Abstract
Background
Despite Sweden's strong entomological tradition, large portions of its insect fauna remain poorly known. As part of the Swedish Taxonomy Initiative, launched in 2002 to document all multi-cellular species occurring in the country, the first taxonomically-broad inventory of the country's insect fauna was initiated, the Swedish Malaise Trap Project (SMTP). In total, 73 Malaise traps were deployed at 55 localities representing a wide range of habitats across the country. Most traps were run continuously from 2003 to 2006 or for a substantial part of that time period. The total catch is estimated to contain 20 million insects, distributed over 1,919 samples (Karlsson et al. 2020). The samples have been sorted into more than 300 taxonomic units, which are made available for expert identification. Thus far, more than 100 taxonomists have been involved in identifying the sorted material, recording the presence of 4,000 species. One third of these had not been recorded from Sweden before and 700 have tentatively been identified as new to science.
New information
Here, we describe the SMTP dataset, published through the Global Biodiversity Information Facility (GBIF). Data on the sorted material are available in the "SMTP Collection Inventory" dataset. It currently includes more than 130,000 records of taxonomically-sorted samples. Data on the identified material are published using the Darwin Core standard for sample-based data. That information is divided up into group-specific datasets, as the sample set processed for each group is different and in most cases non-overlapping. The current data are divided into 79 taxonomic datasets, largely corresponding to taxonomic sorting fractions. The orders Diptera and Hymenoptera together comprise about 90% of the specimens in the material and these orders are mainly sorted to family or subfamily. The remaining insect taxa are mostly sorted to the order level. In total, the 79 datasets currently available comprise around 165,000 specimens, that is, about 1% of the total catch. However, the data are now accumulating rapidly and will be published continuously. The SMTP dataset is unique in that it contains a large proportion of data on previously poorly-known taxa in the Diptera and Hymenoptera.
Keywords: Malaise trap, insect fauna, inventory, survey, all-taxa biodiversity inventory, ATBI, Diptera , Hymenoptera
Introduction
Sweden has a long entomological tradition, starting even before Linnaeus's groundbreaking work on the Swedish fauna and flora. Nevertheless, large portions of the insect fauna remain poorly known to this day. The Swedish distribution of numerous species is documented only by scattered occurrence records and their ecology is poorly documented or unknown. It has also been clear for some time that further research on neglected insect groups would expand the known Swedish species stock significantly and lead to the discovery of a number of species new to science. These neglected groups include many taxa in the orders Diptera and Hymenoptera; it also includes the lice (Phthiraptera) and even some select groups in more well-known insect orders.
To address these knowledge gaps, an ambitious national insect inventory was started in 2003, the Swedish Malaise Trap Project (Karlsson et al. 2005, Karlsson et al. 2020). Malaise traps were chosen as the trapping method because of their efficiency in catching many of the poorly-known taxa of Diptera and Hymenoptera. A total of 73 Malaise traps were deployed at 55 localities spread out over the country and representing a diverse set of habitats. Most of the traps were run continuously from 2003 to 2006 or a significant portion of that time period. A detailed description of all accounts of the project, from collection through transfer to taxonomic experts and collection storage, was recently published (Karlsson et al. 2020).
The total SMTP catch is estimated to contain 20 million insects, distributed over 1,919 samples (Karlsson et al. 2020). The samples have been sorted into more than 300 taxonomic units, suitable for further processing by experts. Thus far, more than 100 taxonomists have been involved in identifying the sorted material, recording the presence of 4,000 species. One third of these had not been recorded from Sweden before and 700 have tentatively been identified as new to science (Ronquist et al. 2020).
The SMTP data are published through the Global Biodiversity Information Facility (GBIF). Data are published both on the sorted taxonomic fractions (the "SMTP Collection Inventory" dataset) and on the identified material of individual groups. The latter data are published using the Darwin Core standard for sample-based data to facilitate biodiversity analyses. The SMTP datasets are unique in that they contain a large proportion of data on previously poorly-known, but species-rich taxa in the Diptera and Hymenoptera. Due to this and the fact that the Swedish insect fauna was better known than most other insect fauna before the start of the inventory, the SMTP data offer biologists one of the best opportunities currently for detailed analysis of the size and composition of temperate insect fauna (Ronquist et al. 2020).
In this paper, we provide background data on the SMTP and describe the rationale behind the data publication strategy. We also provide an overview of the currently available datasets, comprising information on more than 130,000 taxonomically-sorted samples and on the species identity of around 165,000 specimens, that is, about 1% of the total SMTP catch. The species-level data are now accumulating rapidly; more than 600,000 specimens are currently on loan to experts for identification. The data will be published continuously through GBIF as they become available over the coming years.
General description
Purpose
The purpose of this paper is to provide an overview of the SMTP data published through GBIF and to describe the data publication strategy.
Additional information
The SMTP data fall into two different categories: data on the material sorted to taxonomic fractions and data on the specimens identified to species. The data on the sorted material are available in the "SMTP Collection Inventory" dataset (Holston et al. 2020). It currently includes more than 130,000 records of taxonomically-sorted samples. As the taxonomic order sorting of the SMTP material is now complete, the changes to this dataset over the coming year or two will reflect only the finer levels of sorting as they occur. As samples are processed by taxonomists, however, they will disappear from the sorted material dataset so that it will continuously reflect the SMTP material that is currently available to taxonomic experts.
Data on the identified material are published using the Darwin Core standard for sample-based data. The goal is to make the data easy to use for biodiversity analyses, such as species richness estimation. Such analyses typically require that all species in a specific taxon set are recorded for the same set of samples. As the set of SMTP samples processed for each taxonomic unit is different, the information is divided into group-specific datasets. The taxon coverage of each dataset usually corresponds to one of the taxonomic fractions used in the sorting process. In some cases, several related taxonomic fractions are combined into a single dataset, but only if the same set of samples have been processed for all fractions. The circumscription of SMTP datasets is subject to change, based on discussions with the taxonomists involved in identifying the material, amongst other things.
The taxonomic coverage of each dataset is described in its metadata, in the taxonomic coverage section. The field generalTaxonomicCoverage is used to provide information about subtaxa that may be excluded. For instance, a genus may be excluded from the dataset of a family because it is very difficult to identify to species or because it is so numerous that identification to species would be too time-consuming. As appropriate for sample-based data, the absence of a species from the Occurrence table (the Extension table of the Darwin Core Archive) is significant. Provided that the species belongs to the covered taxon set, it means that the species was not encountered in the processed samples.
The samples processed for the taxa covered by the dataset are listed in the Event table (the Core table of the Darwin Core Archive). The sampling site location and sampling effort (time period in days) of each sample are specified, as well as the associated TrapID and EventID identifiers that are used consistently throughout the SMTP project. These identifiers facilitate analyses that look at patterns across SMTP datasets. For instance, one may be interested in the overlap in spatial or temporal coverage amongst SMTP datasets. The absence of one of the 1,919 samples from the Event table is significant: it means that the sample has not been processed for the taxa covered in the dataset. If a sample is listed in the Event table, but there are no occurrence records tied to it, it means that it has been verified that there are no specimens of the covered taxa in that sample. Note that the SMTP samples processed for the taxa in the dataset are usually an arbitrary subset of all the available SMTP samples. If the subset has been chosen according to a principled method, this will be noted in the metadata fields Methods:sampling and Methods:samplingDescription.
