Key Points
Questions
Does universal genetic testing in patients with cancer identify more inherited cancer predisposition variants than a guideline-based approach, and what is the association between universal genetic testing and clinical management?
Findings
In this multicenter cohort study of 2984 patients with cancer, 1 in 8 patients had a pathogenic germline variant, half of which would not have been detected using a guideline-based approach. Nearly 30% of patients with a high-penetrance variant had modifications in their treatment based on the finding.
Meaning
Universal genetic testing detected more clinically actionable variants than a guideline-based approach, with a significant association with clinical management for the patients and their families.
Abstract
Importance
Hereditary factors play a key role in the risk of developing several cancers. Identification of a germline predisposition can have important implications for treatment decisions, risk-reducing interventions, cancer screening, and germline testing.
Objective
To examine the prevalence of pathogenic germline variants (PGVs) in patients with cancer using a universal testing approach compared with targeted testing based on clinical guidelines and the uptake of cascade family variant testing (FVT).
Design, Setting, and Participants
This prospective, multicenter cohort study assessed germline genetic alterations among patients with solid tumor cancer receiving care at Mayo Clinic cancer centers and a community practice between April 1, 2018, and March 31, 2020. Patients were not selected based on cancer type, disease stage, family history of cancer, ethnicity, or age.
Exposures
Germline sequencing using a greater than 80-gene next-generation sequencing platform.
Main Outcomes and Measures
Proportion of PGVs detected with a universal strategy compared with a guideline-directed approach and uptake of cascade FVT in families.
Results
A total of 2984 patients (mean [SD] age, 61.4 [12.2] years; 1582 [53.0%] male) were studied. Pathogenic germline variants were found in 397 patients (13.3%), including 282 moderate- and high-penetrance cancer susceptibility genes. Variants of uncertain significance were found in 1415 patients (47.4%). A total of 192 patients (6.4%) had incremental clinically actionable findings that would not have been detected by phenotype or family history–based testing criteria. Of those with a high-penetrance PGV, 42 patients (28.2%) had modifications in their treatment based on the finding. Only younger age of diagnosis was associated with presence of PGV. Only 70 patients (17.6%) with PGVs had family members undergoing no-cost cascade FVT.
Conclusions and Relevance
This prospective, multicenter cohort study found that universal multigene panel testing among patients with solid tumor cancer was associated with an increased detection of heritable variants over the predicted yield of targeted testing based on guidelines. Nearly 30% of patients with high-penetrance variants had modifications in their treatment. Uptake of cascade FVT was low despite being offered at no cost.
This cohort study examines the prevalence of pathogenic germline variants in patients with cancer using a universal testing approach compared with targeted testing based on clinical guidelines and the subsequent uptake of cascade family variant testing.
Introduction
Hereditary factors play a key role in the risk of developing several cancers.1,2 Identification of a germline predisposition can have important implications for treatment decisions, risk-reducing interventions, cancer screening, and germline testing for affected patients and their close relatives. Prior studies3,4,5 have estimated the prevalence of germline cancer susceptibility in patients with breast, prostate, and colorectal cancer. However, most of these estimates have been from registry populations, genetic testing companies, and high-risk cancer clinics, likely producing a referral and ascertainment bias. Selection for genetic testing has traditionally been based on pathologic features of the cancer, age at diagnosis, family history of cancer, and other factors stipulated in clinical practice guidelines.6 Few studies7,8 have compared the prevalence of germline findings in patients with cancer unselected by practice guidelines. It remains unclear what the impact of broad-based testing for inherited germline variants is in patients with cancer compared with more traditional approaches of selection for genetic testing.
After the identification of a germline cancer predisposition syndrome in a symptomatic proband, cascade family variant testing (FVT) in disease-free relatives affords the opportunity for genetically targeted primary disease prevention.9,10 The degree of cascade FVT uptake significantly impacts the cost-effectiveness of genetic testing programs11,12; however, its uptake remains low, with cost likely being a significant barrier.13 The goals of this study were to examine the incremental yield of pathogenic and likely pathogenic germline variants (PGVs) in inherited susceptibility genes detected by a broad universal testing strategy compared with a targeted strategy based on clinical guidelines, the association of universal genetic testing with clinical management, and the proportion of families who would undergo cascade FVT when offered at no cost.
Methods
Patient Selection
We undertook a prospective, multisite study of germline genetic alterations among patients with solid tumor cancer receiving care at Mayo Clinic Cancer Centers in Rochester, Minnesota; Jacksonville, Florida; and Phoenix, Arizona, and in a community oncology practice (Mayo Clinic Health System, Eau Claire, Wisconsin) between April 1, 2018, and March 31, 2020 (the Interrogating Cancer Etiology Using Proactive Genetic Testing [INTERCEPT] program). Patients were unselected for cancer type, stage of disease, family history of cancer, race/ethnicity, or age and underwent germline testing using a greater than 80-gene next-generation sequencing platform. This study was approved by the Mayo Clinic Institutional Review Board. All patients provided written informed consent. Data were deidentified except to the study investigators. The study followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline.
