Abstract
Ciliates are a highly divergent group of unicellular eukaryotes with separate somatic and germline genomes found in distinct dimorphic nuclei. This characteristic feature is tightly linked to extremely laborious developmentally regulated genome rearrangements in the development of a new somatic genome/nuclei following sex. The transformation from germline to soma genome involves massive DNA elimination mediated by non-coding RNAs, chromosome fragmentation, as well as DNA amplification. In this review, we discuss the similarities and differences in the genome reorganization processes of the model ciliates Paramecium and Tetrahymena (class Oligohymenophorea), and the distantly related Euplotes, Stylonychia, and Oxytricha (class Spirotrichea).
Keywords: Ciliates, Nuclear dimorphism, Genome rearrangement, DNA elimination, Small non-coding RNAs
Introduction
Developmentally regulated genome rearrangements (DRGRs) involve the elimination of specific DNA sequences (from the germline) somatic cell lineages. In most cases, this phenomenon is associated with two forms of DNA elimination either: (a) chromosome elimination where the entire chromosome is lost [1] or (b) chromosome diminution, a process characterized by loss of chromosome portions through chromosome breakage and repair during the developmental transformation from germline to soma [2, 3].
Programmed DNA elimination was first described in 1887 by Theodor Boveri [4] in the horse parasitic nematode, Parascaris univalens. Since then, DRGRs have been identified in diverse multicellular organisms including nematodes, arthropods, hagfish, lampreys [3] and lymphoid lineages of vertebrates [5]. However, it appears most pervasive in ciliates, an ancient clade of microbial eukaryotes (> 1 Gya; [6]), where genome rearrangements lead to the elimination of 30–95% of the germline genome [7–9]. This review will focus on genome rearrangements in the two best-studied classes of ciliates: the Oligohymenophorea (including Paramecium and Tetrahymena), and members of the Spirotrichea (including Euplotes, Oxytricha and Stylonychia).
Ciliates as a model organism
Ciliates are unicellular eukaryotes found in diverse environments (fresh/saltwater as well as soil) across the globe that emerged more than 1 billion years ago [6]. Due to their morphological and morphogenetic characters, the taxonomy of ciliates has been ambiguous for a long time. Numerous studies have improved the phylogenetic relationship between ciliates with the rest of the eukaryotic tree of life, being members of the Alveolata (along with apicomplexans and dinoflagellates) [10]. Similarly, phylogenomic studies within Ciliophora illustrate the great diversity and deep evolutionary history despite limited taxon sampling [11]. Even though just a handful of the ~ 4500 described ciliate species have been studied in-depth [12], they share complex cytoskeletal structures, well-developed ciliary structures at the cell surface (for swimming, food uptake and sensing environmental signals), the separation of germline and somatic genomes into distinct nuclei (nuclear dimorphism), as well as DRGRs (reviewed in [13]). Although the majority of studies are limited to the model genera Tetrahymena, Paramecium, and Oxytricha, these models have greatly contributed to our understanding of biological mechanisms and phenomena present in diverse eukaryotic lineages. This includes discovery and description of ribozymes [14], the discovery of the first histone-modifying enzyme [15], variant nuclear genetic codes [16–20], the initial identification of telomerase and telomere structure [21, 22], numerous examples of small RNA-mediated heterochromatin formation [23, 24], as well as mechanisms enabling the transcription of short DNA fragments [25].
Nuclear dimorphism in ciliates
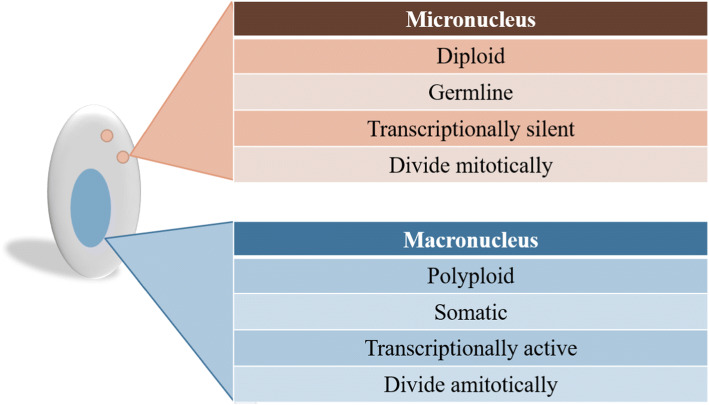
In multicellular eukaryotes, germline and somatic functions are separated into distinct cell types, (e.g. pollen versus leaf in plants, spore versus hyphae in fungi, or egg versus skin in humans). However, in ciliates, both germline and somatic genomes co-exist within a single cell, providing each cell with at least one somatic nucleus used for gene expression and one germline to propagate the genome across sexual generations (Fig. 1).
Fig. 1.
Representative differences between germline and somatic nuclei ciliates
Each ciliate cell possesses at least one micronucleus (MIC) and one macronucleus (MAC); however, their number varies between the species (reviewed in [26]). Interestingly, all micronuclei present in the cell possess features of typical eukaryotic nuclei, (i.e. diploid [27–29], centromeres [30] and are transposon rich [30, 31]). The MAC is transcriptionally active throughout the entire life cycle and possesses highly processed chromosomes. These MAC chromosomes are gene-rich, lack centromeres and can range in ploidy from ~ 2 N in the Karyorelictea to > 13,000 N in the Heterotrichea [32–34]. During asexual growth, the hyper-polyploid MACs divide amitotically, which lacks mitotic spindles and chromatin condensation, separating chromosomes in bulk as large masses, which can result in daughter nuclei with unequal amounts of DNA. The degree of inequality in the segregation of DNA to the two MACs during amitosis can be exacerbated under environmental stressors [35, 36]. Moreover, MAC chromosomes are amplified to elevated copy numbers [26]. For instance, in Oligohymenophorea, each MAC chromosome is present in the equal copy number, namely, ~ 45 copies of each 225 chromosomes in T. thermophila [37, 38] and ~ 800 copies of each chromosome in P. tetraurelia [39, 40]. In contrast, the Oxytricha MAC harbours thousands of unique gene-sized nanochromosomes, which are amplified to ~ 1900 copies [31]. However, unlike Paramecium and Tetrahymena, these nanochromosomes are maintained at unique copy numbers, varying between a few hundred to 106 copies [26, 41–43].
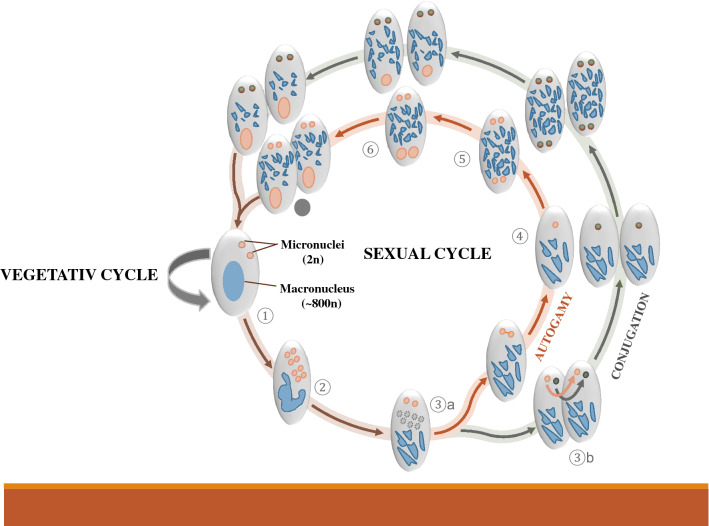
During vegetative (or asexual) growth, the germline remains transcriptionally inactive and divides mitotically [44]. This changes during sex or self-fertilisation/autogamy (Fig. 2) [45]. At the onset of sex and development, micronuclei undergo meiosis and are fused with a partner haploid MIC that gives rise to the zygotic nucleus from which new micro- and macronuclei are formed.
Fig. 2.
The Paramecium sexual cycle (autogamy and conjugation). (1) A vegetative cell with diploid micronuclei and polyploid macronucleus. (2) Meiosis of the micronuclei and beginning of old macronucleus fragmentation. (3a) Mitotic division of one remaining micronucleus (seven out of eight micronuclei degenerates) leading to the production of two identical gametic nuclei. (3b) Alternatively, during conjugation, exchange of haploid nuclei occurs. (4) Zygotic nuclei formation through the fusion of two haploid products. (5) Two subsequent mitotic divisions of the zygotic nuclei. (6) Differentiation of the two mitotic products into new macronuclei. (7) Caryonidal division/separation leading to the formation of two cells each containing two micronuclei and new macronucleus as well as fragments of old macronucleus
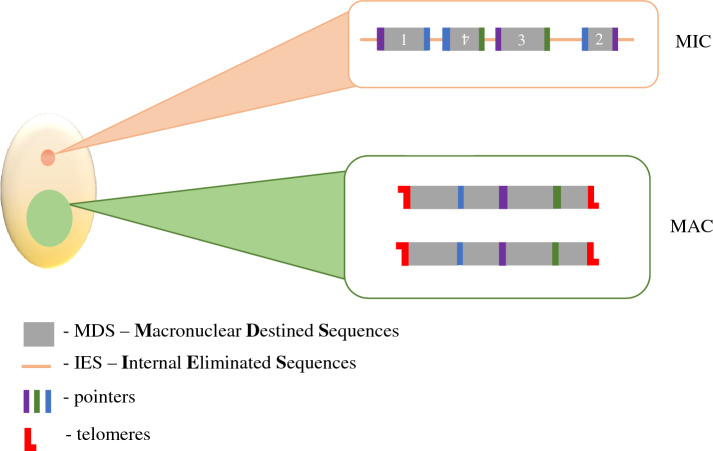
Features and origin of internal eliminated sequences
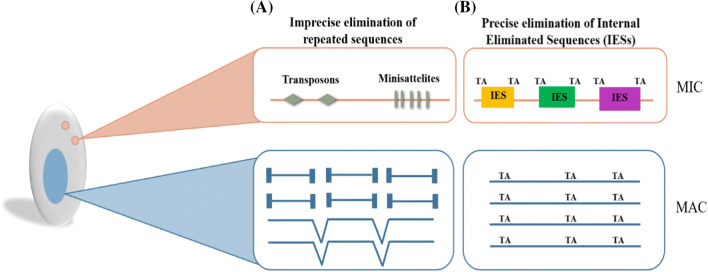
The discovery that germline-limited internal eliminated sequences (IESs) resemble transposon sequences brought a new challenge for scientists trying to understand the mechanisms underlying their elimination. Jacobs and Klobutcher [46] observed that IESs in Euplotes crassus possess the consensus sequence 5′-TATrGCRN-3′ (Y = pyrimidine, R = purine), which resembles terminal inverted repeats (TIRs) at the end of their Tec family transposable elements [46–48] and Paramecium’s Tc1/Mariner transposons [46–49]. All IESs found to date in Euplotes and Paramecium possess 5′-TA-3′ dinucleotide repeats at each boundary, where a single copy of the dinucleotide remains in the new macronucleus after excision. Based on these observations, Klobutcher and Herrick [47, 50] developed a model for the origin of IESs, where transposons initially invade the germline genome, then spread throughout, and ultimately decay over time into the identifiable IESs currently found in ciliate germline genomes.
