Summary
Trained innate immunity, induced via modulation of mature myeloid cells or their bone marrow progenitors, mediates sustained increased responsiveness to secondary challenges. Here, we investigated whether anti-tumor immunity can be enhanced through induction of trained immunity. Pre-treatment of mice with β-glucan, a fungal-derived prototypical agonist of trained immunity, resulted in diminished tumor growth. The anti-tumor effect of β-glucan-induced trained immunity was associated with transcriptomic and epigenetic rewiring of granulopoiesis and neutrophil reprogramming toward an anti-tumor phenotype; this process required type I interferon signaling irrespective of adaptive immunity in the host. Adoptive transfer of neutrophils from β-glucan-trained mice to naive recipients suppressed tumor growth in the latter in a ROS-dependent manner. Moreover, the anti-tumor effect of β-glucan-induced trained granulopoiesis was transmissible by bone marrow transplantation to recipient naive mice. Our findings identify a novel and therapeutically relevant anti-tumor facet of trained immunity involving appropriate rewiring of granulopoiesis.
Keywords: inflammation, trained immunity, innate immune memory, cancer immunotherapy, granulopoiesis, neutrophils, interferon, β-glucan
Graphical Abstract
Highlights
-
•
Trained innate immunity (TII) promotes anti-tumor activity
-
•
TII is linked to transcriptomic and epigenetic rewiring of granulopoiesis
-
•
Trained granulopoiesis promotes an anti-tumor phenotype in neutrophils
-
•
Trained granulopoiesis might bear potential for cancer immunotherapy
Pre-treatment of mice with β-glucan, an agonist of trained immunity responses, leads to epigenetic changes in granulopoiesis and neutrophil function to drive anti-tumor immune responses.
Introduction
Infiltration of solid tumors by immune cells is a hallmark of cancer and plays a critical role in disease progression (Hanahan and Weinberg, 2011). The tumor microenvironment might reprogram tumor-infiltrating immune cells, which thereby acquire pro-tumor functions that facilitate tumor growth (Coussens et al., 2013). Cancer-induced alterations in myelopoiesis drive increased generation of myeloid cells, including monocytes-macrophages and neutrophils, which accumulate at the tumor and often shift toward a specific tumor-promoting phenotype (Gabrilovich et al., 2012; Rice et al., 2018). However, the landscape of tumor-infiltrating myeloid cells is complex and dynamic (Zilionis et al., 2019) and probably specific to the tumor type and innate immune cells might also exert anti-tumor activities (Devlin et al., 2020; Galdiero et al., 2018; Granot et al., 2011; Murray, 2018; Oberg et al., 2019; Ponzetta et al., 2019; Powell and Huttenlocher, 2016). Neutrophils that infiltrate solid tumors, known as tumor-associated neutrophils (TANs), can acquire a phenotype that displays cytotoxic and anti-tumorigenic properties (broadly designated TAN1) or a phenotype associated with tumor progression (TAN2) (Fridlender et al., 2009; Powell and Huttenlocher, 2016). TAN1 and TAN2 can be distinguished by distinct cytokine and chemokine production patterns and other specific molecular signatures, such as related to phagocytosis (Shaul et al., 2016). Type I interferons (IFNs) promote TAN1 (Andzinski et al., 2016), whereas transforming growth factor-β (TGF-β) is linked to TAN2 differentiation (Fridlender et al., 2009). Together, altered granulopoiesis and neutrophil function in the context of cancer (Patel et al., 2018; Powell and Huttenlocher, 2016) are important players shaping tumor immunity and cancer progression.
Fungal-derived polysaccharide β-glucan or the Bacillus Calmette-Guėrin vaccine (BCG) promote a sustained enhanced response of myeloid cells to secondary infectious or inflammatory challenges; this process has been coined “trained innate immunity” or “innate immune memory” and is mediated via transcriptomic, epigenetic, and metabolic reprogramming (Bekkering et al., 2018; Chavakis et al., 2019; Netea et al., 2016; Netea and van der Meer, 2017; Penkov et al., 2019; Saeed et al., 2014). In this context, we and others recently established that the long-term effects of trained immunity are explained by modulation of progenitors of myeloid cells in the bone marrow (BM) (Christ et al., 2018; Kaufmann et al., 2018; Mitroulis et al., 2018). Agents that were recently identified as trained immunity agonists have been also known to exert anti-tumor activities. For instance, BCG is used in the treatment of bladder cancer (Hersh et al., 1977), whereas β-glucan is linked to tumor immunotherapy efficacy (Alexander et al., 2018; Cheung et al., 2002; Li et al., 2010; Masuda et al., 2015; Xu et al., 2016). However, the processes underlying the action of trained immunity agonists in cancer are not understood at mechanistic depth. More importantly, whether the potential tumor-modulating effects of agents, like β-glucan, involve induction of innate immune memory has not been hitherto addressed. Here, we show that innate immune training of granulopoiesis results in potent anti-tumor activity. Specifically, trained immunity induced by pre-treatment of mice with β-glucan resulted in diminished tumor growth. The observed anti-tumor effects of β-glucan-induced trained immunity were associated with transcriptomic and epigenetic rewiring of granulopoietic progenitors, resulting in neutrophil reprogramming toward an anti-tumor phenotype. Therefore, trained immunity enhances host immunity to cancer by anti-tumor rewiring of granulopoiesis.
Results
Trained Innate Immunity Suppresses Tumor Growth Independently of Adaptive Immunity
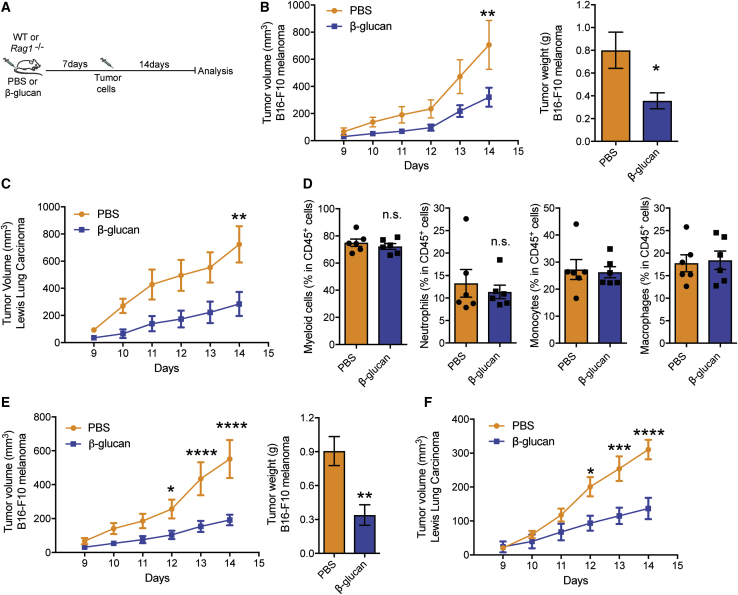
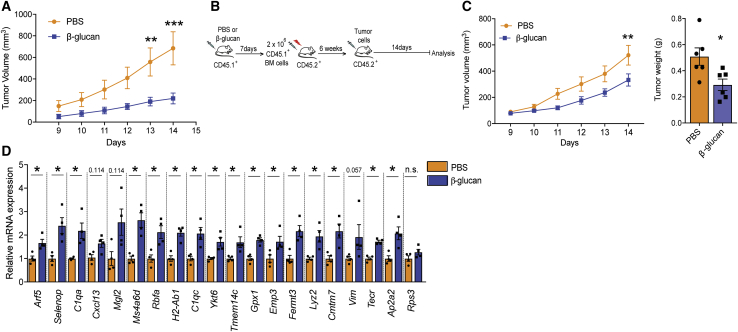
To examine the role of trained immunity on tumor development, we injected wild-type (WT) mice intraperitoneally (i.p.) with a single dose of β-glucan (Mitroulis et al., 2018); 7 days thereafter, mice were subcutaneously inoculated with B16-F10 melanoma cells and tumor growth was monitored (Figure 1A). Training with β-glucan resulted in significantly diminished tumor growth, as assessed by measuring tumor volume and weight 14 days after tumor challenge, as compared with that of control mice that were pre-treated with vehicle control (PBS) (Figure 1B). The tumor-suppressive effect of β-glucan-induced trained immunity was verified by using another ectopic tumor model, the Lewis lung carcinoma (LLC) model (Figure 1C).
Figure 1.
Induction of Trained Immunity Inhibits Tumor Growth
(A) Experimental scheme.
(B) C57BL/6 WT mice received a single i.p. injection of β-glucan or PBS and 7 days thereafter, mice were subcutaneously inoculated with B16-F10 melanoma cells. Shown on the left, tumor volume was monitored for another 14 days after tumor inoculation. Shown on the right, tumor weight at the end of the experiment (n = 6 mice in the PBS group; n = 7 mice in the β-glucan group).
(C) C57BL/6 WT mice received a single i.p. injection of β-glucan or PBS and 7 days thereafter, mice were inoculated with LLC cells. Tumor volume is shown (n = 7 mice in the PBS group; n = 5 mice in the β-glucan group).
(D) C57BL/6 WT mice received β-glucan or PBS and 7 days thereafter, mice were inoculated with B16-F10 melanoma cells. Flow-cytometric analysis for immune cells that are infiltrated in the B16-F10 melanoma tumors was performed at the end of the experiment. Frequencies of myeloid cells (CD45+CD11b+), neutrophils (CD45+CD11b+Ly6g+Ly6c−), monocytes (CD45+CD11b+Ly6g−Ly6c+), and macrophages (CD45+CD11b+F4/80+) within leukocytes (CD45+) are shown (n = 6 mice per group).
(E) Rag1−/− mice received a single i.p. injection of β-glucan or PBS and 7 days thereafter, mice were inoculated with B16-F10 melanoma cells. Shown on the left is tumor volume; on the right is tumor weight at the end of the experiment (n = 8 mice in the PBS group; n = 5 mice in the β-glucan group).
(F) Rag1−/− mice received β-glucan or PBS and 7 days thereafter, mice were inoculated with LLC cells. Tumor volume is shown (n = 16 mice in the PBS group; n = 12 mice in the β-glucan group).
Data are presented as mean ± SEM; n.s., non-significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See also Figure S1.
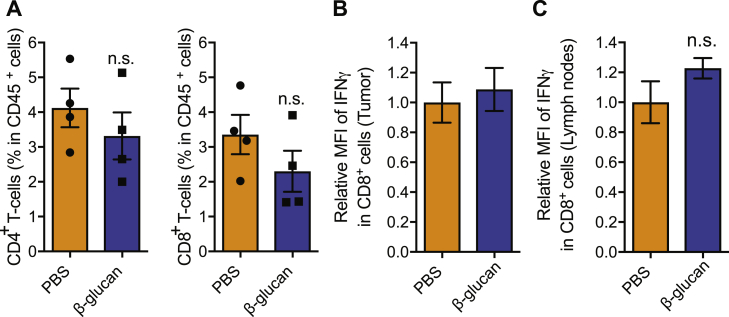
Given that several immune cell types can modulate tumor progression (Gabrilovich et al., 2012), we next examined whether β-glucan-induced trained immunity affects the intra-tumoral immune cell composition. Flow-cytometric analysis of the tumor-infiltrating cells revealed that the frequencies of myeloid cells, neutrophils, monocytes, and macrophages (Figure 1D), and of CD4+ T cells and CD8+ T cells (Figure S1A) were comparable between mice pre-treated with β-glucan or PBS control.
Figure S1.
The Effect of Trained Immunity on Immune Cell Composition in the Tumors and IFNγ Levels in CD8+ Cells of Tumor-Bearing Mice, Related to Figure 1
WT mice received a single intraperitoneal injection with β-glucan or control PBS, and 7 days later received subcutaneous injection of B16-F10 melanoma cells; mice were sacrificed 14 days after tumor inoculation.
(A) Frequencies of CD4+ T cells (CD45+CD4+CD8−) and CD8+ T cells (CD45+CD4−CD8+) within leukocytes (CD45+) in the tumor.
(B and C) Expression of interferon gamma (IFNγ) in CD8+ T cells from tumor tissue (B) and draining lymph nodes (C) was assessed by flow cytometry. Data are shown as relative Mean fluorescence intensity (MFI); the MFI of IFNγ in CD8+ T cells from PBS-treated mice was set as 1 in each case.
Data are presented as mean ± SEM (A and C: n = 4 mice / group; B: n = 5 mice in the PBS group and n = 4 mice in the β-glucan group). n.s., non-significant.
Cells of adaptive immunity are central players in tumor immunity (Chen and Mellman, 2017; Fritz and Lenardo, 2019). To determine whether adaptive immunity is involved in the anti-tumor effect induced by β-glucan, Rag1−/− mice that lack B and T cells were treated with β-glucan prior to the secondary tumor challenge (Figure 1A). Pre-treatment with β-glucan decreased both B16-F10 and LLC tumor burden also in Rag1−/− mice (Figures 1E and 1F), thus showing that the anti-tumor effect of β-glucan-induced trained immunity does not require adaptive immunity. Along the same line, amounts of IFN-γ in cytotoxic CD8+ T cells obtained from tumors and draining lymph nodes were comparable between WT mice pre-treated with β-glucan and control (Figures S1B and S1C).
