Abstract
Objectives
To investigate whether radiomics features extracted from MRI of BRCA-positive patients with sub-centimeter breast masses can be coupled with machine learning to differentiate benign from malignant lesions using model-free parameter maps.
Methods
In this retrospective study, BRCA-positive patients who had an MRI from November 2013 to February 2019 that led to a biopsy (BI-RADS 4) or imaging follow-up (BI-RADS 3) for sub-centimeter lesions were included. Two radiologists assessed all lesions independently and in consensus according to BI-RADS. Radiomics features were calculated using open-source CERR software. Univariate analysis and multivariate modeling were performed to identify significant radiomics features and clinical factors to be included in a machine learning model to differentiate malignant from benign lesions.
Results
Ninety-six BRCA mutation carriers (mean age at biopsy = 45.5 ± 13.5 years) were included. Consensus BI-RADS classification assessment achieved a diagnostic accuracy of 53.4%, sensitivity of 75% (30/40), specificity of 42.1% (32/76), PPV of 40.5% (30/74), and NPV of 76.2% (32/42). The machine learning model combining five parameters (age, lesion location, GLCM-based correlation from the pre-contrast phase, first-order coefficient of variation from the 1st post-contrast phase, and SZM-based gray level variance from the 1st post-contrast phase) achieved a diagnostic accuracy of 81.5%, sensitivity of 63.2% (24/38), specificity of 91.4% (64/70), PPV of 80.0% (24/30), and NPV of 82.1% (64/78).
Conclusions
Radiomics analysis coupled with machine learning improves the diagnostic accuracy of MRI in characterizing sub-centimeter breast masses as benign or malignant compared with qualitative morphological assessment with BI-RADS classification alone in BRCA mutation carriers.
Key Points
• Radiomics and machine learning can help differentiate benign from malignant breast masses even if the masses are small and morphological features are benign.
• Radiomics and machine learning analysis showed improved diagnostic accuracy, specificity, PPV, and NPV compared with qualitative morphological assessment alone.
Electronic supplementary material
The online version of this article (10.1007/s00330-020-06991-7) contains supplementary material, which is available to authorized users.
Keywords: Machine learning, Breast neoplasms, Artificial intelligence, Magnetic resonance imaging
Introduction
Women who inherit BRCA1 and BRCA2 mutations lack tumor suppressor proteins that repair damaged DNA [1]. These women have an increased risk of developing breast cancer at a younger age compared with women who do not have these mutations. MRI is the most sensitive imaging modality for breast cancer detection and therefore, the American Cancer Society and the American College of Radiology recommend yearly mammography in BRCA mutation carriers starting at age 30 years and yearly MRI beginning at age 25 [2–7].
A significant proportion (45%) of BRCA1-related cancers are seen only on MRI [8] where they tend to be cellular with round pushing margins rather than scirrhous with irregular infiltrating margins as seen in other breast cancers. Therefore, early/small tumors may not exhibit classic malignant features but rather may exhibit a benign imaging appearance [9]. As these cancers are also more likely to be high grade and frequently triple negative (hormone receptor and HER-2 negative), the threshold for the recommendation of a biopsy should be low [10, 11]. Prior studies [12, 13] showed how benign morphology is common in invasive cancers of less than 5 mm in diameter regardless of BRCA mutation status and suggested that all masses representing an interval change as well as lesions increasing in size should lead to a biopsy. Unfortunately, BRCA carriers are also more prone to developing benign tumors of the breast [14, 15], resulting in numerous benign biopsies during their life unless prophylactic mastectomy is performed.
To avoid missing significant cancers as well as exposing women to unnecessary biopsies, additional tools to help discriminate benign from malignant lesions should be used to predict the likelihood of malignancy. Radiomics analysis involves the quantitative assessment of the pixel intensity arrangement within specific regions of interest (ROIs) and extracts quantitative features that can be used for further disease characterization. Initial results in women at average risk of breast cancer indicate that radiomics analysis and machine learning (ML) are of value in distinguishing benign and malignant small breast masses [16].
The purpose of our study was to investigate whether radiomics features extracted from MRI of BRCA-positive patients with sub-centimeter breast masses can be coupled with machine learning to differentiate benign from malignant lesions using model-free parameter maps.
Materials and methods
Study population
This was a retrospective Health Insurance Portability and Accountability Act–compliant study conducted at Memorial Sloan Kettering Cancer Center. The study was approved by the Institutional Review Board (protocol number 19-119) and the need for written informed consent was waived.
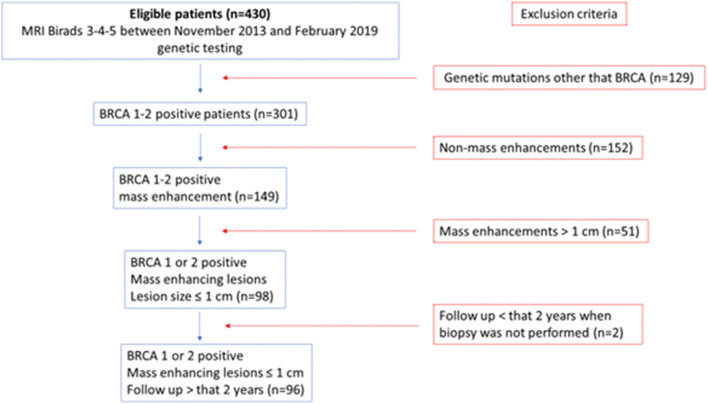
A review of the Department of Radiology database was performed to identify consecutive patients with genetic testing results available and who had an MRI from November 2013 to February 2019 that led to a biopsy or a short-term follow-up. We identified 430 patients. Our inclusion criteria were as follows: BRCA 1– or BRCA 2–positive patients; breast masses with the longest diameter ≤ 10 mm; and BI-RADS 3, 4, or 5 on MRI further assessed with follow-up or vacuum-assisted breast biopsy (MRI or ultrasound-guided) yielding benign or malignant histology. Findings described as non-mass enhancements on MRI were not included. We excluded patients with mutations other than BRCA 1 and 2 and those with a follow-up of less than 2 years when biopsy was not performed (BI-RADS 3 and BI-RADS 4 when target was not visualized at the time of biopsy).
