Abstract
Nearly all retroviruses selectively package two copies of their unspliced RNA genomes from a cellular milieu that contains a substantial excess of non-viral and spliced viral RNAs. Over the past four decades, combinations of genetic experiments, phylogenetic analyses, nucleotide accessibility mapping, in silico RNA structure predictions, and biophysical experiments were employed to understand how retroviral genomes are selected for packaging. Genetic studies provided early clues regarding the protein and RNA elements required for packaging, and nucleotide accessibility mapping experiments provided insights into the secondary structures of functionally important elements in the genome. Three-dimensional structural determinants of packaging were primarily derived by nuclear magnetic resonance (NMR) spectroscopy. A key advantage of NMR, relative to other methods for determining biomolecular structure (such as X-ray crystallography), is that it is well suited for studies of conformationally dynamic and heterogeneous systems—a hallmark of the retrovirus packaging machinery. Here, we review advances in understanding of the structures, dynamics, and interactions of the proteins and RNA elements involved in retroviral genome selection and packaging that are facilitated by NMR.
Keywords: retrovirus, genome, packaging, NMR, cyro-EM, SAXS, RNA, protein, structure
1. Introduction
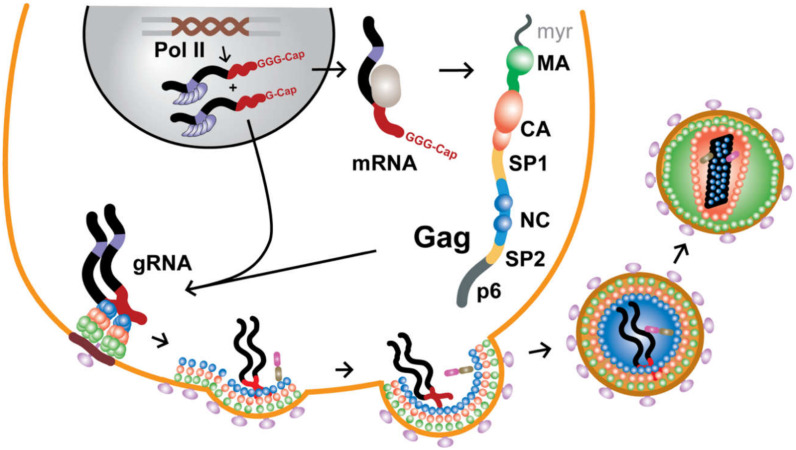
During the late phase of the retroviral replication cycle (after the viral genome is reverse transcribed and the proviral DNA is integrated into the genome of the host cell), two copies of the unspliced, 5′-capped, and 3′-polyadenylated RNA transcript are specifically selected for packaging into assembling virions. Genomes are selected against a background that includes a substantial excess of non-genomic RNA, including cellular mRNAs [1], monomeric unspliced viral transcripts that encode for the viral Gag and Gag-Pol polyproteins, and spliced viral mRNAs that encode for accessory and envelope proteins—all of which are excluded during virus assembly. Over the past ~40 years, considerable effort was made to understand the mechanisms through which retroviruses achieve such exquisite selectivity (for reviews, see [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17]). RNA packaging and virus assembly are both mediated by the Gag polyproteins, which contain three independently folded domains [matrix (MA), capsid (CA), and nucleocapsid (NC)] and additional domains that are intrinsically disordered (for HIV-1, these are: SP1, SP2, p6), Figure 1. The MA domain functions in the selective targeting of Gag to virus assembly sites on the plasma membrane, the CA domain promotes Gag self-association, and the NC domain recognizes and binds the cognate RNA genome. In addition, the conformationally dynamic SP1 domain [18] that bridges the CA and NC domains, adopts a 6-helix bundle structure that stabilizes Gag:Gag interactions in immature virus particles [19,20,21,22]. During or shortly after budding, retroviruses undergo a maturation process in which these domains are liberated by proteolytic cleavage of Gag, leading to large changes in viral morphology; Figure 1. The liberated CA proteins condense to form a conical capsid core particle that encapsidates the viral genome, along with the essential enzymes needed for reverse transcription and subsequent integration of the proviral DNA.
Figure 1.
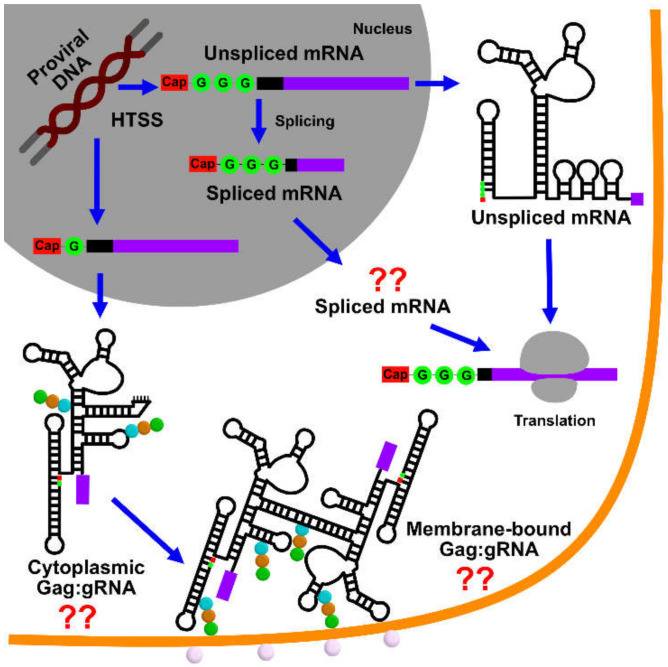
Late phase of the HIV-1 replication cycle. Heterogeneous transcriptional start site usage affords 5′-capped transcripts beginning with a single guanosine that function as genomes (gRNA) and are packaged as dimers, as well as 5′-capped transcripts beginning with two (minor species, not shown) or three guanosines that function as mRNAs (5ʹ-leader, coding region, and Rev-binding sites of the viral RNA shown in black, red, and gray, respectively).
The Retroviridae family includes two subfamilies encompassing eleven genera, categorized primarily by sequence similarity within the pol gene [23]. The Spumaretrovirinae subfamily (Bovispumavirus, Equispumavirus, Felispumavirus, Prosimiispumavirus, Simiispumavirus) diverged early and is evolutionarily distant from the Orthoretrovirinae subfamily (Alpharetrovirus, Betaretrovirus, Deltaretrovirus, Epsilonretrovirus, Gammaretrovirus, Lentivirus). Evolutionarily related retroviruses within a given genus can preferentially package each other’s viral genomes [24,25,26], and the packaging assays indicate that this property is conserved within lentiviruses (HIV and simian immunodeficiency virus (SIV)) [24], as well as gammaretroviruses (Moloney murine sarcoma virus and spleen necrosis virus) [25]. Retroviruses in distinct genera are also found to maintain this property. HIV-1 chimeric virions that contain the Moloney murine leukemia virus (MoMuLV) NC domain, preferentially package the MoMuLV RNA [27], whereas the MoMuLV chimeric virions that contain the HIV-1 NC domain, package the HIV-1 genome [28], indicating that it is the NC domain of Gag that is responsible for genome selection. In fact, the MoMuLV chimeric virions packaged the full-length unspliced HIV-1 genome, indicating that chimeric precursor Gag proteins containing the HIV-1 NC domain can also distinguish between unspliced and spliced viral RNAs, during genome selection [28]. Although it is unclear where in the cell NC–RNA interactions initially occur, confocal studies showed that virus assembly is nucleated by the binding of a small number of Gag proteins (possibly a dozen or fewer [29]) to the viral genome, at plasma membrane assembly sites [30], which leads to the subsequent recruitment of several thousand additional Gag proteins [29,31] and cellular factors that promote virus budding [32,33]. Although virions can assemble in the absence of their genomes by incorporating an equivalent amount of cellular RNAs [34,35,36,37,38,39], vector RNAs containing the authentic packaging signal can efficiently out-compete these more abundant RNAs for packaging [40,41], as long as the non-native downstream vector residues do not interfere with proper folding of the packaging signal [42].
Retroviruses contain two copies of their genomes [43], both of which are utilized for strand transfer-mediated recombination during reverse transcription [44,45,46]. As only one DNA allele is generated, retroviruses are considered “pseudodiploid.” In virions, the genomes form non-covalently linked dimers that become increasingly stable towards thermal denaturation, with increasing virus age [47,48,49,50,51,52]. Dimerization might occur co-transcriptionally [53] but most likely after the genomes are exported from the nucleus [54] and possibly not until they reach the plasma membrane [30]. Dimerization, packaging, and other RNA-dependent functions (including transcriptional activation, splicing, and initiation of reverse transcription) are promoted by the elements located within the 5′-leader of the RNA [1,2,3,4,5,6,7,8,9,10,11,12,13,14,16,17], which is among the most conserved regions of the genome [55,56], (http://www.hiv.lanl.gov/). Although some studies with mutant genomes suggested that residues in the gag open reading frame might also be important for packaging, it now appears that packaging defects were caused by misfolding of the 5′-leader due to interactions with non-native downstream sequences [42]. Studies identified regions within the 5′-leaders of HIV-1 [40], MoMuLV [57,58], and Rous Sarcoma Virus (RSV) [59,60,61,62,63,64], which are independently capable of directing heterologous RNAs into assembling virus-like particles (VLPs). These “core encapsidation signals” incorporate or reside near residues that promote RNA dimerization [3,4,6,8] and there is considerable evidence that genome packaging is critically dependent on dimerization [1,2,11,65,66,67,68]. Although dimerization could be modulated by a riboswitch-like mechanism promoted by the chaperone activity of NC [69,70,71], recent studies indicate that RNA fates are instead controlled at the level of transcription, through the heterogeneous start site usage [72,73,74,75]. In this mechanism, 5′-capped RNAs transcribed with a single 5′-guanosine preferentially form dimers that are packaged into virions, where they function as genomes (gRNA). In contrast, 5′-capped transcripts that begin with two or three guanosines form monomers that promote splicing, are retained in cells, and function as mRNAs [72,73,74,75]; Figure 1.
Solution-state nuclear magnetic resonance (NMR) contributed substantially to current knowledge of the structures and mechanisms that contribute to retroviral genome selection and packaging. NMR offers a number of advantages for structural characterization of biomolecules, including the ability to characterize structures and dynamics of conformationally heterogeneous samples, and to obtain data without the need to prepare crystalline samples. Due to these advantages, seven of the first twelve HIV-1 protein domain structures that were solved during the first decade of HIV-1 structural biology were determined by NMR [76]. However, NMR also has a number of technical disadvantages that confound the studies of the RNA components that are important for genome packaging. Since most RNAs contain only four types of ribonucleotides (guanosine, G; cytidine, C; adenosine, A; uridine, U), chemical shift dispersion is relatively low. Proton density is also lower than that of proteins, particularly for non-exchangeable protons that are found mainly within the major grooves of A-form helices. Interproton distances between different secondary structure elements are typically greater than 5 Å [77], which limits the utility of nuclear Overhauser effect (NOE) experiments for establishing the overall RNA folds [78,79]. In addition, H–C dipolar coupling can severely limit the sensitivity and resolution of the H–C correlation NMR spectra obtainable for larger RNAs with longer rotational correlation times. For these reasons, high-resolution NMR-based structural studies were generally limited to relatively small RNAs [80,81]. The average size of NMR-derived RNA structures deposited in public databases is 30 nucleotides and only six structures were reported for RNAs comprising more than 100 nucleotides (http://www.rcsb.org). However, new 2H-edited NMR methods recently enabled structural studies of RNA elements within the intact, dimeric 5′-leaders of two strains of HIV-1 (HIV-1NL4-3 and HIV-1MAL) (>700 nucleotide dimers; >230 kDa) [71,75,81]. Hybrid approaches that combine high-resolution local structural information provided by NMR with lower-resolution global structural information from cryogenic electron microscopy (cryo-EM), or small angle X-ray scattering (SAXS) could enable structural studies of even larger RNAs and protein–RNA complexes that are important for packaging. Here, we review how NMR studies contributed to the current understanding of the structural and mechanistic determinants of retroviral genome selection and packaging.
2. Protein Components Important for Genome Packaging
2.1. Nucleocapsid Domain of Gag
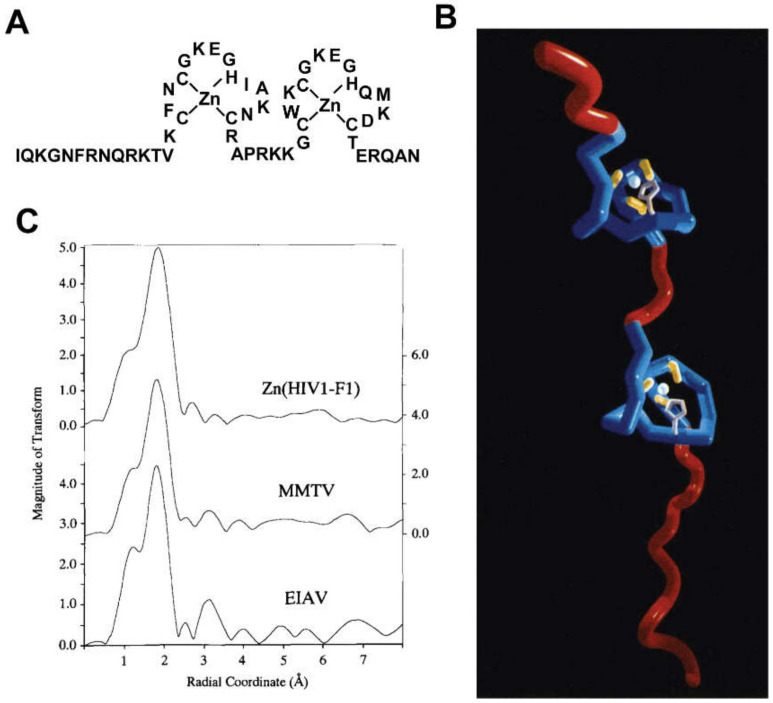
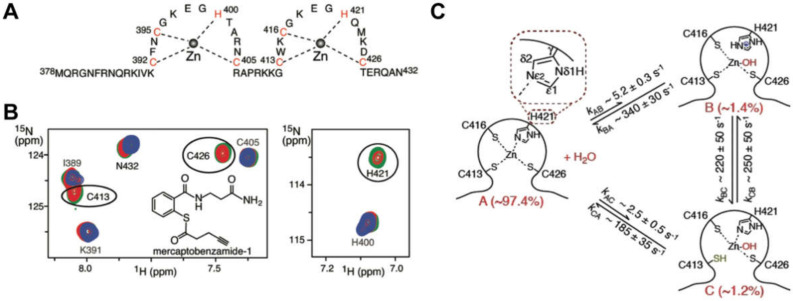
The NC domain of the HIV-1 Gag protein was the first retroviral constituent studied by NMR. Except for the spumaviruses, all retroviral NC proteins contain one or two copies of a conserved cysteine/histidine rich sequence (CCHC; C = cysteine, H = histidine) [82] that was originally discovered by Henderson et al. [83]. After the subsequent discovery of CCHH arrays in Xenopus protein transcription factors and proposals that these arrays function as nucleic acid-binding “zinc fingers” [84], Berg et al. proposed that the retroviral CCHC arrays might also function as zinc binding sites [85]; Figure 2A. Although this proposal was initially controversial [86,87], mutagenesis studies showed that the substitution of a single Cys by Ser (i.e., a single atom S to O substitution) completely ablated viral replication [88]. Furthermore, Goff et al. showed that mutation of several conserved (or conservatively substituted) hydrophobic residues within the CCHC arrays can significantly reduce RNA packaging specificity [89]. Berg et al. showed that an 18-residue CCHC array is capable of binding both Co2+ and Zn2+ with high affinity, likely via 3S/1N coordination [90]. 1H-113Cd heteronuclear spin echo difference NMR applied to a 113Cd-substituted HIV-1 CCHC peptide, revealed that the cysteine-S and histidine-Nε atoms were coordinated to the metal [91], and NOE-based structural studies showed that the peptide adopts a mini-globular “zinc knuckle” structure, upon coordination of zinc [92]. Subsequent NMR studies showed that the CCHC arrays of virus-isolated NC form independently folded zinc knuckle domains that behave like “beads on a string” [93]; Figure 2B. Zinc-edge extended X-ray absorption fine structure spectroscopy confirmed that the CCHC arrays are populated with zinc in mature particles [94]; Figure 2C.
Figure 2.
HIV-1 NC is a zinc metalloprotein. (A) Amino acid sequence and zinc binding mode of the HIV-1NL4-3 NC protein. (B) NMR structure of HIV-1NL4-3 NC. The independently folded CCHC “zinc knuckle” domains behave like “beads on a string.” (C) Similarities of zinc-edge EXAFS spectra obtained for an isolated HIV-1 CCHC peptide (top) and for intact retroviruses [Mouse Mammary Tumor Virus (center) and Equine Infectious Anemia Virus (bottom)]. Panels (B,C) reproduced from [82], with permission.
NMR structures were reported for several isolated retroviral zinc knuckle domains [92,93,95,96,97,98,99], for intact NC proteins [82,100,101,102,103,104,105,106], and NC–RNA complexes [66,104,107,108,109,110,111,112,113,114] (see sections below for more details). Weak NOEs between aromatic residues of the N- and C-terminal HIV-1 NC zinc knuckles [102] and NMR relaxation and chemical shift data [115] were consistent with a model in which the zinc knuckles transiently interact with each other like “beads on a string” [82,115]; Figure 2B. However, the zinc knuckles of HIV-2 and the simian immunodeficiency virus (SIV) NC proteins appear to interact tightly with each other, in the absence of nucleic acids [103,104], and tight inter-knuckle packing was observed for HIV-1 NC when bound to RNAs with high affinity binding sites (see sections below). Residual dipolar couplings (RDCs) of an isolated NC domain interacting with DNA indicate that the two zinc knuckles of NC tightly bind with DNA, and tumble as a singular unit [109,110,114,116]. The NC protein from the mouse mammary tumor virus (MMTV) also contains two CCHC zinc knuckle domains, and NMR studies indicate that these domains do not interact with each other when free in solution [98]. Interestingly, while the proximal zinc knuckle of MMTV NC adopts the same folding observed for all other known retroviral zinc knuckles, the distal knuckle contains an additional C-terminal β-hairpin [98]. A similar reverse turn-like structure was observed for the distal zinc knuckle in the NMR structure of the Mason-Pfizer monkey virus NC protein [106].
2.2. MA Domain of Gag
NMR is a powerful tool in structural characterization of Gag’s MA domain [117,118,119,120,121], elucidating its roles in cytosolic trafficking [121,122,123,124,125], and unveiling MA–RNA interactions [126]. Assembly of HIV particles is facilitated by targeting the viral genome and ~12 copies of Gag to the assembly sites on the inner leaflet of the plasma membrane [29,30,31,127,128]. Roughly 2000 Gag molecules are subsequently anchored to the plasma membrane through Gag’s N-terminally myristoylated MA domain [129]. Given the challenges to prepare an in vitro myristoylation system, the initial structural characterization focused on unmyristoylated MA [myr(-)MA]. In 1994, Matthews et al. used multidimensional solution NMR to report the first structure of an HIV-1 myr(-)MA construct. This structure featured four ⍺-helices, as well as an irregular β-sheet arranged such that several basic residues were available on the protein surface to interact with plasma membrane assembly sites [117]. Structural similarity to the immune modulator interferon-γ led to the suggestion that the MA protein might operate as a dimer [117]. However, the NMR-derived solution structure of a similar construct presented by Massiah et al. in the same year countered this proposal, based on steric hindrance at the dimer interface due to an additional helix at the N-terminus [130]. Matthews et al. also observed this helix following further refinement [131]. Aside from this difference, the NMR-derived structures showed a similar globular fold, comprising five ⍺-helices, a short 310 helical stretch, and a three-strand mixed β-sheet [130]. NMR studies of the 283 N-terminal residues of the Gag-precursor, which include full-length myr(-)MA [118], also revealed a similar fold. X-ray crystallography performed by the Sundquist group, again showed a similar globular fold, but unexpectedly indicated that MA trimerized in three different crystal lattices [132]. Comparison of the crystal and NMR structures revealed a roughly 6 Å displacement of the short 310 helix participating in the trimer interface and ultimately led to the proposal that MA undergoes this structural rearrangement during assembly [133]. Recent virology work suggests that MA trimerization is necessary for recognition of the envelope glycoprotein for incorporation to new viruses [134,135,136]; therefore, this might also be the functional cause for multimerization.
The MA–plasma membrane interaction is dependent not only on the region of basic residues located on MA’s surface but also on N-terminal myristoylation [121,122,137,138], and efforts to characterize the structure of the native, myristoylated MA were realized by Tang et al., using solution NMR [119]. They found that MA adopts both myristoyl exposed and sequestered conformations, but unexpectedly showed that sequestration of the myristoyl group within the protein requires only minor conformational changes and does not alter its tertiary fold. Concentration-dependent chemical shift perturbations, combined with sedimentation equilibrium data, demonstrated a monomer–trimer equilibrium and proposed a model for MA trimerization via intermolecular myr–myr interactions [119] in a manner consistent with the earlier crystal structures [132]. Several subsequent NMR studies demonstrated that myristoylated MA proteins of other retroviruses, including HIV-2 [121] and feline immunodeficiency virus (FIV) [139,140], adopt similar structures to HIV-1 MA. Furthermore, NMR is used to demonstrate that myristoyl exposure is modulated by pH [141], with stabilization of a salt bridge between the protonated side chain of H89 and the acidic side chain of E12 promoting exposure.
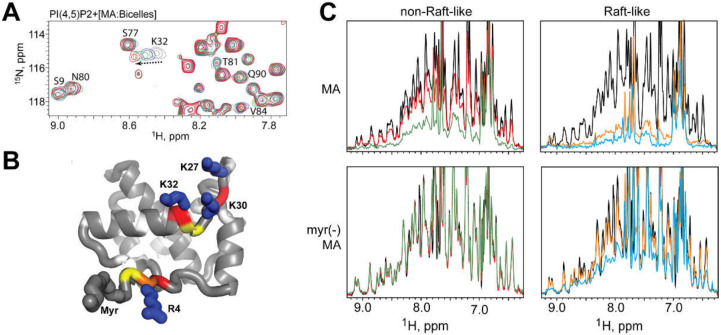
NMR studies also played a key role in characterizing the targeting of MA to the plasma membrane during assembly. MA specifically targets phosphatidylinositol-(4,5)-bisphosphate (PI(4,5)P2) [123,142,143] in lipid rafts, on the inner leaflet of the plasma membrane [144,145,146]. Saad et al. first reported the NMR structure of HIV-1 MA bound to water-soluble truncated PI(4,5)P2 lipids [123], in which the 2′-fatty acid chain of di-C4-PI(4,5)P2 is buried in MA’s hydrophobic cleft in an “extended lipid” conformation, promoting myristoyl exposure [123]. A variety of other phospholipids were proposed to promote membrane binding in a similar manner, based on NMR studies, with lipid analogs containing truncated acyl chains [147]. Development of new NMR techniques employing mimetic membranes allowed re-examination in the context of full-length lipids; Figure 3 [122]. NMR titration experiments employing liposomes and bicelles designed to mimic plasma and viral membranes revealed that, while PI(4,5)P2 promotes MA–liposome interactions, no PI(4,5)P2-dependent chemical shift perturbation are seen in the hydrophobic cleft, calling into question the validity of the extended lipid binding model; Figure 3A. This work prompted consideration of a molecular-dynamics-derived membrane-binding model, in which several MA surfaces interact with PI(4,5)P2 head groups but the acyl chains of PI(4,5)P2 do not extend from the membrane; Figure 3B [122]. NMR was also used to characterize and explain the action of several MA point mutations that inhibit plasma membrane targeting by disruption of the tertiary structure [148], stabilization of the myr-sequestered conformation [120,122], or through removal of basic residues important for both PI(4,5)P2 discrimination and membrane binding, Figure 3C [122].
Figure 3.
NMR studies of HIV-1 MA structure and membrane targeting. (A) 1H-15N HSQC spectra of 15N-labeled MA titrated into bicelles containing increasing mol% native PI(4,5)P2, reveal the binding mode of MA to plasma membrane mimetics. (B) Solution structure of MA determined by NMR. Residues showing large chemical shift perturbations in (A) are highlighted. (C) 1H NMR spectra of MA and myr(-) MA upon addition of liposomes with varying phospholipid compositions. Signal loss indicates binding. MA can bind non-Raft-like membranes due to myristoyl group interactions. Binding to Raft-like membranes is significantly enhanced. Adapted from [122] with permission.
Recent studies demonstrated that MA interacts with cytosolic tRNAs [149], including the tRNALys3 isoform that is present in virions and serves as the primer for reverse transcription [149,150,151,152]. Chukkapalli et al. monitored the influence of MA–RNA interactions on assembly, using liposome flotation assays, which indicated that MA’s basic patch played a role in MA–RNA interactions and that this complex formation prevents MA from binding to PI(4,5)P2 [153]. Gaines et al. applied NMR and isothermal titration calorimetry (ITC) to probe the MA–tRNA interaction and determined that tRNALys3 interacts with residues in the basic patch region of MA. Interestingly, the affinity of MA for tRNALys3 was roughly one order of magnitude stronger than PI(4,5)P2-containing liposomes, and the NMR-based liposome binding assays revealed that tRNALys3 inhibited MA–PI(4,5)P2 interactions [126]. The binding affinity of MA to tRNALys3 is diminished by sample conditions that promote myristate exposure [123], which could also be promoted by Gag self-association [116]. Taken together, these studies point to a model suggesting that Gag–Gag interactions induced by NC binding to the RNA packaging signal might regulate premature binding to the plasma membrane and potentially support MA trimerization [126]. Thus, it is conceivable that genome recognition and Gag–Gag interactions might influence MA’s role in recruiting viral envelope glycoproteins to virions, further shaping the mechanism of HIV assembly [131].