In general, the Occurrence table lists abundance data for the observed species. This may be recorded separately for each sex or as a total number of specimens regardless of sex. In a few cases, the Occurrence table instead lists incidence data or a mix of incidence and abundance data. If so, this is noted in the metadata of the dataset, in the Dataset:additionalInfo field. This field may also contain annotations about the determinations. For instance, in the Phoridae dataset, only the males are determined to species; females are usually determined only to genus.
As far as possible, the species-level taxonomy used for the SMTP data follows the national Swedish checklist Dyntaxa, also available as a checklist through GBIF (https://doi.org/10.15468/j43wfc). Deviations may occur for several reasons. Manuscript names are used in the SMTP datasets for new species; the ambition is to update these records and match them to new entries in Dynyaxa when the taxa are described. In some cases, specialists involved in identifying SMTP material provide corrections of Dyntaxa species names or concepts. These corrections are forwarded to Dyntaxa curators for review and possible action. The ambition is to synchronise the content in the SMTP datasets and Dyntaxa as soon as the issues have been resolved, but this is currently a manual process and time lags may occur.
The sample-based SMTP datasets are linked through their names; they are named "SMTP X", where "X" refers to the taxon set. For instance, the dataset covering Coleoptera is named "SMTP Coleoptera". The metadata of each dataset include standardised fields describing the SMTP project, further facilitating collective retrieval of all SMTP datasets.
Project description
Title
The Swedish Malaise Trap Project
Personnel
The project was initially conceived by Fredrik Ronquist, then at Uppsala University and Thomas Pape, then at the Swedish Museum of Natural History. Project managers have been, from the beginning up to now: Johan Liljeblad, Kjell Arne Johanson, Kajsa Glemhorn and Dave Karlsson. The project was initially hosted by the Swedish Museum of Natural History, but the practical coordination of the project was moved to the field station Station Linné in 2006 and the project has been officially hosted by Station Linné since 2010. Questions about the project can be directed to Dave Karlsson, current SMTP project manager and corresponding author of this paper.
Traps were run largely by volunteers. Sorting has been carried out by staff, students and volunteers at Station Linné (for more details, see Karlsson et al. 2020). A large number of taxonomists (and students) have provided species identifications (see http://www.stationlinne.se/en/research/the-swedish-malaise-trap-project-smtp/taxonomic-units-in-the-smtp/ for updated lists). Crucial for handling the data on behalf of Station Linné have been Dave Karlsson, Pelle Magnusson, Mattias Forshage, Jessica Mo and Marina Karlsson and important for making them public on behalf of the Swedish Museum of Natural History have been Kevin Holston, Ida Li, Anders Telenius, Veronika Johansson, Manash Shah and Markus Skyttner.
Study area description
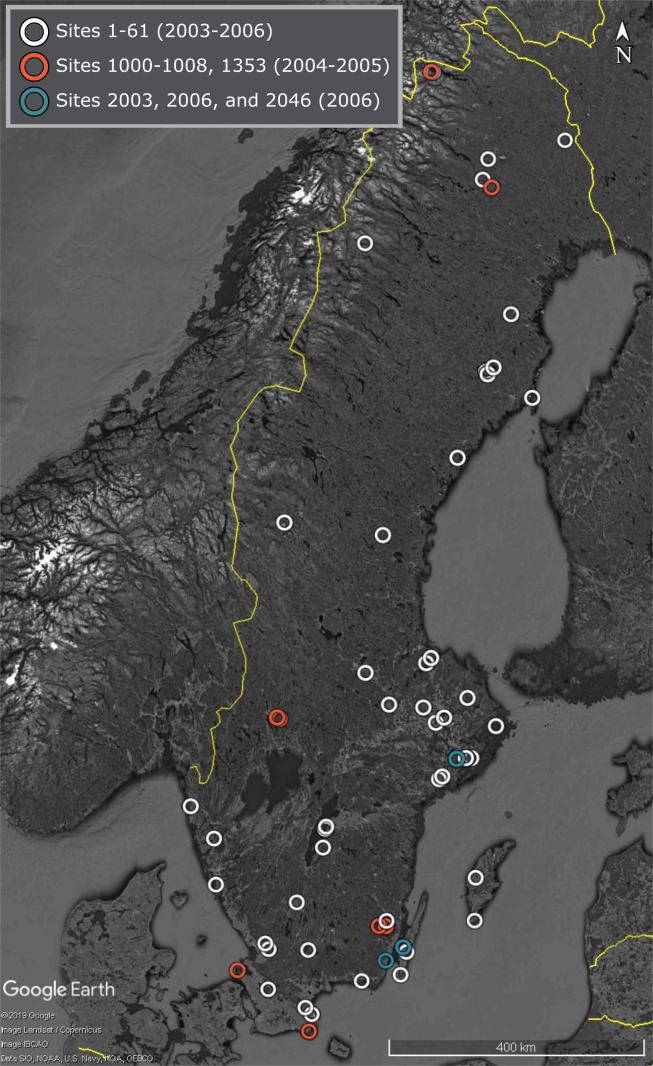
A wide range of representative terrestrial habitats, distributed over Sweden (Fig. 1; Suppl. material 1)
Figure 1.
Map of SMTP trap sites colour coded by trapping period
Design description
Sites were chosen to represent a broad range of habitats across the country of Sweden (Fig. 1, for more details, see Karlsson et al. 2020).
Funding
The SMTP has been funded continuously since 2003 by the Swedish Taxonomy Initiative (Miller 2005, Ronquist and Gärdenfors 2003). Additional support for laboratory technicians has been obtained through the Swedish Public Employment Service and the Swedish Social Insurance Agency.
Sampling methods
Sampling description
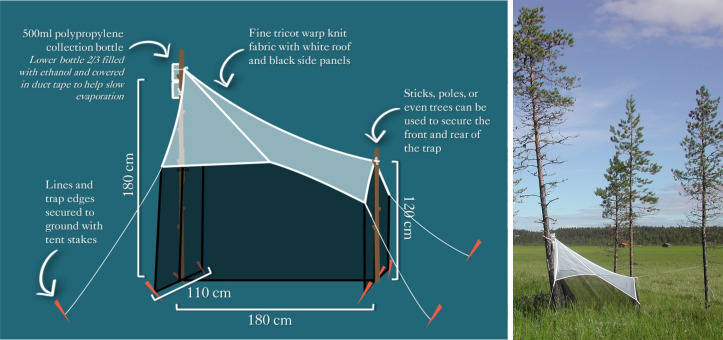
The project used a standard Townes-style Malaise trap obtained from Sante Traps, Lexington, KY, USA (Fig. 2).
Figure 2.
Left: Illustration of the Townes-style Malaise trap used by the SMTP. Right: A trap in operation at Norrbotten, Pajala Kommun (Site 49).
