Abstract
Spotted seal (Phoca largha) is a critically endangered pinniped in China and South Korea. The conventional method to protect and maintain the P. largha population is to keep them captive in artificially controlled environments. However, little is known about the physiological differences between wild and captive P. largha. To generate a preliminary protein expression profile for P. largha, whole blood from wild and captive pups were subjected to a label-free comparative proteomic analysis. According to the results, 972 proteins were identified and predicted to perform functions related to various metabolic, immune, and cellular processes. Among the identified proteins, the expression level of 51 were significantly different between wild and captive P. large pups. These differentially expressed proteins were enriched in a wide range of cellular functions, including cytoskeleton, phagocytosis, proteolysis, the regulation of gene expression, and carbohydrate metabolism. The abundances of proteins involved in phagocytosis and ubiquitin-mediated proteolysis were significantly higher in the whole blood of wild P. largha pups than in captive individuals. In addition, heat shock protein 90-beta, were determined as the key protein associated with the differences in the wild and captive P. largha pups due to the most interactions of it with various differentially expressed proteins. Moreover, wild P. largha pups could be more nutritionally stressed and have more powerful immune capacities than captive pups. This study provides the first data on the protein composition of P. largha and provides useful information on the physiological characteristics for research in this species.
Subject terms: Proteomic analysis, Animal physiology
Introduction
Spotted seals (Phoca largha) are small-bodied pinnipeds that are generally distributed in the cold sea area of the North Pacific Ocean. P. largha has been listed as one of the most endangered species in China and South Korea due to the destruction of their habitat by anthropogenic impacts1. For many wild animals, captivity in artificially-controlled environments is one of the most effective ways to ensure their conservation. During the past decades, China has continuously implemented artificial breeding and rearing activities for the maintenance of the P. largha population. At present, more than 1000 spotted seals are in captivity in the aquarium of China, accounting for 50% of the total population2. Despite advances in species conservation, captivity has also been shown to affect genetic and physiological characteristics in a variety of animals. For example, significant differences in genetics and morphology were observed between wild and captive Leon Springs pupfish, Cyprinodon bovines3. In addition, wild Indian leopards, Panthera pardus fascia, showed higher nucleotide diversity and amino acid polymorphisms in major histocompatibility complex genes and proteins compared to captive individuals4. Moreover, differences in the concentration of plasma cortisol between captive and wild harbor seals (P. vitulina) of the same sex and during the same season were highly significant5. Nonetheless, no research has been performed to describe the potential physiological differences between wild and captive spotted seals.
Previous investigations in spotted seals mainly focused on their distribution6,7, development8, and genetic diversity9,10. To date, only a few studies explored the physiological characteristics of spotted seals at the molecular level. Gao et al.1 assembled the transcriptome in liver and spleen of spotted seals and identified 193 unigenes associated with defense mechanisms. In addition, the normal levels of hematology and serum biochemistry indices in the captive spotted seals were measured, and the age- and gender-related differences in those indices were obtained11. Moreover, the relationships between sexual maturation and the concentrations of serum testosterone, progesterone, and estradiol in captive spotted seals were also reported12. The physiological functions of all organisms are achieved through the “gene-mRNA-protein” pathway. Studies at the gene and mRNA levels do not completely reflect the physiological functions of organisms due to pre- and post-transcriptional regulation13. Proteins are the direct performers of biological functions, and thus, measuring protein expression profiles is a powerful way to understand the physiological characteristics of spotted seals.
Proteomics technologies evaluate the complete protein composition expressed by a genome, cell or tissue, and provide powerful tools to examine the physiological functions of animals14. Over the years, qualitative proteomics techniques based on mass spectrometry (MS), such as 2D-gel-MS, have developed into the most direct and accurate methods for identifying the proteins in animal samples15. However, such traditional techniques have many shortcomings, including their inability to quantitatively recognize the differentially expressed proteins (DEPs) and their poor detection of low-abundance proteins. Hence, a quantitative proteomics technology, named label-free shotgun proteomics, was developed to determine DEPs with extreme accuracy, sensitivity, discrimination, and high-throughput16. At present, label-free shotgun proteomics has been widely used in humans17, plants18, and microorganisms19. Therefore, comparative proteomics research based on label-free shotgun proteomics is suitable for a more comprehensive comparison of the physiological functions between wild and captive spotted seals.
In the present study, preliminary whole blood protein expression profiles for wild and captive P. largha pups were determined using the label-free shotgun proteomics technology. The objectives of this study were to (1) describe the preliminary whole blood protein composition patterns of P. largha pups; (2) provide an overview of the differences in the whole blood proteomes between wild and captive P. largha pups; and (3) identify the key proteins that may potentially alter the physiological functions of P. largha pups due to captivity. To the best of our knowledge, this study is the first application of proteomics technology for the evaluation of spotted seals.
Results
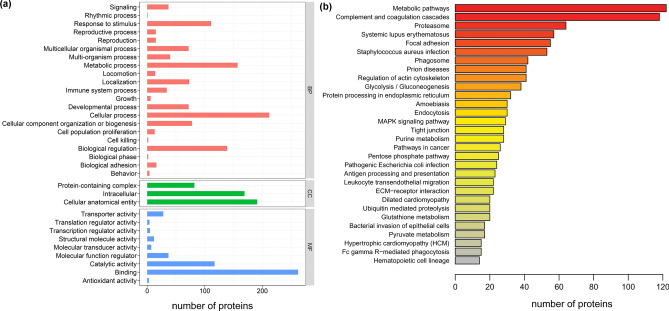
A total of six whole blood samples from P. largha pups (three from wild pups and three from captive pups) were measured using label-free proteomic analysis. After data filtering and protein identification, a total of 4562 unique peptides were obtained and 972 proteins were identified from P. largha pup blood (Supplementary file 1). To investigate the physiological functions of P. largha pups, annotations from the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were extracted based on the reference transcripts that matched the identified P. largha proteins. According to the results of GO annotations, the major protein functions included cellular process, metabolic process, biological regulation, and response to stimulus belonging to the biological process aspect, intracellular and cellular anatomical entity in the cellular component aspect, and binding and catalytic activity attribute to the molecular function aspect (Fig. 1a). For KEGG pathway analyses, the identified proteins exhibited a broad functional distribution, among which the dominant pathways were metabolic pathways, and the complement and coagulation cascades (Fig. 1b).
