Abstract
BACKGROUND
The placenta is the active interface between mother and foetus, bearing the molecular marks of rapid development and exposures in utero. The placenta is routinely discarded at delivery, providing a valuable resource to explore maternal-offspring health and disease in pregnancy. Genome-wide profiling of the human placental transcriptome provides an unbiased approach to study normal maternal–placental–foetal physiology and pathologies.
OBJECTIVE AND RATIONALE
To date, many studies have examined the human placental transcriptome, but often within a narrow focus. This review aims to provide a comprehensive overview of human placental transcriptome studies, encompassing those from the cellular to tissue levels and contextualize current findings from a broader perspective. We have consolidated studies into overarching themes, summarized key research findings and addressed important considerations in study design, as a means to promote wider data sharing and support larger meta-analysis of already available data and greater collaboration between researchers in order to fully capitalize on the potential of transcript profiling in future studies.
SEARCH METHODS
The PubMed database, National Center for Biotechnology Information and European Bioinformatics Institute dataset repositories were searched, to identify all relevant human studies using ‘placenta’, ‘decidua’, ‘trophoblast’, ‘transcriptome’, ‘microarray’ and ‘RNA sequencing’ as search terms until May 2019. Additional studies were found from bibliographies of identified studies.
OUTCOMES
The 179 identified studies were classifiable into four broad themes: healthy placental development, pregnancy complications, exposures during pregnancy and in vitro placental cultures. The median sample size was 13 (interquartile range 8–29). Transcriptome studies prior to 2015 were predominantly performed using microarrays, while RNA sequencing became the preferred choice in more recent studies. Development of fluidics technology, combined with RNA sequencing, has enabled transcript profiles to be generated of single cells throughout pregnancy, in contrast to previous studies relying on isolated cells. There are several key study aspects, such as sample selection criteria, sample processing and data analysis methods that may represent pitfalls and limitations, which need to be carefully considered as they influence interpretation of findings and conclusions. Furthermore, several areas of growing importance, such as maternal mental health and maternal obesity are understudied and the profiling of placentas from these conditions should be prioritized.
WIDER IMPLICATIONS
Integrative analysis of placental transcriptomics with other ‘omics’ (methylome, proteome and metabolome) and linkage with future outcomes from longitudinal studies is crucial in enhancing knowledge of healthy placental development and function, and in enabling the underlying causal mechanisms of pregnancy complications to be identified. Such understanding could help in predicting risk of future adversity and in designing interventions that can improve the health outcomes of both mothers and their offspring. Wider collaboration and sharing of placental transcriptome data, overcoming the challenges in obtaining sufficient numbers of quality samples with well-defined clinical characteristics, and dedication of resources to understudied areas of pregnancy will undoubtedly help drive the field forward.
Keywords: placenta, decidua, transcriptome, microarray, RNA sequencing, pregnancy, trophoblast, development, pre-eclampsia
Introduction
The human placenta undergoes rapid growth and development usually over a span of 9 months. Serving as the maternal–foetal interface, the placenta facilitates communication between mother and child throughout gestation. Therefore, investigating the placenta provides a window into how the pregnancy has progressed and an insight into the potential health trajectory of the child. To better understand maternal–placental–foetal health, especially in the context of pregnancy complications, numerous microarrays and RNA-sequencing studies have been performed to profile the placental transcriptome. Recent rapid technological advancements in ‘omics’ have enabled parallel gathering of large datasets from maternal, placental and foetal tissues across gestation.
This review summarizes genome-wide human placental transcriptome studies performed over the last two decades. We provide an overview of transcriptome study methods, outline the important considerations for study design and interpretation, discuss past study results and highlight knowledge gaps that should be addressed in future studies. As the review is focussed on human placental studies, animal studies will not be referred to. Studies in other mammals (Barreto et al., 2011; Buckberry et al., 2017; Carter, 2018) have undoubtedly provided additional valuable perspectives on placental health and disease and have added to knowledge on comparative placentation across species.
Transcript profiling methods
Two methods used to obtain genome-wide placental transcript profiles are microarrays and RNA sequencing. These high-throughput technologies generate large amounts of data and offer a means to analyse the placenta in an unbiased manner.
Microarrays
Microarrays utilize short oligonucleotide probes embedded on a chip, which when hybridized to specific RNA or DNA sequences present in the sample emits fluorescence (Cox et al., 2015), allowing simultaneous quantitation of many gene transcripts. Commercially available chips from Affymetrix, Agilent and Illumina are commonly used in placental research, although a few studies have custom-made theirs. The main drawback of microarray chips is that a gene-specific probe must be present for a gene to be detected. As the earliest placental microarray studies were performed around the time the first human genome was sequenced, not all transcripts were detectable by gene probes available at that time. Moreover, RNA biology was not as well understood as it is now with the current knowledge of non-coding RNA and RNA gene silencing. Additionally, microarray probes are species-specific. Nevertheless, given established bioinformatics pipelines and readily available statistical tools to analyse microarray data, with publically available datasets for comparison from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) and the European Bioinformatics Institute (EBI) ArrayExpress repositories, microarrays continue to be widely used (Cox et al., 2015).
RNA sequencing
Next-generation RNA sequencing is more sensitive than conventional microarray and generates a fuller picture of the placental transcriptome. The Illumina Hiseq and Genome Analyzer II systems are the most popular platforms in placental research and are increasingly used as the price of sequencing falls. Sequencing can detect rare and novel RNA transcripts, identify single-nucleotide variants in both coding and non-coding RNA of all lengths, and is not species-dependent. Furthermore, recent development of microfluidics technology with RNA sequencing allows transcripts of individual cells to be determined, which was not previously possible (Hu et al., 2018b). However, single-cell RNA sequencing of the syncytiotrophoblast, which forms the placental cellular barrier, remains a challenge since its large size and multi-nucleated nature does not permit its isolation with microfluidics technology. The targeted sequencing depth or number of reads sequenced per sample is dependent on experimental aims and design. For instance, the ENCODE guidelines recommend a minimum sequencing depth of 30 million reads for bulk RNA sequencing that is commonly used for differential gene expression analysis, while 10 000 to 50 000 reads per cell in single-cell sequencing are sufficient to classify cells in an unbiased manner (Haque et al., 2017). Another question-dependent experimental design consideration is whether to sequence library preparations from ribosomal RNA (rRNA)-depleted total RNA (including non-coding RNA) or those enriched for mRNA transcripts by poly A+ selection. Given the vast potential of RNA sequencing, it is unsurprising that its use is growing exponentially in placental research.
Why profile the human placental transcriptome?