These sequences must be excised during the development of a MAC to produce a functional somatic genome. IESs are present in all the ciliate germline genomes studied to date [30, 31, 48, 51–53], although in varying amounts (~ 12,000 IESs in Tetrahymena thermophila [54], ~ 45,000 IES in Paramecium tetraurelia [48, 51] and > 200,000 IESs in Oxytricha trifallax [31]. IESs are typically AT rich (70–100%) and bounded by pairs of short direct repeats (most are 1-8 bp) that help identify the boundaries between Macronuclear Destined Sequences (MDSs) and IESs [30, 31, 48, 52]. Recently, Maurer-Alcalá et al. [53] demonstrated that MDS–IES boundaries are identifiable by sharp changes in GC content. For instance, in P. tetraurelia where IES excision is precise, GC content was decreased in close proximity to its MDS–IES boundaries. On the other hand, in Tetrahymena where almost all IESs are excised imprecisely, GC contents are characterized by the great variability associated with MDS–IES boundaries within the inferred MDS itself [53]. The length and genomic distribution of IESs in germline genomes are very diverse, with most IESs in Tetrahymena being intergenic and “long” (> 100 bp to over 10 Kbp) [55, 56], whereas the IESs in Paramecium and Oxytricha often interrupt protein-coding sequences and are comparatively short (most < 100 bp) [31, 48].
As IESs often disrupt coding regions for most ciliates, they must be accurately excised during development to enable expression of the functional genes in the newly developed MAC [48, 57–59]. However, in Tetrahymena thermophila nearly all of the 7350 well-described IESs (of the ~ 12,000 total IESs) are excised imprecisely, due to variable MDS–IES junction sites. The potential deleterious impacts of this imprecision are likely mitigated by the genomic distribution of these IESs, which are predominantly found in intergenic (6182; 82%) or intronic (1168; 16%) regions [30, 60–62]. In contrast, only the excision of transposon-like sequences and minisatellites in Paramecium is imprecise [63], whereas the ~ 45,000 IESs nestled within or near protein-coding sequences are precisely excised during macronuclear development, although small numbers of IESs are excised at alternative MDS–IES boundaries [40, 48, 64]. Overall, imprecise elimination results either in the fragmentation of micronuclear chromosomes into shorter acentromeric macronuclear chromosomes to which telomeric repeats are added, or to the imprecise re-joining of flanking sequences [63].
Previous work in Paramecium and Tetrahymena have shown that the pointer sequences present at both ends of IESs, influences the efficacy of IES excision. Analyses of Paramecium’s IESs have demonstrated that single basepair-mutations in the conserved terminal repeat of IESs lead to their retention during development [65–67]. Additionally, for some Paramecium IESs, flanking sequences are necessary for excision [68]. For example, the removal of a portion of the 72 bp flanking region of one end of a small 28 bp IES in Paramecium reduced the efficiency of excision, and complete removal of all wild-type sequences adjacent to the TA abolished excision [68]. Recently, it has been shown a small subset of Paramecium’s IESs shares a common 5 bp motif that is implicated in their sRNA-independent excision [69]. In Tetrahymena, flanking sequences are known to have a significant role in the elimination of a number of IESs [70, 71]. Together, Lia3p and Lia3-Like 1 (LTL1) regulatory proteins interact with flanking regulatory sequences to determine MDS–IES boundaries for several IESs for excision [72–74]. These data highlight the importance of both IES pointer sequences and their flanking regions in identifying MDS–IES boundaries.
Another variable that influences IES excision/recognition in Paramecium is its length. Swart et al. [75] indicated that the frequencies of IES sub-terminal bases change with IES length. Moreover, it has been shown that small IESs (shorter than 150 bp) are less sensitive to sRNAs depletion [76], suggesting that some IESs are more difficult to recognize/excise and require additional information (from the sRNAs) for their accurate excision.
Developmentally regulated genome rearrangements
Although the mechanistic details behind ciliate DRGRs differ between even closely related taxa (i.e. P. tetraurelia and T. thermophila), the basic principles of this phenomenon are conserved (Table 1). In the developing macronucleus, rapid DNA synthesis takes place and interstitial DNA sequences such as transposons, minisatellites and IESs are excised. Afterward, the hundreds to thousands of broken chromosome ends created during excision are rejoined through non-homologous end-joining mechanisms [77–79], followed by de novo telomere addition, and finally chromosome amplification (Fig. 3). The end result is the production of a new functional somatic nucleus that contains the streamlined transcriptionally active chromosomes that maintain cell. This section is devoted to describing these phenomena in more detail.
Table 1.
Differences between the genome reorganization processes among ciliates
| Paramecium tetraurelia | Tetrahymena thermophila | Oxytricha trifallax | |
|---|---|---|---|
| MIC chromosomes | ? | 5 [37] | ? |
| MAC chromosomes | 188 [39, 40] | 225 [38] | 15,600 [43] |
| Nanochromosomes | No | No | Yes [43] |
| Unscrambling | No | Noa | Yes [178] |
| IES percentage | 30% [48] | 30% [179] | 90% [31] |
| IES location | Genic and intergenic regions [48] | Intergenic Regions [90] | Genic and intergenic regions [180] |
| Small RNA source | MDS and IES [25, 81] | Biased towards IES [111] | MDS [83] |
| Small RNA target | IES [81] | IES [80] | MDS [82, 83] |
| DNA methylation | 6 mA (P. aurelia) [132] | 6 mA [131] | 5mC, 6 mA [127, 135] |
aOne locus showing unscrambling [181]
Fig. 3.
Macronuclear differentiation process is shown on Paramecium example. a Imprecise elimination of repeated sequences like minisatellites and transposons followed by re-joining of the flanking sequences or de novo telomere addition. b Precise excision of internal eliminated sequences (IESs) possessing two TA repeats at each boundary one copy of which remains after excision
Role of small noncoding RNA in programmed genome rearrangements
An important breakthrough in our understanding of the regulation of DNA elimination was unravelling the involvement of noncoding RNA (ncRNA) in this complex epigenetic process. Briefly, the MIC is bi-directionally transcribed producing long transcripts that are processed into small RNAs. In Paramecium and Tetrahymena (cl: Oligophymenophorea), these sRNAs then “scan” the parental MAC. Those sRNAs corresponding to the parental MAC are lost, leading to the enrichment of MIC-matching sRNAs [23, 76, 80, 81]. However, in Oxytricha and Stylonychia these sRNAs are produced from transcripts derived from the parental MAC genome and are putatively involved in protecting MDSs in the MIC rather than identifying IESs for elimination (as in Paramecium and Tetrahymena) [82–84]. This reflects the extreme differences in germline genome content as ~ 30% of the germline genome is eliminated in Oligohymenophorean ciliates versus ≥ 95% of the germline in spirotrichs. These sRNAs pools ultimately help delineate MDSs and IESs, although details differ among even “closely” related species (e.g. Paramecium and Tetrahymena).
Small RNA-mediated programmed genome rearrangements in Oligohymenophorea
The first insights that DNA elimination relies on homologous RNA molecules originated from work in Oligohymenophorean ciliates (i.e. Paramecium and Tetrahymena). During prophase of meiosis, the MIC is bi-directionally transcribed [85] by RNA polymerase II [86], generating large MIC-based transcripts. These transcripts are then processed into small “scan” RNAs (scnRNAs are 25 nt in Paramecium and 25–29 nt in Tetrahymena), by Dicer-like ribonucleases (Dcl1 in Tetrahymena and Dcl2 and Dcl3 in Paramecium), that “scan” for homologous sequences in the parental MAC genome (Fig. 4a) [76, 81, 87–89]. Initial experiments suggested that scnRNA production from MIC-derived transcripts, represented relatively equal representation of IES and MDS regions [23]. However, recent work in Tetrahymena shows two “pulses” of sRNA production, which are associated with enriched transcription and processing of IES sequences [90]. Interestingly, the biased production of Tetrahymena scnRNAs is predominantly from type-A IESs (per-centromeric and telomeric regions of the MIC chromosomes) [91].
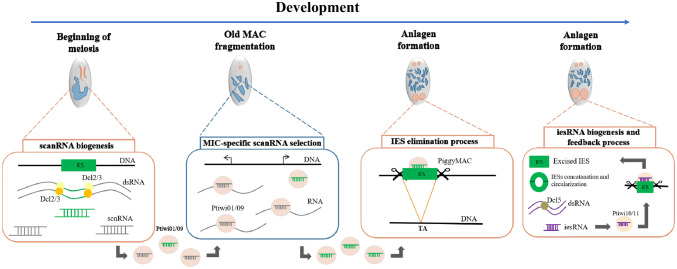
Fig. 4.
