Abstract
This study aimed to verify noteworthy findings between genetic risk factors and autism spectrum disorder (ASD) by employing the false positive report probability (FPRP) and the Bayesian false-discovery probability (BFDP). PubMed and the Genome-Wide Association Studies (GWAS) catalog were searched from inception to 1 August, 2019. We included meta-analyses on genetic factors of ASD of any study design. Overall, twenty-seven meta-analyses articles from literature searches, and four manually added articles from the GWAS catalog were re-analyzed. This showed that five of 31 comparisons for meta-analyses of observational studies, 40 out of 203 comparisons for the GWAS meta-analyses, and 18 out of 20 comparisons for the GWAS catalog, respectively, had noteworthy estimations under both Bayesian approaches. In this study, we found noteworthy genetic comparisons highly related to an increased risk of ASD. Multiple genetic comparisons were shown to be associated with ASD risk; however, genuine associations should be carefully verified and understood.
Keywords: autism spectrum disorder, false positive report probability (FPRP), Bayesian false-discovery probability (BFDP), meta-analysis, Genome-Wide Association Studies (GWAS)
1. Introduction
Autism spectrum disorder (ASD) is a brain-based neurodevelopmental disorder characterized by pervasive impairments in reciprocal social communication, social interaction, and restricted and repetitive behaviors or interests, resulting in a substantial burden of individuals, families, and society [1,2]. The repeated reports of recent increase in the prevalence of ASD have raised substantial public concerns. For example, in large, nationwide population-based studies, the estimated ASD prevalence was reported to be 2.47% among U.S. children and adolescents in 2014–2016 [3,4,5].
Although the full range of etiologies underlying ASD remain largely unexplained, progress has been made in the past decade in identifying some neurobiological and genetic risk factors, and it has been well established that combination of genetic and environmental factors is involved in the etiopathogenesis of autism [1,6]. There is a strong genetic background of ASD, which was demonstrated by the fact that heritability is as high as 80–90% [7,8]. It is possible to estimate the heritability of ASD by taking into the account its covariance within twins, as twins are matched for many characteristics, including in utero and family environment, as well as other developmental aspects [7,9,10].
ASD is polygenic and genetic variants contribute to ASD risk and phenotypic variability. The results of previous studies showed genome-wide genetic links between ASD [11,12]. They indicated that typical variation in social behavior and adaptive functioning and multiple types of genetic risk for ASD influence a continuum of behavioral and developmental traits.
To the best of our knowledge, this is the comprehensive study to summarize the loci that are associated with ASD among the several known loci reported to be related with ASD. We have synthesized all available susceptibility loci for ASD retrieved from meta-analyses regarding the association between the individual polymorphisms and ASD. For the study, we reviewed observational studies, Genome-Wide Association Studies (GWAS) meta-analyses, the combined analysis of GWAS discovery and replication cohorts, the GWAS catalog and GWAS data from GWAS meta-analyses [13]. Furthermore, we applied a Bayesian approaches including false positive report probability (FPRP) and Bayesian false discovery probability (BFDP) to estimate the noteworthiness of the evidence [14,15]. Using these popular Bayesian statistics (i.e., FPRP and BFDP), our study shows that the results of genotype associations between the gene variant and disease were found to be noteworthy (genuine associations). Through these methods, we selected only statistically meaningful values excluding false-positive values and analyzed them again. We aimed to provide an overview to interpret the statistical significance of reported findings and discuss the identified associations in the suggested genetic risk factors for ASD.
2. Materials and Methods
This review was conducted following a registered protocol. The specified methods are available on the PROSPERO database with the registration number CRD42018091704. The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines of this review are shown in Supplementary Table S1.
2.1. Experimental Section
2.1.1. Inclusion and Exclusion Criteria
Studies were included if they satisfied the following conditions: (1) estimated the risk of ASD in humans using meta-analyses in terms of odds ratio (OR) and 95% confidence interval (CI); (2) published in English. Articles were excluded if (1) they did not cover the subject of genetic polymorphism or ASD; (2) did not have individual results for ASD; (3) did not use statistical methods of meta-analysis.
2.1.2. Search Strategy
A PubMed search was performed to extract data from meta-analyses regarding the gene polymorphisms of ASD published until 1 August, 2019. Two of the authors (MJ Son and CY Son) used the search terms (autism AND meta OR meta-analysis) and obtained relevant articles, first, by scanning the titles and abstracts and, second, by reviewing the full-text (Figure 1). During the selection process, all genetic, gen*, and related terms were included in the relevant articles. Any disagreements were resolved by discussion and consensus. In the case of GWAS, the GWAS catalog was additionally used, as well as PubMed, for a more precise search.
Figure 1.
Flow chart of literature search.
2.1.3. Data Extraction
From each article, we extracted the first author, year of publication, the number of individual studies included, the number of cases and controls, and the number of families if a meta-analysis included family-based studies, the type of statistical model (fixed or random) and study design. We also recorded gene name, gene variants, genotypic comparison, OR with 95% CI, and the corresponding p-value. We retrieved all the main data (preferably adjusted), and, for comprehensiveness we additionally extracted subgroup analysis data if the main data were not statistically significant. When data were incomplete, we contacted the corresponding authors for additional information.
Reported association was considered statistically significant if p-value < 0.05 for meta-analyses of observational studies, and <5 × 10−8 for GWAS or meta-analyses of GWAS. Meanwhile, genetic associations with a 5 × 10−8 < p-value < 0.05 were defined as being of borderline significance in GWAS or meta-analyses of GWAS. In addition, we recorded genetic comparisons with p-value < 5 × 10−8 for our gene network, even when they were not re-analyzable due to insufficient raw data.
2.2. Statistical Analysis
Evaluations of the statistical significance of studies about genetic polymorphisms too often inferred false positives, when the evaluations were solely based on p-value [15]. Therefore, to clarify “noteworthy” association between re-analyzable genetic variants and ASD, we employed the two Bayesian approaches: FPRP and BFDP [15]. We used the Excel spreadsheets created by Wacholder et al. [15] and Wakefield [14] to calculate FPRP and BFDP, respectively. We computed FPRP at two prior probability levels of 10−3 and 10−6 and used statistical power to detect two OR levels, 1.2 and 1.5, so that readers can make their own judgment about the evidence for each genetic variant. BFDP is similar to FPRP but uses more information than FPRP [14]. Both prior probability levels were chosen as one of the low and very low values of levels, respectively. We computed BFDP at two prior probabilities levels, 10-3 and 10−6. We set the thresholds of noteworthiness of FPRP and BFDP to be <0.2 and <0.8, respectively, as recommended by the original papers and highlighted corresponding results in bold type [14,15]. Gene variants were determined to have a noteworthy association with ASD if they satisfied both thresholds.
2.3. Construction of Protein-Protein Interaction (PPI) Network
We collected genetic comparisons either with noteworthy results under both FPRP and BFDP or with p-value < 5 × 10−8 to establish a network of genes using STRING 9.1 (protein-protein interaction network, PPI network) related to ASD [16]. Genetic comparison results, which show genome-wide significance (p-value < 5 × 10−8) or borderline significance (p-value < 0.05) with a noteworthy association under both Bayesian approaches, were included. Any results with a p-value < 5 × 10−8 that were not re-analyzable were also added in the network analysis. PPI networks provide a critical assessment of protein function on ASD including direct (physical) as well as indirect (functional) associations.
3. Results
3.1. Study Characteristics
The initial PubMed literature search yielded 747 articles. Out these, 656 articles were excluded after screening the title and abstract, and 64 articles were omitted after reviewing the full-text. Twenty-seven studies were finally included for the re-analysis of observational studies, GWAS, and meta-analyses of GWAS (Figure 1).
Additionally, 25 articles were searched on the GWAS catalog, but 14 articles did not meet the criteria were excluded. Among the remaining 11 articles, five articles were not re-analyzable due to insufficient raw data. Moreover, five articles were already included in our dataset from the PubMed search. However, we retained three of the non-re-analyzable articles [17,18,19] since they satisfied the cut-off value of statistical significance for our PPI network (p-value < 5 × 10−8). Out of the remaining six articles, two were already in our dataset from the literature search from PubMed. Finally, four articles from the GWAS catalog were manually added to 27 articles previously screened from PubMed, leading to a total of 31 eligible articles [17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47] being included in the systematic review (Figure 1).
3.2. Re-Analysis of Meta-Analyses
This paper is divided into two parts: (1) the observational studies part, and (2) the GWAS part. In the observational studies, all statistics were collected considering the overlapping, and results of gene variants with/without statistical significance (Table 1, Supplementary Table S2). Even though genetic variants examined in several studies, we excluded the studies if the data were not significant performed by FPRP or BFDP. In the GWAS part, data from previously published meta-analyses and newly added data from the GWAS catalog were re-analyzed.
Table 1.
Re-analysis results of gene variants with statistical significance (p-value < 0.05) from observational studies.
| Author, Year | Gene/Variant | Comparison | OR (95% CI) | p-Value | Model | No. of Studies | Power OR 1.2 |
Power OR 1.5 |
FPRP Values at Prior Probability | BFDP 0.001 |
BFDP 0.000001 |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR 1.2 | OR 1.5 | |||||||||||||
| 0.001 | 0.000001 | 0.001 | 0.000001 | |||||||||||
| Gene variants with statistically significance (p-value < 0.05), FPRP < 0.2 and BFDP < 0.8 from observational studies | ||||||||||||||
| Rai 2016 [21] | MTHFR C677T | T vs. C | 1.37 (1.25, 1.50) | <0.0001 | Fixed | Overall (13) | 0.002 | 0.975 | 0.000 | 0.005 | 0.000 | 0.000 | 0.000 | 0.001 |
| Mohammad et al., 2016 [23] | MTHFR C677T | T (minor) | 1.47 (1.31, 1.65) | <0.0001 | Fixed | Overall (8) | 0.000 | 0.634 | 0.000 | 0.179 | 0.000 | 0.000 | 0.000 | 0.009 |
| Warrier et al., 2015 [24] | DRD3/rs167771 | G vs. A | 1.822 (1.398, 2.375) | 9.08 × 10−6 | Fixed | Overall (2) | 0.001 | 0.075 | 0.901 | 1.000 | 0.108 | 0.992 | 0.649 | 0.999 |
| Warrier et al., 2015 [24] | RELN/rs362691 | C vs. G | 0.832 (0.763, 0.908) | 3.93 × 10−5 | Fixed | Overall (6) | 0.486 | 1.000 | 0.071 | 0.987 | 0.036 | 0.974 | 0.584 | 0.999 |
| LoParo et al., 2015 [26] | OXTR/rs7632287 | A (minor) | 1.43 (1.23, 1.68) | 0.000005 | Random | Caucasian (2) | 0.016 | 0.720 | 0.451 | 0.999 | 0.018 | 0.950 | 0.432 | 0.999 |
| Gene variants with statistically significance (p-value < 0.05), FPRP > 0.2 or BFDP > 0.8 from observational studies | ||||||||||||||
| Liu et al., 2015 [20] | SLC25A12/rs2056202 | T vs. C | 0.809 (0.713, 0.917) | 0.001 | Fixed | Overall (8) | 0.321 | 0.999 | 0.740 | 1.000 | 0.478 | 0.999 | 0.957 | 1.000 |
| Liu et al., 2015 [20] | SLC25A12/rs2292813 | T vs. C | 0.752 (0.649,0.871) | <0.001 | Fixed | Overall (7) | 0.085 | 0.946 | 0.626 | 0.999 | 0.131 | 0.993 | 0.831 | 1.000 |
| Pu et al., 2013 [22] | MTHFR C677T | TT+CT vs. CC | 1.56 (1.12, 2.18) | 0.009 | Random | Overall (8) | 0.062 | 0.409 | 0.993 | 1.000 | 0.957 | 1.000 | 0.995 | 1.000 |
| Pu et al., 2013 [22] | MTHFR A1298C | CC vs. AA+AC | 0.73 (0.56, 0.97) | 0.03 | Fixed | Overall (5) | 0.181 | 0.734 | 0.994 | 1.000 | 0.976 | 1.000 | 0.997 | 1.000 |
| Warrier et al., 2015 [24] | SLC25A12/rs2292813 | C vs. T | 1.372 (1.161, 1.621) | 1.97 × 10−4 | Fixed | Overall (6) | 0.058 | 0.853 | 0.777 | 1.000 | 0.191 | 0.996 | 0.877 | 1.000 |
| Warrier et al., 2015 [24] | CNTNAP2/rs7794745 | A vs. T | 0.887 (0.828, 0.950) | 1.00 × 10−3 | Fixed | Overall (3) | 0.963 | 1.000 | 0.389 | 0.998 | 0.380 | 0.998 | 0.952 | 1.000 |
| Warrier et al., 2015 [24] | SLC25A12/rs2056202 | T vs. C | 1.227 (1.079, 1.396) | 2.00 × 10−3 | Fixed | Overall (8) | 0.368 | 0.999 | 0.837 | 1.000 | 0.654 | 0.999 | 0.976 | 1.000 |
| Warrier et al., 2015 [24] | OXTR/rs2268491 | T vs. C | 1.31 (1.092, 1.572) | 4.00 × 10−3 | Fixed | Overall (2) | 0.173 | 0.927 | 0.955 | 1.000 | 0.799 | 1.000 | 0.987 | 1.000 |
| Warrier et al., 2015 [24] | EN2/rs1861972 | A vs. G | 1.125 (1.035, 1.224) | 6.00 × 10−3 | Fixed | Overall (8) | 0.933 | 1.000 | 0.869 | 1.000 | 0.861 | 1.000 | 0.993 | 1.000 |
| Warrier et al., 2015 [24] | MTHFR/rs1801133 | T vs. C | 1.370 (1.079, 1.739) | 1.00 × 10−2 | Random | Overall (10) | 0.138 | 0.772 | 0.986 | 1.000 | 0.926 | 1.000 | 0.994 | 1.000 |
| Warrier et al., 2015 [24] | ASMT/rs4446909 | G vs. A | 1.195 (1.038, 1.375) | 1.30 × 10−2 | Fixed | Overall (3) | 0.523 | 0.999 | 0.961 | 1.000 | 0.928 | 1.000 | 0.995 | 1.000 |
| Warrier et al., 2015 [24] | MET/rs38845 | A vs. G | 1.322 (1.013, 1.724) | 1.60 × 10−2 | Random | Overall (3) | 0.237 | 0.824 | 0.994 | 1.000 | 0.979 | 1.000 | 0.998 | 1.000 |
| Warrier et al., 2015 [24] | SLC6A4/rs2020936 | T vs. C | 1.244 (1.036, 1.492) | 1.90 × 10−2 | Fixed | Overall (4) | 0.349 | 0.978 | 0.982 | 1.000 | 0.950 | 1.000 | 0.996 | 1.000 |
| Warrier et al., 2015 [24] | SLC6A4/STin2 VNTR | 12 vs. 9/10 | 1.492 (1.068, 2.083) | 1.90 × 10−2 | Fixed | Caucasian (4) | 0.100 | 0.513 | 0.995 | 1.000 | 0.973 | 1.000 | 0.997 | 1.000 |
| Warrier et al., 2015 [24] | STX1A/rs4717806 | A vs. T | 0.851 (0.741, 0.978) | 2.30 × 10−2 | Fixed | Overall (4) | 0.616 | 1.000 | 0.974 | 1.000 | 0.958 | 1.000 | 0.997 | 1.000 |
| Warrier et al., 2015 [24] | RELN/rs736707 | T vs. C | 1.269 (1.030, 1.563) | 2.50 × 10−2 | Random | Overall (7) | 0.299 | 0.942 | 0.988 | 1.000 | 0.964 | 1.000 | 0.997 | 1.000 |
| Warrier et al., 2015 [24] | PON1/rs662 | A vs. G | 0.794 (0.642, 0.983) | 3.40 × 10−2 | Fixed | Overall (2) | 0.329 | 0.946 | 0.990 | 1.000 | 0.973 | 1.000 | 0.997 | 1.000 |
| Warrier et al., 2015 [24] | OXTR/rs237887 | G vs. A | 1.163 (1.002, 1.349) | 4.70 × 10−2 | Fixed | Overall (2) | 0.660 | 1.000 | 0.986 | 1.000 | 0.979 | 1.000 | 0.998 | 1.000 |
| Warrier et al., 2015 [24] | EN2/rs1861973 | T vs. C | 0.86 (0.791, 0.954) | 3.00 × 10−3 | Fixed | TDT (3) | 0.724 | 1.000 | 0.858 | 1.000 | 0.814 | 1.000 | 0.989 | 1.000 |
| Aoki et al., 2016 [25] | SCL25A12/rs2292813 | G (risk allele) | 1.190 (1.052, 1.346) | 0.006 | Random | Overall (9) | 0.553 | 1.000 | 0.911 | 1.000 | 0.849 | 1.000 | 0.990 | 1.000 |
| Aoki et al., 2016 [25] | SCL25A12/rs2056202 | G (risk allele) | 1.206 (1.035, 1.405) | 0.016 | Random | Overall (10) | 0.474 | 0.997 | 0.972 | 1.000 | 0.942 | 1.000 | 0.996 | 1.000 |
| LoParo et al., 2015 [26] | OXTR/rs237887 | G (minor allele) | 0.89 (0.79, 0.98) | 0.0239 | Random | Overall (3) | 0.910 | 1.000 | 0.951 | 1.000 | 0.947 | 1.000 | 0.997 | 1.000 |
| LoParo et al., 2015 [26] | OXTR/rs2268491 | T (minor allele) | 1.20 (1.05, 1.35) | 0.0075 | Random | Overall (3) | 0.500 | 1.000 | 0.828 | 1.000 | 0.707 | 1.000 | 0.981 | 1.000 |
| Wang et al., 2014 [27] | RELN/rs362691 | R vs. NR | 0.69 (0.56, 0.86) | 0.001 | Fixed | Overall (7) | 0.047 | 0.620 | 0.954 | 1.000 | 0.607 | 0.999 | 0.969 | 1.000 |
| Torrico et al., 2015 [28] | PTCHD1/rs7052177 | T (major allele) | 0.58 (0.45, 0.76) | 6.8 × 10−5 | Fixed | European (4) † | 0.004 | 0.156 | 0.948 | 1.000 | 0.333 | 0.998 | 0.890 | 1.000 |
| Kranz et al., 2016 [29] | OXTR/rs237889 | A vs. G | 1.12 (1.01, 1.24) | 0.0365 | Random | Overall (3) | 0.908 | 1.000 | 0.970 | 1.000 | 0.967 | 1.000 | 0.998 | 1.000 |
Abbreviations: A, Adenine; C, Cytosine; G, Guanine; T, Thymine; R, Risk allele; NR, Non-risk allele; FPRP, false positive rate probability; BFDP, Bayesian false discovery probability; OR, odds ratio; CI, confidence interval; NA, not available; The bold in the table means significant results by FPRP and BFDP. † This article reported only the number of datasets not the number of individual studies included in the meta-analysis. Thus, we wrote the number of datasets in the parenthesis.