Training with β-glucan Promotes an Anti-tumor Phenotype in Neutrophils
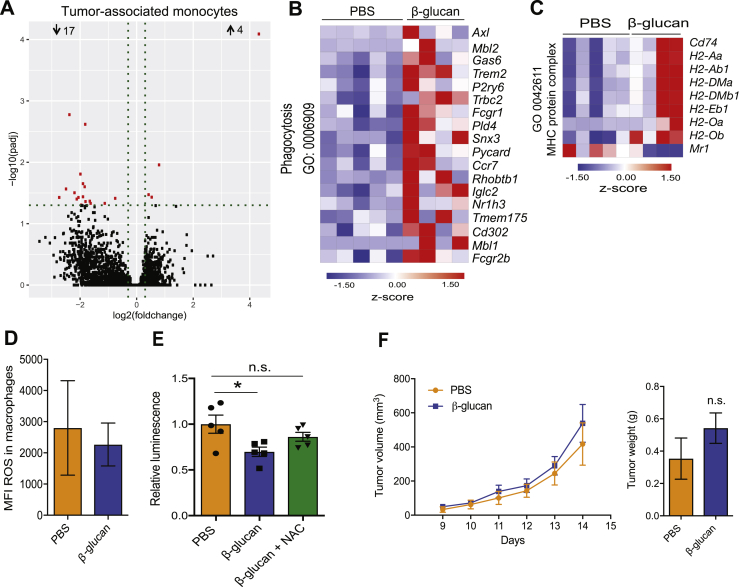
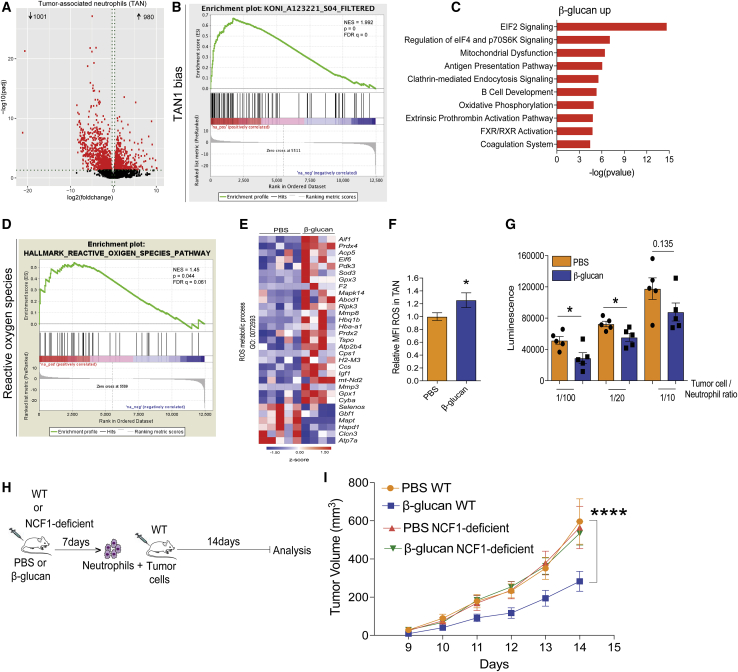
Because analysis of intratumoral innate immune cell populations displayed no quantitative changes by β-glucan pre-treatment, we focused on potential qualitative alterations and thus performed RNA sequencing in TANs (CD45+CD11c−CD11b+Ly6g+Ly6c−) and tumor-associated monocytes (CD45+CD11c−CD11b+Ly6g−Ly6c+) sorted from B16-F10 melanoma tumors of mice that were pre-treated with β-glucan or PBS. Although no major differences were observed between the transcriptome of monocytes from β-glucan-trained and control mice (Figure S2A), TANs from β-glucan-trained mice displayed an altered transcriptomic profile as compared with that of the cells from control-treated mice (Figure 2A). In addition, gene set enrichment analysis (GSEA) using a gene set implicated in the TAN1 phenotype (Shaul et al., 2016), revealed a significant positive correlation of TANs from β-glucan-trained mice with the TAN1 anti-tumor signature (Figure 2B; Table S1). The phagocytic function of innate immune cells is linked to their anti-tumor activity (Feng et al., 2019), and genes related to phagocytosis in neutrophils are upregulated in the TAN1 compared with the TAN2 phenotype (Shaul et al., 2016). Consistently, gene expression linked to the process of phagocytosis was enhanced in TANs from β-glucan-trained mice (Figure S2B). Antigen presentation pathways are also associated with the TAN1 phenotype (Shaul et al., 2016). Consistently, differentially regulated expression of genes linked to the process of antigen presentation was also observed in TANs from trained mice (Figure S2C). These data lend further support to the trained-immunity-induced anti-tumor phenotype of TANs. Ingenuity pathway analysis (IPA) demonstrated that canonical pathways involved in eukaryotic initiation factor 2 (EIF2) as well as in eEIF4 and p70S6K signaling were enriched in TANs from β-glucan-trained mice (Figure 2C). Whereas the p70S6K signaling pathway has been linked to granulocytic differentiation and neutrophil functions, such as chemotaxis (Gomez-Cambronero et al., 2004; Gomez-Cambronero et al., 2003), the EIF2 signaling pathway has been associated with reactive oxygen species (ROS)-induced stress (Liu et al., 2008; Zeeshan et al., 2016; Zhang and Kaufman, 2008). Given that neutrophil ROS are linked to the anti-tumor effects of neutrophils (Granot et al., 2011; Yan et al., 2014), we focused next on the ROS pathway in TANs from β-glucan-trained mice compared with that in TANs from control-treated mice. GSEA using the Molecular Signatures Database (MSigDB) Hallmark gene set collection (Liberzon et al., 2015) displayed a positive correlation with the ROS pathway in TANs from β-glucan-trained mice, as compared with that in TANs from control mice (Figure 2D; Table S2). Moreover, we found increased expression amounts of several genes associated with the ROS metabolic process in TANs from β-glucan-trained mice (Figure 2E). Accordingly, TANs from β-glucan-treated mice displayed increased ROS levels 14 days after tumor inoculation in relation to TANs from mice treated with PBS, whereas no alterations in ROS levels were observed in tumor-associated macrophages (Figures 2F and S2D).
Figure S2.
The Effect of Trained Immunity on the Transcriptomic Profile of Tumor-Associated Monocytes and TAN, Related to Figure 2
(A) WT mice were treated with β-glucan or PBS, and 7 days later received B16-F10 melanoma cells. Mice were sacrificed 14 days after tumor inoculation and monocytes (CD45+CD11c−CD11b+Ly6g−Ly6c+) were sorted from the tumor tissue for RNA sequencing analysis. Differential gene expression in monocytes. Volcano plot showing the distribution of the adjusted p values (−log10(padj)) and fold changes (log2 fold change). FDR ≤ 0.05 (n = 5 mice in the PBS group and n = 4 mice in the β-glucan group).
(B and C) WT mice were treated with β-glucan or PBS, and 7 days later received B16-F10 melanoma cells. Mice were sacrificed 14 days after tumor inoculation and TAN (CD45+CD11c−CD11b+Ly6c−Ly6g+) were sorted from the tumor tissue for RNA sequencing analysis. FDR ≤ 0.05. (B) Heatmap of genes involved in phagocytosis in TAN from β-glucan-treated mice as compared to PBS-treated mice (n = 5 mice in the PBS group and n = 4 mice in the β-glucan group). (C) Heatmap of genes involved in MHC-protein complex in TAN from β-glucan-treated mice as compared to PBS-treated mice (n = 5 mice in the PBS group and n = 4 mice in the β-glucan group).
(D) WT mice were treated with β-glucan or PBS, and 7 days later received B16-F10 melanoma cells. Mice were sacrificed 14 days after tumor inoculation and staining for ROS in tumor-associated macrophages (CD45+CD11b+Ly6g−F4/80+) was performed using flow cytometry. Median fluorescence intensity (MFI) is shown (n = 6 mice / group).
(E) Splenic neutrophils were isolated from mice 7 days after injection with β-glucan or PBS. Neutrophils were co-cultured with luciferase expressing B16-F10 cells at 100:1 neutrophil/tumor cell ratio for 24 h. N-Acetyl-Cysteine (NAC; 5mM) was used to scavenge ROS. Tumor cell survival was assessed by measuring luminescence. Luminescence is expressed relative to the PBS-control group, set as 1 (n = 5 cell isolations per group).
(F) WT mice were injected with β-glucan or PBS and after 7 days splenic monocytes were isolated and were adoptively transferred together with B16-F10 cells into WT mice. Tumor volume (left) and weight of the tumor tissue at the end of the experiment (right) are shown (n = 5 mice in the PBS group; n = 6 mice in the β-glucan group).
Data are presented as mean ± SEM n.s.: non-significant; ∗p < 0.05.
Figure 2.
Trained Immunity Shapes the Transcriptional Profile of TANs and Neutrophils from β-Glucan-Trained Mice Suppress Tumor Growth via ROS Production
(A–E) WT mice were treated with β-glucan or PBS and after 7 days were inoculated with B16-F10 melanoma cells. TANs (CD45+CD11c−CD11b+Ly6c−Ly6g+) were sorted and RNA sequencing analysis was performed (n = 5 mice in the PBS group; n = 4 mice in the β-glucan group) 14 days after the tumor injection. In (A), (C) and (E), false discovery rate (FDR) ≤ 0.05.
(A) Differential gene expression in TANs from mice pre-treated with β-glucan compared with TANs from PBS-treated mice. Volcano plot showing the distribution of the adjusted p values (−log10(padj)) and fold changes (log2 fold change).
(B) GSEA for genes related to a gene set previously implicated in the TAN1 phenotype (Shaul et al., 2016) was used to analyze the transcriptomic effect induced in TANs by β-glucan administration in mice. Abbreviation is as follows: NES, normalized enrichment score.
(C) Top 10 enriched canonical pathways identified by IPA in TANs from β-glucan-treated mice compared to PBS-treated mice.
(D and E ) Shown in (D) is a GSEA for genes related to ROS pathway, and in (E) is a heatmap of genes involved in ROS metabolic process in TANs from β-glucan-treated mice as compared with TANs from PBS-treated mice.
(F) WT mice were treated with β-glucan or PBS and after 7 days were inoculated with B16-F10 melanoma cells. Staining for ROS in TANs (CD45+CD11b+Ly6g+) was performed by flow cytometry 14 days after the tumor cell injection. Relative mean fluorescence intensity (MFI) is shown. MFI of ROS was measured and expressed in relation to the PBS group, set as 1. Data are presented as mean ± SEM (n = 15 per group).
(G) Splenic neutrophils were isolated from mice 7 days after injection with β-glucan or PBS. Neutrophils were co-cultured with luciferase expressing B16-F10 cells for 24 h. Tumor cell survival was assessed by measuring luminescence (n = 5 per group).
(H) Experimental scheme.
(I) As indicated in (H), WT or NCF1-deficient mice were injected with β-glucan or PBS, and after 7 days splenic neutrophils were isolated and were adoptively transferred together with B16-F10 cells into WT recipients. Tumor volume is shown. Data are presented as mean ± SEM (n = 7 mice in the PBS WT group; n = 12 mice in the β-glucan WT group; n = 11 mice in the PBS NCF1-deficient group; n = 14 mice in the β-glucan NCF1-deficient group).
∗p < 0.05, ∗∗∗∗p < 0.0001.
See also Figure S2.
To investigate mechanisms by which neutrophils could exert anti-tumor effects upon induction of trained immunity, we isolated neutrophils from spleens of mice 7 days after injection of β-glucan or PBS control and assessed their anti-tumor function in ex vivo assays. Splenic neutrophils from the β-glucan-trained mice (hereafter designated “trained” neutrophils) or neutrophils from control-treated mice (“non-trained” neutrophils) were co-cultured with B16-F10 melanoma cells expressing a luciferase reporter. Compared with non-trained neutrophils, trained neutrophils displayed enhanced tumor cytotoxicity (Figure 2G). In contrast, no differences were seen in the survival of tumor cells exposed to BM-derived macrophages from β-glucan- or control-treated mice (data not shown). Moreover, the cytotoxic effect of trained neutrophils against tumor cells ex vivo was prevented in the presence of the ROS scavenger N-Acetyl Cysteine (NAC) (Figure S2E), thereby further supporting the involvement of ROS in the anti-tumor activity of trained neutrophils. To provide conclusive in vivo evidence that trained neutrophils exert anti-tumor activity in a ROS-dependent manner, we performed adoptive transfer experiments. To this end, splenic neutrophils were isolated from mice 7 days after pre-treatment with β-glucan or PBS and then were co-injected with B16-F10 melanoma cells into untreated WT mice. Moreover, to interrogate the functional contribution of ROS production to the anti-tumor activity of trained neutrophils, we performed adoptive transfer experiments with neutrophils obtained from mice with impaired nicotinamide adenine dinucleotide phosphate (NADPH) oxidase activity and ROS production resulting from deficiency of neutrophil cytosolic factor 1 (NCF1) protein (Aachoui et al., 2013; Baptista et al., 2016; Huang et al., 2000; Maltez et al., 2015), hereafter designated NCF1-deficient mice. We isolated neutrophils from NCF1-deficient or WT mice that were pre-treated with either β-glucan or PBS control and transferred them, together with B16-F10 melanoma cells, into untreated WT mice (Figure 2H). Tumor growth was significantly suppressed in mice that received neutrophils from the β-glucan-trained WT mice compared with mice that received neutrophils from control-treated mice (Figure 2I). Additionally, neutrophils from β-glucan-treated NCF1-deficient mice did not show enhanced tumor-suppressive activity in recipient WT mice compared with the activity of neutrophils from PBS-treated NCF1-deficient mice (Figure 2I). Adoptive transfer of monocytes from β-glucan-trained or control mice resulted in comparable tumor growth in the respective recipient mice (Figure S2F). These data firmly establish that the anti-tumor activity of trained immunity can be attributed, at least in part, to trained neutrophils and that intact NADPH oxidase-dependent ROS production is an essential feature of the anti-tumor activity of trained neutrophils.
Long-Term Neutrophil-Mediated Anti-tumor Effects of Trained Immunity
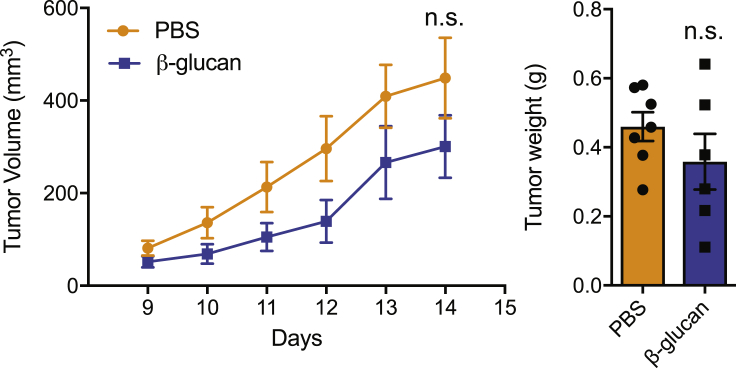
A defining property of trained innate immunity is induction of long-term alterations in innate immune cells (Netea et al., 2020). We therefore next examined whether the anti-tumor effect of trained immunity could be sustained long-term. To this end, inoculation of tumors into WT mice trained with a single injection of β-glucan 28 days earlier resulted in significant inhibition of tumor growth, as compared with that in control-treated non-trained mice (Figure 3A).
Figure 3.
Long-Term Anti-tumor Effects of Trained Granulopoiesis
(A) WT mice were treated with β-glucan or PBS, and after 28 days were subcutaneously inoculated with B16-F10 melanoma cells. Tumor volume was monitored for another 14 days after tumor inoculation (n = 5 mice in the PBS group; n = 6 mice in the β-glucan group).
(B–D) As indicated in the experimental scheme (B), WT CD45.1+ mice were injected with β-glucan or PBS, and after 7 days BM cells were isolated and were transplanted into CD45.2+ mice. Six weeks after transplantation, recipient mice were inoculated with tumors. In (C), (left) tumor volume and (right) the weight of B16-F10 melanoma tumors at the end of the experiment are shown (n = 6 mice per group). Shown in (D), 14 days after the tumor injection in recipient mice, TANs (CD45+CD11c−CD11b+Ly6c−Ly6g+) were sorted and relative mRNA expression of the “trained TAN1-like signature” was performed. Relative mRNA expression was normalized against 18S rRNA and was set as 1 in TANs from recipients that were transplanted with cells from PBS-treated donor mice (n = 4 mice per group).