2.3. Multi-Domain Gag Fragments
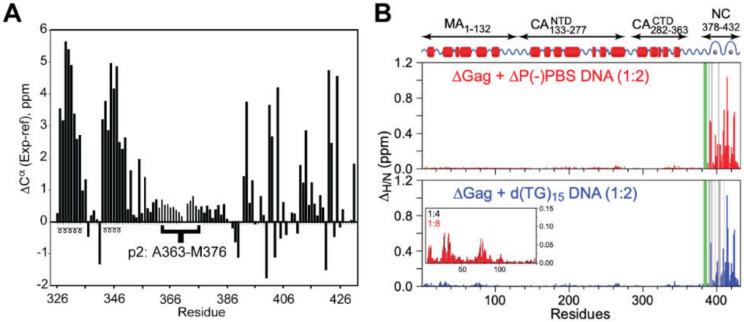
The CA domain of Gag promotes assembly via formation of intermolecular Gag–Gag interactions. The C-terminal domain of CA (CACTD) has a relatively weak propensity to form dimers in solution (dimerization Kd ~10 µM) [154], and both the N- and C-terminal domains participate in intermolecular contacts in assembled CA multimers [32]. To determine the influence of CA multimerization on downstream structure, NMR studies were conducted with a construct comprising CACTD through NC (CACTD-SP1-NC) [18]. These studies revealed that SP1 is conformationally labile and exists as a mixture of unstructured (predominant) and helical (minor) states; Figure 4A. The structure of NC was unaffected by the presence of the upstream CACTD and SP1 domains. An even larger HIV-1 Gag construct (∆Gag) comprising domains spanning unmyristoylated MA through NC (myr(-)MA-CA-SP1-NC; ~100 kDa) was studied by Clore et al. using protein perdeuteration and transverse relaxation optimized spectroscopy (TROSY) NMR [155], to optimize sensitivity and resolution [116]. 1H-15N chemical shift perturbation mapping was used to explore the structural changes that occur as ∆Gag interacts with two different DNA oligonucleotides—∆P(-)PBS and d(TG)15 [116]. Upon addition of either oligonucleotide, some cross-peaks from the MA and NC domains exhibit chemical shift perturbations. Specifically, significant chemical shift perturbations occur within the NC domain as ∆Gag forms a 1:1 complex with the DNA oligonucleotide; Figure 4B. Minor 1H-15N chemical shift perturbations are observed within the MA domain, following saturation of the NC binding site, suggesting weak secondary interactions between the MA and the DNA oligonucleotides [116]; Figure 4B.
Figure 4.
NMR studies of larger Gag constructs. (A) Plot of 13Cα chemical shift indices for SP1 (previously called p2, in braces) and adjacent residues in CACTD-SP1-NC. Consecutive positive values are indicative of α-helical structure. (B) 1H/15N chemical shift perturbation profiles in the presence of DNA [Red—∆P(-)PBS, Blue—d(TG)15]. In the presence of ∆P(-)PBS, chemical shift perturbations are only seen within the NC domain. In the presence of d(TG)15, significant chemical shift perturbations are present within NC but some perturbations are present in regions of MA at higher DNA concentrations (inset). Adapted from [18] (Panel A) and [116] (Panel B), with permission.
3. RNA Components Important for Genome Packaging
3.1. HIV-1
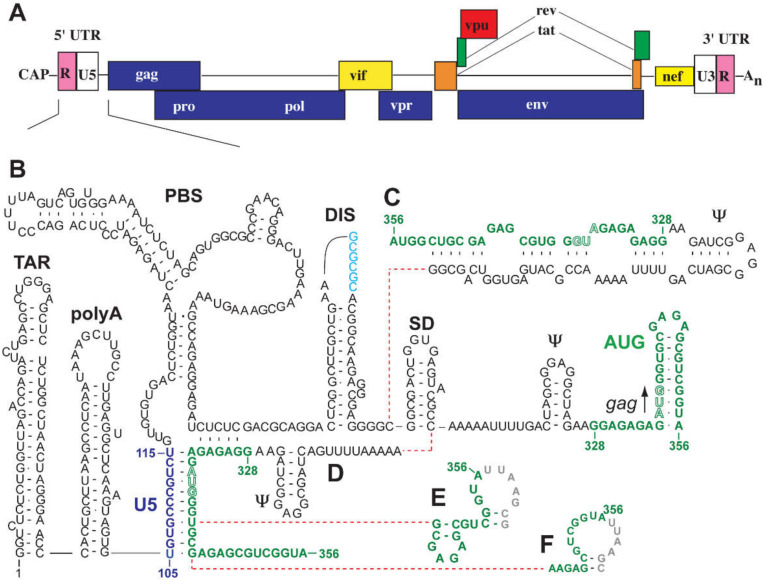
Considerable effort was made over the past 30 years to understand the structure of the HIV-1 5′-leader and its role in genome packaging (reviewed in [12]). A combination of nucleotide accessibility mapping and biophysical studies of recombinant viral genomic RNAs suggest that discrete hairpin structures within the genomic RNA perform specific functions of the genomic RNA, including transcriptional activation, primer binding, dimerization, 5′ splicing activities, and genome packaging [156,157,158,159,160,161,162,163,164,165]. These hairpin structures are, from 5′ to 3′, the trans-activation region (TAR), 5′ polyadenylation signal (polyA), U5 region, primer binding site (PBS), dimer initiation site (DIS; sometimes called SL1), major splice donor (SD; or SL2), Ψ packaging signal (or SL3), and the AUG hairpin (or SL4) [71]; Figure 5. TAR plays an essential role in Tat-mediated transcription activation [166]. The 5′ polyA hairpin contains the polyadenylation signal, AAUAAA, but its function is not as well characterized as the 3′ polyadenylation signal, which recruits cellular machinery to catalyze the addition of a 3′ polyA tail. Some studies suggest that the structure of the 5′ polyA hairpin is involved in translation [167]. The PBS region contains the tRNALys3 primer binding site for initiation of reverse transcription [168,169,170] and might be involved in packaging cellular proteins (see below). The DIS contains a palindromic sequence that allows it to base pair with the DIS of another viral RNA molecule, initiating dimerization (see next section) [71]. The SD hairpin is the major splice donor site for all spliced variants [171], thus all spliced constructs contain a common exon upstream of this site [172]. The Ψ hairpin contains the high affinity binding site for the viral NC protein, and the AUG hairpin (SL4) contains the start codon for Gag and Gag-Pol translation [71].
Figure 5.
(A) Schematic of a representative HIV-1 transcript showing locations of the 5′-UTR and coding regions. (B–F) Five of the more than 20 different secondary structures predicted for the HIV-1 5′-leader, on the basis of nucleotide reactivity probing, phylogenetic analysis, and biochemical studies (highlighting variations in AUG residues, colored green). Adapted from [12], with permission.
Historically, there is little agreement regarding the structures of residues within the 5′-leader that play a role in genome packaging. In fact, more than 20 different structures were proposed for the HIV-1 5′-leader, based on combinations of nucleotide accessibility mapping and phylogenetic analyses [157,158,159,162,173,174,175,176,177,178,179,180]. For example, residues that overlap the gag start codon (which are important for both genome packaging [181] and RNA dimer stability [182]) were proposed to form a hairpin, to base-pair with residues of the U5 element [161,162], or to adopt other conformations [12]; Figure 5. In vivo nucleotide reactivity mapping supported multiple structures for AUG and other elements that are essential for packaging, without consensus [12,163,164,165]. One important advantage of NMR, relative to chemical probing methods, is that it enables direct detection and identification of different structures that might exist within an equilibrium mixture of species. Applications of NMR to different domains of the HIV-1 5′-leader and to the intact leader are described below.
3.1.1. DIS
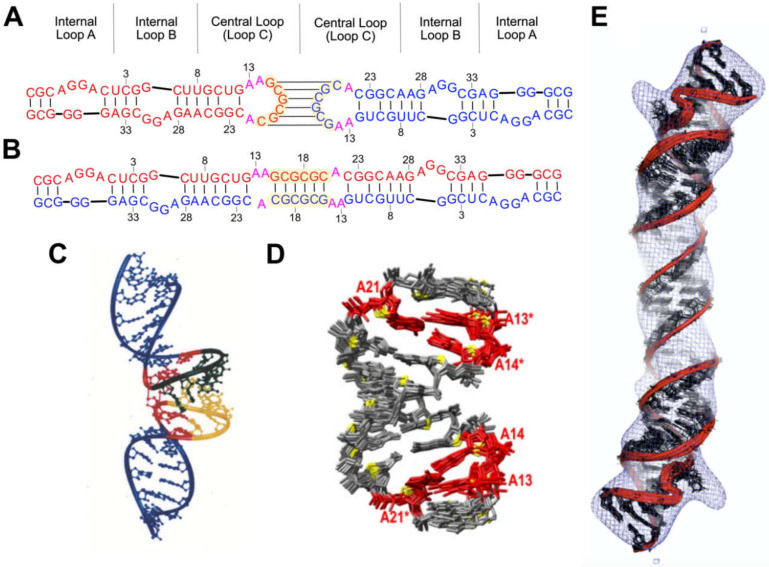
Packaging of the HIV-1 genome is intrinsically dependent on RNA dimerization [6,9,12,54,183,184,185,186,187]. Studies showed that residues of the DIS pseudopalindrome (Figure 5B–F) are required for dimerization of RNAs prepared by in vitro transcription [180,188,189,190,191] or purified from virions [192]. The DIS residues can adopt a hairpin structure with a central loop that contains a six-nucleotide palindrome and is capable of forming an intermolecular “kissing” dimer interface [193,194,195]; Figure 6A. As the DIS is a pseudopalindrome, it is also capable of forming an extended duplex structure (Figure 6B) possibly via formation of an initial kissing species [192,196,197,198,199]. The stem of the hairpin also contains two conservatively substituted bulges (internal loops A and B, Figure 6A,B) [200,201].
Figure 6.
Kissing and extended duplex forms of dimeric DIS. (A,B) Secondary structures of the HIV-1NL4-3 DIS element in kissing (A) and extended duplex (B) conformations. Individual RNAs denoted in red and blue. The palindrome and flanking purines of the apical loop (Loop C) are colored yellow and pink, respectively. Residue numbers correspond to that of a truncated construct utilized in (D). (C) Solution NMR structure of a DIS kissing dimer. (D) Solution NMR structure of the extended dimer. Flanking purines (red) stack to form a zipper motif. (E) Structure of the extended duplex form of DIS, as determined by a hybrid NMR/cryo-EM approach. Panels (C–E) reproduced from [202,203,204], respectively, with permission.
The loop–loop kissing interactions were structurally characterized by both NMR [202,205,206,207,208,209] and X-ray crystallography [210]. All structures exhibited coaxial intermolecular stacking of the helices, with variations in the degree of symmetry between the stacked hairpins [202,205,210]. These discrepancies were attributed to variations in sample conditions and structural restraints, but crystal lattice interactions that favor linear helical structures might also be a factor. HNN-COSY experiments confirmed the presence of intermolecular Watson–Crick G-C base pairs [205], and aromatic and imino proton NOEs were consistent with an underwound A-helical structure at the kissing interface [202,205,206,207]. The NMR data also indicated that the purine residues flanking the palindrome were stacked with residues of the loop residues; Figure 6C [202]. These findings contradict earlier crystal structures in which flanking purines are not stacked within the helix but instead adopt a bulged out conformation [210]. Additional NMR studies revealed that these residues might adopt a dynamic equilibrium between internally stacked and bulged out (minor population) conformations [205,206]. As discussed below, this conformational equilibrium might play a role in the conversion between kissing and extended duplex conformations [208].
Oligonucleotides that adopt the kissing dimer structure can be converted to a more thermodynamically stable extended dimer [180,189,196,211,212] by incubation at 55 °C [203,211,213], or by incubation at lower temperatures in the presence of NC [69,190,214]. Extended duplex DIS oligonucleotides can be prepared by boiling and slow cooling RNA samples, the inclusion of salts, or by use of longer hairpin constructs [204,207]. Three dimensional structures were determined by X-ray crystallography [215], NMR [203,204,207,213,216,217], Figure 6D, and a hybrid NMR/cryo-EM approach [204]; Figure 6E. The extended conformation largely mimics the dimer structure described above, but differs from the kissing species in the region flanking the palindrome, where purines instead exhibit a zipper-like motif with additional stacking contacts that stabilize the extended duplex; Figure 6D [203,207].
Residues of the internal loops (bulges) adopt similar structures in the kissing and extended duplex forms of the DIS dimers [207]. NMR studies defined the non-canonical nature of both internal loops for a number of native and mutant DIS constructs [201,209,217]. Internal loop A adopts a well-defined S-turn structure [201,204], similar to a turn observed in the HIV-1 rev response element (RRE) [218,219]. Internal loop B signals were broadened in some studies, consistent with conformational dynamics and disorder [201,217]. However, the signals were well-resolved in a subsequent NMR study that revealed internally stacked bases, possibly due to intrinsic differences in the constructs studied [207]. These internal loops are thought to serve as potential sites for NC binding [201,217] and helix unwinding, as the RNA converts from the kissing to the extended duplex (see below) [220].
NMR methods are used to monitor the mechanism of DIS conversion from the kissing to the extended duplex structure, as well as the temporal formation and discrimination of intermolecular contacts [212,221,222]. Differential behavior of imino protons identified nucleotide-specific unwinding events upon temperature- and NC-induced structural conversion [206,223]. 1H-15N heteronuclear single quantum coherence (HSQC) studies of wild type and mutant NC constructs identified the N-terminal residues and residues flanking the zinc-knuckle domains as critical to this conversion [224,225]. RDCs, dynamically decoupled spin relaxation, and chemical shift mapping NMR studies enabled direct detection of internal motions within the DIS [220]. These motions were found to be differentially greater for residues “above” internal loop B (closer to the palindrome), and arrested by Mg2+, suggesting a mechanism through which Mg2+ regulates the kissing loop unfolding, to prevent premature conversion to the extended duplex structure, in the absence of NC. Further work also established this mechanism of interconversion to be pH-dependent, upon protonation of a flanking purine residue [208].
Takahashi et al. utilized mixed samples containing unlabeled and 15N-labeled RNAs, to distinguish between kissing and extended duplex dimer interfaces in DIS fragments [212]. 15N splitting patterns observed for Watson–Crick imino protons confirmed that “weak dimers” contain a kissing interface, whereas the dimers that are more thermally stable contain an extended duplex intermolecular interface.
3.1.2. Ψ-hairpin
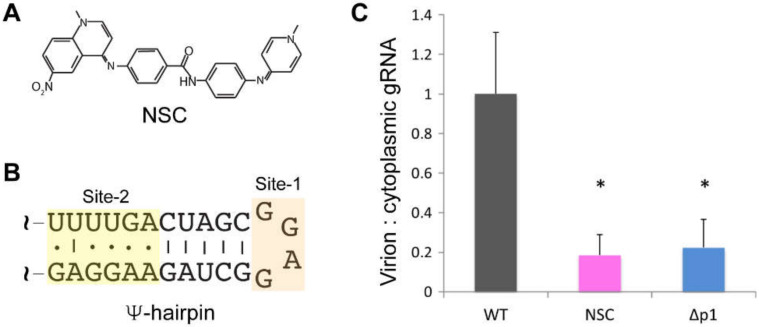
Early mutagenesis studies that deleted residues spanning from the major splice donor site to the gag initiation codon, demonstrated that these residues are essential for wild-type packaging efficiency [226,227,228,229]. RNAs containing this 46-nt region are capable of promoting the packaging of heterologous RNAs into virus like particles [173]. A conserved stretch of residues within this region were predicted to form a conserved hairpin that is historically called the Ψ-hairpin (or SL3) [156,157,159]; Figure 5B–F. The role of Ψ hairpin sequence and structure in genome packaging was probed by mutagenesis. Destabilization of the stem of the Ψ-hairpin significantly reduced dimerization and encapsidation efficiencies, but wild-type efficiencies were rescued by the revertant mutations [175,230,231,232,233]. This finding suggested the structure of the Ψ hairpin, rather than its sequence or downstream GA-rich residues, is critical for genome dimerization and packaging [233]. Other in vitro binding studies with larger Ψ-containing RNA fragments and Gag constructs, support a role for Ψ-NC interactions in Gag binding and packaging [234,235]. Although NC is capable of binding tightly to the conserved GGAG tetraloop of Ψ [109] (see below), substitution of the GGAG loop by GCUA or AAGA did not significantly affect packaging or replication [233]. Recently, NMR and isothermal titration calorimetry (ITC) studies revealed that the weakly base-paired [UUUU]:[GGAG] helical region in the lower stem of the Ψ hairpin, serves as the initial high-affinity NC binding site (Kd ~40 nM) and that structural rearrangements induced by NC binding are required for in vivo RNA packaging [236]. Although considerable effort focused on the structure and NC interactions of the Ψ-hairpin [237,238], it is now clear that competitive RNA packaging requires a much larger portion of the leader (see below) [40].
3.1.3. AUG and U5:AUG
Mutagenesis and other studies showed that the nucleotides surrounding the gag initiation codon are crucial for genome dimerization, Gag binding, and packaging [157,158,175,231,239,240,241]. These residues (AUG) were originally predicted to form a hairpin (Figure 5B) and NMR studies confirmed that oligonucleotides with sequences of AUG form hairpins with structurally well-defined GNRA-type tetraloops (G = guanosine, N = any nucleotide, R = purine, A = adenosine) [242,243]. Phylogenetic studies led to proposals that this stretch of residues instead forms base pairs with residues of an upstream element commonly called U5, thus, forming a U5:AUG helix, Figure 5D [161]. Subsequent structural probing and phylogenetic analyses were also consistent with U5:AUG pairing [162]. Although Paillart et al. did not find evidence for U5:AUG interactions or global RNA rearrangement in cells or in viral particles, using chemical probing [163], Weeks et al. suggested that U5:AUG base pairing does exist in the viral genomic RNA in virions, in transfected cells, and in vitro [164] (although other U5:AUG structures were subsequently proposed by this group [165]).
Long-range U5:AUG base pairing was later supported by NMR studies. 2D NMR analysis showed that an 11-mer U5 RNA can disrupt the AUG stem-loop structure, to form intermolecular U5:AUG duplex [114]. More recently, the U5:AUG interaction was confirmed in the intact leader (712 nt) by NMR using a segmental labeling approach [71]. The unlabeled 5′-leader fragment (residues 1-327) and the 13C-labeled 3′-fragment (residues 328–356) were enzymatically ligated to facilitate the direct probing of the AUG conformations in the context of the large, intact, leader RNA. When the RNA sample was incubated in physiological-like buffers that promote dimerization, NMR spectral changes consistent with an AUG structural conversion from a hairpin to a U5:AUG duplex were observed. The U5:AUG base pairing was then probed directly, using an NMR method for probing long-range interactions through adenosine–interaction detection (lr-AID; see below) [71]. An upfield adenosine signal corresponding to the central adenosine in the UAA triplet of the lr-AID element was observed, providing direct evidence for the presence of U5:AUG base pairing [71]. The three dimensional structure of U5:AUG in a larger fragment of the HIV-1 leader was subsequently reported (see below) [41].
3.1.4. TAR
The TAR element is a 59-nt sequence located at the 5′-end of all HIV-1 nascent viral transcripts and plays an essential role in Tat-mediated transcriptional activation [244,245,246,247,248]. Mutagenesis studies led to proposals that TAR is also involved in a variety of functions, including dimerization [249,250], strand transfer during reverse transcription [251], translation [75], packaging [239,252,253,254], HIV-1 derived microRNA (miRNA) during latency [255], and even the growth and progression of some cancers [256]. Some studies suggested the stability of the lower stem of TAR is important for RNA packaging [252,253]. In these studies, packaging efficiency was reduced by mutations designed to destabilize base pairing in the lower stem of TAR, and was recovered by compensatory mutations that stabilized the lower stem [254]. Das et al. found that viral replication can occur efficiently upon complete deletion of TAR when TAR is not needed for transcription, and that its sequence can be replaced by any sequence that can form a stable stem-loop at the 5′ of the transcript, suggesting the structure rather than sequence of TAR is important for packaging [257]. However, viral packaging studies can be complicated by a variety of factors, such as dominant negative effects [257], lack of viral accessory proteins [258], non-selective packaging [37,38,259,260], high RNA concentration [40], and improper mutation design that leads to misfolding [12,71]. In 2012, Heng et al. showed that the deletion of TAR did not significantly impair leader dimerization or vector RNA packaging, at least in a non-competitive packaging assay [40].
HIV-1 TAR adopts a hairpin structure that consists of two A-form helical domains connected by a three-pyrimidine residue bulge that binds to the Tat protein [261,262,263,264] and an apical 6-nt loop that binds to the cyclin T1 subunit of the cellular positive transcriptional elongation factor (pTEFb) during transcription [265,266]; Figure 5B–F. The first TAR and TAR-Tat complex structures determined by NMR revealed the structural elements that are critical for Tat binding—the most 5′ residue in the bulge (U23), the two base-pairs immediately downstream from the bulge (G26-C39 and A27-U38), and three phosphate groups [267,268]. In the free TAR hairpin, the helix axis is bent by an internal stacked bulge structure [267]. This result was supported by subsequent heteronuclear multi-dimensional NMR studies [269]. TAR undergoes a conformational change when binding to an arginine analog that mimics the TAR recognition site in the Tat protein, where the three bulge nucleotides become unstacked, and the two helix stems are coaxially stacked [267]. In addition, a base-triple forms between U23 and A27-U38, to stabilize the interaction of arginine with G26 and phosphates in the major grooves [267,268]. The base-triple model was supported by later NMR work [270,271,272,273]. NMR-based structural studies revealed that the apical 6-nt loop forms a deep binding pocket with bulge residues, when induced by high affinity Tat binding [271,273]. Conformational shifts in free and bound TAR states show that TAR undergoes extensive dynamic rearrangements related to its functions. More recently, D’Souza et al. used NMR to identify interactions between a large fragment of Tat (the Tat RNA binding domain, RBD) and both TAR and 7SK, a cellular small nuclear RNA that regulates transcriptional elongation [274]. These studies suggest that Tat and TAR evolved dual structural mimicry of cellular HEXIM and 7SK, respectively, providing a mechanism for promoting elongation of stalled HIV-1 transcripts [274]. TAR dynamics were extensively studied for HIV inhibitor development [272,275,276,277].
3.1.5. PBS
The function of the 5′ PBS is well characterized as the annealing site for tRNALys3—the packaged host transfer RNA (see section on matrix) that serves as the primer for viral reverse transcriptase [152]. NC mediates the annealing between the 3′ end of tRNALys3 and a complementary 18-nt PBS fragment (see section on NC–RNA interactions) [278]. An additional eight-nucleotide binding site located upstream of the PBS in the U5 region, called the primer activation site (PAS), anneals with the TΨC arm of the transfer RNA, before reverse transcription is initiated [279]. NMR studies revealed the formation of an intramolecular helix, upstream of the PBS, and significant remodeling of the transfer RNA, to accommodate the base pairing between the PAS and TΨC arm [168,280]. The formation of this structural element was shown to be a kinetic block to reverse transcription [281].
The PBS might also play a role in packaging cellular RNA helicase A (RHA) into virus particles [282,283,284,285]. RHA-knockouts significantly lower reverse transcription efficiencies in viral particles [282,283,284,285], and it appears that RHA might function in coordination with NC to promote reverse transcription elongation, by promoting primer extension through the intramolecular helical structure formed upon tRNALys3 annealing. The recognition mechanism of RHA by the 5′-leader of HIV-1NL4-3 during viral assembly was recently characterized via a combination of NMR and ITC studies, and it was found that RHA and NC noncompetitively interact with the leader RNA [286]. Chemical shift perturbation of A220 in the 5′-leader in NMR titration experiments showed that RHA specifically binds to the PBS region. Additionally, these findings were corroborated through in vivo mutagenesis studies showing that A220-to-cytidine substitution significantly reduced RHA packaging and viral infectivity [286].
3.1.6. Core Encapsidation Signal of HIV-1
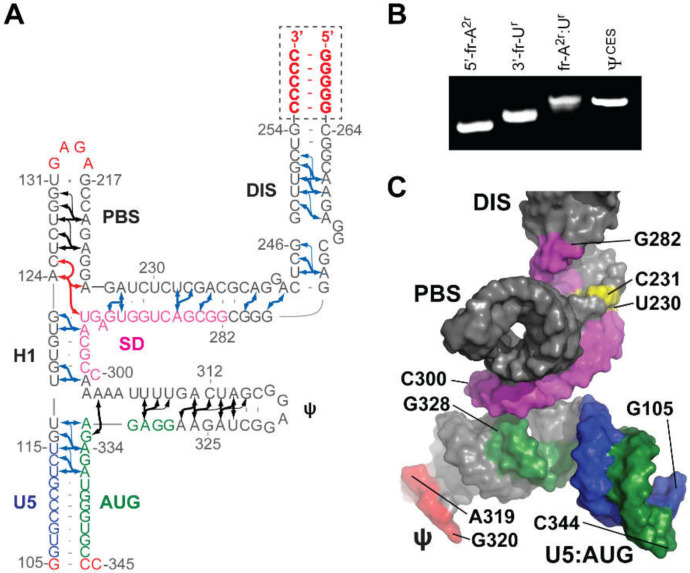
NMR was used to help identify a minimal region of the 5′-leader capable of directing RNA packaging. Deletions and mutations that did not significantly affect NC binding or RNA misfolding in vitro were employed in RNA packaging experiments. These studies revealed that the TAR, polyA, and PBS elements are not required for packaging, and identified a minimal ~260 nt region of the 5′-leader that is capable of efficiently promoting the packaging of heterologous vector RNAs (Core Encapsidation Signal, ΨCES) [40] (see below). Structural studies of ΨCES were facilitated by substituting the dimer-promoting GC-rich loop of the DIS hairpin by a GAGA tetraloop (Figure 7A), effectively cutting the symmetric dimer in half to improve spectral sensitivity and resolution associated with more rapid molecular tumbling [41]. Even with this modification, the size of the resulting RNA construct (155 nt, ΨCESm) was five times larger than the average size of RNA structures previously solved by NMR. Additionally, nucleotide-specific deuterium labeling did not provide sufficient resolution to enable complete assignment of the ΨCESm spectra. Resolution was further improved using a novel approach involving the non-covalent annealing of differentially 2H-labeled RNA fragments [41]; Figure 7B. Assignment and validation of NMR signals were facilitated by comparisons of chemical shifts with average database values, using software that correlates signals based on expected NOE patterns [287,288,289,290]. An improved understanding of the impact of NMR restraints and forcefields on the RNA structure, complemented these tools to help determine the 3D structure of the ΨCESm [41,291]; Figure 7C. Unexpectedly, the RNA adopted a tandem three-way junction structure in which residues of the SD did not form a hairpin but instead were sequestered by long-range base pairing. Several guanosines that were either held in exposed or weakly base-paired conformations were shown by mutagenesis to be essential for high affinity NC binding and RNA packaging. The ΨCESm structure also explains how HIV-1 selectively packages its unspliced genome [55]. Residues immediately downstream of the major splice site are integral to the formation of the tandem three-way junction structure, and differences in spliced mRNA sequences derived from 3′ exons would preclude formation of the packaging, competent, three-way junction structure.
Figure 7.
NMR structure of HIV-1NL4-3 ΨCESm. (A) Fragment-annealed sample used to identify long-range adenosine-H2 detected NOEs (denoted by arrows). Non-native residues are shown in red; U5, blue; AUG, green; and SD, pink. (B) Native polyacrylamide gel electrophoresis showing non-covalent annealing of differentially labeled 5′- and 3′- fragments. (C) Representation of the tandem three-way junction NMR structure adopted by ΨCESm (colors match panel A; conformationally dynamic nucleotides, yellow; Ψ tetraloop, orange). Adapted from [41], with permission.