3.2.1. Re-Analysis of Meta-Analyses of Observational Studies
Among the 31 eligible studies, 19 were meta-analyses of observational studies, which corresponded to 125 genetic comparisons. Thirty one out of 125 genotype comparisons were reported as being statistically significant using the criteria of p-value < 0.05 as listed in Table 1.
Out of the 31 genotype comparisons (Table 1), three (9.7%), and two (6.5%) were verified to be noteworthy (<0.2) using FPRP estimation, at a prior probability of 10−3 and 10−6 with a statistical power to detect an OR of 1.2; seven (22.6%) and two (6.5%) were verified to be noteworthy (<0.2) using FPRP estimation, at a prior probability of 10−3 and 10−6 with a statistical power to detect an OR of 1.5. In terms of BFDP, five (16.1%) and two (6.5%) comparisons had noteworthy findings (<0.8) at a prior probability of 10−3 and 10−6. Two single nucleotide polymorphisms (SNPs) were found to be noteworthy under FPRP estimation only, and not under BFDP (Comparison T vs. C, SLC25A12/rs2292813 [20]; C vs. T, SLC25A12/rs2292813 [24]). In contrast, none of the SNPs were identified to be noteworthy exclusively under BFDP. Consequently, five out of 31 SNPs were found noteworthy using both FPRP and BFDP (T vs. C, MTHFR C677T; T (minor), MTHFR C677T; Comparison G vs. A, DRD3/rs167771; C vs. G, RELN/rs362691; A (minor), OXTR/rs7632287).
3.2.2. Re-Analysis of Meta-Analyses of GWAS
Seven GWAS meta-analyses and one study with a combined analysis of GWAS discovery and replication added up to 203 genetic comparisons [30,31,32,33,34,46,47,48] with statistical or borderline significant results. Out of 277 comparisons, 44 had p-value ≥ 0.05 (Table S2), none of which showed noteworthy estimation of FPRP and BFDP with statistical or borderline significant results. From the 203 comparisons, only one (0.5%), MACROD2/rs4141463 A (minor allele), was statistically significant under the genome-wide significance threshold (p-value < 5 × 10−8), while the remaining 202 comparisons (99.5%) satisfied the criteria of borderline significance (5 × 10−8 < p-value < 0.05) previously defined.
We examined the 203 genetic comparisons with a genome-wide or borderline significance using both FPRP and BFDP estimation. With FPRP estimation, forty-one (20.2%) and four (2.0%) were assessed to be noteworthy at a prior probability of 10−3 and 10−6 with statistical power to detect an OR of 1.2. Moreover, fifty-four (26.6%) and eight (3.9%) were identified as noteworthy at a prior probability of 10−3 and 10−6 with statistical power to detect an OR of 1.5. Overall, forty genetic comparisons (19.7%) were found noteworthy under both Bayesian approaches, which included a single genetic comparison satisfying the conventional significance threshold of p-value < 0.05 (Table 2).
Table 2.
Re-analysis results of gene variants with genome wide statistical significance (p-value < 5 × 10−8) and borderline statistical significance (5 × 10−8 ≤ p-value < 0.05) in GWAS meta-analyses.
| Author, Year | Gene | Variant | Comparison | OR (95% CI) | p-Value | Power OR 1.2 |
Power OR 1.5 |
FPRP Values at Prior Probability | BFDP 0.001 |
BFDP 0.000001 |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR 1.2 | OR 1.5 | ||||||||||||
| 0.001 | 0.000001 | 0.001 | 0.000001 | ||||||||||
| Gene variants with statistically significance (p-value < 5 × 10−8), FPRP < 0.2 and BFDP < 0.8 from meta-analysis of GWAS | |||||||||||||
| Anney et al., 2010 [30] | MACROD2 | rs4141463 | A (minor allele) | 0.73 (0.66–0.82) | 3.7 × 10−8 | 0.013 | 0.937 | 0.009 | 0.898 | 0.000 | 0.107 | 0.008 | 0.891 |
| Gene variants with statistically borderline significance (5 × 10−8 ≤ p-value < 0.05), FPRP < 0.2 and BFDP < 0.8 from meta-analyses of GWAS | |||||||||||||
| Anney et al., 2017 [31] | ALPK3 NMB SCAND2P SEC11A SLC28A1 WDR73 ZNF592 | rs4842996 | T vs. C | 1.08 (1.05–1.12) | 0.00001044 | 1.000 | 1.000 | 0.032 | 0.971 | 0.032 | 0.971 | 0.688 | 1.000 |
| EXOC4 | rs6467494 | T vs. C | 1.07 (1.04–1.09) | 0.0000172 | 1.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| NA | rs13233145 | A vs. C | 1.07 (1.04–1.10) | 0.00002906 | 1.000 | 1.000 | 0.002 | 0.618 | 0.002 | 0.618 | 0.136 | 0.994 | |
| NA | rs7684366 | T vs. C | 0.93 (0.90–0.96) | 0.00003137 | 1.000 | 1.000 | 0.007 | 0.882 | 0.007 | 0.882 | 0.373 | 0.998 | |
| MEGF10 | rs73785549 | C vs. G | 1.15 (1.08–1.21) | 0.0001308 | 0.950 | 1.000 | 0.000 | 0.070 | 0.000 | 0.067 | 0.005 | 0.835 | |
| ANO4 | rs2055471 | A vs. T | 1.07 (1.03–1.10) | 0.0001334 | 1.000 | 1.000 | 0.002 | 0.618 | 0.002 | 0.618 | 0.136 | 0.994 | |
| BNC2 | rs7860276 | A vs. G | 1.10 (1.05–1.15) | 0.0003196 | 1.000 | 1.000 | 0.026 | 0.964 | 0.026 | 0.964 | 0.598 | 0.999 | |
| NA | rs2293280 | C vs. G | 1.12 (1.06–1.18) | 0.0003606 | 0.995 | 1.000 | 0.020 | 0.954 | 0.020 | 0.954 | 0.514 | 0.999 | |
| NA | rs16975940 | T vs. C | 1.07 (1.03–1.10) | 0.0004742 | 1.000 | 1.000 | 0.002 | 0.618 | 0.002 | 0.618 | 0.136 | 0.994 | |
| NA | rs10169115 | C vs. G | 1.06 (1.02–1.09) | 0.004465 | 1.000 | 1.000 | 0.041 | 0.977 | 0.041 | 0.977 | 0.778 | 1.000 | |
| C10orf76 CUEDC2 ELOVL3 FBXL15 GBF1 HPS6 LDB1 MIR146B NFKB2 NOLC1 PITX3 PPRC1 PSD | rs1409313 | T vs. C | 1.10 (1.06–1.14) | 1.467 × 10−6 | 1.000 | 1.000 | 0.000 | 0.145 | 0.000 | 0.145 | 0.014 | 0.936 | |
| ESRRG | rs12725407 | C vs. G | 1.10 (1.06–1.14) | 2.115 × 10−6 | 1.000 | 1.000 | 0.000 | 0.145 | 0.000 | 0.145 | 0.014 | 0.936 | |
| HDAC4 MIR2467 MIR4269 | rs2931203 | A vs. T | 0.92 (0.88–0.95) | 4.243 × 10−6 | 1.000 | 1.000 | 0.000 | 0.261 | 0.000 | 0.261 | 0.031 | 0.970 | |
| Ma et al., 2009 [32] | NA | rs7704909 | C(minor)/T(major) | 1.30 (1.15–1.46) | 1.53 × 10−5 | 0.088 | 0.992 | 0.096 | 0.991 | 0.009 | 0.905 | 0.295 | 0.998 |
| NA | rs1896731 | C(minor)/T(major) | 0.76 (0.67–0.85) | 1.90 × 10−5 | 0.053 | 0.989 | 0.028 | 0.966 | 0.002 | 0.609 | 0.076 | 0.988 | |
| NA | rs12518194 | G(minor)/A(major) | 1.31 (1.16–1.49) | 8.34 × 10−6 | 0.091 | 0.980 | 0.302 | 0.998 | 0.039 | 0.976 | 0.605 | 0.999 | |
| NA | rs4307059 | C(minor)/T(major) | 1.31 (1.16–1.48) | 1.29 × 10−5 | 0.079 | 0.985 | 0.153 | 0.995 | 0.014 | 0.936 | 0.383 | 0.998 | |
| NA | rs4327572 | T(minor)/C(major) | 1.32 (1.17–1.49) | 4.05 × 10−6 | 0.062 | 0.981 | 0.103 | 0.991 | 0.007 | 0.878 | 0.249 | 0.997 | |
| Anney et al., 2010 [30] | NA | rs4078417 | C (minor allele) | 1.19 (1.10–1.30) | 5.6 × 10−5 | 0.574 | 1.000 | 0.167 | 0.995 | 0.103 | 0.991 | 0.795 | 1.000 |
| PPP2R5C | rs7142002 | G (minor allele) | 0.64 (0.53–0.78) | 2.9 × 10−6 | 0.004 | 0.343 | 0.687 | 1.000 | 0.028 | 0.966 | 0.459 | 0.999 | |
| Kuo et al., 2015 [33] | NAALADL2 | rs3914502 | A (minor allele) | 1.4 (1.2–1.6) | 3.5 × 10−6 | 0.012 | 0.844 | 0.062 | 0.985 | 0.001 | 0.482 | 0.051 | 0.982 |
| NAALADL2 | rs2222447 | A (minor allele) | 0.7 (0.6–0.8) | 5.3 × 10−5 | 0.005 | 0.763 | 0.030 | 0.969 | 0.000 | 0.178 | 0.013 | 0.932 | |
| NA | rs12543592 | G (minor allele) | 0.7 (0.6–0.8) | 3.2 × 10−6 | 0.005 | 0.763 | 0.030 | 0.969 | 0.000 | 0.178 | 0.013 | 0.932 | |
| NA | rs7026342 | C (minor allele) | 1.6 (1.2–2.0) | 1.8 × 10−4 | 0.006 | 0.285 | 0.864 | 1.000 | 0.113 | 0.992 | 0.749 | 1.000 | |
| NA | rs7030851 | A (minor allele) | 1.6 (1.3–2.0) | 1.4 × 10−4 | 0.006 | 0.285 | 0.864 | 1.000 | 0.113 | 0.992 | 0.749 | 1.000 | |
| Anney et al., 2012 [34] | RASSF5 | rs11118968 | A | 0.44 (0.32–0.61) | 2.452 × 10−7 | 0.000 | 0.006 | 0.930 | 1.000 | 0.117 | 0.993 | 0.504 | 0.999 |
| DNER | rs6752370 | G | 1.62 (1.33–1.96) | 8.526 × 10−7 | 0.001 | 0.214 | 0.407 | 0.999 | 0.003 | 0.764 | 0.089 | 0.990 | |
| YEATS2 | rs263035 | G | 1.39 (1.22–1.57) | 2.258 × 10−7 | 0.009 | 0.890 | 0.013 | 0.928 | 0.000 | 0.115 | 0.009 | 0.898 | |
| None | rs29456 | A | 1.65 (1.37–1.99) | 1.226 × 10−7 | 0.000 | 0.159 | 0.272 | 0.997 | 0.001 | 0.504 | 0.028 | 0.967 | |
| None | rs1936295 | A | 1.69 (1.37–2.09) | 6.636 × 10−7 | 0.001 | 0.136 | 0.620 | 0.999 | 0.009 | 0.905 | 0.179 | 0.995 | |
| None | rs4761371 | A | 0.46 (0.34–0.63) | 3.914 × 10−7 | 0.000 | 0.010 | 0.924 | 1.000 | 0.111 | 0.992 | 0.521 | 0.999 | |
| None | rs288604 | G | 1.58 (1.32–1.88) | 2.975 × 10−7 | 0.001 | 0.279 | 0.207 | 0.996 | 0.001 | 0.473 | 0.032 | 0.971 | |
| MACROD2 | rs6110458 | A | 1.46 (1.27–1.69) | 1.806 × 10−7 | 0.004 | 0.641 | 0.084 | 0.989 | 0.001 | 0.383 | 0.033 | 0.971 | |
| MACROD2 NCRNA00186 | rs14135 | G | 1.49 (1.28–1.74) | 1.778 × 10−7 | 0.003 | 0.534 | 0.130 | 0.993 | 0.001 | 0.467 | 0.042 | 0.977 | |
| NCRNA00186 MACROD2 | rs1475531 | C | 1.53 (1.30–1.79) | 2.011 × 10−7 | 0.001 | 0.402 | 0.083 | 0.989 | 0.000 | 0.213 | 0.013 | 0.929 | |
| PARD3B | rs4675502 | NA | 1.28 (1.16–1.41) | 4.34 × 10−7 | 0.095 | 0.999 | 0.006 | 0.856 | 0.001 | 0.362 | 0.030 | 0.969 | |
| NA | rs7711337 | NA | 0.82 (0.76–0.89) | 8.25 × 10−7 | 0.350 | 1.000 | 0.006 | 0.854 | 0.002 | 0.672 | 0.091 | 0.990 | |
| NA | rs7834018 | NA | 0.64 (0.53–0.77) | 7.54 × 10−7 | 0.003 | 0.333 | 0.465 | 0.999 | 0.007 | 0.871 | 0.186 | 0.996 | |
| TAF1C | rs4150167 | NA | 0.51 (0.39–0.66) | 2.91 × 10−7 | 0.000 | 0.021 | 0.764 | 1.000 | 0.015 | 0.937 | 0.142 | 0.994 | |
| Gene variants with statistically borderline significance (5 × 10−8 ≤ p-value < 0.05), FPRP > 0.2 or BFDP > 0.2 from meta-analyses of GWAS | |||||||||||||
| Waltes et al., 2014 [46] | CYFIP1c | rs7170637 | G > A | 0.85 (0.75, 0.96) | 0.007 | 0.625 | 1.000 | 0.934 | 1.000 | 0.898 | 1.000 | 0.993 | 1.000 |
