. High-content imaging assay to evaluate Toxoplasma gondii infection and proliferation: A multiparametric assay to screen new compounds. PLoS ONE. 13 (8), e0201678 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thornton LB et al. An ortholog of Plasmodium falciparum chloroquine resistance transporter (PfCRT) plays a key role in maintaining the integrity of the endolysosomal system in Toxoplasma gondii to facilitate host invasion. PLOS Pathogens. 15 (6), e1007775, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larson ET et al. Toxoplasma gondii cathepsin L is the primary target of the invasion-inhibitory compound morpholinurea-leucyl-homophenyl-vinyl sulfone phenyl. The Journal of Biological Chemistry. 284 (39), 26839–26850 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dou Z, McGovern OL, Cristina M, Carruthers VB Toxoplasma gondii Ingests and Digests Host Cytosolic Proteins. mBio. 5 (4), e01188–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cristina M et al. Toxoplasma depends on lysosomal consumption of autophagosomes for persistent infection. Nature Microbiology. 2, 17096 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parussini F, Coppens I, Shah PP, Diamond SL, Carruthers VB Cathepsin L occupies a vacuolar compartment and is a protein maturase within the endo/exocytic system of Toxoplasma gondii. Molecular Microbiology. 76 (6), 1340–1357 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huynh M-H, Carruthers VB Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryotic cell. 8 (4), 530–539 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox BA, Ristuccia JG, Gigley JP, Bzik DJ Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryotic Cell. 8 (4), 520–529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sidik SM, Hackett CG, Tran F, Westwood NJ, Lourido S Efficient Genome Engineering of Toxoplasma gondii Using CRISPR/Cas9. PLoS ONE. 9 (6), e100450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen B, Brown KM, Lee TD, Sibley DL Efficient Gene Disruption in Diverse Strains of Toxoplasma gondii Using CRISPR/CAS9. mBio. 5 (3), e01114–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radke JR et al. Defining the cell cycle for the tachyzoite stage of Toxoplasma gondii. Molecular and Biochemical Parasitology. 115 (2), 165–175 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Ran AF et al. Genome engineering using the CRISPR-Cas9 system. Nature Protocols. 8 (11), 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Research. 44 (W1), W272–W276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heigwer F, Kerr G, Boutros M E-CRISP: fast CRISPR target site identification. Nature Methods. 11 (2), 2812 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Peng D, Tarleton R EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microbial Genomics. 1 (4), e000033 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doench JG et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nature Biotechnology. 32 (12), 1262–1267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sidik SM et al. A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell. 166 (6), 1423–1435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.