3.1.7. NMR Studies of the Intact HIV-1 Leader
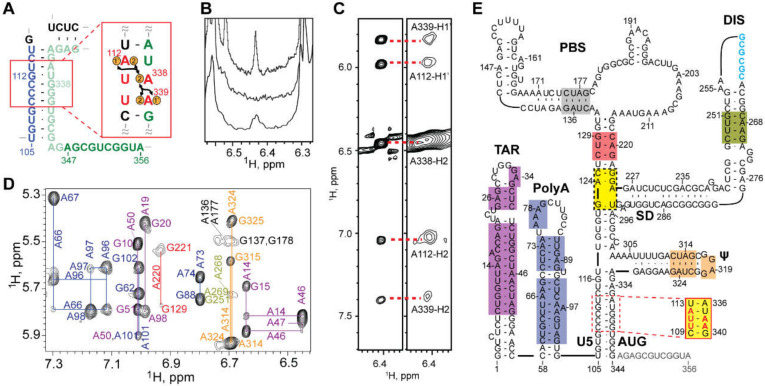
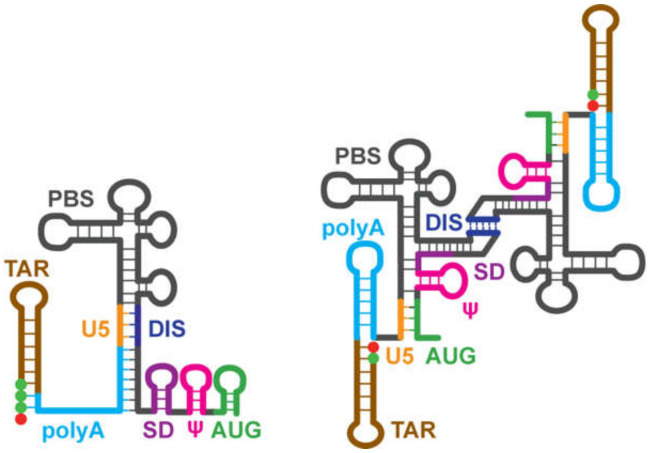
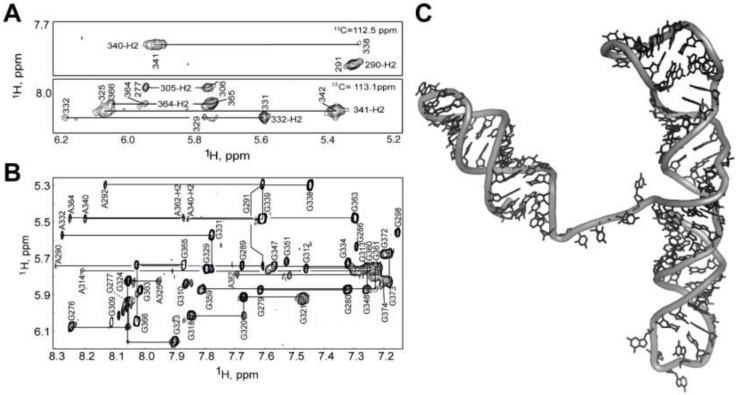
The 5′-leader of the NL4-3 strain of HIV-1 (residues 1-356; Figure 8A) forms a dimer with molecular weight of ~230 kDa, and it was surprising that a system of this size would give rise to interpretable NMR spectra [71]. The unexpectedly narrow NMR signals indicated that the RNA adopts a structure with independently folded subdomains that move with rotational correlation that is normally associated with much smaller molecules. Spectra obtained for nucleotide-specific 2H-labeled samples enabled direct detection of several of the predicted secondary structural elements located within the intact, dimeric 5′-leader; Figure 8A,B [40,71,221]. 1H-13C NMR studies were conducted with a partially 13C labeled leader RNA prepared by enzymatic ligation of an unlabeled 5′ -fragment and a 13C-labeled 3′-fragment [71]. Under low salt conditions where a portion of the RNA exists as a monomer, NMR signals were readily detected for the labeled residues. The 1H-13C NMR correlation spectra matched those of an isolated AUG hairpin, indicating that the AUG element adopts a hairpin structure in the monomeric leader. Under physiological-like buffer conditions that favor the dimer, several 1H-13C NMR signals disappeared due to extensive line broadening, and the remaining signals exhibited linewidths and chemical shifts consistent with an unstructured conformation. These findings confirmed that the AUG residues adopt an alternate (but largely undefined) conformation in the dimer.
Figure 8.
NMR studies of the intact, dimeric HIV-1NL4-3 5′-leader. (A) Illustration depicting mutations in the lr-AID substitution and 1H-1H NOEs. (B) Region of 1D 1H NMR spectra showing (top) the native TAR A46-H2 signal, (middle) A46G substitution, (bottom) A338-H2 signal observed for the lr-AID substitution. (C) 2D 1H-1H NOESY spectra of the same A338-H2 signal observed an isolated U5:AUG hairpin (left) and intact dimer (right) containing the lr-AID substitution. (D) Region of the 2D 1H-1H NOESY spectrum of A2rGrCr-labeled dimeric HIV-1NL4-3 RNA; color-coding matches the elements shown in (E). (E) Secondary structure of the HIV-1NL4-3 in its DIS-exposed, dimer-promoting state. Color-shaded boxes denoted resolved and assigned 2D 1H-1H NOESY signals; yellow denotes sites that were only assignable in mutant constructs using truncated constructs or lrAID substitution. Panels (A–C) adapted from [71] and panels (D,E) from [221], with permission.
The 2H-edited NMR studies led to the discovery that the helical structure adopted by a rare triplet of base pairs, [AiAi+1Ui+2]:[AjUj+1Uj+2], gives rise to well resolved Ai+1-H2 NMR signals. With this knowledge, leader RNAs were engineered to include a [AAU]:[AUU] element in the regions predicted to adopt A-form helical elements. This approach (lr-AID) enabled the direct detection of U5:AUG base pairing in the 230 kDa dimeric HIV-1NL4-3 leader [71]; Figure 8C–E. The approach also revealed that the loop nucleotides of the DIS element form base pairs with an upstream U5 region in the monomeric form of the RNA. This base pairing was also confirmed in NMR studies of the native HIV-1MAL leader (see below) [75]. 2H-edited NOESY NMR spectra were also obtained for HIV-1NL4-3 leader RNAs containing deletions or loop substitutions and compared with spectra of the intact, dimeric leader [40]. Mutations that did not cause global structural remodeling (determined via NMR) were employed for RNA packaging experiments. These studies revealed that the TAR, 5′ polyA, and PBS domains are not required for efficient packaging. Efficient packaging in a non-competitive packaging assay was achieved by a 155-nucleotide region of the leader comprising the U5, DIS, SD, the Ψ-hairpin, and AUG elements [40] (ΨCES, see above).
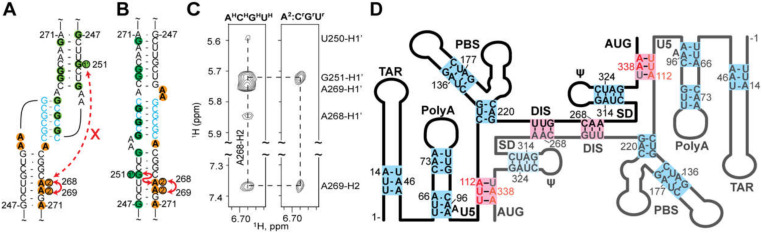
2H-edited NMR was additionally used to probe the nature of the dimer interface in the intact HIV-1NL4-3 leader [221]. Separately prepared HIV-1NL4-3 leader RNAs containing A2-labeling (sample 1) (superscripts denote sites of protonation, all other sites deuterated; e.g., A2 = adenosines protonated only at the C2 position, Gr = guanosines protonated only at ribose carbons) and a Gr-labeling scheme (sample 2) were mixed, and 2D NOE spectroscopy (NOESY) data were obtained. A268-H2-to-G251-H1′ NOE signals were detected for a sample containing an equimolar mixture of A2-leader and CrGrUr-leader RNAs, which is only possible if the leader adopts the extended duplex conformation; Figure 9A–C. Mixed samples containing other Gr labeling schemes showed similar results. In contrast, experiments probing long-range interactions in the TAR, polyA, PBS, and Ψ-hairpins gave results consistent with intramolecular base pairing in these regions of the leader; Figure 9. 1D NMR experiments revealed that the extended interface forms rapidly (within ~10 min) and does not require the presence of NC or other RNA chaperones. More recent single molecule FRET experiments with a 5′-leader fragment containing native U5 and AUG sequences are consistent with the U5: AUG base paired structure observed by NMR, and indicate that the RNA exists as an equilibrium mixture of structures [292].
Figure 9.
Discrimination between kissing and extended duplex base pairing in the HIV-1NL4-3 5′-leader by 2H-edited lr-AID NMR. (A,B) Cross-strand NOEs between A268-H2 and G251-H1′ establish the nature of the intermolecular interface in the kissing (A) versus extended duplex (B) interfaces (green dots = Gr labeling; orange dots = A2 labeling). (C) A268-H2 to G251-H1′ NOEs in both A2Gr and A2:GrCrUr support an extended duplex interface. (D) Summary of intermolecular (pink) (U5:AUG and DIS) and intramolecular (blue) (TAR, PolyA, PBS, Ψ) base pairings detected by NMR. Reproduced from [221], with permission.
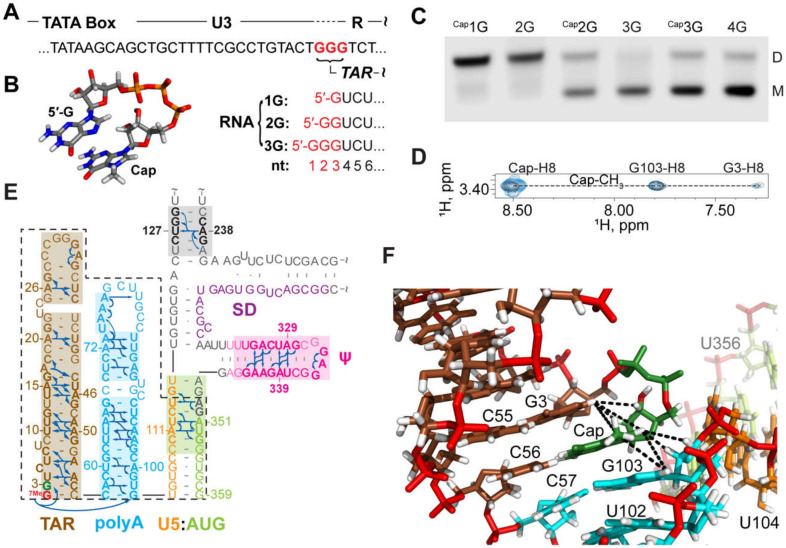
The above studies provided insights into the secondary structures adopted by selected regions within the HIV-1NL4-3 5′-leader. Unfortunately, the high propensity of this leader to form dimers made it difficult to obtain more extensive NMR data for the monomeric form of the RNA, due to the high sample concentrations required for NMR studies (typically > 50 µM). In studies of in vitro transcribed 5′-leader RNAs from different strains of HIV and SIV, it was discovered that the dimerization propensity of the simian immunodeficiency virus (SIVcpzUS strain) leader, which contains a GUGCAC palindrome in the dimer promoting DIS loop, is substantially lower than that of the HIV-1NL4-3 leader [293]. Therefore, studies shifted to the 5′-leader of HIV-1MAL, which also contains a GUGCAC palindrome and exhibits a dimerization propensity similar to that of SIVcpzUS [75]. Thus, the HIV-1MAL 5′-leader could be studied by NMR in its monomeric (low ionic strength) and dimeric (PI buffer) states [75]. In addition, in the course of studying 5′-leader RNAs from other lentiviruses [293], it was discovered that RNAs beginning with more than two guanosines at their 5′ ends exhibit stronger propensities to remain monomeric. Like all eukaryotic cellular mRNAs, HIV-1 transcripts are co-transcriptionally capped by a 5′-5′ triphosphate reverse-linked 7-methylguanosine moiety, shortly after initiation of the RNA polymerase II dependent transcription [294,295,296,297]; Figure 10. Interestingly, the cap moiety influences dimerization in a manner similar to that of an additional guanosine. Thus, non-capped 2G and capped-1G 5′-leader transcripts readily form dimers, whereas non-capped 3G/4G transcripts and capped 2G/3G transcripts, all preferentially form monomers [73].
Figure 10.
Heterogeneous transcriptional start site usage modulates HIV-1MAL RNA dimerization and function. (A) Three guanines (red) can serve as alternative transcription start sites. (B) Transcripts are co-transcriptionally capped by 7-methylguanosine. (C) Effect of 5′-guanosines and capping on 5′-leader dimerization. (D) Portions of 2D NOE spectra showing similarities of Cap-CH3 to Cap-H8, G3, and G108 NOEs observed for G8 intact dimer and the Capped TAR-polyA-U5AUG (Cap1G-leaderTPUA). (E) Assigned A-H2 NOEs and deduced secondary structure; discrete functional elements differentiated by color. Dashed lines denote residues of the Cap1G-leaderTPUA construct used for high-resolution structural studies. (F) Structure of the Cap1G-leaderTPUA showing end-to-end stacking of the TAR and polyA helices and sequestration of the Cap. Adapted from [75], with permission.
A review of published retroviral DNA sequences [73] revealed that the transcripts were predicted to begin with one, two, or three 5′-guanosines (1G, 2G, and 3G, respectively), a consequence of inconsistencies in defining the position of the HIV-1 transcription start site (TSS); Figure 10 [72,296,298,299]. The TSS is genetically defined as the first residue in the Repeat (R) element of the Long Terminal Repeat (LTR), and retroviral proviruses typically contain a conserved stretch of three Gs at the U3/R junction. Early attempts to identify the actual TSS led to inconsistent conclusions [296,298,299], but more recent studies revealed that all three guanosines function as transcriptional start sites during viral replication [72,73]. Importantly, the 1G transcripts are specifically packaged into virions whereas the 2G/3G transcripts are retained in cells and are enriched on polysomes [73]. These findings supported a model in which 1G transcripts adopt a structure that promotes dimerization, NC binding, and packaging, whereas the 2G/3G transcripts adopt an alternative structure that inhibits dimerization, NC binding, and packaging [73].
To understand how transcriptional addition of as few as one or two 5′-guanosines influence the dimerization properties and function of ~9 kilobase HIV-1 transcripts, the secondary structures of 5′-capped HIV-1MAL 1G, 2G, and 3G 5′-leader RNAs were probed by 2H-edited NMR [75]. The dimeric 5′-capped-1G leader RNA adopted a secondary structure consistent with that of the non-capped 2G HIV-1NL4-3 leader; Figure 10. It is important to note that signal diagnostic of the U5:AUG helix were detected in the native HIV-1MAL transcript, unlike the HIV-1NL4-3 transcript that required mutagenesis (lr-AID; see above) for U5:AUG detection. It is also noteworthy that signals were not resolved for the H1 helix observed in the HIV-1NL4-3 ΨCES structure. It is possible these signals were obscured by spectral overlap, and future studies to test for the presence of a tandem three-way junction structure similar to that observed for the HIV-1NL4-3 ΨCES are warranted. The NMR spectra for the intact 5′-capped 1G HIV-1MAL leader also exhibited evidence that the cap residue is sandwiched between the TAR and polyA helices. NMR studies of a leader fragment comprising the capped TAR, polyA, and U5:AUG helices confirmed that the TAR and polyA helices are stacked in an “end-to-end” manner; Figure 10 [75].
2H-edited NMR studies performed for the monomeric forms of the capped 2G/3G MAL leader RNAs, revealed extensive structural remodeling, compared to the capped 1G transcript [75]. Residues of the DIS palindrome were found to base pair with U5 in a manner similar to that observed for the NL4-3 leader (although the HIV-1MAL data were obtained for the native RNA whereas the HIV-1NL4-3 studies were conducted with mutants using the lr-AID NMR method [71]). The remainder of the DIS residues did not form a hairpin but were instead found to from long-range base pairs with upstream residues of polyA, Figure 11. Importantly, the residues of polyA did not adopt a hairpin structure, as observed for the dimeric capped 1G RNA. Instead, a majority of the polyA residues exhibited only sequential NOEs (no long-range NOEs) and chemical shifts consistent with a disordered, non-base paired structure; Figure 11. Furthermore, unlike the dimeric capped 1G leader, in which the cap residue is sandwiched between the TAR and polyA helices, the cap residue of the capped 2G/3G leader RNAs appeared exposed and disordered, a finding confirmed by 3D structural studies of a capped TAR fragment RNA [75]. Biochemical studies revealed that cellular cap binding proteins readily bind the capped 2G/3G MAL leader RNAs, but do not bind to the capped 1G leader, suggesting that cap sequestration/exposure, modulated by heterogeneous TSS usage, might play a role in establishing the function and fate of the HIV-1 transcript [75]; Figure 12.
Figure 11.
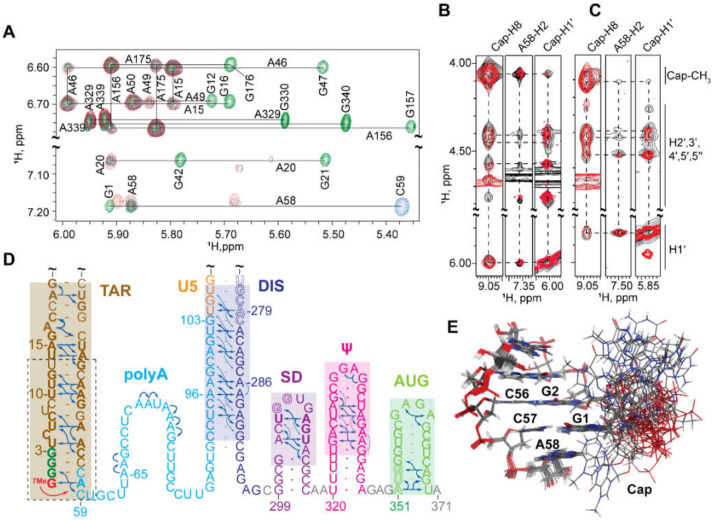
NMR and structural findings for the monomeric Cap3G and Cap2G forms of the HIV-1MAL leader. (A) Regions of the 2D 1H-1H NOESY spectra for the non-capped 3G- leader371 (AH, black; A2rGr, green; A2rCr, blue; G1HA2r, red) used to make secondary structure assignments shown in D. (B) Overlapped 2D 1H-1H NOESY spectra for A2-Cap2G-leader371 (black) and TAR fragment Cap2G-TARm (dashed outlined residues in panel D with U13 and G47 connected by a GAGA tetraloop) (red), showing that the cap methyl group is in close proximity to A58. (C) Similar results were obtained for Cap3G-leader371 (black) and a Cap3G-TARm RNA (red). (D) NOEs (arrows) and secondary structure of monomeric Cap-3G HIV-1MAL leader. (E) Structure of a Cap3G-TARm showing the unstructured Cap. Adapted from [75], with permission.
Figure 12.
Influence of 5′-guanosine number on RNA structure. Capped RNAs containing two or three 5′-guanosines (left) adopt a monomeric structure that exposes the cap and enables RNA processing and metabolism, whereas those with a single capped G (right) adopt a cap-sequestered conformation that promotes dimerization and packaging (Cap = red sphere, guanosines = green spheres). Adapted from [75], with permission.
3.2. Other Retroviruses
3.2.1. Moloney Murine Leukemia Virus (MoMuLV)
MoMuLV is an evolutionarily distant C-type retrovirus that was extensively studied as a model for retroviral genome encapsidation, and as a potential vector for therapeutic gene delivery [300]. A 350-nt “Ψ site” was initially identified by Mann et al., when deletion of the region between the env splice site and gag start codon abrogated the MoMuLV genome packaging [301]. Although the Ψ-site is required for packaging, upstream [302] and downstream [303] residues appear to be important for optimal packaging efficiency. The most efficient packaging was achieved with a region (Ψ+) containing the Ψ site and an additional 474 nt of the gag coding region [304]. We note here that some HIV-1NL4-3 vectors with non-native residues downstream of the leader were also poorly packaged. This was originally attributed to a role of coding residues in packaging; however, more recent studies indicate that the non-native residues might disrupt the structure of the leader and thereby inhibit packaging [42]. It is, thus, possible that the vectors employed in these early MoMuLV packaging experiments might also adversely affect the structure and function of the packaging signal.
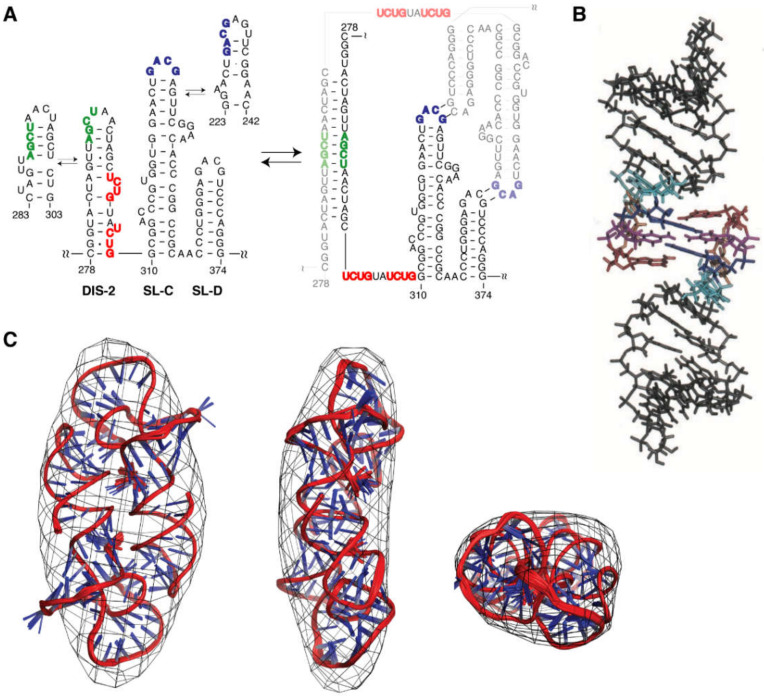
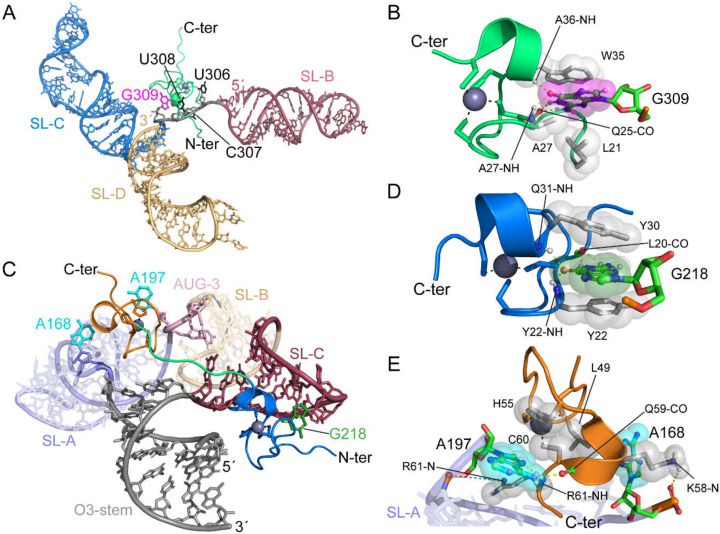
Mutagenesis, chemical accessibility probing, and computational analysis indicated that the Ψ region folds into several stem-loops, which are further condensed by long-range interactions [58,65,305,306,307,308,309,310]. Deletion of conserved residues predicted to form adjacent stem-loop structures (SL-C and SL-D; Figure 13), eliminated the RNA packaging [309,311], and heterologous RNAs containing SL-C, SL-D, and a portion of an upstream pseudopalindrome (SL-B) were efficiently packaged into virions [58] (although SL-C and SL-D alone were insufficient to promote packaging [312]). Native gel electrophoresis and ITC studies also showed that NC binds with a high affinity to an RNA fragment containing all three stem-loops (Kd ~132 nM), but interacts weakly with constructs comprising only one or two stem-loops [313,314]. Thus, this region of the MoMuLV genome [stem-loops DIS-2 (SL-B), SL-C and SL-D] is commonly referred to as the core encapsidation signal (ΨCES).
Figure 13.
Dimerization-dependent structure of the MoMuLV minimal packaging signal. (A) Left: Secondary structure of monomeric mΨ and the base pairings observed at equilibrium for DIS-2 and SL-C. Right: Base pairings in the dimeric form of mΨ. The palindromic sequences required for dimer formation are shown in green (DIS-2) and blue (SL-C and SL-D). NC-binding UCUG elements are shown in red. (B) The NMR structure of the 18-nt SL-D kissing duplex. (C) Superposition of the cryo-electron tomography densities and the NMR ensemble structures of [ΨCD]2. Panels (A,C) reproduced from [315], and panel (B) from [316], with permission.
The MoMuLV ΨCES RNA readily forms dimers under physiological-like ionic strength conditions (as does the entire leader) and undergoes conformational rearrangement upon dimerization [310]. Specifically, DIS-2 (also called SL-B), and an upstream pseudopalindrome called DIS-1 (or SL-A), undergo a register shift in base-pairing [66,310,315,317] upon formation of extended intermolecular duplexes [65,317,318,319,320]; Figure 13. Tinoco et al. were the first to show that SL-D is capable of forming a kissing dimer stabilized by only two intermolecular G-C base pairs [316] (Figure 13B), which was confirmed by subsequent NMR studies [314,315]. The kissing interaction was also characterized by nonequilibrium all-atom molecular dynamics simulation [321]. The tertiary structure of the native SL-CD dimer was determined by NMR using NOE and RDC restraints, and the overall shape of the NMR structure was confirmed by cryo-EM [315]; Figure 13C.
The 3D structure of a mΨ RNA with mutated loops to prevent dimerization was determined by a 2H- and 13C-edited NMR approach [322]; Figure 14. The improved 1H NMR linewidths and spectral quality were readily apparent upon comparison of the 2H- and 13C-edited NMR spectra; Figure 14A,B. Stem-loop SL-B contains five mismatched base-pairs and connects to SL-C via a 4-nt linker. SL-C contains an unusual base-triple platform, and a novel “A-minor K-turn.” SL-D adopts a classical A helical structure and is coaxially stacked with SL-C; Figure 14C. The NMR structure differs significantly from the models proposed on the basis of structural probing, using selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) technology [323], in which the DIS-2 hairpin was proposed to adopt an alternative fold and a large portion of the RNA (residues 283–303) was suggested to be unstructured [324].
Figure 14.
Improved NMR spectral quality afforded by 2H-edited NMR in the studies of MoMuLV minimal packaging signal, with loop residues engineered to prevent dimerization (mΨCES). (A) Representative strips from 3D 1H-13C correlated with the NOESY spectra obtained for 13C-labeled mΨCES. (B) Portion of a 2D NOESY spectrum obtained for an mΨCES sample containing protonated guanosines and with all other predeuterated nucleotides (breakthrough signals from incomplete deuteration of adenosines are also visible). Linewidths are significantly narrower in the 2H-edited spectra. (C) 3D NMR structure of mΨCES. Adapted from [322], with permission.