During the study period, English-speaking adult (18-85 years of age) patients with a new or active cancer diagnosis, with confirmed pathologic diagnosis of carcinoma, and treated in medical oncology, radiation oncology, dermatology, or surgical oncology clinics at any of the 3 Mayo Clinic Cancer Centers or the Mayo Clinic Health System were enrolled. Patients hematologic malignant cancers and those undergoing surveillance after curative cancer were excluded. Patients were recruited using central lists of daily oncology clinic visits by research coordinators at each site. Patients were not selected based on cancer type, stage of disease, family history of cancer, race/ethnicity, age at diagnosis, multifocal tumors, or personal history of multiple malignant neoplasms.
All patients viewed a standardized pretest education video and were offered additional pretest genetic counseling if desired. Germline sequencing using a next-generation sequencing panel of 83 genes (84 genes as of July 2019) on the Invitae Multi-Cancer panel was offered at no cost (eAppendix 1 in the Supplement). All test results were reviewed by a certified genetic counselor (M.K., M.H., S.M.-M.) and disclosed to the patient, and those with PGVs were invited for genetic counseling.
Clinical, demographic, and family history data and pathologic information were collected on all patients from medical records or self-administered electronic questionnaires. Race/ethnicity (ancestry) was determined by patient self-report. Family history information was collected using an electronic pedigree tool (CancerGene Connect).
Sequencing, Germline Variant Interpretation, and Result Reporting
Full gene sequencing, deletion and duplication analysis, and variant interpretation were performed at Invitae as previously described (eAppendix 2 in the Supplement).7,14,15 Pathogenic germline variants were classified as high (relative risk >4), intermediate (relative risk, 2-4), or low (relative risk <2) penetrance or recessive medically actionable variants.
Comparison of Guideline-Based Testing
Guidelines from the National Comprehensive Cancer Network (NCCN) and the National Society of Genetic Counselors (NSGC)6,16,17,18,19,20 and the American College of Medical Genetics (ACMG)21 were used to determine whether genetic testing was indicated for a particular patient. For patients who met the guidelines, it was assumed that the only genes tested were those recommended by the tumor-specific guideline. A PGV was considered incremental if it was detected based on the universal testing performed in this study and would not have been identified in targeted testing approaches recommended by published guidelines.
Family Variant Testing
Cascade FVT was offered at no cost to all blood relatives of affected participants with PGVs within a 90-day window of the patient’s finalized test result report. At the time of genetic counseling, patients were informed of the FVT program offered by Invitae and assisted in communicating this information to their relatives through the use of a standardized template letter and an online video that described the risks and benefits of genetic testing.
Statistical Analysis
Demographic and clinical characteristics of the cohort are presented using descriptive statistics. The prevalences of PGVs and variants of unknown significance (VUSs) are reported in the cohort. Categorical variables were compared using the Pearson χ2 test. Rates of incremental findings were compared between subgroups by the Pearson χ2 test, and proportions of germline findings were compared by stage of disease using the χ2 test. P < .05 was considered to be statistically significant. All statistical tests were 2 sided. Univariate logistic regression models were used to estimate pathogenic germline variants.
Results
Cohort Characteristics
A total of 3095 patients were enrolled in the study; 111 patients were ultimately excluded because (1) no blood sample was obtained for genetic testing (n = 12), (2) consent was withdrawn by the patient (n = 96), or (3) genetic testing was not performed at Invitae (n = 3), leaving a final analytic cohort of 2984 patients (mean [SD] age, 61.4 [12.2] years; 1582 [53.0%] male) (eFigure 1 in the Supplement). The distribution of cancer diagnoses, sex, age, and stage of disease in these patients is given in eTable 1 in the Supplement. A total of 535 patients (18.6%) had stage 0/I disease, 477 (16.7%) had stage II disease, 593 (20.7%) had stage III disease, and 1257 (43.9%) had stage IV disease at the time of genomic analysis. Race/ ethnicity distribution included 159 Hispanic/Latino (5.3%), 110 African American (3.7%), and 53 Asian (1.8%) patients. A family history of cancer in a first-degree relative was reported in 1019 participants (34.1%).