The scanning model in Paramecium. a Bi-directional transcription of the parental micronuclear genome during meiosis and scnRNA production by Dcl2/Dcl3. b Scanning process between scnRNA and transcript of somatic DNA. c scnRNA targeting IES for excision by PiggyMac. d Concatenation and circularization of excised IESs. Transcription of excised and circularized IESs to produce dsRNA. dsRNA cleavage by Dcl5 to produce iesRNA ensuring elimination of all copies of IESs. Old macronucleus degradation and development of the new macronucleus
After production in the micronuclei, scnRNA duplexes are transported to the cytoplasm where they are loaded onto the PIWI family proteins, (Twi1p in Tetrahymena [23, 80] or Ptiwi1/9 complex present in Paramecium [92, 93]) which are then transported to the parental macronucleus (Fig. 4b). There, the genome “scanning” effectively removes the MAC-matching sRNAs, enriching for micronuclear-limited scnRNAs from the initial population. These MIC-enriched scnRNAs are transported to the developing macronucleus to guide DNA excision (Fig. 4c). Interestingly, it has been suggested that Paramecium’s scnRNAs bind to longer RNA transcripts, rather than directly to DNA, in both the old and new MAC [94, 95]. In both Tetrahymena and Paramecium, there is a second wave of sRNAs that aid in ensuring the accurate identification and excision of IESs [76, 91]. In Tetrahymena, these “late scnRNAs” are produced from both types of IESs [Type-A and Type-B (located at the chromosomal arms)] in cis. These late scnRNAs are loaded onto the Twi1p and Twi11p complexes, which further guide heterochromatin formation in trans and ensure the elimination of all IESs copies [91]. However, in Paramecium, excised IESs are eventually circularized, with smaller IESs concatenated together prior to circularization [25]. These IES concatemers act as the transcriptional template for iesRNAs, which further ensure the precise and accurate excision of IESs [25]. These secondary iesRNAs are produced after IES excision in Paramecium, whereas Tetrahymena’s “late” scnRNAs are produced prior to any IES excision [96]. Compared to Tetrahymena where early and late scnRNAs are produced by the same Dicer-like ribonuclease (Dcl1), Paramecium iesRNAs are produced by Dcl5 [76]. As in Tetrahymena the primary scnRNAs and secondary iesRNAs are associated with distinct Piwi proteins, Ptiwi01/09 with scnRNAs and Ptiwi10/11 carries the iesRNAs [93]. Together, the primary and secondary sRNAs ensure the faithful elimination of all copies of IESs present in the developing MAC genome leading to the production of a new functional macronucleus (Fig. 4d).
Small RNA-mediated programmed genome rearrangements in Spirotrichea
DNA elimination in Oxytricha (Spirotrichea) is quite distinct from the distantly related Oligohymenophorea. While Paramecium and Tetrahymena generate scnRNA in the parental MIC, Oxytricha’s, 27-nt-long small RNAs derive from the transcription of the parental macronucleus rather than the germline [82, 83]. In addition to that, these 27mers have been shown to associate with PIWI homologs called Otiwi1 (hence called PIWI-interacting RNAs, piRNAs). The injection of 27 nt piRNAs corresponding to IESs into developing Oxytricha leads to the retention of those IES in the new somatic genome [83]. These data, combined with the apparent parental MAC origin of the piRNAs, suggest that they are responsible for identifying macronuclear destined sequences (MDS) to protect against excision, rather than targeting IESs for excision, as in Tetrahymena and Paramecium. Interestingly, as in Paramecium, Oxytricha also circularizes some excised TE and non-repetitive germline-limited sequences that are also actively transcribed [97, 98]. This presence of development-specific extrachromosomal circular DNA was originally described in Euplotes (cl: Spirotrichea) [99, 100], although the circularization process and content appears to differ between Oxytricha and Paramecium. If these circularized products of excised IES and transposon-like Tec elements in Euplotes lead to the production of small RNAs remains undetermined. Despite the differences in sRNA sources and targets, ciliates have evolved a relatively efficient and low-energy cost means to distinguish soma and germline.
DNA unscrambling
In addition to delineating somatic and germline-limited DNA, macronuclear development in Oxytricha requires a very spectacular form of DNA rearrangement called unscrambling [26, 31, 101]. In the germline, MDSs can also be disordered and/or found on both strands of DNA (i.e. “inverted”) and may even originate from distant germline loci (Fig. 5) [31]. An extreme example is DNA polymerase α in O. nova, O. trifallax [102, 103] and S. lemnae [104], which is broken into more than 40 MDS present at two distinct loci separated by > 3 kbp. Work in Oxytricha has demonstrated that DNA unscrambling is directed by long RNA templates derived from the parental macronucleus [105–107]. These long template RNAs, in conjunction with unique pointer sequences, act as a reference aiding in the accurate reordering of MDSs [108, 109]. The accuracy of DNA unscrambling is incredibly sensitive to these template RNAs. For example, microinjection of alternately unscrambled templates (e.g. swapping the order of MDSs) leads to the production of macronuclear chromosome resembling the introduced template [105]. Furthermore, RNAi knockdown of these long RNA templates results in aberrant or reduced rearrangements of MDSs in the resulting chromosomes found in the new MAC [105].
Fig. 5.
Unscrambling process in Oxytricha
Histone modification in DNA elimination
As in other eukaryotes, histone modifications play an integral role in the effective silencing of transposable elements and germline-limited DNA. In Tetrahymena, heterochromatin-specific marks, H3K9me3 and H3K27me3 are present in the MAC or both MIC and MAC, respectively [24, 87, 110–112]. Accumulation of H3K9me3 and H3K27me3 in the MAC is catalysed by histone methyltransferase Ezl1p, whereas Ezl2p is responsible for H3K27me3 in the MIC [111, 113]. As in other eukaryotes, small RNAs are involved in guiding the deposition of these conserved marks [110]. After deposition, specific marks are subsequently recognized by chromodomain-containing effectors. In particular, Tetrahymana’s Pdd1p (a homolog of HP1) accumulates on IESs, binding to methylated histones [114, 115] and is proposed to aid in recruiting Tetrahymana’s domesticated PiggyBac transposase (Tpb2p) for their excision [116]. Additionally, recent data indicated that RNAi-dependent Polycomb repression pathway is important for controlling transposable elements in Tetrahymena [117]. Disruption of the Polycomb repression pathway (knockout of DCL1, EZL1 and PDD1) results in the activation of TE transcription as well as the germline mobilization of TE [87, 110]. Moreover, numerous other histone modifications in Tetrahymena have been identified and may play roles in its DRGR [118, 119]. As in Tetrahymena, histone-specific marks such as H3K27 and H3K9 trimethylation are mediated by Ezl1 in Paramecium [120] and associated with chromatin assembly factor 1 subunit C-like protein (PtCAF-1) [121]. However, the developmental roles of these marks remain unclear.
As in Oligohymenophorea, heterochromatinization has been observed in Spirotrichea. During development, germline chromosomes are polytenized, with large blocks of observable heterochromatin prior to DNA elimination and fragmentation into thousands of unique gene-sized nanochromosomes [122]. In Stylonychia, this process is linked to the differential expression of a suite of histone H3 variants and subsequent post-translational modifications (PTM) [123–125]. For instance, H3K27me3 was shown to accumulate at the MIC-specific sequences prior to excision [126], while H3.7, acetylated at lysine-32, specifically associate with MDSs [123]. Moreover, knockdown of Piwi impacts the expression of histone H3.3 during macronuclear development in Stylonychia, implicating that H3.3 incorporation into nucleosomes is ncRNA-dependent [123]. Unfortunately, little is known about the roles of histone modifications and variants among other spirotrich ciliates, in Oxytricha and Euplotes.
DNA modification in IES elimination
Besides marking IESs for elimination through histone modifications, chemical modifications of germline DNA may also play a role in ciliate DRGRs. 5-Methylcytosine (5mC) has been identified on some germline limited sequences (e.g. transposons and satellite repeats) in Stylonychia and Oxytricha [127, 128] as well as in aberrant DNA rearrangements and parental DNA undergoing degradation [127]. Additionally, azacytidine and decitabine (DNA methyltransferase-inhibiting drugs) induce demethylation of both somatic and germline DNA during DRGRs, further implicating 5mC as a specific marker for DNA elimination/degradation [127]. Moreover, 5mC in Stylonychia correlates with gene activity as well as with chromatin structure during macronuclear differentiation [129]. However, recent work in Paramecium was unable to detect any evidence for 5mC modifications, suggesting that these modifications may only be involved in the DRGRs of spirotrich ciliates [130].
While the function of 5mC DNA modifications and their phylogenetic distribution in ciliates remains unclear, the only widely conserved DNA modification is 6N-methyladenine (6 mA) [131–135]. Data from Tetrahymena shows that 6 mA is only present in the transcriptionally active MAC and is preferentially enriched in the consensus sequence 5′-AT-3′ [133, 134]. 6 mA modifications also localize to linker DNA regions downstream of the transcription start site (TSS of Polymerase II transcribed genes) and directly influence nucleosome positioning [133, 136–138]. Tetrahymena’s MT-A70 homologue ATM1 (6mA DNA methyltransferase) is required for the normal growth and development of the cell following sex [139]. Interestingly, the enzyme MTA1c (DNA methyltransferase) responsible for 6mA modifications in Oxytricha disfavours nucleosome occupancy, contrary to Tetrahymena [135]. Beh et al. [135] suggest that decreased nucleosome occupancy is due to dA:dT base pair destabilisation by 6mA, which decreases the DNA melting temperature. However, the exact mechanism remains undetermined.
Unfortunately, while DNA modifications during MAC development are present, exactly how they might direct IES excision and/or MDS protection requires further investigation.