3.2.2. Rous Sarcoma Virus (RSV)
RSV is an alpha-retrovirus and was the first oncovirus described that could induce connective tissue tumors in chicken [325]. Unlike other retroviruses, the entire 5′-leader sequence (1-397 nt) of RSV is located upstream of SD and is thus included in all spliced and unspliced viral RNAs [326]. Like other retroviruses, RSV encodes a conserved 5′-leader with PBS, DIS, ψ, and AUG structural elements. The 5′-leaders of avian retroviruses also contain three short open reading frames (ORFs) that were proposed to regulate ribosome scanning [327,328]. One of the ORFs (ORF3) resides within the packaging signal [61]. Some mutagenesis studies suggested that the translation of ORF3 might directly or indirectly increase the packaging efficiency [329,330,331]. A study that competitively assessed Gag- versus ribosome-binding suggested that translation of ORF3 is not required for packaging, but is instead critical for maintaining the balance between translation and packaging [328].
The packaging sequence of RSV was initially discovered by spontaneous deletions near the 5′ end of viral genome, which lead to poor viral infectivity and deficient genome packaging [332,333,334,335,336,337]. All of these deletions contain a 31-nt region between the PBS and SD sequences [338]. A series of studies indicated that the region between the PBS and SD is required for genome packaging and is sufficient to direct the encapsidation of heterogenous RNAs into virus particles [59,60,332,334,335,337,338,339,340], while residues upstream [341,342] and downstream [60,339,343] of this region have little or no effect on packaging efficiency. The packaging signal was further refined to a 160-nt region called MΨ (nucleotides 156–315) [61,63]. Unspliced heterologous RNAs containing MΨ are packaged only about three-fold less efficiently than the wild-type genomic RNA [60,63]. A phylogenetic sequence alignment study predicted the secondary structure of the first 499 nucleotides in 13 strains of avian retroviruses, which consists of leader loops (L1-5), the PBS loop, the O3 loop located just downstream of the ORF3, and G1-3 loops located in the gag coding region [327]. Most of these stem-loop structures are supported by SHAPE experiments [344]. Computational models of MΨ in different strains of avian sarcoma leukosis virus (ASLV) also predicted two major stem-loops: O3 and L3 [61,62,327].
The O3 stem was found to be essential for genome packaging [62,327,345]. Mutations that disrupted the base-pairing of the stem, reduced packaging by 100-fold, compared to wild-type levels [62,327], whereas restoration of the base-pairing restored packaging efficiency [345]. The O3 stem-loop alone was sufficient to direct RNA packaging with efficiency equal to that of MΨ, thus, defining the minimal packaging determinant of the RSV genome (µΨ) [64]. The O3 stem-loop was predicted to consist of three minor stem-loops (O3-SLA, B, and C). Mutations designed to disrupt the minor loop structures greatly reduced packaging, while mutants that maintain RNA structure showed no difference or showed a slightly reduced packaging efficiency compared to the wild-type RNA [61,64], indicating that the structure rather than the sequence of O3 is crucial for genome packaging. Native polyacrylamide gel electrophoresis (PAGE), ITC, and NMR studies revealed that O3 interacts with NC with very high affinity (Kd =1.9 nM). However, isolated stem-loops and multi-stem-loop-fragments bind weakly to NC [346]. These findings set the stage for NMR-based 3D structural studies of the NC: µΨ complex [112] (see below).
It remains to be determined if or how RSV genome dimerization is coupled with packaging [347,348,349,350]. The palindromic sequence in the L3 loop [63,191,351], as well as palindromes in upstream [191] and downstream [347,352] sequences, were proposed to play roles in ALSV dimerization. Mutagenesis studies suggested that the L3 loop might indirectly contribute to packaging, by stabilizing the structure of the packaging signal [62,63]. A SHAPE experiment of the first 636-nt of RSV, mapped out the secondary structure of the entire leader [344]. In this study, L3, and O3-SLA were shown to be required for dimerization, and mutations in the loops of any of these elements abolished dimerization, but did not lead to significant reductions in viral replication. These findings suggested that the dimerization observed in in vitro studies might not be biologically relevant [344]. It is also interesting that the RSV packaging signal is present in both the spliced and unspliced 5′-UTR, even though the unspliced transcripts are preferentially packaged (200-fold enhancement compared with spliced transcripts) [31]. The mechanism through which RSV distinguishes between spliced and unspliced transcripts remains to be understood. One possibility is that the higher order structure of the leader in the unspliced RNA might be different from that in spliced RNAs, which favors a Gag-binding conformation [15].
4. NC–RNA Interactions
4.1. HIV-1
Initial characterizations of the nucleic acid binding activity of HIV-1 NC were performed mainly using deoxyribonucleotides or short polypeptides consisting of one zinc knuckle domain [82,107,111,353,354,355]. NMR titration experiments revealed that the hydrophobic residues Phe16 and Ile24 of the N-terminal zinc knuckle domain (NC-F1) were directly involved in the interactions with single-stranded DNA substrates [107,353,354]. Disruption of the zinc knuckle structure by the addition of a zinc chelating agent (ethylenediaminetetraacetic acid) abolished these interactions [107]. Stacking interactions between NC-F1 aromatic side-chains and nucleic acid bases were suggested to be important for binding [353,355]. Early fluorescent studies showed that NC binding is sequence-specific [82]. NMR titration experiments using DNA oligonucleotides with varied sequences, further revealed that the tight binding to HIV-1 NC-F1 requires the presence of at least one guanosine residue [107]. Two-dimensional NOESY-data-facilitated structural modeling revealed that the packaging site analog, d(ACGCC), binds within the hydrophobic cleft on the NC-F1 surface, and the complex is further stabilized by multiple intermolecular hydrogen bonds involving the guanosine base [107].
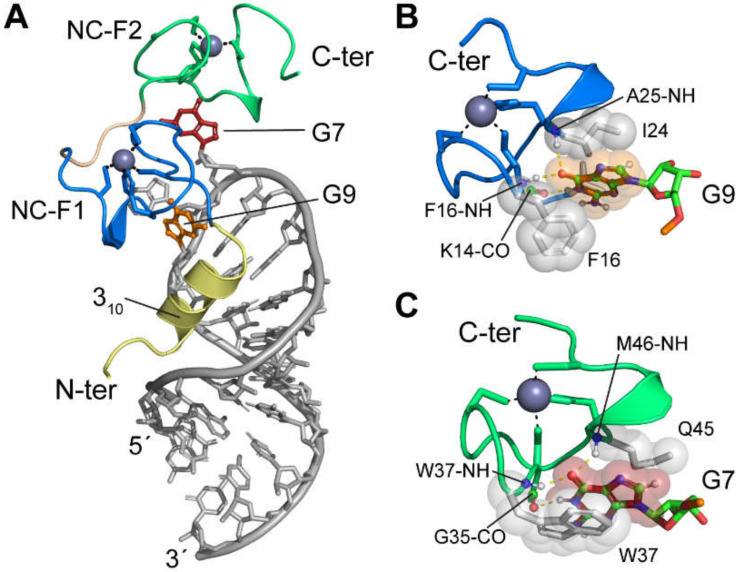
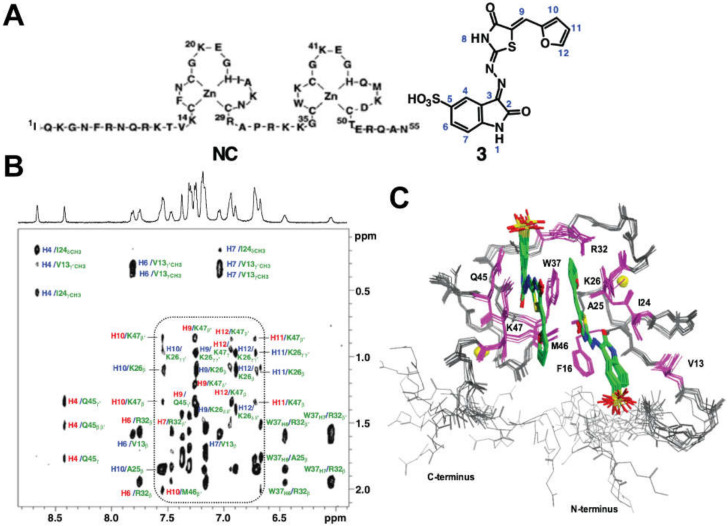
The three-dimensional structure of HIV-1 NC bound to a 20-residue RNA, containing the apical loop of the Ψ-hairpin (originally called “SL3”), was determined by heteronuclear NMR spectroscopy [109]. The specific recognition was mainly mediated by the interactions between the G9 and G7 nucleotide bases of the G6-G7-A8-G9 tetraloop and the hydrophobic clefts of the N- and C-terminal zinc knuckles, respectively (Figure 15A). In the NC-Ψ-hairpin complex structure, the base of G9 is sequestered into the NC-F1 hydrophobic cleft, and directly contacts the bulky side chains of Val13, Phe16, and Ile24; Figure 15B. G9 is further stabilized in the NC-F1 hydrophobic cleft through specific hydrogen bonds between G9-O6 and the backbone NH groups of Phe16 and Ala25, and between G9-H1 and the backbone carbonyl group of Lys14; Figure 15B. G7 interacts with the C-terminal zinc knuckle (NC-F2) hydrophobic cleft in a very similar manner with the nucleotide base packed against the side chains of conservatively substituted Trp37 and Gln45. Additionally, the G7-O6 forms hydrogen bonds with the backbone NH groups of Trp37 and Met46, and G7-H1 forms a hydrogen bond with Gly35-CO; Figure 15C. Extensive non-specific electrostatic interactions also contribute to the tight binding between NC and the Ψ-hairpin [109]. The HIV-1 NC also exhibits structural rearrangements upon binding to the Ψ-hairpin, including the formation of a 310 helix (Lys3 to Arg10) and RNA binding-induced contacts between the two zinc knuckle domains [109], which are hallmarks of adaptive binding.
Figure 15.
NMR structure of HIV-1NL4-3 NC bound to the GGAG loop region of the Ψ-hairpin stem-loop [109]. (A) Overall view showing the relative orientation of the RNA (gray) relative to the 310-helix (yellow), N-terminal zinc knuckle (F1, blue), and C-terminal zinc knuckle (F2, green). Guanosines G7 and G9 are shown in red and orange, respectively. (B) Interactions between G9 and NC-F1. Hydrogen bonds are depicted as yellow dash lines and hydrophobic side chains shown as spheres. (C) Interactions between G7 and NC-F2.
The 3D structure of HIV-1 NC bound to a 19-residue SD-hairpin was also determined by solution NMR [110]. Similar to the specific interactions observed in the NC-Ψ-hairpin structure, the hydrophobic clefts of the N- and C-terminal zinc knuckle domains bind to exposed guanosine bases G11 and G9 of the G8-G9-U10-G11 tetraloop of SL2, respectively. The G11 base is sandwiched between the hydrophobic side chains of Phe16 and Ile24 and stabilized by the hydrogen bonds between G11-O6 and the backbone amide groups of Phe16 and Ala25, as well as by a hydrogen bond from G11-H1 to the backbone carbonyl of Lys14. In the NC-F2 hydrophobic cleft, the G9 nucleobase packs against the side chains of Trp37 and Gln45, and is stabilized by hydrogen bonding from G9-O6 to the backbone amide groups of Trp37 and Met46, and from G9-H1 to the Gly35 backbone carbonyl. Thus, the NC zinc knuckles bind to exposed guanosines of the Ψ-hairpin and SD-hairpin RNAs, in a similar manner. Residues Lys3 to Arg10 also form a 310 helix, upon binding to SD-hairpin, but form different protein-RNA interactions, compared to that of the NC-Ψ-hairpin structure [109,110]. NMR NOE analysis also reveals that the RNA-induced contacts between the NC-F1 and NC-F2 zinc knuckles differ among the SD-hairpin and Ψ-hairpin NC complexes [110]. Thus, it is conceivable that HIV-1 NC exhibits inherent structural flexibility, which allows NC to adaptively bind to different RNA targets with high-affinity.
In addition to the loop region of RNA hairpins, the three-dimensional structure of NC bound to a single-stranded U5 derivative (5′ -UGUGCCCUUCU-3′) was also determined using solution NMR [114]. The two guanosines of this RNA form very similar interactions with the NC-F1 and NC-F2 hydrophobic clefts in the same polarity, as reported for the NC-Ψ-hairpin and NC–SD–hairpin complexes (NC-F1 binding to 3′-guanosine, NC-F2 binding to 5′-guanosine) [114]. Extensive biochemical data demonstrated that the HIV-1 packaging signal contains more than two dozen NC binding sites, with varied affinities and different functional importance [40,41,71]. In particular, NMR-detected NC binding, in conjunction with ITC studies reveal that the [UUUU]:[GGAG] stem region of the Ψ-hairpin contains two very high-affinity binding sites (Kd ~40 nM, compared with Kd ~ 300 nM for the Ψ-hairpin apical loop, under physiologic-like ionic strength) [236]. The structural lability of this [UUUU]:[GGAG] helical region is shown to be required for both tight NC binding in vitro and efficient RNA packaging in transfected cells [236]. However, the molecular mechanism for such high-affinity binding remains unknown, and additional studies of NC bound to other RNA targets in the HIV-1 dimeric 5′-leader are clearly warranted.
4.2. Other Retroviruses
4.2.1. MoMuLV
Initial NMR studies of MoMuLV NC-nucleic acid interactions were conducted with a pentanucleotide, d(ACGCC), which was previously shown to bind to a HIV-1 zinc knuckle peptide [107] and to HIV-1 NC [111,356]. 1H-NMR studies of the complex identified a cluster of hydrophobic residues on the surface of the MoMuLV zinc knuckle that form a binding pocket for a guanosine base (Leu21, Ala27, Ala36). 31P NMR experiments revealed significant chemical shift changes for phosphodiester groups between G3 and C4, and between A1 and C2. These results revealed that the MoMuLV zinc knuckle binds exposed guanosines bases in a manner similar to that observed for HIV-1 zinc knuckles.
Unexpectedly, MoMuLV NC did not exhibit significant affinity for any isolated ΨCES hairpins (DIS-2, SL-C or SL-D), nor to the SL-CD tandem stem-loop RNA [314]. Instead, the protein only bound tightly to RNA constructs containing all three stem-loops (the intact ΨCES), and the binding required (and was shown to induce) RNA dimerization [314]. These data supported a mechanism in which MoMuLV NC targets a binding site that requires the presence of all three hairpins (SL-BCD, ΨCES) [314].
To facilitate 3D structural studies, a ΨCES construct, with loop residue mutations designed to prevent dimerization, while retaining the internal base pairing of the dimer (SL-Bm-Cm-Dm, mΨCES) was examined [66,314]. These studies revealed that the single zinc knuckle domain of MoMuLV NC, binds specifically to the UCUG linker that connects the DIS-2 helix and the SL-C hairpin (residues U306–G309); Figure 16A. The bases of nucleotides U306, C307, and U308 pack against a hydrophobic surface comprising the side chains of Leu21, Ala27, and Tyr28, and binding is further stabilized by a number of direct or water-mediated salt bridges and hydrogen bonds [66]. The G309 base fits into a hydrophobic cleft, lined by the side chains of Leu21, Ala27, Trp35, and Ala36, and is anchored by the following hydrogen bonds—G309-O6–Ala27-NH, G309-O6–Ala36-NH, G309-N1H–Gln25-O, G309-NH21–Gln25-CO; Figure 16B [66]. These hydrophobic and hydrogen bonding interactions are similar to those observed for the guanosine-binding zinc knuckles of HIV-1 NC.
Figure 16.
Complex structures of MoMuLV and RSV NC bound to the recognition elements in their respective packaging signals (reported in [66] and [112], respectively). (A) MoMuLV NC-UCUG complex structure (PDB ID: 1U6P). SL-B, SL-C, and SL-D are colored in red, blue, and gold, respectively. The UCUG linker and binding site is colored gray with G309 in magenta. The zinc knuckle domain of the MoMuLV NC is shown in green, with the black dashes representing the coordination of Zn. (B) The direct interaction between the MoMuLV NC and G309. Hydrogen bonds are shown as yellow dashes and the gray spheres represent hydrophobic interactions. (C) RSV NC bound to the µΨ packaging signal (PDB ID: 2IHX). The O3-stem, SL-A, AUG-3 linker, SL-B, and SL-C are colored gray, purple, pink, gold, and red, respectively. The N-terminal zinc knuckle, linker residues, and C-terminal zinc knuckle are colored blue, green, and orange, respectively. G218 nucleobase is colored green. A168 and A197 nucleobases are colored cyan. (D) The N-terminal zinc knuckle of the RSV NC interacting with G218. (E) The C-terminal zinc knuckle interacting with A168 and A197. Salt bridge interactions are depicted as cyan-colored dashes.
Chemical accessibility experiments conducted with a larger portion of the MoMuLV 5′-leader indicated that multiple UCUG sequences become exposed and accessible, following RNA dimerization, suggesting the presence of additional NC-binding sites for genome recognition and packaging [308,310]. Short oligoribonucleotides were used in a combination of NMR and ITC studies, to characterize the effect of the residues flanking the guanosine on NC-binding and to determine the binding affinity of NC to the other possible recognition elements. Three specific oligoribonucleotides that represented the proposed binding sites were of specific interest—AACAGU, UUUUGCU, and CCUCCGU. Based on 2D-NOESY data, the guanosine at position i gave rise to intermolecular NOEs between Gi-H8 and Trp35 aromatic, Ala27 methyl, Ala36 methyl, and Leu21 side-chain protons, and between Gi-H1′ and Leu21, Trp35, and Arg23 side-chain protons [113]. Additional NOEs were observed between the nucleobase and ribose protons of residues at positions i-1, i-2, and i-3, and the side-chain protons of Ala27, Leu21, Arg18, Tyr28, Ala36, Lys42, and Lys30 [113]. These findings suggest a form of cooperative recognition mechanism where the initial NC-binding might promote RNA dimerization and concomitant exposure of additional NC-binding sites.
4.2.2. Rous Sarcoma Virus
The structure of the RSV NC bound to the minimal packaging signal, µΨ, is also solved by NMR. Unlike other retroviruses, the 5′ UTR of alpha-retroviruses contain three distinct open reading frames or AUG motifs—AUG-1, AUG-2, and AUG-3, and there is controversy over their functional differences [328,329,330,331]. The proposed structure of the minimal packaging signal consists of a central stem, O3 stem, with a 3-way junction between the three stem-loops—SL-A, SL-B, and SL-C where SL-A and SL-B are linked via AUG-3; Figure 16C [64,112] (see above). NMR studies of the NC:µΨ complex suggest that the AUG-3 in the µΨ region is a binding site for NC [112,328,357]. The RSV NC protein has distinct N-terminal and C-terminal zinc knuckle motifs that bind viral RNA through different mechanisms.
NMR analysis showed that the SL-C tetraloop gave rise to distinct NOEs with the aromatic residues in the N-terminal zinc knuckle [112]. Similar to the zinc knuckle domains of HIV-1 and MoMuLV [9,92], the N-terminal zinc knuckle of the RSV NC contains a hydrophobic pocket defined by Tyr22, Tyr30, Leu20, and Gln30, which fits the nucleobase of G218 [112]. The complex is further stabilized via intermolecular hydrogen bonding between backbone amides and oxygens—G218-O6–Tyr22-NH, G218-O6–Gln31-NH, G218-N1H–Leu20-O, and G218-NH21–Leu20-CO; Figure 16D. NOE analyses also indicated that linker residues U224, A225, and G226 fold into the minor groove of SL-C [112].
The C-terminal zinc knuckle structure lacked a hydrophobic pocket and did not give rise to NOEs to guanosine residues, suggesting that it functions differently than other characterized zinc knuckle domains. However, there were NOEs to the linker residues adjacent to the 5′ and 3′ bases of SL-A [112]. On the 3′-end, the exocyclic amino group and N7 of A197 (the adenosine in AUG-3) formed hydrogen bonds with the carbonyl group of Gln59 and the amide proton of Arg61, respectively. The nucleobase of A197 is positioned against His55, Cys60, and Arg61, and is further stabilized by a salt-bridge between the 5′-phosphate of A197 and Arg61; Figure 16E. On the 5′ base of SL-A, the nucleobase of A168 is stacked against Leu49, Cys60, and Lys58 side-chains, and it is further stabilized via hydrogen bonding between the A168 phosphate oxygen and the Lys58 side-chain amino group; Figure 16E [112]. The significance of these interactions was further supported by in vivo mutagenesis studies, where inserting a non-native GAGA SL-C tetraloop or substituting the AUG-3 linker residues significantly reduced viral infectivity [112].
4.3. Chaperone Activity of NC
In addition to its role in genome packaging, the NC domain of Gag (or the mature NC protein) functions as an RNA chaperone to catalyze conformational rearrangements [358]. NC stimulates reverse transcription by annealing the tRNALys3 primer to the primer binding site (PBS) [359,360], promotes the processing of reverse transcriptase by reducing polymerization pausing [361,362], and increases the efficiency of both minus- and plus-strand transfers during reverse transcription [363,364,365,366,367,368,369,370]. NC chaperone activity was also proposed to help catalyze the kissing to extended dimer conversion (see above). The chaperone property of retroviral NC appears to rely on both the highly basic N-terminal residues and the zinc knuckle structures of NC, which involves weak and non-specific NC–RNA interactions [214,371,372]. NMR studies showed that NC performs its chaperone activity by lowering the energy barrier required to break base pairs or by facilitating the formation of new base-pairs [169,358,373,374,375,376,377,378].
5. Inhibition of Genome Packaging
Efforts to develop inhibitors of HIV-1 genome packaging involved approaches that targeted both the NC domain of Gag and the RNA packaging signal. Small molecules that bound to both viral targets and interfered with NC-RNA binding in vitro, were identified. Although these compounds exhibited antiviral activity in cell culture assays, none exhibited the appropriate efficacy and toxicity required for advancement to the clinic. The current progress is summarized below.
5.1. Inhibitors that Target NC
Current anti-retroviral therapies involve a combination of drugs that suppress HIV-1 replication, but resistance can still occur, and this life-long treatment regimen can have adverse side effects. Therefore, there is a need to develop antivirals that target different components of HIV that are essential for replication. The zinc knuckles of NC are essential for replication and, therefore, are attractive potential targets. One class of antiviral agents was shown by NMR to function by a novel “zinc ejection” mechanism, in which the cysteine sulfur atoms that are required for binding zinc and maintaining proper folding are selectively oxidized [379,380,381,382,383]. Mercaptobenzamide thioesters initially oxidize a specific cysteine of the C-terminal zinc knuckle; Figure 17. NMR chemical exchange saturation transfer (CEST) and relaxation experiments showed that the C-terminal zinc knuckle undergoes conformational exchange between one major species and two minor species, which likely explains the enhanced reactivity of the C-terminal zinc knuckle; Figure 17 [384].
Figure 17.
(A) NC treated with mercaptobenzamide-1 targets the sparsely populated states of the C-terminal zinc knuckle of NC. (B) 1H-15N spectrum of NC alone (green) treated with 20-fold molar excess of mercaptobenzamide-1. Three-hour treatment (red) and 6-h treatment (blue) of merceptaobenzamide-1 (Black circles highlight the C-terminal zinc knuckle cross-peaks). (B,C) 15N-CPMG relaxation dispersion (B) and CEST experiments reveal the sparsely populated states (C) of the C-terminal zinc knuckle of NC. Adapted from [384], with permission.
More recently, a series of compounds were identified that could block interactions between NC and TG-rich deoxynucleotides. NMR-based structural studies revealed that two inhibitor molecules interact with the N-terminal and C-terminal zinc knuckles by mimicking exposed guanosine residues [385] (Figure 18), which are known to be important for binding nucleic acids [66,107,109,110].
Figure 18.
Small molecule inhibitors of NC-oligonucleotide binding. (A) NC protein and small molecule (Inhibitor-3) used for structural studies. (B) 2D NOESY data for the NC:Inhibitor-3 complex. (C) NMR structure of the NC:Inhibitor-3 complex (carbon atoms in green). Adapted from [385], with permission.
5.2. Inhibitors that Target the RNA Packaging Signal
In addition to the viral proteins, the highly conserved and structured regions of the RNA genome are also attractive therapeutic targets, as exemplified by the Ψ-hairpin of the HIV-1 RNA packaging signal [386]. Multiple small molecules were identified to inhibit NC-Ψ interactions [387,388]. Among them, a quinolinium derivative NSC260594 (NSC, Figure 19A), was shown to be a specific HIV-1 RNA packaging inhibitor [389] and exhibited potent antiviral activity [388,389]. NSC treatment caused a similar packaging defect as that of the Δp1 mutation—a 19 nt deletion that historically led to the identification of the Ψ-hairpin as a major packaging determinant; Figure 19C [228,389]. NSC was initially identified as an inhibitor of the interaction between Gag and a 20-nt construct containing the upper stem and the apical tetraloop of the Ψ-hairpin [388]. In the same study, 1H NMR spectroscopy showed that NSC specifically bound to the GGAG tetraloop (site-1 in Figure 19B). However, a recent NMR study demonstrated that NSC targeted the structurally labile [UUUU]:[GGAG] stem region of the Ψ-hairpin, in the context of a larger construct of the HIV-1 packaging signal (site-2 in Figure 19B) [236]. Using the full-length HIV-1 5′-leader, SHAPE analysis suggested that the function of NSC is to stabilize the Ψ-hairpin and subsequently stabilize a larger region of the RNA packaging signal [389]. These studies further confirmed the critical function of the [UUUU]:[GGAG] region of the Ψ-hairpin in HIV-1 selective genome packaging, which could serve as a novel specific therapeutic target.
Figure 19.
Small molecule inhibitor of HIV-1 genome packaging. (A) Structure of packaging inhibitor NSC. (B) Binding sites of NSC in the Ψ-hairpin of the HIV-1 packaging signal. (C) NSC treatment reduces HIV-1 selective genome packaging. Asterisks represent statistically significant from wild-type (WT) by Student’s t test, p < 0.05. Panel (C) reproduced from [389], with permission.
In addition to small molecule inhibitors, 1H NMR analysis showed that polypeptides rich in tryptophan also interact with the GGAG tetraloop of the Ψ-hairpin, and possibly block NC binding [390]. The DIS region might also be targeted, since dimerization is a prerequisite for retroviral genome packaging [391].
6. NMR-Based Hybrid Approaches
Many of the challenges associated with NMR studies of large RNAs and protein-RNA complexes are natural targets for hybrid approaches that provide complementary structural information. Commonly employed NMR methods provide high-resolution local structural information but do not inform on overall molecular shape. In contrast, methodologies such as SAXS and cryo-EM, provide information on global structure and overall shape, but typically do not afford information regarding high-resolution local structural features. Hybrid approaches that combine NMR restraints with global shape restraints derived from SAXS or cryo-EM, offer an attractive approach for the study of large RNAs and protein:RNA systems that are difficult to study by any one method.