The trap sites are described in more detail in Suppl. material 1. Trap locations were chosen to maximise habitat diversity (for more details, see Karlsson et al. 2020). To facilitate analyses, the sites have been classified into different habitat types (Suppl. material 1). A somewhat more detailed characterisation of habitat has been done by noting the dominant plants on each site. Suppl. material 2 provides a list of the 1,919 sampling events, each corresponding to a particular range of dates for a particular trap. Within the SMTP project, we maintain a unique set of identifiers for the traps and another set of identifiers for the collecting events. These identifiers are used consistently in all SMTP datasets to facilitate analyses that combine information from several datasets.
Geographic coverage
Description
The trap sites are spread throughout Sweden (Fig. 1; Suppl. material 1). Individual datasets of identified specimens span different subsets of the 1,919 samples and typically only include some of the trapping sites. Therefore, the geographic coverage varies considerably amongst datasets. As the geographic coverage will increase as data are added to each individual dataset, the actual geographic coverage of the dataset has to be computed from the Event table (the Core table in the Darwin Core Archive). The geographic coverage specified in the metadata of the dataset matches that of the entire project.
Taxonomic coverage
Description
The taxonomic coverage of the entire SMTP catch is quite broad. Malaise traps principally target flying insects (especially Hymenoptera and Diptera), but SMTP material also includes large numbers of other terrestrial arthropods (Araneae, Acari, Collembola) and scattered specimens of other invertebrates (Pulmonata, Lumbricidae etc). There are also single examples of unwanted bycatch of vertebrates (several lizards, a bird, a bat), but these specimens have not been preserved as part of the SMTP material.
The taxonomic coverage of each sample-based SMTP dataset is given as part of the metadata published with the dataset. The current data (Table 1, Suppl. material 3) comprise 79 different data datasets. The datasets are dominated by groups belonging to the orders Diptera and Hymenoptera, mostly at the family or subfamily level. Together, these orders comprise about 90% of the specimens in the SMTP material and they also dominate amongst the datasets that cover large numbers of samples and specimens (Table 1, Suppl. material 3). The remaining insect taxa in the SMTP material are mostly sorted to the order level; there are also substantial datasets for some of these. In total, the 79 data datasets currently available cover around 165,000 specimens, that is, about 1% of the total catch. The data are now accumulating rapidly and will be published continuously, so these numbers are likely to increase considerably over the coming years.
Table 1.
The 79 current datasets of the SMTP. Full version is available as a supplementary file.
| Order | Dataset | Link to Dataset | Included Taxa | Order |
| 1 | Odonata | https://doi.org/10.15468/5gtjxx | Odonata: all families | Odonata |
| 2 | Plecoptera | https://doi.org/10.15468/cvcqeb | Plecoptera: all families | Plecoptera |
| 3 | Dermaptera | https://doi.org/10.15468/w5xcbu | Dermaptera: all families | Dermaptera |
| 4 | Psocoptera | https://doi.org/10.15468/37b3pz | Psocoptera: all Psocodea except Phthiraptera | Psocoptera |
| 5 | Thysanoptera | https://doi.org/10.15468/8qm8rd | Thysanoptera: all families | Thysanoptera |
| 6 | Auchenorrhyncha excl Delphacidae | https://doi.org/10.15468/b48ufy | Hemiptera: Auchenorrhyncha: Cicadidae, Cicadellidae, Ulopidae, Membracidae, Aphrophoridae, Achilidae, Caliscelidae, Cixiidae, Issidae | Hemiptera |
| 7 | Psylloidea | https://doi.org/10.15468/cdw57a | Hemiptera: Sternorhyncha: Psylloidea | Hemiptera |
| 8 | Coleoptera | https://doi.org/10.15468/87hk5a | Coleoptera: all families | Coleoptera |
| 9 | Trichoptera | https://doi.org/10.15468/b3yrfu | Trichoptera: all families | Trichoptera |
| 10 | Microlepidoptera | https://doi.org/10.15468/wxyx7e | Lepidoptera: Micropteridoidea, Eriocranioidea, Hepialoidea, Nepticuloidea, Adeloidea, Tischerioidea, Tineoidea, Gracillarioidea, Douglasioidea, Yponomeutoidea, Gelechioidea, Tortricoidea, Schreckensteinioidea, Epermenioidea, Urodoidea, Choreutoidea, Alucitoidea, Pterophoroidea, Pyraloidea, Cossoidea | Lepidoptera |
| 11 | Macrolepidoptera | https://doi.org/10.15468/55t2ay | Lepidoptera: Zygaenoidea, Papilionoidea, Drepanoidea, Geometroidea, Lasiocampoidea, Bombycoidea, Sphingoidea, Noctuoidea | Lepidoptera |
| 12 | Strepsiptera | https://doi.org/10.15468/5ag9hn | Strepsiptera: all families | Strepsiptera |
| 13 | Mycetophilidae, Keroplatidae | https://doi.org/10.15468/dpa676 | Mycetophilidae, Keroplatidae | Diptera |
| 14 | Small Sciaroidea families | https://doi.org/10.15468/stg5z9 | Diadocidiidae, Mycetobiidae, Ditomyiidae, Sciarosoma | Diptera |
| 15 | Dixidae | https://doi.org/10.15468/u7dd6f | Dixidae | Diptera |
| 16 | Pachyneuridae | https://doi.org/10.15468/5epjpb | Pachyneuridae | Diptera |
| 17 | Cecidomyiidae: Porricondylinae (s lat) | https://doi.org/10.15468/hqpbmf | Cecidomyiidae: Porricondylinae, Winnertziinae | Diptera |
| 18 | Phoridae | https://doi.org/10.15468/ws6uh3 | Phoridae | Diptera |
| 19 | Dolichopodidae | https://doi.org/10.15468/rs389z | Dolichopodidae | Diptera |
| 20 | Drosophilidae | https://doi.org/10.15468/9sjvbp | Drosophilidae | Diptera |
| 21 | Sepsidae | https://doi.org/10.15468/a6ugn5 | Sepsidae | Diptera |
| 22 | Heleomyzidae, Odiniidae | https://doi.org/10.15468/gm4esc | Heleomyzidae | Diptera |
| 23 | Chloropidae | https://doi.org/10.15468/xqyhd8 | Chloropidae | Diptera |
| 24 | Piophilidae | https://doi.org/10.15468/wchd9a | Piophilidae | Diptera |
| 25 | Sciomyzidae | https://doi.org/10.15468/bdzv7j | Sciomyzidae | Diptera |
| 26 | Lonchaeidae | https://doi.org/10.15468/8hsehm | Lonchaeidae | Diptera |