Transposases required for DNA excision
Transposase domestication has occurred throughout the eukaryotic tree of life, and can be linked to important DRGRs, such as those in ciliates and those involved in V(D)J recombination in animals [140, 141]. In Paramecium and Tetrahymena, IES excision is performed by a domesticated PiggyBac (PB) transposase (PiggyMac (Pgm) and TPB2 in Paramecium and Tetrahymena, respectively) [142, 143]. Excision of IESs in Paramecium by Pgm generates a 4 bp overhang at 5′ends centred around the "TA" dinucleotide pointer sequence showing the same geometry as those catalyzed in vitro by PB transposases [142, 144]. While Pgm is believed to carry out the physical excision, Paramecium possesses five accessory Pgm-like domesticated transposases (PgmL1-PgmL5) that interact with Pgm individually, with PgmL1 and PgmL3 directly involved in Pgm’s ability for precise IES excision [145]. Compared to Paramecium’s PiggyBac transposase, that possesses a sequence specificity (5′-TTAA-3′) [142], Tpb2p in Tetrahymena possess less stringent sequence specificity, as most of the IESs it excises are not flanked by any common motif [50, 62]. In Tetrahymena excision of the > 10,000 Tpb2p-dependent IESs is imprecise [116, 143]. Tetrahymena also possesses multiple transposase proteins, such as Lia5p (domesticated PB transposase) which localizes on IESs and facilitates Tpb2p-dependent IES elimination [114, 146] and is required for chromosome fragmentation in Tetrahymena [146]. In contrast to the most abundant IESs in Tetrahymena, the excision of 12 particular IESs (possessing TE features, such as terminal inverted repeats and the 5′-TTAA-3′ cutting site) is precise and depends on Tpb1p and Tpb6p [147, 148].
In Oxytricha, IES excision is triggered by telomere-bearing element (TBE) family transposases [149], which belong to a superfamily of transposase genes that possess a common DDE catalytic motif [150]. Analyses of the Oxytricha MIC genome found that complete and partial copies of Tc1/Mariner transposons constitute around 13% of the germline genome [151]. Compared to the Paramecium’s and Tetrahymena’s transposase-related proteins, TBEs in Oxytricha are encoded in the MIC genome itself, rather than the somatic genome [152], and cut with a 3 nt 5′ overhang at an ANT recognition site [98]. Similar to members of the Oligohymenophorea, Oxytricha possesses multiple transposases, all of which have a necessary role in its development [149].
Chromosome fragmentation
Chromosome fragmentation is one of the major events that occur during macronuclear development in ciliates. The most extreme chromosome fragmentation takes place in ciliates with gene-sized chromosomes (e.g. Euplotes, Stylonychia, and Oxytricha) where their MIC chromosomes are fragmented into > 15,000 unique MAC chromosomes [43, 153]. Given the incredibly short size of these nanochromosomes (averaging ~ 2.8–3.2 Kbp in Oxytricha and Stylonychia [43, 153]), most (~ 90%) encode just a single open reading frame (ORF) [43, 154]. While less dramatic, the five MIC chromosomes present in T. thermophila are fragmented into ~ 225 multigene MAC chromosomes (from < 100–1500 Kbp) [155], whereas in P. primaurelia, MIC chromosome fragmentation gives rise to 50–1000 Kbp MAC chromosomes [156].
For some ciliates, the fragmentation of the MIC genome does not occur randomly, but at specific chromosome breakage sequences (CBS) in the germline [157]. In Euplotes crassus, a conserved 10 bp consensus sequence (Euplotes-chromosome breakage sequence; E-CBS: 5′-HATTGAAaHH’, H = A, C or T) directs a staggered double-strand break (DSB) at a precise distance and orientation, which provides the substrates for telomere addition [158–160]. In Tetrahymena, a conserved 15 bp chromosome breakage sequence (CBS: 5′-WAAACCAACCYCNHW-3′, W = A/T; Y = T/C; H = A/T/C; N = G/A/T/C) is necessary for chromosome fragmentation and telomere addition [30, 161–164]. While the E-CBSs in Euplotes are ultimately retained in the MAC [158–160], in Tetrahymena the CBSs themselves are germline-limited and are eliminated with 4–34 bp of flanking DNA on both sides [165]. Interestingly, in Tetrahymena, fragmentation of the germline at the CBSs generates 33 non-maintained chromosomes (NMCs) [30, 164, 166, 167]. Unlike the typical “large” (> 100 Kbp) somatic chromosomes in the MAC, these NMCs are generally short, ranging from 30 to 80 Kbp and have a limited life-span, either being degraded prior to de novo telomere addition or lost by ~ 120 asexual divisions [166]. While a majority of the NMCs harbour functional ORFs (some of which are actively transcribed) [30], it remains unclear what their role, if any, might be in the post-sexual life cycle or the transition from sexual immaturity to maturity.
De novo telomere addition
The presence of DNA double-strand breaks (DSBs) is mostly known to be associated with the induction of repair machinery [Non-Homologous End Joining (NHEJ) or Homologous Recombination (HR)]. The relatively large number of DSBs associated with chromosome fragmentation during development in ciliates generates a large number of chromosomes whose broken ends are “healed” through de novo telomere addition. As in other eukaryotes, ciliate telomeres consist of tandem repeats at the 5′and 3′ends of their chromosomes, such as 5′-GGGGTTTT-3′ (G4T4) in Oxytricha and Euplotes [168], 5′-GGGGTT-3′ (G4T2) in Tetrahymena [22] and 5′-GGGGTT-3′ or 5′-GGGTTT-3′ in Paramecium [169, 170].
Although chromosome fragmentation is a very reproducible and relatively precise event, de novo telomere addition does not typically occur at precise nucleotide position (the exception being E. crassus [158]), generating micro-heterogeneity among the amplified chromosome copies in the developing macronucleus. In Oxytricha and Tetrahymena, telomere addition sites have been found to be clustered within regions ≤ 30 bp, [165, 171–173], whereas this is often ~ 1 to 2 Kbp in Paramecium [169, 170]. However, in E. crassus, there is no heterogeneity and telomeres are added at the same nucleotide positions in all macronuclear copies [158]. Additional heterogeneity can arise from the use of alternative chromosome fragmentation sites in Paramecium and Oxytricha. In Paramecium, the ends of some MAC chromosomes can be generated at alternative telomere addition sites separated by 2–13 Kbp. Each of the regions shows heterogeneity in the telomere’s positions [170, 174, 175]. While most of the chromosome fragmentation in Oxytricha results in gene-sized chromosomes, the use of alternative fragmentation sites (or failure to fragment) can result in macronuclear chromosomes encoding additional ORFs [154, 172, 173].
The exact mechanism of de novo telomere addition remains poorly understood. Data performed on Tetrahymena suggested the involvement of the telomere end binding homologue Pot2p in de novo telomere addition that exclusively localizes to CBSs during chromosome fragmentation [176]. Recent work from Stylonychia has shown that microinjection of RNA templates carrying variable telomeric repeats into the developing macronucleus leads to modified telomeres in vegetative cells suggesting that de novo telomere addition depends on a telomere-containing transcript derived from the parental macronucleus [177]. However, to understand this process in more detail, further work needs to be done.
Conclusions
Ciliates are a diverse group of organisms that have deeply contributed to our recent knowledge about the regulatory role of epigenetics in development. Identification of sRNA pathways as well as histone modifications that mediate DNA elimination is providing a greater understanding of the genome reorganization process in ciliates while shedding new insight into the evolution of epigenetic processes across eukaryotes. While we have a basic understanding of the overall genome reorganization process, numerous outstanding questions remain open. How are IES regions identified and preferentially transcribed to produce sRNAs in the meiotic MIC? What, if any, role is there for retaining IESs in the MIC? How has this process evolved across the ciliate phylogeny? Further comparative analyses of somatic and germline genomes, and the associated DNA elimination process, will be instrumental in answering these questions and will ultimately shed light on the variety of RNA-mediated epigenetic pathways and the dynamic regulation of genome function and structure.
Acknowledgements
This work was supported by the funding 500-3-11-110. We would like to thank Fabian Wieland for help with Fig. 2 as well as the reviewers for valuable comments.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Iwona Rzeszutek, Email: iwona.rzeszutek89@gmail.com.
Mariusz Nowacki, Email: mariusz.nowacki@izb.unibe.ch.