In most pregnancies, the villous placenta is genetically identical to the foetus and is the first foetal-placental tissue in direct contact with the maternal exposome. The placental transcriptome may, therefore, represent to an extent both inherent foetal characteristics and the foetal response to the intrauterine environment. Data generated from genome-wide profiling of the human placenta have several uses. Firstly, analyses of normal placentas throughout gestation enhances understanding of healthy development, which serves as a reference point for studies of how the placenta responds and adapts to various exposures and challenges in complicated pregnancies. Secondly, by analysing placentas from compromised pregnancies, pathological changes linked with different clinical phenotypes can be identified and utilized for developing biomarkers or targets for prediction, diagnosis and therapeutic interventions. For instance, placental-specific gene products that are secreted into the maternal circulation can serve as a non-invasive direct readout of placental function and an indirect measure of foetal wellbeing (Cox et al., 2015). One such success story of transcript profiling is sFLT1, which was first identified to be up-regulated in pre-eclamptic placentas by microarray (Maynard et al., 2003) and is now being trialled in clinical screening to predict if a pregnant woman is at risk of developing pre-eclampsia (Zeisler et al., 2016). Additionally, knowledge of the dysfunctional molecular processes at the maternal–foetal interface provides novel insights into the potential causal mechanisms underlying placental pathologies, which may open up new avenues for developing preventative measures for pregnancy complications. Moreover, the placenta functions as the intermediary between mother and child, participating in the normal programming of the developing foetus to face the prevailing environmental conditions of ex utero life (Burton et al., 2016). Understanding these programming mechanisms and deviations in pathological conditions opens up the possibility of modifying offspring growth and health trajectories arising from compromised intrauterine environments, through interventions that target the placenta, or identifying at-risk children who will benefit from close follow-up and early childhood interventions.
Placental transcriptome: search methods and study themes
The PubMed database and the NCBI GEO and EBI ArrayExpress repositories were used to identify relevant studies and human placental transcriptome datasets, respectively up to May 2019. Our search included terms such as ‘placenta’, ‘decidua’, ‘trophoblast’, ‘transcriptome’, ‘microarray’, ‘RNA sequencing’ and was refined by restricting only to human studies and genome-wide datasets. Additional studies were identified by references within articles.
We identified a total of 179 unique genome-wide datasets related to human placenta. These datasets collectively represent the genome-wide transcriptomes of around 3000 placentas since 2004. Most of these datasets were generated within the last five years, highlighting the rapid expansion of human placental transcriptome studies, with a clear preferential shift towards RNA sequencing from conventional microarrays in the field.
Placental transcriptome studies can broadly be divided into four main study themes: healthy placental development (Table I), pregnancy complications (Table II), exposures during pregnancy (Table III) and in vitro placental cultures (Table IV). The tables list studies in chronological order and studies with fewer than five samples are presented separately in Supplementary Table SI. Studies in each theme are summarized and discussed in the context of the following considerations.
Table I.
Healthy placental development.
| Year | Accession ID | Platform | Type | Tissue/cell | Sampling site | Total no. | Gestation | Purpose | Associated publications |
|---|---|---|---|---|---|---|---|---|---|
|
Gestation-specific effects | |||||||||
| 2008 | NCBI GEO GSE9984 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Tissue | Chorionic villi |
|
1st, 2nd and 3rd trimester | Profile placenta across gestation | Mikheev et al. (2008) |
| 2011 | NCBI GEO GSE28551 | ABI Human Genome Survey Microarray Version 2 | Microarray | Tissue | Chorionic villi |
|
1st and 3rd trimester | Assess placental development across gestation | Sitras et al. (2012) |
| 2015 | NCBI GEO GSE66302 |
|
RNA-seq | Tissue | Amnion, chorion, chorionic villi, decidua, umbilical cord |
|
1st and 2nd trimester | Development from early to mid-pregnancy | Roost et al. (2015) |
| 2016 | NCBI GEO GSE75010 | Affymetrix Human Gene 1.0 ST Array | Microarray | Tissue | Placenta biopsy (basal plate + chorionic villi) | 157 | 3rd trimester | Identify markers of villous maturation (with histological data) | Leavey et al. (2017) |
| 2017 | NCBI GEO GSE98752 |
|
RNA-seq | Tissue | Chorionic villi |
|
1st and 3rd trimester | Placental development across gestation (with linked methylation data) | Lim et al. (2017b) |
| 2018 | NCBI GEO GSE100051 | Illumina HumanHT-12 V4.0 Expression Beadchip | Microarray | Tissue | Chorionic villi |
|
1st, 2nd and 3rd trimester | Profile placenta across gestation | Soncin et al. (2018) |
| Not publically available | Affymetrix Human Genome U133A Array | Microarray | Tissue | Chorionic villi |
|
1st and 3rd trimester | Identify placental-specific transcripts in maternal blood | Tsui et al. (2004) | |
|
| |||||||||
|
Cellular characterization and differentiation | |||||||||
| 2008 | NCBI GEO GSE10612 | Affymetrix Human Genome U133A Array; Affymetrix Human Genome U133A 2.0 Array | Microarray | Sorted cell (flow cytometry/ magnetic bead) | Decidua | 11 | 1st trimester | Characterize decidual macrophages | Gustafsson et al. (2008) |
| 2008 | NCBI GEO GSE11510 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Primary cell cultures | Amnion, chorionic plate, chorionic villi, decidua, umbilical cord | 10 | 3rd trimester | Placental cell taxonomy | Kawamichi et al. (2010) |