Breast MRI technique
Breast MRI was performed on either a 1.5-T or a 3-T magnet (Sigma; GE) using an 8-channel or 16-channel dedicated surface breast coil. The imaging sequences are included in Table 1.
Table 1.
Summary of imaging sequences and acquisition parameters used for the study
| MR sequences | Acquisition parameters |
|---|---|
| Axial fat-suppressed 2D T2-weighted imaging | TR, 5000–6000 ms; TE, 90–110 ms; refocusing flip angle, “auto”; slice thickness, 3 mm; gap, 0 mm; field of view, 34–38 cm; matrix size, 320 × 320; bandwidth, 125 kHz for 1.5 T and 83 kHz for 3.0 T; parallel imaging, “ASSET” |
| Axial non-fat-suppressed 3D T1-weighted imaging | |
| Axial fat-suppressed 3D T1-weighted imaging using a Volume Image Breast Assessment (VIBRANT) gradient echo. One sequence before and 3 sequences after intravenous administration of a gadolinium-based contrast agent | TR, 4–4.5 ms; TE, 2.1 ms; flip angle, 10°; bandwidth, 62 kHz; field of view, 34–38 cm; matrix size, 320 × 192 (for 1.5 T) and 300 × 300 (for 3.0 T); slice thickness, 1.1 mm; gap, 0 mm; parallel imaging, “ASSET” |
| Axial DWI using single-shot with echo-planar imaging (EPI) | 2 b-values (b = 0, 800); TR, 6000 ms; TE, “minimum”; flip angle, 90°; field of view, 34–38 cm, matrix size, 128 × 128 (for 1.5 T), 256 × 256 (for 3 T); fat suppression, “special”; dual shims, “on”; slice thickness, 4–5 mm; parallel imaging, “ASSET” |
| ADC mapping available in 65 lesions |
ASSET, array spatial sensitivity encoding technique; TR, repetition time; TE, echo time
Imaging assessment by radiologists
All images were independently assessed by two dedicated fellowship-trained breast radiologists in one session (R1: R.L., and R2: I.D., both with 4 years of experience in breast imaging and interpreting breast MRI) blinded to the final histopathological diagnoses and prior or subsequent conventional and MRI imaging. For each lesion, the following morphological features were assessed according to the BI-RADS lexicon on post-contrast-enhanced T1-weighted images: lesion shape, margin, and internal enhancement characteristics. Readers also assigned a BI-RADS classification. Lesion size was measured as the single largest diameter. On T2-weighted and DW images, signal intensity, morphology, background parenchymal enhancement (BPE), and fibroglandular tissue (FGT) for each breast were also assessed. Time–intensity kinetic curve analysis (signal enhancement in relation to time after contrast injection) was performed on a dedicated workstation with a commercially available computer-aided diagnosis system (OsiriX, OsiriX Foundation) by R1. The reader qualitatively measured the kinetic curve pattern described as washout, plateau, or persistent, according to the BI-RADS lexicon. The location of lesions within the breast (anterior, middle, or posterior depth) was also assessed by R1.
After independent review was conducted, the cases in which there was disagreement between the two readers were re-reviewed in consensus to generate an overall consensus assessment.
Reference standard
Preferentially, histopathology was used as the reference standard established by either image-guided needle biopsy or surgery. In two patients who had benign high-risk lesions on biopsy, the histological report from the surgical biopsy was recorded to confirm the benign nature of the lesion. When biopsy was not performed, stability of more than 2 years on follow-up MRI was considered benign.
Radiomics analysis
Digital Imaging and Communications in Medicine (DICOM) images from the DCE-MRI and non-contrast-enhanced T1-weighted MRI were loaded into the open-source image processing tool OsiriX. Both radiologists reviewed the images in consensus before delineating the ROIs and R1manually delineated the ROIs, tracing the borders of each lesion to include the entire enhancing lesion.
Given the small size of the lesions sampled yielding a small number of pixels per slice, an in-house code written in MATLAB (The MathWorks, Inc.) was used to input the ROIs into the open-source CERR software environment (freely available through GitHub) which calculated the radiomics features [17]. Data was reduced to 16 gray levels and only an interpixel distance of one was considered (for small lesions, higher interpixel distances are not appropriate and would reduce counting statistics drastically). CERR analysis resulted in 102 radiomics features sub-divided into six categories: 22 first-order features, 26 features based on the gray level co-occurrence matrix (GLCM), 16 features based on the run length matrix (RLM), 16 features based on the size zone matrix (SZM), 17 features based on the neighborhood gray level dependence matrix, and 5 features based on the neighborhood gray tone difference matrix. Since patients were scanned at either 1.5 T (27 benign cases and 17 malignant cases) or 3 T (49 benign cases and 23 malignant cases), ComBat harmonization (Supplemental Info A1) was employed prior to statistical analysis to remove center effects [18].
Univariate analysis was initially performed to select significant radiomics features able to differentiate between benign and malignant lesions. An AUC cutoff of ≥ 0.65 was used to reduce the number of features of interest. Correlation analysis was then employed to further remove redundant features. For any significant correlations in which the Spearman rank correlation coefficient > 0.9, the feature with the lowest AUC was removed from consideration. This resulted in a more manageable number of features for subsequent multivariate modeling. Using a fine Gaussian support vector machine, perfect separation of benign and malignant cases was obtained. To limit data overfitting, a fivefold cross-validation was employed to develop a robust ML model which should produce similar results for new data.