| CAMK4c | rs25925 | C > G | 1.31 (1.04, 1.64) | 0.021 | 0.222 | 0.881 | 0.988 | 1.000 | 0.954 | 1.000 | 0.996 | 1.000 | |
| Anney et al., 2017 [31] | NA | rs1436358 | T vs. C | 0.86 (0.79–0.93) | 0.00001473 | 0.785 | 1.000 | 0.168 | 0.995 | 0.137 | 0.994 | 0.844 | 1.000 |
| MACROD2 MACROD2-AS1 | rs6079556 | A vs. C | 0.94 (0.91–0.97) | 0.00001731 | 1.000 | 1.000 | 0.102 | 0.991 | 0.102 | 0.991 | 0.887 | 1.000 | |
| LINC00535 | chr8_94389815_I | I vs. D | 0.92 (0.89–0.96) | 0.00002102 | 1.000 | 1.000 | 0.109 | 0.992 | 0.109 | 0.992 | 0.867 | 1.000 | |
| LINCR-0001 PRSS55 | rs4840484 | T vs. C | 1.07 (1.04–1.11) | 0.00002307 | 1.000 | 1.000 | 0.232 | 0.997 | 0.232 | 0.997 | 0.945 | 1.000 | |
| Anney et al., 2017 (continued) | ADTRP | rs10947543 | C vs. G | 0.94 (0.91–0.97) | 0.000031 | 1.000 | 1.000 | 0.102 | 0.991 | 0.102 | 0.991 | 0.887 | 1.000 |
| LRRC4 MIR593 SND1 SND1-IT1 | chr7_127644308_D | D vs. I | 0.93 (0.90–0.97) | 0.00003235 | 1.000 | 1.000 | 0.422 | 0.999 | 0.422 | 0.999 | 0.972 | 1.000 | |
| CCDC93 DDX18 INSIG2 | chr2_118616767_D | I vs. D | 0.85 (0.78–0.93) | 0.00003531 | 0.667 | 1.000 | 0.374 | 0.998 | 0.285 | 0.997 | 0.921 | 1.000 | |
| NA | chr14_99235398_I | I vs. D | 0.87 (0.81–0.94) | 0.00003765 | 0.862 | 1.000 | 0.327 | 0.998 | 0.296 | 0.998 | 0.930 | 1.000 | |
| TTBK1 | rs2756174 | A vs. C | 0.94 (0.91–0.97) | 0.00005245 | 1.000 | 1.000 | 0.102 | 0.991 | 0.102 | 0.991 | 0.887 | 1.000 | |
| HCG4B HLA-A HLA-H | rs115254791 | T vs. G | 0.94 (0.90–0.97) | 0.00005321 | 1.000 | 1.000 | 0.102 | 0.991 | 0.102 | 0.991 | 0.887 | 1.000 | |
| MIR2113 | rs9482120 | A vs. C | 0.94 (0.91–0.97) | 0.00009513 | 1.000 | 1.000 | 0.102 | 0.991 | 0.102 | 0.991 | 0.887 | 1.000 | |
| CRTAP SUSD5 | chr3_33191013_D | I vs. D | 0.93 (0.89–0.97) | 0.0000957 | 1.000 | 1.000 | 0.422 | 0.999 | 0.422 | 0.999 | 0.972 | 1.000 | |
| NA | rs9285005 | A vs. G | 0.91 (0.86–0.96) | 0.0001147 | 0.999 | 1.000 | 0.354 | 0.998 | 0.354 | 0.998 | 0.956 | 1.000 | |
| LOC100505609 | rs73065342 | T vs. C | 0.89 (0.83–0.95) | 0.0001169 | 0.976 | 1.000 | 0.322 | 0.998 | 0.317 | 0.998 | 0.941 | 1.000 | |
| DCAF4 DPF3 PAPLN PSEN1 RBM25 ZFYVE1 | rs1203311 | A vs. C | 0.86 (0.79–0.94) | 0.0001394 | 0.756 | 1.000 | 0.540 | 0.999 | 0.470 | 0.999 | 0.960 | 1.000 | |
| MACROD2 | rs192259652 | A vs. T | 0.91 (0.85–0.96) | 0.0001438 | 0.999 | 1.000 | 0.354 | 0.998 | 0.354 | 0.998 | 0.956 | 1.000 | |
| FOXP1 | rs76188283 | T vs. C | 1.09 (1.05–1.14) | 0.0002093 | 1.000 | 1.000 | 0.142 | 0.994 | 0.142 | 0.994 | 0.892 | 1.000 | |
| CCDC38 NTN4 SNRPF | chr12_96221819_D | I vs. D | 0.94 (0.91–0.97) | 0.0002128 | 1.000 | 1.000 | 0.102 | 0.991 | 0.102 | 0.991 | 0.887 | 1.000 | |
| NA | chr3_182308608_I | D vs. I | 0.94 (0.90–0.97) | 0.0002755 | 1.000 | 1.000 | 0.102 | 0.991 | 0.102 | 0.991 | 0.887 | 1.000 | |
| ASTN2 PAPPA PAPPA-AS1 | rs7026354 | A vs. G | 1.05 (1.03–1.08) | 0.0003018 | 1.000 | 1.000 | 0.407 | 0.999 | 0.407 | 0.999 | 0.979 | 1.000 | |
| NA | rs2368140 | A vs. G | 0.94 (0.91–0.98) | 0.0003049 | 1.000 | 1.000 | 0.783 | 1.000 | 0.783 | 1.000 | 0.993 | 1.000 | |
| NA | rs13016472 | T vs. C | 0.94 (0.91–0.98) | 0.0003629 | 1.000 | 1.000 | 0.783 | 1.000 | 0.783 | 1.000 | 0.993 | 1.000 | |
| DSCAM | rs62235658 | T vs. C | 0.92 (0.87–0.97) | 0.0004132 | 1.000 | 1.000 | 0.668 | 1.000 | 0.668 | 1.000 | 0.986 | 1.000 | |
| NA | rs3113169 | C vs. G | 0.93 (0.90–0.97) | 0.0004234 | 1.000 | 1.000 | 0.422 | 0.999 | 0.422 | 0.999 | 0.972 | 1.000 | |
| CASKIN2 GGA3 GRB2 LOC100287042 MIF4GD MIR3678 MIR6785 MRPS7 NUP85 SLC25A19 TMEM94 TSEN54 | rs12950709 | A vs. G | 0.92 (0.87–0.97) | 0.0004387 | 1.000 | 1.000 | 0.668 | 1.000 | 0.668 | 1.000 | 0.986 | 1.000 | |
| CAMP CDC25A CSPG5 DHX30 MAP4 MIR1226 MIR4443 SMARCC1 ZNF589 | rs7429990 | A vs. C | 0.94 (0.91–0.97) | 0.0004525 | 1.000 | 1.000 | 0.102 | 0.991 | 0.102 | 0.991 | 0.887 | 1.000 | |
| NA | chr8_84959513_D | D vs. I | 0.89 (0.83–0.96) | 0.0004634 | 0.956 | 1.000 | 0.728 | 1.000 | 0.718 | 1.000 | 0.985 | 1.000 | |
| ACTN2 | rs4659712 | A vs. G | 0.95 (0.92–0.98) | 0.0004976 | 1.000 | 1.000 | 0.550 | 0.999 | 0.550 | 0.999 | 0.986 | 1.000 | |
| ASB4 | rs113706540 | T vs. C | 0.93 (0.88–0.97) | 0.0005006 | 1.000 | 1.000 | 0.422 | 0.999 | 0.422 | 0.999 | 0.972 | 1.000 | |
| GJD4 | rs7897060 | C vs. G | 0.95 (0.91–0.98) | 0.0005789 | 1.000 | 1.000 | 0.550 | 0.999 | 0.550 | 0.999 | 0.986 | 1.000 | |
| AK5 DNAJB4 FAM73A FUBP1 GIPC2 MGC27382 NEXN NEXN-AS1 USP33 ZZZ3 |
rs12126604 | T vs. C | 0.92 (0.87–0.97) | 0.0006161 | 1.000 | 1.000 | 0.668 | 1.000 | 0.668 | 1.000 | 0.986 | 1.000 | |
| SEMA6D | rs17387110 | T vs. G | 0.95 (0.92–0.98) | 0.0006996 | 1.000 | 1.000 | 0.550 | 0.999 | 0.550 | 0.999 | 0.986 | 1.000 | |
| NA | chr16_62649826_D | D vs. I | 0.87 (0.80–0.95) | 0.0007369 | 0.831 | 1.000 | 0.697 | 1.000 | 0.657 | 0.999 | 0.979 | 1.000 | |
| NA | rs4239875 | A vs. G | 1.06 (1.03–1.10) | 0.0008018 | 1.000 | 1.000 | 0.672 | 1.000 | 0.672 | 1.000 | 0.990 | 1.000 | |
| CTNNA3 DNAJC12 HERC4 MYPN POU5F1P5 SIRT1 | chr10_69763783_D | I vs. D | 0.91 (0.86–0.97) | 0.0008401 | 0.997 | 1.000 | 0.792 | 1.000 | 0.791 | 1.000 | 0.991 | 1.000 | |
| CLIC5 ENPP4 ENPP5 | rs7762549 | A vs. G | 0.95 (0.92–0.98) | 0.00085 | 1.000 | 1.000 | 0.550 | 0.999 | 0.550 | 0.999 | 0.986 | 1.000 | |
| NA | chr18_76035713_D | D vs. I | 0.93 (0.88–0.97) | 0.000884 | 1.000 | 1.000 | 0.422 | 0.999 | 0.422 | 0.999 | 0.972 | 1.000 | |
| BRICD5 CASKIN1 DNASE1L2 E4F1 MIR3180-5 MIR4516 MLST8 PGP PKD1 RAB26 SNHG19 SNORD60 TRAF7 | rs2078282 | A vs. G | 0.94 (0.91–0.98) | 0.0009187 | 1.000 | 1.000 | 0.783 | 1.000 | 0.783 | 1.000 | 0.993 | 1.000 | |
| OPCML | rs7952100 | C vs. G | 1.06 (1.03–1.10) | 0.0009399 | 1.000 | 1.000 | 0.672 | 1.000 | 0.672 | 1.000 | 0.990 | 1.000 | |
| LOC101927907 LRRTM4 | rs58500924 | A vs. G | 0.90 (0.84–0.96) | 0.0009721 | 0.990 | 1.000 | 0.581 | 0.999 | 0.579 | 0.999 | 0.977 | 1.000 | |
| RNGTT | rs35675874 | A vs. G | 0.94 (0.91–0.98) | 0.001031 | 1.000 | 1.000 | 0.783 | 1.000 | 0.783 | 1.000 | 0.993 | 1.000 | |
| LOC101928505 LOC101928539 | chr5_57079215_I | D vs. I | 1.07 (1.03–1.11) | 0.001076 | 1.000 | 1.000 | 0.232 | 0.997 | 0.232 | 0.997 | 0.945 | 1.000 | |
| DPP4 SLC4A10 | rs2909451 | T vs. C | 0.94 (0.90–0.98) | 0.001078 | 1.000 | 1.000 | 0.783 | 1.000 | 0.783 | 1.000 | 0.993 | 1.000 | |
| ERAP2 LNPEP | rs55767008 | T vs. C | 0.89 (0.82–0.96) | 0.001182 | 0.956 | 1.000 | 0.728 | 1.000 | 0.718 | 1.000 | 0.985 | 1.000 | |
| C2orf15 KIAA1211L LIPT1 LOC101927070 TSGA10 | rs10202643 | A vs. T | 0.95 (0.92–0.98) | 0.001269 | 1.000 | 1.000 | 0.550 | 0.999 | 0.550 | 0.999 | 0.986 | 1.000 | |
| AUTS2 | rs2293507 | T vs. G | 0.88 (0.81–0.96) | 0.001337 | 0.890 | 1.000 | 0.817 | 1.000 | 0.799 | 1.000 | 0.989 | 1.000 | |
| NA | rs138457704 | A vs. G | 1.07 (1.03–1.11) | 0.001357 | 1.000 | 1.000 | 0.232 | 0.997 | 0.232 | 0.997 | 0.945 | 1.000 | |
| GLDC | rs13288399 | C vs. G | 0.95 (0.91–0.98) | 0.001357 | 1.000 | 1.000 | 0.550 | 0.999 | 0.550 | 0.999 | 0.986 | 1.000 | |
| MTFR1 PDE7A | rs1513723 | C vs. G | 0.95 (0.92–0.98) | 0.001447 | 1.000 | 1.000 | 0.550 | 0.999 | 0.550 | 0.999 | 0.986 | 1.000 | |
| ASTN2 ASTN2-AS1 PAPPA TRIM32 | rs146737360 | T vs. G | 0.95 (0.92–0.98) | 0.001534 | 1.000 | 1.000 | 0.550 | 0.999 | 0.550 | 0.999 | 0.986 | 1.000 | |
| NA | chr6_45726254_D | D vs. I | 0.90 (0.83–0.96) | 0.001606 | 0.990 | 1.000 | 0.581 | 0.999 | 0.579 | 0.999 | 0.977 | 1.000 | |
| NA | rs6742513 | C vs. G | 1.07 (1.03–1.11) | 0.001611 | 1.000 | 1.000 | 0.232 | 0.997 | 0.232 | 0.997 | 0.945 | 1.000 | |
| NA | rs73204738 | A vs. C | 0.92 (0.88–0.97) | 0.001617 | 1.000 | 1.000 | 0.668 | 1.000 | 0.668 | 1.000 | 0.986 | 1.000 | |
| LINC01553 | rs11817353 | A vs. C | 0.95 (0.92–0.98) | 0.001678 | 1.000 | 1.000 | 0.550 | 0.999 | 0.550 | 0.999 | 0.986 | 1.000 | |
| Anney et al., 2017 (continued) | RAD51B | rs2842330 | A vs. C | 1.10 (1.04–1.16) | 0.001845 | 0.999 | 1.000 | 0.303 | 0.998 | 0.303 | 0.998 | 0.946 | 1.000 |
| RBFOX1 | rs12930616 | C vs. G | 1.05 (1.02–1.09) | 0.001985 | 1.000 | 1.000 | 0.913 | 1.000 | 0.913 | 1.000 | 0.998 | 1.000 | |
| GRID2 | rs6811974 | T vs. C | 0.95 (0.93–0.98) | 0.001995 | 1.000 | 1.000 | 0.550 | 0.999 | 0.550 | 0.999 | 0.986 | 1.000 | |
| NA | rs7135621 | T vs. C | 0.96 (0.93–0.98) | 0.002059 | 1.000 | 1.000 | 0.094 | 0.991 | 0.094 | 0.991 | 0.915 | 1.000 | |
| GFER NOXO1 NPW RNF151 RPS2 SNHG9 SNORA78 SYNGR3 TBL3 ZNF598 | rs55742253 | T vs. C | 0.93 (0.88–0.98) | 0.002075 | 1.000 | 1.000 | 0.868 | 1.000 | 0.868 | 1.000 | 0.995 | 1.000 | |
| PTPRB | rs10784860 | T vs. C | 0.95 (0.91–0.98) | 0.002211 | 1.000 | 1.000 | 0.550 | 0.999 | 0.550 | 0.999 | 0.986 | 1.000 | |
| LOC101927768 | rs9387201 | C vs. G | 1.09 (1.03–1.14) | 0.002427 | 1.000 | 1.000 | 0.142 | 0.994 | 0.142 | 0.994 | 0.892 | 1.000 | |
| BTBD11 LOC101929162 PRDM4 PWP1 | rs4964602 | T vs. G | 0.95 (0.91–0.98) | 0.00256 | 1.000 | 1.000 | 0.550 | 0.999 | 0.550 | 0.999 | 0.986 | 1.000 | |
| NA | rs1376888 | T vs. C | 1.05 (1.02–1.08) | 0.002668 | 1.000 | 1.000 | 0.407 | 0.999 | 0.407 | 0.999 | 0.979 | 1.000 | |
| KLHL29 | rs10182178 | A vs. G | 1.05 (1.02–1.08) | 0.003508 | 1.000 | 1.000 | 0.407 | 0.999 | 0.407 | 0.999 | 0.979 | 1.000 | |
| UBE2H | rs78661858 | A vs. G | 0.91 (0.85–0.97) | 0.003665 | 0.997 | 1.000 | 0.792 | 1.000 | 0.791 | 1.000 | 0.991 | 1.000 | |
| VAPA | rs29063 | A vs. G | 1.04 (1.01–1.07) | 0.004075 | 1.000 | 1.000 | 0.873 | 1.000 | 0.873 | 1.000 | 0.997 | 1.000 | |