Data are presented as mean ± SEM; n.s.. non-significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figure S3.
Given that trained innate immunity mediates long-term effects on myeloid cells via modulation of their progenitors in the BM (Christ et al., 2018; Kaufmann et al., 2018; Mitroulis et al., 2018), we interrogated whether the anti-tumor effects of trained immunity are mediated by sustained adaptations in BM hematopoietic progenitors. To address whether the anti-tumor actions of trained immunity are transmissible by BM transplantation to recipient non-trained mice, donor mice were treated with β-glucan or PBS, and 7 days later their BM cells were transplanted to non-trained irradiated recipient mice. After establishment of hematopoiesis in recipient mice, tumors were implanted (Figure 3B). Tumor burden was significantly decreased in mice that had received BM cells from β-glucan-trained donor mice, as compared with that of mice that received BM cells from control-treated mice (Figure 3C). Therefore, the tumor-suppressive properties of β-glucan-induced trained immunity can be transferred by BM transplantation to recipient non-trained mice.
Multiple gene expression markers associated with the anti-tumor TAN1 phenotype were significantly upregulated in TANs isolated from the tumors of recipient mice transplanted with BM cells from β-glucan-trained mice, as compared with those of TANs of mice that received BM from control-treated donors (Figure 3D). The “trained TAN1-like signature” shown in Figure 3D was selected according to the study by Shaul et al. (2016), and is defined in STAR Methods under “RNA isolation and real time PCR.” An upregulation in the expression of the vast majority of the genes comprising the “trained TAN1-like signature” was present in TANs from mice that received BM cells from β-glucan-trained mice as compared with TANs of mice transplanted with BM cells from control-treated donors (Figure 3D). We also identified the human orthologs of the mouse “trained TAN1-like signature” from Mouse Genome Informatics (http://www.informatics.jax.org/) and used the Gene Expression Profiling Interactive Analysis (GEPIA-2), which dichotomizes patients in datasets from The Cancer Genome Atlas (TCGA) on the basis of median expression of a gene signature (Tang et al., 2019). High expression of the human ortholog “trained TAN1-like signature” was found to be a good prognostic factor for survival in certain cancers, for instance, in human skin cutaneous melanoma (hazard ratio 0.61, p(HR) = 0.00032), but not in other cancers (data not shown). It should be noted that the mouse “trained TAN1-like signature” derives from RNA-sequencing analysis of the sorted neutrophils from B16-F10 tumor-bearing mice, whereas the analysis performed with GEPIA-2 on The Cancer Genome Atlas (TCGA) datasets assesses this signature in RNA-sequencing data from total tumors and thus does not necessarily represent expression of these genes in TANs, limiting the applicability of such analysis.
To gain additional insight into whether the anti-tumor effects of β-glucan-induced trained immunity are mediated through adaptations in BM progenitors rather than direct effects on neutrophils themselves, we performed adoptive transfer of splenic neutrophils from mice that were treated for only one day with β-glucan or vehicle control. Comparable tumor growth was observed between the two recipient groups (Figure S3), probably because the limited training time did not allow the emergence of trained neutrophils in donor mice. Together, these findings are consistent with the conclusion that the anti-tumor effects of β-glucan are initiated by training of BM progenitors and, therefore, require sufficient time to mount.
Figure S3.
Adoptive Transfer of Neutrophils after Short-Term β-Glucan-Treatment in Mice, Related to Figure 3
WT mice were injected with β-glucan or PBS and after 1 day splenic neutrophils were isolated and were adoptively transferred together with B16-F10 cells into WT mice. Tumor volume (left) and weight of the tumor tissue at the end of the experiment (right) is shown (n = 7 mice in the PBS group; n = 6 mice in the β-glucan group).
Data are presented as mean ± SEM. n.s., non-significant.
Innate Immune Training of Neutrophils toward an Anti-tumor Phenotype Is Associated with Transcriptomic and Epigenetic Rewiring of Granulopoiesis
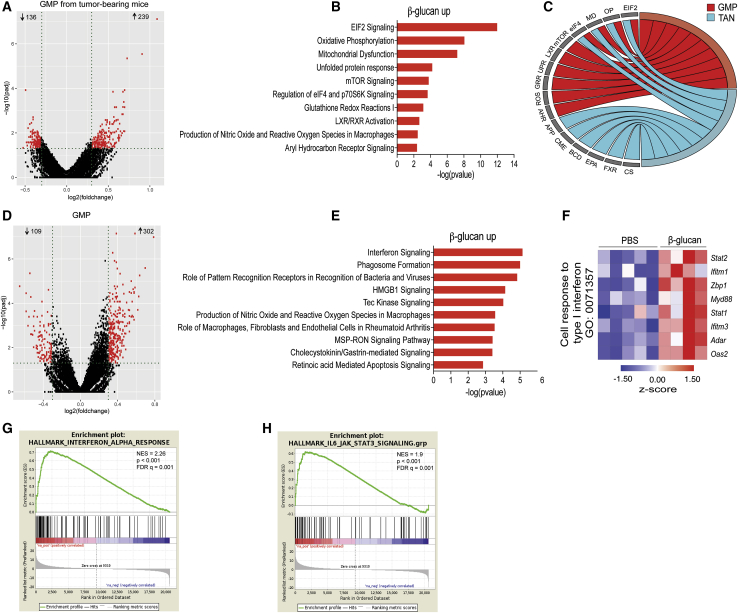
Because our findings so far suggested that the anti-tumor effects of β-glucan involve training of granulopoiesis in the BM, we next focused on the BM precursors of neutrophils, granulocyte-monocyte progenitors (GMPs). To this end, GMPs (Lin−c-Kit+Sca1−CD16/32+CD34+) from control-treated tumor-bearing mice and GMPs from β-glucan-trained tumor-bearing mice were sorted 14 days after tumor inoculation and subjected to RNA sequencing, which revealed that pre-treatment with β-glucan (7 days prior to tumor injection) led to significant differences in gene expression (Figure 4A). IPA displayed a partial overlap between the pathways enriched in GMPs from tumor-bearing and β-glucan-trained mice (Figures 4B and 4C) and the enriched pathways observed in TANs from β-glucan-trained mice (Figure 2D). For instance, oxidative phosphorylation, regulation of eEIF4 and p70S6K signaling and the EIF2 signaling pathways were among the top commonly enriched pathways between GMPs and TANs from tumor-bearing mice pre-treated with β-glucan (Figures 4B and 4C). Moreover, upstream regulator analysis using IPA revealed common upstream regulators in the transcriptomic profile of TANs and BM GMPs from tumor-bearing mice after induction of β-glucan-mediated trained immunity (Figure S4A). These findings suggest that granulopoiesis progenitors are targets of trained immunity that leads to the induction of neutrophils with enhanced anti-tumor activity.
Figure 4.
Transcriptomic Alterations in GMP Due to Trained Immunity
(A–C) WT mice were treated with β-glucan or PBS and after 7 days were inoculated with B16-F10 melanoma cells. BM GMPs (Lin−c-Kit+Sca1−CD16/32+CD34+) were sorted 14 days after tumor cell injection and RNA sequencing analysis was performed (n = 4 mice in the PBS group and n = 3 mice in the β-glucan group).
(A) Differential gene expression in GMPs from tumor-bearing mice pre-treated with β-glucan as compared with GMPs from PBS-treated mice. Volcano plot showing the distribution of the adjusted p values (−log10(padj)) and fold changes (log2 fold change). FDR ≤ 0.05.
(B) Top 10 enriched canonical pathways identified by IPA in GMPs from β-glucan-treated mice, compared with GMPs from PBS-treated mice. FDR ≤ 0.05.
(C) Circos plot showing the commonly enriched pathways (within the top 10 enriched canonical pathways) between TANs and GMPs from tumor-bearing mice. Abbreviations are as follows: EIF2, EIF2 signaling; OP, oxidative phosphorylation; MD, mitochondrial dysfunction; eIF4, regulation of eIF4 and p70S6K signaling; mTOR, mTOR signaling; LXR, LXR/RXR activation; UPR, unfolded protein response; GRR, glutathione redox reactions I; ROS, production of nitric oxide and reactive oxygen species in macrophages; AHR, aryl hydrocarbon receptor signaling; APP, antigen presentation pathway; CME, clathrin-mediated endocytosis signaling; BCD, B cell development; EPA, extrinsic prothrombin activation pathway; FXR, FXR/RXR activation; CS, coagulation system.
(D–H) WT mice were treated with β-glucan or PBS and after 7 days BM GMP were sorted for RNA sequencing analysis (n = 5 mice in the PBS group and n = 4 mice in the β-glucan group). In (D)–(F), FDR ≤ 0.05.
(D) Differential gene expression in GMPs from mice pre-treated with β-glucan compared with GMPs from PBS-treated mice. Volcano plot showing the distribution of the adjusted p values (−log10(padj)) and fold changes (log2 fold change).
(E) Top 10 enriched canonical pathways identified by IPA in GMPs from β-glucan-treated mice as compared with GMPs from PBS-treated mice.
(F and G) In (F) is a heatmap of genes involved in the cell response to type I IFN, and in (G) is a GSEA for genes related to IFN-α response in GMPs from β-glucan-treated mice as compared with GMPs from PBS-treated mice.
(H) GSEA for genes related to the IL6-Jak-Stat3 signaling pathway in GMPs from β-glucan-treated mice as compared with GMPs from PBS-treated mice.
See also Figure S4.
Figure S4.
The Effect of Trained Immunity on the Transcriptomic Profile of TAN and BM GMP and on IFNα Levels in BM pDCs and CD169+ Macrophages, Related to Figure 4
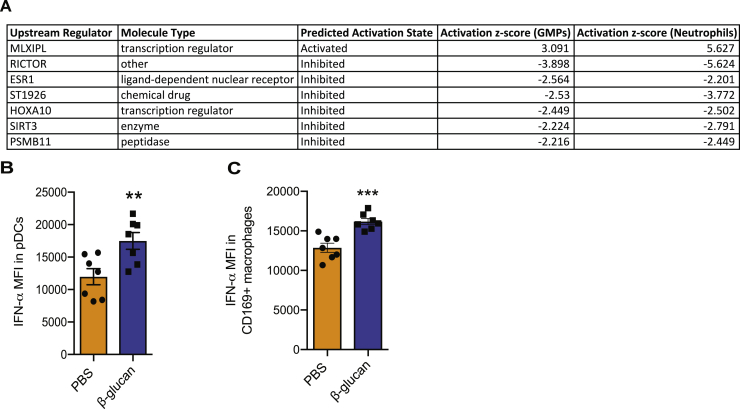
(A) WT mice were treated with β-glucan or PBS and after 7 days were inoculated with B16-F10 melanoma tumors. 14 days after the tumor injection, TAN and BM GMP were sorted and RNA sequencing analysis was performed (RNA sequencing analysis of TANs is shown in Figure 2). Upstream regulator analysis in transcriptomic data using IPA was performed. Common upstream regulators in the transcriptomic profile of TAN and BM GMPs from tumor-bearing mice are listed (TAN: n = 5 mice in the PBS group and n = 4 mice in the β-glucan group; GMP: n = 4 mice in the PBS group and n = 3 mice in the β-glucan group).
(B) Intracellular staining for IFNα in pDCs from the BM of mice 7 days after β-glucan or PBS treatment was performed. Median fluorescence intensity (MFI) (n = 7 mice / group).
(C) Intracellular staining for IFNα in CD169+ macrophages from the BM of mice 7 days after β-glucan or PBS treatment was performed. Median fluorescence intensity (MFI) (n = 7 mice / group).
∗∗p < 0.01; ∗∗∗p < 0.001.
To evaluate potential mechanisms underlying the innate immune training of granulopoiesis toward anti-tumor activity, we determined the transcriptomic profile of BM GMPs 7 days after β-glucan administration to mice in the absence of tumors (Figure 4D). GSEA in GMPs from trained mice revealed a GMPs transcriptome consistent with preferential differentiation to neutrophils compared with that of other myeloid cell types (Table S3). IPA demonstrated that inflammatory pathways with foremost IFN signaling pathway were enriched in upregulated genes of GMPs upon induction of β-glucan-mediated trained immunity (Figure 4E). Type I IFN responses have been previously associated with induction of the TAN1 anti-tumor phenotype (Andzinski et al., 2016; Pylaeva et al., 2016); we therefore focused on this pathway. Indeed, GMPs from β-glucan-trained mice showed increased expression amounts of genes involved in the cell response to type I IFN (Figure 4F). Along the same line, GSEA demonstrated a positive correlation with the IFN-α response and with the JAK-STAT signaling pathway (Nan et al., 2017) in GMPs from β-glucan-trained mice, as compared with GMPs of control-treated mice (Figures 4G and 4H; Table S4). To interrogate the cellular source of IFN-α in the BM, we analyzed CD169+ macrophages, plasmacytoid dendritic cells (pDCs), and classical DCs. IFN-α protein amounts were increased in pDCs and CD169+ macrophages (Figures S4B and S4C), but not in classical DCs (data not shown) upon induction of β-glucan-mediated trained immunity, as compared with IFN-α levels in cells from control-treated mice. Collectively, these findings support the notion that type I IFN signaling might be a key regulator of innate immune training of granulopoiesis.