Statistical analysis
Statistical analysis was conducted using SAS (version 9.4, SAS Institute). Continuous variables were summarized using means (± standard deviation) and medians (range); categorical variables were summarized using proportions. Univariate analysis using the chi-square test or Fisher’s exact test was performed to assess associations between the imaging parameters (from independent and consensus assessment) with disease status (malignant vs. benign). p values < 0.05 were considered significant. To determine inter-observer agreement, weighted Cohen’s κ was used to assess ordinal parameters, while simple Cohen’s κ was used to assess the inter-reader agreement for nominal parameters.
For radiomics data, statistical analysis was performed using SPSS (version 25, IBM Corp.) and MATLAB (R2017b, The MathWorks, Inc.). Univariate analysis was performed to identify radiomics features that were significantly different between malignant and benign lesions. Since the number of patients was not large (especially in the malignant cohort), normality in the malignant and benign cohort distributions was tested using the Shapiro–Wilk test and Q-Q plots. For a minority (21/102) of normally distributed features, a two-tailed independent t test was used to determine the significant features. For the majority of non-normally distributed features (81/102), the Mann–Whitney U test for two independent samples was used to determine the significant features.
Clinical factors considered as potential predictors of malignancy (age, BRCA status, menopausal status, and lesion location) were assessed for statistically significant associations with disease status using the Mann–Whitney U test (for age) and the Pearson chi-square test (for all other clinical factors). Significant clinical factors were incorporated into multivariate modeling along with significant radiomics features to produce a robust ML model for discriminating between benign and malignant lesions. All ML modelling was performed using a predefined Gaussian support vector machine.
Results
Patient population and breast lesion characteristics
The study population included 96 patients (Fig. 1).
Fig. 1.
Flowchart of inclusion and exclusion criteria for the study
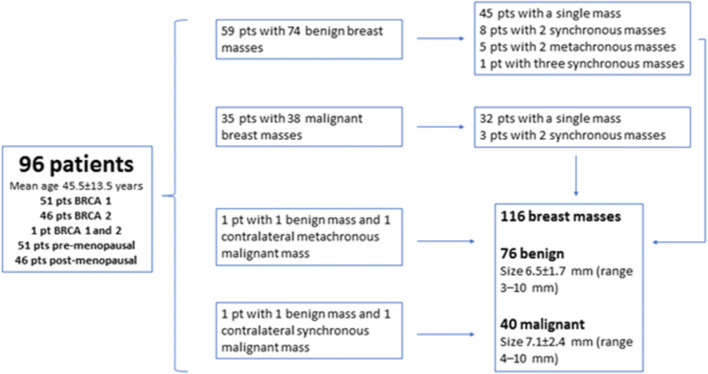
Table 2 and Fig. 2 show the patient and breast lesion characteristics. Figures 3 and 4 are examples of benign and malignant breast masses included in this study. After segmentation, the median benign lesion size was 514.5 pixels (range 85–2425 pixels) and the median malignant lesion size was 816 pixels (range 66–2116 pixels).
Table 2.
Histopathology of the 76 benign and 40 malignant masses
| Benign (n = 76) | Malignant (n = 40) |
|---|---|
| FAD 21; complex FAD 1; FAD with atypia 1 | IDC 29 |
| Ruptured cyst, adenosis, stromal fibrosis and normal breast parenchyma 21 | IDC + DCIS 4 |
| PASH 12 | ILC + DCIS 1 |
| Papilloma 3 | DCIS microinvasive 1 |
| Usual ductal hyperplasia 2 | DCIS 4 |
| Fat necrosis 1 | Metastatic intramammary lymph node 1 |
| LCIS 1; ALH 1 | |
| Columnar changes with atypia 1 | |
| Benign follow-up (BI-RADS 3) 8 | |
| Benign follow-up (BI-RADS 4), not visible at time of biopsy 3 |
FAD, fibroadenoma; PASH, pseudoangiomatous stromal hyperplasia; LCIS, lobular carcinoma in situ; ALH, atypical lobular hyperplasia; IDC, invasive ductal carcinoma; DCIS, ductal carcinoma in situ
Fig. 2.
Patient and breast lesion characteristics
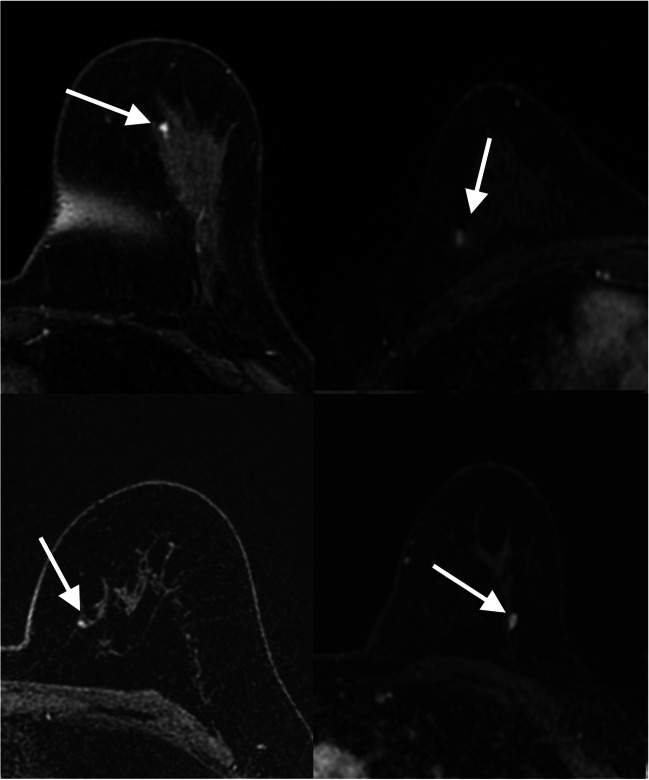
Fig. 3.