| NA | rs190401890 | A vs. T | 1.12 (1.04–1.20) | 0.004114 | 0.975 | 1.000 | 0.568 | 0.999 | 0.562 | 0.999 | 0.975 | 1.000 | |
| LOC102723427 | rs192668887 | T vs. C | 0.91 (0.84–0.97) | 0.004205 | 0.997 | 1.000 | 0.792 | 1.000 | 0.791 | 1.000 | 0.991 | 1.000 | |
| SLC12A7 | rs73031119 | A vs. C | 0.91 (0.84–0.97) | 0.004399 | 0.997 | 1.000 | 0.792 | 1.000 | 0.791 | 1.000 | 0.991 | 1.000 | |
| ADGRL2 | rs75695875 | A vs. G | 0.93 (0.87–0.98) | 0.004715 | 1.000 | 1.000 | 0.868 | 1.000 | 0.868 | 1.000 | 0.995 | 1.000 | |
| NA | rs1943999 | C vs. G | 0.96 (0.92–0.99) | 0.004915 | 1.000 | 1.000 | 0.903 | 1.000 | 0.903 | 1.000 | 0.998 | 1.000 | |
| DNAH6 | rs2222734 | A vs. G | 0.92 (0.87–0.98) | 0.005058 | 0.999 | 1.000 | 0.906 | 1.000 | 0.906 | 1.000 | 0.996 | 1.000 | |
| OR8A1 OR8B12 | rs2226753 | T vs. C | 0.96 (0.93–0.99) | 0.005074 | 1.000 | 1.000 | 0.903 | 1.000 | 0.903 | 1.000 | 0.998 | 1.000 | |
| TUSC5 | rs35713482 | A vs. G | 1.05 (1.01–1.08) | 0.005154 | 1.000 | 1.000 | 0.407 | 0.999 | 0.407 | 0.999 | 0.979 | 1.000 | |
| C5orf15 VDAC1 | rs67120295 | T vs. C | 1.06 (1.02–1.10) | 0.005745 | 1.000 | 1.000 | 0.672 | 1.000 | 0.672 | 1.000 | 0.990 | 1.000 | |
| NA | rs76010911 | A vs. G | 1.11 (1.04–1.19) | 0.006255 | 0.986 | 1.000 | 0.769 | 1.000 | 0.767 | 1.000 | 0.989 | 1.000 | |
| MTMR9 SLC35G5 TDH | rs6601581 | T vs. C | 1.06 (1.02–1.11) | 0.006463 | 1.000 | 1.000 | 0.930 | 1.000 | 0.930 | 1.000 | 0.998 | 1.000 | |
| HSDL2 MIR3134 PTBP3 SUSD1 | rs7024761 | A vs. G | 1.05 (1.02–1.09) | 0.00648 | 1.000 | 1.000 | 0.913 | 1.000 | 0.913 | 1.000 | 0.998 | 1.000 | |
| CRTC3 GABARAPL3 IQGAP1 ZNF774 | rs2601187 | A vs. G | 1.05 (1.01–1.08) | 0.006859 | 1.000 | 1.000 | 0.407 | 0.999 | 0.407 | 0.999 | 0.979 | 1.000 | |
| LOC101927189 LRRC1 | rs4715431 | A vs. G | 1.04 (1.01–1.08) | 0.007007 | 1.000 | 1.000 | 0.977 | 1.000 | 0.977 | 1.000 | 0.999 | 1.000 | |
| NA | rs646680 | A vs. G | 0.95 (0.92–0.99) | 0.00723 | 1.000 | 1.000 | 0.937 | 1.000 | 0.937 | 1.000 | 0.998 | 1.000 | |
| CCNE1 | rs12609867 | A vs. G | 0.95 (0.91–0.99) | 0.00743 | 1.000 | 1.000 | 0.937 | 1.000 | 0.937 | 1.000 | 0.998 | 1.000 | |
| NOS1AP OLFML2B | rs75192393 | T vs. C | 1.07 (1.02–1.12) | 0.007697 | 1.000 | 1.000 | 0.787 | 1.000 | 0.787 | 1.000 | 0.993 | 1.000 | |
| KDM4A KDM4A-AS1 LOC101929592 MIR6079 PTPRF ST3GAL3 |
rs79857083 | T vs. C | 1.04 (1.01–1.08) | 0.007758 | 1.000 | 1.000 | 0.977 | 1.000 | 0.977 | 1.000 | 0.999 | 1.000 | |
| NA | rs142968358 | T vs. G | 1.04 (1.01–1.07) | 0.007789 | 1.000 | 1.000 | 0.873 | 1.000 | 0.873 | 1.000 | 0.997 | 1.000 | |
| C3orf30 IGSF11 IGSF11-AS1 UPK1B | rs1102586 | A vs. G | 1.06 (1.02–1.10) | 0.007844 | 1.000 | 1.000 | 0.672 | 1.000 | 0.672 | 1.000 | 0.990 | 1.000 | |
| NA | chr11_98107192_D | D vs. I | 1.04 (1.01–1.08) | 0.00785 | 1.000 | 1.000 | 0.977 | 1.000 | 0.977 | 1.000 | 0.999 | 1.000 | |
| C9orf135 | rs76014157 | A vs. G | 0.90 (0.82–0.98) | 0.007946 | 0.962 | 1.000 | 0.941 | 1.000 | 0.939 | 1.000 | 0.997 | 1.000 | |
| NA | rs6437449 | A vs. G | 1.07 (1.02–1.11) | 0.008708 | 1.000 | 1.000 | 0.232 | 0.997 | 0.232 | 0.997 | 0.945 | 1.000 | |
| MYO5A | chr15_52811815_D | I vs. D | 0.90 (0.81–0.98) | 0.008799 | 0.962 | 1.000 | 0.941 | 1.000 | 0.939 | 1.000 | 0.997 | 1.000 | |
| NA | rs9466619 | A vs. G | 0.95 (0.92–0.99) | 0.009071 | 1.000 | 1.000 | 0.937 | 1.000 | 0.937 | 1.000 | 0.998 | 1.000 | |
| NA | rs6117854 | A vs. G | 0.96 (0.93–0.99) | 0.01012 | 1.000 | 1.000 | 0.903 | 1.000 | 0.903 | 1.000 | 0.998 | 1.000 | |
| C7orf33 | rs6955951 | A vs. T | 1.04 (1.01–1.07) | 0.01015 | 1.000 | 1.000 | 0.873 | 1.000 | 0.873 | 1.000 | 0.997 | 1.000 | |
| LHX6 | rs72767788 | A vs. C | 0.95 (0.91–0.99) | 0.01093 | 1.000 | 1.000 | 0.937 | 1.000 | 0.937 | 1.000 | 0.998 | 1.000 | |
| NA | rs2028664 | A vs. C | 1.04 (1.01–1.07) | 0.01095 | 1.000 | 1.000 | 0.873 | 1.000 | 0.873 | 1.000 | 0.997 | 1.000 | |
| ELAVL2 | rs180861134 | A vs. T | 1.05 (1.01–1.09) | 0.01104 | 1.000 | 1.000 | 0.913 | 1.000 | 0.913 | 1.000 | 0.998 | 1.000 | |
| RASGEF1C | rs12659560 | T vs. C | 1.04 (1.01–1.07) | 0.0112 | 1.000 | 1.000 | 0.873 | 1.000 | 0.873 | 1.000 | 0.997 | 1.000 | |
| MIR548AZ SYNE2 | rs2150291 | T vs. C | 1.05 (1.01–1.09) | 0.0113 | 1.000 | 1.000 | 0.913 | 1.000 | 0.913 | 1.000 | 0.998 | 1.000 | |
| WDFY4 | rs118059975 | A vs. C | 0.95 (0.91–0.99) | 0.01146 | 1.000 | 1.000 | 0.937 | 1.000 | 0.937 | 1.000 | 0.998 | 1.000 | |
| LINC01525 MAN1A2 | rs3820500 | A vs. G | 1.04 (1.01–1.07) | 0.0116 | 1.000 | 1.000 | 0.873 | 1.000 | 0.873 | 1.000 | 0.997 | 1.000 | |
| GALNT10 | rs17629195 | T vs. C | 1.04 (1.01–1.07) | 0.012 | 1.000 | 1.000 | 0.873 | 1.000 | 0.873 | 1.000 | 0.997 | 1.000 | |
| MIR597 TNKS | rs78853604 | T vs. C | 1.05 (1.01–1.08) | 0.01256 | 1.000 | 1.000 | 0.407 | 0.999 | 0.407 | 0.999 | 0.979 | 1.000 | |
| EXT1 | rs7835763 | A vs. T | 1.04 (1.01–1.08) | 0.01283 | 1.000 | 1.000 | 0.977 | 1.000 | 0.977 | 1.000 | 0.999 | 1.000 | |
| NA | rs4652928 | A vs. G | 0.96 (0.92–0.99) | 0.01384 | 1.000 | 1.000 | 0.903 | 1.000 | 0.903 | 1.000 | 0.998 | 1.000 | |
| PDE1C | rs11976985 | T vs. C | 0.95 (0.92–0.99) | 0.0141 | 1.000 | 1.000 | 0.937 | 1.000 | 0.937 | 1.000 | 0.998 | 1.000 | |
| BAX FTL GYS1 | rs2230267 | T vs. C | 1.04 (1.01–1.07) | 0.01429 | 1.000 | 1.000 | 0.873 | 1.000 | 0.873 | 1.000 | 0.997 | 1.000 | |
| Anney et al., 2017 (continued) | GRID2 | rs6854329 | C vs. G | 0.92 (0.86–0.99) | 0.01486 | 0.996 | 1.000 | 0.963 | 1.000 | 0.963 | 1.000 | 0.998 | 1.000 |
| NA | rs1926229 | C vs. G | 1.05 (1.01–1.08) | 0.01496 | 1.000 | 1.000 | 0.407 | 0.999 | 0.407 | 0.999 | 0.979 | 1.000 | |
| NA | rs261351 | T vs. C | 0.96 (0.93–0.99) | 0.01498 | 1.000 | 1.000 | 0.903 | 1.000 | 0.903 | 1.000 | 0.998 | 1.000 | |
| RAPGEF2 | rs4440173 | A vs. G | 1.04 (1.01–1.07) | 0.01564 | 1.000 | 1.000 | 0.873 | 1.000 | 0.873 | 1.000 | 0.997 | 1.000 | |
| MIR4650-1 MIR4650-2 POM121 SBDSP1 SPDYE7P TYW1B | rs4392770 | T vs. C | 1.05 (1.01–1.09) | 0.01564 | 1.000 | 1.000 | 0.913 | 1.000 | 0.913 | 1.000 | 0.998 | 1.000 | |
| NA | rs138493916 | C vs. G | 1.08 (1.02–1.14) | 0.01783 | 1.000 | 1.000 | 0.840 | 1.000 | 0.840 | 1.000 | 0.994 | 1.000 | |
| NA | rs615512 | A vs. G | 1.08 (1.02–1.14) | 0.01811 | 1.000 | 1.000 | 0.840 | 1.000 | 0.840 | 1.000 | 0.994 | 1.000 | |
| EP400 EP400NL PUS1 SNORA49 | rs11608890 | T vs. G | 0.94 (0.88–0.99) | 0.0187 | 1.000 | 1.000 | 0.951 | 1.000 | 0.951 | 1.000 | 0.998 | 1.000 | |
| DIAPH3 | chr13_60161890_I | I vs. D | 1.05 (1.01–1.09) | 0.01984 | 1.000 | 1.000 | 0.913 | 1.000 | 0.913 | 1.000 | 0.998 | 1.000 | |
| ADAM12 | rs1674923 | T vs. C | 0.96 (0.93–0.99) | 0.0203 | 1.000 | 1.000 | 0.903 | 1.000 | 0.903 | 1.000 | 0.998 | 1.000 | |
| ATP2B2 GHRL GHRLOS IRAK2 LINC00852 MIR378B MIR885 SEC13 TATDN2 |
rs7619385 | A vs. G | 1.04 (1.01–1.07) | 0.02102 | 1.000 | 1.000 | 0.873 | 1.000 | 0.873 | 1.000 | 0.997 | 1.000 | |
| UNC13C | rs75099274 | A vs. G | 1.08 (1.01–1.14) | 0.02123 | 1.000 | 1.000 | 0.840 | 1.000 | 0.840 | 1.000 | 0.994 | 1.000 | |
| ZSWIM6 | rs10053166 | A vs. G | 0.95 (0.90–0.99) | 0.02226 | 1.000 | 1.000 | 0.937 | 1.000 | 0.937 | 1.000 | 0.998 | 1.000 | |
| HIVEP3 | rs2786484 | T vs. C | 0.93 (0.86–0.99) | 0.0237 | 1.000 | 1.000 | 0.958 | 1.000 | 0.958 | 1.000 | 0.998 | 1.000 | |
| FJX1 TRIM44 | rs76847144 | T vs. C | 0.93 (0.86–0.99) | 0.02643 | 1.000 | 1.000 | 0.958 | 1.000 | 0.958 | 1.000 | 0.998 | 1.000 | |
| WBSCR17 | rs148521358 | C vs. G | 0.94 (0.88–0.99) | 0.02731 | 1.000 | 1.000 | 0.951 | 1.000 | 0.951 | 1.000 | 0.998 | 1.000 | |
| MIR3134 SUSD1 | rs2564899 | T vs. C | 0.97 (0.94–1.00) | 0.02735 | 1.000 | 1.000 | 0.980 | 1.000 | 0.980 | 1.000 | 0.999 | 1.000 | |
| NA | chr8_138837351_I | I vs. D | 1.05 (1.01–1.09) | 0.0284 | 1.000 | 1.000 | 0.913 | 1.000 | 0.913 | 1.000 | 0.998 | 1.000 | |
| LINC01393 MDFIC | rs7799732 | A vs. G | 1.03 (1.00–1.06) | 0.03114 | 1.000 | 1.000 | 0.978 | 1.000 | 0.978 | 1.000 | 0.999 | 1.000 | |
| TBX18 TBX18-AS1 | rs76397051 | A vs. G | 1.05 (1.01–1.10) | 0.034 | 1.000 | 1.000 | 0.975 | 1.000 | 0.975 | 1.000 | 0.999 | 1.000 | |
| NA | rs171794 | T vs. C | 1.06 (1.01–1.12) | 0.03587 | 1.000 | 1.000 | 0.974 | 1.000 | 0.974 | 1.000 | 0.999 | 1.000 | |
| GDA | rs4327921 | A vs. G | 0.97 (0.94–1.00) | 0.03938 | 1.000 | 1.000 | 0.980 | 1.000 | 0.980 | 1.000 | 0.999 | 1.000 | |
| NA | rs2167341 | T vs. G | 1.05 (1.00–1.10) | 0.04203 | 1.000 | 1.000 | 0.975 | 1.000 | 0.975 | 1.000 | 0.999 | 1.000 | |
| EVA1C | rs62216215 | A vs. C | 1.04 (1.00–1.08) | 0.04598 | 1.000 | 1.000 | 0.977 | 1.000 | 0.977 | 1.000 | 0.999 | 1.000 | |
| LINC01036 | rs17589281 | T vs. C | 0.95 (0.89–1.00) | 0.04716 | 1.000 | 1.000 | 0.980 | 1.000 | 0.980 | 1.000 | 0.999 | 1.000 | |
| LOC283585 | rs61979775 | T vs. C | 0.97 (0.93–1.00) | 0.04813 | 1.000 | 1.000 | 0.980 | 1.000 | 0.980 | 1.000 | 0.999 | 1.000 | |
| CHMP4A GMPR2 MDP1 NEDD8 NEDD8-MDP1 TM9SF1 TSSK4 |
rs72694312 | T vs. G | 1.06 (1.00–1.11) | 0.04814 | 1.000 | 1.000 | 0.930 | 1.000 | 0.930 | 1.000 | 0.998 | 1.000 | |
| Ma et al., 2009 [32] | NA | rs10065041 | T(minor)/C(major) | 1.21 (1.08–1.36) | 3.24 × 10−4 | 0.445 | 1.000 | 0.757 | 1.000 | 0.581 | 0.999 | 0.970 | 1.000 |
| NA | rs10038113 | C(minor)/T(major) | 0.75 (0.70–0.90) | 3.40 × 10−6 | 0.129 | 0.897 | 0.939 | 1.000 | 0.688 | 1.000 | 0.979 | 1.000 | |
| NA | rs6894838 | T(minor)/C(major) | 1.26 (1.12–1.42) | 8.00 × 10−5 | 0.212 | 0.998 | 0.416 | 0.999 | 0.131 | 0.993 | 0.827 | 1.000 | |