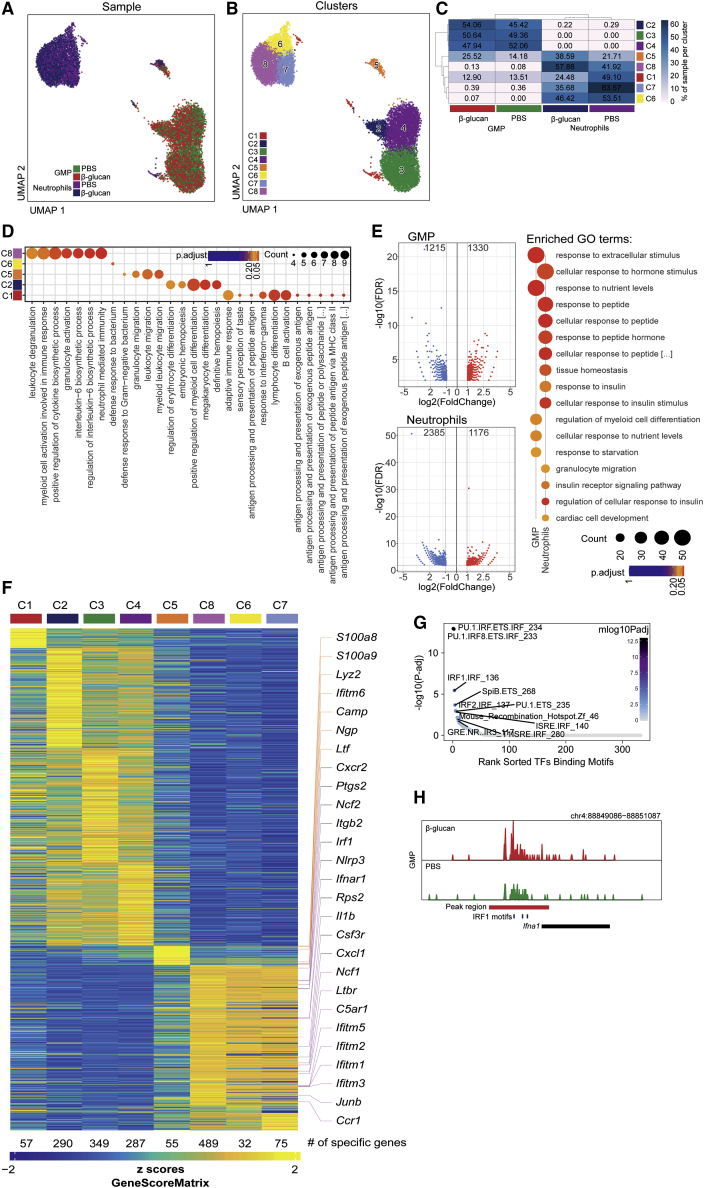
To determine whether epigenetic rewiring is involved in trained granulopoiesis, we performed single-cell assay for transposase-accessible chromatin with high-throughput sequencing (scATACseq) to determine chromatin accessibility in splenic neutrophils and BM GMPs from mice that were trained by pre-treatment with β-glucan, or treated with PBS control 7 days earlier. Two-dimensional Uniform Manifold Approximation and Projection (UMAP) of 3,702, 4,433, 2,256, and 2,992 cells in the PBS-GMP, β-glucan-GMP, PBS-neutrophil and β-glucan-neutrophil groups, respectively, on the basis of genome-wide tile matrices using 500 bp bins and Louvain clustering, partitioned GMPs and neutrophils into 8 clusters (C1–C8): C2–C4 comprised GMPs, C6–C8 comprised neutrophils, and C1 and C5 included both neutrophils and GMPs (Figures 5A–5C). C5 and C8 were enriched upon induction of β-glucan-mediated trained immunity (Figure 5C). Gene ontology (GO) enrichment results of cluster-specific marker genes based on gene activity scores revealed that the C5 and C8 subpopulations were characterized by enrichment of several pathways related to granulocyte activation (Figure 5D). Differential accessibility analysis, based on MACS2-defined peak regions for GMPs and neutrophils from β-glucan-treated mice compared with the respective cells from control-treated mice, revealed differentially accessible regions (DARs) in GMPs and neutrophils upon induction of trained immunity (Figure 5E). The top 10 significantly enriched GO terms in GMPs and neutrophils, identified on the basis of genes annotated to regions more accessible due to β-glucan-induced training, not only revealed terms such as “regulation of myeloid cell differentiation,” but also cell metabolism-related terms (Figure 5E), consistent with the findings of the RNA sequencing results from TANs (Figure 2C) and with the previously described involvement of immunometabolic pathways in the induction of trained immunity (Arts et al., 2016; Ieronymaki et al., 2019).
Figure 5.
Epigenetic Rewiring of Trained Granulopoiesis
Splenic neutrophils and BM GMPs were sorted from mice that were treated with β-glucan or with PBS 7 days earlier and scATACseq was performed.
(A and B) Two-dimensional UMAP representation of 13,383 cells, on the basis of genome-wide tile matrices of 500 bp bins colored, according to (A) sample origin and (B) results of Louvain clustering.
(C) Heatmap visualization of the distribution of cells from the four different samples (GMPs and neutrophils from PBS-treated or β-glucan-treated mice) within each of the identified clusters, normalized for the number of cells per sample in the dataset.
(D) GO enrichment results of cluster-specific marker genes determined on the basis of gene activity scores (Bonferroni-corrected p value cut-off = 0.1).
(E) Volcano plots displaying differential accessibility analysis results based on MACS2-defined peak regions for GMP and neutrophils from β-glucan-treated mice as compared with the respective cells from PBS-treated mice (FDR ≤ 0.01 and abs(Log2FC) ≥ 1). Top 10 significantly enriched GO terms sorted by GeneRatio identified on the basis of genes annotated to regions more accessible due to β-glucan treatment are shown on the side (Bonferroni-corrected p value cut-off = 0.1).
(F) Heatmap visualization of gene activity scores of cluster-specific marker genes (FDR ≤ 0.01, log2FC ≥ 1). Selected genes are indicated.
(G) Visualization of transcription factor (TF) binding motif enrichment analysis results for the β-glucan specifically accessible regions in GMPs by using the homer TF motif database.
(H) Genome browser track showing a DAR in proximity to the Ifna1 gene locus and the IRF1 binding motifs within this region.
See also Figure S5.
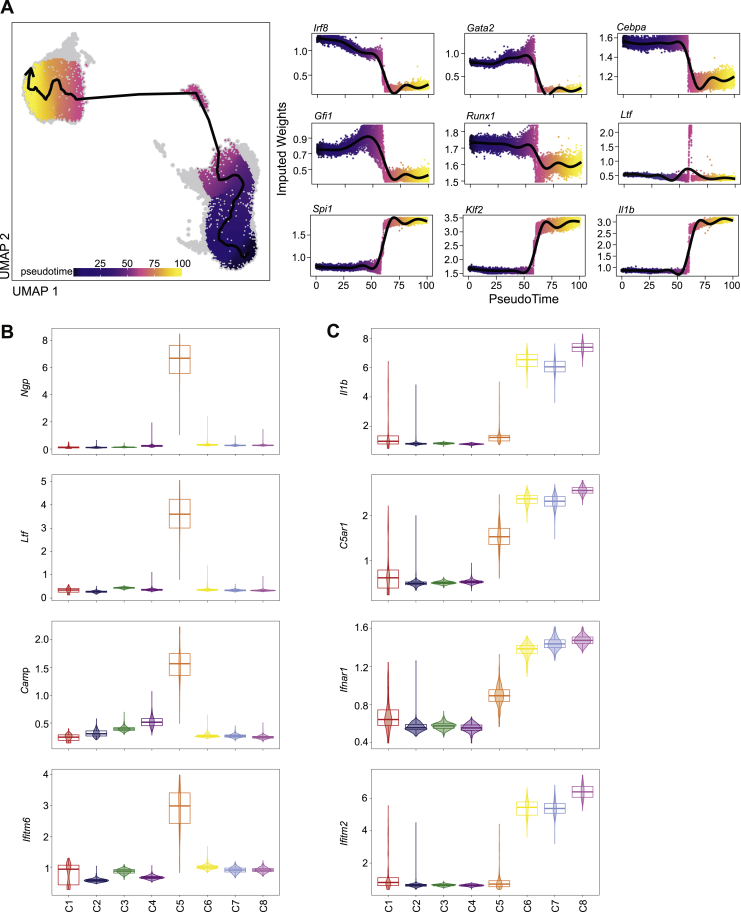
Cells clustering to C5, enriched by trained immunity, comprised both GMPs and neutrophils, suggesting that this cluster represents a transition cluster associated with trained granulopoiesis. A cellular trajectory spanning from GMPs over C5 to neutrophils revealed corresponding changes in the accessibility of key genes of the respective cellular stages in granulopoiesis, as recently identified by Evrard et al. (2018) (Figure S5A). C5 comprises cells at the transition of GMPs to neutrophils, as indicated by analysis of GMP-specific transcription factors such as Irf8 and Gata2 and of neutrophil-specific genes such as Spi1 (PU.1) and Il1b (Figure S5A). C5 revealed enhanced chromatin accessibility of secondary granule genes Ngp (neutrophilic granule protein), encoding NGP protein, which was previously identified to exert anti-metastastic activity in Gr1+CD11b+ cells (Boutté et al., 2011), Camp (cathelicidin anti-microbial peptide) and Ltf (lactoferrin) (Figures 5F and S5B). C5 is therefore reminiscent of differentiating neutrophils, as recently identified (Xie et al., 2020).
Figure S5.
scATACseq Analysis, Related to Figure 5
(A) Two-dimensional UMAP representation of 13383 cells, based on genome-wide tile matrices of 500 bp bins, colored by pseudo-time values, as determined by trajectory analysis (left). The arrow indicates the approximated differentiation path. Gene activity scores of selected key genes of granulopoiesis (‘GMP-genes’: Irf8, Gata2, Cebpa; ‘pre-neutrophils / immature neutrophils-genes’: Gfi1, Runx1, Ltf; ‘mature neutrophils-genes’: Spi1, Klf2, Il1b, as previously defined (Evrard et al., 2018) are visualized as a function of the pseudo-time as dot plots (right).
(B) Violin plots showing gene activity scores of Ngp, Ltf, Camp, and Ifitm6 across the identified clusters.
(C) Violin plots showing gene activity scores of Il1b, C5ar1, Ifnar1, and Ifitm2 across the identified clusters.
The neutrophil cluster C8 displayed enhanced chromatin accessibility in the genes of the ROS-producing factors Ncf1 and Ncf2 (Figure 5F), consistent with the role of Ncf1 and ROS production in mediating the anti-tumor effect of trained neutrophils (Figure 2I). Focusing on C8 cluster marker genes revealed a pro-inflammatory signature (enhanced chromatin accessibility in regions, such as Il1b, Csf3r, C5ar1, and Cxcr1) as well as a type I IFN signaling-related signature (enhanced accessibility of Ifnar1, Irf1, Ifitm1, Ifitm2, and Ifitm3) (Figures 5F and S5C). Transcription factor motif enrichment analysis for the open regions mediated by β-glucan-induced trained immunity revealed IRF1 motifs among the top enriched motifs in the DARs of the GMPs (Figure 5G). A genome browser track showing a DAR in proximity to the Ifna1 gene locus and the IRF1 binding motifs within this region in trained and non-trained GMPs is shown in Figure 5H. Together, transcriptomic and single-cell epigenomic analysis revealed that β-glucan-induced training is associated with enhanced IFN-related signaling in granulopoiesis.
Type I IFN Signaling Mediates the Anti-tumor Activity of Trained Neutrophils
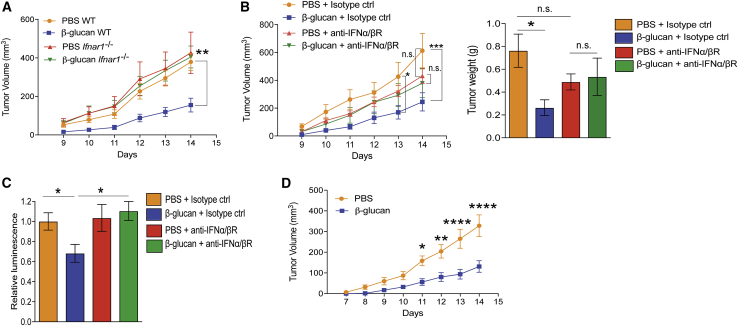
Our findings so far suggested the involvement of type I IFN in inducing trained granulopoiesis. To investigate whether type I IFN signaling contributes to the anti-tumor neutrophil phenotype that is elicited upon induction of trained innate immunity, we isolated neutrophils from Ifnar1 deficient (Ifnar1−/−) mice (i.e., mice lacking type I IFN receptor function) that were pre-treated with either β-glucan or PBS control and transferred them, together with B16-F10 melanoma cells, into untreated WT mice. The tumor-suppressive effect of trained neutrophils was abolished when neutrophils from Ifnar1−/− mice were transferred (Figure 6A), suggesting that cell-intrinsic responsiveness to type I IFN signaling is critical for trained granulopoiesis in the context of cancer.
Figure 6.
Type I IFN Signaling Promotes the Anti-tumor Activity of Trained Granulopoiesis
(A) WT or Ifnar1−/− mice were injected with β-glucan or PBS, and after 7 days splenic neutrophils were isolated and adoptively transferred together with B16-F10 cells into WT recipients. Tumor volume was determined (n = 9–13 mice per group).
(B and C) WT mice were injected with a monoclonal antibody against the receptor for IFNα/β (anti-IFNα/βR) or isotype control one day before and on the same day that β-glucan or PBS was administered. Seven days after the second injection, splenic neutrophils were isolated and were (B) adoptively transferred together with B16-F10 cells into WT recipients, or (C) were co-cultured with luciferase-expressing B16-F10 cells at 100/1 neutrophil/tumor cell ratio for 24 h. In (B), (left) tumor volume and (right) tumor weight at the end of the experiment are shown (n = 7–12 mice per group). In (C), survival of tumor cells was assessed by measuring luminescence. Luminescence is expressed in relation to the PBS + isotype control group, set as 1 (n = 10 per group).
(D) WT mice were injected with β-glucan or PBS, and after 7 days splenic neutrophils were isolated and systemically administered to mice that were inoculated with B16-F10 tumors 5 days earlier. Tumor volume is shown (n = 10 mice per group).
Data are presented as mean ± SEM. n.s., non-significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Additionally, WT mice were treated with a neutralizing antibody against the receptor for IFNα/β, (IFNα/βR) or with isotype control one day before and on the same day of β-glucan or PBS administration. Seven days after the second injection, splenic neutrophils were isolated and adoptively transferred, together with B16-F10 melanoma cells, into WT mice. Although neutrophils from β-glucan-trained mice that received isotype control antibody mediated enhanced inhibition of tumor growth in recipient mice, as compared with neutrophils from non-trained mice that received isotype control antibody, the anti-tumor effect of trained neutrophils was abrogated by anti-IFNα/βR treatment. Specifically, upon IFNα/βR blockade, compared with non-trained neutrophils, trained neutrophils did not show enhanced tumor-suppressive activity in recipient mice (Figure 6B). Consistently, the enhanced capacity of trained neutrophils from mice treated with β-glucan to kill tumor cells, as compared with that of non-trained neutrophils, was abrogated by inhibition of IFNα/βR signaling (Figure 6C).
To explore the anti-tumor potential of trained granulopoiesis in a therapeutic setting, we systemically administered trained neutrophils after tumor inoculation. Neutrophils from donor mice that were trained by pre-treatment with β-glucan or control-treated mice were transferred retro-orbitally into tumor-bearing recipient mice 5 days after tumor inoculation. Systemic administration of trained neutrophils resulted in inhibition of tumor growth compared with that in mice that received non-trained neutrophils (Figure 6D), thus establishing the therapeutic potential of trained immunity.