Variants Detected
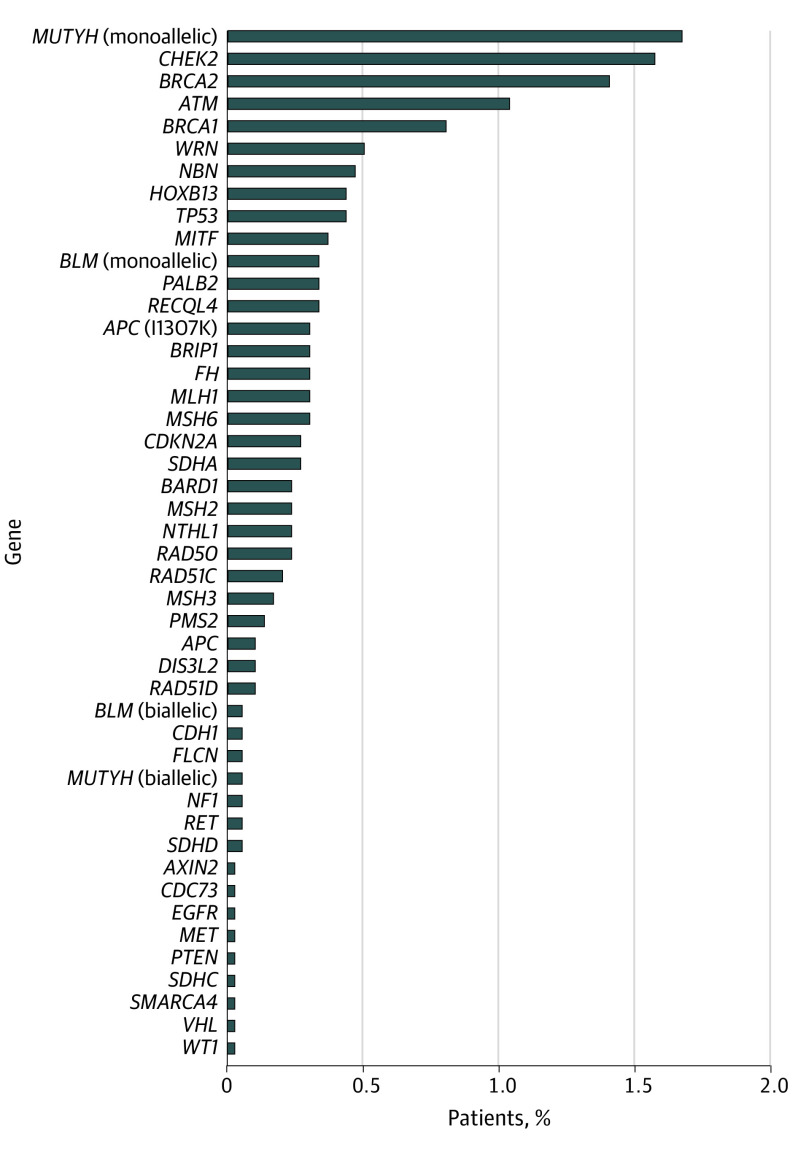
Among a total of 2984 patients, 397 (13.3%) harbored 426 PGVs. More than 1 PGV variant was detected in 27 patients, of whom 2 had 3 variants detected. Pathogenic germline variants could be stratified into those with high (n = 149), moderate (n = 133), or low (n = 65) penetrance, and 50 patients were carriers of variants associated with recessive syndromes. The PGV rates ranged from a low of 7.8% at the Mayo Clinic Health System and rates of 12% to 17% at the 3 Mayo Clinic Cancer Centers (P = .03). The 6 most common PGVs were found in BRCA1 (OMIM 113705) and BRCA2 (OMIM 600185) (66 [2.2%]), monoallelic MUTYH (OMIM 604933) (50 [1.7%]), CHEK2 (OMIM 604373) (47 [1.6%]), Lynch mismatch repair genes (29 [1.0%]), and ATM (OMIM 607585) (31 [1.0%]) (Figure 1). A total of 1415 patients (47.4%) had VUSs. The incidence of germline PGVs in various tumors ranged from 7.3% for melanoma to more than 13% for several cancer types (ovarian, 20.6%; pancreas, 15.9%; colorectal, 15.3%; prostate, 13.7%; lung, 14.7%; cholangiocarcinoma, 14.5%; endometrial, 13.3%; and bladder, 14.2%). The distribution of sex, age, ancestry, and cancer type and stage stratified by variant type is given in eTable 2 in the Supplement. Of importance, the variant rate by stage of cancer was similar: 13.7% in stage 0 to 2 disease and 13.1% in stage 3 to 4 disease (P = .24). Those with a family history of cancer in the same organ system had a high rate of PGVs (21.8%). A total of 13.8% of the recruited population had non-White ancestry, and the prevalence of PGVs was 10.9% and VUSs was 62.2% (vs 53.4% for White ancestry, P < .001). Figure 2 shows the distribution of genetic variants seen in each cancer type listed by penetrance class (high, moderate, low, or recessive) and variant classification (pathogenic/likely pathogenic, VUS, or benign). eFigure 2 in the Supplement shows the specific type of PGV (deletion, insertion, duplication, or missense) listed by gene and tumor type. Most PGVs were missense variants (250 [58.7%]), followed by deletion (131 [30.8%]), duplication (33 [7.7%]), and insertion (12 [2.8%]).