| 2013 | NCBI GEO GSE44368 | Affymetrix Human Gene 1.0 ST Array | Microarray | Primary cell cultures | Chorionic villi |
|
3rd trimester | Sexual dimorphism of trophoblast and endothelial cells | Cvitic et al. (2013) |
| 2013 | NCBI GEO GSE24268 | Agilent-014850 Whole Human Genome Microarray 4 × 44K G4112F | Microarray | Sorted cell (flow cytometry) | Decidua | Pooled | 1st trimester | Characterize decidual natural killer cells | Wang et al. (2014) |
| 2015 | NCBI GEO GSE59126 | Affymetrix Human Genome U133A Array | Microarray | Primary cell cultures | Chorionic villi (1st trimester), chorionic plate (3rd trimester) |
|
1st and 3rd trimester | Compare 1st trimester trophoblast vs 3rd trimester endothelial degradome | Ghaffari-Tabrizi-Wizsy et al. (2014) |
| 2015 | NCBI GEO GSE57834 | Affymetrix Human Gene Expression Array | Microarray | Sorted cell (flow cytometry) | Chorionic villi | 5 | 1st trimester | Characterize trophoblast subpopulations in early pregnancy | James et al. (2015) |
| 2017 | EBI ArrayExpress E-MTAB-5517 | Affymetrix Human Transcriptome Array 2.0 | Microarray | Sorted cell (flow cytometry) | Decidua | 6 | 3rd trimester | Characterize decidual T-cell populations | Powell et al. (2017) |
| 2017 | EBI EGA EGAD00001003705 (access requires approval) |
|
RNA-seq | Single cell | Chorionic villi | 8 | 3rd trimester | Characterize cells from term and early-onset pre-eclampsia-affected placentas | Tsang et al. (2017) |
| 2018 | NCBI GEO GSE79879 |
|
RNA-seq | Sorted cell (flow cytometry) | Decidua | 5 (pooled) | 1st trimester | Profile decidual natural killer cell population with expanded memory in subsequent pregnancy | Gamliel et al. (2018) |
| 2018 | NCBI GEO GSE89497 |
|
RNA-seq | Single cell with pre-sorting (magnetic bead) | Chorionic villi (1st trimester), basal plate (2nd trimester) |
|
1st and 2nd trimester | Characterize placental cell subpopulations | Liu et al. (2018) |
| 2018 | NCBI GEO GSE107824 | Illumina HumanHT-12 V4.0 Expression Beadchip | Microarray | Primary trophoblast cell culture | Chorionic villi |
|
1st, 2nd and 3rd trimester | Assess trophoblast differentiation across gestation | Soncin et al. (2018) |
| 2018 | NCBI SRA PRJNA492324 |
|
RNA-seq | Single cell and tissue | Chorionic villi, decidua | 9 | 1st trimester | Characterize cells from the first-trimester maternal–foetal interface | Suryawanshi et al. (2018) |
| 2018 | EBI ArrayExpress E-MTAB-6678 |
|
RNA-seq | Single cell with pre-sorting (flow cytometry) | Decidua | 5 | 1st trimester | Characterize cells from the first-trimester maternal–foetal interface | Vento-Tormo et al. (2018) |
| 2018 | EBI ArrayExpress E-MTAB-6701 |
|
RNA-seq | Single cell with pre-sorting (flow cytometry) | Chorionic villi, decidua | 7 | 1st trimester | Characterize cells from the first-trimester maternal–foetal interface | Vento-Tormo et al. (2018) |
| 2018 | NCBI GEO GSE124282 |
|
RNA-seq | Primary trophoblast cell culture | Chorionic villi |
|
2nd and 3rd trimester | Assess trophoblast differentiation across gestation (with linked ChIP sequencing data) | Wang et al. (2019a) |
|
| |||||||||
|
Gene expression variation and regulation | |||||||||
| 2006 | NCBI GEO GSE4421 | Stanford Functional Genomics Human cDNA Microarray | Microarray | Tissue | Amnion, basal plate, chorion, chorionic plate, chorionic villi, umbilical cord | 19 | 3rd trimester | Examine gene expression variation within the placenta and between closely associated tissues | Sood et al. (2006) |
| 2012 | NCBI GEO GSE36828 | Illumina HumanHT-12 V3.0 Expression Beadchip | Microarray | Tissue | Chorionic villi | 48 | 3rd trimester | Examine relationship of placental gene expression with methylation | Turan et al. (2012) |
| 2014 | NCBI GEO GSE56524 |
|
RNA-seq | Tissue | Whole placenta biopsy (basal plate + chorionic plate + chorionic villi) | 10 | 3rd trimester | Identify imprinted genes in the placenta | Metsalu et al. (2014) |
| 2015 | NCBI GEO GSE66622 |
|
RNA-seq | Tissue | Chorionic villi | 40 | 3rd trimester | Assess inter and intra-variation in placental transcriptome (African-American, European-American, South Asian and East Asian ancestry) | Hughes et al. (2015) |
| 2017 | NCBI GEO GSE77085 |
|
RNA-seq | Tissue | Chorionic villi | 16 | 3rd trimester | Identify gene co-expression patterns in normal term placenta | Buckberry et al. (2017) |
| 2018 | EBI ArrayExpress E-MTAB-6683 | Affymetrix Human Gene 2.1 ST Array | Microarray | Tissue | Chorionic villi | 8 | 1st trimester | Validate source and source-derived organoid | Turco et al. (2018) |
| 2019 | dbGaP Study Accession: phs001717.v1.p1 (access requires approval) |
|
RNA-seq | Tissue | Chorionic villi | 80 | 3rd trimester | Determine placental gene regulation (with linked genotyping and methylation data) | Delahaye et al. (2018) |
| 2019 | NCBI GEO GSE109120 |
|
RNA-seq | Tissue | Chorionic villi | 39 | 1st trimester | Sex: males vs females | Gonzalez et al. (2018) |
| Not publically available |
|
RNA-seq | Tissue | Amnion, chorion, basal plate | 5 (pooled) | 3rd trimester | Examine gene expression variation between placental-associated tissues | Kim et al. (2012) | |
| Not publically available |
|
RNA-seq | Tissue | Placenta biopsy (no details provided) | 20 | 3rd trimester | Identify placental-enriched genes compared to other tissue types | Saben et al. (2014b) | |
| Not publically available |
|
RNA-seq | Tissue | Chorionic villi | 159 | 3rd trimester | Determine expression quantitative trait loci in the placenta (with linked genotyping data) | Peng et al. (2017) | |
ChIP, chromatin immunoprecipitation; dbGaP, database for Genotypes and Phenotypes; EBI, European Bioinformatics Institute; EGA, European Genome-phenome Archive; GEO, Gene Expression Omnibus; M, million reads; NCBI, National Center for Biotechnology Information; SRA, sequence read archive.
Table II.
Pregnancy complications.