Figure 1.
Functional distribution of the proteins identified in the whole blood of wild and captive Phoca largha pups based on the GO (a) and KEGG (b) databases.
Based on the identified proteins, partial least squares discrimination analysis (PLS-DA) was used to comprehensively investigate the divergence of the preliminary protein expression profiles between wild and captive P. largha pups (Fig. 2). The first two components explained 57.8% of the total protein expression variations, and samples from the wild and captive individuals were observed in isolated clusters, separated from one another. Those results suggested that captivity in artificial environments could significantly affect the protein composition and abundance in the whole blood of P. largha pups. Furthermore, the expression of 51 proteins exhibited significant variations between the whole blood of wild and captive P. largha pups (Supplementary File 2). Compared to the samples from captive pups, the number of up-regulated DEPs in the wild-pup samples was 26 (Table 1), while seven DEPs were down-regulated (up-regulated in captive, Table 2). Moreover, there were eight and ten proteins unique to the wild- and captive-pup samples, respectively (Table 3). Thus, those results revealed the dissimilarity in the protein expression profiles in whole blood from wild and captive P. largha pups.
Figure 2.
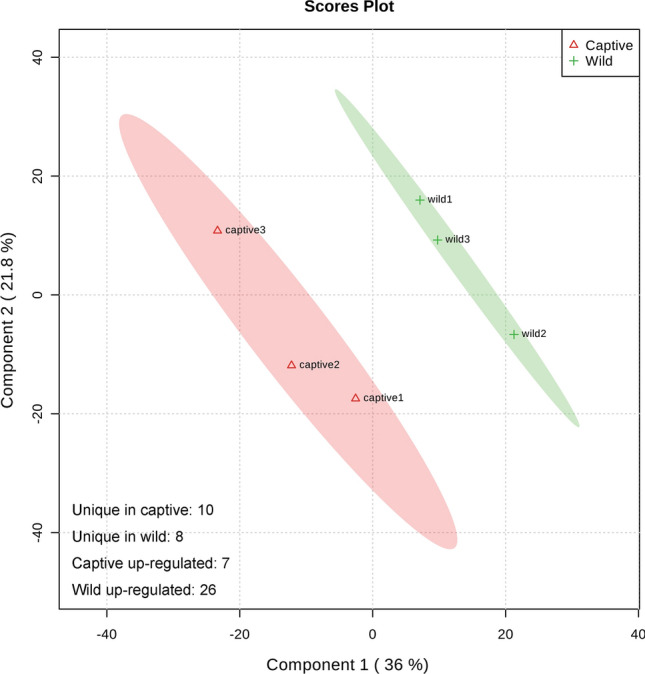
PLS-DA of proteins in the whole blood from wild and captive Phoca largha pups.
Table 1.
Up-regulated proteins in the whole blood of wild Phoca largha pups compared to captive pups.
| Protein identity | Fold change | P-value |
|---|---|---|
| Filamin A (FLNA) | 9.48 | 0.001 |
| Tubulin beta chain (TUBB) | 6.76 | 0.006 |
| Cathepsin S (CTSS) | 4.22 | 0.039 |
| Ubiquitin-conjugating enzyme E2-230K (UBE2) | 3.59 | 0.024 |
| Barrier to autointegration factor 1 (BANF1) | 2.90 | 0.045 |
| Alpha-2-HS-glycoprotein (AHSG) | 2.78 | 0.034 |
| Regulator of G-protein signaling 10 (RGS10) | 2.56 | 0.019 |
| Vinculin (VCL) | 2.54 | 0.027 |
| Ubiquitin-conjugating enzyme E2 O (UBE2O) | 2.52 | 0.026 |
| Plasma kallikrein (KLKB1) | 2.34 | 0.039 |
| NIF3-like protein 1 (NIF3L1) | 2.26 | 0.011 |
| S-100P (S100P) | 2.26 | 0.025 |
| Nucleosome assembly protein 1 (NAP1) | 2.13 | 0.026 |
| Protein phosphatase 1A (PP1A) | 2.07 | 0.033 |
| CD59 glycoprotein (CD59) | 1.98 | 0.009 |
| Protein phosphatase type 2A (PP2A) | 1.89 | 0.006 |
| Phosphatidylcholine-sterol acyltransferase precursor (LCAT) | 1.86 | 0.025 |
| Glucose-6-phosphate 1-dehydrogenase X (G6PDX) | 1.84 | 0.042 |
| Heat shock protein HSP 90-beta (HSP90AB1) | 1.80 | 0.034 |
| Ras suppressor protein 1 (RSU1) | 1.68 | 0.035 |
| Ubiquitin-like-conjugating enzyme (ATG3) | 1.68 | 0.021 |
| Calpain-1 catalytic subunit (CAPN1) | 1.68 | 0.027 |
| Calcium-regulated heat stable protein 1 (CARHSP1) | 1.63 | 0.043 |
| Ubiquitin-like modifier-activating enzyme 1 (UBA1) | 1.62 | 0.048 |
| Eukaryotic translation initiation factor 5 (EIF5) | 1.60 | 0.035 |
| Exportin-1 (XPO1) | 1.55 | 0.039 |
Table 2.
Down-regulated proteins in the whole blood of wild Phoca largha pups compared to captive pups.
| Protein identity | Fold change | P-value |
|---|---|---|
| Adenosylhomocysteinase (AHCY) | 0.65 | 0.040 |
| Proteasome subunit alpha type-2 (PSMA2) | 0.62 | 0.031 |
| Protein DDI1 homolog 2 (DDI2) | 0.62 | 0.002 |
| C10C5.4 | 0.53 | 0.038 |
| Aldehyde dehydrogenase family 1 member A3 (ALDH1A3) | 0.45 | 0.030 |
| Vitamin K-dependent protein S (PROS1) | 0.37 | 0.032 |
| Glutathione S-transferase theta-1 (GSTT1) | 0.32 | 0.037 |
Table 3.
