Abstract
Posttranscriptional modifications of anticodon loops contribute to the decoding efficiency of tRNAs by supporting codon recognition and loop stability. Consistently, strong synthetic growth defects are observed in yeast strains simultaneously lacking distinct anticodon loop modifications. These phenotypes are accompanied by translational inefficiency of certain mRNAs and disturbed protein homeostasis resulting in accumulation of protein aggregates. Different combinations of anticodon loop modification defects were shown to affect distinct tRNAs but provoke common transcriptional changes that are reminiscent of the cellular response to nutrient starvation. Multiple mechanisms may be involved in mediating inadequate starvation response upon loss of critical tRNA modifications. Recent evidence suggests protein aggregate induction to represent one such trigger.
Keywords: tRNA modification, Protein aggregation, Decoding, Starvation response
Background
During decoding of mRNA, codons are recognized by the tRNA anticodon. For efficient decoding, the tRNA must be correctly folded into an L-shaped structure and the anticodon presented in an unpaired open loop. Posttranscriptional modifications in the anticodon loop are thought to improve codon recognition and contribute to anticodon loop stability by promoting base stacking interactions, reducing the flexibility of the sugar phosphate backbone and preventing unwanted across-the-loop base pairing (Agris 2008; Sokołowski et al. 2017; Väre et al. 2017; Vendeix et al. 2012). For example, tRNALysUUU contains mcm5s2U34 (5-methoxycarbonylmethyl-2-thiouridine at position 34) and ct6A37 (cyclic N6-threonylcarbamoyladenosine at position 37) modifications which each fulfill one or more of these tasks (Johansson et al. 2018; Miyauchi et al. 2013; Schaffrath and Leidel 2017; Thiaville et al. 2014). Both, mcm5s2U and ct6A are formed by multiple biosynthetic enzymes and steps. Completion of mcm5s2U synthesis is abolished at distinct steps in elp3 and urm1 mutants, while ct6A formation from the t6A (N6-threonylcarbamoyladenosine) precursor requires TCD1 (Huang et al. 2005; Leidel et al. 2009; Miyauchi et al. 2013). Hence, in elp3, urm1 and tcd1 mutants, distinct pathway intermediates are formed at the target nucleosides U34 and A37. Consistent with functional redundancy, joint abrogation of mcm5s2U synthesis at different steps and prevention of t6A to ct6A conversion results in a functional defect of tRNALysUUU normally carrying these modifications (Klassen et al. 2016). A similar functional redundancy exists in the tRNAGlnUUG anticodon loop which naturally carries mcm5s2U and Ψ38 (pseudouridine at position 38) (Han et al. 2015; Klassen et al. 2016). Combined absence of mcm5s2U and Ψ38 in elp3 deg1 or urm1 deg1 double mutants causes a severe functional impairment of this tRNA. When formation of mcm5s2U is completely abolished by combining elp3 and urm1 or elp6 and ncs2 modifications, both, tRNAGlnUUG and tRNALysUUU are functionally impaired (Björk et al. 2007; Klassen et al. 2015; Nedialkova and Leidel 2015; Xu et al. 2019).