| Year | Accession ID | Platform | Type | Tissue/cell | Sampling location | Total no. | Gestation | Purpose | Associated publications |
|---|---|---|---|---|---|---|---|---|---|
|
Pre-eclampsia | |||||||||
| 2006 | NCBI GEO GSE4707 | Agilent-012391 Whole Human Genome Oligo Microarray G4112A | Microarray | Tissue | Chorionic villi |
|
3rd trimester | EOPE or LOPE vs controls | Nishizawa et al. (2007) |
| 2007 | EBI ArrayExpress E-MEXP-1050 | Affymetrix GeneChip Human Genome Focus Array | Microarray | Tissue | Decidua |
|
3rd trimester | PE or IUGR vs controls | Eide et al. (2008) |
| 2007 | NCBI GEO GSE6573 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Tissue | Decidua, placenta biopsy (no details provided) |
|
3rd trimester | PE vs controls | Herse et al. (2007) |
| 2008 | NCBI GEO GSE12767 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Tissue | Chorionic villi |
|
1st trimester | Subsequent PE vs controls | Founds et al. (2009) |
| 2008 | NCBI GEO GSE10588 | Applied Biosystems Human Genome Survey Microarray Version 2 | Microarray | Tissue | Chorionic villi |
|
3rd trimester | PE vs controls | Sitras et al. (2009b) |
| 2009 | EBI ArrayExpress E-TABM-682 | Illumina Human-6 v2 Expression BeadChip | Microarray | Tissue | Decidua |
|
3rd trimester | PE or SGA vs controls | Loset et al. (2011) |
| 2009 | NCBI GEO GSE14722 | Affymetrix Human Genome U133A Array; Affymetrix Human Genome U133B Array | Microarray | Tissue | Basal plate |
|
3rd trimester | PE vs preterm controls | Winn et al. (2009) |
| 2010 | NCBI GEO GSE24129 | Affymetrix Human Gene 1.0 ST Array | Microarray | Tissue | Chorionic villi |
|
3rd trimester | PE or IUGR vs controls | Nishizawa et al. (2011) |
| 2010 | NCBI GEO GSE25906 | Illumina humanWG-6 V2.0 Expression Beadchip | Microarray | Tissue | Chorionic villi |
|
3rd trimester | PE vs controls | Tsai et al. (2011) |
| 2011 | NCBI GEO GSE22526 | Operon Human Genome Array Ready Oligo Set version 2.1 | Microarray | Tissue | Placenta biopsy (basal plate + chorionic villi) |
|
3rd trimester | EOPE vs LOPE | Junus et al. (2012) |
| 2011 | NCBI GEO GSE30186 | Illumina HumanHT-12 V4.0 Expression Beadchip | Microarray | Tissue | Chorionic villi |
|
3rd trimester | PE vs controls | Meng et al. (2012) |
| 2012 | NCBI GEO GSE25861 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Sorted cells (magnetic beads) | Chorionic villi |
|
3rd trimester | Placental endothelial cells in PE with IUGR vs preterm controls | Dunk et al. (2012) |
| 2012 | NCBI GEO GSE35574 | Illumina HumanWG-6 V2.0 Expression Beadchip | Microarray | Tissue | Chorionic villi |
|
3rd trimester | PE or IUGR vs controls | Guo et al. (2013) |
| 2013 | NCBI GEO GSE44711 | Illumina HumanHT-12 V4.0 Expression Beadchip | Microarray | Tissue | Chorionic villi |
|
3rd trimester | EOPE vs preterm controls (with linked methylation data) | Blair et al. (2013) |
| 2013 | NCBI GEO GSE47187 | Agilent-028004 SurePrint G3 Human GE 8 × 60K Microarray | Microarray | Tissue | Chorionic villi |
|
3rd trimester | PE vs preterm controls | Song et al. (2013) |
| 2013 | NCBI GEO GSE43942 | NimbleGen Homo sapiens HG18 090828 opt expr HX12 (12 × 135k) | Microarray | Tissue | Placenta biopsy (basal plate + chorionic villi) |
|
3rd trimester | PE vs controls | Xiang et al. (2013) |
| 2014 | NCBI GEO GSE54618 | Illumina HumanHT-12 V4.0 Expression Beadchip | Microarray | Tissue | Chorionic villi |
|
3rd trimester | PE vs controls | Jebbink et al. (2015) |
| 2015 | EBI ArrayExpress E-MTAB-3265 | Illumina HumanHT-12 V3.0 Expression BeadChip | Microarray | Tissue | Chorionic villi |
|
1st trimester | High risk vs low risk of PE (based on uterine artery Doppler resistance index) | Leslie et al. (2015) |
| 2015 | NCBI GEO GSE74341 | Agilent-039494 SurePrint G3 Human GE v2 8 × 60K | Microarray | Tissue | Chorionic villi |
|
3rd trimester | EOPE or LOPE vs controls | Liang et al. (2016) |
| 2015 | NCBI GEO GSE73374 | Affymetrix Human Gene 2.0 ST Array | Microarray | Tissue | Whole placenta biopsy (basal plate + chorionic plate + chorionic villi) |
|
3rd trimester | PE vs controls (with linked methylation data) | Martin et al. (2015) |
| 2015 | NCBI GEO GSE60438 | Illumina HumanWG-6 V3.0 Expression Beadchip; Illumina HumanHT-12 V4.0 Expression Beadchip | Microarray | Tissue | Decidua |
|
3rd trimester | PE vs controls | Yong et al. (2015) |
| 2016 | NCBI GEO GSE75010 | Affymetrix Human Gene 1.0 ST Array | Microarray | Tissue | Placenta biopsy (basal plate + chorionic villi) |
|
3rd trimester | PE vs controls (with linked methylation and histological data) | Leavey et al. (2016); Christians et al. (2017); Leavey et al. (2018); Benton et al. (2018); Gibbs et al. (2019) |
| 2017 | NCBI GEO GSE94643 | Affymetrix Human Gene 2.0 ST Array | Microarray | Tissue | Decidua |
|
3rd trimester | Decidualization in PE vs preterm controls | Garrido-Gomez et al. (2017) |
| 2017 | NCBI GEO GSE93839 | Affymetrix Human Gene 2.0 ST Array | Microarray | Tissue (laser microdissected) | Placenta biopsy (basal plate + chorionic villi) |
|
3rd trimester | Trophoblast populations in PE vs preterm controls | Gormley et al. (2017) |
| 2017 | NCBI GEO GSE99007 | Agilent SurePrint G3 Human GE v3 8 × 60K Microarray | Microarray | Primary cell culture | Chorionic villi |
|
3rd trimester | Fibroblasts from PE vs controls | Ohmaru-Nakanishi et al. (2018) |
| 2017 | EBI EGA EGAD00001003705 (access requires approval) | Illumina NextSeq 500 -mean sequencing depth ∼21 471 reads | RNA-seq | Single cell | Chorionic villi |
|
Late 2nd and 3rd trimester | EOPE vs controls | Tsang et al. (2017) |
| 2018 | NCBI GEO GSE96984 | Agilent-078298 human ceRNA array V1.0 4X180K | Microarray | Tissue | Basal plate |
|
3rd trimester | PE vs controls | Unpublished |
| 2018 | NCBI GEO GSE66273 | Agilent-014850 Whole Human Genome Microarray 4 × 44K G4112F | Microarray | Tissue | Chorionic villi |
|
3rd trimester | EOPE vs preterm controls | Than et al. (2018); Varkonyi et al. (2011) |
| 2019 | NCBI GEO GSE102897 | Agilent-078298 human ceRNA array V1.0 4X180K | Microarray | Tissue | Chorionic villi |
|
3rd trimester | PE vs controls | Hu et al. (2018a) |
| Not publically available, but available on request | CapitalBio Genomewide Microarray | Microarray | Tissue | Chorionic villi |