Unique proteins in the whole blood of wild and captive Phoca largha pups.
| Unique in wild spotted seal | Unique in captive spotted seal |
|---|---|
| 60S acidic ribosomal protein P1 (RPLP1) | Aldo–keto reductase family 7 (AKR7) |
| Ankyrin-3 (ANK3) | Creatine kinase B (CKB) |
| LZIC-like isoform 2 (LZIC2) | Fibrinogen alpha chain (FGA) |
| Methylosome protein 50 (MEP50) | Flotillin-1 (FLOT1) |
| Translationally-controlled tumor protein 1 (TPT1) | Galectin-3-binding protein (LGALS3BP) |
| Protein argonaute-2 (AGO2) | Immunoglobulin alpha heavy chain (IGHA) |
| F-actin-capping protein subunit alpha-1 (CAPZA1) | Kell blood group glycoprotein (KEL) |
| Vasodilator-stimulated phosphoprotein (VASP) | Kynureninase (KYNU) |
| Myc box-dependent-interacting protein 1 (BIN1) | |
| Tumor protein D54 (TPD54) |
As illustrated in the DEP tables, some important immune-related proteins, such as Cathepsin S (CTSS), Protein phosphatase 1A (PP1A), CD59 glycolicprotein (CD59), Protein phosphatase type 2A (PP2A), Heat shock protein HSP 90-beta (HSP90AB1), and Calpain-1 catalytic subunit (CAPN1) were up-regulated in the blood of wild P. largha. In contrast, five proteins related to immune response were up-regulated or uniquely expressed in captive pups, including Glutathione S-transferase theta-1 (GSTT1), Vitamin K-dependent protein S (PROS1), Protein DDI1 homolog 2 (DDI2), Immunoglobulin alpha heavy chain (IGHA), and Galectin-3-binding protein (LGALS3BP). Moreover, several proteins involved in the regulation of gene expression were significantly up-regulated in the blood of wild P. largha pups, including Barrier to autointegration factor 1 (BANF1), NIF3-like protein 1 (NIF3L1), Calcium-regulated heat stable protein 1 (CARHSP1), Eukaryotic translation initiation factor 5 (EIF5), and Exportin-1 (XPO1). It is also worth noting that the expressions of multiple proteins related to cell adhesion were differentially expressed in the blood of wild P. largha pups, including Filamin A (FLNA), Tubulin beta chain (TUBB), Vinculin (VCL), Ankyrin-3 (ANK3), Translationally-controlled tumor protein 1 (TPT1), Protein argonaute-2 (AGO2), F-actin-capping protein (CAPZA1), Vasodilator-stimulated phosphoprotein (VASP), and Fibrinogen alpha chain (FGA).
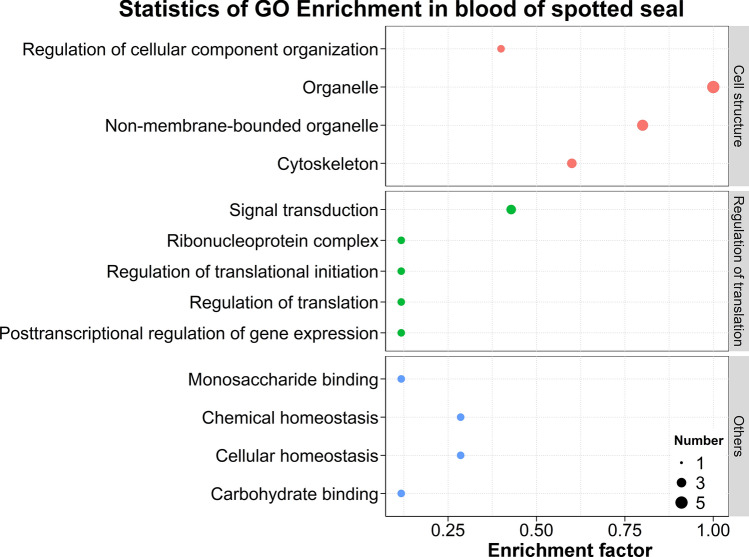
We also performed GO and KEGG enrichment analyses to explore the biological functions and pathways that could be significantly impacted due to captivity. The results showed that 13 GO terms were significantly enriched in the DEPs between wild and captive P. largha pups (Fig. 3). The enriched GO terms were assigned to three classes: (1) cell structure, including different kinds of organelle and cytoskeleton functions; (2) regulation of translation, which involves a broad range of post transcriptional and translational regulations; and (3) others, containing monosaccharide and carbohydrate binding, and cellular and chemical homeostasis.
Figure 3.
GO terms were significantly enriched (p adjusted by FDR < 0.05, performed by DAVID) in the whole blood of wild and captive Phoca largha pups based on the DEP dataset.
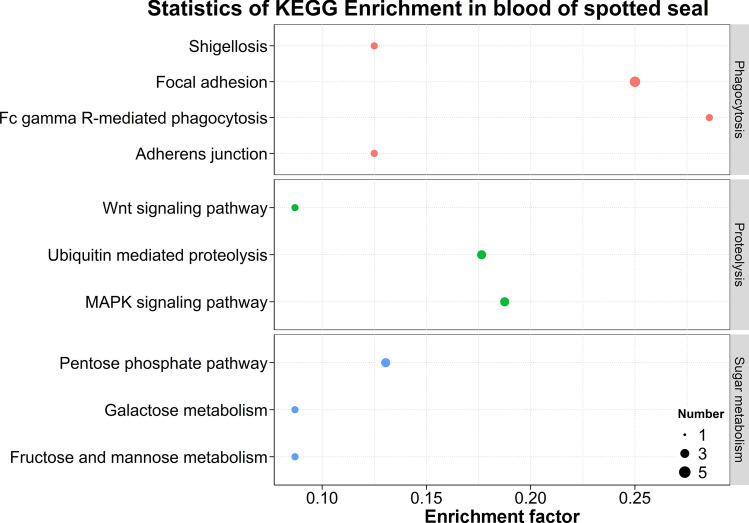
KEGG pathway enrichment analyses based on the DEPs revealed that ten pathways were significantly enriched in the blood of wild and captive P. largha pups (Fig. 4). According to the function of the enriched pathways, they were classified into three categories, including phagocytosis, proteolysis, and carbohydrate metabolism. The phagocytosis category included Fc gamma R-mediated phagocytosis and its related cell adhesion pathways. The proteolysis category included ubiquitin-mediated proteolysis and its accompanying signaling pathways. The carbohydrate metabolism category included the pentose, galactose, fructose and mannose metabolism pathways.