| Anney et al., 2010 [30] | HAT1 | rs6731562 | G (minor allele) | 1.25 (1.11–1.41) | 2.0 × 10−4 | 0.253 | 0.998 | 0.527 | 0.999 | 0.220 | 0.996 | 0.891 | 1.000 |
| POU6F2 | rs10258862 | G (minor allele) | 1.09 (1.00–1.18) | 4.6 × 10−2 | 0.991 | 1.000 | 0.971 | 1.000 | 0.971 | 1.000 | 0.998 | 1.000 | |
| NA | rs6557675 | A (minor allele) | 0.84 (0.76–0.93) | 1.0 × 10−3 | 0.561 | 1.000 | 0.583 | 0.999 | 0.440 | 0.999 | 0.953 | 1.000 | |
| MYH11 | rs17284809 | A (minor allele) | 0.63 (0.50–0.79) | 5.7 × 10−5 | 0.008 | 0.312 | 0.891 | 1.000 | 0.168 | 0.995 | 0.821 | 1.000 | |
| GSG1L | rs205409 | G (minor allele) | 0.91 (0.84–0.99) | 2.8 × 10−2 | 0.980 | 1.000 | 0.966 | 1.000 | 0.966 | 1.000 | 0.998 | 1.000 | |
| TAF1C | rs4150167 | A (minor allele) | 0.54 (0.40–0.73) | 2.1 × 10−5 | 0.002 | 0.085 | 0.963 | 1.000 | 0.420 | 0.999 | 0.905 | 1.000 | |
| Kuo et al., 2015 [33] | GLIS1 | rs12082358 | C (minor allele) | 1.3 (1.1–1.5) | 2.2 × 10−4 | 0.136 | 0.975 | 0.705 | 1.000 | 0.251 | 0.997 | 0.906 | 1.000 |
| GLIS1 | rs12080993 | A (minor allele) | 1.3 (1.1–1.5) | 1.5 × 10−4 | 0.136 | 0.975 | 0.705 | 1.000 | 0.251 | 0.997 | 0.906 | 1.000 | |
| GPD2 | rs3916984 | A (minor allele) | 1.3 (1.1–1.5) | 3.1 × 10−4 | 0.136 | 0.975 | 0.705 | 1.000 | 0.251 | 0.997 | 0.906 | 1.000 | |
| LRP2/BBS5 | rs13014164 | C (minor allele) | 1.7 (1.3–2.3) | 8.6 × 10−5 | 0.012 | 0.209 | 0.980 | 1.000 | 0.735 | 1.000 | 0.974 | 1.000 | |
| PDGFRA | rs7697680 | G (minor allele) | 1.5 (1.2–1.9) | 9.2 × 10−4 | 0.032 | 0.500 | 0.960 | 1.000 | 0.607 | 0.999 | 0.967 | 1.000 | |
| FSTL4 | rs11741756 | A (minor allele) | 1.3 (1.1–1.5) | 1.2 × 10−2 | 0.136 | 0.975 | 0.705 | 1.000 | 0.251 | 0.997 | 0.906 | 1.000 | |
| NA | rs13211684 | G (minor allele) | 1.3 (1.1–1.5) | 2.5 × 10−3 | 0.136 | 0.975 | 0.705 | 1.000 | 0.251 | 0.997 | 0.906 | 1.000 | |
| NA | rs10966205 | T (minor allele) | 1.3 (1.2–1.5) | 2.9 × 10−5 | 0.136 | 0.975 | 0.705 | 1.000 | 0.251 | 0.997 | 0.906 | 1.000 | |
| C10orf68 | rs10763893 | A (minor allele) | 1.6 (1.2–2.2) | 6.1 × 10−4 | 0.038 | 0.346 | 0.990 | 1.000 | 0.917 | 1.000 | 0.992 | 1.000 | |
| NA | rs12366025 | A (minor allele) | 1.3 (1.1–1.6) | 3.8 × 10−3 | 0.225 | 0.912 | 0.983 | 1.000 | 0.936 | 1.000 | 0.995 | 1.000 | |
| NA | rs11030597 | G (minor allele) | 1.3 (1.1–1.6) | 4.1 × 10−3 | 0.225 | 0.912 | 0.983 | 1.000 | 0.936 | 1.000 | 0.995 | 1.000 | |
| NA | rs7933990 | A (minor allele) | 1.3 (1.1–1.6) | 2.5 × 10−3 | 0.225 | 0.912 | 0.983 | 1.000 | 0.936 | 1.000 | 0.995 | 1.000 | |
| NA | rs11030606 | A (minor allele) | 1.3 (1.1–1.6) | 5.6 × 10−3 | 0.225 | 0.912 | 0.983 | 1.000 | 0.936 | 1.000 | 0.995 | 1.000 | |
| MACROD2 | rs17263514 | A (minor allele) | 1.2 (1.0–1.4) | 1.4 × 10−2 | 0.500 | 0.998 | 0.976 | 1.000 | 0.953 | 1.000 | 0.996 | 1.000 | |
| BCAS1/CYP24A1 | rs12479663 | C (minor allele) | 1.5 (1.3–1.9) | 4.0 × 10−5 | 0.032 | 0.500 | 0.960 | 1.000 | 0.607 | 0.999 | 0.967 | 1.000 | |
Abbreviations: A, Adenine; C, Cytosine; G, Guanine; T, Thymine; D, Deletion; I, Insertion; R, Risk allele; NR, Non-risk allele; FPRP, false positive rate probability; BFDP, Bayesian false discovery probability; OR, odds ratio; CI, confidence interval; NA, not available.
3.2.3. Re-Analysis of Results from the GWAS Catalog and GWAS Datasets Included in the GWAS Meta-Analyses
Genetic comparisons additionally extracted from the GWAS catalog were also re-analyzed (Table 3). Among the 20 included comparisons, two (10.0%) genotype comparisons, MACROD2/rs4141463 and LOCI105370358-LOCI107984602/rs4773054, extracted from the GWAS catalog were reported to be significant with a p-value < 5 × 10−8. The remaining 18 comparisons were of borderline statistical significance (p-value between 0.05 and 5 × 10−8).
Table 3.
Re-analysis results of gene variants with genome wide statistical significance (p-value < 5 × 10−8) and borderline statistical significance (5 × 10−8 ≤ p-value < 0.05) in the genome-wide association studies (GWAS) catalog.
| Author, Year | Gene | Variant | Comparison | OR (95% CI) | p-Value | Power OR 1.2 |
Power OR 1.5 |
FPRP Values at Prior Probability | BFDP 0.001 |
BFDP 0.000001 |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR 1.2 | OR 1.5 | ||||||||||||
| 0.001 | 0.000001 | 0.001 | 0.000001 | ||||||||||
| Gene variants with statistically significance (p-value < 5 × 10−8), FPRP < 0.2 and BFDP < 0.8 from GWAS catalog | |||||||||||||
| Anney et al., 2010 [30] | MACROD2 | rs4141463 | NA | 1.37 (1.22–1.52) | 4.00 × 10−8 | 0.006 | 0.956 | 0.000 | 0.316 | 0.000 | 0.003 | 0.000 | 0.208 |
| Chaste et al., 2014 [35] | AL163541.1 | rs4773054 | NA | 2.66 (1.83–3.86) | 5.00 × 10−8 | 0.000 | 0.001 | 0.949 | 1.000 | 0.169 | 0.995 | 0.526 | 0.999 |
| Gene variants with statistically borderline significance (5 × 10−8 ≤ p-value < 0.05), FPRP < 0.2 and BFDP < 0.8 from GWAS catalog | |||||||||||||
| Anney et al., 2010 [30] | PPP2R5C | rs7142002 | NA | 1.56 (1.28–1.89) | 3.00 × 10−6 | 0.004 | 0.344 | 0.602 | 0.999 | 0.016 | 0.942 | 0.338 | 0.998 |
| Anney et al., 2012 [34] | TAF1C | rs4150167 | NA | 1.96 (1.52–2.56) | 3.00 × 10−7 | 0.000 | 0.025 | 0.832 | 1.000 | 0.031 | 0.969 | 0.269 | 0.997 |
| Anney et al., 2012 [34] | PARD3B | rs4675502 | NA | 1.28 (1.16–1.41) | 4.00 × 10−7 | 0.095 | 0.999 | 0.006 | 0.856 | 0.001 | 0.362 | 0.030 | 0.969 |
| Anney et al., 2012 [34] | AC113414.1 | rs7711337 | NA | 1.22 (1.12–1.32) | 8.00 × 10−7 | 0.340 | 1.000 | 0.002 | 0.689 | 0.001 | 0.429 | 0.038 | 0.975 |
| Anney et al., 2012 [34] | AC009446.1, EYA1 | rs7834018 | NA | 1.56 (1.3–1.89) | 8.00 × 10−7 | 0.004 | 0.344 | 0.602 | 0.999 | 0.016 | 0.942 | 0.338 | 0.998 |
| Anney et al., 2017 [31] | AL133270.1, AL139093.1 | rs142968358 | T (risk allele) | 1.1 (1.06–1.14) | 1.00 × 10−6 | 1.000 | 1.000 | 0.000 | 0.145 | 0.000 | 0.145 | 0.014 | 0.936 |
| Anney et al., 2017 [31] | EXT1 | rs7835763 | A (risk allele) | 1.1 (1.06–1.14) | 2.00 × 10−6 | 1.000 | 1.000 | 0.000 | 0.145 | 0.000 | 0.145 | 0.014 | 0.936 |
| Chaste et al., 2014 [35] | INHCAP | rs1867503 | NA | 1.55 (1.30–1.84) | 4.00 × 10−7 | 0.002 | 0.354 | 0.241 | 0.997 | 0.002 | 0.608 | 0.058 | 0.984 |
| Chaste et al., 2014 [35] | CUEDC2 | rs1409313 | NA | 1.75 (1.40–2.18) | 4.00 × 10−7 | 0.000 | 0.085 | 0.610 | 0.999 | 0.007 | 0.876 | 0.121 | 0.993 |
| Chaste et al., 2014 [35] | CTU2 | rs11641365 | NA | 2.06 (1.54–2.76) | 3.00 × 10−7 | 0.000 | 0.017 | 0.897 | 1.000 | 0.071 | 0.987 | 0.433 | 0.999 |
| Chaste et al., 2014 [35] | AC067752.1, AC024598.1, ZNF365 | rs93895 | NA | 1.91 (1.48–2.47) | 2.00 × 10−7 | 0.000 | 0.033 | 0.804 | 1.000 | 0.024 | 0.961 | 0.241 | 0.997 |
| Kuo et al., 2015 [33] | LINC01151, AC108136.1 | rs12543592 | G (risk allele) | 1.43 (1.25–1.67) | 3.00 × 10−6 | 0.013 | 0.727 | 0.318 | 0.998 | 0.008 | 0.895 | 0.275 | 0.997 |
| Kuo et al., 2015 [33] | NAALADL2 | rs3914502 | A (risk allele) | 1.4 (1.20–1.60) | 4.00 × 10−6 | 0.012 | 0.844 | 0.062 | 0.985 | 0.001 | 0.482 | 0.051 | 0.982 |
| Kuo et al., 2015 [33] | OR2M4 | rs10888329 | NA | 1.82 (1.39–2.33) | 8.00 × 10−6 | 0.000 | 0.062 | 0.809 | 1.000 | 0.031 | 0.970 | 0.338 | 0.998 |
| Kuo et al., 2015 [33] | SGSM2 | rs2447097 | A (risk allele) | 1.53 (1.27–1.85) | 9.00 × 10−6 | 0.006 | 0.419 | 0.652 | 0.999 | 0.026 | 0.965 | 0.467 | 0.999 |
| Ma et al., 2009 [32] | Intergenic (RNU6-374P - MSNP1) | rs10038113 | T (risk allele) | 1.33 (1.11–1.43] | 3.00 × 10−6 | 0.003 | 0.999 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Gene variants with statistically borderline significance (5 × 10-8≤ p-value < 0.05), FPRP > 0.2 or BFDP > 0.8 from GWAS catalog | |||||||||||||
| Chaste et al., 2014 [35] | AL163541.1 | rs4773054 | NA | 2.9 (1.91–4.39) | 7.00 × 10−8 | 0.000 | 0.001 | 0.970 | 1.000 | 0.345 | 0.998 | 0.741 | 1.000 |
| Anney et al., 2017 [31] | HLA-A, AL671277.1 | rs115254791 | G (risk allele) | 1.0869565 (1.05–1.14) | 4.00 × 10−6 | 1.000 | 1.000 | 0.376 | 0.998 | 0.376 | 0.998 | 0.963 | 1.000 |
Abbreviations: A, Adenine; G; Guanine; T, Thymine; FPRP, false positive rate probability; BFDP, Bayesian false discovery probability; OR, odds ratio; CI, confidence interval; F, fixed effects model; R, random effects model; NA, not available; ASD, autism spectrum disorder.
While assessing noteworthiness, five (25.0%) and three (15.0%) were verified as being noteworthy using FPRP estimation, at a prior probability of 10−3 and 10−6, respectively, with the statistical power to detect a 1.2 OR. In addition, eighteen (90.0%) and four (25.0%) showed noteworthiness at a prior probability of 10−3 and 10−6 with the statistical power to detect a 1.5 OR, respectively. In the BFDP estimation, nineteen (95.0%) and two (10.0%) were assessed as being noteworthy at a prior probability of 10−3 and 10−6, respectively. Finally, 18 genetic associations (95%) of both significant and borderline statistically significant results were verified as being noteworthy under both the FPRP and BFDP approaches. The total number of associations included two comparisons with genome-wide significance (p-value < 5 × 10−8) and sixteen comparisons with borderline significance (p-value between 0.05 and 5 × 10−8).
In order to develop the analysis further, we extracted the GWAS data that was both statistically significant and noteworthy under both Bayesian approaches, from the GWAS meta-analysis and GWAS catalog. They were extracted from five articles [30,31,32,33,34], with 70 of the GWAS data being noteworthy under both FPRP and BFDP. Results with noteworthy association are summarized in Table 4.
Table 4.