|
3rd trimester | PE vs controls | Zhou et al. (2006) | |
| Not publically available | Affymetrix Human Genome U133 Plus 2 | Microarray | Tissue | Placenta biopsy (no details provided) |
|
3rd trimester | PE with IUGR vs controls | Jarvenpaa et al. (2007) | |
| Not publically available | Operon Human Genome Array Ready Oligo Set version 2.1 | Microarray | Tissue | Placenta biopsy (basal plate + chorionic villi) |
|
3rd trimester | PE vs controls | Enquobahrie et al. (2008) | |
| Not publically available | Affymetrix Human Genome U133A Array | Microarray | Tissue | Placenta biopsy (basal plate + chorionic villi) |
|
3rd trimester | PE vs controls | Hoegh et al. (2010) | |
| Not publically available | Agilent Human Genome Microarray 4 × 44K | Microarray | Tissue | Chorionic villi |
|
3rd trimester | PE vs controls | Lee et al. (2010) | |
| Not publically available | Operon Human Genome Array Ready Oligo Set versions 2.1 and 2.1.1 | Microarray | Tissue | Chorionic villi |
|
3rd trimester | PE vs controls (with Doppler findings) | Centlow et al. (2011) | |
| Not publically available | GE Healthcare Codelink Human Whole Genome Bioarrays | Microarray | Tissue | Placenta biopsy (basal plate + chorionic plate + chorionic villi) |
|
3rd trimester | PE vs controls | Kang et al. (2011) | |
| Not publically available | Illumina HumanRef-12 v3 Expression BeadChip | Microarray | Tissue | Chorionic villi |
|
3rd trimester | PE or PTL vs TNL (with linked miRNA data) | Mayor-Lynn et al. (2011) | |
| Not publically available |
|
RNA-seq | Tissue | Placental biopsy (basal plate + chorionic plate + chorionic villi) |
|
3rd trimester | LOPE, GDM, SGA or LGA vs controls | Sober et al. (2015) | |
| Not publically available |
|
RNA-seq | Tissue | Chorionic villi |
|
3rd trimester | LOPE or GDM vs controls | Lekva et al. (2016) | |
| Not publically available |
|
RNA-seq | Tissue | Decidua |
|
3rd trimester | EOPE or LOPE vs controls | Tong et al. (2018) | |
|
| |||||||||
|
Intrauterine growth restriction/small for gestational age | |||||||||
| 2007 | EBI ArrayExpress E-MEXP-1050 | Affymetrix GeneChip Human Genome Focus Array | Microarray | Tissue | Decidua |
|
3rd trimester | IUGR or PE vs controls | Eide et al. (2008) |
| 2008 | NCBI GEO GSE12216 | ABI Human Genome Survey Microarray Version 2 | Microarray | Tissue | Chorionic villi |
|
3rd trimester | IUGR with placental insufficiency vs controls | Sitras et al. (2009a) |
| 2009 | EBI ArrayExpress E-TABM-682 | Illumina Human-6 v2 Expression BeadChip | Microarray | Tissue | Decidua |
|
3rd trimester | SGA or PE vs controls | Loset et al. (2011) |
| 2010 | NCBI GEO GSE24129 | Affymetrix Human Gene 1.0 ST Array | Microarray | Tissue | Chorionic villi |
|
3rd trimester | IUGR or PE vs controls | Nishizawa et al. (2011) |
| 2012 | NCBI GEO GSE25861 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Sorted cells (magnetic bead) | Chorionic villi |
|
3rd trimester | Placental endothelial cells in IUGR with PE vs preterm controls | Dunk et al. (2012) |
| 2012 | NCBI GEO GSE35574 | Illumina HumanWG-6 V2.0 Expression Beadchip | Microarray | Tissue | Chorionic villi |
|
3rd trimester | IUGR or PE vs controls | Guo et al. (2013) |
| 2014 | EBI ArrayExpress E-MTAB-1956 | NimbleGen Human HG18 60mer 4 × 72K Expression Array | Microarray | Tissue | Chorionic villi |
|
3rd trimester | IUGR vs control | Madeleneau et al. (2015) |
| 2017 | NCBI SRA PRJNA358255 |
|
RNA-seq | Tissue | Placenta biopsy (chorionic plate + chorionic villi) |
|
3rd trimester | SGA or LGA vs controls | Deyssenroth et al. (2017) |
| 2018 | NCBI GEO GSE100415 | Affymetrix Human Gene 1.0 ST Array | Microarray | Tissue | Placenta biopsy (basal plate + chorionic villi) |
|
3rd trimester | Normotensive suspected IUGR only (extension of earlier study comparing healthy controls and hypertensive suspected IUGR) | Gibbs et al. (2019) (extension of GSE75010) |
| 2018 | Available as supplementary data within journal article |
|
RNA-seq | Tissue | Placenta biopsy (basal plate + chorionic villi) |
|
3rd trimester | IUGR vs controls in women with and without chemotherapy during pregnancy | Verheecke et al. (2018) |
| 2019 | EBI ENA PRJEB30656 |
|
RNA-seq | Tissue | Placenta biopsy (no details provided) |
|
3rd trimester | IUGR vs controls | Majewska et al. (2019) |
| Not publically available | Affymetrix U95A Microarray | Microarray | Tissue | Placenta biopsy (no details provided) |
|
3rd trimester | IUGR vs controls | Roh et al. (2005) | |
| Not publically available | Affymetrix Human Genome U133 Plus 2 | Microarray | Tissue | Placenta biopsy (no details provided) |
|
3rd trimester | IUGR with PE vs controls | Jarvenpaa et al. (2007) | |
| Not publically available | Affymetrix Human Genome U133A Array | Microarray | Tissue | Placenta biopsy (basal plate + chorionic villi) |
|
3rd trimester | IUGR vs controls | McCarthy et al. (2007) | |
| Not publically available | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Tissue | Placenta biopsy (chorionic plate + chorionic villi) |
|
3rd trimester | IUGR vs preterm controls | Struwe et al. (2010) | |
| Not publically available | Affymetrix Human Genome U219 Array | Microarray | Tissue | Chorionic villi |
|
3rd trimester | IUGR or macro vs controls | Sabri et al. (2014) | |
| Not publically available |
|
RNA-seq | Tissue | Placental biopsy (basal plate + chorionic plate + chorionic villi) |
|
3rd trimester | SGA, LGA or PE or GDM vs controls | Sober et al. (2015) | |
|
| |||||||||
|
Macrosomia/large for gestational age | |||||||||
| 2017 | NCBI SRA PRJNA358255 |
|
RNA-seq | Tissue | Placenta biopsy (chorionic plate + chorionic villi) |
|
3rd trimester | LGA or SGA vs controls | Deyssenroth et al. (2017) |
| Not publically available | Affymetrix Human Genome U219 Array | Microarray | Tissue | Chorionic villi |
|
3rd trimester | Macro or IUGR vs controls | Sabri et al. (2014) | |
| Not publically available |
|
RNA-seq | Tissue | Placental biopsy (basal plate + chorionic plate + chorionic villi) |
|
3rd trimester | LGA, SGA, PE or GDM vs controls | Sober et al. (2015) | |
| Not publically available | Arraystar Human LncRNA Microarray V3.0 | Microarray | Tissue | Placenta biopsy (basal plate + chorionic villi) |
|