References
- 1.Streit A, Wang J, Kang Y, Davis RE. Gene silencing and sex determination by programmed DNA elimination in parasitic nematodes. Curr Opin Microbiol. 2016;32:120–127. doi: 10.1016/j.mib.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kloc M, Zagrodzinska B. Chromatin elimination—an oddity or a common mechanism in differentiation and development? Differentiation. 2001;68:84–91. doi: 10.1046/j.1432-0436.2001.680202.x. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Davis RE. Programmed DNA elimination in multicellular organisms. Curr Opin Genet Dev. 2014;27:26–34. doi: 10.1016/j.gde.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boveri T. Ueber Differenzierung der Zellkerne wahrend der Furchung des Eies von Ascaris megalocephala. The first discovery and description of a DNA elimination, chromatin diminution, in the parasitic nematode Parascaris. Anat Anz. 1887;2:688–693. [Google Scholar]
- 5.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109:45–55. doi: 10.1016/S0092-8674(02)00675-X. [DOI] [PubMed] [Google Scholar]
- 6.Parfrey LW, Lahr DJG, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalker DL, Yao M-C. DNA elimination in ciliates: transposon domestication and genome surveillance. Annu Rev Genet. 2011;45:227–246. doi: 10.1146/annurev-genet-110410-132432. [DOI] [PubMed] [Google Scholar]
- 8.Bracht JR, Fang W, Goldman AD, et al. Genomes on the edge: programmed genome instability in ciliates. Cell. 2013;152:406–416. doi: 10.1016/j.cell.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zufall RA, Robinson T, Katz LA. Evolution of developmentally regulated genome rearrangements in eukaryotes. J Exp Zool B Mol Dev Evol. 2005;304:448–455. doi: 10.1002/jez.b.21056. [DOI] [PubMed] [Google Scholar]
- 10.Grant JR, Katz LA (2014) Building a phylogenomic pipeline for the eukaryotic tree of life—addressing deep phylogenies with genome-scale data. PLoS Curr 6:. Doi: 10.1371/currents.tol.c24b6054aebf3602748ac042ccc8f2e9 [DOI] [PMC free article] [PubMed]
- 11.Gao F, Katz LA. Phylogenomic analyses support the bifurcation of ciliates into two major clades that differ in properties of nuclear division. Mol Phylogenet Evol. 2014;70:240–243. doi: 10.1016/j.ympev.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foissner W, Chao A, Katz LA. Diversity and geographic distribution of ciliates (Protista: Ciliophora) Biodivers Conserv. 2008;17:345–363. doi: 10.1007/s10531-007-9254-7. [DOI] [Google Scholar]
- 13.Simon M, Plattner H. Unicellular eukaryotes as models in cell and molecular biology: critical appraisal of their past and future value. Int Rev Cell Mol Biol. 2014;309:141–198. doi: 10.1016/B978-0-12-800255-1.00003-X. [DOI] [PubMed] [Google Scholar]
- 14.Kruger K, Grabowski PJ, Zaug AJ, et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 15.Brownell JE, Zhou J, Ranalli T, et al. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/S0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 16.Caron F, Meyer E. Does Paramecium primaurelia use a different genetic code in its macronucleus? Nature. 1985;314:185–188. doi: 10.1038/314185a0. [DOI] [PubMed] [Google Scholar]
- 17.Preer JR, Preer LB, Rudman BM, Barnett AJ. Deviation from the universal code shown by the gene for surface protein 51A in Paramecium. Nature. 1985;314:188–190. doi: 10.1038/314188a0. [DOI] [PubMed] [Google Scholar]
- 18.Lozupone CA, Knight RD, Landweber LF. The molecular basis of nuclear genetic code change in ciliates. Curr Biol. 2001;11:65–74. doi: 10.1016/S0960-9822(01)00028-8. [DOI] [PubMed] [Google Scholar]
- 19.Swart EC, Serra V, Petroni G, Nowacki M. Genetic codes with no dedicated stop codon: context-dependent translation termination. Cell. 2016;166:691–702. doi: 10.1016/j.cell.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heaphy SM, Mariotti M, Gladyshev VN, et al. Novel ciliate genetic code variants including the reassignment of all three stop codons to sense codons in condylostoma magnum. Mol Biol Evol. 2016;33:2885–2889. doi: 10.1093/molbev/msw166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 22.Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- 23.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/S0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 24.Taverna SD, Coyne RS, Allis CD. Methylation of histone H3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/S0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- 25.Allen SE, Hug I, Pabian S, et al. Circular Concatemers of Ultra-Short DNA Segments Produce Regulatory RNAs. Cell. 2017;168:990–999. doi: 10.1016/j.cell.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1016/0022-2836(72)90095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barid SE, Klobutcher LA. Genetic characterization and use of a restriction fragment length variant in the hypotrichous ciliate euplotes crassus. J Protozool. 1988;35:459–465. doi: 10.1111/j.1550-7408.1988.tb04130.x. [DOI] [PubMed] [Google Scholar]
- 28.Godiska R, Aufderheidet KJ, Gilley D, et al. Transformation of paramecium by microinjection of a cloned serotype gene. Proc Natl Acad Sci USA. 1987;84:7590–7594. doi: 10.1073/pnas.84.21.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martindale D, Mardtindale H, BP, (1986) Nucleic acids research. Nucleic Acids Res 14:1341–1354. 10.1093/nar/gkh411 [DOI] [PMC free article] [PubMed]
- 30.Hamilton EP, Kapusta A, Huvos PE, et al. Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome. Elife. 2016;5:e19090. doi: 10.7554/eLife.19090.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Bracht JR, Goldman AD, et al. The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development. Cell. 2014;158:1187–1198. doi: 10.1016/j.cell.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan Y, Rogers AJ, Gao F, Katz LA. Unusual features of non-dividing somatic macronuclei in the ciliate class Karyorelictea. Eur J Protistol. 2017;61:399–408. doi: 10.1016/j.ejop.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wancura MM, Yan Y, Katz LA, Maurer-Alcalá XX. Nuclear features of the heterotrich ciliate blepharisma americanum: genomic amplification, life cycle, and nuclear inclusion. J Eukaryot Microbiol. 2018;65:4–11. doi: 10.1111/jeu.12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juranek SA, Lipps HJ. New Insights into the Macronuclear Development in Ciliates. Int Rev Cytol. 2007;262:219–251. doi: 10.1016/S0074-7696(07)62005-1. [DOI] [PubMed] [Google Scholar]
- 35.Doerder FP, De Bault LE. Cytofluorimetric analysis of nuclear DNA during meiosis, fertilization and macronuclear development in the ciliate \textit{Tetrahymena pyriformis}, syngen 1. J Cell Sci. 1975;17:471–493. doi: 10.1242/jcs.17.3.471. [DOI] [PubMed] [Google Scholar]
- 36.Seyfert HM, Cleffmann G. Mean macronuclear DNA contents are variable in the ciliate Tetrahymena. J Cell Sci. 1982;58:211–223. doi: 10.1242/jcs.58.1.211. [DOI] [PubMed] [Google Scholar]
- 37.Doerder FP, Deak JC, Lief JH. Rate of phenotypic assortment in Tetrahymena thermophila. Dev Genet. 1992;13:126–132. doi: 10.1002/dvg.1020130206. [DOI] [PubMed] [Google Scholar]
- 38.Eisen JA, Coyne RS, Wu M, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aury JM, Jaillon O, Duret L, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 40.Duret L, Cohen J, Jubin C, et al. Analysis of sequence variability in the macronuclear DNA of Paramecium tetraurelia: A somatic view of the germline. Genome Res. 2008;18:585–596. doi: 10.1101/gr.074534.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nowacki M, Haye JE, Fang W, et al. RNA-mediated epigenetic regulation of DNA copy number. Proc Natl Acad Sci. 2010;12:367–389. doi: 10.1073/pnas.1012236107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu K, Doak TG, Lipps HJ, et al. Copy number variations of 11 macronuclear chromosomes and their gene expression in Oxytricha trifallax. Gene. 2012;505:75–80. doi: 10.1016/j.gene.2012.05.045. [DOI] [PubMed] [Google Scholar]
- 43.Swart EC, Bracht JR, Magrini V, et al. The Oxytricha trifallax Macronuclear Genome: A Complex Eukaryotic Genome with 16,000 Tiny Chromosomes. PLoS Biol. 2013;11:e1001473. doi: 10.1371/journal.pbio.1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonneborn TM (1974) Paramecium aurelia. In: Handbook of Genetics. pp 469–594.
- 45.Sonneborn TM. Sex, Sex Inheritance and Sex Determination in Paramecium Aurelia. Proc Natl Acad Sci. 1937;23:378–385. doi: 10.1073/pnas.23.7.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobs ME, Klobutcher LA (1996) The long and the short of developmental dna deletion in Euplotes crassus. In: Journal of Eukaryotic Microbiology. pp 442–452. [DOI] [PubMed]
- 47.Klobutcher LA, Herrick G. Consensus inverted terminal repeat sequence of Paramecium lESs: Resemblance to termini of Tc1 -related and Euplotes Tec transposons. Nucleic Acids Res. 1995;23:2006–2013. doi: 10.1093/nar/23.11.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnaiz O, Mathy N, Baudry C, et al. The Paramecium Germline Genome Provides a Niche for Intragenic Parasitic DNA: Evolutionary Dynamics of Internal Eliminated Sequences. PLoS Genet. 2012;8:e1002984. doi: 10.1371/journal.pgen.1002984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jaraczewski JW, Jahn CL. Elimination of Tec elements involves a novel excision process. Genes Dev. 1993;7:95–105. doi: 10.1101/gad.7.1.95. [DOI] [PubMed] [Google Scholar]
- 50.Klobutcher LA, Herrick G (1997) Developmental Genome Reorganization in Ciliated Protozoa: The Transposon Link. pp 1–62. [DOI] [PubMed]
- 51.Guérin F, Arnaiz O, Boggetto N, et al. Flow cytometry sorting of nuclei enables the first global characterization of Paramecium germline DNA and transposable elements. BMC Genomics. 2017;18:327. doi: 10.1186/s12864-017-3713-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maurer-Alcalá XX, Yan Y, Pilling OA, et al. Twisted Tales: Insights into Genome Diversity of Ciliates Using Single-Cell ’Omics. Genome Biol Evol. 2018;10:1927–1939. doi: 10.1093/gbe/evy133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maurer-Alcalá XX, Knight R, Katz LA. Exploration of the germline genome of the ciliate Chilodonella uncinata through single-cell omics (Transcriptomics and genomics) MBio. 2018;9:e01836–e1917. doi: 10.1128/mBio.01836-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Callahan RC, Shalke G, Gorovsky MA. Developmental rearrangements associated with a single type of expressed α-tubulin gene in tetrahymena. Cell. 1984;36:441–445. doi: 10.1016/0092-8674(84)90237-X. [DOI] [PubMed] [Google Scholar]
- 55.Yao MC, Gorovsky MA. Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma. 1974;48:1–18. doi: 10.1007/BF00284863. [DOI] [PubMed] [Google Scholar]
- 56.Yao MC, Choi J, Yokoyama S, et al. DNA elimination in tetrahymena: A developmental process involving extensive breakage and rejoining of DNA at defined sites. Cell. 1984;36:433–440. doi: 10.1016/0092-8674(84)90236-8. [DOI] [PubMed] [Google Scholar]
- 57.Gratias A, Betermier M. Processing of Double-Strand Breaks Is Involved in the Precise Excision of Paramecium Internal Eliminated Sequences. Mol Cell Biol. 2003;23:7152–7162. doi: 10.1128/mcb.23.20.7152-7162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eder C, Maercker C, Meyer J, Lipps HJ. The processing of macronuclear DNA sequences during macronuclear development of the hypotrichous ciliate Stylonychia lemnae. Int J Dev Biol. 1993;37:473–477. doi: 10.1387/ijdb.8292542. [DOI] [PubMed] [Google Scholar]
- 59.Prescott DM, DuBois ML (1996) Internal eliminated segments (IESs) of oxytrichidae. In: Journal of Eukaryotic Microbiology. pp 432–441. [DOI] [PubMed]