Figure 1. Distribution of Pathogenic and Likely Pathogenic Variants in 397 Patients With Cancer.
Figure 2. Germline Variant and Pathogenicity Shown by Tumor Type.
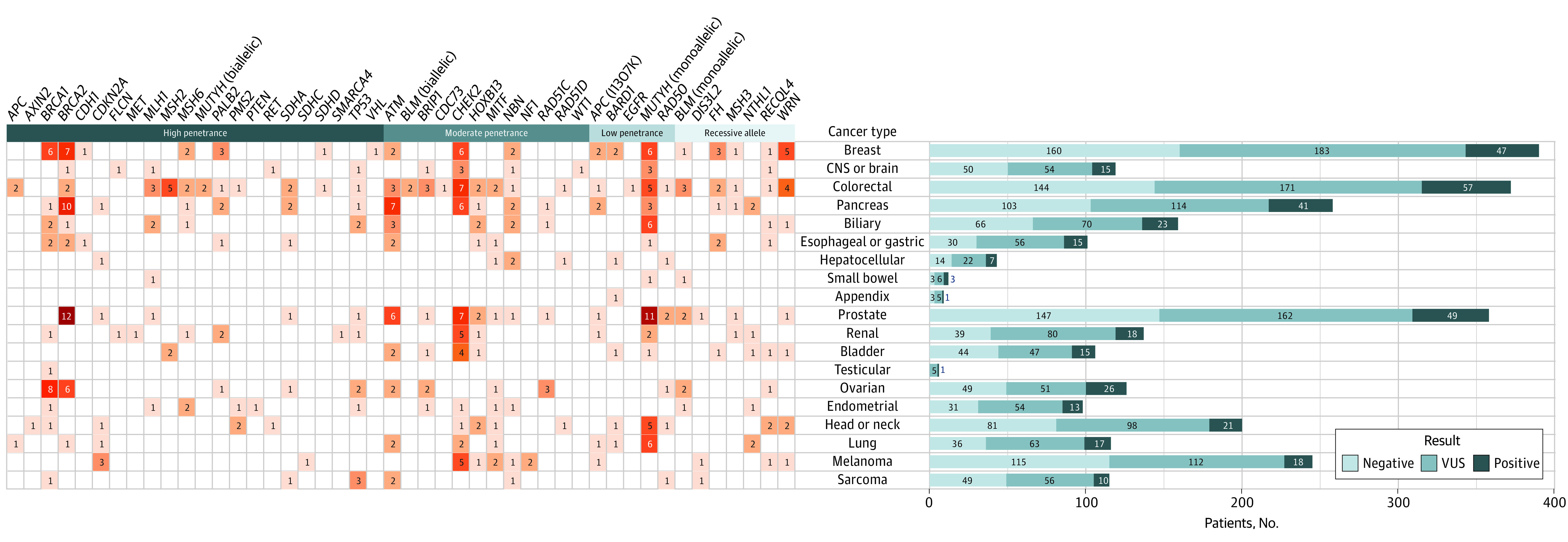
CNS indicates central nervous system; VUS, variant of unknown significance. Shades of color correspond to the number of PGVs in the particular genes, with light to dark representing the fewest to the most.
Application of Clinical Genetic Referral Criteria
A total of 192 cases had incremental findings that would not have been detected by phenotype or family history–based testing criteria using the 2018 NCCN, NSGC, or ACMG guidelines (eTable 3 in the Supplement). This finding represents 6.4% of the 2984 cases overall and 48.4% of the 397 patients with PGVs. Of the 192 patients with incremental findings, 35 (18.2%) carried high-penetrance PGVs, 83 (43.2%) carried moderate-penetrance PGVs, 41 (21.4%) carried low-penetrance PGVs, and 33 (17.2%) carried recessive-penetrance PGVs. The incremental detection rate only changed minimally (42.6%) when using current (2020) guidelines (P = .10 comparing 2018 and 2020 miss rates). Patients with incremental PGVs based on 2020 criteria (n = 169) are described in eTable 4 in the Supplement. These patients are older, less likely to have a BRCA or Lynch mismatch repair gene variant, and less likely to have breast or gynecologic cancer compared with those with nonincremental PGVs.
The ACMG has identified 59 genes that are considered medically actionable. With the use of the panel recommended by the ACMG, the prevalence of PGVs would be 5.8% (n = 174), and VUSs would be present in 11.0% (n = 329). However, the panel would not have identified 223 patients with PGVs identified using the larger panel in this study, including 168 with PGVs in high- or moderate-penetrance genes.
Clinical Implications of PGVs
With few exceptions, the genes in which high-penetrance PGVs were identified have published management recommendations, including options for precision therapy or enrollment in clinical trials. Of the 149 patients with a high-penetrance PGV, 42 (28.2%) had clinically actionable management and treatment changes that were indicated by the detected PGVs (eTable 5 in the Supplement). These changes can be broadly categorized as need for surgery (n = 18), targeted therapy (n = 21), or enrollment in a clinical trial (n = 2).