Re-analysis results of gene variants with genome wide statistical significance (p-value < 5 × 10−8) and borderline statistical significance (5 × 10−8 ≤ p-value < 0.05) in the GWAS datasets included in GWAS meta-analyses (results of FPRP < 0.2 and BFDP < 0.8).
| Author, Year | Trait | Gene(s) | Variant | Comparison | OR (95% CI) | p-Value | Power OR 1.2 | Power OR 1.5 | FPRP Values at Prior Probability | BFDP 0.001 |
BFDP 0.000001 |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR 1.2 | OR 1.5 | |||||||||||||
| 0.001 | 0.000001 | 0.001 | 0.000001 | |||||||||||
| Anney et al., 2012 [34] | ASD (European) | ERBB4 | rs1879532 | A | 2.02 (1.57–2.59) | 1.55 × 10−8 | 0.000 | 0.009 | 0.595 | 0.999 | 0.003 | 0.757 | 0.026 | 0.964 |
| Anney et al., 2012 [34] | Autism (European) | None | rs289932 | A | 0.49 (0.38–0.64) | 5.04 × 10−8 | 0.000 | 0.012 | 0.772 | 1.000 | 0.014 | 0.932 | 0.114 | 0.992 |
| Anney et al., 2012 [34] | ASD | TMEM132B | rs16919315 | A | 0.53 (0.42–0.67) | 5.12 × 10−8 | 0.000 | 0.028 | 0.589 | 0.999 | 0.004 | 0.800 | 0.049 | 0.981 |
| Anney et al., 2012 [34] | Autism (European) | ERBB4 | rs1879532 | A | 1.72 (1.39–2.11) | 1.66 × 10−7 | 0.000 | 0.095 | 0.416 | 0.999 | 0.002 | 0.676 | 0.044 | 0.979 |
| Anney et al., 2010 [30] | Autism | NA | rs6557675 | A (minor allele) | 0.61 (0.51–0.71) | 2.20 × 10−7 | 0.000 | 0.126 | 0.006 | 0.861 | 0.000 | 0.001 | 0.000 | 0.048 |
| Anney et al., 2012 [34] | Autism (European) | None | rs289858 | A | 0.52 (0.40–0.67) | 2.81 × 10−7 | 0.000 | 0.027 | 0.762 | 1.000 | 0.015 | 0.940 | 0.161 | 0.995 |
| Anney et al., 2012 [34] | ASD | SYNE2 | rs2150291 | A | 1.72 (1.40–2.13) | 2.83 × 10−7 | 0.000 | 0.105 | 0.579 | 0.999 | 0.006 | 0.864 | 0.119 | 0.993 |
| Anney et al., 2012 [34] | ASD (European) | RPH3AL | rs7207517 | A | 1.97 (1.51–2.57) | 3.05 × 10−7 | 0.000 | 0.022 | 0.817 | 1.000 | 0.025 | 0.963 | 0.226 | 0.997 |
| Anney et al., 2012 [34] | Autism (European) | None | rs4761371 | A | 0.46 (0.34–0.63) | 3.91 × 10−7 | 0.000 | 0.010 | 0.924 | 1.000 | 0.111 | 0.992 | 0.521 | 0.999 |
| Anney et al., 2012 [34] | ASD (European) | PRAMEF12 | rs1812242 | A | 1.44 (1.25–1.66) | 4.29 × 10−7 | 0.006 | 0.713 | 0.077 | 0.988 | 0.001 | 0.411 | 0.038 | 0.975 |
| Anney et al., 2012 [34] | ASD | None | rs10904487 | G | 0.63 (0.52–0.75) | 4.29 × 10−7 | 0.001 | 0.262 | 0.198 | 0.996 | 0.001 | 0.440 | 0.028 | 0.966 |
| Anney et al., 2012 [34] | Autism (European) | None | rs289932 | A | 0.67 (0.57–0.79) | 5.42 × 10−7 | 0.005 | 0.524 | 0.286 | 0.998 | 0.004 | 0.784 | 0.135 | 0.994 |
| Anney et al., 2010 [30] | Autism | MACROD2 | rs4141463 | A (minor allele) | 0.62 (0.52–0.73) | 5.50 × 10−7 | 0.000 | 0.192 | 0.047 | 0.980 | 0.000 | 0.048 | 0.002 | 0.655 |
| Anney et al., 2012 [34] | Autism | None | rs9608521 | A | 1.46 (1.25–1.69) | 7.62 × 10−7 | 0.004 | 0.641 | 0.084 | 0.989 | 0.001 | 0.383 | 0.033 | 0.971 |
| Anney et al., 2012 [34] | ASD | None | rs1408744 | A | 0.65 (0.54–0.77) | 8.06 × 10−7 | 0.002 | 0.385 | 0.235 | 0.997 | 0.002 | 0.618 | 0.062 | 0.985 |
| Anney et al., 2017 [31] | ASD | LINC00535 | chr8_94389815_I | I vs. D | 1.14 (1.09–1.19) | 9.47 × 10−7 | 0.990 | 1.000 | 0.000 | 0.002 | 0.000 | 0.002 | 0.686 | 1.000 |
| Anney et al., 2012 [34] | ASD (European) | PC | rs7122539 | A | 0.60 (0.49–0.74) | 9.64 × 10−7 | 0.001 | 0.162 | 0.628 | 0.999 | 0.011 | 0.917 | 0.213 | 0.996 |
| Anney et al., 2010 [30] | Autism | MACROD2 | rs4814324 | A (minor allele) | 1.58 (1.34–1.86) | 9.80 × 10−7 | 0.000 | 0.266 | 0.076 | 0.988 | 0.000 | 0.128 | 0.006 | 0.859 |
| Anney et al., 2010 [30] | Autism | MACROD2 | rs6079544 | A (minor allele) | 1.57 (1.33–1.84) | 1.20 × 10−6 | 0.000 | 0.287 | 0.053 | 0.982 | 0.000 | 0.081 | 0.004 | 0.797 |
| Anney et al., 2017 [31] | ASD | EXOC4 | rs6467494 | T vs. C | 1.12 (1.07–1.16) | 1.43 × 10−6 | 1.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.197 | 0.996 |
| Anney et al., 2010 [30] | Autism | MACROD2 | rs6079536 | A (minor allele) | 0.64 (0.54–0.75) | 1.60 × 10−6 | 0.001 | 0.307 | 0.059 | 0.984 | 0.000 | 0.102 | 0.005 | 0.837 |
| Anney et al., 2010 [30] | ASD | MYH11 | rs17284809 | A (minor allele) | 0.52 (0.39–0.69) | 1.70 × 10−6 | 0.001 | 0.043 | 0.915 | 1.000 | 0.121 | 0.993 | 0.636 | 0.999 |
| Anney et al., 2010 [30] | Autism | MACROD2 | rs6079553 | A (minor allele) | 1.55 (1.31–1.82) | 2.10 × 10−6 | 0.001 | 0.344 | 0.090 | 0.990 | 0.000 | 0.204 | 0.011 | 0.920 |
| Anney et al., 2010 [30] | Autism | MACROD2 | rs6074798 | A (minor allele) | 1.56 (1.32–1.84) | 2.10 × 10−6 | 0.001 | 0.321 | 0.123 | 0.993 | 0.000 | 0.287 | 0.017 | 0.945 |
| Anney et al., 2017 [31] | ASD | OPCML | rs7952100 | C vs.G | 1.14 (1.09–1.19) | 2.49 × 10−6 | 0.990 | 1.000 | 0.000 | 0.002 | 0.000 | 0.002 | 0.686 | 1.000 |
| Anney et al., 2010 [30] | Autism | MACROD2 | rs10446030 | G (minor allele) | 1.54 (1.30–1.81) | 3.20 × 10−6 | 0.001 | 0.375 | 0.116 | 0.992 | 0.000 | 0.301 | 0.019 | 0.951 |
| Kuo et al., 2015 [33] | ASD | STYK1 | rs16922945 | C (minor allele) | 1.86 (1.43–2.43) | 3.43 × 10−6 | 0.001 | 0.057 | 0.891 | 1.000 | 0.085 | 0.989 | 0.572 | 0.999 |
| Anney et al., 2010 [30] | ASD | POU5F2 | rs10258862 | G (minor allele) | 1.41 (1.23–1.61) | 3.70 × 10−6 | 0.009 | 0.820 | 0.043 | 0.978 | 0.000 | 0.319 | 0.027 | 0.966 |
| Anney et al., 2010 [30] | Autism | MACROD2 | rs6079540 | A (minor allele) | 0.65 (0.55–0.77) | 3.70 × 10−6 | 0.002 | 0.385 | 0.235 | 0.997 | 0.002 | 0.618 | 0.062 | 0.985 |
| Anney et al., 2010 [30] | Autism | MACROD2 | rs6074787 | A (minor allele) | 1.53 (1.30–1.80) | 4.10 × 10−6 | 0.002 | 0.406 | 0.147 | 0.994 | 0.001 | 0.418 | 0.031 | 0.970 |
| Anney et al., 2010 [30] | ASD | MACROD2 | rs6074798 | A (minor allele) | 1.38 (1.22–1.56) | 4.80 × 10−6 | 0.013 | 0.909 | 0.020 | 0.954 | 0.000 | 0.224 | 0.018 | 0.948 |
| Anney et al., 2010 [30] | Autism | MACROD2 | rs980319 | G (minor allele) | 1.52 (1.29–1.79) | 5.10 × 10−6 | 0.002 | 0.437 | 0.184 | 0.996 | 0.001 | 0.543 | 0.050 | 0.981 |
| Anney et al., 2010 [30] | Autism | MACROD2 | rs6079537 | G (minor allele) | 1.52 (1.29–1.79) | 6.00 × 10−6 | 0.002 | 0.437 | 0.184 | 0.996 | 0.001 | 0.543 | 0.050 | 0.981 |
| Kuo et al., 2015 [33] | ASD | NA | rs10966205 | A (minor allele) | 1.52 (1.27–1.83) | 6.25 × 10−6 | 0.006 | 0.444 | 0.609 | 0.999 | 0.022 | 0.957 | 0.426 | 0.999 |
| Kuo et al., 2015 [33] | ASD | OR2M4 | rs10888329 | T (minor allele) | 0.55 (0.43–0.72) | 8.05 × 10−6 | 0.001 | 0.081 | 0.916 | 1.000 | 0.144 | 0.994 | 0.718 | 1.000 |
| Anney et al., 2010 [30] | ASD | MACROD2 | rs6079536 | A (minor allele) | 0.73 (0.65–0.83) | 8.50 × 10−6 | 0.022 | 0.917 | 0.067 | 0.986 | 0.002 | 0.628 | 0.084 | 0.989 |
| Anney et al., 2010 [30] | ASD | NA | rs6557675 | A (minor allele) | 0.72 (0.63–0.82) | 8.70 × 10−6 | 0.014 | 0.877 | 0.051 | 0.982 | 0.001 | 0.457 | 0.047 | 0.980 |
| Kuo et al., 2015 [33] | ASD | NA | rs7933990 | A (minor allele) | 1.72 (1.35–2.19) | 9.40 × 10−6 | 0.002 | 0.133 | 0.861 | 1.000 | 0.075 | 0.988 | 0.606 | 0.999 |
| Kuo et al., 2015 [33] | ASD | MNT | rs2447097 | A (minor allele) | 1.53 (1.27–1.85) | 9.45 × 10−6 | 0.006 | 0.419 | 0.652 | 0.999 | 0.026 | 0.965 | 0.467 | 0.999 |
| Anney et al., 2010 [30] | ASD | GSG1L | rs205409 | G (minor allele) | 0.72 (0.64–0.82) | 9.60 × 10−6 | 0.014 | 0.877 | 0.051 | 0.982 | 0.001 | 0.457 | 0.047 | 0.980 |
| Kuo et al., 2015 [33] | ASD | OR2M4 | rs6672981 | C (minor allele) | 0.55 (0.42–0.72) | 9.64 × 10−6 | 0.001 | 0.081 | 0.916 | 1.000 | 0.144 | 0.994 | 0.718 | 1.000 |
| Kuo et al., 2015 [33] | ASD | OR2M4 | rs4397683 | C (minor allele) | 0.55 (0.42–0.72) | 9.86 × 10−6 | 0.001 | 0.081 | 0.916 | 1.000 | 0.144 | 0.994 | 0.718 | 1.000 |
| Anney et al., 2010 [30] | ASD | MACROD2 | rs980319 | G (minor allele) | 1.36 (1.20–1.54) | 1.00 × 10−5 | 0.024 | 0.939 | 0.049 | 0.981 | 0.001 | 0.570 | 0.068 | 0.987 |
| Kuo et al., 2015 [33] | ASD | BCAS1/CYP24A1 | rs12479663 | G (minor allele) | 1.81 (1.38–2.36) | 1.08 × 10−5 | 0.001 | 0.083 | 0.907 | 1.000 | 0.124 | 0.993 | 0.687 | 1.000 |
| Anney et al., 2010 [30] | ASD | MACROD2 | rs4814324 | A (minor allele) | 1.36 (1.20–1.54) | 1.10 × 10−5 | 0.024 | 0.939 | 0.049 | 0.981 | 0.001 | 0.570 | 0.068 | 0.987 |
| Kuo et al., 2015 [33] | ASD | KRR1 | rs3741496 | C (minor allele) | 1.49 (1.24–1.78) | 1.15 × 10−5 | 0.009 | 0.529 | 0.565 | 0.999 | 0.020 | 0.954 | 0.430 | 0.999 |
| Kuo et al., 2015 [33] | ASD | OR2M4 | rs4642918 | C (minor allele) | 0.56 (0.43–0.73) | 1.24 × 10−5 | 0.002 | 0.099 | 0.917 | 1.000 | 0.155 | 0.995 | 0.745 | 1.000 |
| Anney et al., 2010 [30] | ASD | MACROD2 | rs6079544 | A (minor allele) | 1.35 (1.20–1.53) | 1.30 × 10−5 | 0.033 | 0.951 | 0.074 | 0.988 | 0.003 | 0.733 | 0.124 | 0.993 |
| Kuo et al., 2015 [33] | ASD | NA | rs13211684 | G (minor allele) | 1.56 (1.28–1.91) | 1.36 × 10−5 | 0.006 | 0.352 | 0.750 | 1.000 | 0.045 | 0.979 | 0.572 | 0.999 |
| Kuo et al., 2015 [33] | ASD | MNT | rs2447095 | A (minor allele) | 1.52 (1.26–1.84) | 1.45 × 10−5 | 0.008 | 0.446 | 0.695 | 1.000 | 0.038 | 0.975 | 0.552 | 0.999 |
| Kuo et al., 2015 [33] | ASD | NA | rs12543592 | G (minor allele) | 0.67 (0.56–0.81) | 1.63 × 10−5 | 0.012 | 0.521 | 0.744 | 1.000 | 0.063 | 0.985 | 0.678 | 1.000 |
| Anney et al., 2010 [30] | ASD | MACROD2 | rs6079553 | A (minor allele) | 1.35 (1.19–1.52) | 1.70 × 10−5 | 0.026 | 0.959 | 0.027 | 0.965 | 0.001 | 0.424 | 0.041 | 0.977 |
| Kuo et al., 2015 [33] | ASD | KRR1 | rs1051446 | C (minor allele) | 1.47 (1.23–1.76) | 1.77 × 10−5 | 0.014 | 0.587 | 0.669 | 1.000 | 0.045 | 0.979 | 0.614 | 0.999 |
| Anney et al., 2010 [30] | ASD | NA | rs4078417 | C (minor allele) | 1.38 (1.21–1.57) | 1.90 × 10−5 | 0.017 | 0.897 | 0.055 | 0.983 | 0.001 | 0.524 | 0.059 | 0.984 |
| Anney et al., 2010 [30] | ASD | MACROD2 | rs10446030 | G (minor allele) | 1.34 (1.19–1.52) | 2.20 × 10−5 | 0.043 | 0.960 | 0.110 | 0.992 | 0.006 | 0.847 | 0.210 | 0.996 |
| Kuo et al., 2015 [33] | ASD | GPD2 | rs3916984 | T (minor allele) | 0.62 (0.49–0.77) | 2.25 × 10−5 | 0.004 | 0.256 | 0.804 | 1.000 | 0.056 | 0.984 | 0.595 | 0.999 |
| Kuo et al., 2015 [33] | ASD | NA | rs12366025 | T (minor allele) | 1.67 (1.31–2.11) | 2.49 × 10−5 | 0.003 | 0.184 | 0.860 | 1.000 | 0.086 | 0.989 | 0.662 | 0.999 |
| Ma et al., 2009 [32] | Autism | NA | rs10038113 | C(minor)/T(major) | 0.67 (0.56–0.81) | 2.75 × 10−5 | 0.012 | 0.521 | 0.744 | 1.000 | 0.063 | 0.985 | 0.678 | 1.000 |
| Anney et al., 2010 [30] | ASD | MACROD2 | rs6079540 | A (minor allele) | 0.75 (0.66–0.84) | 2.90 × 10−5 | 0.034 | 0.979 | 0.019 | 0.950 | 0.001 | 0.399 | 0.037 | 0.975 |
| Anney et al., 2010 [30] | Autism | HAT1 | rs6731562 | G (minor allele) | 1.51 (1.27–1.81) | 3.30 × 10−5 | 0.006 | 0.471 | 0.562 | 0.999 | 0.017 | 0.946 | 0.383 | 0.998 |
| Anney et al., 2010 [30] | ASD | MACROD2 | rs6074787 | A (minor allele) | 1.33 (1.18–1.50) | 3.40 × 10−5 | 0.047 | 0.975 | 0.067 | 0.986 | 0.003 | 0.776 | 0.147 | 0.994 |
| Kuo et al., 2015 [33] | ASD | GLIS1 | rs12080933 | A (minor allele) | 1.48 (1.23–1.78) | 3.57 × 10−5 | 0.013 | 0.557 | 0.707 | 1.000 | 0.053 | 0.983 | 0.648 | 0.999 |
| Kuo et al., 2015 [33] | ASD | FSTL4 | rs11741756 | T (minor allele) | 1.67 (1.31–2.13) | 3.64 × 10−5 | 0.004 | 0.194 | 0.903 | 1.000 | 0.157 | 0.995 | 0.785 | 1.000 |
| Kuo et al., 2015 [33] | ASD | STYK1 | rs7953930 | G (minor allele) | 1.65 (1.30–2.09) | 3.83 × 10−5 | 0.004 | 0.215 | 0.888 | 1.000 | 0.133 | 0.994 | 0.761 | 1.000 |
| Anney et al., 2010 [30] | Autism | NA | rs4078417 | C (minor allele) | 1.50 (1.26–1.79) | 4.10 × 10−5 | 0.007 | 0.500 | 0.509 | 0.999 | 0.014 | 0.933 | 0.339 | 0.998 |
| Anney et al., 2010 [30] | ASD | MACROD2 | rs4141463 | A (minor allele) | 0.75 (0.66–0.85) | 4.30 × 10−5 | 0.049 | 0.967 | 0.118 | 0.993 | 0.007 | 0.873 | 0.243 | 0.997 |
| Kuo et al., 2015 [33] | ASD | OR2M3 | rs11204613 | G (minor allele) | 0.58 (0.45–0.75) | 4.60 × 10−5 | 0.003 | 0.144 | 0.920 | 1.000 | 0.185 | 0.996 | 0.799 | 1.000 |
| Anney et al., 2010 [30] | ASD | MACROD2 | rs6079537 | G (minor allele) | 1.32 (1.17–1.49) | 5.40 × 10−5 | 0.062 | 0.981 | 0.103 | 0.991 | 0.007 | 0.878 | 0.249 | 0.997 |
| Anney et al., 2010 [30] | Autism | GSG1L | rs205409 | G (minor allele) | 0.69 (0.58–0.81) | 1.10 × 10−4 | 0.011 | 0.663 | 0.353 | 0.998 | 0.009 | 0.896 | 0.271 | 0.997 |
| Anney et al., 2010 [30] | Autism | POU5F2 | rs10258862 | G (minor allele) | 1.43 (1.21–1.71) | 1.80 × 10−4 | 0.027 | 0.700 | 0.764 | 1.000 | 0.112 | 0.992 | 0.799 | 1.000 |
Abbreviations: ASD, Autism spectrum disorders; A, Adenine; C, Cytosine; G, Guanine; T, Thymine; D, Deletion; I, Insertion; FPRP, false positive rate probability; BFDP, Bayesian false discovery probability; OR, odds ratio; CI, confidence interval; GWAS, Genome-Wide Association Studies; NA, not available.