SAXS has many advantages in this regard, because (i) small-angle scattering profiles do not require sample crystallization, (ii) there are no size limitations, (ii) the electron-rich backbone of RNA scatters X-rays strongly, (iii) the 3D RNA structures are typically non-spherical, and (iv) procedures for implementing sparse NMR restraints and SAXS data could be accomplished with existing NMR structure refinement software packages [392,393,394,395,396]. A disadvantage of SAXS is that data analysis can be complicated if the sample exists as a mixture of multiple conformers or contains even small amounts of higher-order species. An attempt to use SAXS to analyze the structure of a tandem hairpin RNA structure that is important for MoMuLV genome packaging was unsuccessful due to the fact that the RNA existed in solution as a mixture of monomeric, dimeric, and higher-order multimeric species, each of which was directly detectable in a single sample, through cryo-EM [315]. Although there are presently no examples of hybrid SAXS/NMR methods applied to the studies of retroviral genome packaging, these approaches provided important insights into HIV-1 RNA structures that contribute to transcriptional activation [274], splicing [397], and nuclear export [398]. For example, Tolbert and co-workers used a hybrid SAXS/NMR approach to determine the structure adopted by residues of a phylogenetically conserved intron splice silencer stem-loop. A handful of cellular RNAs were also examined by this approach (for examples, see [394,399,400,401,402]).
Cryo-EM is another complementary technique traditionally used to determine the tertiary structures of large protein complexes [403,404]. Unlike SAXS and NMR, samples are frozen in a thin layer of vitrified ice and density maps generated from the scattering data were obtained for a large number of individual particles, thereby producing a 3D envelope for an ensemble average of particles [405]. A particular advantage of cryo-EM is that densities for individual molecules that exist in different conformations or oligomerization states can be “binned,” enabling studies of samples containing structurally heterogeneous molecules. Resolution is limited by several factors, including flexibility, symmetry, and size [404]. Cryo-EM is historically used to study the structures of large macromolecular assemblies, and it was initially surprising that high-quality electron density maps could be obtained for the structures of RNAs as small as the SL-C/SL-D tandem hairpin of the MoMuLV packaging signal (see Figure 13, above) [315]. At the time, this RNA was simultaneously among the largest RNAs to be structurally characterized by NMR, and the smallest to be characterized by cryo-EM [315]. These studies revealed that the helical regions of the tandem hairpin pack side-by-side, a finding that was inferred by the NMR-directed calculations but not experimentally verifiable using the NMR methods employed. Perhaps the most important point of this early work was that the low-resolution global structural information obtainable by cryo-EM was highly complementary to the high-resolution local structural information readily obtainable by NMR [315].
The combination of NMR and cryo-EM was further utilized in the structural characterization of the extended duplex form of the 30 kDa HIV-1 DIS [204]; Figure 6E. The cryo-EM map, refined to 9 Å resolution, revealed the major groove of the RNA and the presence of an overall super-helical twist. The cryo-EM data also revealed bulges in the electron density maps, indicative of bulged nucleotides. The combination of NMR and cryo-EM in this study clearly demonstrated the utility of merging global and local restraints in structure determination [204].
7. Summary and Future Directions
During the late 1980’s and early 90’s, NMR was used to study relatively small retroviral proteins and RNAs were believed to be important for packaging. These studies provided insights into the role of zinc in viral replication, characterized the structure of NC, and its intrinsic CCHC zinc knuckle domains, and compared and contrasted the NC domains of different retroviruses. These studies were complemented by studies with small nucleic acids, which identified sequences that bind the NC zinc knuckles with high affinity, established the mechanism of DIS-promoted RNA dimerization, and led to the structural characterization of nucleic acid complexes with isolated zinc knuckles and intact NC proteins. NMR studies also provided insights into the structures of other regions of the 5′-leader that are important for viral replication, and led to the identification of novel reagents and mechanisms for inhibiting viral replication by promoting zinc ejection from the CCHC zinc knuckles or binding competitively to NC or to RNA elements within the 5′-leader. In addition, NMR studies of a minimal packaging signal from the MoMuLV 5′-leader, led to the proposal for a dimerization-dependent RNA structural switch packaging mechanism, in which nucleotides important for NC binding are sequestered by base pairing in the monomeric form of the RNA and become exposed to promote NC-binding upon dimerization. It remains to be determined if MoMuLV or other retroviruses utilizes heterogeneous transcriptional start site usage as a means of controlling transcript dimerization, as appears to be the case for HIV-1. Interestingly, more than 40 years ago, Levin and Rosenak proposed that MuLV-producing cells likely contain two non-interconverting pools of viral transcripts that function separately as viral mRNA and gRNA, based on the sensitivity of gRNA incorporation into virions produced in the presence of the transcription inhibitor, actinomycin D [406], and more recently Mougel et al. showed that nuclear export of MuLV gRNA and mRNAs occurs via different pathways [407]. Although HIV evolved a Rev-dependent mechanism to promote nuclear export [408], it remains plausible that the evolutionarily distant HIV and MoMuLV retroviruses could both utilize heterogenous transcriptional start site usage as a means of producing distinct pools of messenger and genomic RNA transcripts.
RNA dimerization-dependent packaging mechanisms were proposed for HIV, based primarily on biochemical and nucleotide accessibility mapping studies of in vitro transcribed RNAs, but applications of NMR to test structural and mechanistic hypotheses were not performed until the past decade or so, due to the large size of the HIV 5′-leader (~350 nucleotides). The first reported NMR studies of the intact HIV-1 5′-leader were performed using both chemical ligation of differentially labeled (13C-labeled and un-labeled) fragments and nucleotide specific 2H editing. Both strategies aimed to enhance spectral sensitivity and resolution and enable detection of NMR chemical shifts and NOE signals for the specific nucleotides of interest [409]. Although the 1H-13C signals were detectable for the regions of the leader undergoing rapid rotational motions (e.g., the 3′-terminal AUG hairpin), they could not be detected for residues in larger, more globular regions of the RNA undergoing more restricted rotational motions (e.g., the AUG residues when base paired with the upstream U5 element). Thus, although the 1H-13C correlated NMR methods that are widely used to study protein structure were able to characterize smaller, structurally independent regions of the leader, they had a limited utility for the characterization of large globular regions. For example, 1H-13C correlated NMR methods showed that the AUG residues adopt a hairpin in the monomer and an alternate (undetermined) structure in the dimer, likely due to their participation in a larger, globular structure in the dimer.
In contrast, the signals observed for the adenosine-H2 protons in highly deuterated samples exhibited good resolution and sensitivity for a majority of the adenosines, enabling a more complete analysis of the RNA structure involving these residues (and nearby protonated nucleotides). Initial studies focused on the non-capped 2G HIV-1NL4-3 leader that adopts a stable dimer structure under physiological-like conditions, and the NMR studies confirmed several aspects of secondary structures that were proposed on the basis of chemical probing and phylogenetic analyses. For example, signals in the 2H-edited NOESY NMR spectra obtained for the dimeric leader, were consistent with a structure in which the residues of TAR and polyA form hairpins and residues of AUG base pair, with an upstream U5 segment.
Attempts to study the monomeric form of the 2G HIV-1NL4-3 leader were confounded by the inability to obtain homogeneous samples of monomeric species, at the high concentrations required for NMR measurements. Nevertheless, studies of samples with measurable populations of the monomer revealed that residues of AUG form a hairpin, and residues of the DIS palindrome form base pairs with U5, in the monomeric form of the HIV-1NL4-3 leader. Based on the observation of A-H2 signals that are unambiguously diagnostic of the DIS hairpin, those studies suggested that the DIS retains its hairpin structure in the monomer. Subsequent studies shifted focus to the leader of HIV-1MAL because the samples containing higher percentages of the monomer could be obtained. These studies clearly showed that the DIS does not adopt a hairpin structure in the monomer, and it now appears that the DIS hairpin signals observed in the HIV-1NL4-3 spectra were associated with an equilibrium presence of the dimeric species. The NMR data clearly showed that the monomeric form of the HIV-1MAL leader adopted a structure in which the DIS palindrome formed base pairs with U5, as observed by NMR for the NL4-3 leader, and the remaining residues form long-range base pairs with residues of the upstream polyA and U5 elements. This topology has similarities to the “Long Distance Interactive” (LDI) model proposed by Abbink and Berkhout (although aspects of the LDI and NMR-derived models differ). NMR also helped shed light on the factors that modulate RNA dimerization and thereby modulate the transcript function and fate. 5′-Capped transcripts predominantly begin with either one or three guanosines, resulting from “twinned” start site usage. Transcripts that begin with a single guanosine, adopt a dimeric structure that sequesters the 5′-cap and exposes high-affinity NC binding sites, whereas 5′-capped transcripts that begin with two (less common) or three guanosines, form monomers that are substantially remodeled, compared to the dimeric RNA. The energetics are exquisitely balanced, with remodeling occurring at the energetic cost of a single G-C base pair (~3 kCal/mol).
Future studies of retroviral packaging mechanisms will benefit from studies of ever larger RNAs and protein–RNA complexes, including spliced RNA transcripts and unspliced transcripts bound to Gag, both within the cytoplasm and associated with the plasma membrane; Figure 20. Confocal imaging studies indicate that the ribonucleoprotein complex targeted to the plasma membrane assembly sites comprise about a dozen Gag proteins bound to the RNA genome, and this complex is too large to study using the presently available solution-based NMR methods. Solution-state NMR studies of larger complexes would require the development of improved methodologies for enhancing sensitivity and resolution, in addition to the development of improved labeling methods for rapid and economical preparation of segment- and site-specifically labeled RNAs. In this regard, Wang et al. developed a method for site-specific isotopic labeling of RNAs [410], but this potentially powerful technology requires specialized equipment and is yet to be widely adopted. Expanded use of 1H-15N correlated NMR methods for detecting the H2 protons of 15N-labeled adenosines [411] (e.g., for detection of pseudo-contact shifts in RNAs with site-specifically bound proteins that contain paramagnetic tags) should prove useful for studies of larger systems. The continued development of hybrid approaches that combine high-resolution, short-range NMR restraints, with lower resolution global structural restraints provided by cryo-EM or SAXS, could lead to the derivation of more accurate structures for systems of greater size and complexity. In addition, solid-state NMR methods could prove useful for studying large protein-RNA assemblies, if appropriately homogeneous samples can be prepared.
Figure 20.
Open questions regarding the structural biology of HIV-1 genome packaging. Structures of spliced mRNAs that avoid packaging are unknown, and structures of gRNA-Gag complexes that anchor the genome to the plasma membrane and nucleate virus assembly are also unknown.
In summary, NMR contributed considerably to our understanding of the atomic-level structural determinants of retroviral genome selection and packaging, more so than any other structural methodology. Since the first reported NMR structure of a retroviral zinc knuckle 30 years ago, all 3D structural information for retroviral zinc knuckle domains, NC proteins, and NC–RNA complexes were provided by NMR. Challenges associated with the crystallization of structurally flexible polycationic proteins such as NC, or structurally heterogeneous RNA polyanions, makes NMR an ideal methodology for studying retroviral RNA packaging. The NMR techniques developed over the course of these studies should be broadly applicable to the rapidly expanding field of non-coding cellular RNAs. As questions about genome packaging shift to larger systems, new approaches involving hybrid methodologies, advancements in labeling techniques, and possibly solid-state NMR will likely make important future contributions.
Acknowledgments
We thank Cindy Finch and Holly Summers (Howard Hughes Medical Institute at the University of Maryland, Baltimore County) for assistance with preparation of the manuscript.
Author Contributions
All authors contributed to the writing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by research grants from the NIH (National Institute of Allergy and Infectious Diseases [NIAID] Grant 8R01 AI150498 to M.F.S. and NIAID Grant U54 AI150470 to J.M.). J.D.B. was supported by NIH predoctoral fellowship F31 GM123803, and J.D.B., J.B.B., C.S., P.S.B. were supported by an NIH grant that promotes doctoral diversity (IMSD 5R25 GM055036-24), and S.Y. was supported by an NIH T32 training grant (5T32 HL007698-26).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Berkowitz R., Fisher J., Goff S.P. RNA packaging. Curr. Top. Microbiol. Immun. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 2.Rein A. Retroviral RNA packaging: A review. Arch. Virol. 1994;9:513–522. doi: 10.1007/978-3-7091-9326-6_49. [DOI] [PubMed] [Google Scholar]
- 3.Greatorex J., Lever A. Retroviral RNA dimer linkage. J. Gen. Virol. 1998;79:2877–2882. doi: 10.1099/0022-1317-79-12-2877. [DOI] [PubMed] [Google Scholar]
- 4.Paillart J.-C., Marquet R., Skripkin E., Ehresmann C., Ehresmann B. Dimerization of retroviral genomic RNAs: Structural and functional implications. Biochimie. 1996;78:639–653. doi: 10.1016/S0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 5.Jewell N.A., Mansky L.M. In the beginning: Genome recognition, RNA encapsidation and the initiation of complex retrovirus assembly. J. Gen. Virol. 2000;81:1889–1899. doi: 10.1099/0022-1317-81-8-1889. [DOI] [PubMed] [Google Scholar]
- 6.Paillart J.-C., Shehu-Xhilaga M., Marquet R., Mak J. Dimerization of retroviral RNA genomes: An inseparable pair. Nat. Rev. Microbiol. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 7.Russell R.S., Liang C., Wainberg M.A. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology. 2004;1:23. doi: 10.1186/1742-4690-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greatorex J. The retroviral RNA dimer linkage: Different structures may reflect different roles. Retrovirology. 2004;1:22. doi: 10.1186/1742-4690-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Souza V., Summers M.F. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 10.Johnson S.F., Telesnitsky A. Retroviral RNA dimerization and packaging: The what, how, when, where, and why. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzembayeva M., Dilley K., Sardo L., Hu W.-S. Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology. 2014;454:362–370. doi: 10.1016/j.virol.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu K., Heng X., Summers M.F. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 2011;410:609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellmund C., Lever A.M. Coordination of genomic RNA packaging with viral assembly in HIV-1. Viruses. 2016;8:192. doi: 10.3390/v8070192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mailler E., Bernacchi S., Marquet R., Paillart J.C., Vivet-Boudou V., Smyth R.P. The life-cycle of the HIV-1 Gag-RNA complex. Viruses. 2016;8:248. doi: 10.3390/v8090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maldonado R.K., Parent L.J. Orchestrating the selection and packaging of genomic RNA by retroviruses: An ensemble of viral and host factors. Viruses. 2016;8:257. doi: 10.3390/v8090257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bieniasz P., Telesnitsky A. Multiple, Switchable protein: RNA interactions regulate human immunodeficiency virus type 1 assembly. Annu. Rev. Virol. 2018;5:165–183. doi: 10.1146/annurev-virology-092917-043448. [DOI] [PubMed] [Google Scholar]
- 17.Rein A. RNA Packaging in HIV. Trends Microbiol. 2019;27:715–723. doi: 10.1016/j.tim.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman J.L., Butcher E.W., Patel D.T., Mikhaylenko Y., Summers M.F. Flexibility in the P2 domain of the HIV-1 Gag polyprotein. Protein Sci. 2004;13:2101–2107. doi: 10.1110/ps.04614804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright E.R., Schooler J.B., Ding H.J., Kieffer C., Fillmore C., Sundquist W.I., Jensen G.J. Electron crytomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schur F.K., Hagen W.J., Rumlova M., Ruml T., Muller B., Krausslich H.G., Briggs J.A. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nature. 2015;517:505–508. doi: 10.1038/nature13838. [DOI] [PubMed] [Google Scholar]
- 21.Schur F.K., Obr M., Hagen W.J., Wan W., Jakobi A.J., Kirkpatrick J.M., Sachse C., Krausslich H.G., Briggs J.A. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science. 2016;353:506–508. doi: 10.1126/science.aaf9620. [DOI] [PubMed] [Google Scholar]
- 22.Wagner J.M., Zadrozny K.K., Chrustowicz J., Purdy M.D., Yeager M., Ganser-Pornillos B.K., Pornillos O. Crystal structure of an HIV assembly and maturation switch. ELife. 2016;5:5. doi: 10.7554/eLife.17063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker P.J., Siddell S.G., Lefkowitz E.J., Mushegian A.R., Dempsey D.M., Dutilh B.E., Harrach B., Harrison R.L., Hendrickson R.C., Junglen S., et al. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2019;164:2417–2429. doi: 10.1007/s00705-019-04306-w. [DOI] [PubMed] [Google Scholar]
- 24.Rizvi T.A., Panganiban A.T. Simian immunodeficiency Virus-Rna is efficiently encapsidated by human-immunodeficiency-virus type-1 particles. J. Virol. 1993;67:2681–2688. doi: 10.1128/JVI.67.5.2681-2688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin P.D., Hu W.-S. RNAs from genetically distinct retroviruses can copackage and exchange genetic information in vivo. J. Virol. 1997;71:6237–6242. doi: 10.1128/JVI.71.8.6237-6242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaye J.F., Lever A.M. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: A possible role for the p2 domain of gag in RNA encapsidation. J. Virol. 1998;72:5877–5885. doi: 10.1128/JVI.72.7.5877-5885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berkowitz R.D., Ohagen A., Hoglund S., Goff S.P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 1995;69:6445–6456. doi: 10.1128/JVI.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J. Virol. 1995;69:5716–5722. doi: 10.1128/JVI.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jouvenet N., Simon S.M., Bieniasz P.D. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. USA. 2009;106:19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J., Rahman S.A., Nikolaitchik O.A., Grunwald D., Sardo L., Burdick R.C., Plisov S., Liang E., Tai S., Pathak V.K., et al. HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein. Proc. Natl. Acad. Sci. USA. 2016;113:E201–E208. doi: 10.1073/pnas.1518572113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jouvenet N., Bieniasz P.D., Simon S.M. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganser-Pornillos B.K., Yeager M., Sundquist W.I. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundquist W.I., Krausslich H.G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levin J.G., Grimley P.M., Ramseur J.M., Berezesky I.K. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J. Virol. 1974;14:152–161. doi: 10.1128/JVI.14.1.152-161.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poon D.T.K., Wu J., Aldovini A. Charged amino acid residues of human immunodeficiency virus type-1 nucleocapsid P7 protein involved in RNA pacakaging and infectivity. J. Virol. 1996;70:6607–6617. doi: 10.1128/JVI.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cimarelli A., Sandin S., Hoglund S., Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 2000;74:3046–3057. doi: 10.1128/JVI.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S.-W., Aldovini A. RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J. Virol. 2002;76:11853–11865. doi: 10.1128/JVI.76.23.11853-11865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muriaux D., Mirro J., Harvin D., Rein A. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA. 2001;98:5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muriaux D., Mirro J., Nagashima K., Harvin D., Rein A. Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J. Virol. 2002;76:11405–11413. doi: 10.1128/JVI.76.22.11405-11413.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heng X., Kharytonchyk S., Garcia E.L., Lu K., Divakaruni S.S., LaCotti C., Edme K., Telesnitsky A., Summers M.F. Identification of a minimal region of the HIV-1 5’-leader required for RNA dimerization, NC binding, and packaging. J. Mol. Biol. 2012;417:224–239. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keane S.C., Heng X., Lu K., Kharytonchyk S., Ramakrishnan V., Carter G., Barton S., Hosic A., Florwick A., Santos J., et al. Structure of the HIV-1 RNA packaging signal. Science. 2015;348:917–921. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kharytonchyk S., Brown J.D., Stilger K., Yasin S., Iyer A.S., Collins J., Summers M.F., Telesnitsky A. Influence of gag and RRE sequences on HIV-1 RNA packaging signal structure and function. J. Mol. Biol. 2018;430:2066–2079. doi: 10.1016/j.jmb.2018.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogt V.M. Retroviral virions and genomes. In: Coffin J.M., Hughes S.H., Varmus H.E., editors. Retroviruses. Volume 1. Cold Spring Harbor Laboratory Press; Plainview, NY, USA: 1997. pp. 27–69. [PubMed] [Google Scholar]
- 44.Hu W.-S., Temin H.M. Genetic consequences of packaging two RNA genomes in one retroviral particle: Pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu W.S., Temin H.M. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 46.Onafuwa-Nuga A., Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol. Mol. Biol. Rev. 2009;73:451–480. doi: 10.1128/MMBR.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bender W., Chien Y.-H., Chattopadhyay S., Vogt P.K., Gardner M.B., Davidson N. High-molecular-weight RNAs of AKR, NZB and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J. Virol. 1978;25:888–896. doi: 10.1128/JVI.25.3.888-896.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kung H.-J., Hu S., Bender W., Baily J.M., Davidson N., Nicolson M.O., McAllister R.M. RD-114, baboon and wolly monkey viral RNAs compared in size and structure. Cell. 1976;7:609–620. doi: 10.1016/0092-8674(76)90211-7. [DOI] [PubMed] [Google Scholar]
- 49.Maisel J., Bender W., Hu S., Duesberg P.H., Davidson N. Structure of 50 to 70S RNA from Moloney sarcoma viruses. J. Virol. 1978;25:384–394. doi: 10.1128/JVI.25.1.384-394.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murti K.G., Bondurant M., Tereba A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J. Virol. 1981;37:411–419. doi: 10.1128/JVI.37.1.411-419.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu W., Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 1993;67:5443–5449. doi: 10.1128/JVI.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oritz-Conde B.A., Hughes S.H. Studies of the genomic RNA of leukosis viruses: Implications for RNA dimerization. J. Virol. 1999;73:7165–7174. doi: 10.1128/JVI.73.9.7165-7174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flynn J.A., Telesnitsky A. Two distinct Moloney murine leukemia virus RNAs produced from a single locus dimerize at random. Virology. 2006;344:391–400. doi: 10.1016/j.virol.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moore M.D., Nikolaitchik O.A., Chen J., Hammarskjold M.L., Rekosh D., Hu W.S. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 2009;5:e1000627. doi: 10.1371/journal.ppat.1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coffin J.M., Hughes S.H., Varmus H.E. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview, NY, USA: 1997. [PubMed] [Google Scholar]
- 56.Lever A.M. HIV-1 RNA packaging. Adv. Pharmacol. 2007;55:1–32. doi: 10.1016/S1054-3589(07)55001-5. [DOI] [PubMed] [Google Scholar]
- 57.Adam M.A., Miller A.D. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J. Virol. 1988;62:3802–3806. doi: 10.1128/JVI.62.10.3802-3806.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mougel M., Barklis E. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J. Virol. 1997;71:8061–8065. doi: 10.1128/JVI.71.10.8061-8065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aronoff R., Linial M. Specificity of retroviral RNA packaging. J. Virol. 1991;65:71–80. doi: 10.1128/JVI.65.1.71-80.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aronoff R., Hajjar A.M., Linial M.L. Avian retroviral RNA encapsidation: Reexamination of functional 5’ RNA sequences and the role of nucleocapsid Cys-His motifs. J. Virol. 1993;67:178–188. doi: 10.1128/JVI.67.1.178-188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Banks J.D., Yeo A., Green K., Cepeda F., Linial M.L. A minimal avian retroviral packaging sequence has a complex structure. J. Virol. 1998;72:6190–6194. doi: 10.1128/JVI.72.7.6190-6194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doria-Rose N.A., Vogt V.M. In vivo selection of Rous sarcoma virus mutants with randomized sequences in the packaging signal. J. Virol. 1998;72:8073–8082. doi: 10.1128/JVI.72.10.8073-8082.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banks J.D., Kealoha B.O., Linial M.L. An MY containing heterologous RNA, but not env mRNA, is efficiently packaged into avian retroviral particles. J. Virol. 1999;73:8926–8933. doi: 10.1128/JVI.73.11.8926-8933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Banks J.D., Linial M.L. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J. Virol. 2000;74:456–464. doi: 10.1128/JVI.74.1.456-464.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prats A.-C., Roy C., Wang P., Erard M., Housset V., Gabus C., Paoletti C., Darlix J.-L. Cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J. Virol. 1990;64:774–783. doi: 10.1128/JVI.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.D’Souza V., Summers M.F. Structural basis for packaging the dimeric genome of Moloney Murine Leukaemia Virus. Nature. 2004;431:586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 67.Miyazaki Y., Garcia E., King S.R., Iyalla K., Loeliger K., Starck P., Syed S., Telesnitsky A., Summers M.F. An RNA structural switch regulates diploid genome packaging by Moloney Murine leukemia virus. J. Mol. Biol. 2010;396:141–152. doi: 10.1016/j.jmb.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gherghe C., Lombo T., Leonard C.W., Datta S.A., Bess J.W., Gorelick R.J., Rein A., Weeks K.M. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc. Natl. Acad. Sci. USA. 2010;107:19248–19253. doi: 10.1073/pnas.1006897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Darlix J.-L., Gabus C., Nugeyre M.-T., Clavel F., Barre-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 70.Ooms M., Huthoff H., Russell R., Liang C., Berkhout B. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 2004;78:10814–10819. doi: 10.1128/JVI.78.19.10814-10819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu K., Heng X., Garyu L., Monti S., Garcia E., Kharytonchyk S., Dorjsuren B., Kulandaivel G., Jones S., Hiremath A., et al. NMR detection of structures in the HIV-1 5´-leader RNA that regulate genome packaging. Science. 2011;344:242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Masuda T., Sato Y., Huang Y.L., Koi S., Takahata T., Hasegawa A., Kawai G., Kannagi M. Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci. Rep. 2015;5:17680. doi: 10.1038/srep17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kharytonchyk S., Monti S., Smaldino P.J., Van V., Bolden N.C., Brown J.D., Russo E., Swanson C., Shuey A., Telesnitsky A., et al. Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc. Natl. Acad. Sci. USA. 2016;113:13378–13383. doi: 10.1073/pnas.1616627113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Esquiaqui J.M., Kharytonchyk S., Drucker D., Telesnitsky A. HIV-1 spliced RNAs display transcription start site bias. RNA. 2020;26:708–714. doi: 10.1261/rna.073650.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brown J.D., Kharytonchyk S., Chaudry I., Iyer A.S., Carter H., Becker G., Desai Y., Glang L., Choi S.H., Singh K., et al. Structural basis for transcriptional start site control of HIV-1 RNA fate. Science. 2020;368:413–417. doi: 10.1126/science.aaz7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Turner B.G., Summers M.F. Structural Biology of HIV. J. Mol. Biol. 1999;285:1–32. doi: 10.1006/jmbi.1998.2354. [DOI] [PubMed] [Google Scholar]