Cascade FVT
No-cost FVT was offered to all blood relatives of affected participants. Seventy patients (17.6%) with PGVs had relatives who underwent FVT within a 3-month window of their test result. Of the 176 family members who received FVT, 79 (45%) had positive results . The characteristics of patients whose families underwent FVT vs those whose families did not are summarized in eTable 6 in the Supplement. The median number of family members tested per proband was 2.0 (range, 1-14). Similarly, the number of FVTs was low in families in which a PGV was present in the BRCA1, BRCA2, or Lynch mismatch repair gene (range, 16%-20%).
Factors Associated With PGVs
Logistic regression analyses found that younger age at cancer diagnosis (<50 years) was associated with an increased likelihood of having a PGV (odds ratio, 1.38; 95% CI, 1.06-1.78). Patients with colorectal, pancreatic, and prostate cancer had similar prevalences of PGVs compared with patients with breast cancer, whereas patients with melanoma had a significantly lower likelihood of PGVs (odds ratio, 0.58; 95% CI, 0.32-1.00). Sex, stage of cancer, and family history of cancer were not associated with a higher likelihood of having a PGV (eTable 7 in the Supplement). Results were similar in the adjusted logistic regression model.
Discussion
In this multisite, prospective cohort study of unselected patients with cancer, universal germline genetic testing found that approximately 1 in 8 patients (13.3%) harbored a PGV, and 48% of those would not have been detected using standard guidelines. Nearly 30% of patients with a high-penetrance PGV had modifications in their treatment based on the finding, and cascade FVT was underused even when cost was not a barrier.
The overall PGV prevalence in this study was 13.3%. A wide range of PGV prevalence has been reported in the literature (range, 3%-17.5%).22,23,24 Our results are consistent with those from a multicenter study of patients recruited from specialized cancer genetics clinics in California.8 Although a study from New York23 reported a much higher PGV prevalence of 17.5%, this study was restricted to patients with advanced-stage disease. Our study has the advantage of having enrollment from multiple cancer care sites across 4 US states, including a community cancer program, and a broad representation of cancer types and stages compared with prior publications.8,23
A striking result was that nearly 1 in 2 identified PGVs would have not been detected using the 2018 or 2020 NCCN guidelines, including identification of 124 high- or moderate-penetrance PGVs. This finding highlights the limitations of clinical and guideline-based risk assessment and is consistent with what other studies have reported.23,25,26,27,28,29
In 28% of patients with PGV in high-penetrance genes, clinical management and treatment changes were made based on the detected PGVs, including 15.4% who receiving targeted therapies or participated in clinical trials. This finding is consistent with a prior study23 from New York that reported discussion of initiation of targeted therapy in 18% of patients with PGVs.
There are limited data on PGV rates among non-White populations undergoing multigene panel testing. In our cohort, 413 patients (13.8%) reported non-White ancestry, and the rate of PGV was 10.9% (n = 45 of 413). Our data are consistent with reports24,30,31 from several other centers at which PGV rates in Hispanic populations have ranged from 6.7% to 14.1%, depending on the size of gene panel used. The rate of VUSs in non-White populations was also much higher than in the White population in our study, which may reflect the historical emphasis of genetic research on White European populations, leading to an association with variant interpretation in minority populations.32,33 This finding indicates the importance of investigating the use and implication of multipanel genetic testing in underrepresented populations.
Concerns have been raised about high rates of VUSs identified in multigene panel testing. Consistent with prior studies,3,8,15,22,23 we report a VUS rate of 47%. Several studies34,35,36 have described the limited confidence that oncologists have with the interpretation and correct management of VUS results. A related concern is when genes with unknown or unclear clinical relevance may prompt invasive procedures or morbid prophylactic operations. There were also genes for which no PGVs were identified, including some recently introduced to guidelines and those involved with rare cancers. Although one might argue that these genes should not be tested, decreasing costs of testing allow broader application of comprehensive panels, which enables identification of clinically relevant PGVs that might otherwise be missed because of limited family history or a nonclassic phenotype. These issues will be important to address as broader genetic testing is incorporated into practice.
Equally important to the discovery of PGVs in a patient with cancer is the potential to share these findings with their relatives, allowing for earlier disease detection and cancer prevention. In our study, the traditional barrier of cost was removed, and efforts were made to ensure accurate communication of the results to family members with a letter and an easily accessible video that explained the benefits and limitations of genetic testing. Surprisingly, FVT was pursued in less than 20% of families of probands with a PGV. This result is consistent with several other studies37,38 that have found low rates of cascade FVT, including a low-cost online initiative from Color Genomics39 and a study40 of free FVT conducted in Singapore, in which uptake was 21%. It is possible that other barriers led to low uptake of FVT, such as limited comprehension by at-risk relatives after notification, the technical nature of genetic test reports, concerns regarding genetic discrimination and insurance coverage, and emotional burden of a cancer diagnosis. Although it is possible that FVT was conducted through other laboratories, the number of such patients would likely be small because of the financial advantage (no cost) of completing the testing through the same laboratory used in the study.