- 60.Heinonen TYK, Pearlman RE. A germ line-specific sequence element in an intron in Tetrahymena thermophila. J Biol Chem. 1994;269:17428–17433. [PubMed] [Google Scholar]
- 61.Chilcoat ND, Turkewitz AP. In vivo analysis of the major exocytosis-sensitive phosphoprotein in Tetrahymena. J Cell Biol. 1997;139:1197–1207. doi: 10.1083/jcb.139.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fass JN, Joshi NA, Couvillion MT, et al (2011) Genome-Scale Analysis of Programmed DNA Elimination Sites in Tetrahymena thermophila. G3|Genes|Genomes|Genetics 1:515–522. Doi: 10.1534/g3.111.000927 [DOI] [PMC free article] [PubMed]
- 63.Le Mouël A, Butler A, Caron F, Meyer E. Developmentally regulated chromosome fragmentation linked to imprecise elimination of repeated sequences in Paramecia. Eukaryot Cell. 2003;2:1076–1090. doi: 10.1128/EC.2.5.1076-1090.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Catania F, McGrath CL, Doak TG, Lynch M. Spliced DNA sequences in the Paramecium germline: Their properties and evolutionary potential. Genome Biol Evol. 2013;5:1200–1211. doi: 10.1093/gbe/evt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mayer KM, Mikami K, Forney JD. A mutation in Paramecium tetraurelia reveals functional and structural features of developmentally excised DNA elements. Genetics. 1998;148:139–149. doi: 10.1093/genetics/148.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mayer KM, Forney JD. A mutation in the flanking 5’-TA-3’ dinucleotide prevents excision of an internal eliminated sequence from the Paramecium tetraurelia genome. Genetics. 1999;151:597–694. doi: 10.1093/genetics/151.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gratias A, Lepère G, Garnier O, et al. Developmentally programmed DNA splicing in Paramecium reveals short-distance crosstalk between DNA cleavage sites. Nucleic Acids Res. 2008;36:3244–3251. doi: 10.1093/nar/gkn154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ku M, Mayer K, Forney JD. Developmentally regulated excision of a 28-base-pair sequence from the Paramecium genome requires flanking DNA. Mol Cell Biol. 2000;20:8390–8396. doi: 10.1128/MCB.20.22.8390-8396.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bhullar S, Wilkes CD, Arnaiz O, et al. A mating-type mutagenesis screen identifies a zinc-finger protein required for specific DNA excision events in Paramecium. Nucleic Acids Res. 2018;46:9550–9562. doi: 10.1093/nar/gky772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chalker DL, La Terza A, Wilson A, et al. Flanking regulatory sequences of the Tetrahymena R deletion element determine the boundaries of DNA rearrangement. Mol Cell Biol. 1999;19:5631–5641. doi: 10.1128/MCB.19.8.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Godiska R, James C, Yao MC. A distant 10-bp sequence specifies the boundaries of a programmed DNA deletion in Tetrahymena. Genes Dev. 1993;7:2357–2365. doi: 10.1101/gad.7.12a.2357. [DOI] [PubMed] [Google Scholar]
- 72.Lin C-YG, Chao J-L, Tsai H-K, et al. Setting boundaries for genome-wide heterochromatic DNA deletions through flanking inverted repeats in Tetrahymena thermophila. Nucleic Acids Res. 2019;47:5181–5192. doi: 10.1093/nar/gkz209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jaspan VN, Taye ME, Carle CM, et al. Boundaries of eliminated heterochromatin of Tetrahymena are positioned by the DNA-binding protein Ltl1. Nucleic Acids Res. 2019;14:7348–7362. doi: 10.1093/nar/gkz504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carle CM, Zaher HS, Chalker DL. A Parallel G Quadruplex-Binding Protein Regulates the Boundaries of DNA Elimination Events of Tetrahymena thermophila. PLoS Genet. 2016;12:e1005842. doi: 10.1371/journal.pgen.1005842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Swart EC, Wilkes CD, Sandoval PY, et al. Genome-wide analysis of genetic and epigenetic control of programmed DNA deletion. Nucleic Acids Res. 2014;42:8970–8983. doi: 10.1093/nar/gku619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sandoval P, Swart E, Arambasic M, Nowacki M. Functional Diversification of Dicer-like Proteins and Small RNAs Required for Genome Sculpting. Dev Cell. 2014;28:174–188. doi: 10.1016/j.devcel.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 77.Kapusta A, Matsuda A, Marmignon A, et al. Highly precise and developmentally programmed genome assembly in paramecium requires ligase IV-dependent end joining. PLoS Genet. 2011;7:e1002049. doi: 10.1371/journal.pgen.1002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin I-T, Chao J-L, Yao M-C. An essential role for the DNA breakage-repair protein Ku80 in programmed DNA rearrangements in Tetrahymena thermophila. Mol Biol Cell. 2012;23:2213–2225. doi: 10.1091/mbc.e11-11-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marmignon A, Bischerour J, Silve A, et al. Ku-Mediated Coupling of DNA Cleavage and Repair during Programmed Genome Rearrangements in the Ciliate Paramecium tetraurelia. PLoS Genet. 2014;10:e1004552. doi: 10.1371/journal.pgen.1004552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mochizuki K, Gorovsky MA. Small RNAs in genome rearrangement in Tetrahymena. Curr Opin Genet Dev. 2004;14:181–187. doi: 10.1016/j.gde.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 81.Lepère G, Nowacki M, Serrano V, et al. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic Acids Res. 2009;37:903–915. doi: 10.1093/nar/gkn1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zahler AM, Neeb ZT, Lin A, Katzman S. Mating of the stichotrichous Ciliate Oxytricha trifallax induces production of a class of 27 nt small RNAs derived from the parental macronucleus. PLoS ONE. 2012;7:e42371. doi: 10.1371/journal.pone.0042371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fang W, Wang X, Bracht JR, et al. Piwi-interacting RNAs protect DNA against loss during oxytricha genome rearrangement. Cell. 2012;151:1243–1255. doi: 10.1016/j.cell.2012.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Postberg J, Jönsson F, Weil PP, et al. 27nt-RNAs guide histone variant deposition via “RNA-induced DNA replication interference” and thus transmit parental genome partitioning in Stylonychia. Epigenetics and Chromatin. 2018;11:31. doi: 10.1186/s13072-018-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chalker DL, Yao MC. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 2001;15:1287–1298. doi: 10.1101/gad.884601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mochizuki K, Gorovsky MA. RNA polymerase II localizes in Tetrahymena thermophila meiotic micronuclei when micronuclear transcription associated with genome rearrangement occurs. Eukaryot Cell. 2004;3:1233–1240. doi: 10.1128/EC.3.5.1233-1240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Malone CD, Anderson AM, Motl JA, et al. Germ Line Transcripts Are Processed by a Dicer-Like Protein That Is Essential for Developmentally Programmed Genome Rearrangements of Tetrahymena thermophila. Mol Cell Biol. 2005;25:9151–9164. doi: 10.1128/MCB.25.20.9151-9164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hoehener C, Hug I, Nowacki M. Dicer-like Enzymes with Sequence Cleavage Preferences. Cell. 2018;173:234–247.e7. doi: 10.1016/j.cell.2018.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schoeberl UE, Kurth HM, Noto T, Mochizuki K. Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena. Genes Dev. 2012;26:1729–1742. doi: 10.1101/gad.196493.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Noto T, Kataoka K, Suhren JH, et al. Small-RNA-Mediated Genome-wide trans-Recognition Network in Tetrahymena DNA Elimination. Mol Cell. 2015;59:229–242. doi: 10.1016/j.molcel.2015.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bouhouche K, Gout JF, Kapusta A, et al. Functional specialization of Piwi proteins in Paramecium tetraurelia from post-transcriptional gene silencing to genome remodelling. Nucleic Acids Res. 2011;39:4249–4264. doi: 10.1093/nar/gkq1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Furrer DI, Swart EC, Kraft MF, et al. Two Sets of Piwi Proteins Are Involved in Distinct sRNA Pathways Leading to Elimination of Germline-Specific DNA. Cell Rep. 2017;20:505–520. doi: 10.1016/j.celrep.2017.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maliszewska-Olejniczak K, Gruchota J, Gromadka R, et al. TFIIS-Dependent Non-coding Transcription Regulates Developmental Genome Rearrangements. PLoS Genet. 2015;10:e1004552. doi: 10.1371/journal.pgen.1005383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lepère G, Bétermier M, Meyer E, Duharcourt S. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 2008;22:1501–1512. doi: 10.1101/gad.473008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mutazono M, Noto T, Mochizuki K. Diversification of small RNA amplification mechanisms for targeting transposon-related sequences in ciliates. Proc Natl Acad Sci U S A. 2019;116:14639–14644. doi: 10.1073/pnas.1903491116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yerlici VT, Lu MW, Hoge CR, et al. Programmed genome rearrangements in Oxytricha produce transcriptionally active extrachromosomal circular DNA. Nucleic Acids Res. 2019;47:9741–9760. doi: 10.1093/nar/gkz725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Williams K, Doak TG, Herrick G. Developmental precise excision of Oxytricha trifallax telomere-bearing elements and formation of circles closed by a copy of the flanking target duplication. EMBO J. 1993;12:4593–4601. doi: 10.1002/j.1460-2075.1993.tb06148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tausta SL, Klobutcher LA. Detection of circular forms of eliminated DNA during macronuclear development in E. crassus. Cell. 1989;59:1019–1026. doi: 10.1016/0092-8674(89)90758-7. [DOI] [PubMed] [Google Scholar]
- 100.Klobutcher LA, Turner LR, LaPlante J. Circular forms of developmentally excised DNA in Euplotes crassus have a heteroduplex junction. Genes Dev. 1993;7:84–94. doi: 10.1101/gad.7.1.84. [DOI] [PubMed] [Google Scholar]
- 101.Chang W-J, Bryson PD, Liang H, et al. The evolutionary origin of a complex scrambled gene. Proc Natl Acad Sci. 2005;102:15149–15154. doi: 10.1073/pnas.0507682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hoffman DC, Prescott DM. Phylogenetic relationships among hypotrichous ciliates determined with the macronuclear gene encoding the large, catalytic subunit of DNA polymerase α. J Mol Evol. 1997;45:301–310. doi: 10.1007/PL00006234. [DOI] [PubMed] [Google Scholar]
- 103.Hoffman D. The germline gene encoding DNA polymerase alpha in the hypotrichous ciliate Oxytricha nova is extremely scrambled. Nucleic Acids Res. 1996;45:301–310. doi: 10.1093/nar/24.17.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Landweber LF, Kuo TC, Curtis EA. Evolution and assembly of an extremely scrambled gene. Proc Natl Acad Sci U S A. 2000;97:3298–3303. doi: 10.1073/pnas.97.7.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nowacki M, Vijayan V, Zhou Y, et al. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lindblad KA, Bracht JR, Williams AE, Landweber LF. Thousands of RNA-cached copies of whole chromosomes are present in the ciliate Oxytricha during development. RNA. 2017;23:1200–1208. doi: 10.1261/rna.058511.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Khurana JS, Wang X, Chen X, et al. Transcription-independent functions of an RNA polymerase II subunit, Rpb2, during genome rearrangement in the ciliate, Oxytricha trifallax. Genetics. 2014;197:839–849. doi: 10.1534/genetics.114.163279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kari L, Landweber LF (1999) Computational Power of Gene Rearrangement. Proceedings of DNA Based Computers V, E. Winfree, D. Gifford eds., MIT, Boston 0000:207–216.