| 27 | Milichiidae excl Phyllomyza | https://doi.org/10.15468/ajpx9r | Milichiidae excl. Phyllomyza | Diptera |
| 28 | Muscidae | https://doi.org/10.15468/s5y2mb | Muscidae | Diptera |
| 29 | Anthomyzidae | https://doi.org/10.15468/zhb2jn | Anthomyzidae | Diptera |
| 30 | Tephritidae, Ulidiidae | https://doi.org/10.15468/y6dzym | Tephritidae, Ulidiidae | Diptera |
| 31 | Acartophthalmidae | https://doi.org/10.15468/nw85cf | Acartophthalmidae | Diptera |
| 32 | Lauxaniidae | https://doi.org/10.15468/q6653a | Lauxaniidae | Diptera |
| 33 | Trixoscelididae | https://doi.org/10.15468/7v7ybr | Trixoscelididae | Diptera |
| 34 | Pallopteridae | https://doi.org/10.15468/z244v6 | Pallopteridae | Diptera |
| 35 | Asteiidae | https://doi.org/10.15468/z244v6 | Asteiidae | Diptera |
| 36 | Syrphidae | https://doi.org/10.15468/4km26f | Syrphidae | Diptera |
| 37 | Dryomyzidae | https://doi.org/10.15468/9s6ktn | Dryomyzidae | Diptera |
| 38 | Conopidae | https://doi.org/10.15468/2denvh | Conopidae | Diptera |
| 39 | Aulacigastridae | https://doi.org/10.15468/n9qrv4 | Aulacigastridae | Diptera |
| 40 | Clusiidae | https://doi.org/10.15468/2ng6hg | Clusiidae | Diptera |
| 41 | Asilidae | https://doi.org/10.15468/6d663p | Asilidae | Diptera |
| 42 | Therevidae | https://doi.org/10.15468/3n7h7p | Therevidae | Diptera |
| 43 | Symphyta | https://doi.org/10.15468/fag596 | Xyelidae, Cephidae, Pamphiliidae, Siricidae, Xiphydriidae, Argidae, Blasticotomidae, Cimbicidae, Diprionidae, Tenthredinidae, Heptamelidae, Orussidae | Hymenoptera |
| 44 | Platygastridae (s str) | https://doi.org/10.15468/jh6duy | Platygastridae | Hymenoptera |
| 45 | Figitidae excl Charipinae | https://doi.org/10.15468/es2jyy | Figitidae: Anacharitinae, Aspicerinae, Eucoilinae, Figitinae | Hymenoptera |
| 46 | Ismaridae | https://doi.org/10.15468/zx6dxp | Ismaridae | Hymenoptera |
| 47 | Heloridae | https://doi.org/10.15468/wkxgbq | Heloridae | Hymenoptera |
| 48 | Evaniidae | https://doi.org/10.15468/t2nc3b | Evaniidae | Hymenoptera |
| 49 | Gasteruptiidae, Aulacidae | https://doi.org/10.15468/xknzj2 | Gasteruptiidae, Aulacidae | Hymenoptera |
| 50 | Eupelmidae | https://doi.org/10.15468/y7ndas | Eupelmidae | Hymenoptera |
| 51 | Omphale | https://doi.org/10.15468/yvwnpu | Eulophidae: Entedoninae: Omphale | Hymenoptera |
| 52 | Mymar | https://doi.org/10.15468/sdgpvp | Mymaridae: Mymar | Hymenoptera |
| 53 | Ichneumoninae excl Phaeogenini | https://doi.org/10.15468/mer99q | Ichneumonidae: Ichneumoninae: Eurylabini, Goedartiinae, Heresiarchini, Ichneumonini, Listrodromini, Oedicephalini, Platylabini, Zimmeriini | Hymenoptera |
| 54 | Adelognathinae | https://doi.org/10.15468/f46vf7 | Ichneumonidae: Adelognathinae | Hymenoptera |
| 55 | Diplazontinae | https://doi.org/10.15468/f9u8kc | Ichneumonidae: Diplazontinae | Hymenoptera |
| 56 | Pimplinae | https://doi.org/10.15468/8pu4y5 | Ichneumonidae: Pimplinae | Hymenoptera |
| 57 | Rhyssinae | https://doi.org/10.15468/at6azc | Ichneumonidae: Rhyssinae | Hymenoptera |
| 58 | Poemeniinae | https://doi.org/10.15468/kq78ja | Ichneumonidae: Poemeniinae | Hymenoptera |
| 59 | Xoridinae | https://doi.org/10.15468/btczj6 | Ichneumonidae: Xoridinae | Hymenoptera |
| 60 | Phrudus group | https://doi.org/10.15468/bt7w4p | Ichneumonidae: Tersilochinae: Phrudus group | Hymenoptera |
| 61 | Neorhacodes | https://doi.org/10.15468/saparq | Ichneumonidae: Neorhacodinae | Hymenoptera |
| 62 | Banchini | https://doi.org/10.15468/h8jaj2 | Ichneumonidae: Banchinae: Banchini | Hymenoptera |
| 63 | Netelia | https://doi.org/10.15468/p3r2rb | Ichneumonidae: Tryphoninae: Netelia | Hymenoptera |
| 64 | Anomaloninae | https://doi.org/10.15468/4smbc7 | Ichneumonidae: Anomaloninae | Hymenoptera |
| 65 | Brachycyrtinae | https://doi.org/10.15468/5zhs46 | Ichneumonidae: Brachycyrtinae | Hymenoptera |
| 66 | Diacritinae | https://doi.org/10.15468/zycqbx | Ichneumonidae: Diacritinae | Hymenoptera |
| 67 | Cheloninae excl Adelius | https://doi.org/10.15468/68s836 | Braconidae: Cheloninae excl. Adelius | Hymenoptera |
| 68 | Meteorini | https://doi.org/10.15468/c5fqds | Braconidae: Euphorinae: Meteorini | Hymenoptera |
| 69 | Rogadinae | https://doi.org/10.15468/jmn2xn | Braconidae: Rogadinae | Hymenoptera |
| 70 | Adelius | https://doi.org/10.15468/wg5p5e | Braconidae: Cheloninae: Adelius | Hymenoptera |
| 71 | Pompilidae | https://doi.org/10.15468/62qwjj | Pompilidae | Hymenoptera |
| 72 | Dryinidae, Embolemidae | https://doi.org/10.15468/3k4v3x | Dryinidae, Embolemidae | Hymenoptera |
| 73 | Small families of parasitic aculeates | https://doi.org/10.15468/d8xe2j | Mutillidae, Myrmosidae, Tiphiidae, Methochidae, Sapygidae, Pompilidae | Hymenoptera |
| 74 | Sphecidae (s lat) | https://doi.org/10.15468/bwhfe6 | Crabronidae, Sphecidae, Ampulicidae | Hymenoptera |
| 75 | Vespinae | https://doi.org/10.15468/qck78f | Vespidae: Vespinae | Hymenoptera |
| 76 | Eumeninae | https://doi.org/10.15468/x656zx | Vespidae: Eumeninae | Hymenoptera |
| 77 | Formicidae | https://doi.org/10.15468/hd5npb | Formicidae | Hymenoptera |
| 78 | Chrysididae | https://doi.org/10.15468/g44yc3 | Chrysididae | Hymenoptera |
| 79 | Bethylidae | https://doi.org/10.15468/633y44 | Bethylidae | Hymenoptera |
The datasets only cover a portion of the available taxonomic fractions from the SMTP material. About 330 taxonomic sorting fractions are currently used (Table 2; see the list at the Station Linné webpage for more details, including older sorting fractions). Around half of these fractions are at present actively being worked on. As new experts become involved in the project, we expect that the taxonomic coverage of the identified material will increase. The taxonomic fractions have changed slightly through the years, reflecting how experts want to work with the material at a given point in time and according to the sorting expertise available in the SMTP lab. These types of changes are likely to continue over the coming years. Similarly, we expect that the taxonomic circumscription of published datasets will change slowly over time.