3rd trimester | Non-diabetes macro vs controls | Song et al. (2018) | |
|
| |||||||||
|
Gestational diabetes mellitus | |||||||||
| 2005 | NCBI GEO GSE2956 | Affymetrix Human U133 Array | Microarray | Tissue | Chorionic villi | 15 −8 C −7 GDM |
3rd trimester | GDM vs controls | Radaelli et al. (2003) |
| 2015 | NCBI GEO GSE70493 | Affymetrix Human Transcriptome Array 2.0 | Microarray | Tissue | Placenta biopsy (basal plate + chorionic villi) |
|
3rd trimester | GDM vs controls (with linked methylation data) | Binder et al. (2015) |
| Not publically available | Operon Human Genome Array Ready Oligo Set version 2.1 | Microarray | Tissue | Chorionic villi |
|
3rd trimester | GDM vs controls | Enquobahrie et al. (2009) | |
| Not publically available | Affymetrix Human U133 Array | Microarray | Tissue | Chorionic villi |
|
3rd trimester | GDM or T1D vs controls | Radaelli et al. (2009) | |
| Not publically available |
|
RNA-seq | Tissue | Placental biopsy (basal plate + chorionic plate + chorionic villi) |
|
3rd trimester | GDM, PE, SGA or LGA vs controls | Sober et al. (2015) | |
| Not publically available | Affymetrix HuGene ST 1.0 Array | Microarray | Tissue (laser microdissected) | Placenta biopsy (no details provided) |
|
3rd trimester | GDM vs obese non-diabetic vs lean controls | Bari et al. (2016) | |
| Not publically available |
|
RNA-seq | Tissue | Placenta biopsy (no details provided) |
|
3rd trimester | GDM vs controls (with linked miRNA data) | Ding et al. (2018) | |
| Not publically available |
|
RNA-seq | Tissue | Chorionic villi |
|
3rd trimester | GDM or LOPE vs controls | Lekva et al. (2016) | |
| Not publically available, but available on request |
|
RNA-seq | Tissue | Placenta biopsy (chorionic plate + chorionic villi) |
|
3rd trimester | Diabetes during pregnancy (GDM and T2D) vs controls (with linked methylation data) | Alexander et al. (2018) | |
|
| |||||||||
|
Antenatal infections and inflammation | |||||||||
| 2007 | NCBI GEO GSE7586 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Tissue | Chorionic villi |
|
3rd trimester | Active placental malaria vs controls | Muehlenbachs et al. (2007) |
| 2009 | EBI ArrayExpress E-TABM-577 | Affymetrix Human Genome U133 Plus 2.0 | Microarray | Tissue | Chorionic villi |
|
3rd trimester | Idiopathic PI vs controls | Kim et al. (2009) |
| 2015 | NCBI GEO GSE68474 | Illumina HumanHT-12 WG V4.0 Expression Beadchip | Microarray | Tissue | Placental biopsy (basal plate + chorionic villi) |
|
3rd trimester | Chronic PI vs controls | Raman et al. (2015) |
| 2016 | NCBI GEO GSE73712 |
|
RNA-seq | Tissue | Chorionic villi, decidua |
|
3rd trimester | Preterm with PI or preterm without PI vs controls | Ackerman et al. (2016) |
| 2018 | NCBI GEO GSE107376 |
|
RNA-seq | Tissue | Placenta biopsy (basal plate + chorionic plate + chorionic villi) |
|
3rd trimester | TrypC+ vs controls | Juiz et al. (2018) |
|
Labour | |||||||||
| 2009 | NCBI GEO GSE18809 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Tissue | Chorionic villi |
|
3rd trimester | PTSL vs TSL | Chim et al. (2012) |
| 2009 | NCBI GEO GSE18850 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Tissue | Chorionic villi |
|
3rd trimester | TSL vs TNL | Chim et al. (2012) |
| 2016 | NCBI GEO GSE73685 | Affymetrix Human Gene 1.0 ST Array | Microarray | Tissue | Amnion, chorion, decidua, placenta biopsy (no details provided) |
|
3rd trimester | Preterm vs term with and without labour | Bukowski et al. (2017) |
| 2017 | EBI ArrayExpress E-MTAB-5353 | Illumina Human HT-12 WG V4.0 Expression Beadchip | Microarray | Tissue | Decidua |
|
3rd trimester | Preterm vs term with and without labour | Rinaldi et al. (2017) |
| Not publically available | Illumina HumanRef-12 v3 Expression BeadChip | Microarray | Tissue | Chorionic villi |
|
3rd trimester | PTL or PE vs TNL (with linked miRNA data) | Mayor-Lynn et al. (2011) | |
|
| |||||||||
|
Recurrent miscarriage | |||||||||
| 2010 | NCBI GEO GSE22490 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Tissue | Placenta biopsy (chorionic villi + basal plate) |
|
1st and 2nd trimester | RM vs controls | Rull et al. (2013) |
| 2016 | NCBI GEO GSE76862 | Affymetrix GeneChip Human Transcriptome Array 2.0 | Microarray | Primary cell culture | Chorionic villi | Not stated (pooled into 6 groups of 3 to 5 samples) | 1st trimester | Placental trophoblast cells from RM vs controls | Tian et al. (2016) |
| 2018 | NCBI GEO GSE121950 |
|
RNA-seq | Tissue | Chorionic villi |
|
1st trimester | RM vs controls | Huang et al. (2018) |
| 2018 | NCBI GEO GSE113790 |
|
RNA-seq | Tissue | Decidua |
|
1st trimester | RM vs controls (with linked methylation data) | Yu et al. (2018) |
| Not publically available |
|
RNA-seq | Tissue | Chorionic villi |
|
1st trimester | RM vs controls | Sober et al. (2016) | |
|
| |||||||||
|
Chromosomal abnormalities | |||||||||
| 2015 | NCBI GEO GSE70102 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Tissue | Basal plate |
|
2nd trimester | Aneupoidy vs controls | Bianco et al. (2016) |
| Not publically available | Affymetrix GeneChip Human Genome U133 Plus 2.0 Array | Microarray | Tissue | Chorionic villi |
|
1st trimester | T21 vs controls | Lim et al. (2017a) | |
|
| |||||||||
|
Intrahepatic cholestasis of pregnancy | |||||||||
| 2013 | NCBI GEO GSE46157 | Agilent-026652 Whole Human Genome Microarray 4 × 44K v2 | Microarray | Tissue | Chorionic villi |
|
3rd trimester | IHCP vs controls | Du et al. (2014) |
C, control; ENA, European Nucleotide Archive; EOPE, early-onset pre-eclampsia; GDM, gestational diabetes mellitus; HELLP, haemolysis, elevated liver enzymes, low-platelet count syndrome; IHCP, intrahepatic cholestasis of pregnancy; iPTB, idiopathic preterm birth; IUGR, intrauterine growth restriction; LGA, large for gestational age; LOPE, late-onset pre-eclampsia; macro, macrosomia; miRNA, microRNA; PE, pre-eclampsia; PI – placental inflammation; PTL, preterm labour; PTNL, preterm no labour; PTSL, preterm spontaneous labour; RM, recurrent miscarriage; SGA, small for gestational age; T13, trisomy 13; T18, trisomy 18; T21, trisomy 21; T1D, type 1 diabetes; T2D, type 2 diabetes; TL, term labour; TNL, term no labour; TSL, term spontaneous labour; TrypC+, Trypanosoma cruzi seropositive.