Effects of modification loss on decoding and protein homeostasis
In the mutants carrying combinations of tRNA modification defects, negative phenotypes and translational incompetence are routinely suppressed by overexpression of the functionally impaired tRNAs (Björk et al. 2007; Han et al. 2015; Klassen et al. 2015, 2016; Nedialkova and Leidel 2015). Elevated abundance of the hypomodified tRNA is thought to counteract the translational deficiency, which may result from increased rejection rate during the codon recognition process (Ranjan and Rodnina 2017; Rezgui et al. 2013). Another cellular consequence of such specific tRNA defects is a severe protein homeostasis disturbance, resulting in the accumulation of protein aggregates (Fig. 1) (Nedialkova and Leidel 2015). The exact mechanism how combined tRNA modification defects trigger protein aggregation is not known, but it can be assumed that ribosomal pausing is an important factor for this effect. Ribosomal pausing at CAA (Gln) and AAA (Lys) codons has been indeed demonstrated for yeast strains lacking mcm5s2U (Nedialkova and Leidel 2015), and mcm5s2U deficiency in combination with either loss of ct6A or Ψ38 likely aggravates pausing at CAA or AAA codons, respectively (Bruch et al. 2020; Klassen et al. 2016; Pollo-Oliveira et al. 2020). Such pause during translational elongation may as well increase the occurrence of ribosomal errors including + 1 frameshifts and, potentially, mistranslation due to misreading by near- or non-cognate tRNAs. An increase in + 1 frameshift rates has in fact been detected in yeast strains lacking mcm5s2U alone and in combination with ct6A defects (Klassen et al. 2017; Pollo-Oliveira et al. 2020; Tükenmez et al. 2015). While in such mutants, the efficiency of near/non-cognate tRNA misincorporation at the AAA or CAA codons has not been investigated, a similar effect was observed for misreading of CGC (Arg) codons by tRNAHisGUG (Khonsari and Klassen 2020). Here, absence of Pus1 dependent Ψ in the normal CGC decoder tRNAArgICG increased misreading by tRNAHisGUG, which is not naturally modified by Pus1. Thus, in general, impairment of a cognate tRNA upon loss of critical modifications may increase near cognate misreading by a competitor tRNA that does not rely on the same modification. In addition to effects potentially associated with ribosomal pausing, the ability of the hypomodified tRNA itself to engage in misreading might also be affected by loss of critical anticodon loop modifications. Some specific errors indeed increased in the absence of mcm5s2U (Joshi et al. 2018). However, when misreading of near cognate codons with wobble base mismatches to tRNALysUUU was studied, mcm5s2U promoted rather than inhibited these types of errors (Joshi et al. 2018).
Fig. 1.
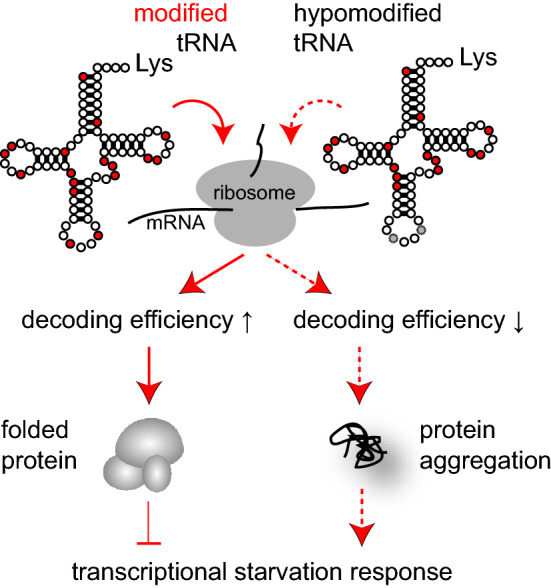
Model for the induction of a transcriptional starvation response in combined absence of anticodon loop modifications. tRNALysUUU is depicted with modified positions (indicated in red). In combined elp3 tcd1 or urm1 tcd1 mutants, anticodon loop modifications mcm5s2U and ct6A are missing (indicated in grey), causing decreased decoding efficiency of cognate AAA (Lys) codons. Multiple mechanisms are discussed how such decoding defect may cause accumulation of cellular protein aggregates. New results suggest that protein aggregates are involved in triggering a subsequent transcriptional response reminiscent of nutrient starvation
An alternative mechanism how protein aggregation might be linked to tRNA modification defects causing ribosomal pausing lies with disturbance of co-translational protein folding (Nedialkova and Leidel 2015). Support for this assumption stems from the observation of similarities in protein aggregate induction in a mcm5s2U-deficient yeast strain and a mutant lacking the ribosome-associated chaperones Ssb1/2, which are important for co-translational protein folding (Nedialkova and Leidel 2015). Thus, multiple mechanisms might link tRNA modification defects to the production of faulty proteins, which may be relevant for the common observation of impaired protein homeostasis in different tRNA modification mutants (Klassen et al. 2016; Nedialkova and Leidel 2015; Pollo-Oliveira et al. 2020; Thiaville et al. 2016; Xu et al. 2019).
Starvation responses of tRNA modification mutants
Interestingly, different tRNA modification defects also evoke major transcriptome changes and part of these are reminiscent of the transcriptomic response to nutrient depletion. Several yeast tRNA modification mutants including those lacking mcm5s2U and ct6A induce GCN4-dependent amino acid biosynthesis genes despite the presence of amino acids in the medium (Daugeron et al. 2011; Zinshteyn and Gilbert 2013). In absence of either mcm5s2U or ct6A, GCN4 induction occurred independent of the Gcn2 kinase which is activated upon binding of uncharged tRNA (Daugeron et al. 2011; Zinshteyn and Gilbert 2013). The GCN2-independent GCN4 induction in these mutants suggested a non-canonical mechanism is involved in expression of general amino acid control (GAAC) genes in different tRNA modification mutants (Daugeron et al. 2011; Zinshteyn and Gilbert 2013). The recent characterization of transcriptomic changes after combined loss of mcm5s2U and either ct6A or Ψ38/39 revealed additional facets of a common starvation in response to loss of different tRNA modifications (Bruch et al. 2020).