Table 2.
Sorting fractions in the SMTP.
| First Tier Sorting Fraction | Second Tier Sorting Fraction | Third Tier Sorting Fraction |
| ACARI | ||
| AMPHIBIA | ||
| AMPHIPODA | ||
| ANNELIDA | ||
| ARANEAE | Agelenidae | |
| ARANEAE | Amaurobiidae | |
| ARANEAE | Anyphaenidae | |
| ARANEAE | Araneidae | |
| ARANEAE | Argyronetidae | |
| ARANEAE | Atypidae | |
| ARANEAE | Clubionidae | |
| ARANEAE | Corinnidae (Syn: Phrurolithidae) | |
| ARANEAE | Dictynidae | |
| ARANEAE | Dysderidae | |
| ARANEAE | Eresidae | |
| ARANEAE | Gnaphosidae | |
| ARANEAE | Hahniidae | |
| ARANEAE | Linyphiidae | |
| ARANEAE | Liocranidae | |
| ARANEAE | Lycosidae | |
| ARANEAE | Mimetidae (Syn: Eutichuridae) | |
| ARANEAE | Miturgidae | |
| ARANEAE | Nesticidae | |
| ARANEAE | Oecobiidae | |
| ARANEAE | Oonopidae | |
| ARANEAE | Oxyopidae | |
| ARANEAE | Philodromidae | |
| ARANEAE | Pholcidae | |
| ARANEAE | Pisauridae | |
| ARANEAE | Salticidae | |
| ARANEAE | Segestriidae | |
| ARANEAE | Sparassidae (Heteropodidae) | |
| ARANEAE | Tetragnathidae | |
| ARANEAE | Theridiidae | |
| ARANEAE | Theridiosomatidae | |
| ARANEAE | Thomisidae | |
| ARANEAE | Titanoecidae | |
| ARANEAE | Uloboridae | |
| ARANEAE | Zoridae (Syn: Miturgidae) | |
| ARCHAEOGNATHA | ||
| AUCHENORRHYNCHA | ||
| AVES | ||
| BLATTODEA | ||
| BRACHYCERA | Acartophthalmidae | |
| BRACHYCERA | Acroceridae | |
| BRACHYCERA | Agromyzidae | |
| BRACHYCERA | Anthomyiidae | |
| BRACHYCERA | Anthomyzidae | |
| BRACHYCERA | Asilidae | |
| BRACHYCERA | Asteiidae | |
| BRACHYCERA | Athericidae | |
| BRACHYCERA | Aulacigasteridae | |
| BRACHYCERA | Bombyliidae | |
| BRACHYCERA | Braulidae | |
| BRACHYCERA | Calliphoridae | |
| BRACHYCERA | Camillidae | |
| BRACHYCERA | Campichoetidae | |
| BRACHYCERA | Carnidae | |
| BRACHYCERA | Chamaemyiidae | |
| BRACHYCERA | Chiropteromyzidae | |
| BRACHYCERA | Chloropidae | |
| BRACHYCERA | Chyromyidae | |
| BRACHYCERA | Clusiidae | |
| BRACHYCERA | Coelopidae | |
| BRACHYCERA | Coenomyiidae | |
| BRACHYCERA | Conopidae | |
| BRACHYCERA | Diastatidae | |
| BRACHYCERA | Dolichopodidae | |
| BRACHYCERA | Drosophilidae | |
| BRACHYCERA | Dryomyzidae | |
| BRACHYCERA | Empididae sensu lato (incl. Hybotidae, Atelestidae, Microphoridae, Brachystomatidae) | |
| BRACHYCERA | Empididae sensu stricto | |
| BRACHYCERA | Ephydridae | |
| BRACHYCERA | Fanniidae | |
| BRACHYCERA | Gasterophilidae | |
| BRACHYCERA | Helcomyzidae | |
| BRACHYCERA | Heleomyzidae (incl Borboropsidae) | |
| BRACHYCERA | Heterocheilidae (note: junior homonym) | |
| BRACHYCERA | Hippoboscidae | |
| BRACHYCERA | Hybotidae | |
| BRACHYCERA | Hypodermatidae | |
| BRACHYCERA | Lauxaniidae | |
| BRACHYCERA | Lonchaeidae | |
| BRACHYCERA | Lonchopteridae | |
| BRACHYCERA | Megamerinidae | |
| BRACHYCERA | Micropezidae | |
| BRACHYCERA | Milichiidae | |
| BRACHYCERA | Muscidae | |
| BRACHYCERA | Mythicomyiidae | |
| BRACHYCERA | Nycteribiidae | |
| BRACHYCERA | Odiniidae | |
| BRACHYCERA | Oestridae | |
| BRACHYCERA | Opetiidae | |
| BRACHYCERA | Opomyzidae | |
| BRACHYCERA | Pallopteridae | |
| BRACHYCERA | Periscelididae | |
| BRACHYCERA | Phaeomyiidae | |
| BRACHYCERA | Piophilidae | |
| BRACHYCERA | Pipunculidae | |
| BRACHYCERA | Platypezidae | |
| BRACHYCERA | Platystomatidae | |
| BRACHYCERA | Pseudopomyzidae | |
| BRACHYCERA | Psilidae | |
| BRACHYCERA | Rhagionidae | |
| BRACHYCERA | Rhinophoridae | |
| BRACHYCERA | Sarcophagidae | |
| BRACHYCERA | Scathophagidae | |
| BRACHYCERA | Scenopinidae | |
| BRACHYCERA | Sciomyzidae | |
| BRACHYCERA | Sepsidae | |
| BRACHYCERA | Sphaeroceridae | |
| BRACHYCERA | Stenomicridae | |
| BRACHYCERA | Stratiomyidae | |
| BRACHYCERA | Strongylophthalmyiidae | |
| BRACHYCERA | Syrphidae | |
| BRACHYCERA | Tabanidae | |
| BRACHYCERA | Tachinidae | |
| BRACHYCERA | Tanypezidae | |
| BRACHYCERA | Tephritidae | |
| BRACHYCERA | Tethinidae | |
| BRACHYCERA | Therevidae | |
| BRACHYCERA | Trixoscelididae | |
| BRACHYCERA | Ulidiidae | |
| BRACHYCERA | Xylomyidae | |
| BRACHYCERA | Xylophagidae | |
| BRACHYCERA : Phoridae | ||
| COLEOPTERA | ||
| COLLEMBOLA | ||
| DERMAPTERA | ||
| DIPLURA | ||
| EPHEMEROPTERA | ||
| GASTROPODA | ||
| HETEROPTERA | ||
| HYMENOPTERA | Apiformes (Anthophila) | |
| HYMENOPTERA | Aulacidae | |
| HYMENOPTERA | Bethylidae | |
| HYMENOPTERA | Braconidae | Adeliinae (Adelius) |
| HYMENOPTERA | Braconidae | Agathidinae |
| HYMENOPTERA | Braconidae | Alysiinae: Alysiini |
| HYMENOPTERA | Braconidae | Alysiinae: Dacnusini |
| HYMENOPTERA | Braconidae | Aphidiinae |
| HYMENOPTERA | Braconidae | Brachistinae: Blacini |
| HYMENOPTERA | Braconidae | Brachistinae:Brachistini |
| HYMENOPTERA | Braconidae | Braconinae |
| HYMENOPTERA | Braconidae | Cardiochilinae |