Figure 4.
KEGG pathways were significantly enriched (p adjusted by FDR < 0.05, performed by DAVID) in the whole blood of wild and captive Phoca largha pups based on the DEP dataset.
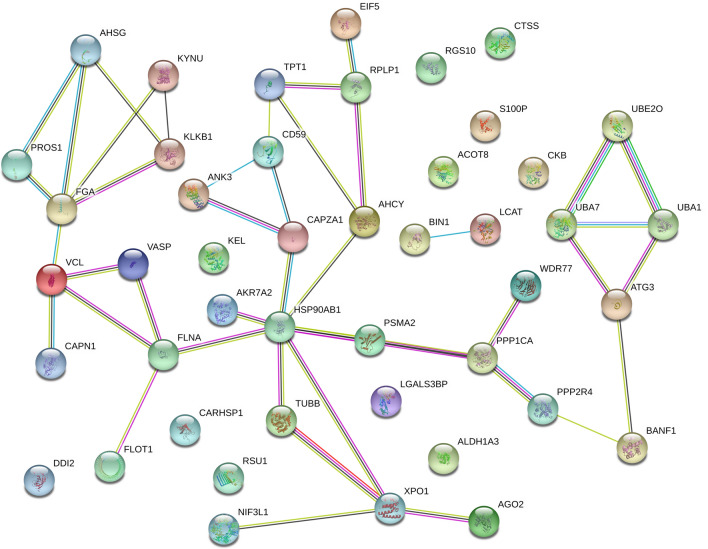
The protein–protein interaction (PPI) networks of the identified DEPs were analyzed using STRING software and the results are shown in Fig. 5. Based on the search results, HSP90AB1 had the most predicted interactions with the other DEPs. A variety of cell adhesion and cytoskeleton proteins (VASP, VCL, and FGA, among others) was predicted to interact with HSP90AB1 via FLNA. Additionally, HSP90AB1 was predicted to associate with a series of post-transcriptional (XPO1, AGO2 and NIF3L1) and translational regulation (RPLP1, EIF5, TPT1, etc.) proteins through its interactions with AHCY and TUBB, respectively. Moreover, the interaction between HSP90AB1 and PSMA2 was associated with proteins involved in ubiquitin-mediated proteolysis (UBE2O, UBA1 and UBA7).
Figure 5.
Protein–protein interaction (PPI) networks for differentially-expressed proteins between the whole blood of wild and captive Phoca largha pups. The PPI network had an average node degree of 1.35 and clustering coefficient of 0.491. The PPI enrichment p-value was 0.00532, which indicated that the network had significantly more interactions than expected. The colors of edges specify different types of interaction evidences and the thickness indicates the confidence of data support. STRING20 (https://string-db.org/) was used to construct the PPI networks of the DEPs to identify the key proteins that changed the physiological functions of P. largha due to captivity.
Discussion
Until now, no studies have been performed on the variations in protein composition and expression in P. largha. One of the aims of the current study was to determine the preliminary proteomic profile of P. largha. A total of 972 proteins were identified using label-free proteomics analyses. Regarding other marine mammals, a previous study of kidney diseases in wild animals identified a total of 2694 proteins in the urine of 19 California sea lions (Zalophus californianus)21. However, only 206 proteins were identified from the cerebrospinal fluid of 11 California sea lions22. Conversely, two previous studies of plasma samples from bottlenose dolphins (Tursiops truncates) only identified 58 and 196 unique proteins, respectively23,24. Thus, those results suggested that the complexity of the protein compositions of marine mammals varies between different tissues and species.
Another aim of this study was to identify the proteins that were differentially expressed between the whole blood of wild and captive P. largha. In the present study, the Fc gamma R-mediated phagocytosis pathway was enriched in the blood of wild and captive P. largha pups. Phagocytosis is an important innate immune defense mechanism in animals, and is accompanied by a chain of cellular processes as diverse as cytoskeletal rearrangement, alterations in membrane trafficking, and the activation of microbial killing mechanisms25.
Several cytoskeleton-related proteins were found to be up-regulated in the blood of wild P. largha pups compared to captive pups, including VASP, FLNA, TUBB, VCL, and CAPZA1. VASP is an actin-associated protein that promotes actin filament elongation26. The activation of Fc gamma receptors during phagocytosis triggers the recruitment of VASP to phagosomes27. FLNA is an actin-binding protein that anchors various transmembrane proteins to the actin cytoskeleton to promote cell–cell contact28. FLNA has been reported to play a role in neutrophil phagocytosis in humans29, and the deletion of FLNA resulted in a twofold reduction in collagen phagocytosis in mice30. The rate-limiting step during phagocytosis is the binding of specific adhesion receptors, such as integrins31. TUBB and VCL are intracellular adaptor proteins that link actin filaments and integrins to construct the cytoskeleton32,33. CAPZA1 performed to the fast-growing ends of actin filaments and its mutation resulted in actin accumulation, thereby blocking cytoskeletal rearrangement34. Conversely, FGA, an extracellular protein that interacts with integrins for cell–cell adhesion35, was uniquely detected in the blood of captive P. largha. It has been shown that the binding of FGA by Streptococcus progenies was involved in their resistance to phagocytosis in human blood36. Thus, all of the above findings suggest that the blood of wild P. largha pups contained more phagosomes than that of captive pups.
Moreover, during phagocytosis, macrophages alter the plasma membrane to form the phagosome, which is the principal site for killing microorganisms through proteolysis37. In the present study, proteins involved in the ubiquitin-mediated proteolysis pathway were enriched in the blood of P. largha pups, including UBE2, UBE2O, UBA1 and ATG3. The activation of ubiquitin-mediated proteolysis requires ATP to degrade microbial membrane proteins38. G6PDX, which catalyzes the rate-limiting step of the oxidative phosphate pathway to produce ATP39, was up-regulated in the blood of wild P. largha pups.