Table III.
Pregnancy exposures.
| Year | Accession ID | Platform | Type | Tissue/cell | Sampling location | Total no. | Gestation | Purpose | Associated publications |
|---|---|---|---|---|---|---|---|---|---|
|
Obesity | |||||||||
| 2013 | EBI ArrayExpress E-MTAB-4541 | Affymetrix Human Gene 2.0 ST Array | Microarray | Tissue | Chorionic villi |
|
3rd trimester | Obese vs lean before pregnancy | Altmae et al. (2017) |
| 2018 | NCBI SRA PRJNA478464 | Illumina NextSeq 500 -mean sequencing depth ∼31M | RNA-seq | Tissue | Placenta biopsy (no details provided) |
|
3rd trimester | Obese vs lean before pregnancy | Sureshchandra et al. (2018) |
| 2019 | NCBI GEO GSE128381 | Agilent-039494 SurePrint G3 Human GE v2 8 × 60K 039381 | Microarray | Tissue | Chorionic villi |
|
3rd trimester | Across a continuum of maternal pre-pregnancy BMI including 41% overweight or obese | Cox et al. (2019) |
| Not publically available |
|
RNA-seq | Tissue | Placenta biopsy (no details provided) |
|
3rd trimester | Obese vs lean before pregnancy | Saben et al. (2014a) | |
| Not publically available | Affymetrix HuGene ST 1.0 Array | Microarray | Tissue (laser microdissected) | Placenta biopsy (no details provided) |
|
3rd trimester | GDM vs obese non-diabetic vs lean controls | Bari et al. (2016) | |
|
| |||||||||
|
Smoking | |||||||||
| 2007 | NCBI GEO GSE7434 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Tissue | Placenta biopsy (no details provided) |
|
3rd trimester | Smoking vs non-smoking controls | Huuskonen et al. (2008) |
| 2009 | NCBI GEO GSE18044 | Illumina humanRef-8 v2.0 Expression Beadchip | Microarray | Tissue | Chorionic villi |
|
3rd trimester | Smoking vs non-smoking controls | Bruchova et al. (2010) |
| 2011 | NCBI GEO GSE27272 | Illumina HumanRef-8 V3.0 Expression Beadchip | Microarray | Tissue | Chorionic villi |
|
3rd trimester | Smoking vs non-smoking controls | Votavova et al. (2011) |
| 2011 | NCBI GEO GSE30032 | Illumina HumanRef-8 V3.0 Expression Beadchip | Microarray | Tissue | Chorionic villi |
|
3rd trimester | Passive smokers vs non-smoking controls | Votavova et al. (2012) |
|
Clinical trials | |||||||||
| 2012 | NCBI GEO GSE39290 | Agilent-014850 Whole Human Genome Microarray 4 × 44K G4112F | Microarray | Tissue | Placenta biopsy (basal plate + chorionic plate + chorionic villi) |
|
3rd trimester | High vs low choline intake in third trimester | Jiang et al. (2013) |
| 2014 | NCBI GEO GSE53291 | Affymetrix NuGO array (human) NuGO_Hs1a520180 | Microarray | Tissue | Chorionic villi |
|
3rd trimester | Omega-3 supplementation vs placebo | Sedlmeier et al. (2014) |
| 2015 | EBI ArrayExpress E-MTAB-6418 | Illumina HumanHT-12 V4.0 Expression Beadchip | Microarray | Tissue | Placenta biopsy (basal plate + chorionic plate + chorionic villi) |
|
3rd trimester | Obese women treated with metformin vs placebo | Chiswick et al. (2016) |
|
| |||||||||
|
IVF | |||||||||
| 2018 | NCBI GEO GSE122214 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Tissue | Chorionic villi |
|
1st trimester | IVF vs controls | Zhao et al. (2019) |
| Not publically available | Affymetrix GeneChip Human Gene 1.0 ST Array | Microarray | Tissue | Chorionic villi |
|
3rd trimester | IVF vs controls | Nelissen et al. (2014) | |
| Not publically available |
|
RNA-seq | Tissue | Chorionic villi |
|
1st trimester | IVF or non-IVF fertility treated vs controls | Lee et al. (2019) | |
|
| |||||||||
|
Antenatal depression | |||||||||
| 2014 | Available as supplementary data within journal article | Affymetrix GeneChip Human Gene 1.0 ST Array | Microarray | Tissue | Placenta biopsy (chorionic plate + chorionic villi) |
|
3rd trimester | Depressed women with and without SSRI treatment vs controls | Olivier et al. (2014) |
IVF, in vitro fertilisation; SSRI, selective serotonin reuptake inhibitor.
Table IV.
In vitro placental cultures.