- 109.Ehrenfeucht A, Harju T, Rozenberg G. Gene assembly through cyclic graph decomposition. Theor Comput Sci. 2002;281:325–349. doi: 10.1016/S0304-3975(02)00019-1. [DOI] [Google Scholar]
- 110.Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci U S A. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu Y, Taverna SD, Muratore TL, et al. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Frapporti A, Miró Pina C, Arnaiz O, et al. The Polycomb protein Ezl1 mediates H3K9 and H3K27 methylation to repress transposable elements in Paramecium. Nat Commun. 2019;10:2710. doi: 10.1038/s41467-019-10648-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Papazyan R, Voronina E, Chapman JR, et al. Methylation of histone H3K23 blocks DNA damage in pericentric heterochromatin during meiosis. Elife. 2014;3:e02996. doi: 10.7554/eLife.02996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Suhren JH, Noto T, Kataoka K, et al. Negative Regulators of an RNAi-Heterochromatin Positive Feedback Loop Safeguard Somatic Genome Integrity in Tetrahymena. Cell Rep. 2017;18:2494–2507. doi: 10.1016/j.celrep.2017.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kataoka K, Mochizuki K. Phosphorylation of an HP1-like Protein Regulates Heterochromatin Body Assembly for DNA Elimination. Dev Cell. 2015;35:775–788. doi: 10.1016/j.devcel.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cheng CY, Vogt A, Mochizuki K, Yao MC. A Domesticated piggyBac Transposase Plays Key Roles in Heterochromatin Dynamics and DNA Cleavage during Programmed DNA Deletion in Tetrahymena thermophila. Mol Biol Cell. 2010;21:1753–1762. doi: 10.1091/mbc.E09-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhao X, Xiong J, Mao F, et al. RNAi-dependent polycomb repression controls transposable elements in Tetrahymena. Genes Dev. 2019;33:348–364. doi: 10.1101/gad.320796.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Taverna SD, Ueberheide BM, Liu Y, et al. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc Natl Acad Sci U S A. 2007;104:2086–2091. doi: 10.1073/pnas.0610993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Garcia BA, Hake SB, Diaz RL, et al. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 120.Lhuillier-Akakpo M, Frapporti A, Denby Wilkes C, et al. Local Effect of Enhancer of Zeste-Like Reveals Cooperation of Epigenetic and cis-Acting Determinants for Zygotic Genome Rearrangements. PLoS Genet. 2014;10:e1004665. doi: 10.1371/journal.pgen.1004665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ignarski M, Singh A, Swart EC, et al. Paramecium tetraurelia chromatin assembly factor-1-like protein PtCAF-1 is involved in RNA-mediated control of DNA elimination. Nucleic Acids Res. 2014;42:11952–11964. doi: 10.1093/nar/gku874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ammermann D. Morphology and development of the macronuclei of the ciliates Stylonychia mytilus and Euplotes aediculatus. Chromosoma. 1971;33:209–238. doi: 10.1007/BF00285634. [DOI] [PubMed] [Google Scholar]
- 123.Forcob S, Bulic A, Jönsson F, et al (2014) Differential expression of histone H3 genes and selective association of the variant H3.7 with a specific sequence class in Stylonychia macronuclear development. Epigenetics and Chromatin. Doi: 10.1186/1756–8935–7–4 [DOI] [PMC free article] [PubMed]
- 124.Postberg J, Heyse K, Cremer M, et al. Spatial and temporal plasticity of chromatin during programmed DNA-reorganization in Stylonychia macronuclear development. Epigenetics Chromatin. 2008;1:3. doi: 10.1186/1756-8935-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Juranek SA, Rupprecht S, Postberg J, Lipps HJ. snRNA and heterochromatin formation are involved in DNA excision during macronuclear development in stichotrichous ciliates. Eukaryot Cell. 2005;4:1934–1941. doi: 10.1128/EC.4.11.1934-1941.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Meyer GF, Lipps HJ. Chromatin elimination in the hypotrichous ciliate Stylonychia mytilus. Chromosoma. 1980;77:285–297. doi: 10.1007/BF00286054. [DOI] [PubMed] [Google Scholar]
- 127.Bracht JR, Perlman DH, Landweber LF. Cytosine methylation and hydroxymethylation mark DNA for elimination in Oxytricha trifallax. Genome Biol. 2012;13:R99. doi: 10.1186/gb-2012-13-10-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Juranek S, Wieden HJ, Lipps HJ. De novo cytosine methylation in the differentiating macronucleus of the stichotrichous ciliate Stylonychia lemnae. Nucleic Acids Res. 2003;31:1387–1391. doi: 10.1093/nar/gkg233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bulic A, Postberg J, Fischer A, et al. A permissive chromatin structure is adopted prior to site-specific DNA demethylation of developmentally expressed genes involved in macronuclear differentiation. Epigenetics and Chromatin. 2013;6:5. doi: 10.1186/1756-8935-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Singh A, Vancura A, Woycicki RK, et al. Determination of the presence of 5-methylcytosine in Paramecium tetraurelia. PLoS ONE. 2018;13:e0206667. doi: 10.1371/journal.pone.0206667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Harrison GS, FindlyKarrer RCKM. Site-specific methylation of adenine in the nuclear genome of a eucaryote. Tetrahymena thermophila Mol Cell Biol. 1986;6:2364–2370. doi: 10.1128/MCB.6.7.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cummings DJ, Tait A, Goddard JM. Methylated bases in DNA from Paramecium aurelia. BBA Sect Nucleic Acids Protein Synth. 1974;374:1–11. doi: 10.1016/0005-2787(74)90194-4. [DOI] [PubMed] [Google Scholar]
- 133.Wang Y, Chen X, Sheng Y, et al. N6-adenine DNA methylation is associated with the linker DNA of H2A.Z-containing well-positioned nucleosomes in Pol II-transcribed genes in Tetrahymena. Nucleic Acids Res. 2017;45:11594–11606. doi: 10.1093/nar/gkx883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gorovsky MA, Hattman S, Pleger GL. [6N]methyl adenine in the nuclear DNA of a eucaryote. Tetrahymena Pyriformis J Cell Biol. 1973;56:697–701. doi: 10.1083/jcb.56.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Beh LY, Debelouchina GT, Clay DM, et al. identification of a DNA N6-adenine methyltransferase complex and its impact on chromatin organization. Cell. 2019;177:1781–1796. doi: 10.1016/j.cell.2019.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Luo GZ, Hao Z, Luo L, et al. N 6 -methyldeoxyadenosine directs nucleosome positioning in Tetrahymena DNA. Genome Biol. 2018;19:200. doi: 10.1186/s13059-018-1573-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang Y, Sheng Y, Liu Y, et al. N6-methyladenine DNA modification in the unicellular eukaryotic organism Tetrahymena thermophila. Eur J Protistol. 2017;58:94–102. doi: 10.1016/j.ejop.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 138.Karrer KM. Methylation of adenine in the nuclear DNA of Tetrahymena is internucleosomal and independent of histone H1. Nucleic Acids Res. 2002;30:1364–1370. doi: 10.1093/nar/30.6.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang Y, Sheng Y, Liu Y, et al. A distinct class of eukaryotic MT-A70 methyltransferases maintain symmetric DNA N6-adenine methylation at the ApT dinucleotides as an epigenetic mark associated with transcription. Nucleic Acids Res. 2019;47:11771–11789. doi: 10.1093/nar/gkz1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pamcer Z, Amemiya CT, Ehrhardt GRA, et al. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–180. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]
- 141.Flajnik MF. Another manifestation of GOD. Nature. 2004;430:157–158. doi: 10.1038/430157a. [DOI] [PubMed] [Google Scholar]