In summary, the DEPs involved in phagocytosis and proteolytic activities detected in the present study suggest that wild P. largha pups may be more resistant to pathogen infection than captive pups. The heightened level of immunity may be due to the result of ontogenetic differences between the two groups. In a species with such rapid post-natal development, animals that are < 1 month of age (wild pups in this study) are going to have dramatically different proteome profiles than animals that are 4 months of age (captive pups in this study). Similar to humans, newborn pups will acquire more powerful protection via maternal passive immunity from their mothers to ensure their survival40, and the immunity inherited from the mother could not have been completely degraded in the detected wild pups. Moreover, captive pups lived in relatively stable environments and were cared for by humans; thus, they were likely to be less environmentally stressed than wild pups. Such factors could be additional reasons for the higher expression of proteins associated with immune function of wild P. largha pups compared to their captive counterparts.
The biological roles of the DEPs identified in the present study were further examined based on their PPI networks to understand differences between wild and captive P. largha pups. HSP90AB1, a type of heat shock protein was upregulated in the wild P. largha pups, and was predicted to interact with a variety of DEPs of different functions, including those involved in cell adhesion, regulation of gene expression, and proteolysis. This result illustrated that HSP90AB1 could be the key protein related to the differences in the protein expression profiles between wild and captive P. largha pups. The realization of HSP90AB1 physiological functions depends on the consumption of ATP. Upon ATP binding, HSP90 undergoes significant conformational changes to become active and the ATP was hydrolyzed to ADP after the activation of client proteins41. This phenomenon was consistent with the up-regulation of proteins involved in carbohydrate metabolism in the blood of wild P. largha pups; thus, indicating that there might be more energy demand. Spotted seal pups in the wild are born in pack ice, which exposes them to higher predation than other animals. The pack ice breeding strategy resulted in the evolution of shorter lactation times and higher daily energy outputs in spotted seals42. In captive environments, newborn spotted seal pups are weaned and regularly fed. However, even if they have begun foraging, wild pups are likely to be more nutritionally-stressed than captive pups. On the other hand, wild pups are likely to respond differently to presence of and handling by researchers than pups in captivity. Sampling may induce a more robust physiological stress response in wild pups than captive pups, resulting the alteration of protein expression. Thus, the potentially higher nutritional and physiological stresses could be the underlying reason for the upregulation of proteins involved in carbohydrate metabolism and immunity in the blood of wild P. largha pups.
Due to ecological destruction and poaching, the number of spotted seals in Liaodng Bay, China is very low. Field rescue and captive breeding are the dominant programs for maintaining the Liaodong Bay population of spotted seals. The simultaneous occurrence of on-ice births, weaning pups, and ice-melting are important characteristics for the reproductive biology of these seals43. Increased ice melting due to global warming and harsh sea conditions have caused high death rates among spotted seal pups42. Therefore, higher expression of proteins associated with carbohydrate metabolism in wild compared to captive pups may be the result of higher metabolic demands of postnatal development in wild seals. Conversely, spotted seal pups born in aquariums are susceptible to diarrhea and pneumonia, both of which are caused by pathogenic bacteria44,45. The diseases of spotted seal pups in captive environments can lead to anorexia and death in severe cases. The downregulation of proteins associated with phagocytosis and proteolysis detected in the present study was consistent with high incidence of disease in captive pups. Therefore, it is necessary to improve the immunity of captive spotted seal pups through food additives or other methods. In addition, while proteomes of spotted seal adults were not measured currently, the results of spotted seal pups in this study are likely to differ from the adults. The data described here only provide preliminary insights into the physiology of this species, specifically in pups, but are by no means a comprehensive explanation of the impact of captivity on ringed seal physiology. In conclusion, the information provided herein not only expands the understanding of protein expression profiles in spotted seals, but also provides information for the conservation of this species.
Methods
The wild P. largha pups investigated in this study were sampled from Liaodong Bay, China, during the Spotted seal Rescue Survey in 2019. Based on the white lanugo on their skin, and the fact that they were independent of their mother indicated that they ended lactation, but were less than one month of age. The captive P. largha pups used in this study were from the Dalian Sun Asia Aquarium (DSAA), China, and were all newborn under human care in 2019. The captive pups were weaned and were regularly fed prior to sampling. The diet of the captive spotted seals was a 1:1 ratio of capelin Mallotus villosus and Atlantic herring, Clupea harengus (m:m). The amount of feed was approximately 8% of the pup’s weight per day. The ontogenetic differences of wild and captive P. largha pups are provided in Table 4. Approximately 3 mL of blood was collected from the veins in the hind flippers when the animals were restrained on a V-shaped bench. The handling protocol was the same for wild and captive pups. Blood samples were stored in medical biochemical tubes with anti-coagulation gel at − 80 °C prior to protein extraction. All operations were performed in accordance with the relevant guidelines and regulations.
Table 4.
The ontogenetic differences between wild and captive P. largha pups used in this study.
| Sample ID | Birth/rescue date | Sampling date | Body weight (kg) | Body size (cm) | Age (months) | Gender | Captive/discovery location |
|---|---|---|---|---|---|---|---|
| Captive1 | 2019.02.19 | 2019.06.15 | 39.6 | 105 | 4 | Male | DSAA, China |
| Captive2 | 2019.02.23 | 2019.06.15 | 43.5 | 105 | 4 | Female | DSAA, China |
| Captive3 | 2019.02.13 | 2019.06.15 | 46.8 | 108 | 4 | Male | DSAA, China |
| Wild1 | 2019.02.11 | 2019.02.14 | 12.0 | 75 | < 1 | Male | Dalian, China |
| Wild2 | 2019.02.11 | 2019.02.14 | 11.2 | 80 | < 1 | Female | Dalian, China |
| Wild3 | 2019.03.04 | 2019.03.05 | 13.5 | 81 | < 1 | Female | Panjin, China |
Label-free proteomics technology was used to generate the preliminary whole blood protein expression profiles of wild and captive P. largha pups. Proteins from the whole blood samples were extracted using the Mammalian Tissue and Cell Protein Extraction Kit (Bangfei Bioscience Co., Ltd, Beijing, China). The concentrations of protein in each sample were determined using a protein quantification kit (Dingguo Changsheng, Beijing, China), according to the manufacturer's instructions. The extracted proteins were then excised from the preparative tube and destained with NH4HCO3. Following reduction and alkylation with dl-dithiothreitol and iodoacetamide, respectively, the samples were digested with trypsin using the Filter Aided Sample Preparation protocol46. All digested peptide samples were stored at − 80 °C prior to MS analysis.