Strengths and Limitations
This study has several strengths. This is a prospective, multicenter study of a diverse population of patients with cancer across the southwestern, southeastern, and midwestern US, including a community oncology practice. This study implemented a model of pretest genetic education via video that would be practical for a high volume practice. The recruitment of patients regardless of cancer type, stage, age of diagnosis, or family history of cancer allows for generalization of the findings to a wide variety of cancer practices. This study was uniquely able to examine the association of the PGV finding with clinical management and to ascertain cascade FVT within the integrated multistate health system. To our knowledge, this is the first study of universal guideline agnostic multigene panel testing in a large, prospective, multicenter cohort of nearly 3000 patients with cancer.
This study also has limitations. For example, there is a lack of long-term follow-up to assess cancer-related mortality and the morbidity associated with targeted therapy, preventive screening, or prophylactic surgery. The interpretation of family history and need for testing was performed by expert reviewers using guidelines that changed during the study. The cascade FVT portion of the study relied on communication of genetic test results to family members solely by proband-mediated disclosure, which is suboptimal and complicated by a range of complex personal, social, and cultural factors. Finally, the demographic characteristics and case mix of patients seen at the multiple Mayo Clinic sites that participated in this study may not mirror those in all regions of the country and thus may limit generalizability.
Conclusions
In this large, prospective, multisite cohort study with a broad mixture of cancer types and stages, a variety of academic and community cancer practices, and patients unselected for family cancer history, universal multigene panel testing was associated with increased detection of clinically actionable, heritable variants over the predicted yield by targeted testing based on clinical guidelines. Nearly 30% of patients with high-penetrance variants had modifications in their treatment. The rate of FVT remained low, although the financial barrier of such testing was not present in this study, suggesting the importance of removing financial barriers from this process. This study offers significant insight into the performance of multigene panel testing and has broad implications for its wide clinical implementation and acceptance in oncology practice.
eAppendix 1. Invitae Multigene Panel List by Penetrance Category
eFigure 1. Patient Flow Diagram
eFigure 2. Germline Mutation Type Shown by Tumor Type
eTable 1. Participant Characteristics by Clinical Site
eTable 2. Participant and Tumor Characteristics by Genetic Test Result
eTable 3. Incidence of Findings Not Predicted by Clinical Guidelines (Incremental Findings)
eTable 4. Participant and Tumor Characteristics by Incremental Vs Nonincremental PGM
eTable 5. Clinical Implications of Those With High Penetrance PGM
eTable 6. Characteristics of Participants Whose Families Did or Did Not Undergo Cascade Family Variant Testing
eTable 7. Logistic Regression ORs and 95% CI of Patient and Tumor Predictors of Pathogenic/Likely Pathogenic Mutation
eReferences
References
- 1.Samadder NJ, Baffy N, Giridhar KV, Couch FJ, Riegert-Johnson D. Hereditary cancer syndromes-a primer on diagnosis and management, part 2: gastrointestinal cancer syndromes. Mayo Clin Proc. 2019;94(6):1099-1116. doi: 10.1016/j.mayocp.2019.01.042 [DOI] [PubMed] [Google Scholar]
- 2.Samadder NJ, Giridhar KV, Baffy N, Riegert-Johnson D, Couch FJ. Hereditary cancer syndromes: a primer on diagnosis and management, part 1: breast-ovarian cancer syndromes. Mayo Clin Proc. 2019;94(6):1084-1098. doi: 10.1016/j.mayocp.2019.02.017 [DOI] [PubMed] [Google Scholar]
- 3.Yurgelun MB, Kulke MH, Fuchs CS, et al. Cancer susceptibility gene mutations in individuals with colorectal cancer. J Clin Oncol. 2017;35(10):1086-1095. doi: 10.1200/JCO.2016.71.0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desmond A, Kurian AW, Gabree M, et al. Clinical actionability of multigene panel testing for hereditary breast and ovarian cancer risk assessment. JAMA Oncol. 2015;1(7):943-951. doi: 10.1001/jamaoncol.2015.2690 [DOI] [PubMed] [Google Scholar]
- 5.Nicolosi P, Ledet E, Yang S, et al. Prevalence of germline variants in prostate cancer and implications for current genetic testing guidelines. JAMA Oncol. 2019;5(4):523-528. doi: 10.1001/jamaoncol.2018.6760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta S, Provenzale D, Llor X, et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Colorectal, Version 2.2019. J Natl Compr Canc Netw. 2019;17(9):1032-1041. doi: 10.6004/jnccn.2019.0044 [DOI] [PubMed] [Google Scholar]