| HYMENOPTERA | Braconidae | Cenocoeliinae |
| HYMENOPTERA | Braconidae | Charmontinae |
| HYMENOPTERA | Braconidae | Cheloninae |
| HYMENOPTERA | Braconidae | Doryctinae |
| HYMENOPTERA | Braconidae | Euphorinae sensu stricto |
| HYMENOPTERA | Braconidae | Exothecinae |
| HYMENOPTERA | Braconidae | Gnamptodontinae |
| HYMENOPTERA | Braconidae | Helconinae sensu stricto |
| HYMENOPTERA | Braconidae | Histeromerinae |
| HYMENOPTERA | Braconidae | Homolobinae sensu stricto |
| HYMENOPTERA | Braconidae | Hormiinae |
| HYMENOPTERA | Braconidae | Ichneutinae |
| HYMENOPTERA | Braconidae | Lysiterminae |
| HYMENOPTERA | Braconidae | Macrocentrinae |
| HYMENOPTERA | Braconidae | Euphorinae: Meteorini |
| HYMENOPTERA | Braconidae | Microgastrinae |
| HYMENOPTERA | Braconidae | Microtypinae |
| HYMENOPTERA | Braconidae | Miracini |
| HYMENOPTERA | Braconidae | Euphorinae: Neoneurini |
| HYMENOPTERA | Braconidae | Opiinae |
| HYMENOPTERA | Braconidae | Orgilinae |
| HYMENOPTERA | Braconidae | Pambolinae |
| HYMENOPTERA | Braconidae | Proteropini |
| HYMENOPTERA | Braconidae | Rhysipolinae |
| HYMENOPTERA | Braconidae | Rhyssalinae |
| HYMENOPTERA | Braconidae | Rogadinae sensu stricto |
| HYMENOPTERA | Braconidae | Sigalphinae |
| HYMENOPTERA | Ceraphronidae | |
| HYMENOPTERA | Chalcidoidea sensu lato | Aphelinidae sensu lato (incl. Azotidae & Eriaporidae) |
| HYMENOPTERA | Chalcidoidea sensu lato | Chalcididae |
| HYMENOPTERA | Chalcidoidea sensu lato | Encyrtidae |
| HYMENOPTERA | Chalcidoidea sensu lato | Eulophidae |
| HYMENOPTERA | Chalcidoidea sensu lato | Eupelmidae |
| HYMENOPTERA | Chalcidoidea sensu lato | Eurytomidae |
| HYMENOPTERA | Chalcidoidea sensu lato | Mymaridae |
| HYMENOPTERA | Chalcidoidea sensu lato | Ormyridae |
| HYMENOPTERA | Chalcidoidea sensu lato | Perilampidae |
| HYMENOPTERA | Chalcidoidea sensu lato | Pteromalidae |
| HYMENOPTERA | Chalcidoidea sensu lato | Signiphoridae |
| HYMENOPTERA | Chalcidoidea sensu lato | Tetracampidae |
| HYMENOPTERA | Chalcidoidea sensu lato | Torymidae |
| HYMENOPTERA | Chalcidoidea sensu lato | Trichogrammatidae |
| HYMENOPTERA | ChalcidoideaMymaridae: Mymar | |
| HYMENOPTERA | Chrysididae | |
| HYMENOPTERA | Cynipoidea | Charipinae |
| HYMENOPTERA | Cynipoidea | Cynipidae |
| HYMENOPTERA | Cynipoidea | Figitidae (excl. Charipinae) |
| HYMENOPTERA | Cynipoidea | Ibaliidae |
| HYMENOPTERA | Diapriidae: Belytinae | |
| HYMENOPTERA | Diapriidae: Diapriinae | |
| HYMENOPTERA | Dryinidae | |
| HYMENOPTERA | Embolemidae | |
| HYMENOPTERA | Evaniidae | |
| HYMENOPTERA | Formicidae | |
| HYMENOPTERA | Gasteruptiidae | |
| HYMENOPTERA | Heloridae | |
| HYMENOPTERA | Ichneumonidae | Acaenitinae |
| HYMENOPTERA | Ichneumonidae | Adelognathinae |
| HYMENOPTERA | Ichneumonidae | Agriotypinae Haliday |
| HYMENOPTERA | Ichneumonidae | Alomyinae |
| HYMENOPTERA | Ichneumonidae | Anomaloninae |
| HYMENOPTERA | Ichneumonidae | Atrophini |
| HYMENOPTERA | Ichneumonidae | Banchinae |
| HYMENOPTERA | Ichneumonidae | Banchini |
| HYMENOPTERA | Ichneumonidae | Brachycyrtinae |
| HYMENOPTERA | Ichneumonidae | Campopleginae (Porizontinae) |
| HYMENOPTERA | Ichneumonidae | Collyriinae |
| HYMENOPTERA | Ichneumonidae | Cremastinae |
| HYMENOPTERA | Ichneumonidae | Cryptini Kirby |
| HYMENOPTERA | Ichneumonidae | Ctenopelmatini |
| HYMENOPTERA | Ichneumonidae | Cylloceriinae |
| HYMENOPTERA | Ichneumonidae | Diacritinae |
| HYMENOPTERA | Ichneumonidae | Diplazontinae |
| HYMENOPTERA | Ichneumonidae | Ephialtini excl. Polysphincta group (incl. Delomeristini and Perithoini) |
| HYMENOPTERA | Ichneumonidae | Eucerotinae |
| HYMENOPTERA | Ichneumonidae | Euryproctini |
| HYMENOPTERA | Ichneumonidae | Exenterini (incl. Eclytini) |
| HYMENOPTERA | Ichneumonidae | Glyptini |
| HYMENOPTERA | Ichneumonidae | Helictes group (≈Plectiscinae) |
| HYMENOPTERA | Ichneumonidae | Hemigastrini |
| HYMENOPTERA | Ichneumonidae | Hybrizontinae (Paxylommatinae) |
| HYMENOPTERA | Ichneumonidae | Ichneumonini sensu lato |
| HYMENOPTERA | Ichneumonidae | Idiogrammatini |
| HYMENOPTERA | Ichneumonidae | Lycorininae |
| HYMENOPTERA | Ichneumonidae | Mesochorinae |
| HYMENOPTERA | Ichneumonidae | Mesoleiini |
| HYMENOPTERA | Ichneumonidae | Metopiinae |
| HYMENOPTERA | Ichneumonidae | Microleptinae |
| HYMENOPTERA | Ichneumonidae | Neorhacodinae |
| HYMENOPTERA | Ichneumonidae | Oedemopsini |
| HYMENOPTERA | Ichneumonidae | Olethrodotini |
| HYMENOPTERA | Ichneumonidae | Ophioninae |
| HYMENOPTERA | Ichneumonidae | Orthocentrus group (Orthocentrini) |
| HYMENOPTERA | Ichneumonidae | Orthopelmatinae |
| HYMENOPTERA | Ichneumonidae | Oxytorinae |
| HYMENOPTERA | Ichneumonidae | Perilissini |
| HYMENOPTERA | Ichneumonidae | Phaeogenini |