In these strains, when grown to early exponential phase, premature transcriptional activation of genes occurred that are normally expressed only upon entry into stationary phase or nutrient depletion. This includes a loss of glucose repression and induction of nitrogen catabolite-repressed (NCR) genes in addition to the activation of different amino acid biosynthesis genes (Bruch et al. 2020). Also, autophagy (another cellular starvation response) was induced as judged from studying loss of Atg13 phosphorylation and degradation of a GFP-Atg8 fusion protein. Since NCR and autophagy are controlled by the TORC1 complex in budding yeast, these cellular responses to combined tRNA modification defects might be caused by loss or suppression of TORC1 activity (Bruch et al. 2020). Additional evidence for a role of Elp3-dependent tRNA modification in reciprocal regulation of TORC1 and TORC2 activities was obtained in a recent fission yeast study (Candiracci et al. 2019). In budding yeast, TORC1 activity also appears to be influenced by the level of uncharged tRNAs (Kamada 2017). These results suggest that the TOR complex, which represents a master regulator of growth and metabolism (Loewith and Hall 2011) might monitor the modification and charging status of tRNA. Loss of mcm5U or s2U modifications not only influences nutrient sensitive gene expression signatures, but also results in robust changes in cellular metabolism, and some of these are again reminiscent of cellular responses to nutrient starvation (Gupta et al. 2019; Karlsborn et al. 2016). Thus, apart from tRNA aminoacylation, multiple lines of evidence support an emerging role for tRNA anticodon loop modifications in the cellular signaling of nutrient availability.
Potential mediators of nutrient signaling defects in tRNA modification mutants
Several tRNA modification defects in yeast are known to trigger GCN4 expression in the absence of amino acid starvation. This includes not only the mcm5s2U and ct6A defective mutants described above, but was also observed in deg1, pus7, rit1, trm1, trm7, mod5 and tyw3 mutants lacking various other tRNA modifications (Chou et al. 2017; Han et al. 2018). While such amino acid starvation response appeared to be independent of the Gcn2 kinase responding to uncharged tRNA in mcm5s2U and ct6A defective strains, it was shown to be Gcn2 dependent in trm7 mutants (Daugeron et al. 2011; Han et al. 2018; Zinshteyn and Gilbert 2013). In these mutants, which lack 2′-O-methylation of C32 and G34 in tRNAPhe, reduced charging of the hypomodified tRNA was observed (Han et al. 2018). Hence, the GAAC starvation response in tRNA modification mutants can be triggered in some cases by effects on the tRNA aminoacylation efficiency.
In s2U-deficient strains, robust metabolic changes involve increased storage carbohydrate synthesis, which normally occurs after glucose depletion (Gupta et al. 2019). Interestingly, these effects were linked to a disturbance of phosphate homeostasis. Increased trehalose synthesis likely occurs to counteract reduced intracellular phosphate levels since trehalose generation from trehalose phosphate can replenish intracellular phosphate levels. The phosphate shortage in s2U-deficient mutants is thought to be triggered by transcriptional and translational downregulation of PHO genes involved in phosphate uptake (Gupta et al. 2019). A similar mechanism might be involved in starvation like responses in other tRNA modification mutants, including those required for formation of mcm5U and ct6A, since transcriptional downregulation of PHO genes was observed (Chou et al. 2017). In the s2U-deficient strain, however, no robust transcriptional starvation response was triggered (Gupta et al. 2019), which is in contrast to the changes seen in combined mutants. Since the combined mutants exhibit growth defects exceeding those of the s2U-deficient strain, more robust changes might occur also at the metabolic level (Bruch et al. 2020; Klassen et al. 2016). It remains unknown, however, how exactly the transcriptional response is mediated.