- 7.Beitsch PD, Whitworth PW, Hughes K, et al. Underdiagnosis of hereditary breast cancer: are genetic testing guidelines a tool or an obstacle? J Clin Oncol. 2019;37(6):453-460. doi: 10.1200/JCO.18.01631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Idos G Multicenter prospective cohort study of the diagnostic yield and patient experience of multiplex gene panel testing for hereditary cancer risk. JCO Precis Oncol. 2019;3:1-12. doi: 10.1200/PO.18.00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samimi G, Bernardini MQ, Brody LC, et al. Traceback: a proposed framework to increase identification and genetic counseling of BRCA1 and BRCA2 mutation carriers through family-based outreach. J Clin Oncol. 2017;35(20):2329-2337. doi: 10.1200/JCO.2016.70.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knowles JW, Rader DJ, Khoury MJ. Cascade screening for familial hypercholesterolemia and the use of genetic testing. JAMA. 2017;318(4):381-382. doi: 10.1001/jama.2017.8543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eccleston A, Bentley A, Dyer M, et al. A cost-effectiveness evaluation of germline BRCA1 and BRCA2 testing in UK women with ovarian cancer. Value Health. 2017;20(4):567-576. doi: 10.1016/j.jval.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mvundura M, Grosse SD, Hampel H, Palomaki GE. The cost-effectiveness of genetic testing strategies for Lynch syndrome among newly diagnosed patients with colorectal cancer. Genet Med. 2010;12(2):93-104. doi: 10.1097/GIM.0b013e3181cd666c [DOI] [PubMed] [Google Scholar]
- 13.Roberts MC, Dotson WD, DeVore CS, et al. Delivery of cascade screening for hereditary conditions: a scoping review of the literature. Health Aff (Millwood). 2018;37(5):801-808. doi: 10.1377/hlthaff.2017.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakane T, Chiba S. Constrictor responses of isolated and perfused monkey coronary arteries to pindolol. Jpn J Pharmacol. 1988;47(1):91-94. doi: 10.1254/jjp.47.91 [DOI] [PubMed] [Google Scholar]
- 15.Kurian AW, Hare EE, Mills MA, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol. 2014;32(19):2001-2009. doi: 10.1200/JCO.2013.53.6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daly MB, Pilarski R, Yurgelun MB, et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 1.2020. J Natl Compr Canc Netw. 2020;18(4):380-391. doi: 10.6004/jnccn.2020.0017 [DOI] [PubMed] [Google Scholar]
- 17.Daly MB, Pilarski R, Berry M, et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast and Ovarian, Version 2.2017. J Natl Compr Canc Netw. 2017;15(1):9-20. doi: 10.6004/jnccn.2017.0003 [DOI] [PubMed] [Google Scholar]
- 18.Gupta S, Provenzale D, Regenbogen SE, et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Colorectal, Version 3.2017. J Natl Compr Canc Netw. 2017;15(12):1465-1475. doi: 10.6004/jnccn.2017.0176 [DOI] [PubMed] [Google Scholar]
- 19.Mohler JL, Antonarakis ES, Armstrong AJ, et al. Prostate Cancer, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2019;17(5):479-505. doi: 10.6004/jnccn.2019.0023 [DOI] [PubMed] [Google Scholar]
- 20.Mohler JL, Armstrong AJ, Bahnson RR, et al. Prostate Cancer, Version 1.2016. J Natl Compr Canc Netw. 2016;14(1):19-30. doi: 10.6004/jnccn.2016.0004 [DOI] [PubMed] [Google Scholar]
- 21.Hampel H, Bennett RL, Buchanan A, Pearlman R, Wiesner GL; Guideline Development Group, American College of Medical Genetics and Genomics Professional Practice and Guidelines Committee and National Society of Genetic Counselors Practice Guidelines Committee . A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet Med. 2015;17(1):70-87. doi: 10.1038/gim.2014.147 [DOI] [PubMed] [Google Scholar]
- 22.Tung N, Lin NU, Kidd J, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol. 2016;34(13):1460-1468. doi: 10.1200/JCO.2015.65.0747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandelker D, Zhang L, Kemel Y, et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA. 2017;318(9):825-835. doi: 10.1001/jama.2017.11137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenthal ET, Bernhisel R, Brown K, Kidd J, Manley S. Clinical testing with a panel of 25 genes associated with increased cancer risk results in a significant increase in clinically significant findings across a broad range of cancer histories. Cancer Genet. 2017;218-219:58-68. doi: 10.1016/j.cancergen.2017.09.003 [DOI] [PubMed] [Google Scholar]
- 25.Neben CL, Zimmer AD, Stedden W, et al. Multi-gene panel testing of 23,179 individuals for hereditary cancer risk identifies pathogenic variant carriers missed by current genetic testing guidelines. J Mol Diagn. 2019;21(4):646-657. doi: 10.1016/j.jmoldx.2019.03.001 [DOI] [PubMed] [Google Scholar]