| HYMENOPTERA | Ichneumonidae | Phrudini |
| HYMENOPTERA | Ichneumonidae | Phygadeuontini |
| HYMENOPTERA | Ichneumonidae | Phytodietini |
| HYMENOPTERA | Ichneumonidae | Pimplini |
| HYMENOPTERA | Ichneumonidae | Pionini |
| HYMENOPTERA | Ichneumonidae | Poemeniinae |
| HYMENOPTERA | Ichneumonidae | Polysphincta group (Polysphinctini) |
| HYMENOPTERA | Ichneumonidae | Rhyssinae |
| HYMENOPTERA | Ichneumonidae | Scolobatini |
| HYMENOPTERA | Ichneumonidae | Sphinctini |
| HYMENOPTERA | Ichneumonidae | Stilbopinae |
| HYMENOPTERA | Ichneumonidae | Tersilochinae sensu stricto |
| HYMENOPTERA | Ichneumonidae | Tryphonini |
| HYMENOPTERA | Ismaridae | |
| HYMENOPTERA | Megaspilidae | |
| HYMENOPTERA | Mutillidae | |
| HYMENOPTERA | Mymarommatoidea | |
| HYMENOPTERA | Myrmosidae | |
| HYMENOPTERA | Platygastridae | |
| HYMENOPTERA | Pompilidae | |
| HYMENOPTERA | Proctotrupidae | |
| HYMENOPTERA | Sapygidae | |
| HYMENOPTERA | Scelionidae | |
| HYMENOPTERA | Scoliidae | |
| HYMENOPTERA | Sparasionidae | |
| HYMENOPTERA | Spheciformes (Sphecidae sensu lato) | |
| HYMENOPTERA | Symphyta | Argidae |
| HYMENOPTERA | Symphyta | Blasticotomidae |
| HYMENOPTERA | Symphyta | Cephidae |
| HYMENOPTERA | Symphyta | Cimbicidae |
| HYMENOPTERA | Symphyta | Diprionidae |
| HYMENOPTERA | Symphyta | Heptamelidae |
| HYMENOPTERA | Symphyta | Tenthredinidae: Nematinae |
| HYMENOPTERA | Symphyta | Orussidae |
| HYMENOPTERA | Symphyta | Pamphiliidae |
| HYMENOPTERA | Symphyta | Siricidae |
| HYMENOPTERA | Symphyta | Tenthredinidae (excl. Nematinae) |
| HYMENOPTERA | Symphyta | Xiphydriidae |
| HYMENOPTERA | Symphyta | Xyelidae |
| HYMENOPTERA | Thynnidae | |
| HYMENOPTERA | Tiphiidae | |
| HYMENOPTERA | Vanhorniidae | |
| HYMENOPTERA | Vespidae | |
| ISOPODA Latreille, 1817 | ||
| LEPIDOPTERA | ||
| LEPIDOSAURIA | ||
| MAMMALIA | ||
| "MECOPTERA" | ||
| MEGALOPTERA | ||
| MYRIAPODA | ||
| "NEMATOCERA" | Anisopodidae | |
| "NEMATOCERA" | Bibionidae (incl Pleciidae) | |
| "NEMATOCERA" | Bolitophilidae | |
| "NEMATOCERA" | Canthyloscelidae (incl Synneuridae) | |
| "NEMATOCERA" | Cecidomyiidae | |
| "NEMATOCERA" | Ceratopogonidae | |
| "NEMATOCERA" | Chaoboridae | |
| "NEMATOCERA" | Chironomidae | |
| "NEMATOCERA" | Culicidae | |
| "NEMATOCERA" | Cylindrotomidae | |
| "NEMATOCERA" | Diadocidiidae | |
| "NEMATOCERA" | Ditomyiidae | |
| "NEMATOCERA" | Dixidae | |
| "NEMATOCERA" | Keroplatidae | |
| "NEMATOCERA" | Limoniidae | |
| "NEMATOCERA" | Mycetobiidae | |
| "NEMATOCERA" | Mycetophilidae | |
| "NEMATOCERA" | Pachyneuridae | |
| "NEMATOCERA" | Pediciidae | |
| "NEMATOCERA" | Psychodidae | |
| "NEMATOCERA" | Ptychopteridae | |
| "NEMATOCERA" | Scatopsidae | |
| "NEMATOCERA" | Sciaridae | |
| "NEMATOCERA" | Simuliidae | |
| "NEMATOCERA" | Thaumaleidae | |
| "NEMATOCERA" | Tipulidae | |
| "NEMATOCERA" | Trichoceridae | |
| ODONATA | ||
| OPILIONES | ||
| ORTHOPTERA sensu stricto (Saltatoria) | ||
| PHTHIRAPTERA | ||
| NEUROPTERA sensu stricto (Planipennia) | ||
| PLECOPTERA | ||
| PROTURA | ||
| PSEUDOSCORPIONES | ||
| "PSOCOPTERA" | ||
| RAPHIDIOPTERA | ||
| SIPHONAPTERA | ||
| STERNORRHYNCHA | ||
| STREPSIPTERA | ||
| THYSANOPTERA | ||
| TRICHOPTERA | ||
| ZYGENTOMA (Thysanura sensu stricto) | ||
Taxa included
| Rank | Scientific Name | |
|---|---|---|
| class | Arachnida | |
| class | Entognatha | |
| class | Insecta |
Temporal coverage
Data range: 2003-6-08 – 2006-11-23; 2007-4-11 – 2009-1-01.
Notes
The primary collection phase started in the summer of 2003 and ended in late 2006 and involves 71 of the 73 traps at 53 locations. Single traps were run continuously throughout this period or through substantial parts of it (Karlsson et al. 2020). Two complementary traps were run at two additional sites from 11 April 2007 until 1 January 2009 (Suppl. material 2).
Trap managers were instructed to empty the traps every two weeks. However, during the most intense summer period, some traps had to be emptied more often. Conversely, during the winter, sample periods were often much longer (Suppl. material 2). Individual datasets of identified material span only a subset of the 1,919 samples and the temporal coverage therefore varies.
The temporal coverage of individual sample-based datasets varies considerably, depending on which samples have been processed. The temporal coverage will increase over time as data are added to the dataset. The actual temporal coverage has to be computed from the Event table (the Core table of the Darwin Core Archive); the metadata specify the temporal coverage of the entire project.
Usage rights
Use license
Creative Commons Public Domain Waiver (CC-Zero)
IP rights notes
All SMTP datasets are released under the most permissive licence possible to facilitate use of the data.