MS analysis was performed according to a previous study with only one technical replicate for each sample47. Digested peptide mixtures were first pressure-loaded onto a fused silica capillary column packed with 3 μm dionex C18 material (Phenomenex, USA). The column was washed with buffer A (water, 0.1% formic acid) and buffer B (acetonitrile, 0.1% formic acid), and an Agilent 1100 quaternary high-performance liquid chromatography (HPLC) was applied to analyzed. The first step of HPLC measure was consisted of a 5 min gradient from 0 to 2% buffer B, followed by a 45 min gradient to 40% buffer B. Following this, a 3 min gradient from 40 to 80% buffer B was performed followed by a 10 min hold at 80% buffer B. A 2 min gradient of buffer B from 80 to 2% was performed, and approximately 100 μg of the tryptic peptide mixture was loaded onto the columns and was separated at a flow rate of 0.5 μL/min using a linear gradient. As peptides were eluted from the micro-capillary column, they were electrosprayed directly into a micrOTOF-Q II mass spectrometer (BRUKER Scientific, Beijing, China) with the application of a distal 180 °C source temperature. The mass spectrometer was operated in the MS/MS mode. Survey MS scans were acquired in the TOF-Q II with the resolution set to a value of 20,000. Each survey scan (50–2500) was followed by five data-dependent tandem mass scans at a normalized scan speed of 2 Hz.
Tandem mass spectra were searched against the reference transcriptome of spotted seals (NCBI Sequence Read Archive: SRA050171)1 using the Proteome Discoverer 2.1 software by the Mascot search engine (Matrix Science, London, UK; version 2.3.02) based on the standard LFQ module. The following options were used to identify the proteins: Peptide mass tolerance = ± 15 ppm, MS/MS tolerance = 0.02 Da, enzyme = trypsin, missed cleavage = 2, fixed modification: carbamidomethyl, variable modification: oxidation, database pattern = decoy. The results were then filtered using a cutoff of 0.01 for the false discovery rate. The minimum number of peptides required to identify a protein was set to 1 and proteins with at least two unique peptides were used for abundance quantification. The quantification of peptides was based on MS1-level data. For differential expression analysis, proteins with missing abundance values in more than one biological replicate of wild or captive groups were filtered out. If a protein with more than one abundance values in one groups and missing abundance values in all three replicates in another group, it was identified as unique in this group. Then, remaining proteins with fold changes > 1.5 and p-values < 0.05 (t-test) between the wild and captive P. largha pups were considered to be significantly differentially expressed.
To determine the biological functions of the preliminary whole blood proteome of P. largha pups, the GO and KEGG annotations of the identified proteins were extracted from the annotation results of the reference transcriptome. Such annotations were obtained from our previous study by aligning the unigenes to the GO and KEGG databases48 using BLASTx with an E-value cutoff of less than 10–51.
PLS-DA based on the identified proteins and their expression levels in all samples was performed using the “mixOmics” package v6.12.149 in the R v4.0.0 platform to evaluate the differences in whole blood protein expression profiles between the wild and captive P. largha pups. The GO and KEGG enrichment analyses were performed using the DAVID tool based on the GO and KEGG annotations of DEPs50, and the p-values were adjusted using the FDR method. STRING20 was used to construct the PPI networks of the DEPs to identify the key proteins that changed the physiological functions of P. largha due to captivity. All other charts were drawn using the “ggplot2” package in R v4.0.0 software.
Ethic approval
The protocols for samples collection of spotted seals was/were approved by the Ministry of Agriculture and Rural Affairs of the People’s Republic of China, permit number: 1376.
Supplementary information
Acknowledgements
We thank the staff from Dalian Sun Asia Aquarium for their assistance in collecting the blood samples of spotted seals.
Author contributions
Conceptualization: J.T., J.H. and Z.L.; Methodology: J.T. and J.D.; Formal analysis and investigation: J.T., J.D. and X.B.; Writing—original draft preparation: J.T.; Writing—review and editing: J.D., J.H. and Z.L.; Funding acquisition: J.H. and Z.L.; Resources: J.T., and X.S.; Supervision: J.H. and Z.L.
Funding
This work was supported by the Foundation of Liaoning Province Department of Ocean and Fisheries [Grant numbers 201812, 201822]; and the China Environment and Zoology Protection for Offshore Oil and Ocean Foundation [Grant number CF-MEEC/ER/2019-10].
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD020112.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-75759-2.