- 77.Wüthrich K. NMR of Proteins and Nucleic Acids. John Wiley & Sons; New York, NY, USA: 1986. [Google Scholar]
- 78.Allain F.H.-T., Varani G. How accurately and precisely can RNA structure be determined by NMR? J. Mol. Biol. 1997;267:338–351. doi: 10.1006/jmbi.1996.0855. [DOI] [PubMed] [Google Scholar]
- 79.Lukavsky P.J., Puglisi J.D. Structure determination of large biological RNAs. Methods Enzymol. 2005;394:399–415. doi: 10.1016/S0076-6879(05)94016-0. [DOI] [PubMed] [Google Scholar]
- 80.Lu K., Miyazaki Y., Summers M.F. Isotope labeling strategies for NMR studies of RNA. J. Biomol. NMR. 2010;46:113–125. doi: 10.1007/s10858-009-9375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kotar A., Foley H.N., Baughman K.M., Keane S.C. Advanced approaches for elucidating structures of large RNAs using NMR spectroscopy and complementary methods. Methods. 2020 doi: 10.1016/j.ymeth.2020.01.009. [DOI] [PubMed] [Google Scholar]
- 82.Summers M.F., Henderson L.E., Chance M.R., Bess J.W.J., South T.L., Blake P.R., Sagi I., Perez-Alvarado G., Sowder R.C.I., Hare D.R., et al. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Henderson L.E., Copeland T.D., Sowder R.C., Smythers G.W., Oroslzan S. Primary structure of the low molecular weight nucleic acid-binding proteins of murine leukemia viruses. J. Biol. Chem. 1981;256:8400–8406. [PubMed] [Google Scholar]
- 84.Miller J., McLachlan A.D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berg J.M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232:485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- 86.Schiff L.A., Nibert M.L., Fields B.N. Characterization of a zinc blotting technique: Evidence that a retroviral gag protein binds zinc. Proc. Natl. Acad. Sci. USA. 1988;85:4195–4199. doi: 10.1073/pnas.85.12.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith L.M., Jentoft J.E. Conserved cysteine and histidine residues in avian myeloblastosis virus nucleocapsid protein pp12 are not zinc binding ligands. Biophys. J. 1988;53:295a. doi: 10.1073/pnas.85.19.7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gorelick R.J., Henderson L.E., Hanser J.P., Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: Evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. USA. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Méric C., Goff S.P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J. Virol. 1989;63:1558–1568. doi: 10.1128/JVI.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Green L.M., Berg J.M. A retroviral Cys-Xaa2-Cys-Xaa4-His-Xaa4-Cys peptide binds metal ions: Spectroscopic studies and a proposed three-dimensional structure. Proc. Natl. Acad. Sci. USA. 1989;86:4047–4051. doi: 10.1073/pnas.86.11.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.South T.L., Kim B., Summers M.F. 113Cd NMR studies of a 1:1 Cd adduct with an 18-residue zinc finger peptide from HIV-1 nucleic acid binding protein, p7. J. Am. Chem. Soc. 1989;111:395–396. doi: 10.1021/ja00183a074. [DOI] [Google Scholar]
- 92.Summers M.F., South T.L., Kim B., Hare D.R. High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach. Biochemistry. 1990;29:329–340. doi: 10.1021/bi00454a005. [DOI] [PubMed] [Google Scholar]
- 93.South T.L., Blake P.R., Sowder R.C., 3rd, Arthur L.O., Henderson L.E., Summers M.F. The nucleocapsid protein isolated from HIV-1 particles binds zinc and forms retroviral-type zinc fingers. Biochemistry. 1990;29:7786–7789. doi: 10.1021/bi00486a002. [DOI] [PubMed] [Google Scholar]
- 94.Chance M.R., Sagi I., Wirt M.D., Frisbie S.M., Scheuring E., Chen E., Bess J.W., Henderson L.E., Arthur L.O., South T.L. Extended x-ray absorption fine structure studies of a retrovirus: Equine infectious anemia virus cysteine arrays are coordinated to zinc. Proc. Natl. Acad. Sci. USA. 1992;89:10041–10045. doi: 10.1073/pnas.89.21.10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.South T.L., Blake P.R., Hare D.R., Summers M.F. C-terminal retroviral-type zinc finger domain from the HIV-1 nucleocapsid protein is structurally similar to the N-terminal zinc finger domain. Biochemistry. 1991;30:6342–6349. doi: 10.1021/bi00239a036. [DOI] [PubMed] [Google Scholar]
- 96.Omichinski J.G., Clore G.M., Sakaguchi K., Appella E., Gronenborn A.M. Structural characterization of a 39-residue synthetic peptide containing the two zinc binding domains from the HIV-1 p7 nucleocapsid protein by CD and NMR spectroscopy. Fed. Eur. Biochem. Soc. Lett. 1991;292:25–30. doi: 10.1016/0014-5793(91)80825-n. [DOI] [PubMed] [Google Scholar]
- 97.Déméné H., Dong C.Z., Ottmann M., Rouyez M.C., Jullian N., Morellet N., Mely Y., Darlix J.L., Fournié-Zaluski M.C., Saragosti S., et al. 1H NMR structure and biological studies of the His23 to Cys mutant nucleocapsid protein of HIV-1 indicate that the conformation of the first zinc finger is critial for virus infectivity. Biochemistry. 1994;33:11707–11716. doi: 10.1021/bi00205a006. [DOI] [PubMed] [Google Scholar]
- 98.Klein D., Johnson P.E., Zollars E.S., de Guzman R.N., Summers M.F. The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle. Biochemistry. 2000;39:1604–1612. doi: 10.1021/bi9922493. [DOI] [PubMed] [Google Scholar]
- 99.Bertola F., Manigand C., Picard P., Belghazi M., Precigoux G. Human T-lymphotrophic virus type I nucleocapsid protein NCp15: Structural study and stability of the N-terminal zinc-finger. Biochem. J. 2000;352:293–300. doi: 10.1042/bj3520293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mély Y., Piémont E., Sorinas-Jimeno M., de Rocquigny H., Julian N., Morellet N., Roques B.P., Gérard D. Structural and dynamic characterization of the aromatic amino acids of the human immunodeficiency virus type 1 nucleocapsid protein zinc fingers and their involvement in heterologous tRNAPhe binding: A steady-state and time resolved flourescence study. Biophys. J. 1993;65:1513–1522. doi: 10.1016/S0006-3495(93)81222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Déméné H., Jullian N., Morellet N., de Rocquigny H., Cornille F., Maigret B., Roques B.P. Three-dimensional 1H NMR structure of the nucleocapsid protein NCp10 of Moloney murine leukemia virus. J. Biomol. NMR. 1994;4:153–170. doi: 10.1007/BF00175244. [DOI] [PubMed] [Google Scholar]
- 102.Morellet N., Jullian N., de Rocquigny H., Maigret B., Darlix J.-L., Roques B.P. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Morellet N., Meudal H., Bouaziz S., Roques B.P. Structure of the zinc finger domain encompassing residues 13–51 of the nucleocapsid protein from simian immunodeficiency virus. Biochem. J. 2006;393:725–732. doi: 10.1042/BJ20051203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kodera Y., Sato K., Tsukahara T., Ksmatsu H., Maeda T., Kohno T. High-resolution solution NMR structure of the minimal active domain of the human immunodeficiency virus type-2 nucleocapsid protein. Biochemistry. 1998;37:17704–17713. doi: 10.1021/bi981818o. [DOI] [PubMed] [Google Scholar]
- 105.Gao Y., Boyd J., Pielak G.J., Williams R.J.P. Proton nuclear magnetic resonance as a probe of differences in structure between the C102T and F82S, C102T Variants of Iso-1-cytochrome c from the Yeast Saccharomyces cerevisiae. Biochemistry. 1991;30:7033–7040. doi: 10.1021/bi00242a032. [DOI] [PubMed] [Google Scholar]
- 106.Gao Y., Kaluarachchi K., Giedroc D.P. Solution structure and backbone dynamics of Mason-Pfizer monkey virus (MPMV) nucleocapsid protein. Protein Sci. 1998;7:2265–2280. doi: 10.1002/pro.5560071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.South T.L., Summers M.F. Zinc- and sequence-dependent binding to nucleic acids by the N-terminal zinc finger of the HIV-1 nucleocapsid protein: NMR structure of the complex with the Psi-site analog, dACGCC. Protein Sci. 1993;2:3–19. doi: 10.1002/pro.5560020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.De Guzman R.N., Turner R.B., Summers M.F. Protein-RNA recognition. Biopolymers. 1999;48:181–195. doi: 10.1002/(SICI)1097-0282(1998)48:2<181::AID-BIP7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 109.De Guzman R.N., Wu Z.R., Stalling C.C., Pappalardo L., Borer P.N., Summers M.F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Y-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 110.Amarasinghe G.K., De Guzman R.N., Turner R.B., Chancellor K., Wu Z.-R., Summers M.F. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the Y-RNA packaging signal. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 111.Morellet N., Demene H., Teilleux V., Huynh-Dinh T., de Rocquigny H., Fournie-Zaluski M.C., Roques B.P. Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC) J. Mol. Biol. 1998;283:419–434. doi: 10.1006/jmbi.1998.2098. [DOI] [PubMed] [Google Scholar]
- 112.Zhou J., Bean R.L., Vogt V.M., Summers M.F. Solution structure of the Rous sarcoma virus nucleocapsid protein:uY RNA packaging signal complex. J. Mol. Biol. 2007;365:453–467. doi: 10.1016/j.jmb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dey A., York D., Smalls-Mantey A., Summers M.F. Composition and sequence dependent binding of RNA to the nucleocapsid protein of Moloney Murine Leukemia Virus. Biochemistry. 2005;44:3735–3744. doi: 10.1021/bi047639q. [DOI] [PubMed] [Google Scholar]
- 114.Spriggs S., Garyu L., Connor R., Summers M.F. Potential intra- and intermolecular interactions involving the Unique-5′ Region of the HIV-1 5′-UTR. Biochemistry. 2008;46:13064–13073. doi: 10.1021/bi8014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee B.M., de Guzman R.N., Turner B.G., Tjandra N., Summers M.F. Dynamical behavior of the HIV-1 nucleocapsid protein. J. Mol. Biol. 1998;279:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- 116.Deshmukh L., Ghirlando R., Clore G.M. Conformation and dynamics of the Gag polyprotein of the human immunodeficiency virus 1 studied by NMR spectroscopy. Proc. Natl. Acad. Sci. USA. 2015;112:3374–3379. doi: 10.1073/pnas.1501985112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Matthews S., Barlow P., Boyd J., Barton G., Russell R., Mills H., Cunningham M., Meyers N., Burns N., Clark N., et al. Structural similarity between the p17 matrix protein of HIV-1 and interferon-g. Nature. 1994;370:666–668. doi: 10.1038/370666a0. [DOI] [PubMed] [Google Scholar]
- 118.Tang C., Ndassa Y., Summers M.F. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 2002;9:537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- 119.Tang C., Loeliger E., Luncsford P., Kinde I., Beckett D., Summers M.F. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA. 2004;101:517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Saad J.S., Loeliger E., Luncsford P., Liriano M., Tai J., Kim A., Miller J., Joshi A., Freed E.O., Summers M.F. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J. Mol. Biol. 2007;366:574–585. doi: 10.1016/j.jmb.2006.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Saad J.S., Ablan S.D., Ghanam R.H., Kim A., Andrews K., Nagashima K., Soheilian F., Freed E.O., Summers M.F. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J. Mol. Biol. 2008;382:434–447. doi: 10.1016/j.jmb.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mercredi P.Y., Bucca N., Loeliger B., Gaines C.R., Mehta M., Bhargava P., Tedbury P.R., Charlier L., Floquet N., Muriaux D., et al. Structural and molecular determinants of membrane binding by the HIV-1 matrix protein. J. Mol. Biol. 2016;428:1637–1655. doi: 10.1016/j.jmb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saad J.S., Miller J., Tai J., Kim A., Ghanam R.H., Summers M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Saad J.S., Kim A., Ghanam R.H., Dalton A.K., Vogt M.V., Wu Z., Lu W., Summers M.F. Mutations that mimic phosphorylation of the HIV-1 matrix protein: Implications for trafficking. Protein Sci. 2007;16:1793–1797. doi: 10.1110/ps.072987607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Murphy R.E., Smal A.B., Blach J., Mas V., Prevelige P.E., Saad J.S. Structural and biophysical characterizations of HIV-1 matrix trimer binding to lipid nanodiscs shed light on virus assembly. J. Biol. Chem. 2019;294:18600–18612. doi: 10.1074/jbc.RA119.010997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gaines C.R., Tkacik E., Rivera-Oven A., Somani P., Achimovich A., Alabi T., Zhu A., Getachew N., Yang A.L., McDonough M., et al. HIV-1 Matrix protein interactions with tRNA: Implications for membrane targeting. J. Mol. Biol. 2018;430:2113–2127. doi: 10.1016/j.jmb.2018.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jouvenet N., Simon S.M., Bieniasz P.D. Visualizing HIV-1 assembly. J. Mol. Biol. 2011;410:501–511. doi: 10.1016/j.jmb.2011.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Freed E.O. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015;13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Carlson L.A., Briggs J.A., Glass B., Riches J.D., Simon M.N., Johnson M.C., Muller B., Grunewald K., Krausslich H.G. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Massiah M.A., Starich M.R., Paschall C., Summers M.F., Christensen A.M., Sundquist W.I. Three dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 131.Matthews S., Barlow P., Clark N., Kingsman S., Kingsman A., Campbell I. Refined solution structure of p17, the HIV matrix protein. Biochem. Soc. Trans. 1995;23:725–728. doi: 10.1042/bst0230725. [DOI] [PubMed] [Google Scholar]
- 132.Hill C.P., Worthylake D., Bancroft D.P., Christensen A.M., Sundquist W.I. Crystal Structures of the Trimeric HIV-1 Matrix Protein: Implications for Membrane Association. Proc. Natl. Acad. Sci. USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Massiah M.A., Worthylake D., Christensen A.M., Sundquist W.I., Hill C.P., Summers M.F. Comparison of the NMR and X-ray structures of the HIV-1 matrix protein: Evidence for conformational changes during viral assembly. Protein Sci. 1996;5:2391–2398. doi: 10.1002/pro.5560051202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Freed E.O., Martin A.M. Domains of the human immonodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 1996;70:341–351. doi: 10.1128/JVI.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tedbury P.R., Novikova M., Alfadhli A., Hikichi Y., Kagiampakis I., KewalRamani V.N., Barklis E., Freed E.O. HIV-1 Matrix trimerization-impaired mutants are rescued by matrix substitutions that enhance envelope glycoprotein incorporation. J. Virol. 2019;94:1. doi: 10.1128/JVI.01526-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tedbury P.R., Novikova M., Ablan S.D., Freed E.O. Biochemical evidence of a role for matrix trimerization in HIV-1 envelope glycoprotein incorporation. Proc. Natl. Acad. Sci. USA. 2016;113:E182–E190. doi: 10.1073/pnas.1516618113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Freed E.O., Englund G., Martin A.M. Role of the basic domain of human immondeficiency virus type 1 matrix in macrophage infection. J. Virol. 1995;69:3949–3954. doi: 10.1128/JVI.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Deichaite I., Casson L.P., Ling H.P., Resh M.D. In vitro synthesis of pp60v-src: Myristylation in a cell-free system. Mol. Cell Biol. 1988;8:4295–4301. doi: 10.1128/MCB.8.10.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Brown L.A., Cox C., Baptiste J., Summers H., Button R., Bahlow K., Spurrier V., Kyser J., Luttge B.G., Kuo L., et al. NMR structure of the myristylated feline immunodeficiency virus matrix protein. Viruses. 2015;7:2210–2229. doi: 10.3390/v7052210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Brown J.B., Summers H.R., Brown L.A., Marchant J., Canova P.N., O’Hern C.T., Abbott S.T., Nyaunu C., Maxwell S., Johnson T., et al. Structural and Mechanistic studies of the rare myristoylation signal of the feline immunodeficiency virus. J. Mol. Biol. 2020;432:4076–4091. doi: 10.1016/j.jmb.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fledderman E.L., Fujii K., Ghanam R.H., Waki K., Prevelige P.E., Freed E.O., Saad J.S. Myristate exposure in the human immunodeficiency virus type 1 matrix protein is modulated by pH. Biochemistry. 2010;49:9551–9562. doi: 10.1021/bi101245j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ono A., Ablan S.D., Lockett S.J., Nagashima K., Freed E.O. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zimmerman C., Klein K.C., Kiser P.K., Singh A.R., Bonnie F.L., Riba S.C., Lingappa J.R. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature. 2002;415:88–92. doi: 10.1038/415088a. [DOI] [PubMed] [Google Scholar]
- 144.Behnia R., Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- 145.Brugger B., Glass B., Haberkant P., Leibrecht I., Wieland F.T., Krausslich H.G. The HIV lipidome: A raft with an unusual composition. Proc. Natl. Acad. Sci. USA. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nguyen D.H., Hildreth J.E. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 2000;74:3264–3272. doi: 10.1128/JVI.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vlach J., Saad J.S. Trio engagement via plasma membrane phospholipids and the myristoyl moiety governs HIV-1 matrix binding to bilayers. Proc. Natl. Acad. Sci. USA. 2013;110:3525–3530. doi: 10.1073/pnas.1216655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cannon P.M., Matthews S., Clark N., Byles E.D., Iourin O., Hockley D.J., Kingsman S., Kingsman A. Structure-Function studies of the Human immunodefeciency virus type 1 matrix protein, p17. J. Virol. 1997;71:3474–3483. doi: 10.1128/JVI.71.5.3474-3483.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kutluay S.B., Zang T., Blanco-Melo D., Powell C., Jannain D., Errando M., Bieniasz P.D. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell. 2014;159:1096–1109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bénas P., Bec G., Keith G., Marquet R., Ehresmann C., Ehresmann B., Dumas P. The crystal structure of HIV reverse-transcription primer tRNA(Lys,3) shows a canonical anticodon loop. RNA. 2000;6:1347–1355. doi: 10.1017/S1355838200000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kleiman L., Jones C., Musier-Forsyth K. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 2010;584:359–365. doi: 10.1016/j.febslet.2009.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kleiman L. tRNA(Lys3): The primer tRNA for reverse transcription in HIV-1. IUBMB Life. 2002;53:107–114. doi: 10.1080/15216540211469. [DOI] [PubMed] [Google Scholar]
- 153.Chukkapalli V., Oh S.J., Ono A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. USA. 2010;107:1600–1605. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gamble T.R., Yoo S., Vajdos F.F., von Schwedler U.K., Korthylake D.K., Wang H., McCutcheon J.P., Sundquist W.I., Hill C.P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 155.Pervushin K., Riek R., Wider G., Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Harrison G.P., Lever A.M.L. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J. Virol. 1992;66:4144–4153. doi: 10.1128/JVI.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Baudin F., Marquet R., Isel C., Darlix J.-L., Ehresmann B., Ehresmann C. Functional sites in the 5’ region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 158.Clever J., Sassetti C., Parslow T.G. RNA secondary structure and binding sites for gag gene products in the 5’ packaging signal of human immunodeficiency virus type 1. J. Virol. 1995;69:2101–2109. doi: 10.1128/JVI.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.McBride M.S., Panganiban A.T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 1996;70:2963–2973. doi: 10.1128/JVI.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Clever J.L., Miranda J.D., Parslow T.G. RNA structure and packaging signals in the 5’ leader region of the human immunodeficiency virus type 1 genome. J. Virol. 2002;76:12381–12387. doi: 10.1128/JVI.76.23.12381-12387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Abbink T.E.M., Berkhout B. A novel long distance base-pairing interaction in Human Immunodeficiency Virus Type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003;278:11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 162.Damgaard C.K., Andersen E.S., Knudsen B., Gorodkin J., Kjems J. RNA interactions in the 5’ region of the HIV-1 genome. J. Mol. Biol. 2004;336:369–379. doi: 10.1016/j.jmb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 163.Paillart J.C., Dettenhofer M., Yu X.-F., Ehresmann C., Ehresmann B., Marquet R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004;279:48397–48403. doi: 10.1074/jbc.M408294200. [DOI] [PubMed] [Google Scholar]
- 164.Wilkinson K.A., Gorelick R.J., Vasa S.M., Guex N., Rein A., Mathews D.H., Giddings M.C., Weeks K.M. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:883–899. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Watts J.M., Dang K.K., Gorelick R.J., Leonard C.W., Bess J.W., Jr., Swanstrom R., Burch C.L., Weeks K.M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Carroll R., Peterlin B.M., Derse D. Inhibition of human immunodeficiency virus type 1 Tat activity by coexpression of heterologous trans activators. J. Virol. 1992;66:2000–2007. doi: 10.1128/JVI.66.4.2000-2007.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Boeras I., Seufzer B., Brady S., Rendahl A., Heng X., Boris-Lawrie K. The basal translation rate of authentic HIV-1 RNA is regulated by 5’UTR nt-pairings at junction of R and U5. Sci. Rep. 2017;7:6902. doi: 10.1038/s41598-017-06883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sleiman D., Barraud P., Brachet F., Tisne C. The Interaction between tRNA(Lys) 3 and the primer activation signal deciphered by NMR spectroscopy. PLoS ONE. 2013;8:e64700. doi: 10.1371/journal.pone.0064700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sleiman D., Goldschmidt V., Barraud P., Marquet R., Paillart J.C., Tisne C. Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions. Virus Res. 2012;169:324–339. doi: 10.1016/j.virusres.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 170.Rhim H., Park J., Morrow C.D. Deletion in the tRNALys primer-binding site of human immunodeficiency virus type 1 identity essential regions for reverse transcription. J. Virol. 1991;65:4555–4564. doi: 10.1128/JVI.65.9.4555-4564.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Abbink T.E.M., Berkhout B. RNA structure modulates splicing efficiency at the human immunodeficiency virus type 1 major splice donor. J. Virol. 2008;82:3090–3098. doi: 10.1128/JVI.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Plank T.D., Whitehurst J.T., Cencic R., Pelletier J., Kieft J.S. Internal translation initiation from HIV-1 transcripts is conferred by a common RNA structure. Translation. 2014;2:e27694. doi: 10.4161/trla.27694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hayashi T., Shioda T., Iwakura Y., Shibuta H. RNA packaging signal of human immunodeficiency virus type 1. Virology. 1992;188:590–599. doi: 10.1016/0042-6822(92)90513-O. [DOI] [PubMed] [Google Scholar]
- 174.Hayashi T., Ueno Y., Okamoto T. Elucidation of a conserved RNA stem-loop structure in the packaging signal of human immunodeficiency virus type 1. FEBS Lett. 1993;327:213–218. doi: 10.1016/0014-5793(93)80172-Q. [DOI] [PubMed] [Google Scholar]
- 175.McBride M.S., Panganiban A.T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J. Virol. 1997;71:2050–2058. doi: 10.1128/JVI.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Harrison G.P., Miele G., Hunter E., Lever A.M.L. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J. Virol. 1998;72:5886–5896. doi: 10.1128/JVI.72.7.5886-5896.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Clever J.L., Parslow T.G. Mutant Human Immunodeficiency Virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 1997;71:3407–3414. doi: 10.1128/JVI.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Berkhout B. Structural features in TAR RNA of human and simian immunodeficiency viruses: A phylogenetic analysis. Nucleic Acids Res. 1992;20:27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Berkhout B., Klaver B., Das A.T. A conserved hairpin structure predicted for the poly(A) signal of human and simian immunodeficiency viruses. Virology. 1995;207:276–281. doi: 10.1006/viro.1995.1077. [DOI] [PubMed] [Google Scholar]
- 180.Skripkin E., Paillart J.C., Marquet R., Ehresmann B., Ehresmann C. Identification of the primary site of the human immunodefieiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nikolaitchik O., Rhodes T.D., Ott D., Hu W.-S. Effects of mutations in the Human Immunodeficiency Virus Type 1 gag gene on RNA packaging and recombination. J. Virol. 2006;80:4691–4697. doi: 10.1128/JVI.80.10.4691-4697.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Song R., Kafaie J., Laughrea M. Role of the 5’ TAR stem–loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA. Biochemistry. 2008;47:3283–3293. doi: 10.1021/bi7023173. [DOI] [PubMed] [Google Scholar]