- 142.Baudry C, Malinsky S, Restituito M, et al. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev. 2009;23:2478–2483. doi: 10.1101/gad.547309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vogt A, Mochizuki K. A domesticated piggybac transposase interacts with heterochromatin and catalyzes reproducible DNA elimination in tetrahymena. PLoS Genet. 2013;9:e1004032. doi: 10.1371/journal.pgen.1004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mitra R, Fain-Thornton J, Craig NL. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008;27:1097–1109. doi: 10.1038/emboj.2008.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bischerour J, Bhullar S, Wilkes CD, et al. Six domesticated piggybac transposases together carry out programmed DNA elimination in paramecium. Elife. 2018;7:e37927. doi: 10.7554/eLife.37927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shieh AWY, Chalker DL. LIA5 Is Required for nuclear reorganization and programmed DNA rearrangements occurring during tetrahymena macronuclear differentiation. PLoS ONE. 2013;8:e75337. doi: 10.1371/journal.pone.0075337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cheng CY, Young JM, Lin CYG, et al. The piggyBac transposon-derived genes TPB1 and TPB6 mediate essential transposon-like excision during the developmental rearrangement of key genes in Tetrahymena thermophila. Genes Dev. 2016;30:2724–2736. doi: 10.1101/gad.290460.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Feng L, Wang G, Hamilton EP, et al. A germline-limited piggyBac transposase gene is required for precise excision in Tetrahymena genome rearrangement. Nucleic Acids Res. 2017;45:9481–9502. doi: 10.1093/nar/gkx652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nowacki M, Higgins BP, Maquilan GM, et al. A functional role for transposases in a large eukaryotic genome. Science (80- ) 2009;324:935–938. doi: 10.1126/science.1170023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Doak TG, Doerder FP, Jahn CL, Herrick G. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc Natl Acad Sci USA. 1994;91:942–946. doi: 10.1073/pnas.91.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen X, Landweber LF. Phylogenomic analysis reveals genome-wide purifying selection on TBE transposons in the ciliate Oxytricha. Mob DNA. 2016;7:2. doi: 10.1186/s13100-016-0057-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hunter DJ, Williams K, Cartinhour S, Herrick G. Precise excision of telomere-bearing transposons during Oxytricha fallax macronuclear development. Genes Dev. 1989;3:2101–2112. doi: 10.1101/gad.3.12b.2101. [DOI] [PubMed] [Google Scholar]
- 153.Aeschlimann SH, Jönsson F, Postberg J, et al. The draft assembly of the radically organized Stylonychia lemnae macronuclear genome. Genome Biol Evol. 2014;6:1707–1723. doi: 10.1093/gbe/evu139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Seegmiller A, Williams KR, Herrick G. Two two-gene macronuclear chromosomes of the hypotrichous ciliates Oxytricha fallax and O. trifallax generated by alternative processing of the 81 locus. Dev Genet. 1997;20:348–357. doi: 10.1002/(SICI)1520-6408(1997)20:4<348::AID-DVG6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 155.Conover RK, Brunk CF. Macronuclear DNA molecules of Tetrahymena thermophila. Mol Cell Biol. 1986;6:900–905. doi: 10.1128/MCB.6.3.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Phan HL, Forney J, Blackburn EH. Analysis of paramecium macronuclear DNA using pulsed field gel electrophoresis. J Protozool. 1989;36:402–408. doi: 10.1111/j.1550-7408.1989.tb05535.x. [DOI] [PubMed] [Google Scholar]
- 157.Yao MC, Yao CH, Monks B. The controlling sequence for site-specific chromosome breakage in tetrahymena. Cell. 1990;63:763–772. doi: 10.1016/0092-8674(90)90142-2. [DOI] [PubMed] [Google Scholar]
- 158.Baird SE, Klobutcher LA. Characterization of chromosome fragmentation in two protozoans and identification of a candidate fragmentation sequence in Euplotes crassus. Genes Dev. 1989;3:585–597. doi: 10.1101/gad.3.5.585. [DOI] [PubMed] [Google Scholar]
- 159.Klobutcher LA. Characterization of in vivo developmental chromosome fragmentation intermediates in E. crassus. Mol Cell. 1999;4:695–704. doi: 10.1016/S1097-2765(00)80380-9. [DOI] [PubMed] [Google Scholar]
- 160.Klobutcher LA, Gygax SE, Podoloff JD, et al. Conserved DNA sequences adjacent to chromosome fragmentation and telomere addition sites in Euplotes crassus. Nucleic Acids Res. 1998;26:4230–4240. doi: 10.1093/nar/26.18.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yao MC, Yao CH. Accurate processing and amplification of cloned germ line copies of ribosomal DNA injected into developing nuclei of Tetrahymena thermophila. Mol Cell Biol. 1989;9:1092–1099. doi: 10.3928/1081597X-20101214-01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fan Q, Yao MC. A long stringent sequence signal for programmed chromosome breakage in Tetrahymena thermophila. Nucleic Acids Res. 2000;28:895–900. doi: 10.1093/nar/28.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hamilton EP, Williamson S, Dunn S, et al. The highly conserved family of Tetrahymena thermophila chromosome breakage elements contains an invariant 10-base-pair core. Eukaryot Cell. 2006;5:771–780. doi: 10.1128/EC.5.4.771-780.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lin CYG, Lin IT, Yao MC. Programmed Minichromosome elimination as a mechanism for somatic genome reduction in Tetrahymena thermophila. PLoS Genet. 2016;12:e1006403. doi: 10.1371/journal.pgen.1006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fan Q, Yao M. New telomere formation coupled with site-specific chromosome breakage in Tetrahymena thermophila. Mol Cell Biol. 1996;16:1267–1274. doi: 10.1128/MCB.16.3.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cassidy-Hanley D, Bisharyan Y, Fridman V, et al. Genome-wide characterization of Tetrahymena thermophila chromosome breakage sites. II Phys Genet Mapping Genet. 2005;170:1623–1631. doi: 10.1534/genetics.104.031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hamilton E, Bruns P, Lin C, et al. Genome-wide characterization of Tetrahymena thermophila chromosome breakage sites. I. Cloning and identification of functional sites. Genetics. 2005;170:1611–1621. doi: 10.1534/genetics.104.031401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Klobutcher LA, Swanton MT, Donini P, Prescott DM. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3’ terminus. Proc Natl Acad Sci. 1981;78:3015–3019. doi: 10.1073/pnas.78.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Baroin A, Prat A, Caron F. Telomeric site position heterogeneity in macronuclear DNA of Paramecium primaurelia. Nucleic Acids Res. 1987;15:1717–1728. doi: 10.1093/nar/15.4.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Forney JD, Blackburn EH. Developmentally controlled telomere addition in wild-type and mutant paramecia. Mol Cell Biol. 1988;8:251–258. doi: 10.1128/MCB.8.1.251.Updated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Baird SE, Fino GM, Tausta SL, Klobutcher LA. Micronuclear genome organization in Euplotes crassus: a transposonlike element is removed during macronuclear development. Mol Cell Biol. 1989;9:3793–3807. doi: 10.1128/MCB.9.9.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Herrick G, Hunter D, Williams K, Kotter K. Alternative processing during development of a macronuclear chromosome family in Oxytricha fallax. Genes Dev. 1987;1:1047–1058. doi: 10.1101/gad.1.10.1047. [DOI] [PubMed] [Google Scholar]
- 173.Williams KR, Doak TG, Herrick G. Telomere formation on macronuclear chromosomes of Oxytricha trifallax and O. fallax: Alternatively processed regions have multiple telomere addition sites. BMC Genet. 2002;3:16. doi: 10.1186/1471-2156-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Amar L. Chromosome end formation and internal sequence elimination as alternative genomic rearrangements in the ciliate paramecium. J Mol Biol. 1994;236:421–426. doi: 10.1006/jmbi.1994.1154. [DOI] [PubMed] [Google Scholar]
- 175.Caron F. A high degree of macronuclear chromosome polymorphism is generated by variable DNA rearrangements in Paramecium primaurelia during macronuclear differentiation. J Mol Biol. 1992;225:661–678. doi: 10.1016/0022-2836(92)90393-X. [DOI] [PubMed] [Google Scholar]
- 176.Cranert S, Heyse S, Linger BR, et al. Tetrahymena Pot2 is a developmentally regulated paralog of Pot1 that localizes to chromosome breakage sites but not to telomeres. Eukaryot Cell. 2014;13:1519–1529. doi: 10.1128/ec.00204-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fuhrmann G, Jönsson F, Weil PP, et al. RNA-template dependent de novo telomere addition. RNA Biol. 2016;13:733–739. doi: 10.1080/15476286.2015.1134414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Greslin AF, Prescott DM, Oka Y, et al. Reordering of nine exons is necessary to form a functional actin gene in Oxytricha nova. Proc Natl Acad Sci U S A. 1989;86:6264–6268. doi: 10.1073/pnas.86.16.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Coyne RS, Stover NA, Miao W. Whole Genome studies of Tetrahymena. Methods Cell Biol. 2012;109:53–81. doi: 10.1016/B978-0-12-385967-9.00004-9. [DOI] [PubMed] [Google Scholar]
- 180.Prescott DM, Prescott JD, Prescott RM. Coding properties of macronuclear DNA molecules in Sterkiella nova (Oxytricha nova) Protist. 2002;153:71–77. doi: 10.1078/1434-4610-00084. [DOI] [PubMed] [Google Scholar]
- 181.Cervantes MD, Hamilton EP, Xiong J, et al. Selecting one of several mating types through gene segment joining and deletion in Tetrahymena thermophila. PLoS Biol. 2013;13:e1002284. doi: 10.1371/journal.pbio.1001518. [DOI] [PMC free article] [PubMed] [Google Scholar]