Discussion
The recognition of the important role of tumor immunity in cancer has led to the introduction of immunotherapeutic strategies (Fridman et al., 2017; Weiden et al., 2018). Cells of the myeloid lineage are a major component of the tumor immune cell infiltrate (Balkwill et al., 2005; Broz et al., 2014; Gabrilovich et al., 2012); yet, current immunotherapies target exclusively the adaptive arm of immunity (Fridman et al., 2017). The tumor and its microenvironment usually alter the phenotype of myeloid cells toward a phenotype that promotes tumor growth (Gabrilovich et al., 2012). Although the default scenario is that cancer hijacks innate immunity and granulopoiesis to promote tumor growth, innate immune cells do have the potential to exert anti-tumor activities (Galdiero et al., 2018; Murray, 2018; Ponzetta et al., 2019; Powell and Huttenlocher, 2016). Therefore, immunotherapies aiming at reversing the pro-tumor phenotype or promoting the anti-tumor phenotype in innate immune cells, represent an attractive anti-tumor approach that could act synergistically to current immunotherapies targeting adaptive immunity. We demonstrate that β-glucan-induced trained immunity might represent a novel approach to cancer immunotherapy by modulating granulopoiesis toward anti-tumor activity. The anti-tumor effects of trained immunity were transmissible to recipient naive mice not only via adoptive transfer of trained neutrophils, but also via transplantation of BM cells from trained mice to non-trained recipients. Thus, the anti-tumor effects of trained granulopoiesis are initiated by alterations of long-lived BM progenitors, in line with recent studies that trained immunity is initiated in BM progenitor cells (Christ et al., 2018; Kaufmann et al., 2018; Mitroulis et al., 2018).
To date, cells of the monocytic lineage (monocytes/macrophages) were assumed as the main targets of trained immunity (Kleinnijenhuis et al., 2012; Netea et al., 2016; Netea and van der Meer, 2017; Saeed et al., 2014). Here, we demonstrate that, at least in the context of anti-tumor immunity, neutrophils, and granulopoietic progenitors are major cellular effectors of β-glucan-induced trained immunity. Indeed, the anti-tumor effect of β-glucan-induced trained immunity was mediated by qualitative changes in neutrophils, rather than in cells of the monocytic lineage. Moreover, the GMP transcriptome upon β-glucan-induced trained immunity was consistent with skewed differentiation to neutrophils compared with that of other myeloid cell types. We also showed previously that hematopoietic stem cell transplantation from β-glucan-trained mice to naive recipients gives rise to increased frequency of neutrophils in the circulation of the latter (Mitroulis et al., 2018). Our findings together with a recent study showing that changes in hematopoietic progenitors and in granulocytes are integral to BCG-induced trained immunity in humans (Cirovic et al., 2020) point to the emerging and hitherto underappreciated role of neutrophils as effectors of trained immunity.
The trained-immunity-mediated induction of a tumor-suppressive phenotype in neutrophils was associated with training of granulopoiesis mediated by type I IFN signaling. This is in line with previous studies showing that type I IFNs play a central role in granulopoiesis and promote an anti-tumor phenotype in neutrophils (Andzinski et al., 2016). Mice lacking IFN-β displayed defective maturation of BM hematopoietic progenitors and reduced blood neutrophil counts, accompanied by more aggressive tumor growth (Deonarain et al., 2003). Our scATACseq analysis provided additional evidence for trained immunity-induced epigenetic rewiring of granulopoiesis toward an anti-tumor phenotype and corroborated the experimentally demonstrated IFN- and ROS-related mechanisms therein. It should be noted that our findings only show a correlation between these two major mechanisms (type I IFN signaling and ROS production) involved in the anti-tumor effects of trained granulopoiesis; however, we cannot exclude a causal link, given that type I IFN priming was previously shown to enhance ROS production in neutrophils (Wright et al., 2008). Furthermore, the neutrophil heterogeneity observed by our single-cell analysis is in agreement with the existence of different neutrophil subsets, as recently identified; additionally, the existence of a distinct neutrophil subset exhibiting a type I IFN signature was recently shown (Xie et al., 2020; Zilionis et al., 2019).
Neutrophil ROS might exert either pro-tumorigenic (immunosuppressive) or anti-tumorigenic (tumor-killing) effects of neutrophils (Granot et al., 2011; Rice et al., 2018; Yan et al., 2014). Our present findings suggest an important role of ROS production in neutrophils in the anti-tumor effect of trained immunity, given that NCF1 deficiency abrogated the anti-tumor effect of β-glucan-trained neutrophils. Thus, ROS can account, at least in part, for the anti-tumor activity of trained neutrophils, although we cannot exclude that ROS might additionally contribute to the initial training of granulopoiesis. Moreover, it is conceivable that, besides a potential direct anti-tumor effect, adoptively transferred trained neutrophils might exert further indirect actions on other immune or stromal cells within the tumor microenvironment. Although the anti-tumor effect of β-glucan-induced training could be exerted in the absence of adaptive immunity, it is conceivable that trained immunity might also potentiate the crosstalk between neutrophils and adaptive immune cells in anti-tumor immunity in WT mice. In keeping with this notion, we found that the pathway of antigen presentation was upregulated in TANs from trained mice. Moreover, a recent paper demonstrated that neutrophils mediate a novel IFN-γ-dependent anti-tumor pathway, which is associated with a subset of CD4−CD8− unconventional αβ T cells (Ponzetta et al., 2019).
We observed inhibition of tumor growth by systemic transfer of trained neutrophils into already tumor-bearing mice. As granulocyte transfusion is currently considered as a therapy in humans with neutropenia (Estcourt et al., 2016), it is conceivable that cancer patients could receive as an adjuvant immunotherapy granulocytes from normal donors after induction of trained immunity in the latter. This scenario should be addressed in a future clinical study. In this regard, neutrophils from some healthy donors (but not from cancer patients) naturally have potent cancer-killing activity against different human cancer cell lines (Yan et al., 2014), although the trained state of these neutrophils was not addressed. Future studies should also address the possibility that therapeutically administered β-glucan or trained granulocytes can be used synergistically with checkpoint inhibitors in cancer therapy, as performed in a current clinical trial (Uhlik et al., 2020).
Although agonists of trained immunity, such as BCG and β-glucan, have been used against cancer in preclinical studies and in selected patient groups (Hersh et al., 1977; Liu et al., 2009; Xiang et al., 2012; Zhang et al., 2018), the underlying mechanisms were incompletely understood. Intriguingly, neutrophils have been implicated as mediators of the anti-tumor effect of BCG in a murine model of bladder cancer (Suttmann et al., 2006). Our study is the first to link the anti-tumor actions of β-glucan to trained immunity. We show here that the innate immune training and rewiring of granulopoiesis underlies the anti-tumor effect of β-glucan and perhaps other agonists of trained immunity. Our findings clearly suggest that harnessing trained granulopoiesis represents a promising avenue for neutralizing the hijacking of innate immunity by cancer. Therefore, innate immune training merits further investigation as an adjuvant tumor immunotherapy.
Limitations of the Study
Our study shows that β-glucan-induced trained immunity inhibits tumor growth and suggests that type I IFN signaling is involved in trained granulopoiesis and in mediating the rewiring of neutrophils toward an anti-tumor phenotype. Neutrophil-derived ROS were integral to the anti-tumor effects of β-glucan-induced trained immunity. However, our study also has limitations. At this point, we cannot exclude that ROS might also contribute to the initial training of granulopoiesis, besides promoting the tumor-killing activity of trained neutrophils (i.e., ROS might constitute both an outcome of trained granulopoiesis and a reinforcing stimulus for its maintenance). In addition, it is currently uncertain whether type I IFN signaling and ROS are causally linked in mediating the anti-tumor activity of trained granulopoiesis and, specifically, whether type I IFN signaling is involved in ROS upregulation in trained neutrophils; this interesting possibility requires additional mechanistic investigation in future studies. Our translational findings, derived from injectable tumor models, clearly suggest that trained granulopoiesis might bear potential as an adjuvant tumor immunotherapy. Addressing the role of trained innate immunity in autochthonous cancer models could provide complementary fundamental and applied insights pertinent to the role of trained immunity in carcinogenesis and therefore merits future investigation. In conclusion, future studies in pre-clinical models might further expand our current mechanistic and translational understanding of the anti-tumor potential of trained immunity.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Cell Lines | ||
| Lewis lung carcinoma (LLC1) | ATCC | Cat#CRL-1642; RRID: CVCL_4358 |
| B16-F10 | ATCC | Cat#CRL-6475; RRID: CVCL_0159 |
| B16-F10-Luc | Creative Biogene | Cat#CSC-RR0234 |
| Antibodies | ||
| Rat anti-mouse CD11b | Biolegend | Cat#101216; RRID: AB_312799 |
| Rat anti-mouse Sca1 (Ly6-A/E) | Biolegend | Cat#122514; RRID: AB_756199 |
| Mouse Lineage Antibody Cocktail | BD Biosciences | Cat#558074; RRID: AB_1645213 |
| Biotin Mouse Lineage Panel | BD Biosciences | Cat#559971; RRID: AB_10053179 |
| Rat anti-mouse cKit (CD117) | BD Biosciences | Cat#553355; RRID: AB_394806 |
| Rat anti-mouse CD16/CD32 | Biolegend | Cat#101332; RRID: AB_2650889 |
| Rat anti-mouse CD34 | Thermo Fisher Scientific | Cat#11-0341-82; RRID: AB_465021 |
| Rat anti-mouse CD45 | Biolegend | Cat#103132; RRID: AB_893340 |
| Rat anti-mouse CD4 | Biolegend | Cat#116006; RRID: 313691 |
| Rat anti-mouse CD8 | Biolegend | Cat#100722; RRID: AB_312761 |
| Rat anti-mouse Ly6g | Biolegend | Cat#127624; RRID: AB_10640819 |
| Biotin anti-mouse Ly6g | Biolegend | Cat# 127604; RRID: AB_1186108 |
| Rat anti-mouse anti-Ly6c | Biolegend | Cat#128032; RRID: AB_2562178 |
| Biotin anti-mouse Ly6c | Biolegend | Cat#128004; RRID: AB_1236553 |
| Rat anti-mouse F4/80 | Biolegend | Cat#123109 and 123116; RRID: AB_893498 and 893481 |
| Rat anti-mouse Gr-1 | Biolegend | Cat#108445; RRID: AB_2562903 |
| Rat anti-mouse CD115 | Biolegend | Cat#135523; RRID: AB_2566459 |
| Rat anti-mouse CD169 | Biolegend | Cat#142417; RRID: AB_2565640 |
| Rat anti-mouse B220 | Biolegend | Cat#103207; RRID: AB_312992 |
| Armenian hamster anti-mouse CD11c | Biolegend | Cat#117310; RRID: AB_313779 |
| Rat anti-mouse CD317 | Biolegend | Cat#127023; RRID: AB_2687109 |
| Rat anti-mouse CD90.2 | Biolegend | Cat#105319; RRID: AB_493724 |
| Rat anti-mouse CD19 | Biolegend | Cat#115527; RRID: AB_493734 |
| Mouse anti-mouse NK1.1 | Biolegend | Cat#108729; RRID: AB_2074426 |
| Rat anti-mouse anti-IFNα | R&D Systems | Cat#22100-3; RRID: AB_884210 |
| Rat anti-mouse IFNγ | Biolegend | Cat#505808; RRID: AB_315402 |
| Ultra-LEAF Purified anti-mouse IFNAR-1 Antibody | Biolegend | Cat#127321; RRID: AB_11150409 |
| Ultra-LEAF™ Purified Mouse IgG1, κ Isotype Ctrl Antibody | Biolegend | Cat#400166; RRID:AB_11146992 |
| Chemicals, Enzymes, and Buffers | ||
| Beta-glucan peptide (BGP) | Invivogen | Cat#tlrl-bgp |
| PMA | Sigma | Cat#P1585 |
| Ionomycin | Sigma | Cat#I0634 |
| Brefeldin A | Biolegend | Cat#420601 |
| Deoxyribonuclease I from bovine pancreas | Sigma-Aldrich | Cat#D5025 |
| Collagenase D | Roche | Cat#11088866001 |
| 10X RBC Lysis Buffer | eBioscience | Cat#00-4300-54 |
| Puromycin | Invivogen | Cat#ant-pr1-1 |
| N-Acetyl-L-cysteine | Sigma | Cat#A7250 |
| Nonidet P40 Substrate | Sigma | Cat#74385 |
| Nuclei Buffer | 10x Genomics | Cat#2000153 |
| Dynabeads MyOne SILANE | 10x Genomics | Cat#2000048 |
| SPRIselect | Beckman Coulter | Cat#B23318 |
| Critical Commercial Assays | ||
| CellROX™ Green Flow Cytometry Assay Kit | Thermo Fisher | Cat#C10492 |
| Foxp3/Transcription Factor Buffer Set | eBioscience | Cat#00-5523-00 |
| Intracellular Fixation & Permeabilization Buffer | eBioscience | Cat#88-8824-00 |
| iScript cDNA Synthesis Kit | BioRad | Cat#1708891 |
| High-Capacity RNA-to-cDNA Kit | Applied Biosystems | Cat#4387406 |
| SsoFast EvaGreen Supermix | BioRad | Cat#1725202 |
| Fast SYBR Green Master Mix | Applied Biosystems | Cat#4385612 |
| Anti-Biotin MicroBeads | Miltenyi Biotec | Cat#130-090-485 |
| RNeasy Plus Micro Kit | QIAGEN | Cat#74034 |
| MycoAlertTM Mycoplasma Detection Kit | Lonza | Cat#LT07-318 |
| SMARTer Ultra Low Input RNA for Illumina Sequencing | Takara Bio | Cat#634828 |
| NEBNext Ultra DNA Library Prep Kit for Illumina | New England Biolabs | Cat#E7370L |
| Luciferase assay system | Promega | Cat#E1500 |
| Chromium Next GEM Chip H Single Cell Kit v1.1 | 10x Genomics | Cat#1000162 |
| Chromium Next GEM Single Cell ATAC Library & Gel Bead Kit v1.1 | 10x Genomics | Cat#1000176 |
| NGS High Sensitivity Fragment Analysis Kit | Agilent | Cat#DNF-474 |
| NextSeq 500/550 High Output Kit v2.5;150 Cycles | Illumina | Cat#20024907 |
| Deposited Data | ||
| RNA sequencing data | This paper | GEO: GSE139450, GSE139451, GSE139452, and GSE139456 |
| Single cell ATAC sequencing data | This paper | GEO: GSE152353 |
| Experimental models: Organisms/Strains | ||
| Mouse: C57BL/6 | Janvier Labs | C57BL/6JRj |
| Mouse: C57BL/6 | The Jackson Laboratory | Stock#000664 |
| Mouse: C57BL/6-CD45.1 B6.SJL-PtprcaPepcb/BoyJ | The Jackson Laboratory | Stock#002014 |
| Mouse: B6(Cg)-Ncf1m1J/J | The Jackson Laboratory | Stock#004742 |
| Mouse: B6.129S7-Rag1tm1Mom/J | The Jackson Laboratory | Stock#002216 |
| Mouse: B6(Cg)-Ifnar1tm1.2Ees/J | The Jackson Laboratory | Stock#028288 |
| Oligonucleotides | ||
| qPCR primers | This paper | See Table S5 |
| Software, Algorithms and Protocols | ||
| GraphPad Prism 6 | Graphpad Software | N/A |
| Ingenuity Pathway Analysis | QIAGEN | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/ |
| GSEA software | Broad Institute | http://software.broadinstitute.org/gsea/index.jsp |
| Morpheus | Broad Institute | https://software.broadinstitute.org/morpheus/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| GSNAP | Wu and Nacu, 2010 | http://research-pub.gene.com/gmap/ |
| Ensembl gene annotation version 81 | EMBL-EBI | https://www.ebi.ac.uk/ |
| Ensembl Gene IDs using Ensembl Biomart (Release 96) | EMBL-EBI | https://www.ebi.ac.uk/ |
| featureCounts | Liao et al., 2014 | http://bioinf.wehi.edu.au/featureCounts/ |
| FlowJo version 10 | Tree Star | https://www.flowjo.com/solutions/flowjo |
| NovoExpress® software | ACEA Biosciences | https://www.aceabio.com |
| Cell Ranger ATAC version 1.2.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-atac/software/overview/welcome |
| R package ChIPseeker v1.22.1 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/ChIPseeker.html |
| R package clusterProfiler v3.14.3 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| Chromium Single Cell ATAC Reagent Kits protocol | 10xGenomics | Cat#CG000168 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Triantafyllos Chavakis (Triantafyllos.Chavakis@uniklinikum-dresden.de).