- 26.Brand R, Borazanci E, Speare V, et al. Prospective study of germline genetic testing in incident cases of pancreatic adenocarcinoma. Cancer. 2018;124(17):3520-3527. doi: 10.1002/cncr.31628 [DOI] [PubMed] [Google Scholar]
- 27.Lowery MA, Wong W, Jordan EJ, et al. Prospective evaluation of germline alterations in patients with exocrine pancreatic neoplasms. J Natl Cancer Inst. 2018;110(10):1067-1074. doi: 10.1093/jnci/djy024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlo MI, Mukherjee S, Mandelker D, et al. Prevalence of germline mutations in cancer susceptibility genes in patients with advanced renal cell carcinoma. JAMA Oncol. 2018;4(9):1228-1235. doi: 10.1001/jamaoncol.2018.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Idos G, Gupta S. When should patients undergo genetic testing for hereditary colon cancer syndromes? Clin Gastroenterol Hepatol. 2018;16(2):181-183. doi: 10.1016/j.cgh.2017.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ricker C, Culver JO, Lowstuter K, et al. Increased yield of actionable mutations using multi-gene panels to assess hereditary cancer susceptibility in an ethnically diverse clinical cohort. Cancer Genet. 2016;209(4):130-137. doi: 10.1016/j.cancergen.2015.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caswell-Jin JL, Gupta T, Hall E, et al. Racial/ethnic differences in multiple-gene sequencing results for hereditary cancer risk. Genet Med. 2018;20(2):234-239. doi: 10.1038/gim.2017.96 [DOI] [PubMed] [Google Scholar]
- 32.Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell. 2019;177(4):1080. doi: 10.1016/j.cell.2019.04.032 [DOI] [PubMed] [Google Scholar]
- 33.Ndugga-Kabuye MK, Issaka RB. Inequities in multi-gene hereditary cancer testing: lower diagnostic yield and higher VUS rate in individuals who identify as Hispanic, African or Asian and Pacific Islander as compared to European. Fam Cancer. 2019;18(4):465-469. doi: 10.1007/s10689-019-00144-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pal T, Cragun D, Lewis C, et al. A statewide survey of practitioners to assess knowledge and clinical practices regarding hereditary breast and ovarian cancer. Genet Test Mol Biomarkers. 2013;17(5):367-375. doi: 10.1089/gtmb.2012.0381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurian AW, Friese CR, Bondarenko I, et al. Second opinions from medical oncologists for early-stage breast cancer: prevalence, correlates, and consequences. JAMA Oncol. 2017;3(3):391-397. doi: 10.1001/jamaoncol.2016.5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurian AW, Li Y, Hamilton AS, et al. Gaps in incorporating germline genetic testing into treatment decision-making for early-stage breast cancer. J Clin Oncol. 2017;35(20):2232-2239. doi: 10.1200/JCO.2016.71.6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffin NE, Buchanan TR, Smith SH, et al. Low rates of cascade genetic testing among families with hereditary gynecologic cancer: an opportunity to improve cancer prevention. Gynecol Oncol. 2020;156(1):140-146. doi: 10.1016/j.ygyno.2019.11.005 [DOI] [PubMed] [Google Scholar]
- 38.Stoffel EM, Ford B, Mercado RC, et al. Sharing genetic test results in Lynch syndrome: communication with close and distant relatives. Clin Gastroenterol Hepatol. 2008;6(3):333-338. doi: 10.1016/j.cgh.2007.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caswell-Jin JL, Zimmer AD, Stedden W, Kingham KE, Zhou AY, Kurian AW. Cascade genetic testing of relatives for hereditary cancer risk: results of an online initiative. J Natl Cancer Inst. 2019;111(1):95-98. doi: 10.1093/jnci/djy147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Courtney E, Chok AK, Ting Ang ZL, et al. Impact of free cancer predisposition cascade genetic testing on uptake in Singapore. NPJ Genom Med. 2019;4:22. doi: 10.1038/s41525-019-0096-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix 1. Invitae Multigene Panel List by Penetrance Category
eFigure 1. Patient Flow Diagram
eFigure 2. Germline Mutation Type Shown by Tumor Type
eTable 1. Participant Characteristics by Clinical Site
eTable 2. Participant and Tumor Characteristics by Genetic Test Result
eTable 3. Incidence of Findings Not Predicted by Clinical Guidelines (Incremental Findings)
eTable 4. Participant and Tumor Characteristics by Incremental Vs Nonincremental PGM
eTable 5. Clinical Implications of Those With High Penetrance PGM
eTable 6. Characteristics of Participants Whose Families Did or Did Not Undergo Cascade Family Variant Testing
eTable 7. Logistic Regression ORs and 95% CI of Patient and Tumor Predictors of Pathogenic/Likely Pathogenic Mutation
eReferences