Transverse first post-contrast bilateral dynamic MR images (TR/TE, 4.5/2.1 ms; flip angle, 10°) of four patients with benign-appearing small breast masses (white arrows) in which biopsy yielded invasive ductal carcinoma
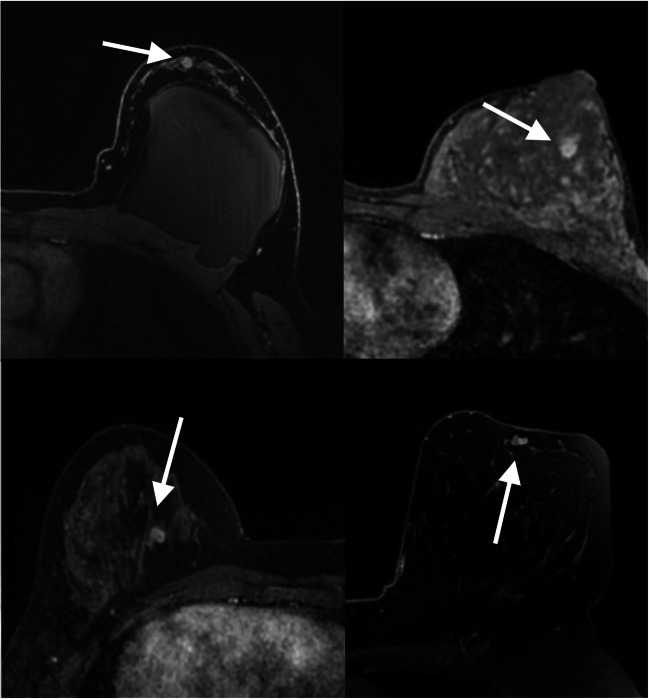
Fig. 4.
Transverse first post-contrast bilateral dynamic MR images (TR/TE, 4.5/2.1 ms; flip angle, 10°) of four patients with suspicious-appearing small breast masses categorized as BI-RADS 4 in which biopsy results yielded fibroadenoma (white arrows) and pseudoangiomatous stromal hyperplasia (white arrow)
Imaging assessment by radiologists
Consensus BI-RADS classification achieved a sensitivity of 75%, specificity of 42.1%, PPV of 40.5%, NPV of 76.2%, and accuracy of 53.4%. Time–intensity kinetic curve analysis was performed of 109/116 lesions; 7 lesions were not analyzed due to motion-related artifacts. Progressive contrast enhancement was present in 54.2% of patients with benign lesions (38/70) and in 23% of patients with malignant lesions (9/39); there was a statistically significant association with disease status based on kinetic analysis (p = 0.01).
Table 3 shows the results from univariate analysis according to independent assessments by the two radiologists.
Table 3.
Univariate analysis according to independent radiologist assessment
| Reader 1 | Reader 2 | |||||
|---|---|---|---|---|---|---|
| Imaging feature | Malign. | Benign | p value | Malign. | Benign | p value |
| BI-RADS | 0.003 | 0.002 | ||||
| 2 | 1 (1) | 1 (1) | 3 (3) | 12 (10) | ||
| 3 | 8 (7) | 29 (25) | 8 (7) | 21(18) | ||
| 4 | 26 (22) | 46 (40) | 22 (19) | 43 (37) | ||
| 5 | 5 (4) | 0 (0) | 7 (6) | 0 (0) | ||
| BPE | 0.047 | 0.33 | ||||
| Minimal | 20 (17) | 20 (17) | 23 (20) | 35 (30) | ||
| Mild | 13(11) | 33 (29) | 10 (9) | 25 (22) | ||
| Moderate | 5 (4) | 14 (12) | 5 (4) | 8 (7) | ||
| Marked | 1 (1) | 9 (8) | 1 (1) | 8 (7) | ||
| Bilateral mastectomy | 1 | 1 | ||||
| Contrast enhancement | 0.07 | 0.04 | ||||
| Homogeneous | 11 (10) | 25 (22) | 10 (9) | 17 (15) | ||
| Heterogeneous | 15(13) | 14 (12) | 17 (15) | 16 (14) | ||
| Rim enhancement | 10 (9) | 18 (16) | 7 (6) | 16 (14) | ||
| Dark internal septations | 4 (4) | 19 (16) | 6 (5) | 27 (23) | ||
| DWI signal | 0.22 | 0.91 | ||||
| Homogeneous | 12 (19) | 19 (30) | 9 (14) | 16 (25) | ||
| Heterogeneous | 0 (0) | 3 (5) | 1 (2) | 5 (8) | ||
| Rim | 2 (3) | 1 (2) | 1 (2) | 3 (5) | ||
| No correlation | 7 (11) | 21 (32) | 10 (15) | 20 (31) | ||
| Margins | 0.06 | 0.01 | ||||
| Circumscribed | 22 (19) | 54 (47) | 18 (16) | 48 (41) | ||
| Irregular | 16 (14) | 22 (19) | 18 (16) | 28 (24) | ||
| Spiculated | 2 (2) | 0 (0) | 4 (3) | 0 (0) | ||
| Shape | 0.19 | 0.03 | ||||
| Oval | 11 (10) | 34 (29) | 13 (11) | 36 (31) | ||
| Round | 15 (13) | 21 (18) | 11 (10) | 27 (23) | ||
| Irregular | 14 (12) | 21 (18) | 16 (14) | 13 (11) | ||
| T2 signal intensity | 0.02 | 0.17 | ||||
| Hypointense | 6 (5) | 4 (4) | 3 (3) | v6 (5) | ||
| Isointense | 6 (5) | 13 (11) | 11 (10) | 17 (15) | ||
| Hyperintense | 24 (21) | 35 (30) | 23 (20) | 35 (30) | ||
| No correlation | 4 (4) | 24 (21) | 3 (3) | 18 (16) | ||
| FGT breast with mass | 0.07 | 0.07 | ||||
| Almost entirely fat | 5 (4) | 2 (2) | 5 (4) | 9 (8) | ||
| Scattered FGT | 13 (11) | 24 (21) | 21 (18) | 25 (22) | ||
| Heterogeneous FGT | 13 (11) | 21 (18) | 8 (7) | 17 (15) | ||
| Extreme FGT | 8 (7) | 29 (25) | 5 (4) | 25 (22) | ||
| Mastectomy | 1 | 0 | 1 | 0 | ||
| FGT contralateral breast | 0.13 | 0.07 | ||||
| Almost entirely fat | 3 (3) | 1 (1) | 3 (4) | 8 (7) | ||
| Scattered FGT | 12 (11) | 24 (22) | 20 (18) | 24 (22) | ||