Materials Availability
This study did not generate any unique reagents.
Data and Code Availability
Data are available upon request to the Lead Contact. Sequencing data are available at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under the accession numbers GSE139450, GSE139451, GSE139452, GSE139456 and GSE152353.
Experimental Model and Subject Details
Cell lines
The Lewis lung carcinoma cell line (LLC1) and the melanoma cell line B16-F10 were purchased from the ATCC. The B16-F10 melanoma cell line that expresses the luciferase reporter (B16-F10-Luc) was obtained from Creative Biogene.
Mice
C57BL/6 mice were purchased from Jackson Laboratories and Janvier Laboratories. Ncf1m1j/m1j mice (Stock# 004742) with deficiency in NCF1 due to a mutation in the Ncf1 gene (Aachoui et al., 2013; Baptista et al., 2016; Huang et al., 2000; Maltez et al., 2015) referred to as NCF1-deficient mice hereafter, Ifnar1−/− mice (Stock# 028288) and respective WT C57BL/6 mice, B6/SJL (CD45.1) mice (Stock# 002014) and Rag1−/− mice (Stock# 002216) were from Jackson Laboratories. Mice were housed under specific pathogen-free conditions on a standard 12/12 h light/dark cycle and were used at the age of 9-11 weeks. Food and water were provided ad libitum. Animal experiments were approved by the Landesdirektion Sachsen, Dresden Germany, the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania and the Institutional Committee of Protocol Evaluation of the Biomedical Research Foundation of the Academy of Athens in conjunction with the related veterinary authority of the Region of Attika, Greece.
Method Details
Mouse experiments
Induction of solid tumors was performed as previously described (Alissafi et al., 2018). Briefly, mice were injected subcutaneously (s.c.) with 3 × 105 B16-F10 melanoma or LLC1 cells into the right flank. To examine the role of trained innate immunity in tumor development, mice were pre-treated with a single dose of 1 mg of β-glucan from Trametes versicolor (Invivogen) in PBS or with PBS vehicle alone (control) i.p., followed by tumor cell injection 7 or 28 days later. Volume of palpable tumors was monitored and was calculated using the equation (length x width2) / 2. Tumor weight and flow cytometry analysis were performed at the end of the experiment (14 days after tumor inoculation). For adoptive transfer experiments, mice were injected s.c. with 3 × 105 B16-F10 cells together with 106 magnetically isolated splenic neutrophils or monocytes from donor mice (Alissafi et al., 2018; Shaul et al., 2016; Sierra et al., 2017); donor mice received β-glucan or PBS i.p. 1 or 7 days before neutrophil isolation or 7 days before monocyte isolation. Inhibition of type I IFN signaling was performed by i.p. injection of mice with a neutralizing anti-mouse antibody against the receptor for IFNα/β (clone MAR1-5A3, Biolegend) or an isotype control (clone MOPC-21, Biolegend) one day before and on the same day (500 μg / dose) of treatment of mice with β-glucan or PBS. Seven days after the latter injection, neutrophils were isolated and used in adoptive transfer experiments, as described above. In another experiment, mice were injected s.c. with 3 × 105 B16-F10 cells and 5 days later they received retro-orbitally 106 magnetically isolated splenic neutrophils from donor mice that had received β-glucan or PBS i.p. 7 days before neutrophil isolation. To generate BM chimeras, a total of 2 × 106 CD45+ BM cells from B6/SJL (CD45.1) mice that were pre-treated with a single i.p. injection of control vehicle (PBS) or β-glucan 7 days earlier, were transplanted into lethally irradiated (9 Gy) WT mice (CD45.2). Six weeks after transplantation, recipient mice were inoculated with B16-F10 melanoma cells and mice were followed for 14 days after tumor inoculation. Mice were excluded from experiments if pre-established exclusion criteria were fulfilled, for instance, tumor ulceration as an animal protocol-defined endpoint or, in cases that no tumor growth was observed (Chen et al., 2019; Michod et al., 2009; Voltarelli et al., 2017).
To isolate neutrophils for adoptive transfer and co-culture experiments, splenocytes were incubated with a biotin anti-mouse Ly6g antibody (clone 1A8; Biolegend) followed by anti-biotin microbeads (Miltenyi Biotec). Neutrophils were then positively selected on a magnetic field according to the manufacturer’s instructions (MACS separation columns, Miltenyi Biotec). To isolate splenic monocytes, neutrophils were removed from splenocyte preparation by negative selection for Ly6g+ cells and then positive selection for Ly6c+ (clone HK1.4; Biolegend) cells was performed. Purity of the magnetically isolated populations was >90%.
Flow cytometry and sorting
Cell analysis was performed by FACS Canto II (BD, Heidelberg, Germany) and cell sorting was performed using a FACS Aria cell sorter (BD, Heidelberg, Germany) (Chung et al., 2017; Kourtzelis et al., 2019; Mitroulis et al., 2017). Cell purity was above 95%. Single cell suspensions from tumors were prepared after incubation of the dissected tissue for 45 min at 37°C in RPMI culture medium containing 0.25 mg/mL DNase I (Sigma-Aldrich) and 1 mg/mL collagenase D (Roche). Mouse spleens and lymph nodes were homogenized and splenic single cell suspensions from spleen were prepared after erythrocyte lysis with red blood cell lysis buffer (eBioscience). For cell surface phenotype analysis, a lineage cocktail (Lin: anti-CD3e (clone 145-2C11), anti-CD11b (clone M1/70), anti-Gr-1 (anti-Ly6g/Ly6c; clone RB6-8C5), anti-B220 (clone RA3-6B2) and anti-TER119 (clone TER-119)), anti-Sca1 (clone E13-161.7), anti-cKit (clone 2B8), anti-CD16/CD32 (clone 93), anti-CD34 (clone RAM34), anti-CD45 (clone 30-F11), anti-CD4 (clone RM4-4), anti-CD8 (clone 53-6.7) anti-CD11b (clone M1/70), anti-Ly6g (clone 1A8), anti-Ly6c (clone HK1.4), anti-F4/80 (clone BM8), anti-Gr-1 (anti-Ly6g/Ly6c; clone RB6-8C5), anti-CD115 (clone AFS98), anti-CD169 (clone 3D6.112), anti-B220 (clone RA3-6B2), anti-CD11c (clone N418), anti-CD317 (BST2, PCDA-1; clone 927), anti-CD90.2 (clone 30-H12), anti-CD19 (clone 6D5), anti- NK1.1 (clone PK136), anti-IFNα (clone RMMA-1) and anti-IFNγ (clone XMG1.2) were used. Data analysis was performed using FlowJo (Tree Star) software or NovoExpress software (ACEA Biosciences).
Enrichment of Lin− cells was performed prior to sorting of GMP from BM by magnetic negative selection using MidiMACS Separator (Miltenyi Biotec). To this end, cells were incubated with a biotin mouse lineage panel (Biotin-Conjugated Mouse Lineage Panel, BD PharMingen) and subsequently with anti-Biotin MicroBeads (Miltenyi Biotec). GMP were sorted as Lin−c-Kit+Sca1−CD16/32+CD34+ cells and splenic neutrophils were sorted as CD11b+Ly6c−Ly6g+ cells.
To detect the presence of IFNγ in lymphocytes, single cell suspensions from tumors and lymph nodes were stained for cell surface markers, fixed and permeabilized using fixation / permeabilization buffer (Foxp3/Transcription Factor Buffer Set; eBioscience) and then stained with the respective antibodies. Cells were incubated with 50 ng/mL phorbol-12-myristate-13-acetate (Sigma), 1 μg/mL ionomycin (Sigma) and 5 μg/mL brefeldin A (Biolegend) for 3 h at 37°C prior to the addition of antibodies.
To detect the levels of IFNα in BM dendritic cells and macrophages, BM single-cell suspensions were stained for cell surface markers, fixed and permeabilized with eBioscience Intracellular Fixation & Permeabilization Buffer and stained for IFNα. Staining for pDCs (CD45+NK1.1−CD90.2−CD19−CD11c+CD11b−B220+BST2+) and CD169+ macrophages (CD45+Gr-1−CD115intF4/80+SSClowCD169+) was performed, as described (Chow et al., 2011; Macal et al., 2016). To determine intracellular levels of reactive oxygen species (ROS), single cell suspensions from tumors were stained with antibodies against surface markers and then were stained with Cell Rox Green flow cytometry assay kit (Thermo Fisher).
Cytotoxicity assay
Splenic neutrophils (isolated as described under ‘Mouse experiments’) were co-cultured with B16-F10 melanoma cells that express a luciferase reporter in culture medium containing 1 μg/mL puromycin for 24 h. Cells were then washed and lysed, and luminenscence was measured using a luciferase reporter assay system (Promega) on a Synergy HT multi-mode microplate reader (Biotek Instruments) according to the manufacturer’s instructions. To block reactive oxygen species, co-culture experiments were performed in the presence of 5mM N-acetyl-cysteine (Sigma).
RNA isolation and real time PCR
RNA isolation was performed using RNeasy Plus Micro Kit (QIAGEN) or TRIzol, according to manufacturer’s instructions. Complementary DNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad) or High-Capacity RNA-to-cDNA Kit (Applied Biosystems). qPCR was performed by using the SsoFast EvaGreen Supermix (BioRad) or Fast SYBR Green Master Mix (Applied Biosystems) and gene-specific primers for the ‘trained TAN1-like signature’ (Table S5) in a CFX384 Real time PCR detection system (BioRad). 18S was used as an internal control for normalization (Kourtzelis et al., 2019). Data were analyzed using the comparative (ΔΔCt) method. The ‘trained TAN1-like signature’ was defined as follows: Based on the RNaseq analysis, we identified genes that were significantly upregulated in TAN from β-glucan–trained as compared to the control-treated mice. The ‘trained TAN1-like signature’ represented the top 20 genes from the intersection of the upregulated genes in TANs from trained mice and the previously described TAN1-like gene dataset (Shaul et al., 2016).
RNA sequencing
RNA isolation was performed as previously described (Mitroulis et al., 2018). Briefly, isolated RNA was subjected to the amplification workflow of the SMARTer Ultra HV v2 kit (Takara Bio). Amplified cDNA was successively converted into short read sequencing libraries using the NEBnext Ultra DNA library preparation chemistry (New England Biolabs). Libraries were equimolarly pooled and sequenced on an Illumina HiSeq 2500, resulting in 27.9 – 59.9 million single end reads per library.
Single-cell ATAC sequencing
Nuclei isolation for single-cell ATAC sequencing was performed according to the protocol of 10x Genomics. Briefly, cells were sorted into V bottom plate, centrifuged at 300 rcf for 5 min at 4°C. Chilled Lysis buffer (2.5mM Tris-HCl (pH 7.4), 2.5mM NaCl, 0.9mM MgCl2, Tween-20 0.025%, Nonidet P40 Substrate (Sigma) 0.025%, Digitonin 0.0025%, BSA 0.25%, nuclease free water) was added to the pellet and cells were incubated on ice for 2 min. Cells were washed once with Wash buffer (2.5mM Tris-HCl (pH 7.4), 2.5mM NaCl, 0.9mM MgCl2, Tween-20 0.025%) and concentrated by centrifugation at 500 rcf for 5 min at 4°C; after removing the supernatant, 7 μl 1x Nuclei Buffer (10x Genomics) was added. 1 μl of the nuclei suspension was taken for evaluation (nuclei quality control and determining nuclei concentration). Nuclei suspension was then used according to the Chromium Single Cell ATAC Reagent Kits protocol (10x Genomics). 5 μl of the nuclei suspension was mixed with the tagmentation mix and incubated for one h at 37°C. After finishing the tagmentation, the still intact nuclei were mixed with a barcoding mix and loaded into a 10x chip H (10x Genomics, Chromium Next GEM Chip H Single Cell Kit v1.1; Chromium Next GEM Single Cell ATAC Library & Gel Bead Kit v1.1) together with barcoded beads and partitioning oil and encapsulated using the Chromium controller. The gel emulsion was transferred into a PCR tube to perform 12 cycles of amplification in a thermocycler. The gel emulsion containing barcoded DNA was broken, purified (10x Genomics, Dynabeads MyOne SILANE) and subjected to a final index PCR for 11 cycles. After size selection (0.4x/1.2x volume of beads) (Beckman Coulter SPRIselect), the library was examined on a fragment analyzer (Agilent, NGS High Sensitivity Fragment Analysis Kit) for its quality and quantity and sequenced on an Illumina NextSeq 500 (Illumina NextSeq 500/550 High Output Kit v2.5;150 Cycles) in 72 PE mode (R1 72, R2 72, I1 8, I2 16) at an average depth of 5400 fragments/cell. More information on the library prep can be found at the site https://www.10xgenomics.com/resources/support-documentation/.