Intriguingly, when studying the transcriptional induction of nutrient responsive genes in combined tRNA modification mutants, their expression was dampened upon overexpression of the very same tRNAs that conferred a suppression of growth defects (Bruch et al. 2020; Klassen et al. 2016). As outlined above, the overexpressed tRNA presumably directly counteracts the inefficiency in decoding. At the same time, the propensity to accumulate protein aggregates (see above) is significantly lowered by the tRNA overexpression constructs. Hence, protein aggregates are linked to the decoding defect and are potentially involved in the observed gene expression changes (Fig. 1). Further support for this hypothesis was obtained from studying a mutant (zuo1) accumulating protein aggregates independent of a tRNA modification defect (Bruch et al. 2020). In zuo1 mutants, the ribosome-associated chaperone system is severely compromised, leading to accumulation of protein aggregates (Bruch et al. 2020). At the same time, marker genes that are subject to glucose repression or NCR become transcriptionally induced despite the presence of glucose and ammonia in the medium. Thus, protein aggregates might be mechanistically involved in mediating transcriptional changes in response to combined loss of tRNA modifications. Possibly, the proteasome-mediated turnover of normally short-lived transcription factors is altered upon cellular accumulation of protein aggregates, ultimately leading to the observed changes in gene expression signatures. Further work will be required to test this hypothesis and other potentially involved mechanisms.
Acknowledgements
We gratefully acknowledge support from the Deutsche Forschungsgemeinschaft (DFG) to R.S. (SCHA750/15-2) and their Priority Program SPP1784 Chemical Biology of Native Nucleic Acid Modifications to R.S. (SCHA750/20-2) and R.K. (KL2937/1-2).
Funding
This work was supported by Deutsche Forschungsgemeinschaft (DFG) Grants SCHA750/15-2, SCHA750/20-2 to R.S. and KL2937/1-2 to R.K. Open Access funding provided by Projekt DEAL.
Compliance with ethical standards
Conflict of interest
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008;9:629–635. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk GR, Huang B, Persson OP, Byström AS. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 2007;13:1245–1255. doi: 10.1261/rna.558707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruch A, Laguna T, Butter F, Schaffrath R, Klassen R. Misactivation of multiple starvation responses in yeast by loss of tRNA modifications. Nucleic Acids Res. 2020 doi: 10.1093/nar/gkaa455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candiracci J, Migeot V, Chionh Y-H, Bauer F, Brochier T, Russell B, Shiozaki K, Dedon P, Hermand D. Reciprocal regulation of TORC signaling and tRNA modifications by Elongator enforces nutrient-dependent cell fate. Sci Adv. 2019 doi: 10.1126/sciadv.aav0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H-J, Donnard E, Gustafsson HT, Garber M, Rando OJ. Transcriptome-wide analysis of roles for tRNA modifications in translational regulation. Mol Cell. 2017;68:978–992.e4. doi: 10.1016/j.molcel.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron M-C, Lenstra TL, Frizzarin M, El Yacoubi B, Liu X, Baudin-Baillieu A, Lijnzaad P, Decourty L, Saveanu C, Jacquier A, Holstege FCP, de Crécy-Lagard V, van Tilbeurgh H, Libri D. Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res. 2011;39:6148–6160. doi: 10.1093/nar/gkr178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Walvekar AS, Liang S, Rashida Z, Shah P, Laxman S. A tRNA modification balances carbon and nitrogen metabolism by regulating phosphate homeostasis. Elife. 2019 doi: 10.7554/eLife.44795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Kon Y, Phizicky EM. Functional importance of Ψ38 and Ψ39 in distinct tRNAs, amplified for tRNAGln(UUG) by unexpected temperature sensitivity of the s2U modification in yeast. RNA. 2015;21:188–201. doi: 10.1261/rna.048173.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Guy MP, Kon Y, Phizicky EM. Lack of 2′-O-methylation in the tRNA anticodon loop of two phylogenetically distant yeast species activates the general amino acid control pathway. PLoS Genet. 2018;14:e1007288. doi: 10.1371/journal.pgen.1007288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Johansson MJO, Byström AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJO, Xu F, Byström AS. Elongator-a tRNA modifying complex that promotes efficient translational decoding. Biochim Biophys Acta Gene Regul Mech. 2018;1861:401–408. doi: 10.1016/j.bbagrm.2017.11.006. [DOI] [PubMed] [Google Scholar]