| Heterogeneous FGT | 12 (11) | 20 (18) | 7 (6) | 17 (16) | ||
| Extreme FGT | 8 (7) | 29 (27) | 5 (5) | 25 (23) | ||
| Mastectomy | 5 | 2 | 5 | 2 | ||
Values represent number of patients (percentages)
BI-RADS, Breast Imaging and Reporting and Data System; BPE, background parenchymal enhancement; DWI, diffusion-weighted imaging; FGT, fibroglandular tissue
Table 4 shows the results from univariate analysis according to overall consensus assessment as well as according to singular assessment performed for kinetics and lesion location, BRCA mutation status, and menopausal status. In consensus reading, there was no significant association with disease status based on margin (p = 0.11), shape (p = 0.97), enhancement pattern (p = 0.05), T2 signal intensity (p = 0.16), DWI (p = 0.54), BPE (p = 0.32), and BRCA mutation status (BRCA1 vs. BRCA2, p = 0.79). There was a statistically significant association with disease status based on lesion location within the breast (p = 0.03), menopausal status (p = 0.0001), and BI-RADS classification (p < 0.001).
Table 4.
Consensus analysis according to independent radiologist assessment
| Disease status | |||
|---|---|---|---|
| Imaging feature | Malignant | Benign | p value |
| BI-RADS | < 0.001 | ||
| 2 | 1 (1) | 5 (4) | |
| 3 | 9 (8) | 27 (23) | |
| 4 | 22 (19) | 44 (38) | |
| 5 | 8 (7) | 0 (0) | |
| BPE | 0.33 | ||
| Minimal | 23 (20) | 35 (30) | |
| Mild | 10 (9) | 25 (22) | |
| Moderate | 5 (4) | 8 (7) | |
| Marked | 1 (1) | 8 (7) | |
| BRCA | 0.80 | ||
| 1 | 20 (18) | 38 (33) | |
| 2 | 18 (16) | 38 (33) | |
| Contrast enhancement | 0.05 | ||
| Homogeneous | 9 (8) | 18 (16) | |
| Heterogeneous | 20 (17) | 21(18) | |
| Rim enhancement | 7 (6) | 16 (14) | |
| Dark internal septation | 4 (4) | 21 (18) | |
| DCE (kinetics)* | 0.01 | ||
| Progressive | 9 (8) | 38 (35) | |
| Plateau | 22 (20) | 23 (21) | |
| Washout | 8 (7) | 9 (8) | |
| DWI signal | 0.54 | ||
| Homogeneous | 10 (16) | 19 (30) | |
| Heterogeneous | 0 (0) | 3 (5) | |
| Rim | 2 (3) | 2 (3) | |
| No correlation | 8 (13) | 19 (30) | |
| Location* | 0.03 | ||
| Anterior | 5 (4) | 20 (17) | |
| Middle | 14 (12) | 34 (29) | |
| Posterior | 21 (18) | 22 (19) | |
| Margins | 0.11 | ||
| Circumscribed | 23 (20) | 51 (44) | |
| Irregular | 15 (13) | 25 (22) | |
| Spiculated | 2 (2) | 0 (0.0) | |
| Menopausal status | < 0.001 | ||
| Fertile | 12 (10) | 51 (44) | |
| Menopause | 28 (24) | 25 (22) | |
| Shape | 0.97 | ||
| Oval | 14 (12) | 28 (24) | |
| Round | 14 (12) | 25 (22) | |
| Irregular | 12 (10) | 23 (20) | |
| T2 Signal intensity | 0.16 | ||
| Hypointense | 5 (4) | 4 (4) | |
| Isointense | 8 (7) | 12 (10) | |
| Hyperintense | 22 (19) | 38 (33) | |
| No correlation | 5 (4) | 22 (19) | |
Values represent number of patients (percentages)
BI-RADS, Breast Imaging and Reporting and Data System; BPE, background parenchymal enhancement; DCE, dynamic contrast-enhanced; DWI, diffusion-weighted imaging. *Evaluated by reader 1
Radiomics analysis
ML Model using only the first post-contrast phase
At univariate analysis, 37/102 radiomics features were found to be significantly different between benign and malignant lesions (Supplemental Table S1). The AUC cutoff of ≥ 0.65 reduced the number of features of interest to 21/102. Correlation analysis resulted in 11 features (from 5 classes) for subsequent multivariate modeling (Supplemental Table S2). Using a fine Gaussian support vector machine with all 11 parameters, a perfect separation of benign and malignant cases was obtained, demonstrating 100% accuracy. However, this ML model undoubtedly overfitted the data (Supplemental Table S3).