Data resources
Data package title
SMTP DATA
Resource link
SMTP Collection Inventory: https://doi.org/10.15468/5becml; SMTP Taxonomic Datasets (available as 79 datasets as published by Station Linné): https://www.gbif.org/publisher/bee30abf-e3a6-4e4e-b626-7ef57800b027
Number of data sets
2
Data set 1.
Data set name
SMTP Collection Inventory
Number of columns
20
Data set 1.
| Column label | Column description |
|---|---|
| Scientific name | Latin name of species |
| Country or Area | Location of specimen collection |
| Coordinates | Latitude and Longitude |
| Month & Year | Month and year of specimen collection |
| Basis of Record | All records based on preserved specimens |
| Dataset | The dataset containing the record |
| Individual count | The number of individual specimens recorded |
| Recorded by | All records by the Swedish Malaise Trap Project |
| Collection code | Unique identifier for collection |
| Institution code | Code of institution owning record/specimen |
| Identified by | Taxonomic expert who made identification |
| Publisher | All records published by the Swedish Malaise Trap Project |
| Rank | Rank of identification known |
| Kingdom | Level of identification |
| Phylum | Level of identification |
| Class | Level of identification |
| Order | Level of identification |
| Family | Level of identification |
| Genus | Level of identification |
| Species | Level of identification |
Data set 2.
Data set name
SMTP Taxonomic Dataset (broken into 79 subsets)
Number of columns
20
Data set 2.
| Column label | Column description |
|---|---|
| Scientific name | Latin name of species |
| Country or Area | Location of specimen collection |
| Coordinates | Latitude and Longitude |
| Month & Year | Month and year of specimen collection |
| Basis of Record | All records based on preserved specimens |
| Dataset | The dataset containing the record |
| Individual count | The number of individual specimens recorded |
| Recorded by | All records by the Swedish Malaise Trap Project |
| Collection code | Unique identifier for collection |
| Institution code | Code of institution owning record/specimen |
| Identified by | Taxonomic expert who made identification |
| Publisher | All records published by the Swedish Malaise Trap Project |
| Rank | Rank of identification known |
| Kingdom | Level of identification |
| Phylum | Level of identification |
| Class | Level of identification |
| Order | Level of identification |
| Family | Level of identification |
| Genus | Level of identification |
| Species | Level of identification |
Additional information
Availability of material
The SMTP collection is maintained and curated as part of the insect collections of the Swedish Museum of Natural History (NRM) in Stockholm, Sweden. Most of the collection is kept in 95% ethanol at -20°C in modern storage facilities at Station Linné (see Karlsson et al. 2020 for details). Reference material identified by experts, as well as any type specimens, are deposited in the insect collections at the NRM.
Data availability
The SMTP data will be continuously published through GBIF and we refer readers to GBIF (https://gbif.org) and the national Swedish hubs for natural history collections (https://naturarv.se) and biodiversity data (https://bioatlas.se), for the most recent versions of the datasets. General project information will be available from the Station Linné web site (https://stationlinne.se).
Supplementary Material
Trapping site information
Karlsson, D; Forshage, M; Holston, K; Ronquist, F
Data type
Site
File: oo_452514.csv
Sampling events
Karlsson, D; Forshage, M; Holston, K; Ronquist, F
Data type
Unique identifiers
Brief description
Each event corresponding to a particular range of dates for a particular trap
File: oo_452524.xlsx
The current 79 datasets of the SMTP
Karlsson, D; Forshage, M; Holston, K; Ronquist, F
Data type
Occurrences
Brief description
Full version.
File: oo_455180.xlsx
Acknowledgements
A final thanks to the hundreds, at present and in the past, who have contributed to the SMTP. A special thanks to Emily Hartop who made the illustrations and to Harald Havnås who helped us with the tables in this paper.
Funding Statement
Swedish Taxonomy Initiative
References
- Holston K, Karlsson D, Forshage M, Telenius A, Saha M, Johansson V. Swedish Museum of Natural History via GBIF.org; 2020. Swedish Malaise Trap Project (SMTP) Collection Inventory. [DOI] [Google Scholar]
- Karlsson D, Pape T, Johanson KA, Liljeblad J, Ronquist F. Svenska Malaisefälleprojektet, eller hur många arter steklar, flugor och myggor finns i Sverige? http://www.sef.nu/download/entomologisk_tidskrift/et_2005/ET2005%2043-53.pdf Entomologisk Tidskrift. 2005;126:43–53. [Google Scholar]
- Karlsson Dave, Hartop Emily, Forshage Mattias, Jaschhof Mathias, Ronquist Fredrik. The Swedish Malaise Trap Project: A 15 year retrospective on a countrywide insect inventory. Biodiversity Data Journal. 2020;8:e47255. doi: 10.3897/BDJ.8.e47255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G. TAXONOMY: Linnaeus's legacy carries on. Science. 2005;307(5712):1038–1039. doi: 10.1126/science.307.5712.1038a. [DOI] [PubMed] [Google Scholar]
- Ronquist Fredrik, Gärdenfors Ulf. Taxonomy and biodiversity inventories: time to deliver. Trends in Ecology & Evolution. 2003;18(6):269–270. doi: 10.1016/s0169-5347(03)00098-3. [DOI] [Google Scholar]
- Ronquist Fredrik, Forshage Mattias, Häggqvist Sibylle, Karlsson Dave, Hovmöller Rasmus, Bergsten Johannes, Holston Kevin, Britton Tom, Abenius Johan, Andersson Bengt, Buhl Peter Neerup, Coulianos Carl-Cedric, Fjellberg Arne, Gertsson Carl-Axel, Hellqvist Sven, Jaschhof Mathias, Kjærandsen Jostein, Klopfstein Seraina, Kobro Sverre, Liston Andrew, Meier Rudolf, Pollet Marc, Riedel Matthias, Roháček Jindřich, Schuppenhauer Meike, Stigenberg Julia, Struwe Ingemar, Taeger Andreas, Ulefors Sven-Olof, Varga Oleksandr, Withers Phil, Gärdenfors Ulf. Completing Linnaeus's inventory of the Swedish insect fauna: Only 5,000 species left? PloS one. 2020;15(3):e0228561. doi: 10.1371/journal.pone.0228561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Trapping site information
Karlsson, D; Forshage, M; Holston, K; Ronquist, F
Data type
Site
File: oo_452514.csv
Sampling events
Karlsson, D; Forshage, M; Holston, K; Ronquist, F
Data type
Unique identifiers
Brief description
Each event corresponding to a particular range of dates for a particular trap
File: oo_452524.xlsx
The current 79 datasets of the SMTP
Karlsson, D; Forshage, M; Holston, K; Ronquist, F
Data type
Occurrences
Brief description
Full version.
File: oo_455180.xlsx
Data Availability Statement
The SMTP data will be continuously published through GBIF and we refer readers to GBIF (https://gbif.org) and the national Swedish hubs for natural history collections (https://naturarv.se) and biodiversity data (https://bioatlas.se), for the most recent versions of the datasets. General project information will be available from the Station Linné web site (https://stationlinne.se).