- 183.Sakuragi J.-I., Shioda T., Panganiban A.T. Duplication of the primary encapsidation and dimer linkage region of Human immunodeficiency virus type 1 RNA results in the appearance of monomeric RNA in virions. J. Virol. 2001;75:2557–2565. doi: 10.1128/JVI.75.6.2557-2565.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Laughrea M., Jette L., Mak J., Kleiman L., Liang C., Wainberg M.A. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J. Virol. 1997;71:3397–3406. doi: 10.1128/JVI.71.5.3397-3406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Moore M.D., Fu W., Nikolaitchik O., Chen J., Ptak R.G., Hu W.-S. Dimer initiation signal of human immunodeficiency virus type 1: Its role in partner selection during RNA copackaging and its effects on recombination. J. Virol. 2007;81:4002–4011. doi: 10.1128/JVI.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shen N., Jette L., Liang C., Wainberg M.A., Laughrea M. Impact of human immunodeficiency virus type 1 RNA dimerization on viral infectivity and of stem-loop B on RNA dimerization and reverse transcription and dissociation of dimerization from packaging. J. Virol. 2000;74:5729–5735. doi: 10.1128/JVI.74.12.5729-5735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Paillart J.C., Berthoux L., Ottmann M., Darlix J.L., Marquet R., Ehresmann B., Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J. Virol. 1996;70:8348–8354. doi: 10.1128/JVI.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Laughrea M., Jette L. A 19-nucleotide sequence upstream of the 5’ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 189.Paillart J.C., Marquet R., Skripkin E., Ehresmann B., Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- 190.Muriaux D., de Rocquigny H., Roques B.P., Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J. Biol. Chem. 1996;271:33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 191.Fosse P., Motte N., Roumer A., Gabus C., Muriaux D., Darlix J.-L., Paoletti J. A short autocomplementary sequence plays an essential role in avian sarcoma-leukosis virus RNA dimerization. Biochemistry. 1996;35:16601–16609. doi: 10.1021/bi9613786. [DOI] [PubMed] [Google Scholar]
- 192.Song R., Kafaie J., Yang L., Laughrea M. HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. J. Mol. Biol. 2007;371:1084–1096. doi: 10.1016/j.jmb.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 193.Paillart J.-C., Skripkin E., Ehresmann B., Ehresmann C., Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Clever J.L., Wong M.L., Parslow T.G. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 1996;70:5902–5908. doi: 10.1128/JVI.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chen J., Nikolaitchik O., Singh J., Wright A., Bencsics C.E., Coffin J.M., Ni N., Lockett S., Pathak V.K., Hu W.S. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. USA. 2009;106:13535–13540. doi: 10.1073/pnas.0906822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Muriaux D., Girard P.-M., Bonnet-Mathoniere B., Paoletti J. Dimerization of HIV-1Lai RNA at low ionic strength: An autocomplementary sequence in the 5’ leader region is evidence dby an antisense oligonucleotide. J. Biol. Chem. 1995;270:8209–8216. doi: 10.1074/jbc.270.14.8209. [DOI] [PubMed] [Google Scholar]
- 197.Windbichler N., Werner M., Schroeder R. Kissing complex-mediated dimerisation of HIV-1 RNA: Coupling extended duplex formation to ribozyme cleavage. Nucleic Acids Res. 2003;31:6419–6427. doi: 10.1093/nar/gkg873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Laughrea M., Jette L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248–271 are dispensable for dimer formation. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 199.Laughrea M., Jette L. HIV-1 genome dimerization: Kissing-loop hairpin dictates whether nucleotides downstream of the 5’ splice junction contribute to loose and tight dimerization of human immunodeficiency virus RNA. Biochemistry. 1997;36:9501–9508. doi: 10.1021/bi970862l. [DOI] [PubMed] [Google Scholar]
- 200.Sakuragi S., Yokoyama M., Shioda T., Sato H., Sakuragi J.-I. SL1 revisited: Functional analysis of the structure and conformation of HIV-1 genome RNA. Retrovirology. 2016;13:1–13. doi: 10.1186/s12977-016-0310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Greatorex J., Gallego J., Varani G., Lever A. Structure and stability of wild-type and mutant RNA internal loops from the SL1 domain of the HIV-1 packaging signal. J. Mol. Biol. 2002;322:543–557. doi: 10.1016/S0022-2836(02)00776-3. [DOI] [PubMed] [Google Scholar]
- 202.Mujeeb A., Clever J.L., Billeci T.M., James T.L., Parslow T.G. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat. Struct. Biol. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 203.Ulyanov N.B., Mujeeb A., Du Z., Tonelli M., Parslow T.G., James T.L. NMR structure of the full-length linear dimer of stem-loop-1 RNA in the HIV-1 dimer initiation site. J. Biol. Chem. 2006;281:16168–16177. doi: 10.1074/jbc.M601711200. [DOI] [PubMed] [Google Scholar]
- 204.Zhang K., Keane S.C., Su Z., Irobalieva R.N., Chen M., Van V., Sciandra C.A., Marchant J., Heng X., Schmid M.F., et al. Structure of the 30 kDa HIV-1 RNA dimerization signal by a hybrid Cryo-EM, NMR, and molecular dynamics approach. Structure. 2018;26:490–498. doi: 10.1016/j.str.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kieken F., Paquet F., Brule F., Paoletti J., Lancelot G. A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer. Nucleic Acids Res. 2006;34:343–352. doi: 10.1093/nar/gkj427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Dardel F., Marquet R., Ehresmann C., Ehresmann B., Blanquet S. Solution studies of the dimerization initiation site of HIV-1 genomic RNA. Nucleic Acids Res. 1998;26:3567–3571. doi: 10.1093/nar/26.15.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Baba S., Takahashi K., Noguchi S., Takaku H., Koyanagi Y., Yamamoto N., Kawai G. Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers. J. Biochem. 2005;138:583–592. doi: 10.1093/jb/mvi158. [DOI] [PubMed] [Google Scholar]
- 208.Mihailescu M.-R., Marino J.P. A proton-coupled dynamic conformational switch in the HIV-1 dimerization initiation site kissing complex. Proc. Natl. Acad. Sci. USA. 2004;101:1189–1194. doi: 10.1073/pnas.0307966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lawrence D.C., Stover C.C., Noznitsky J., Wu Z.-R., Summers M.F. Structure of the intact stem and bulge of HIV-1 ψ-RNA stem loop SL1. J. Mol. Biol. 2003;326:529–542. doi: 10.1016/S0022-2836(02)01305-0. [DOI] [PubMed] [Google Scholar]
- 210.Ennifar E., Walter P., Ehresmann B., Ehresmann C., Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 2001;8:1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 211.Muriaux D., Fosse P., Paoletti J. A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry. 1996;35:5075–5082. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 212.Takahashi K.-I., Baba S., Hayashi Y., Koyanagi Y., Yamamoto N., Takaku H., Kawai G. NMR analysis of intra- and inter-molecular stems in dimerization initiation site. J. Biochem. 2000;127:681–686. doi: 10.1093/oxfordjournals.jbchem.a022657. [DOI] [PubMed] [Google Scholar]
- 213.Mujeeb A., Parslow T.G., Zarrinpar A., Das C., James T.L. NMR structure of the mature dimer initiation complex of HIV-1 genomic RNA. FEBS Lett. 1999;458:387–392. doi: 10.1016/S0014-5793(99)01183-7. [DOI] [PubMed] [Google Scholar]
- 214.Feng Y.-X., Copeland T.D., Henderson L.E., Gorelick R.J., Bosche W.J., Levin J.G., Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ennifar E., Yusupov M., Walter P., Marquet R., Ehresmann B., Ehresmann C., Dumas P. The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges. Structure. 1999;7:1439–1449. doi: 10.1016/S0969-2126(00)80033-7. [DOI] [PubMed] [Google Scholar]
- 216.Girard F., Barbault F., Gouyett C., Huynh-dinh T., Paoletti J., Lamcelot G. Dimer initiation sequence of HIV-1 Lai genomic RNA: NMR solution structure of the extended nucleus. J. Biomol. Struct. Dyn. 1999;16:1145–1157. doi: 10.1080/07391102.1999.10508323. [DOI] [PubMed] [Google Scholar]
- 217.Yuan Y., Kerwood D.J., Paoletti A.C., Shubsda M.F., Borer P.N. Stem of SL1 RNA in HIV-1: Structure and nucleocapsid protein binding for a 1 × 3 internal loop. Biochemistry. 2003;42:5259–5269. doi: 10.1021/bi034084a. [DOI] [PubMed] [Google Scholar]
- 218.Battiste J.L., Mao H., Rao N.S., Tan R., Muhandiram D.R., Kay L.E., Frankel A.D., Williamson J.R. Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 219.Peterson R.D., Feigon J. Structural change in Rev responsive element RNA of HIV-1 on binding Rev peptide. J. Mol. Biol. 1996;264:863–877. doi: 10.1006/jmbi.1996.0683. [DOI] [PubMed] [Google Scholar]
- 220.Sun X., Zhang Q., Al-Hashimi H.M. Resolving fast and slow motions in the internal loop containing stem-loop 1 of HIV-1 that are modulated by Mg2+ binding: Role in the kissing-duplex structural transition. Nucleic Acids Res. 2007;35:1698–1713. doi: 10.1093/nar/gkm020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Keane S.C., Van V., Frank H.M., Sciandra C.A., McCowin S., Santos J., Heng X., Summers M.F. NMR detection of intermolecular interaction sites in the dimeric 5’-leader of the HIV-1 genome. Proc. Natl. Acad. Sci. USA. 2016;113:13033–13038. doi: 10.1073/pnas.1614785113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ganser L.R., Al-Hashimi H.M. HIV-1 leader RNA dimeric interface revealed by NMR. Proc. Natl. Acad. Sci. USA. 2016;113:13263–13265. doi: 10.1073/pnas.1615789113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mujeeb A., Ulyanov N.B., Georgantis S., Smirnov I., Chung J., Parslow T.G., James T.L. Nucleocapsid protein-mediated maturation of dimer initiation complex of full-length SL1 stemloop of HIV-1: Sequence effects and mechanism of RNA refolding. Nucleic Acids Res. 2007;35:2026–2034. doi: 10.1093/nar/gkm097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Takahashi K., Baba S., Koyanagi Y., Yamamoto N., Takaku H., Kawai G. Two basic regions of NCp7 are sufficient for conformational conversion of HIV-1 dimerization initiation site from kissing-loop dimer to extended-duplex dimer. J. Biol. Chem. 2001;276:31274–31278. doi: 10.1074/jbc.M104577200. [DOI] [PubMed] [Google Scholar]
- 225.Aduri R., Briggs K.T., Gorelick R.J., Marino J.P. Molecular determinants of HIV-1 NCp7 chaperone activity in maturation of the HIV-1 dimerization initiation site. Nucleic Acids Res. 2013;41:2565–2580. doi: 10.1093/nar/gks1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Aldovini A., Young R.A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 1990;64:1920–1926. doi: 10.1128/JVI.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Poznansky M., Lever A.M.L., Bergeron L., Haseltine W., Sodroski J. Gene transfer into human lymphocytes by a defective human immunodeficiency virus type 1 vector. J. Virol. 1991;65:532–536. doi: 10.1128/JVI.65.1.532-536.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Lever A.M.L., Göttlinger H.G., Haseltine W.A., Sodroski J.G. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 1989;63:4085–4087. doi: 10.1128/JVI.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Clavel F., Orenstein J.M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J. Virol. 1990;64:5230–5234. doi: 10.1128/JVI.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sakuragi J., Sakuragi S., Shioda T. Minimal region sufficient for genome dimerization in the human immunodeficiency virus type 1 virion and its potential roles in the early stages of viral replication. J. Virol. 2007;81:7985–7992. doi: 10.1128/JVI.00429-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sakaguchi K., Zambrano N., Baldwin E.T., Shapiro B.A., Erickson J.W., Omichinski J.G., Clore G.M., Gronenborn A.M., Appella E. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proc. Natl. Acad. Sci. USA. 1993;90:5219–5223. doi: 10.1073/pnas.90.11.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Rong L., Russell R.S., Hu J., Laughrea M., Wainberg M.A., Liang C. Deletion of stem-loop 3 is compensated by second-site mutations within the Gag protein of human immunodeficiency virus type 1. Virology. 2003;314:221–228. doi: 10.1016/S0042-6822(03)00405-7. [DOI] [PubMed] [Google Scholar]
- 233.Russell R.S., Hu J., Beriault V., Mouland A.J., Kleiman L., Wainberg M.A., Liang C. Sequences downstream of the 5’ splice donor site are required for both packaging and dimerization of human immunodeficiency virus type-1 RNA. J. Virol. 2003;77:84–96. doi: 10.1128/JVI.77.1.84-96.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Webb J.A., Jones C.P., Parent L.J., Rouzina I., Musier-Forsyth K. Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: Implications for viral genomic RNA packaging. RNA. 2013;19:1078–1088. doi: 10.1261/rna.038869.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Comas-Garcia M., Datta S.A., Baker L., Varma R., Gudla P.R., Rein A. Dissection of specific binding of HIV-1 Gag to the “packaging signal” in viral RNA. Elife. 2017;6:6. doi: 10.7554/eLife.27055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ding P., Kharytonchyk S., Waller A., Mbaekwe U., Basappa S., Kuo N., Frank H.M., Quasney C., Kidane A., Swanson C., et al. Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal. Proc. Natl. Acad. Sci. USA. 2020;117:17737–17746. doi: 10.1073/pnas.2008519117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Pappalardo L., Kerwood D.J., Pelczer I., Borer P.N. Three-dimensional folding of an RNA hairpin required for packaging HIV-1. J. Mol. Biol. 1998;282:801–818. doi: 10.1006/jmbi.1998.2046. [DOI] [PubMed] [Google Scholar]
- 238.Zeffman A., Hassard S., Varani G., Lever A. The major HIV-1 packaging signal is an extended bulged stem loop whose structure is altered on interaction with the Gag polyprotein. J. Mol. Biol. 2000;297:877–893. doi: 10.1006/jmbi.2000.3611. [DOI] [PubMed] [Google Scholar]
- 239.McBride M.S., Schwartz M.D., Panganiban A.T. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 1997;71:4544–4554. doi: 10.1128/JVI.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Luban J., Goff S.P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 1994;68:3784–3793. doi: 10.1128/JVI.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Parolin C., Dorfman T., Palu G., Gottlinger H.G., Sodroski J. Analysis in human immunodeficiency virus type 1 vectors of cis-acting sequences that affect gene transfer into human lymphocytes. J. Virol. 1994;68:3888–3895. doi: 10.1128/JVI.68.6.3888-3895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kerwood D.J., Cavaluzzi M.J., Borer P.N. Structure of SL4 RNA from the HIV-1 packaging signal. Biochemistry. 2001;40:14518–14529. doi: 10.1021/bi0111909. [DOI] [PubMed] [Google Scholar]
- 243.Amarasinghe G.K., Zhou J., Miskimon M., Chancellor K.J., McDonald J.A., Matthews A.G., Miller R.A., Rouse M.D., Summers M.F. Stem-loop SL4 of the HIV-1 Ψ-RNA packaging signal exhibits weak affinity for the nucleocapsid protein. Structural studies and implications for genome recognition. J. Mol. Biol. 2001;314:961–969. doi: 10.1006/jmbi.2000.5182. [DOI] [PubMed] [Google Scholar]
- 244.Wimmer J., Fujinaga K., Taube R., Cujec T.P., Zhu Y., Peng J., Price D.H., Peterlin B.M. Interactions between Tat and TAR and human immunodeficiency virus replication are facilitated by human cyclin T1 but not cyclins T2a or T2b. Virology. 1999;255:182–189. doi: 10.1006/viro.1998.9589. [DOI] [PubMed] [Google Scholar]
- 245.Brady J., Kashanchi F. Tat gets the “green” light on transcription initiation. Retrovirology. 2005;2:69. doi: 10.1186/1742-4690-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Laspia M.F., Rice A.P., Mathews M.B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989;59:283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- 247.Chulze-Gahmen U., Hurley J.H. Structural mechanism for HIV-1 TAR loop recognition by Tat and the super elongation complex. Proc. Natl. Acad. Sci. USA. 2018;115:12973–12978. doi: 10.1073/pnas.1806438115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Chavali S.S., Bonn-Breach R., Wedekind J.E. Face-time with TAR: Portraits of an HIV-1 RNA with diverse modes of effector recognition relevant for drug discovery. J. Biol. Chem. 2019;294:9326–9341. doi: 10.1074/jbc.REV119.006860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Jalalirad M., Saadatmand J., Laughrea M. Dominant role of the 5’ TAR bulge in dimerization of HIV-1 genomic RNA, but no evidence of TAR-TAR kissing during in vivo virus assembly. Biochemistry. 2012;51:3744–3758. doi: 10.1021/bi300111p. [DOI] [PubMed] [Google Scholar]
- 250.Andersen E.S., Contera S.A., Knudsen B., Damgaard C.K., Besenbacher F., Kjems J. Role of the trans-activation response element in dimerization of HIV-1 RNA. J. Biol. Chem. 2004;279:22243–22249. doi: 10.1074/jbc.M314326200. [DOI] [PubMed] [Google Scholar]
- 251.Berkhout B., Vastenhouw N.L., Klasens B.I., Huthoff H. Structural features in the HIV-1 repeat region facilitate strand transfer during reverse transcription. RNA. 2001;7:1097–1114. doi: 10.1017/S1355838201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Helga-Maria C., Hammarskjold M.-L., Rekosh D. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type-1 genomic RNA. J. Virol. 1999;73:4127–4135. doi: 10.1128/JVI.73.5.4127-4135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Clever J.L., Eckstein D.A., Parslow T.G. Genetic dissociation of the encapsidation and reverse transcription functions in the 5’ R region of human immunodeficiency virus type 1. J. Virol. 1999;73:101–109. doi: 10.1128/JVI.73.1.101-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Das A.T., Klaver B., Berkhout B. The 5´ and 3´ TAR elements of human immunodeficiency virus exert effects at several points in the virus life cycle. J. Virol. 1998;72:9217–9223. doi: 10.1128/JVI.72.11.9217-9223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Bennasser Y., Le S.Y., Yeung M.L., Jeang K.T. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology. 2004;1:43. doi: 10.1186/1742-4690-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Chen L., Feng Z., Yue H., Bazdar D., Mbonye U., Zender C., Harding C.V., Bruggeman L., Karn J., Sieg S.F., et al. Exosomes derived from HIV-1-infected cells promote growth and progression of cancer via HIV TAR RNA. Nat. Commun. 2018;9:4585. doi: 10.1038/s41467-018-07006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Das A.T., Harwig A., Vrolijk M.M., Berkhout B. The TAR hairpin of human immunodeficiency virus Type 1 can be deleted when not required for Tat-mediated activation of transcription. J. Virol. 2007;81:7742–7748. doi: 10.1128/JVI.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Brandt S., Blissenbach M., Grewe B., Konietzny R., Grunwald T., Uberla K. Rev proteins of human and simian immunodeficiency virus enhance RNA encapsidation. PLoS Pathog. 2007;3:e54. doi: 10.1371/journal.ppat.0030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.O’Carroll I.P., Soheilian F., Kamata A., Nagashima K., Rein A. Elements in HIV-1 Gag contributing to virus particle assembly. Virus Res. 2013;171:341–345. doi: 10.1016/j.virusres.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Rulli J.S.J., Hibbert C.S., Mirro J., Pederson T., Biswal S., Rein A. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 2007;81:6623–6631. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Aboul-Ela F., Karn J., Varani G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J. Mol. Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 262.Hauber J., Malim M.H., Cullen B.R. Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J. Virol. 1989;63:1181–1187. doi: 10.1128/JVI.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Delling U., Reid L.S., Barnett R.W., Ma M.Y., Climie S., Sumner-Smith M., Sonenberg N. Conserved nucleotides in the TAR RNA stem of human immunodeficiency virus type 1 are critical for Tat binding and trans activation: Model for TAR RNA tertiary structure. J. Virol. 1992;66:3018–3025. doi: 10.1128/JVI.66.5.3018-3025.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Dingwall C., Ernberg I., Gait M.J., Green S.M., Heaphy S., Karn J., Lowe A.D., Singh M., Skinner M.A. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 1990;9:4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wei P., Garber M.E., Fang S.-M., Fischer W.H., Jones K.A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/S0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 266.Richter S., Ping Y.H., Rana T.M. TAR RNA loop: A scaffold for the assembly of a regulatory switch in HIV replication. Proc. Natl. Acad. Sci. USA. 2002;99:7928–7933. doi: 10.1073/pnas.122119999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Puglisi J.D., Tan R., Calnan B.J., Frankel A.D., Williamson J.R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 268.Puglisi J.D., Chen L., Frankel A.D., Williamson J.R. Role of RNA structure in arginine recognition of TAR RNA. Proc. Natl. Acad. Sci. USA. 1993;90:3680–3684. doi: 10.1073/pnas.90.8.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Aboul-Ela F., Karn J., Varani G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Long K.S., Crothers D.M. Characterization of the solution conformations of unbound and Tat peptide-bound forms of HIV-1 TAR RNA. Biochemistry. 1999;38:10059–10069. doi: 10.1021/bi990590h. [DOI] [PubMed] [Google Scholar]
- 271.Davidson A., Leeper T.C., Athanassiou Z., Patora-Komisarska K., Karn J., Robinson J.A., Varani G. Simultaneous recognition of HIV-1 TAR RNA bulge and loop sequences by cyclic peptide mimics of Tat protein. Proc. Natl. Acad. Sci. USA. 2009;106:11931–11936. doi: 10.1073/pnas.0900629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Pitt S.W., Majumdar A., Serganov A., Patel D.J., Al-Hashimi H.M. Argininamide binding arrests global motions in HIV-1 TAR RNA: Comparison with Mg2+-induced conformational stabilization. J. Mol. Biol. 2004;338:7–16. doi: 10.1016/j.jmb.2004.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Shortridge M.D., Wille P.T., Jones A.N., Davidson A., Bogdanovic J., Arts E., Karn J., Robinson J.A., Varani G. An ultra-high affinity ligand of HIV-1 TAR reveals the RNA structure recognized by P-TEFb. Nucleic Acids Res. 2019;47:1523–1531. doi: 10.1093/nar/gky1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Pham V.V., Salguero C., Khan S.N., Meagher J.L., Brown W.C., Humbert N., de Rocquigny H., Smith J.L., D’Souza V.M. HIV-1 Tat interactions with cellular 7SK and viral TAR RNAs identifies dual structural mimicry. Nat. Commun. 2018;9:4266. doi: 10.1038/s41467-018-06591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Dethoff E.A., Petzold K., Chugh J., Casiano-Negroni A., Al-Hashimi H.M. Visualizing transient low-populated structures of RNA. Nature. 2012;491:724–728. doi: 10.1038/nature11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Ganser L.R., Kelly M.L., Patwardhan N.N., Hargrove A.E., Al-Hashimi H.M. Demonstration that small molecules can bind and stabilize Low-abundance Short-lived RNA excited conformational states. J. Mol. Biol. 2020;432:1297–1304. doi: 10.1016/j.jmb.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zhang Q., Stelzer A.C., Fisher C.K., Al-Hashimi H.M. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 278.Tisne C., Roques B.P., Dardel F. The annealing mechanism of HIV-1 reverse transcription primer onto the viral genome. J. Biol. Chem. 2004;279:3588–3595. doi: 10.1074/jbc.M310368200. [DOI] [PubMed] [Google Scholar]
- 279.Beerens N., Groot F., Berkhout B. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J. Biol. Chem. 2001;276:31247–31256. doi: 10.1074/jbc.M102441200. [DOI] [PubMed] [Google Scholar]
- 280.Puglisi E.V., Puglisi J.D. Secondary structure of the HIV reverse transcription initiation complex by NMR. J. Mol. Biol. 2011;410:863–874. doi: 10.1016/j.jmb.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Goldschmidt V., Rigourd M., Ehresmann C., Le Grice S.F., Ehresmann B., Marquet R. Direct and indirect contributions of RNA secondary structure elements to the initiation of HIV-1 reverse transcription. J. Biol. Chem. 2002;277:43233–43242. doi: 10.1074/jbc.M205295200. [DOI] [PubMed] [Google Scholar]
- 282.Xing L., Liang C., Kleiman L. Coordinate Roles of Gag and RNA Helicase A in Promoting the Annealing of tRNALys3 to HIV-1 RNA. J. Virol. 2011;85:1847–1860. doi: 10.1128/JVI.02010-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Jeang K.-T., Yedavalli V. Role of RNA helicases in HIV-1 replication. Nucleic Acids Res. 2006;34:4198–4205. doi: 10.1093/nar/gkl398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Roy B.B., Hu J., Guo X., Russell R.S., Guo F., Kleiman L., Liang C. Association of RNA Helicase A with human immunodeficiency virus type 1 particles. J. Biol. Chem. 2006;281:12625–12635. doi: 10.1074/jbc.M510596200. [DOI] [PubMed] [Google Scholar]
- 285.Bolinger C., Sharma A., Singh D., Yu L., Boris-Lawrie K. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res. 2010;38:1686–1696. doi: 10.1093/nar/gkp1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Boeras I., Song Z., Moran A., Franklin J., Brown W.C., Johnson M., Boris-Lawrie K., Heng X. DHX9/RHA Binding to the PBS-Segment of the genomic RNA during HIV-1 assembly bolsters virion infectivity. J. Mol. Biol. 2016;428:2418–2429. doi: 10.1016/j.jmb.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Barton S., Heng X., Johnson B.A., Summers M.F. Database proton NMR chemical shifts for RNA signal assignment and validation. J. Biomol. NMR. 2013;55:33–46. doi: 10.1007/s10858-012-9683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Brown J.D., Summers M.F., Johnson B.A. Prediction of hydrogen and carbon chemical shifts from RNA using database mining and support vector regression. J. Biomol. NMR. 2015;63:39–52. doi: 10.1007/s10858-015-9961-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Johnson B.A., Blevins R.A. NMRview: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 290.Marchant J., Summers M.F., Johnson B.A. Assigning NMR spectra of RNA, peptides and small organic molecules using molecular network visualization software. J. Biomol. NMR. 2019;73:525–529. doi: 10.1007/s10858-019-00271-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Tolbert B.S., Miyazaki Y., Barton S., Kinde B., Starck P., Singh R., Bax A., Case D.A., Summers M.F. Major groove width variations in RNA structures determined by NMR and impact of 13C residual chemical shift anisotropy and 1H-13C residual dipolar coupling on refinement. J. Biomol. NMR. 2010;47:205–219. doi: 10.1007/s10858-010-9424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Brigham B.S., Kitzrow J.P., Reyes J.C., Musier-Forsyth K., Munro J.B. Intrinsic conformational dynamics of the HIV-1 genomic RNA 5’UTR. Proc. Natl. Acad. Sci. USA. 2019;116:10372–10381. doi: 10.1073/pnas.1902271116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Tran T., Liu Y., Marchant J., Monti S., Seu M., Zaki J., Yang A.L., Bohn J., Ramakrishnan V., Singh R., et al. Conserved determinants of lentiviral genome dimerization. Retrovirology. 2015;12:83. doi: 10.1186/s12977-015-0209-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Chiu Y.L., Coronel E., Ho C.K., Shuman S., Rana T.M. HIV-1 Tat protein interacts with mammalian capping enzyme and stimulates capping of TAR RNA. J. Biol. Chem. 2001;276:12959–12966. doi: 10.1074/jbc.M007901200. [DOI] [PubMed] [Google Scholar]
- 295.Zhou M., Deng L., Kashanchi F., Brady J.N., Shatkin A.J., Kumar A. The Tat/TAR-dependent phosphorylation of RNA polymerase II C-terminal domain stimulates cotranscriptional capping of HIV-1 mRNA. Proc. Natl. Acad. Sci. USA. 2003;100:12666–12671. doi: 10.1073/pnas.1835726100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Menees T.M., Muller B., Krausslich H.G. The major 5’ end of HIV type 1 RNA corresponds to G456. AIDS Res. Hum. Retrovir. 2007;23:1042–1048. doi: 10.1089/aid.2006.0275. [DOI] [PubMed] [Google Scholar]
- 297.Sharma A., Yilmaz A., Marsh K., Cochrane A., Boris-Lawrie K. Thriving under stress: Selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 2012;8:e1002612. doi: 10.1371/journal.ppat.1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Muesing M.A., Smith D.H., Cabradilla C.D., Benton C.V., Lasky L.A., Capon D.J. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature. 1985;313:450–458. doi: 10.1038/313450a0. [DOI] [PubMed] [Google Scholar]
- 299.Starcich B., Ratner L., Josephs S.F., Okamoto T., Gallo R.C., Wong-Staal F. Characterization of long terminal repeat sequences of HTLV-III. Science. 1985;227:538–540. doi: 10.1126/science.2981438. [DOI] [PubMed] [Google Scholar]