References
- 1.Gao XG, Han JB, Lu ZC, Zhang PJ, He CBD. novo assembly and characterization of spotted seal Phoca largha transcriptome using Illumina paired-end sequencing. Comp. Biochem. Phys. D. 2013;8(2):103–110. doi: 10.1016/j.cbd.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Gao XG, Han JB, Lu ZC, Zhang PJ, He CB. Sequence variation and gene duplication at the MHC DRB loci of the spotted seal Phoca largha. Genet. Mol. Res. 2015;14(1):2055–2062. doi: 10.4238/2015.March.20.15. [DOI] [PubMed] [Google Scholar]
- 3.Black AN, Seears HA, Hollenbeck CM, Samollow PB. Rapid genetic and morphologic divergence between captive and wild populations of the endangered Leon Springs pupfish Cyprinodon bovinus. Mol. Ecol. 2017;26(8):2237–2256. doi: 10.1111/mec.14028. [DOI] [PubMed] [Google Scholar]
- 4.Parmar DR, Mitra S, Bhadouriya S, Rao T, Kunteepuram V, Gaur A. Characterization of major histocompatibility complex class I, and class II DRB loci of captive and wild Indian leopards (Panthera pardus fusca) Genetica. 2017;145(1):541–558. doi: 10.1007/s10709-017-9979-5. [DOI] [PubMed] [Google Scholar]
- 5.Gardiner KJ, Hall AJ. Diel and annual variation in plasma cortisol concentrations among wild and captive harbor seals (Phoca vitulina) Can. J. Zool. 1997;75(11):1773–1780. doi: 10.1139/z97-806. [DOI] [Google Scholar]
- 6.Wang PL, Han JB, Ma ZQ. Status survey of spotted Seal (Phoca largha) in Bohai and Yellow Sea. Chin. J. Wildlife. 2008;29(1):29–31. [Google Scholar]
- 7.Katin IO, Nesterenko VA. Inshore associations of the spotted seal (Phoca largha Pallas, 1811) Contemp. Probl. Ecol. 2010;3(1):127–132. doi: 10.1134/S1995425510010200. [DOI] [Google Scholar]
- 8.Mizuno AW, Ohtaishi N. Cranial features of the spotted seal, Phoca largha, in the Nemuro Strait, considering age effects. J. Vet. Med. Sci. 2002;64(2):137–144. doi: 10.1292/jvms.64.137. [DOI] [PubMed] [Google Scholar]
- 9.Mizuno AW, Onuma M, Takahashi M, Ohtaishi N. Population genetic structure of the spotted seal Phoca largha along the Coast of Hokkaido, based on mitochondrial DNA Sequences. Zool. Sci. 2003;20(6):783–788. doi: 10.2108/zsj.20.783. [DOI] [PubMed] [Google Scholar]
- 10.Han JB, Sun FY, Gao XG, He CB, Wang PL, Ma ZQ, Wang ZH. Low microsatellite variation in spotted seal (Phoca largha) shows a decrease in population size in the Liaodong Gulf Colony. Ann. Zool. Fenn. 2010;47(1):15–27. doi: 10.5735/086.047.0102. [DOI] [Google Scholar]
- 11.Song XR, Wang QG, Zhang PJ, Yang Y, Han JB, Yu CZ, Chu SH, Zhang SJ. Normal values of hematology and serum biochemistry in cultured spotted seal Phoca largha. Fish. Sci. 2014;33(7):427–432. [Google Scholar]
- 12.Zhang P, Yang Y, Han J, Lu Z, Wang L, Tian J, Wang Q. Serum testosterone, progesterone, and estradiol concentrations and sexual maturation in spotted seals (Phoca largha) Theriogenology. 2014;82(3):475–480. doi: 10.1016/j.theriogenology.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Johnson PF, Mcknight SL. Eukaryotic transcriptional regulatory proteins. Annu. Rev. Biochem. 1989;58(1):799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- 14.Thakur S, Geiger T, Chatterjee B, Bandilla P, Frohlich F, Cox J, Mann M. Deep and highly sensitive proteome coverage by LC–MS/MS without prefractionation. Mol. Cell. Proteomics. 2011;10:M110.003699. doi: 10.1074/mcp.M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabilloud T, Chevallet M, Luche S, Lelong C. Two-dimensional gel electrophoresis in proteomics: Past, present and future. J. Proteome. 2010;73:2064–2077. doi: 10.1016/j.jprot.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 16.Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, Hettich RL, Samatova NF. Detecting differential and correlated protein expression in label-free shotgun proteomics. J. Proteome. Res. 2006;5(11):2909–2918. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- 17.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics. 2005;4(10):1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Matros A, Kaspar S, Witzel K, Mock HP. Recent progress in liquid chromatography-based separation and label-free quantitative plant proteomics. Phytochemistry. 2011;72(10):963–974. doi: 10.1016/j.phytochem.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Porteus B, Kocharunchitt C, Nilsson RE, Ross T, Bowman JP. Utility of gel-free, label-free shotgun proteomics approaches to investigate microorganisms. Appl. Microbiol. Biotechnol. 2011;90(2):407–416. doi: 10.1007/s00253-011-3172-z. [DOI] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Morris J, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva N, Roth A, Bork P, Jensen L, Mering C. The string database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neely BA, Prager KC, Bland AM, Fontaine C, Gulland FM, Janech MG. Proteomic analysis of urine from California Sea Lions (Zalophus californianus): A resource for urinary biomarker discovery. J. Proteome Res. 2018;17:3281–3291. doi: 10.1021/acs.jproteome.8b00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neely BA, Soper JL, Gulland FMD, Bell PD, Kindy M, Arthur JM, Janech MG. Proteomic analysis of cerebrospinal fluid in California Sea Lions (Zalophus Californianus) With domoic acid toxicosis identifies proteins associated with neurodegeneration. Proteomics. 2015;15:4051–4063. doi: 10.1002/pmic.201500167. [DOI] [PubMed] [Google Scholar]
- 23.Sobolesky P, Parry C, Boxall B, Wells R, Venn-Watson S, Janech MG. Proteomic analysis of non-depleted serum proteins from bottlenose dolphins uncovers a high vanin-1 phenotype. Sci. Rep. 2016;6:33879. doi: 10.1038/srep33879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller BA, Nanni P, Fortes C, Grossmann J, Arreoia MR, Vences M, Canales R, Sanchez-Okrucku R, de Almeida AM, Bergfelt DR. Plasma proteome and clinical biochemistry associated with performance-based physical activity in Bottlenose Dolphins (Tursiops truncatus) Aquat. Mamm. 2017;43:453–464. doi: 10.1578/AM.43.5.2017.453. [DOI] [Google Scholar]
- 25.Underhill DM, Ozinsky A. Phagocytosis of microbes: Complexity in action. Annu. Rev. Immunol. 2002;20(1):825–852. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 26.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertier FB. Ena/VASP proteins: Regulators of the actin cytoskeleton and cell migration. Annu. Rev. Cell. Dev. Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- 27.Coppolino MG, Krause M, Hagendorff P, Monner DA, Trimble W, Grinstein S, Wehland J, Sechi AS. Evidence for a molecular complex consisting of Fyb/SLAP, SLP-76, Nck, VASP and WASP that links the actin cytoskeleton to Fcgamma receptor signalling during phagocytosis. J. Cell. Sci. 2011;114(Pt 23):4307–4318. doi: 10.1242/jcs.114.23.4307. [DOI] [PubMed] [Google Scholar]
- 28.Adams M, Simms RJ, Abdelhamed Z, Dawe HR, Szymanska K, Logan CV, Wheway G, Pitt E, Gull K, Knowles MA, Blair E, Cross SH, Sayer JA, Johnson CA. A meckelin-filamin A interaction mediates ciliogenesis. Hum. Mol. Geneti. 2012;21(6):1272–1286. doi: 10.1093/hmg/ddr557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth H, Samereier M, Begandt D, Pick R, Salvermoser M, Brechtefeld D, Schleicher M, Walzog B, Muller-Taubenberger A. Filamin A promotes efficient migration and phagocytosis of neutrophil-like HL-60 cells. Eur. J. Cell. Biol. 2017;96(6):553–566. doi: 10.1016/j.ejcb.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Mezawa M, Pinto VI, Kazembe MP, Lee WS, McCulloch CA. Filamin A regulates the organization and remodeling of the pericellular collagen matrix. FASEB J. 2016;30(10):3613–3627. doi: 10.1096/fj.201600354RR. [DOI] [PubMed] [Google Scholar]
- 31.Staudinger, L. A. Functional interactions between the discoidin domain receptor 1 and β1 integrins. University of Toronto. (Accessed 05 August 2020); https://tspace.library.utoronto.ca/bitstream/1807/35138/3/Staudinger_Lisa_A_201303_MSc_Thesis.pdf (2013).