Quantification and Statistical Analysis
RNA sequencing analysis
Analysis was performed as previously described (Mitroulis et al., 2018). Briefly, FastQC (http://www.bioinformatics.babraham.ac.uk/) was used to perform a basic quality control on the resulting reads. As an additional control, library diversity was assessed by redundancy investigation in the reads. Alignment of the reads to the mouse reference (mm10) was done with GSNAP (Wu and Nacu, 2010) and Ensembl gene annotation version 81 was used to detect splice sites. The uniquely aligned reads were counted with featureCounts (Liao et al., 2014) and the same Ensembl annotation. Normalization of the raw read counts based on the library size and testing for differential expression between conditions was performed with the DESeq2 R package (Love et al., 2014). Genes, which have an adjusted p value (padj) < 0.05 and counts > 50 were considered as differentially expressed. Pathway and upstream regulator analysis of gene lists containing significantly differentially expressed genes (padj < 0.05, log2FC < -0.3 and > 0.3) was done with Ingenuity Pathway Analysis (IPA, QIAGEN Redwood City, www.qiagen.com/ingenuity). Top canonical pathways derived from IPA are shown. Morpheus software (Broad Institute) was used to generate heatmaps. ggplot2_v1.0.1 (Wickham, 2009) and GOplot (Walter et al., 2015) were used to generate volcano plots and circos plot, respectively. To perform gene set enrichment analysis (GSEA), gene sets were ranked by taking the -log10 transform of the p value and signed as positive or negative based on the direction of fold change. GSEA pre-ranked analysis (1000 permutations, minimum term size of 15, maximum term size of 500) was then performed using the GSEA software (Broad Institute) (Musso et al., 2015). Annotated gene sets from Molecular Signatures Database (MSigDB) were used as input. Shown in heatmaps (Figures 2E, S2B, S2C, and 4F) are selected genes that were significantly regulated in our datasets, as assessed by our RNA sequencing analyses, and also included in the GO terms ‘ROS metabolic process’, ‘phagocytosis’, ‘MHC protein complex’ and ‘Cell response to type I IFN’. Significantly regulated genes that were present in both ‘ROS metabolic process’ and ‘phagocytosis’ GO terms or in both ‘ROS metabolic process’ and ‘MHC protein complex’ GO terms are presented only in the heatmap of Figure 2E as ‘ROS metabolic process’.
For TAN1 pathway overrepresentation analysis, data was extracted from GSE101584 (Shaul et al., 2016). In particular, Shaul et al. show in Table S4 of their paper genes with a greater than 10-fold change in expression between TAN1 and TAN2. The gene symbols listed by the study were first mapped to ILMN microarray probe IDs using the array design file GPL6885 that was provided as part of the GEO dataset GSE101584. These ILMN probe IDs were then mapped to Ensembl Gene IDs using Ensembl Biomart (Release 96) with ambiguously and non-mapping probes excluded, leading to retaining 92 Ensembl Gene IDs from the original gene set of Table S4 from Shaul et al. that were downregulated in TAN2 compared to TAN1. These 92 genes implicated as playing a role in the functionality of TAN1 were used as a gene set for GSEA. The GSEA analysis was performed as described above using the ranked-list of differentially expressed genes.
Single cell ATAC sequencing analysis
For data pre-processing Cell Ranger ATAC version 1.2.0 (https://support.10xgenomics.com/single-cell-atac/software/overview/welcome) was used to process raw sequencing data. ‘cellranger-atac count’ pipeline was used to align reads and generate single-cell accessibility counts for the cells. Reference genome file refdata-cellranger-atac-mm10-1.2.0 corresponding to mm10 was downloaded from the 10X Genomics website (https://support.10xgenomics.com/single-cell-atac/software/downloads/). This file was used as reference genome file for alignment and generating single-cell accessibility counts. Downstream analysis of the scATACseq data was performed using the R package ArchR v0.9.3 (Granja et al., 2020), following the developers' default recommendations, unless otherwise indicated. Arrow files were generated by reading in accessible read fragments produced by the ‘cellranger-atac count’ pipeline for each sample. After quality control to remove the contribution of low-quality cells and doublet removal, a tile matrix was created using 500-bp bins. A layered dimensionality reduction approach using Latent Semantic Indexing (LSI) and Singular Value Decomposition (SVD) on the previously created genome-wide tile matrix followed by Uniform Manifold Approximation and Projection (UMAP) was performed to visualize the data structure in the two-dimensional space. Subsequently, single-cell accessibility profiles were clustered using the Louvain clustering approach as implemented in the R package Seurat (Stuart et al., 2019). Gene activity scores were calculated based on the local accessibility of gene regions, which includes the promoter and gene body, and cluster-specific marker genes were identified based on these using the following cut-offs: FDR ≤ 0.01 & Log2FC ≥ 1 and presented in a heatmap. We used the R package clusterProfiler v3.14.3 to determine Gene Ontology (GO) terms enriched for the cluster-specific gene sets (Yu et al., 2012). Based on the gene activity scores, we further constructed a cellular trajectory spanning from GMP over cluster C5 to neutrophils using the addTrajectory function implemented in ArchR. Subsequently, open chromatin peaks were called based on pseudo-bulk replicates of the GMP and neutrophils of control (PBS)- or β-glucan-treated mice (designated PBS-GMP, β-glucan-GMP, PBS-neutrophil and β-glucan-neutrophil groups) using MACS2 and differentially accessible regions (DAR) were called between cells from β-glucan-treated and control-treated mice using a pairwise binomial test after binarization of the peak matrix (FDR ≤ 0.01 & abs(Log2FC) ≥ 1). Peak regions were annotated using the R package ChIPseeker v1.22.1 (Yu et al., 2015) and GO terms enriched for the genes annotated to the treatment-specific DAR of GMP and neutrophils were identified using the R package clusterProfiler v3.14.3. Additionally, we analyzed the set of treatment-specific DAR in GMP for enrichment of transcription factor binding motifs using the peakAnnoEnrichment function implemented in ArchR. The script written for the analysis of the scATAC-seq data can be found at https://github.com/schultzelab/Kalafati_2020.
Statistical analysis
Results are presented as mean ± SEM. Data were analyzed by two-tailed Student’s t test or Mann-Whitney U-test as appropriate. Multiple-group comparisons were performed using one-way or two-way ANOVA followed by Tukey’s or Sidak's multiple comparison tests. Statistical analyses were performed with GraphPad Prism software (GraphPad Inc., La Jolla, CA), unless otherwise stated in STAR METHODS, and statistical significance was set at p < 0.05.
Acknowledgments
This work was supported by grants from the European Research Council (DEMETINL to T.C.) and the US National Institutes of Health (DE024716 to G.H. and DE026152 and DE028561 to G.H. and T.C.). M.G.N. is supported by a Spinoza grant of the Netherlands Organisation for Scientific Research and an ERC Advanced Grant (#833247). I.M. is supported by the National Center for Tumor Diseases, Partner Site Dresden. I.K. is supported by start-up funding from the Hull York Medical School of University of York. We thank K. Bär and S. Grossklaus (Institute for Clinical Chemistry and Laboratory Medicine, Faculty of Medicine, Technische Universität Dresden, Dresden, Germany) and P. Alexakos (Biomedical Research Foundation, Academy of Athens, Athens, Greece) for technical assistance.
Author Contributions
L.K. designed and performed research, analyzed data, and contributed to writing the manuscript; I.K. designed the study, performed experiments, analyzed, and interpreted data and wrote the manuscript; J.S.S. analyzed and interpreted data and edited the manuscript; A.S., C.H., S.D., A.M.J.D., M.L., A.D., and I.H. analyzed data; X.L., A.H., T.G., S.S., E.H., M.N., A.N., A.C., A.Z., P.S., P.Mir., and K.-J.C. performed experiments and analyzed data; B.W., P.Mur., J.L.S., and M.G.N. interpreted data, contributed to study design, and edited the manuscript; G.H., P.V., and I.M. co-designed the study, interpreted data, supervised research, and contributed to writing the paper; T.C. designed the study, interpreted data, supervised research, and wrote the manuscript.
Declaration of interests
M.G.N. is scientific founder of Trained Therapeutics and Discovery (TTxD).
Published: October 29, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.09.058.
Contributor Information
Ioannis Kourtzelis, Email: ioannis.kourtzelis@york.ac.uk.
Triantafyllos Chavakis, Email: triantafyllos.chavakis@uniklinikum-dresden.de.
Supplemental Information
References
- Aachoui Y., Leaf I.A., Hagar J.A., Fontana M.F., Campos C.G., Zak D.E., Tan M.H., Cotter P.A., Vance R.E., Aderem A., Miao E.A. Caspase-11 protects against bacteria that escape the vacuole. Science. 2013;339:975–978. doi: 10.1126/science.1230751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M.P., Fiering S.N., Ostroff G.R., Cramer R.A., Mullins D.W. Beta-glucan-induced inflammatory monocytes mediate antitumor efficacy in the murine lung. Cancer Immunol. Immunother. 2018;67:1731–1742. doi: 10.1007/s00262-018-2234-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alissafi T., Hatzioannou A., Mintzas K., Barouni R.M., Banos A., Sormendi S., Polyzos A., Xilouri M., Wielockx B., Gogas H., Verginis P. Autophagy orchestrates the regulatory program of tumor-associated myeloid-derived suppressor cells. J. Clin. Invest. 2018;128:3840–3852. doi: 10.1172/JCI120888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andzinski L., Kasnitz N., Stahnke S., Wu C.F., Gereke M., von Köckritz-Blickwede M., Schilling B., Brandau S., Weiss S., Jablonska J. Type I IFNs induce anti-tumor polarization of tumor associated neutrophils in mice and human. Int. J. Cancer. 2016;138:1982–1993. doi: 10.1002/ijc.29945. [DOI] [PubMed] [Google Scholar]
- Arts R.J., Joosten L.A., Netea M.G. Immunometabolic circuits in trained immunity. Semin. Immunol. 2016;28:425–430. doi: 10.1016/j.smim.2016.09.002. [DOI] [PubMed] [Google Scholar]
- Balkwill F., Charles K.A., Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell. 2005;7:211–217. doi: 10.1016/j.ccr.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Baptista M.A., Keszei M., Oliveira M., Sunahara K.K., Andersson J., Dahlberg C.I., Worth A.J., Liedén A., Kuo I.C., Wallin R.P. Deletion of Wiskott-Aldrich syndrome protein triggers Rac2 activity and increased cross-presentation by dendritic cells. Nat. Commun. 2016;7:12175. doi: 10.1038/ncomms12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkering S., Arts R.J.W., Novakovic B., Kourtzelis I., van der Heijden C., Li Y., Popa C.D., Ter Horst R., van Tuijl J., Netea-Maier R.T. Metabolic Induction of Trained Immunity through the Mevalonate Pathway. Cell. 2018;172:135–146. doi: 10.1016/j.cell.2017.11.025. [DOI] [PubMed] [Google Scholar]
- Boutté A.M., Friedman D.B., Bogyo M., Min Y., Yang L., Lin P.C. Identification of a myeloid-derived suppressor cell cystatin-like protein that inhibits metastasis. FASEB J. 2011;25:2626–2637. doi: 10.1096/fj.10-180604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz M.L., Binnewies M., Boldajipour B., Nelson A.E., Pollack J.L., Erle D.J., Barczak A., Rosenblum M.D., Daud A., Barber D.L. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell. 2014;26:638–652. doi: 10.1016/j.ccell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavakis T., Mitroulis I., Hajishengallis G. Hematopoietic progenitor cells as integrative hubs for adaptation to and fine-tuning of inflammation. Nat. Immunol. 2019;20:802–811. doi: 10.1038/s41590-019-0402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- Chen J., Cao Y., Markelc B., Kaeppler J., Vermeer J.A., Muschel R.J. Type I IFN protects cancer cells from CD8+ T cell-mediated cytotoxicity after radiation. J. Clin. Invest. 2019;129:4224–4238. doi: 10.1172/JCI127458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung N.K., Modak S., Vickers A., Knuckles B. Orally administered beta-glucans enhance anti-tumor effects of monoclonal antibodies. Cancer Immunol. Immunother. 2002;51:557–564. doi: 10.1007/s00262-002-0321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow A., Lucas D., Hidalgo A., Méndez-Ferrer S., Hashimoto D., Scheiermann C., Battista M., Leboeuf M., Prophete C., van Rooijen N. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J. Exp. Med. 2011;208:261–271. doi: 10.1084/jem.20101688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ A., Gunther P., Lauterbach M.A.R., Duewell P., Biswas D., Pelka K., Scholz C.J., Oosting M., Haendler K., Bassler K. Western Diet Triggers NLRP3-Dependent Innate Immune Reprogramming. Cell. 2018;172:162–175. doi: 10.1016/j.cell.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K.J., Chatzigeorgiou A., Economopoulou M., Garcia-Martin R., Alexaki V.I., Mitroulis I., Nati M., Gebler J., Ziemssen T., Goelz S.E. A self-sustained loop of inflammation-driven inhibition of beige adipogenesis in obesity. Nat. Immunol. 2017;18:654–664. doi: 10.1038/ni.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirovic B., de Bree L.C.J., Groh L., Blok B.A., Chan J., van der Velden W., Bremmers M.E.J., van Crevel R., Handler K., Picelli S. BCG Vaccination in Humans Elicits Trained Immunity via the Hematopoietic Progenitor Compartment. Cell host & microbe. 2020;28:322–334. doi: 10.1016/j.chom.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens L.M., Zitvogel L., Palucka A.K. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339:286–291. doi: 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deonarain R., Verma A., Porter A.C., Gewert D.R., Platanias L.C., Fish E.N. Critical roles for IFN-beta in lymphoid development, myelopoiesis, and tumor development: links to tumor necrosis factor alpha. Proc. Natl. Acad. Sci. USA. 2003;100:13453–13458. doi: 10.1073/pnas.2230460100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin J.C., Zwack E.E., Tang M.S., Li Z., Fenyo D., Torres V.J., Ruggles K.V., Loke P. Distinct Features of Human Myeloid Cell Cytokine Response Profiles Identify Neutrophil Activation by Cytokines as a Prognostic Feature during Tuberculosis and Cancer. J. Immunol. 2020;204:3389–3399. doi: 10.4049/jimmunol.1901133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estcourt L.J., Stanworth S.J., Hopewell S., Doree C., Trivella M., Massey E. Granulocyte transfusions for treating infections in people with neutropenia or neutrophil dysfunction. Cochrane Database Syst. Rev. 2016;4:CD005339. doi: 10.1002/14651858.CD005339.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard M., Kwok I.W.H., Chong S.Z., Teng K.W.W., Becht E., Chen J., Sieow J.L., Penny H.L., Ching G.C., Devi S. Developmental Analysis of Bone Marrow Neutrophils Reveals Populations Specialized in Expansion, Trafficking, and Effector Functions. Immunity. 2018;48:364–379. doi: 10.1016/j.immuni.2018.02.002. [DOI] [PubMed] [Google Scholar]