After fivefold cross-validation, LASSO (least absolute shrinkage and selection operator) was used to further reduce the number of parameters. The final ML model utilized three parameters (GLCM-based correlation, SZM-based gray level non-uniformity normalized, and SZM-based zone emphasis). This ML model achieved a diagnostic accuracy of 75% but it can be regarded as a robust ML model which should produce similar results for new data (Supplemental Table S4). This ML model achieved a sensitivity of 55.0% (22/40), specificity of 85.5% (65/76), PPV of 66.7% (22/33), and NPV of 78.3% (65/83).
ML model combining radiomics features from the first post-contrast phase and clinical factors
We included clinical factors in multivariate modeling to further improve the model. Multivariate results showed that disease status was associated with menopausal status (χ2 = 11.86, p = 0.001), age (p < 0.0005), and lesion location (χ2 = 6.84, p = 0.03). There was no association with BRCA status (χ2 = 0.17, p = 0.68). A fivefold cross-validation was again employed to develop a robust ML model. The final ML model utilized six parameters (age, first-order coefficient of variation, GLCM-based joint entropy, GLCM-based correlation, GLCM-based cluster prominence, and RLM-based run emphasis). This robust ML model resulted in a diagnostic accuracy of 79.3% (Supplemental Table S5). This ML model achieved a sensitivity of 52.5% (21/40), specificity of 93.4% (71/76), PPV of 80.8% (21/26), and NPV of 78.9% (71/90).
ML model combining radiomics features from all dynamic phases and clinical factors
The results for the ML model using all dynamic phases and clinical factors are provided in the Supplemental Data (Supplemental Info A2, Table S6, Table S7, Table S8). This ML model resulted in a diagnostic accuracy of 81.5% and can be regarded as a robust model. The results from all radiomics models are illustrated in Table 5.
Table 5.
Summary of radiomics features model results
| Accuracy | Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|---|
| 1st PC phase (no validation) | 90.5% (83.7–95.2) | 75.0% (58.8–87.3) | 98.7% (92.9–100.0) | 96.8% (80.9–99.5) | 88.2% (81.4–92.8) |
| 1st PC phase (fivefold validation) | 75.0% (66.1–82.6) | 55.0% (38.5–70.7) | 85.5% (75.6–92.6) | 66.7% (52.0–78.7) | 78.3% (71.7–83.7) |
| 1st PC phase and clinical factors (fivefold validation) | 79.3% (70.8–86.3) | 52.5% (36.1–68.5) | 93.4% (85.3–97.8) | 80.8% (63.1–91.2) | 78.9 (72.9–83.9) |
| All phases and clinical factors (fivefold validation) | 81.5% (72.9–88.3) | 63.2% (46.0–78.2) | 91.4% (82.3–96.8) | 80.0% (64.2–89.9) | 82.1% (75.0–87.5) |
Confidence intervals are in parenthesis
PC, post-contrast; PPV, positive predictive value; NPV, negative predictive value
Discussion
In this study, we investigated whether radiomics analysis and ML with MRI can accurately differentiate sub-centimeter benign from malignant lesions in BRCA mutation carriers using model-free parameter maps. We demonstrated that radiomics analysis coupled with ML aids in the differentiation of benign and malignant enhancing sub-centimeter masses in these patients. The T2-weighted signal intensity and DW imaging did not help to differentiate benign from malignant lesions. While larger cancers have been well-described and characterized on MRI, sub-centimeter lesions, particularly those less than 0.5 cm, have traditionally been regarded as being too small to characterize according to morphological descriptors, negatively impacting accuracy. With advancements in hardware and software, the spatial resolution of MRI has improved, allowing not only the detection but also the morphologic characterization of small enhancing lesions [19].
Meissnitzer et al [13] showed that sub-centimeter invasive breast cancers often present with benign morphologic features such as persistent enhancement (30%) and high T2 signal (17%). Raza et al [20] demonstrated that breast cancers smaller than 5 mm tend to present with circumscribed margins (71%), benign shape (67%), and benign kinetic characteristics (41%). The presence of a BRCA mutation is an additional confounding factor as breast cancers in this population often present with benign morphologic features (e.g., oval shape and well-defined margins) on MRI and can resemble a fibroadenoma or a cyst in 23–38% of cases [12, 20]. Yet, these cancers are more aggressive with fast growth rates and a short lead time [20].
Our results confirmed that for sub-centimeter masses in BRCA mutation carriers, morphologic BI-RADS descriptors are not particularly useful for breast cancer diagnosis; there was only moderate inter-rater agreement for morphology although there was at least substantial inter-rater agreement for the BI-RADS assessment categories. Compared with Ha et al [21] who concluded that any T2 hypointense enhancing focus representing an interval change should be biopsied rather than undergo short-term follow-up, we found no significant difference in T2 signal intensity between benign and malignant lesions. This is in agreement with Zhang et al who also showed that T2-weighted imaging does not significantly contribute to differentiating benign from malignant lesions [22]. In addition, we found that DWI signal analysis did not contribute to the accuracy of assessing these lesions, which can in part be explained by its limited spatial resolution which makes it challenging to accurately evaluate sub-centimeter masses.
Several studies have shown that radiomics and machine learning can be used as adjuvant tools to support radiologist image interpretation in differentiating benign from malignant lesions using mammography [23], digital breast tomosynthesis [24], and MRI [16, 25]. A study by Truhn et al [26] demonstrated that radiomics and CNN were superior compared with radiomics analysis in differentiating benign from malignant breast masses but both were inferior to the assessment performed by the radiologist. However, for this study, the authors included lesions with overall average diameter of 22.4 ± 20.3; thus, their results could be due to the fact that when lesions are larger in size, they are easier to be characterized as benign or malignant by just analyzing BI-RADS descriptors.