- Joshi K, Bhatt MJ, Farabaugh PJ. Codon-specific effects of tRNA anticodon loop modifications on translational misreading errors in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y. Novel tRNA function in amino acid sensing of yeast Tor complex1. Genes Cells. 2017;22:135–147. doi: 10.1111/gtc.12462. [DOI] [PubMed] [Google Scholar]
- Karlsborn T, Mahmud AKMF, Tükenmez H, Byström AS. Loss of ncm5 and mcm5 wobble uridine side chains results in an altered metabolic profile. Metabolomics. 2016;12:177. doi: 10.1007/s11306-016-1120-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khonsari B, Klassen R. Impact of Pus1 pseudouridine synthase on specific decoding events in Saccharomyces cerevisiae. Biomolecules. 2020 doi: 10.3390/biom10050729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen R, Grunewald P, Thüring KL, Eichler C, Helm M, Schaffrath R. Loss of anticodon wobble uridine modifications affects tRNA(Lys) function and protein levels in Saccharomyces cerevisiae. PLoS ONE. 2015;10:e0119261. doi: 10.1371/journal.pone.0119261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen R, Ciftci A, Funk J, Bruch A, Butter F, Schaffrath R. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res. 2016;44:10946–10959. doi: 10.1093/nar/gkw705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen R, Bruch A, Schaffrath R. Independent suppression of ribosomal +1 frameshifts by different tRNA anticodon loop modifications. RNA Biol. 2017;14:1252–1259. doi: 10.1080/15476286.2016.1267098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Pedrioli PGA, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi K, Kimura S, Suzuki T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat Chem Biol. 2013;9:105–111. doi: 10.1038/nchembio.1137. [DOI] [PubMed] [Google Scholar]
- Nedialkova DD, Leidel SA. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015;161:1606–1618. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollo-Oliveira L, Klassen R, Davis N, Ciftci A, Bacusmo JM, Martinelli M, DeMott MS, Begley TJ, Dedon PC, Schaffrath R, de Crécy-Lagard V. Loss of elongator- and KEOPS-dependent tRNA modifications leads to severe growth phenotypes and protein aggregation in yeast. Biomolecules. 2020 doi: 10.3390/biom10020322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan N, Rodnina MV. Thio-Modification of tRNA at the wobble position as regulator of the kinetics of decoding and translocation on the ribosome. J Am Chem Soc. 2017;139:5857–5864. doi: 10.1021/jacs.7b00727. [DOI] [PubMed] [Google Scholar]
- Rezgui VAN, Tyagi K, Ranjan N, Konevega AL, Mittelstaet J, Rodnina MV, Peter M, Pedrioli PGA. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc Natl Acad Sci U S A. 2013;110:12289–12294. doi: 10.1073/pnas.1300781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffrath R, Leidel SA. Wobble uridine modifications—a reason to live, a reason to die?! RNA Biol. 2017;14:1209–1222. doi: 10.1080/15476286.2017.1295204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokołowski M, Klassen R, Bruch A, Schaffrath R, Glatt S. Cooperativity between different tRNA modifications and their modification pathways. Biochim Biophys Acta. 2017 doi: 10.1016/j.bbagrm.2017.12.003. [DOI] [PubMed] [Google Scholar]
- Thiaville PC, Iwata-Reuyl D, de Crécy-Lagard V. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t(6)A), a universal modification of tRNA. RNA Biol. 2014;11:1529–1539. doi: 10.4161/15476286.2014.992277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville PC, Legendre R, Rojas-Benítez D, Baudin-Baillieu A, Hatin I, Chalancon G, Glavic A, Namy O, de Crécy-Lagard V. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb Cell. 2016;3:29–45. doi: 10.15698/mic2016.01.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tükenmez H, Xu H, Esberg A, Byström AS. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res. 2015;43:9489–9499. doi: 10.1093/nar/gkv832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Väre VYP, Eruysal ER, Narendran A, Sarachan KL, Agris PF. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules. 2017 doi: 10.3390/biom7010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeix FAP, Murphy FV, Cantara WA, Leszczyńska G, Gustilo EM, Sproat B, Malkiewicz A, Agris PF. Human tRNA(Lys3)(UUU) is pre-structured by natural modifications for cognate and wobble codon binding through keto-enol tautomerism. J Mol Biol. 2012;416:467–485. doi: 10.1016/j.jmb.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Byström AS, Johansson MJO. SSD1 suppresses phenotypes induced by the lack of Elongator-dependent tRNA modifications. PLoS Genet. 2019;15:e1008117. doi: 10.1371/journal.pgen.1008117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinshteyn B, Gilbert WV. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 2013;9:e1003675. doi: 10.1371/journal.pgen.1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]