3.3. Protein-Protein Interaction (PPI) Network
We established PPI networks related to the risk of ASD by filtering genes noteworthy under both FPRP and BFDP or genes with a p-value < 5 × 10−8. We included the results of both re-analyzed and non-re-analyzable genetic comparisons from meta-analyses of observational studies and GWAS, GWAS included in meta-analyses of GWAS, and the GWAS catalog. The statistically significant results of non-re-analyzable studies are presented in the Supplement Table S3.
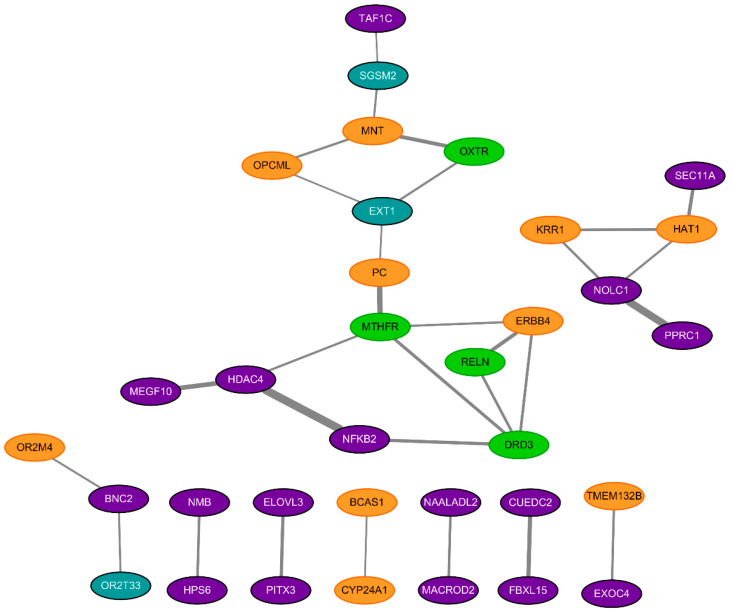
The major genes that included a strong genetic connection were the myc-associated factor X (MAX) network transcriptional repressor (MNT), oxytocin receptor (OXTR), nucleolar and coiled-body phosphoprotein (NOLC1), peroxisome proliferator-activated receptor gamma related coactivator-related 1 (PPRC1), pyruvate carboxylase (PC), methylenetetrahydrofolate reductase (MTHFR), multiple epidermal growth factor like domains 10 (MEGF10), nuclear factor kappa B subunit 2 (NFKB2), histone deacetylase 4 (HDAC4), etc. (Figure 2 and Table 5).
Figure 2.
Protein-protein interaction network of ASD. There were 34 distinct genes with about 30 genetic connections among them. The thickness of the line connecting genes represents the score of PPI interaction using STRING9.1 and the color of each gene represents the source of the data; orange, GWAS data: green, GWAS catalog: purple, meta-analysis of GWAS: light green, meta-analysis of observational studies.
Table 5.
Lists of genes involved in the PPI network.
| Gene | Function of the Encoding Proteins |
|---|---|
| OXTR | Receptor for oxytocin associated with social recognition and emotion processing |
| MTHFR | Influences susceptibility to neural tube defect by changing folate metabolism |
| RELN | Control cell positioning and neural migration during brain development |
| DRD3 | D3 subtype of the five dopamine receptors; localized to the limbic areas of the brain |
| MNT | Protein member of the Myc/Max/Mad network; transcriptional repressor and an antagonist of Myc-dependent transcriptional activation and cell growth |
| OPCML | Member of the IgLON subfamily in the immunoglobulin protein superfamily of proteins; localized in the plasma membrane; accessory role in opioid receptor function |
| PC | Pyruvate carboxylase; gluconeogenesis, lipogenesis, insulin secretion and synthesis of neurotransmitter glutamate |
| ERBB4 | Tyr protein kinase family and the epidermal growth factor receptor subfamily; binds to and is activated by neuregulins, and induces mitogenesis and differentiation |
| OR2M4 | Members of a large family of GPCR; olfactory receptors initiating a neuronal response that triggers the perception of a smell |
| BCAS1 | Oncogene; highly expressed in three amplified breast cancer cell lines and in one breast tumor without amplification at 20q13.2. |
| CYP24A1 | Cytochrome P450 superfamily of enzymes; drug metabolism and synthesis of cholesterol, steroids and other lipids |
| TMEM132B | The function remains poorly understood despite their mutations associated with non-syndromic hearing loss, panic disorder, and cancer |
| KRR1 | Nucleolar protein; 18S rRNA synthesis and 40S ribosomal assembly |
| HAT1 | Type B histone acetyltransferase; rapid acetylation of newly synthesized cytoplasmic histones; replication-dependent chromatin assembly |
| SGSM2 | GTPase activator; regulators of membrane trafficking |
| EXT1 | Endoplasmic reticulum-resident type II transmembrane glycosyltransferase; involved in the chain elongation step of heparan sulfate biosynthesis |
| OR2T33 | Members of a large family of GPCR; share a 7-transmembrane domain structure with many neurotransmitter and hormone receptors |
| TAF1C | Binds to the core promoter of ribosomal RNA genes to position the polymerase properly; acts as a channel for regulatory signals |
| HDAC4 | Class II of the histone deacetylase/acuc/apha family; represses transcription when tethered to a promoter |
| MEGF10 | Member of the multiple epidermal growth factor-like domains protein family; cell adhesion, motility and proliferation; critical mediator of apoptotic cell phagocytosis; amyloid-beta peptide uptake in brain |
| NFKB2 | Subunit of the transcription factor complex nuclear factor-kappa-B; central activator of genes involved in inflammation and immune function |
| BNC2 | Conserved zinc finger protein; skin color saturation |
| NMB | Member of the bombesin-like family of neuropeptides; negatively regulate eating behavior; regulate colonic smooth muscle contraction |
| HPS6 | Organelle biogenesis associated with melanosomes, platelet dense granules, and lysosomes |
| ELOVL3 | GNS1/SUR4 family; elongation of long chain fatty acids to provide precursors for synthesis of sphingolipids and ceramides |
| PITX3 | Member of the RIEG/PITX homeobox family; transcription factors; lens formation during eye development |
| NAALADL2 | Not well-known, but diseases associated with NAALADL2 include Chromosome 6Pter-P24 Deletion Syndrome and Cornelia De Lange Syndrome. |
| MACROD2 | Deacetylase removing ADP-ribose from mono-ADP-ribosylated proteins; translocate from the nucleus to the cytoplasm upon DNA damage |
| CUEDC2 | CUE domain-containing protein; down-regulate ESR1 protein levels through progesterone-induced and degradation of receptors |
| FBXL15 | Substrate recognition component of SCF E3 ubiquitin-protein ligase complex; mediates the ubiquitination and subsequent proteasomal degradation of SMURF1 |
| EXOC4 | Component of the exocyst complex; targeting exocytic vesicles to specific docking sites on the plasma membrane |
| NOLC1 | Nucleolar protein; act as a regulator of RNA polymerase I; neural crest specification; nucleologenesis |
| PPRC1 | Similar to PPAR-gamma coactivator 1; activate mitochondrial biogenesis through NRF1 in response to proliferative signals |
| SEC11A | Member of the peptidase S26B family; subunit of the signal peptidase complex; cell migration and invasion, gastric cancer and lymph node metastasis |
Abbreviations: OXTR, Oxytocin Receptor; MTHFR, Methylene tetrahydrofolate reductase; RELN, reelin, DRD3, Dopamine Receptor D3; MNT, Myc-associated factor X (MAX) Network Transcriptional Repressor; OPCML, opioid binding protein/cell adhesion molecule-like; PC, Pyruvate carboxylase; ERBB4, Erb-B2 Receptor Tyrosine Kinase 4; OR2M4, olfactory receptor family 2 subfamily M member 4; GPCR, G protein-coupled receptor; BCAS1, Breast Carcinoma Amplified Sequence 1; CYP24A1, Cytochrome P450 Family 24 Subfamily A Member 1; TMEM132B, transmembrane protein 132B; KRR1, KRR1 small subunit processome component homolog; HAT1, histone acetyltransferase 1; SGSM2, small G protein signaling modulator 2; EXT1, Exostosin-1; OR2T33, Olfactory receptor 2T33; TAF1C, TATA-Box Binding Protein Associated Factor, RNA Polymerase I Subunit C; HDAC4, Histone deacetylase 4; MEGF10, Multiple Epidermal Growth Factor Like Domains 10; NFKB2, Nuclear Factor Kappa B Subunit 2; BNC2, basonuclin-2; NMB, Neuromedin B; HPS6, Hermansky–Pudlak syndrome 6; ELOVL3, Elongation Of Very Long Chain Fatty Acids Protein 3, PITX3, Pituitary homeobox 3; NAALADL2, N-Acetylated Alpha-Linked Acidic Dipeptidase Like 2; MACROD2, Mono-ADP Ribosylhydrolase 2; CUEDC2, CUE domain containing 2; FBXL15, F-Box And Leucine Rich Repeat Protein 15; EXOC4, Exocyst Complex Component 4; NOLC1, Nucleolar And Coiled-Body Phosphoprotein 1; PPRC1, peroxisome proliferator-activated receptor gamma, coactivator-related 1; SEC11A, SEC11 Homolog A, Signal Peptidase Complex Subunit.
4. Discussion
To our knowledge, this study is the first study of ASD genetic risk factors, which assessed the levels of evidence of the published meta-analyses showing the association between susceptible loci and ASD. Overall, genetic comparisons with noteworthy results were confirmed as risk factors for ASD. The genetic comparisons highly related to an increased risk of ASD might reflect the implication in neurodevelopment and specific synaptogenesis of ASD.
According to the PPI network, composed of noteworthy results obtained when using both Bayesian approaches, multiple genes were included as a risk factor for ASD. Investigating the lists genes as a risk factor, promising candidates encoded the protein associated with neural development and specification, and also with neurotransmitters and its receptors. These genes were RELN and DRD3 from observational studies, and PC, OPCML, ERBB4, OR2M4, MEGF10, OR2T33, NMB, and NOLC1, from GWAS. In line with our findings, previous reports have supported that the migration and proliferation of neuronal cells is essential to understanding neurodevelopmental disorders such as ASD or schizophrenia [49,50]. In addition, apart from anatomical approaches, genes correlated with neuropeptides and receptors, such as those in the brain or hippocampus, also explain the pathophysiology of the disease at a molecular level [51]. The list of genes included is presented in Table 5.
The present comprehensive re-analyses shows that, although a large number of studies have suggested numerous possible genetic risk factors for ASD, truly significant results are small and a partial part of whole results. For instance, we detected false positive results in 26 out of 31 (83.9%) meta-analyses of observational studies and 163 out of 203 (80.3%) in meta-analyses of GWAS, respectively. However, only a small portion of genetic comparisons with a p-value < 0.05 exhibited noteworthy associations with ASD under both Bayesian approaches (Table 1, Table 2, Table 3 and Table 4).
Moreover, we also detected that genetic comparisons with borderline statistical significance (5 × 10−8 < p-value < 0.05) accounted for 53 out of 126 (42%) noteworthy comparisons from GWAS or meta-analyses of GWAS. These genetic comparisons might have been neglected if the p-value alone was considered to determine noteworthiness. Using the two Bayesian approaches as we did, or relaxing the current GWAS threshold as Panagiotou et al. suggests, might enable better interpretation of GWAS results [48].
Based on the observational studies, out of 31 statistically significant genotype comparisons, five (16.1%) were found noteworthy under both FPRP and BFDP: T vs. C, MTHFR C677T; T (minor), MTHFR C677T; G vs. A, DRD3/rs167771; C vs. G, RELN/rs362691; A (minor), OXTR/rs7632287. From the meta-analyses of GWAS, we could confirm that 34 distinct genes are noteworthy under both Bayesian approaches with about 30 genetic connections. However, the fact that all three comparisons with a p-value < 5 × 10−8—rs1879532 (Table S3), rs4773054 (Table 2), rs4141463 (Table 2)—displayed noteworthiness may indicate that the stringent threshold of p < 5 × 10−8 is a good tool for verification of the true noteworthiness of genetic risk factors.
There are several limitations in our review. First, we did not include studies that have not been meta-analyzed, or meta-analyses that had insufficient data in our review. Secondly, we only included the single findings of a meta-analysis with the lowest p-value per genetic variant. Therefore, we could not consider potentially meaningful subgroup analyses for different ethnicity, location, gender, and type of genotype comparison (i.e., random or fixed) when selecting a certain outcome. We focused on whether the individual genotype variant was truly associated with ASD or not, regardless of the specific type of the genotype comparison or ethnicity.
Our study has several strengths and implications. For example, to our knowledge, this is the first study that simultaneously analyzed a sizeable amount of data about genetic factors including not only GWAS but also the GWAS catalog. Despite the known high heritability of ASD and abundant research in ASD that has focused on the underlying genetic causes, the literature on genetic risk factors for ASD has not fully reached a consensus. This comprehensive review of genetic associations linked to ASD may improve understanding of the strengths and limitations of each form of research, and advance better and novel approaches for examining ASD in the field of genetic research. The findings of this study could provide mechanisms that may be explored for the development of novel neurotherapeutic agents both for the prevention and treatment of ASD.