Our study shows a more accurate means of differentiating benign from malignant lesions in BRCA mutation carriers. Gibbs et al evaluated the utility of radiomics and ML from DCE-based parameter maps to diagnose small breast lesions in the general population [16]. The best AUC was 0.78 ± 0.12 and their results showed that radiomics can potentially improve the evaluation of small, benign-appearing breast masses, with increased PPV (fewer biopsies needed) and NPV (more cancers diagnosed) compared with the currently used BI-RADS classification alone. In our study population of BRCA mutation carriers, our data indicate that radiomics analysis and ML can in fact spare women from unnecessary biopsies for benign-appearing small breast nodules. Three radiomics features (coefficient of variation, cluster prominence, and Haralick correlation) were able to separate benign from malignant masses with a diagnostic accuracy of 79.3% when only the first post-contrast scan, combined with clinical data, was used in a ML model.
Another study by D’Amico et al [27] examined 12 malignant and 33 benign enhancing foci in 45 patients. From these foci, over 200 radiomics features were extracted and performances of selected features were evaluated by means of k-nearest neighbor (kNN). A fast and robust classification algorithm yielded a sensitivity of 27/27 (100%, 95% CI 87–100%), a specificity of 37/41 (90%, 95% CI 77–97%), and an accuracy of 64/68 (94%, 95% CI 86–98%). Compared with D’Amico et al, our study compared machine learning to radiologist’s image interpretation according to BI-RADS from 2 different readers, included a larger sample size of 116 lesions (vs. 45), and included a more homogeneous patient populations with BRCA mutations.
Recently, alternative abbreviated protocols have been proposed for screening women [19, 28] to reduce scan time by acquiring only one pre-contrast and one early post-contrast T1-weighted image set. In agreement with the results of Gibbs et al [16], our results showed that delayed post-contrast phases did not add any significant discriminative value to the analysis. This study therefore provides indirect evidence for the potential use of radiomics analysis in abbreviated protocols which have been recently proposed as an alternative for screening high-risk women with dense breast tissue [19] without concerns regarding a decrease in specificity related to the lack of information of enhancement kinetics in the delayed phases.
This study has limitations. By using only single-center data, it is difficult to predict how the developed models might perform with data acquired under different imaging protocols, especially in the case of poorer spatial resolution and slice thickness. We included only sub-centimeter breast masses which do not constitute many pixels in an image, leading to lower spatial resolution and fewer pixels in the final ROI and an increased proportion of pixels that can be regarded as potentially contaminated by partial volume effects. To ensure adequate counting statistics, we decreased the data to only 16 gray levels (vs. 32 or 64 gray levels that have previously been employed in breast MRI) [29]. Another limitation is the relatively small sample size of 116 breast masses due to our strict inclusion criteria. With only 40 cases in the malignant group, feature selection was performed prior to any cross-validation fold.
In conclusion, radiomics analysis coupled with machine learning improves the diagnostic accuracy in small breast masses in BRCA mutation carriers compared with the qualitative morphological assessment with BI-RADS classification alone. Further studies, preferentially multi-center studies in larger patient cohorts, are needed to confirm these promising results.
Electronic supplementary material
(DOCX 38.7 kb)
Abbreviations
- BI-RADS
Breast Imaging Reporting and Data System
- BPE
Background parenchymal enhancement
- FGT
Fibroglandular tissue
- GLCM
Gray level co-occurrence matrix
- ML
Machine learning
- NPV
Negative predictive value
- PPV
Positive predictive value
- RLM
Run length matrix
- SZM
Size zone matrix
Funding information
Open access funding provided by Medical University of Vienna. This study received funding from the NIH/NCI Cancer Center Support Grant (P30 CA008748), the Breast Cancer Research Foundation, Susan G. Komen, Spanish Foundation Alfonso Martin Escudero, and the European School of Radiology.
Compliance with ethical standards
Guarantor
The scientific guarantor of this publication is Katja Pinker, MD PhD.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
Two of the authors (Peter Gibbs, PhD, and Almir Bitencourt, MD, PhD) have significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• observational
• performed at one institution
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Levy-Lahad E, Friedman E. Cancer risks among BRCA1 and BRCA2 mutation carriers. Br J Cancer. 2007;96:11–15. doi: 10.1038/sj.bjc.6603535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhl C, Weigel S, Schrading S, et al. Prospective multicenter cohort study to refine management recommendations for women at elevated familial risk of breast cancer: the EVA trial. J Clin Oncol. 2010;28:1450–1457. doi: 10.1200/JCO.2009.23.0839. [DOI] [PubMed] [Google Scholar]
- 3.Warner E, Plewes DB, Hill KA, et al. Surveillance of BRCA1 and BRCA2 mutation carriers with magnetic resonance imaging, ultrasound, mammography, and clinical breast examination. JAMA. 2004;292:1317–1325. doi: 10.1001/jama.292.11.1317. [DOI] [PubMed] [Google Scholar]
- 4.Kriege M, Brekelmans CT, Boetes C, et al. Efficacy of MRI and mammography for breast-cancer screening in women with a familial or genetic predisposition. N Engl J Med. 2004;351:427–437. doi: 10.1056/NEJMoa031759. [DOI] [PubMed] [Google Scholar]
- 5.Rijnsburger AJ, Obdeijn IM, Kaas R, et al. BRCA1-associated breast cancers present differently from BRCA2-associated and familial cases: long-term follow-up of the Dutch MRISC Screening Study. J Clin Oncol. 2010;28:5265–5273. doi: 10.1200/JCO.2009.27.2294. [DOI] [PubMed] [Google Scholar]