- 300.Vogt V. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 1996;214:95–132. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 301.Mann R., Mulligan R.C., Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 302.Murphy J.E., Goff S.P. Construction and analysis of deletion mutations in the U5 region of Moloney murine leukemia virus: Effects on RNA packaging and reverse transcription. J. Virol. 1989;63:319–327. doi: 10.1128/JVI.63.1.319-327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Yu S.S., Kim J.-M., Kim S. The 17 nucleotides downstream from the env gene stop codon are important for Murine Leukemia Virus packaging. J. Virol. 2000;74:8775–8780. doi: 10.1128/jvi.74.18.8775-8780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Bender M.A., Palmer T.D., Gelinas R.E., Miller A.D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J. Virol. 1987;61:1639–1646. doi: 10.1128/JVI.61.5.1639-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Alford R.L., Honda S., Lawrence C.B., Belmont J.W. RNA secondary structure analysis of the packing signal for Moloney murine leukemia virus. Virology. 1991;183:611–619. doi: 10.1016/0042-6822(91)90990-S. [DOI] [PubMed] [Google Scholar]
- 306.Fisher J., Goff S.P. Mutational analysis of stem-loops in the RNA packaging signal of the moloney murine leukemia virus. Virology. 1998;244:133–145. doi: 10.1006/viro.1998.9090. [DOI] [PubMed] [Google Scholar]
- 307.Konings D.A.M., Nash M.A., Maizel J.V., Arlinghaus R.B. Novel GACG-hairpin pair motif in the 5’ untranslated region of type C retrovirus related to murine leukemia virus. J. Virol. 1992;66:632–640. doi: 10.1128/JVI.66.2.632-640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Mougel M., Tounekti N., Darlix J.-L., Paoletti J., Ehresmann B., Ehresmann C. Conformational analysis of the 5’ leader and the gag initiation site of Mo-MuLV RNA and allosteric transitions induced by dimerization. Nucleic Acids Res. 1993;21:4677–4684. doi: 10.1093/nar/21.20.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Mougel M., Zhang Y., Barklis E. Cis-active structural motifs involved in specific encapsidation of Moloney Murine leukemia virus RNA. J. Virol. 1996;70:5043–5050. doi: 10.1128/JVI.70.8.5043-5050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Tounekti N., Mougel M., Roy C., Marquet R., Darlix J.-L., Paoletti J., Ehresmann B., Ehresmann C. Effect of dimerization on the conformation of the encapsidation psi domain of Moloney murine leukemia virus RNA. J. Mol. Biol. 1992;223:205–220. doi: 10.1016/0022-2836(92)90726-Z. [DOI] [PubMed] [Google Scholar]
- 311.Shinnick T.M., Lerner R.A., Sutcliffe J.G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 312.Beasley B.E., Hu W.S. Cis-acting elements imporant for retroviral RNA packaging specificity. J. Virol. 2002;76:4950–4960. doi: 10.1128/JVI.76.10.4950-4960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Evans M.J., Bacharach E., Goff S.P. RNA sequences in the Moloney murine leukemia virus genome bound by the Gag precursor protein in the Yeast three-hybrid system. J. Virol. 2004;78:7677–7684. doi: 10.1128/JVI.78.14.7677-7684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.D’Souza V., Melamed J., Habib D., Pullen K., Wallace K., Summers M.F. Identification of a high-affinity nucleocapsid protein binding site within the Moloney Murine Leukemia Virus Y-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2001;314:217–232. doi: 10.1006/jmbi.2001.5139. [DOI] [PubMed] [Google Scholar]
- 315.Miyazaki Y., Irobalieva R.N., Tolbert B.S., Smalls-Manty A., Iyalla K., Loeliger K., D’Souza V., Khant H., Schmid M.F., Garcia E., et al. Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J. Mol. Biol. 2010;404:751–772. doi: 10.1016/j.jmb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Kim C.-H., Tinoco I., Jr. A retroviral RNA kissing complex containing only two G-C base pairs. Proc. Natl. Acad. Sci. USA. 2000;97:9396–9401. doi: 10.1073/pnas.170283697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Oroudjev E.M., Kang P.C.E., Kohlstaedt L.A. An additional dimer linkage structure in Moloney Murine Leukemia Virus RNA. J. Mol. Biol. 1999;291:603–613. doi: 10.1006/jmbi.1999.2984. [DOI] [PubMed] [Google Scholar]
- 318.Girard P.-M., Bonnet-Mathoniere B., Mauriaux D., Paoletti J. A short autocomplementary sequence in the 5’ leader region is responsible for dimerization of MoMuLV genomic RNA. Biochemistry. 1995;34:9785–9794. doi: 10.1021/bi00030a016. [DOI] [PubMed] [Google Scholar]
- 319.De Tapia M., Metzler V., Mougel M., Ehresmann B., Ehresmann C. Dimerization of MoMuLV genomic RNA: Redefinition of the role of the palindromic stem-loop H1 (278–303) and new roles for stem-loops H2 (310-352) and H3 (355-374) Biochemistry. 1998;37:6077–6085. doi: 10.1021/bi9800303. [DOI] [PubMed] [Google Scholar]
- 320.Ly H., Parslow T.G. Bipartite signal for genomic RNA dimerization in the Moloney Murine Leukemia Virus. J. Virol. 2002;76:3135–3144. doi: 10.1128/JVI.76.7.3135-3144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Chen A.A., Garcia A.E. Mechanism of enhanced mechanical stability of a minimal RNA kissing complex elucidated by nonequilibrium molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 2012;109:E1530–E1539. doi: 10.1073/pnas.1119552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.D’Souza V., Dey A., Habib D., Summers M.F. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J. Mol. Biol. 2004;337:427–442. doi: 10.1016/j.jmb.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 323.Merino E.J., Wilkinson K.A., Coughlan J.L., Weeks K.M. RNA structure analysis at single nucleotide resolution by selective 2’-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 324.Badorrek C.S., Weeks K.M. Flexibility in the dimerization domain of a gamma retrovirus. Nat. Chem. Biol. 2005;1:104–111. doi: 10.1038/nchembio712. [DOI] [PubMed] [Google Scholar]
- 325.Brickell P.M. The p60c-src family of protein-tyrosine kinases: Structure, regulation and function. Crit. Rev. Oncog. 1992;3:401–446. [PubMed] [Google Scholar]
- 326.Hackett P.B., Swanstrom R., Varmus H.E., Bishop J.M. The leader sequence of the subgenomic mRNA’s of Rous sarcoma virus is approximately 390 nucleotides. J. Virol. 1982;41:527–534. doi: 10.1128/JVI.41.2.527-534.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Hackett P., Dalton M.W., Johnson D.P., Peterson R.B. Phylogenetic and physical analysis of the 5’ leader RNA sequences of avian retroviruses. Nucleic Acids Res. 1991;19:6929–6934. doi: 10.1093/nar/19.24.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Sonstegard T.S., Hackett P.B. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J. Virol. 1996;70:6642–6652. doi: 10.1128/JVI.70.10.6542-6552.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Donze O., Damay P., Spahr P.-F. The first and third uORFs in RSV leader RNA are efficiently translated: Implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Moustakas A., Sonstegard T.S., Hackett P.B. Alterations of the three short open reading frames in the Rous sarcoma virus leader RNA modulate viral replication and gene expression. J. Virol. 1993;67:4337–4349. doi: 10.1128/JVI.67.7.4337-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Donze O., Spahr P.-F. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992;11:3747–3757. doi: 10.1002/j.1460-2075.1992.tb05460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Anderson D.J., Lee P., Levine K.L., Sang J.S., Shah S.A., Yang O.O., Shank P.R., Linial M.L. Molecular cloning and characterization of the RNA packaging-defective retrovirus SE21Q1b. J. Virol. 1992;66:204–216. doi: 10.1128/JVI.66.1.204-216.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Gallis B., Linial M., Eisenman R. An avian oncovirus mutant deficient in genomic RNA: Characterization of the packaged RNA as cellular messenger RNA. Virology. 1979;94:146–161. doi: 10.1016/0042-6822(79)90445-8. [DOI] [PubMed] [Google Scholar]
- 334.Kawai S., Koyama T. Characterization of a Rouse Sarcoma Virus mutant defective in packaging it’s own genomic RNA: Biological properties of mutant TK15 and mutant-induced transformation. J. Virol. 1984;51:147–153. doi: 10.1128/JVI.51.1.147-153.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Koyama T., Harada F., Kawai S. Characterization of a Rous sarcoma virus mutant defectie in packaging its own genomic RNA: Biochemical properties of mutant TK15 and mutant-induced transformants. J. Virol. 1984;51:154–162. doi: 10.1128/JVI.51.1.154-162.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Linial M., Medeiros E., Hayward W.S. An avian oncovirus mutant (SE 21Q1b) deficient in genomic RNA: Biological and biochemical characterization. Cell. 1978;15:1371–1381. doi: 10.1016/0092-8674(78)90062-4. [DOI] [PubMed] [Google Scholar]
- 337.Nishizawa M., Koyama T., Kawai S. Unusual features of the leader sequence of Rous sarcoma virus packaging mutant TK15. J. Virol. 1985;55:881–885. doi: 10.1128/JVI.55.3.881-885.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Katz R.A., Terry R.W., Skalka A.M. A conserved cis-acting sequence in the 5’ leader of avian sarcoma virus RNA is required for packaging. J. Virol. 1986;59:163–167. doi: 10.1128/JVI.59.1.163-167.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Stoker A.W., Bissell M.J. Development of avian sarcoma and leukosis virus based vector packaging cell lines. J. Virol. 1988;16:1161–1170. doi: 10.1128/JVI.62.3.1008-1015.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Shank P.R., Linial M. Avian oncovirus mutant (SE21Q1b) deficient in genomic RNA: Characterization of a deletion in the provirus. J. Virol. 1980;36:450–456. doi: 10.1128/JVI.36.2.450-456.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Cobrink D., Aiyar A., Ge Z., Katzman M., Huang H., Leis J. Overlapping retrovirus U5 sequence elements are required for efficient integration and initiation of reverse transcriptiuon. J. Virol. 1991;65:3864–3872. doi: 10.1128/JVI.65.7.3864-3872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Cobrink D., Soskey L., Leis J. A retroviral RNA secondary structure required for efficient initiation of reverse transcription. J. Virol. 1988;62:3622–3630. doi: 10.1128/JVI.62.10.3622-3630.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Norton P.A., Coffin J.M. Bacterial b-galactosidase as a marker of Rous sarcoma virus gene expression and replication. Mol. Cell Biol. 1985;5:281–290. doi: 10.1128/MCB.5.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Liu S., Maldonado R.K., Rye-McCurdy T., Binkley C., Bah A., Chen E.C., Rice B.L., Parent L.J., Musier-Forsyth K. Rous Sarcoma Virus genomic RNA Dimerization capability in vitro is not a prerequisite for viral infectivity. Viruses. 2020;12:568. doi: 10.3390/v12050568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Knight J.B., Hai S.Z., Stoltzfus M.C. A base-paired sequence in the avian sarcoma virus 5’ leader is required for efficient encapsidation of RNA. J. Virol. 1994;68:4493–4502. doi: 10.1128/JVI.68.7.4493-4502.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Zhou J., McAllen K., Tailor Y., Summers M.F. High affinity nucleocapsid protein binding to the μΨ RNA packaging signal of Rous sarcoma virus. J. Mol. Biol. 2005;349:976–988. doi: 10.1016/j.jmb.2005.04.046. [DOI] [PubMed] [Google Scholar]
- 347.Lear A.L., Haddrick M., Heaply S. A study of the dimerization of the Rous sarcoma virus RNA in vitro and in vivo. Virology. 1995;212:47–57. doi: 10.1006/viro.1995.1452. [DOI] [PubMed] [Google Scholar]
- 348.Canaani E., Helm K.V.D., Duesberg P. Evidence for a 30–40s RNA as a precursor for the 60-70s RNA of the Rous Sarcoma Virus. Proc. Natl. Acad. Sci. USA. 1973;70:401–405. doi: 10.1073/pnas.70.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Cheung K.S., Smith R.E., Stone M.P., Joklik W.K. Comparison of immature (rapid harvest) and mature Rous Sarcoma Virus. Virology. 1972;50:851–864. doi: 10.1016/0042-6822(72)90439-4. [DOI] [PubMed] [Google Scholar]
- 350.Oertle S., Spahr P.-F. Role of the Gag polyprotein precursor in packaging and maturation of Rous sarcoma virus genomic RNA. J. Virol. 1990;64:5757–5763. doi: 10.1128/JVI.64.12.5757-5763.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Polge E., Darlix J.-L., Paoletti J., Fosse P. Characterization of the loose and tight dimer forms of avian leukosis virus RNA. J. Mol. Biol. 2000;300:41–56. doi: 10.1006/jmbi.2000.3832. [DOI] [PubMed] [Google Scholar]
- 352.Bieth E., Gabus C., Darlix J.-L. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 1990;18:119–127. doi: 10.1093/nar/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Delahunty M., South T.L., Summers M.F., Karpel R.L. Nucleic acid interactive properties of a peptide corresponding to the N-terminal zinc finger domain of HIV-1 nucleocapsid protein. Biochemistry. 1992;31:6461–6469. doi: 10.1021/bi00143a015. [DOI] [PubMed] [Google Scholar]
- 354.Summers M.F. Zinc finger motif for single-stranded nucleic acids? Investigations by nuclear magnetic resonance. J. Cell. Biochem. 1991;45:41–48. doi: 10.1002/jcb.240450110. [DOI] [PubMed] [Google Scholar]
- 355.South T.L., Kim B., Hare D.R., Summers M.F. Zinc fingers and molecular recognition. Structure and nucleic acid binding studies of an HIV zinc finger-like domain. Biochem. Pharm. 1990;40:123–129. doi: 10.1016/0006-2952(90)90187-P. [DOI] [PubMed] [Google Scholar]
- 356.Schuler W., Dong C., Wecker K., Roques B.P. NMR structure of the complex between the zinc finger protein NCp10 of Moloney murine leukemia virus and the single-stranded pentanucleotide d(ACGCC): Comparison with HIV-NCp7 complexes. Biochemistry. 1999;38:12984–12994. doi: 10.1021/bi990378d. [DOI] [PubMed] [Google Scholar]
- 357.Deffaud C., Darlix J.-L. Rous sarcoma virus translation revisited: Characterization of an internal ribosome entry segment in the 5’ leader of the genomic RNA. J. Virol. 2000;74:11581–11588. doi: 10.1128/JVI.74.24.11581-11588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Levin J.G., Guo J., Rouzina I., Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 359.Fu W., Ortiz-Conde B.A., Gorelick R.J., Hughes S.H., Rein A. Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J. Virol. 1997;71:6940–6946. doi: 10.1128/JVI.71.9.6940-6946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Feng Y.-X., Campbell S., Harvin D., Ehresmann B., Ehresmann C., Rein A. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: Possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 1999;73:4251–4256. doi: 10.1128/JVI.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Wu W., Henderson L.E., Copeland T.D., Gorelick R.J., Bosche W.J., Rein A., Levin J.G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J. Virol. 1996;70:7132–7142. doi: 10.1128/JVI.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Tanchou V., Gabus C., Rogemond V., Darlix J.-L. Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J. Mol. Biol. 1995;252:563–571. doi: 10.1006/jmbi.1995.0520. [DOI] [PubMed] [Google Scholar]
- 363.Darlix J.-L., Vincent C., Gabus H., de Rocquigny H., Roques B. Trans-activation of the 5’ to 3’ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV-1. C. R. Acad. Sci. 1993;316:763–771. [PubMed] [Google Scholar]
- 364.Allain B., Lapadat-Tapolsky M., Berlioz C., Darlix J.-L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Peliska J.A., Balasubramanian S., Giedroc D.P., Benkovic S.J. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNaseH activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 366.Kim J.K., Palaniappan C., Wu W., Fay P.J., Bambara R.A. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J. Biol. Chem. 1997;272:16769–16777. doi: 10.1074/jbc.272.27.16769. [DOI] [PubMed] [Google Scholar]
- 367.Tsuchihashi Z., Brown P.O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1994;68:5863–5870. doi: 10.1128/JVI.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Auxilien S., Keith G., Le Grice S.F., Darlix J.-L. Role of post-transcriptional modifications of primer tRNALys, 3 in the fidelity and efficacy of plus strand DNA transfer during HIV-1 reverse transcription. J. Biol. Chem. 1999;274:4412–4420. doi: 10.1074/jbc.274.7.4412. [DOI] [PubMed] [Google Scholar]
- 369.Wu T., Guo J., Bess J., Henderson L.E., Levin J.G. Molecular requirements for human immunodeficiency virus type 1 plus-strand transfer: Analysis in reconstituted and endogenous reverse transcription systems. J. Virol. 1999;73:4794–4805. doi: 10.1128/JVI.73.6.4794-4805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Muthuswami R., Chen J., Burnett B.P., Thimmig R.L., Janjic N., McHenry C.S. The HIV plus-strand transfer reaction: Determination of replication-competent intermediates and identification of a novel lentiviral element, the primer over-extension sequence. J. Mol. Biol. 2002;315:311–323. doi: 10.1006/jmbi.2001.5205. [DOI] [PubMed] [Google Scholar]
- 371.Guo J., Wu T., Anderson J., Kane B.F., Johnson D.G., Gorelick R.J., Henderson L.E., Levin J.G. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 2000;74:8980–8988. doi: 10.1128/JVI.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Williams M.C., Rouzina I., Wenner J.R., Gorelick R.J., Musier-Forsyth K., Bloomfield V.A. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl. Acad. Sci. USA. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Johnson P.E., Turner R.B., Wu Z.-R., Hairston L., Guo J., Levin J.G., Summers M.F. A mechanism for (+) strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry. 2000;39:9084–9091. doi: 10.1021/bi000841i. [DOI] [PubMed] [Google Scholar]
- 374.Barraud P., Gaudin C., Dardel F., Tisne C. New insights into the formation of HIV-1 reverse transcription initiation complex. Biochimie. 2007;89:1204–1210. doi: 10.1016/j.biochi.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 375.Tisne C., Roques B.P., Dardel F. Heteronuclear NMR studies of the interaction of tRNA(Lys)3 with HIV-1 nucleocapsid protein. J. Mol. Biol. 2001;306:443–454. doi: 10.1006/jmbi.2000.4391. [DOI] [PubMed] [Google Scholar]
- 376.Bourbigot S., Ramalanjaona N., Boudier C., Salgado G.F., Roques B.P., Mely Y., Bouaziz S., Morellet N. How the HIV-1 nucleocapsid protein binds and destabilises the (-)primer binding site during reverse transcription. J. Mol. Biol. 2008;383:1112–1128. doi: 10.1016/j.jmb.2008.08.046. [DOI] [PubMed] [Google Scholar]
- 377.Bazzi A., Zargarian L., Chaminade F., Boudier C., de Rocquigny H., Rene B., Mely Y., Fosse P., Mauffret O. Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: Role of sugar deoxyriboses in the binding polarity of NC. Nucleic Acids Res. 2011;39:3903–3916. doi: 10.1093/nar/gkq1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Belfetmi A., Zargarian L., Tisne C., Sleiman D., Morellet N., Lescop E., Maskri O., Rene B., Mely Y., Fosse P., et al. Insights into the mechanisms of RNA secondary structure destabilization by the HIV-1 nucleocapsid protein. RNA. 2016;22:506–517. doi: 10.1261/rna.054445.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Rice W.G., Turpin J.A., Schaeffer C.A., Graham L., Clanton D., Buckheit R.W., Zaharevitz D., Summers M.F., Wallqvist A., Covell D.G. Evaluation of selected chemotypes in coupled cellular and molecular target-based screens identifies novel HIV-1 zinc finger inhibitors. J. Med. Chem. 1996;39:3606–3616. doi: 10.1021/jm960375o. [DOI] [PubMed] [Google Scholar]
- 380.Rice W.G., Schaeffer C.A., Harten B., Villinger F., South T.L., Summers M.F., Henderson L.E., Bess J.W., Jr., Arthur L.O., McDougal J.S., et al. Inhibition of HIV-1 infectivity by zinc-ejecting aromatic C-nitroso compounds. Nature. 1993;361:473–475. doi: 10.1038/361473a0. [DOI] [PubMed] [Google Scholar]
- 381.Rice W.G., Turpin J.A., Huang M., Clanton D., Buckheit R.W., Jr., Covell D.G., Wallqvist A., McDonnell N.B., DeGuzman R.N., Summers M.F., et al. Azodicarbonamide inhibits HIV-1 replication by targeting the nucleocapsid protein. Nat. Med. 1997;3:341–345. doi: 10.1038/nm0397-341. [DOI] [PubMed] [Google Scholar]
- 382.McDonnell N.B., De Guzman R.N., Rice W.G., Turpin J.A., Summers M.F. Zinc ejection as a new rationale for the use of cystamine and related dislufide-containing antiviral agents in the treatment of AIDS. J. Med. Chem. 1997;40:1969–1976. doi: 10.1021/jm970147+. [DOI] [PubMed] [Google Scholar]
- 383.Tummino P.J., Scholten J.D., Harvey P.J., Holler T.P., Maloney L., Gogliotti R., Domagala J., Hupe D. The in vitro ejection of zinc from human immunodeficiency virus (HIV) type 1 nucleocapsid protein by disulfide benzamides with cellular anti-HIV activity. Proc. Natl. Acad. Sci. USA. 1996;93:969–973. doi: 10.1073/pnas.93.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Deshmukh L., Tugarinov V., Appella D.H., Clore G.M. Targeting a dark excited state of HIV-1 nucleocapsid by antiretroviral thioesters revealed by NMR spectroscopy. Angew. Chem. Int. Ed. 2018;57:2687–2691. doi: 10.1002/anie.201713172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Goudreau N., Hucke O., Faucher A.M., Grand-Maitre C., Lepage O., Bonneau P.R., Mason S.W., Titolo S. Discovery and structural characterization of a new inhibitor series of HIV-1 nucleocapsid function: NMR solution structure determination of a ternary complex involving a 2:1 inhibitor/NC stoichiometry. J. Mol. Biol. 2013;425:1982–1998. doi: 10.1016/j.jmb.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 386.Le Grice S.F. Targeting the HIV RNA genome: High-hanging fruit only needs a longer ladder. Curr. Top. Microbiol. Immunol. 2015;389:147–169. doi: 10.1007/82_2015_434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Warui D.M., Baranger A.M. Identification of small molecule inhibitors of the HIV-1 nucleocapsid-stem-loop 3 RNA complex. J. Med. Chem. 2012;55:4132–4141. doi: 10.1021/jm2007694. [DOI] [PubMed] [Google Scholar]
- 388.Bell N.M., L’Hernault A., Murat P., Richards J.E., Lever A.M., Balasubramanian S. Targeting RNA-protein interactions within the human immunodeficiency virus type 1 lifecycle. Biochemistry. 2013;52:9269–9274. doi: 10.1021/bi401270d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Ingemarsdotter C.K., Zeng J., Long Z., Lever A.M.L., Kenyon J.C. An RNA-binding compound that stabilizes the HIV-1 gRNA packaging signal structure and specifically blocks HIV-1 RNA encapsidation. Retrovirology. 2018;15:25. doi: 10.1186/s12977-018-0407-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Dietz J., Koch J., Kaur A., Raja C., Stein S., Grez M., Pustowka A., Mensch S., Ferner J., Moller L., et al. Inhibition of HIV-1 by a peptide ligand of the genomic RNA packaging signal Psi. ChemMedChem. 2008;3:749–755. doi: 10.1002/cmdc.200700194. [DOI] [PubMed] [Google Scholar]
- 391.Chung J., Ulyanov N.B., Guilbert C., Mujeeb A., James T.L. Binding characteristics of small molecules that mimic nucleocapsid protein-induced maturation of stem-loop 1 of HIV-1 RNA. Biochemistry. 2010;49:6341–6351. doi: 10.1021/bi100660r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Tria G., Mertens H.D., Kachala M., Svergun D.I. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ. 2015;2:207–217. doi: 10.1107/S205225251500202X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Fang X., Stagno J.R., Bhandari Y.R., Zuo X., Wang Y.X. Small-angle X-ray scattering: A bridge between RNA secondary structures and three-dimensional topological structures. Curr. Opin. Struct. Biol. 2015;30:147–160. doi: 10.1016/j.sbi.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Grishaev A., Ying J., Canny M.D., Pardi A., Bax A. Solution structure of tRNAVal from refinement of homology model against residual dipolar coupling and SAXS data. J. Biomol. NMR. 2008;42:99–109. doi: 10.1007/s10858-008-9267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Wang J., Zuo X., Yu P., Xu H., Starich M.R., Tiede D.M., Shapiro B.A., Schwieters C.D., Wang Y.X. A method for helical RNA global structure determination in solution using small-angle x-ray scattering and NMR measurements. J. Mol. Biol. 2009;393:717–734. doi: 10.1016/j.jmb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Wang Y.X., Zuo X., Wang J., Yu P., Butcher S.E. Rapid global structure determination of large RNA and RNA complexes using NMR and small-angle X-ray scattering. Methods. 2010;52:180–191. doi: 10.1016/j.ymeth.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Jain N., Morgan C.E., Rife B.D., Salemi M., Tolbert B.S. Solution structure of the HIV-1 Intron splicing silencer and its interactions with the UP1 domain of heterogeneous nuclear Ribonucleoprotein (hnRNP) A1. J. Biol. Chem. 2016;291:2331–2344. doi: 10.1074/jbc.M115.674564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Fang X., Wang J., O’Carroll I.P., Mitchell M., Zuo X., Wang Y., Yu P., Liu Y., Rausch J.W., Dyba M.A., et al. An unusual topological structure of the HIV-1 Rev response element. Cell. 2013;155:594–605. doi: 10.1016/j.cell.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Zuo X., Wang J., Yu P., Eyler D., Xu H., Starich M.R., Tiede D.M., Simon A.E., Kasprzak W., Schwieters C.D., et al. Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3’ UTR of turnip crinkle virus. Proc. Natl. Acad. Sci. USA. 2010;107:1385–1390. doi: 10.1073/pnas.0908140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Burke J.E., Sashital D.G., Zuo X., Wang Y.X., Butcher S.E. Structure of the yeast U2/U6 snRNA complex. RNA. 2012;18:673–683. doi: 10.1261/rna.031138.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Cornilescu G., Didychuk A.L., Rodgers M.L., Michael L.A., Burke J.E., Montemayor E.J., Hoskins A.A., Butcher S.E. Structural Analysis of multi-helical RNAs by NMR-SAXS/WAXS: Application to the U4/U6 di-snRNA. J. Mol. Biol. 2016;428:777–789. doi: 10.1016/j.jmb.2015.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Imai S., Kumar P., Hellen C.U., D’Souza V.M., Wagner G. An accurately preorganized IRES RNA structure enables eIF4G capture for initiation of viral translation. Nat. Struct. Mol. Biol. 2016;23:859–864. doi: 10.1038/nsmb.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Luque D., Caston J.R. Cryo-electron microscopy for the study of virus assembly. Nat. Chem. Biol. 2020;16:231–239. doi: 10.1038/s41589-020-0477-1. [DOI] [PubMed] [Google Scholar]
- 404.Merk A., Bartesaghi A., Banerjee S., Falconieri V., Rao P., Davis M.I., Pragani R., Boxer M.B., Earl L.A., Milne J.L.S., et al. Breaking Cryo-EM resolution barriers to facilitate drug discovery. Cell. 2016;165:1698–1707. doi: 10.1016/j.cell.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Geraets J.A., Pothula K.R., Schroder G.F. Integrating cryo-EM and NMR data. Curr. Opin. Struct. Biol. 2020;61:173–181. doi: 10.1016/j.sbi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 406.Levin J.G., Rosenak M.J. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: Evidence for persistence of viral messenger RNA. Proc. Natl. Acad. Sci. USA. 1976;73:1154–1158. doi: 10.1073/pnas.73.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Mougel M., Akkawi C., Chamontin C., Feuillard J., Pessel-Vivares L., Socol M., Laine S. NXF1 and CRM1 nuclear export pathways orchestrate nuclear export, translation and packaging of murine leukaemia retrovirus unspliced RNA. RNA Biol. 2020;17:528–538. doi: 10.1080/15476286.2020.1713539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Malim M.H., Hauber J., Le S.Y., Maizel J.V., Cullen B.R. The HIV-1 rev transactivator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 409.Zhang H., Keane S.C. Advances that facilitate the study of large RNA structure and dynamics by nuclear magnetic resonance spectroscopy. Wiley Interdiscip. Rev. RNA. 2019;10:e1541. doi: 10.1002/wrna.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Liu Y., Holmstrom E., Zhang J., Yu P., Wang J., Dyba M.A., Chen D., Ying J., Lockett S., Nesbitt D.J., et al. Synthesis and applications of RNAs with position-selective labelling and mosaic composition. Nature. 2015;522:368–372. doi: 10.1038/nature14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Marchant J., Bax A., Summers M.F. Accurate measurement of residual dipolar couplings in large RNAs by Variable flip angle NMR. J. Am. Chem. Soc. 2018;140:6978–6983. doi: 10.1021/jacs.8b03298. [DOI] [PMC free article] [PubMed] [Google Scholar]