5. Conclusions
In conclusion, we synthesized published meta-analyses on risk factors of ASD to acquire noteworthy findings and false positive results by adopting two Bayesian approaches for genetic factors. We attempted to synthesize all meta-analyses on genetic polymorphisms linked to ASD and found noteworthy genetic factors highly related to an increased risk of ASD. We also investigated their validity by discovering false positive results under Bayesian methods. To verify results obtained from genetic analyses, both approaches may have advantages, especially for interpretation of results obtained from observational studies. We found noteworthy results from GWAS, not only with p-value ranging between 0.05 and 5 × 10−8, but also from genetic variants within borderline significance rage which were almost half of the genetic variants. This finding speculates that the genetic variants with borderline significance needs to be further analyzed to determine what associations are genuine.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3425/10/10/692/s1, Supplementary Table S1. PRISMA 2009 Checklist; Supplementary Table S2. Gene variants without statistical significance (p-value ≥ 0.05) in meta-analyses of observational studies; Supplementary Table S3. Non-re-analyzable gene variants with genome wide statistical significance (p-value < 5 × 10−8) from the GWAS catalog, meta-analyses of GWAS and the GWAS datasets included in the GWAS meta-analysis.
Author Contributions
J.L., M.J.S., J.I.S. and P.F.-P. designed the study. J.L., M.J.S., C.Y.S., G.H.J., K.H.L., K.S.L. and J.I.S. collected the data and M.J.S., G.H.J., K.H.L. and Y.K. did the analysis. J.L., M.J.S., C.Y.S., G.H.J., K.H.L., K.S.L., Y.K., J.Y.K., J.Y.L., J.R., M.E., F.G., A.K. (Ai Koyanagi), B.S., M.S., T.B.R., A.K. (Andreas Kronbichler), E.D., D.F.P.V., F.R.P.d.S., K.T., A.R.B., A.F.C., S.C., S.T., A.S., L.S., T.T., J.I.S., and P.F.-P. wrote the first draft of the manuscript and gave critical comments on manuscript draft. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Lyall K., Croen L., Daniels J., Fallin M.D., Ladd-Acosta C., Lee B.K., Park B.Y., Snyder N.W., Schendel D., Volk H., et al. The changing epidemiology of autism spectrum disorders. Annu. Rev. Public Health. 2017;38:81–102. doi: 10.1146/annurev-publhealth-031816-044318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders (dsm-5®) 5th ed. American Psychiatric Pub.; Arlington, VA, USA: 2013. [Google Scholar]
- 3.Xu G., Strathearn L., Liu B., Bao W. Prevalence of autism spectrum disorder among us children and adolescents, 2014–2016. JAMA. 2018;319:81–82. doi: 10.1001/jama.2017.17812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zablotsky B., Black L.I., Maenner M.J., Schieve L.A., Blumberg S.J. Estimated prevalence of autism and other developmental disabilities following questionnaire changes in the 2014 national health interview survey. Natl. Health Stat. Rep. 2015;87:1–20. [PubMed] [Google Scholar]
- 5.Christensen D.L., Baio J., Van Naarden Braun K., Bilder D., Charles J., Constantino J.N., Daniels J., Durkin M.S., Fitzgerald R.T., Kurzius-Spencer M., et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—Autism and developmental disabilities monitoring network, 11 sites, united states, 2012. MMWR Surveill. Summ. 2016;65:1–23. doi: 10.15585/mmwr.ss6503a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J.Y., Son M.J., Son C.Y., Radua J., Eisenhut M., Gressier F., Koyanagi A., Carvalho A.F., Stubbs B., Solmi M., et al. Environmental risk factors and biomarkers for autism spectrum disorder: An umbrella review of the evidence. Lancet Psychiatry. 2019;6:590–600. doi: 10.1016/S2215-0366(19)30181-6. [DOI] [PubMed] [Google Scholar]
- 7.Ronald A., Hoekstra R.A. Autism spectrum disorders and autistic traits: A decade of new twin studies. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2011;156B:255–274. doi: 10.1002/ajmg.b.31159. [DOI] [PubMed] [Google Scholar]
- 8.Bai D., Yip B.H.K., Windham G.C., Sourander A., Francis R., Yoffe R., Glasson E., Mahjani B., Suominen A., Leonard H., et al. Association of genetic and environmental factors with autism in a 5-country cohort. JAMA Psychiatry. 2019 doi: 10.1001/jamapsychiatry.2019.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacGregor A.J., Snieder H., Schork N.J., Spector T.D. Twins: Novel uses to study complex traits and genetic diseases. Trends Genet. 2000;16:131–134. doi: 10.1016/S0168-9525(99)01946-0. [DOI] [PubMed] [Google Scholar]
- 10.Modabbernia A., Velthorst E., Reichenberg A. Environmental risk factors for autism: An evidence-based review of systematic reviews and meta-analyses. Mol. Autism. 2017;8:13. doi: 10.1186/s13229-017-0121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson E.B., St Pourcain B., Anttila V., Kosmicki J.A., Bulik-Sullivan B., Grove J., Maller J., Samocha K.E., Sanders S.J., Ripke S., et al. Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat. Genet. 2016;48:552–555. doi: 10.1038/ng.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.State M.W., Levitt P. The conundrums of understanding genetic risks for autism spectrum disorders. Nat. Neurosci. 2011;14:1499–1506. doi: 10.1038/nn.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. The new nhgri-ebi catalog of published genome-wide association studies (gwas catalog) Nucleic Acids Res. 2016;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wakefield J. A bayesian measure of the probability of false discovery in genetic epidemiology studies. Am. J. Hum. Genet. 2007;81:208–227. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wacholder S., Chanock S., Garcia-Closas M., El Ghormli L., Rothman N. Assessing the probability that a positive report is false: An approach for molecular epidemiology studies. J. Natl. Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. String v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K., Zhang H., Ma D., Bucan M., Glessner J.T., Abrahams B.S., Salyakina D., Imielinski M., Bradfield J.P., Sleiman P.M., et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia K., Guo H., Hu Z., Xun G., Zuo L., Peng Y., Wang K., He Y., Xiong Z., Sun L., et al. Common genetic variants on 1p13.2 associate with risk of autism. Mol. Psychiatry. 2014;19:1212–1219. doi: 10.1038/mp.2013.146. [DOI] [PubMed] [Google Scholar]
- 19.Grove J., Ripke S., Als T.D., Mattheisen M., Walters R.K., Won H., Pallesen J., Agerbo E., Andreassen O.A., Anney R., et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019;51:431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J., Yang A., Zhang Q., Yang G., Yang W., Lei H., Quan J., Qu F., Wang M., Zhang Z. Association between genetic variants in slc25a12 and risk of autism spectrum disorders: An integrated meta-analysis. Am. J. Med Genet. Part B Neuropsychiatr. Genet. 2015;168:236–246. doi: 10.1002/ajmg.b.32304. [DOI] [PubMed] [Google Scholar]
- 21.Rai V. Association of methylenetetrahydrofolate reductase (mthfr) gene c677t polymorphism with autism: Evidence of genetic susceptibility. Metab. Brain Dis. 2016;31:727–735. doi: 10.1007/s11011-016-9815-0. [DOI] [PubMed] [Google Scholar]
- 22.Pu D., Shen Y., Wu J. Association between mthfr gene polymorphisms and the risk of autism spectrum disorders: A meta-analysis. Autism Res. 2013;6:384–392. doi: 10.1002/aur.1300. [DOI] [PubMed] [Google Scholar]
- 23.Shaik Mohammad N., Sai Shruti P., Bharathi V., Krishna Prasad C., Hussain T., Alrokayan S.A., Naik U., Radha Rama Devi A. Clinical utility of folate pathway genetic polymorphisms in the diagnosis of autism spectrum disorders. Psychiatr. Genet. 2016;26:281–286. doi: 10.1097/YPG.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 24.Warrier V., Chee V., Smith P., Chakrabarti B., Baron-Cohen S. A comprehensive meta-analysis of common genetic variants in autism spectrum conditions. Mol. Autism. 2015;6:49. doi: 10.1186/s13229-015-0041-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aoki Y., Cortese S. Mitochondrial aspartate/glutamate carrier slc25a12 and autism spectrum disorder: A meta-analysis. Mol. Neurobiol. 2016;53:1579–1588. doi: 10.1007/s12035-015-9116-3. [DOI] [PubMed] [Google Scholar]
- 26.LoParo D., Waldman I.D. The oxytocin receptor gene (oxtr) is associated with autism spectrum disorder: A meta-analysis. Mol. Psychiatr. 2015;20:640–646. doi: 10.1038/mp.2014.77. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z., Hong Y., Zou L., Zhong R., Zhu B., Shen N., Chen W., Lou J., Ke J., Zhang T., et al. Reelin gene variants and risk of autism spectrum disorders: An integrated meta-analysis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2014;165B:192–200. doi: 10.1002/ajmg.b.32222. [DOI] [PubMed] [Google Scholar]
- 28.Torrico B., Fernandez-Castillo N., Hervas A., Mila M., Salgado M., Rueda I., Buitelaar J.K., Rommelse N., Oerlemans A.M., Bralten J., et al. Contribution of common and rare variants of the ptchd1 gene to autism spectrum disorders and intellectual disability. Eur. J. Hum. Genet. 2015;23:1694–1701. doi: 10.1038/ejhg.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kranz T.M., Kopp M., Waltes R., Sachse M., Duketis E., Jarczok T.A., Degenhardt F., Gorgen K., Meyer J., Freitag C.M., et al. Meta-analysis and association of two common polymorphisms of the human oxytocin receptor gene in autism spectrum disorder. Autism Res. 2016;9:1036–1045. doi: 10.1002/aur.1597. [DOI] [PubMed] [Google Scholar]
- 30.Anney R., Klei L., Pinto D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S., Sykes N., Pagnamenta A.T., et al. A genome-wide scan for common alleles affecting risk for autism. Hum. Mol. Genet. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium Meta-analysis of gwas of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism. 2017;8:21. doi: 10.1186/s13229-017-0137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma D., Salyakina D., Jaworski J.M., Konidari I., Whitehead P.L., Andersen A.N., Hoffman J.D., Slifer S.H., Hedges D.J., Cukier H.N., et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Ann. Hum. Genet. 2009;73:263–273. doi: 10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo P.H., Chuang L.C., Su M.H., Chen C.H., Chen C.H., Wu J.Y., Yen C.J., Wu Y.Y., Liu S.K., Chou M.C., et al. Genome-wide association study for autism spectrum disorder in taiwanese han population. PLoS ONE. 2015;10:e0138695. doi: 10.1371/journal.pone.0138695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anney R., Klei L., Pinto D., Almeida J., Bacchelli E., Baird G., Bolshakova N., Bolte S., Bolton P.F., Bourgeron T., et al. Individual common variants exert weak effects on the risk for autism spectrum disorders. Hum. Mol. Genet. 2012;21:4781–4792. doi: 10.1093/hmg/dds301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaste P., Klei L., Sanders S.J., Hus V., Murtha M.T., Lowe J.K., Willsey A.J., Moreno-De-Luca D., Timothy W.Y., Fombonne E. A genome-wide association study of autism using the simons simplex collection: Does reducing phenotypic heterogeneity in autism increase genetic homogeneity? Biol. Psychiatry. 2015;77:775–784. doi: 10.1016/j.biopsych.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Main P.A., Angley M.T., O’Doherty C.E., Thomas P., Fenech M. The potential role of the antioxidant and detoxification properties of glutathione in autism spectrum disorders: A systematic review and meta-analysis. Nutr. Metab. 2012;9:35. doi: 10.1186/1743-7075-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C.H., Santangelo S.L. Autism and serotonin transporter gene polymorphisms: A systematic review and meta-analysis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147B:903–913. doi: 10.1002/ajmg.b.30720. [DOI] [PubMed] [Google Scholar]
- 38.Curran S., Bolton P., Rozsnyai K., Chiocchetti A., Klauck S.M., Duketis E., Poustka F., Schlitt S., Freitag C.M., Lee I., et al. No association between a common single nucleotide polymorphism, rs4141463, in the macrod2 gene and autism spectrum disorder. Am. J. Med Genet. Part B: Neuropsychiatr. Genet. 2011;156:633–639. doi: 10.1002/ajmg.b.31201. [DOI] [PubMed] [Google Scholar]
- 39.Yang P.Y., Menga Y.J., Li T., Huang Y. Associations of endocrine stress-related gene polymorphisms with risk of autism spectrum disorders: Evidence from an integrated meta-analysis. Autism Res. 2017;10:1722–1736. doi: 10.1002/aur.1822. [DOI] [PubMed] [Google Scholar]
- 40.Song R.R., Zou L., Zhong R., Zheng X.W., Zhu B.B., Chen W., Liu L., Miao X.P. An integrated meta-analysis of two variants in hoxa1/hoxb1 and their effect on the risk of autism spectrum disorders. PLoS ONE. 2011;6:e25603. doi: 10.1371/journal.pone.0025603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen N., Bao Y., Xue Y., Sun Y., Hu D., Meng S., Lu L., Shi J. Meta-analyses of reln variants in neuropsychiatric disorders. Behav. Brain Res. 2017;332:110–119. doi: 10.1016/j.bbr.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 42.Werling A.M., Bobrowski E., Taurines R., Gundelfinger R., Romanos M., Grunblatt E., Walitza S. Cntnap2 gene in high functioning autism: No association according to family and meta-analysis approaches. J. Neural. Transm. 2016;123:353–363. doi: 10.1007/s00702-015-1458-5. [DOI] [PubMed] [Google Scholar]
- 43.Zhang T., Zhang J., Wang Z., Jia M., Lu T., Wang H., Yue W., Zhang D., Li J., Wang L. Association between cntnap2 polymorphisms and autism: A family-based study in the chinese han population and a meta-analysis combined with gwas data of psychiatric genomics consortium. Autism Res. 2019;12:553–561. doi: 10.1002/aur.2078. [DOI] [PubMed] [Google Scholar]
- 44.Noroozi R., Taheri M., Ghafouri-Fard S., Bidel Z., Omrani M.D., Moghaddam A.S., Sarabi P., Jarahi A.M. Meta-analysis of gabrb3 gene polymorphisms and susceptibility to autism spectrum disorder. J. Mol. Neurosci. 2018;65:432–437. doi: 10.1007/s12031-018-1114-2. [DOI] [PubMed] [Google Scholar]
- 45.Mahdavi M., Kheirollahi M., Riahi R., Khorvash F., Khorrami M., Mirsafaie M. Meta-analysis of the association between gaba receptor polymorphisms and autism spectrum disorder (ASD) J. Mol. Neurosci. 2018;65:1–9. doi: 10.1007/s12031-018-1073-7. [DOI] [PubMed] [Google Scholar]
- 46.Waltes R., Duketis E., Knapp M., Anney R.J., Huguet G., Schlitt S., Jarczok T.A., Sachse M., Kampfer L.M., Kleinbock T., et al. Common variants in genes of the postsynaptic fmrp signalling pathway are risk factors for autism spectrum disorders. Hum. Genet. 2014;133:781–792. doi: 10.1007/s00439-013-1416-y. [DOI] [PubMed] [Google Scholar]
- 47.Torrico B., Chiocchetti A.G., Bacchelli E., Trabetti E., Hervas A., Franke B., Buitelaar J.K., Rommelse N., Yousaf A., Duketis E., et al. Lack of replication of previous autism spectrum disorder gwas hits in european populations. Autism Res. 2017;10:202–211. doi: 10.1002/aur.1662. [DOI] [PubMed] [Google Scholar]
- 48.Panagiotou O.A., Ioannidis J.P.A., Genome-Wide Significance Project What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int. J. Epidemiol. 2011;41:273–286. doi: 10.1093/ije/dyr178. [DOI] [PubMed] [Google Scholar]
- 49.Skaar D.A., Shao Y., Haines J.L., Stenger J.E., Jaworski J., Martin E.R., DeLong G.R., Moore J.H., McCauley J.L., Sutcliffe J.S., et al. Analysis of the reln gene as a genetic risk factor for autism. Mol. Psychiatry. 2005;10:563–571. doi: 10.1038/sj.mp.4001614. [DOI] [PubMed] [Google Scholar]
- 50.Glessner J.T., Wang K., Cai G., Korvatska O., Kim C.E., Wood S., Zhang H., Estes A., Brune C.W., Bradfield J.P., et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purcell A.E., Jeon O.H., Zimmerman A.W., Blue M.E., Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001;57:1618–1628. doi: 10.1212/WNL.57.9.1618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.