- 6.Saslow D, Boetes C, Burke W, et al. American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. CA Cancer J Clin. 2007;57:75–89. doi: 10.3322/canjclin.57.2.75. [DOI] [PubMed] [Google Scholar]
- 7.Lee CH, Dershaw DD, Kopans D, et al. Breast cancer screening with imaging: recommendations from the Society of Breast Imaging and the ACR on the use of mammography, breast MRI, breast ultrasound, and other technologies for the detection of clinically occult breast cancer. J Am Coll Radiol. 2010;7:18–27. doi: 10.1016/j.jacr.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 8.Riedl CC, Luft N, Bernhart C, et al. Triple-modality screening trial for familial breast cancer underlines the importance of magnetic resonance imaging and questions the role of mammography and ultrasound regardless of patient mutation status, age, and breast density. J Clin Oncol. 2015;33:1128–1135. doi: 10.1200/JCO.2014.56.8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lakhani SR, Jacquemier J, Sloane JP, et al. Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. J Natl Cancer Inst. 1998;90:1138–1145. doi: 10.1093/jnci/90.15.1138. [DOI] [PubMed] [Google Scholar]
- 10.Krammer J, Pinker-Domenig K, Robson ME, et al. Breast cancer detection and tumor characteristics in BRCA1 and BRCA2 mutation carriers. Breast Cancer Res Treat. 2017;163:565–571. doi: 10.1007/s10549-017-4198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakhani SR, Van De Vijver MJ, Jacquemier J, et al. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20:2310–2318. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 12.Marino MA, Riedl CC, Bernathova M, et al. Imaging phenotypes in women at high risk for breast cancer on mammography, ultrasound, and magnetic resonance imaging using the fifth edition of the Breast Imaging Reporting and Data System. Eur J Radiol. 2018;106:150–159. doi: 10.1016/j.ejrad.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meissnitzer M, Dershaw DD, Feigin K, Bernard-Davila B, Barra F, Morris EA. MRI appearance of invasive subcentimetre breast carcinoma: benign characteristics are common. Br J Radiol. 2017;90:20170102. doi: 10.1259/bjr.20170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kauff ND, Brogi E, Scheuer L, et al. Epithelial lesions in prophylactic mastectomy specimens from women with BRCA mutations. Cancer. 2003;97:1601–1608. doi: 10.1002/cncr.11225. [DOI] [PubMed] [Google Scholar]
- 15.Isern AE, Loman N, Malina J, Olsson H, Ringberg A. Histopathological findings and follow-up after prophylactic mastectomy and immediate breast reconstruction in 100 women from families with hereditary breast cancer. Eur J Surg Oncol. 2008;34:1148–1154. doi: 10.1016/j.ejso.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Gibbs P, Onishi N, Sadinski M, et al. Characterization of sub-1 cm breast lesions using radiomics analysis. J Magn Reson Imaging. 2019;50:1468–1477. doi: 10.1002/jmri.26732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apte AP, Iyer A, Crispin-Ortuzar M et al (2018) Technical note: extension of CERR for computational radiomics: a comprehensive MATLAB platform for reproducible radiomics research. Med Phys. 10.1002/mp.13046 [DOI] [PMC free article] [PubMed]
- 18.Fortin JP, Parker D, Tunc B, et al. Harmonization of multi-site diffusion tensor imaging data. Neuroimage. 2017;161:149–170. doi: 10.1016/j.neuroimage.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhl CK. Abbreviated breast MRI for screening women with dense breast: the EA1141 trial. Br J Radiol. 2018;91:20170441. doi: 10.1259/bjr.20170441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raza S, Sekar M, Ong EM, Birdwell RL. Small masses on breast MR: is biopsy necessary? Acad Radiol. 2012;19:412–419. doi: 10.1016/j.acra.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 21.Ha R, Sung J, Lee C, Comstock C, Wynn R, Morris E. Characteristics and outcome of enhancing foci followed on breast MRI with management implications. Clin Radiol. 2014;69:715–720. doi: 10.1016/j.crad.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Zhang M, Horvat JV, Bernard-Davila B, et al. Multiparametric MRI model with dynamic contrast-enhanced and diffusion-weighted imaging enables breast cancer diagnosis with high accuracy. J Magn Reson Imaging. 2019;49:864–874. doi: 10.1002/jmri.26285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drukker K, Giger ML, Joe BN, et al. Combined benefit of quantitative three-compartment breast image analysis and mammography radiomics in the classification of breast masses in a clinical data set. Radiology. 2019;290:621–628. doi: 10.1148/radiol.2018180608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakai A, Onishi Y, Matsui M et al (2019) A method for the automated classification of benign and malignant masses on digital breast tomosynthesis images using machine learning and radiomic features. Radiol Phys Technol. 10.1007/s12194-019-00543-5 [DOI] [PubMed]
- 25.Sutton EJ, Huang EP, Drukker K, et al. Breast MRI radiomics: comparison of computer- and human-extracted imaging phenotypes. Eur Radiol Exp. 2017;1:22. doi: 10.1186/s41747-017-0025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truhn D, Schrading S, Haarburger C, Schneider H, Merhof D, Kuhl C. Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology. 2019;290:290–297. doi: 10.1148/radiol.2018181352. [DOI] [PubMed] [Google Scholar]
- 27.D'Amico NC, Grossi E, Valbusa G, et al. A machine learning approach for differentiating malignant from benign enhancing foci on breast MRI. Eur Radiol Exp. 2020;4:5. doi: 10.1186/s41747-019-0131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leithner D, Moy L, Morris EA, Marino MA, Helbich TH, Pinker K. Abbreviated MRI of the breast: does it provide value? J Magn Reson Imaging. 2019;49:e85–e100. doi: 10.1002/jmri.26291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrading S, Kuhl CK. Mammographic, US, and MR imaging phenotypes of familial breast cancer. Radiology. 2008;246:58–70. doi: 10.1148/radiol.2461062173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 38.7 kb)