- 32.Ezzell RM, Goldmann WH, Wang N, Parashurama N, Ingbre DE. Vinculin promotes cell spreading by mechanically coupling integrins to the cytoskeleton. Exp. Cell. Res. 1997;231(1):14–26. doi: 10.1006/excr.1996.3451. [DOI] [PubMed] [Google Scholar]
- 33.Zambrano JL, Sorodo O, Alcala A, Vizzi E, Diaz Y, Ruiz MC, Michelangeli F, Liprandi F, Ludert JE. Rotavirus infection of cell in culture induces activation of RhoA and changes in the actin and tubulin cytoskeleton. PLoS ONE. 2012;7(10):e47612. doi: 10.1371/journal.pone.0047612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delalle I. Mutations in the Drosophila Orthologs of the F-actin capping protein alpha- and beta-subunits cause actin accumulation and subsequent retinal degeneration. Genetics. 2005;171(4):1757–1765. doi: 10.1534/genetics.105.049213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Felding-Habermann B, Ruggeri ZM, Cheresh DA. Distinct biological consequences of integral alpha v beta 3-mediated melanoma cell adhesion to fibrinogen and it plasmic fragments. J. Biol. Chem. 1992;267(8):5070–5077. [PubMed] [Google Scholar]
- 36.Li Y, Courtney HS. Promotion of phagocytosis of Streptococcus pyogenes in human blood by a fibrinogen-binding peptide. Microbes. Infect. 2011;13(4):413–418. doi: 10.1016/j.micinf.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 37.Pillay CS, Elliott E, Dennison C. Endolysosomal proteolysis and its regulation. Biochem. J. 2002;363(3):417–429. doi: 10.1042/bj3630417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Debigaré R, Price SR. Proteolysis, the ubiquitin-proteasome system, and renal diseases. Am. J. Physiol. Renal. Physiol. 2003;285(1):F1–F8. doi: 10.1152/ajprenal.00244.2002. [DOI] [PubMed] [Google Scholar]
- 39.Totani M, Hirota K. Glucose-6-phosphate dehydrogenase (G6PD) Nihon. Rinsho. 1999;53(Su Pt 2):206–209. [PubMed] [Google Scholar]
- 40.Saha K, Hollowell D, Wong PKY. Mother-to-baby transfer of humoral immunity against retrovirus-induced neurologic disorders and immunodeficiency. Virology. 1994;198(1):129–137. doi: 10.1006/viro.1994.1015. [DOI] [PubMed] [Google Scholar]
- 41.Richter K, Soroka J, Skalniak L, Leskovar A, Hessling M, Reinstein J, Buchner J. Conserved conformational changes in the ATPase cycle of human Hsp90. J. Biol. Chem. 2008;283(26):17757–17765. doi: 10.1074/jbc.M800540200. [DOI] [PubMed] [Google Scholar]
- 42.Zhang PJ, Song XR, Han JB, Wang LM, Yang Y. Milk composition, milk consumption, and growth rate of a captive spotted seal (Phoca largha) pup from Liaodong Bay, China. Can. J. Zool. 2014;92:449–452. doi: 10.1139/cjz-2013-0295. [DOI] [Google Scholar]
- 43.Atkinson S. Reproductive biology of seals. Rev. Reprod. 1997;2:175–194. doi: 10.1530/ror.0.0020175. [DOI] [PubMed] [Google Scholar]
- 44.Florez ID, Niño-Serna LF, Beltrán-Arroyave CP. Acute infectious diarrhea and gastroenteritis in children. Curr. Infect. Dis. Rep. 2020;22(2):4. doi: 10.1007/s11908-020-0713-6. [DOI] [PubMed] [Google Scholar]
- 45.Yin S, Huang M, Li D, Tang N. Difference of coagulation features between severe pneumonia induced by SARS-Cov2 and non-SARS-Cov2. J. Thromb. Thrombolys. 2020 doi: 10.1007/s11239-020-02105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 47.Jiang J, Zhao Z, Pan Y, Dong Y, Gao S, Jiang B, Xiao Y, Jiang P, Zhang G, Wang X, Zhou Z. Proteomics reveals the gender differences in humoral immunity and physiological characteristics associated with reproduction in the sea cucumber Apostichopus japonicus. J. Proteom. 2020;217:103687. doi: 10.1016/j.jprot.2020.103687. [DOI] [PubMed] [Google Scholar]
- 48.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohart F, Gautier B, Singh A, Le Cao KA. mixOmics: An R package for omics feature selection and multiple data integration. PLoS Comput. Biol. 2017;13(11):e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang D, Sherman B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD020112.