- Feng M., Jiang W., Kim B.Y.S., Zhang C.C., Fu Y.X., Weissman I.L. Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat. Rev. Cancer. 2019;19:568–586. doi: 10.1038/s41568-019-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridlender Z.G., Sun J., Kim S., Kapoor V., Cheng G., Ling L., Worthen G.S., Albelda S.M. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell. 2009;16:183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman W.H., Zitvogel L., Sautès-Fridman C., Kroemer G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017;14:717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- Fritz J.M., Lenardo M.J. Development of immune checkpoint therapy for cancer. J. Exp. Med. 2019;216:1244–1254. doi: 10.1084/jem.20182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich D.I., Ostrand-Rosenberg S., Bronte V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdiero M.R., Varricchi G., Loffredo S., Mantovani A., Marone G. Roles of neutrophils in cancer growth and progression. J. Leukoc. Biol. 2018;103:457–464. doi: 10.1002/JLB.3MR0717-292R. [DOI] [PubMed] [Google Scholar]
- Gomez-Cambronero J., Horn J., Paul C.C., Baumann M.A. Granulocyte-macrophage colony-stimulating factor is a chemoattractant cytokine for human neutrophils: involvement of the ribosomal p70 S6 kinase signaling pathway. J. Immunol. 2003;171:6846–6855. doi: 10.4049/jimmunol.171.12.6846. [DOI] [PubMed] [Google Scholar]
- Gomez-Cambronero J., Frye T., Baumann M. Ribosomal p70S6K basal activity increases upon induction of differentiation of myelomonocytic leukemic cell lines HL60, AML14 and MPD. Leuk. Res. 2004;28:755–762. doi: 10.1016/j.leukres.2003.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granja J.M., Corces M.R., Pierce S.E., Bagdatli S.T., Choudhry H., Chang H.Y., Greenleaf W.J. ArchR: An integrative and scalable software package for single-cell chromatin accessibility analysis. bioRxiv. 2020 doi: 10.1101/2020.04.28.066498. [DOI] [Google Scholar]
- Granot Z., Henke E., Comen E.A., King T.A., Norton L., Benezra R. Tumor entrained neutrophils inhibit seeding in the premetastatic lung. Cancer Cell. 2011;20:300–314. doi: 10.1016/j.ccr.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hersh E.M., Gutterman J.U., Mavligit G.M. BCG as adjuvant immunotherapy for neoplasia. Annu. Rev. Med. 1977;28:489–515. doi: 10.1146/annurev.me.28.020177.002421. [DOI] [PubMed] [Google Scholar]
- Huang C.K., Zhan L., Hannigan M.O., Ai Y., Leto T.L. P47(phox)-deficient NADPH oxidase defect in neutrophils of diabetic mouse strains, C57BL/6J-m db/db and db/+ J. Leukoc. Biol. 2000;67:210–215. doi: 10.1002/jlb.67.2.210. [DOI] [PubMed] [Google Scholar]
- Ieronymaki E., Daskalaki M.G., Lyroni K., Tsatsanis C. Insulin Signaling and Insulin Resistance Facilitate Trained Immunity in Macrophages Through Metabolic and Epigenetic Changes. Front. Immunol. 2019;10:1330. doi: 10.3389/fimmu.2019.01330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann E., Sanz J., Dunn J.L., Khan N., Mendonca L.E., Pacis A., Tzelepis F., Pernet E., Dumaine A., Grenier J.C. BCG Educates Hematopoietic Stem Cells to Generate Protective Innate Immunity against Tuberculosis. Cell. 2018;172:176–190. doi: 10.1016/j.cell.2017.12.031. [DOI] [PubMed] [Google Scholar]
- Kleinnijenhuis J., Quintin J., Preijers F., Joosten L.A., Ifrim D.C., Saeed S., Jacobs C., van Loenhout J., de Jong D., Stunnenberg H.G. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. USA. 2012;109:17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtzelis I., Li X., Mitroulis I., Grosser D., Kajikawa T., Wang B., Grzybek M., von Renesse J., Czogalla A., Troullinaki M. DEL-1 promotes macrophage efferocytosis and clearance of inflammation. Nat. Immunol. 2019;20:40–49. doi: 10.1038/s41590-018-0249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Cai Y., Qi C., Hansen R., Ding C., Mitchell T.C., Yan J. Orally administered particulate beta-glucan modulates tumor-capturing dendritic cells and improves antitumor T-cell responses in cancer. Clin. Cancer Res. 2010;16:5153–5164. doi: 10.1158/1078-0432.CCR-10-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wise D.R., Diehl J.A., Simon M.C. Hypoxic reactive oxygen species regulate the integrated stress response and cell survival. J. Biol. Chem. 2008;283:31153–31162. doi: 10.1074/jbc.M805056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Gunn L., Hansen R., Yan J. Combined yeast-derived beta-glucan with anti-tumor monoclonal antibody for cancer immunotherapy. Exp. Mol. Pathol. 2009;86:208–214. doi: 10.1016/j.yexmp.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macal M., Tam M.A., Hesser C., Di Domizio J., Leger P., Gilliet M., Zuniga E.I. CD28 Deficiency Enhances Type I IFN Production by Murine Plasmacytoid Dendritic Cells. J. Immunol. 2016;196:1900–1909. doi: 10.4049/jimmunol.1501658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltez V.I., Tubbs A.L., Cook K.D., Aachoui Y., Falcone E.L., Holland S.M., Whitmire J.K., Miao E.A. Inflammasomes Coordinate Pyroptosis and Natural Killer Cell Cytotoxicity to Clear Infection by a Ubiquitous Environmental Bacterium. Immunity. 2015;43:987–997. doi: 10.1016/j.immuni.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y., Nawa D., Nakayama Y., Konishi M., Nanba H. Soluble β-glucan from Grifola frondosa induces tumor regression in synergy with TLR9 agonist via dendritic cell-mediated immunity. J. Leukoc. Biol. 2015;98:1015–1025. doi: 10.1189/jlb.1A0814-415RR. [DOI] [PubMed] [Google Scholar]
- Michod D., Annibaldi A., Schaefer S., Dapples C., Rochat B., Widmann C. Effect of RasGAP N2 fragment-derived peptide on tumor growth in mice. J. Natl. Cancer Inst. 2009;101:828–832. doi: 10.1093/jnci/djp100. [DOI] [PubMed] [Google Scholar]
- Mitroulis I., Chen L.S., Singh R.P., Kourtzelis I., Economopoulou M., Kajikawa T., Troullinaki M., Ziogas A., Ruppova K., Hosur K. Secreted protein Del-1 regulates myelopoiesis in the hematopoietic stem cell niche. J. Clin. Invest. 2017;127:3624–3639. doi: 10.1172/JCI92571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitroulis I., Ruppova K., Wang B., Chen L.S., Grzybek M., Grinenko T., Eugster A., Troullinaki M., Palladini A., Kourtzelis I. Modulation of Myelopoiesis Progenitors Is an Integral Component of Trained Immunity. Cell. 2018;172:147–161. doi: 10.1016/j.cell.2017.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P.J. Immune regulation by monocytes. Semin. Immunol. 2018;35:12–18. doi: 10.1016/j.smim.2017.12.005. [DOI] [PubMed] [Google Scholar]
- Musso G., Mosimann C., Panáková D., Burger A., Zhou Y., Zon L.I., MacRae C.A. Generating and evaluating a ranked candidate gene list for potential vertebrate heart field regulators. Genom. Data. 2015;6:199–201. doi: 10.1016/j.gdata.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan Y., Wu C., Zhang Y.J. Interplay between Janus Kinase/Signal Transducer and Activator of Transcription Signaling Activated by Type I Interferons and Viral Antagonism. Front. Immunol. 2017;8:1758. doi: 10.3389/fimmu.2017.01758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., van der Meer J.W. Trained Immunity: An Ancient Way of Remembering. Cell Host Microbe. 2017;21:297–300. doi: 10.1016/j.chom.2017.02.003. [DOI] [PubMed] [Google Scholar]
- Netea M.G., Joosten L.A., Latz E., Mills K.H., Natoli G., Stunnenberg H.G., O’Neill L.A., Xavier R.J. Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Domínguez-Andrés J., Barreiro L.B., Chavakis T., Divangahi M., Fuchs E., Joosten L.A.B., van der Meer J.W.M., Mhlanga M.M., Mulder W.J.M. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 2020;20:375–388. doi: 10.1038/s41577-020-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberg H.H., Wesch D., Kalyan S., Kabelitz D. Regulatory Interactions Between Neutrophils, Tumor Cells and T Cells. Front. Immunol. 2019;10:1690. doi: 10.3389/fimmu.2019.01690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S., Fu S., Mastio J., Dominguez G.A., Purohit A., Kossenkov A., Lin C., Alicea-Torres K., Sehgal M., Nefedova Y. Unique pattern of neutrophil migration and function during tumor progression. Nat. Immunol. 2018;19:1236–1247. doi: 10.1038/s41590-018-0229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkov S., Mitroulis I., Hajishengallis G., Chavakis T. Immunometabolic Crosstalk: An Ancestral Principle of Trained Immunity? Trends Immunol. 2019;40:1–11. doi: 10.1016/j.it.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzetta A., Carriero R., Carnevale S., Barbagallo M., Molgora M., Perucchini C., Magrini E., Gianni F., Kunderfranco P., Polentarutti N. Neutrophils Driving Unconventional T Cells Mediate Resistance against Murine Sarcomas and Selected Human Tumors. Cell. 2019;178:346–360. doi: 10.1016/j.cell.2019.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell D.R., Huttenlocher A. Neutrophils in the Tumor Microenvironment. Trends Immunol. 2016;37:41–52. doi: 10.1016/j.it.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylaeva E., Lang S., Jablonska J. The Essential Role of Type I Interferons in Differentiation and Activation of Tumor-Associated Neutrophils. Front. Immunol. 2016;7:629. doi: 10.3389/fimmu.2016.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C.M., Davies L.C., Subleski J.J., Maio N., Gonzalez-Cotto M., Andrews C., Patel N.L., Palmieri E.M., Weiss J.M., Lee J.M. Tumour-elicited neutrophils engage mitochondrial metabolism to circumvent nutrient limitations and maintain immune suppression. Nat. Commun. 2018;9:5099. doi: 10.1038/s41467-018-07505-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed S., Quintin J., Kerstens H.H., Rao N.A., Aghajanirefah A., Matarese F., Cheng S.C., Ratter J., Berentsen K., van der Ent M.A. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul M.E., Levy L., Sun J., Mishalian I., Singhal S., Kapoor V., Horng W., Fridlender G., Albelda S.M., Fridlender Z.G. Tumor-associated neutrophils display a distinct N1 profile following TGFβ modulation: A transcriptomics analysis of pro- vs. antitumor TANs. OncoImmunology. 2016;5:e1232221. doi: 10.1080/2162402X.2016.1232221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra R.A., Trillo-Tinoco J., Mohamed E., Yu L., Achyut B.R., Arbab A., Bradford J.W., Osborne B.A., Miele L., Rodriguez P.C. Anti-Jagged Immunotherapy Inhibits MDSCs and Overcomes Tumor-Induced Tolerance. Cancer Res. 2017;77:5628–5638. doi: 10.1158/0008-5472.CAN-17-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttmann H., Riemensberger J., Bentien G., Schmaltz D., Stöckle M., Jocham D., Böhle A., Brandau S. Neutrophil granulocytes are required for effective Bacillus Calmette-Guérin immunotherapy of bladder cancer and orchestrate local immune responses. Cancer Res. 2006;66:8250–8257. doi: 10.1158/0008-5472.CAN-06-1416. [DOI] [PubMed] [Google Scholar]
- Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlik M.T., Bose N., Cox J., Mattson P., Gargano M., O’Day S., Borges V., Chmielowski B., Rao R., Abu-Khalaf M. Abstract PD1-02: Response and clinical benefit assessment of the combination of the dectin-1 agonist imprime PGG and anti-PD-1 pembrolizumab in chemotherapy-resistant metastatic triple negative breast cancer (TNBC) Cancer Research. 2020;80 https://www.biothera.com/wp-content/uploads/2019/12/Biothera-2019-SABCS-Spotlight-Poster-PD1-02.pdf [Google Scholar]
- Voltarelli F.A., Frajacomo F.T., Padilha C.S., Testa M.T.J., Cella P.S., Ribeiro D.F., de Oliveira D.X., Veronez L.C., Bisson G.S., Moura F.A., Deminice R. Syngeneic B16F10 Melanoma Causes Cachexia and Impaired Skeletal Muscle Strength and Locomotor Activity in Mice. Front. Physiol. 2017;8:715. doi: 10.3389/fphys.2017.00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter W., Sánchez-Cabo F., Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31:2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- Weiden J., Tel J., Figdor C.G. Synthetic immune niches for cancer immunotherapy. Nat. Rev. Immunol. 2018;18:212–219. doi: 10.1038/nri.2017.89. [DOI] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag New York; 2009. ggplot2. [Google Scholar]
- Wright H.J., Matthews J.B., Chapple I.L., Ling-Mountford N., Cooper P.R. Periodontitis associates with a type 1 IFN signature in peripheral blood neutrophils. J. Immunol. 2008;181:5775–5784. doi: 10.4049/jimmunol.181.8.5775. [DOI] [PubMed] [Google Scholar]
- Wu T.D., Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang D., Sharma V.R., Freter C.E., Yan J. Anti-tumor monoclonal antibodies in conjunction with β-glucans: a novel anti-cancer immunotherapy. Curr. Med. Chem. 2012;19:4298–4305. doi: 10.2174/092986712802884303. [DOI] [PubMed] [Google Scholar]
- Xie X., Shi Q., Wu P., Zhang X., Kambara H., Su J., Yu H., Park S.Y., Guo R., Ren Q. Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat. Immunol. 2020;21:1119–1133. doi: 10.1038/s41590-020-0736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zou S., Xu X., Zhang L. Anti-tumor effect of β-glucan from Lentinus edodes and the underlying mechanism. Sci. Rep. 2016;6:28802. doi: 10.1038/srep28802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Kloecker G., Fleming C., Bousamra M., 2nd, Hansen R., Hu X., Ding C., Cai Y., Xiang D., Donninger H. Human polymorphonuclear neutrophils specifically recognize and kill cancerous cells. OncoImmunology. 2014;3:e950163. doi: 10.4161/15384101.2014.950163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., He Q.-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- Zeeshan H.M., Lee G.H., Kim H.R., Chae H.J. Endoplasmic Reticulum Stress and Associated ROS. Int. J. Mol. Sci. 2016;17:327. doi: 10.3390/ijms17030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Kaufman R.J. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Kim J.A., Huang A.Y. Optimizing Tumor Microenvironment for Cancer Immunotherapy: β-Glucan-Based Nanoparticles. Front. Immunol. 2018;9:341. doi: 10.3389/fimmu.2018.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilionis R., Engblom C., Pfirschke C., Savova V., Zemmour D., Saatcioglu H.D., Krishnan I., Maroni G., Meyerovitz C.V., Kerwin C.M. Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity. 2019;50:1317–1334. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon request to the Lead Contact. Sequencing data are available at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under the accession numbers GSE139450, GSE139451, GSE139452, GSE139456 and GSE152353.