| Year | Accession ID | Platform | Type | Model | Treatment | Source | Purpose | Associated publications | |
|---|---|---|---|---|---|---|---|---|---|
|
Trophoblast cultures | |||||||||
| 2004 | NCBI GEO GSE1302 | Affymetrix Human Genome U95B Array, Affymetrix Human Genome U95C Array, Affymetrix Human Genome U95D Array, Affymetrix Human Genome U95E Array, Affymetrix Human Genome U95 Version 2 Array | Microarray | Primary cytotrophoblast culture | Peroxisome proliferator-activated receptor gamma ligand GW7845 | Term placenta | Assess accuracy of microarray data | Mecham et al. (2004) | |
| 2005 | NCBI GEO GSE2531 | Affymetrix Human Genome U133A Array | Microarray | Cytrophoblast cell lines | None | JEG-3 and BeWo | Compare trophoblast cell lines | Burleigh et al. (2007) | |
| 2008 | NCBI GEO GSE13475 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Cytrophoblast cell lines | Transfected with STOX1 overexpression plasmid | JEG-3 | Determine effect of STOX1 overexpression on trophoblast function | Rigourd et al. (2008) | |
| 2010 | NCBI GEO GSE20510 | Affymetrix Human Genome U133A Array | Microarray | Cytotrophoblast and extravillous trophoblast cell lines | None | SGHPL-5, HTR-8/SVneo, BeWo, JEG-3 and ACH-3P | Compare trophoblast cell lines | Bilban et al. (2010) | |
| 2011 | EBI ArrayExpress E-MEXP-2800 | Agilent Whole Human Genome Microarray 4 × 44K 014850 G4112F | Microarray | Cytotrophoblast cell line | Infected with Coxiella burnetii | BeWo | Determine pathways of Coxiella burnetii (Q fever) placental infection in trophoblast | Ben Amara et al. (2010) | |
| 2011 | NCBI GEO GSE27909 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Extravillous trophoblast cell line | Hoechst dye | HTR-8/SVneo | Compare side population (consists of stem and progenitor cells) and non-side trophoblast population | Takao et al. (2011) | |
| 2011 | NCBI GEO GSE20404 | Affymetrix Human Gene 1.0 ST Array | Microarray | Cytotrophoblast cell line | Heme oxygenase-1 silencing | BeWo | Assess effect of HO-1 silencing on trophoblast cell adhesion | Tauber et al. (2010) | |
| 2011 | NCBI GEO GSE31679 | Illumina HumanHT-12 V3.0 expression beadchip | Microarray | Extravillous trophoblast cell line | Cobalt chloride, interleukin-1-beta, tumour necrosis factor alpha | Swan-71 | Assess effects of chemical hypoxia and pro-inflammatory cytokines on trophoblast function | Unpublished | |
| 2012 | NCBI GEO GSE40182 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Primary cytotrophoblast culture | Exposed to pre-eclampsia in vivo | 3rd trimester placenta | Assess long-term effect of exposure to pre-eclampsia in vivo on trophoblast | Zhou et al. (2013) | |
| 2012 | NCBI GEO GSE30330 | Agilent Whole Human Genome Microarray 4 × 44K 014850 G4112F | Microarray | Cytotrophoblast cell line | Infected with Coxiella burnetii | JEG3 | Determine pathways of Coxiella burnetii (Q fever) placental infection in trophoblast | Unpublished | |
| 2012 | NCBI GEO GSE41441 | Agilent-012391 Whole Human Genome Oligo Microarray G4112A | Microarray | Cytotrophoblast cell line | Valproic acid | JEG-3 | Determine adverse effects of valproic acid on trophoblast | Unpublished | |
| 2013 | NCBI GEO GSE49922 | Agilent-028004 SurePrint G3 Human GE 8 × 60K Microarray | Microarray | Primary cytotrophoblast culture | Irradiation | Term placenta | Determine irradiated trophoblast response | Kanter et al. (2014) | |
| 2013 | NCBI GEO GSE41331 | Illumina HumanHT-12 V4.0 Expression Beadchip | Microarray | Primary cytotrophoblast culture | Hypoxia | Term placenta | Determine hypoxic trophoblast response | Yuen et al. (2013) | |
| 2014 | EBI ArrayExpress E-MTAB-429 | Illumina HumanHT-12 v3.0 Expression BeadChip | Microarray | Cytotrophoblast and extravillous trophoblast primary culture and cell lines | None | First-trimester placenta, JAR and JEG-3 | Compare trophoblast cell lines | Apps et al. (2011) | |
| 2014 | NCBI GEO GSE56564 | Agilent-028004 SurePrint G3 Human GE 8 × 60K Microarray | Microarray | Primary cytotrophoblast culture and extravillous trophoblast cell line | Transfected with plasmid for C19MC miRNA expression | Term placenta; HTR-8/SVneo | Determine effect of C19MC miRNA on trophoblast function | Xie et al. (2014) | |
| 2015 | NCBI GEO GSE60432 | Agilent-028004 SurePrint G3 Human GE 8 × 60K Microarray | Microarray | Primary cytotrophoblast culture | Hypoxia | Term placenta | Determine hypoxic trophoblast response | Unpublished | |
| 2016 | NCBI GEO GSE79333 | Stanford Functional Genomics Facility SHDZ | Microarray | Primary cytotrophoblast and syncytiotrophoblast culture | In vitro differentiation | Term placenta | Characterize in vitro cytotrophoblast differentiation | Rouault et al. (2016) | |
| 2016 | NCBI GEO GSE73016 |
|
RNA-seq | Primary cytrophoblast culture | Time course differentiation | Term placenta | Compare synctytiotrophoblast derived from placenta and human pluripotent stem cells | Yabe et al. (2016) | |
| 2017 | NCBI GEO GSE98523 | Applied Biosystems Human Genome Survey Microarray Version 2 | Microarray | Cytotrophoblast cell line | In vitro differentiation with forskolin | BeWo | Characterize in vitro cytotrophoblast differentiation | Gauster et al. (2018) | |
| 2017 | NCBI GEO GSE86171 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Primary cytotrophoblast culture | In vitro differentiation with Matrigel | 2nd trimester placenta | Characterize in vitro cytotrophoblast differentiation | Robinson et al. (2017) | |
| 2017 | NCBI SRA PRJNA383955 |
|
RNA-seq | Cytotrophoblast cell line | Dexamethasone | Not stated, obtained from cell bank | Determine trophoblast response to dexamethasone | Shang et al. (2018) | |
| 2018 | NCBI SRA PRJNA397241 |
|
RNA-seq | Primary cytotrophoblast culture and cell line | In vitro differentiation by time course or with forskolin | Term placenta and BeWo | Characterize in vitro cytotrophoblast differentiation | Azar et al. (2018) | |
| 2018 | NCBI GEO GSE65866 | Illumina HumanHT-12 V4.0 Expression Beadchip | Microarray | Cytotrophoblast cell line | ZNF554 silencing | BeWo | Determine role of ZNF554 in trophoblast function | Than et al. (2018) | |
| 2018 | NCBI GEO GSE65940 | Illumina HumanHT-12 V4.0 Expression Beadchip | Microarray | Extravillous trophoblast cell line | ZNF554 silencing | HTR-8/SVneo | Determine role of ZNF554 in trophoblast function | Than et al. (2018) | |
| 2018 | NCBI GEO GSE66304 |
|
RNA-seq | Cytotrophoblast cell line | PEG10 silencing | JEG-3 | Determine role of PEG10 in trophoblast function | Unpublished | |
| 2018 | NCBI GEO GSE118351 | Affymetrix Human Gene Expression Array | Microarray | Primary cytotrophoblast culture | Time course differentiation | Term placenta | Characterize in vitro cytotrophoblast differentiation | Unpublished | |
| 2019 | NCBI GEO GSE125778 |
|
RNA-seq | Extravillous trophoblast cell line | Interferon-gamma | HTR-8/SVneo | Assess effect of interferon-gamma on trophoblast invasion | Verma et al. (2018) | |
| 2019 | NCBI GEO GSE124586 |
|
RNA-seq | Extravillous trophoblast cell line | Epidermal growth factor | HTR-8/SVneo | Assess effect of epidermal growth factor on trophoblast invasion | Unpublished | |
| 2019 | NCBI GEO GSE127170 | Affymetrix Human Gene 1.0 ST Array | Microarray | Cytotrophoblast cell line | In vitro differentiation with forskolin | BeWo and JEG-3 | Identify genes associated with trophoblast fusion | Unpublished | |
| Not publically available | Affymetrix U95A array | Microarray | Primary cytotrophoblast culture | Hypoxia | Term placenta | Determine hypoxic trophoblast response | Roh et al. (2005) | ||
| Not publically available |
|
RNA-seq | Cytotrophoblast cell line | In vitro differentiation with forskolin | BeWo | Assess relationship of cell fusion transcriptome with methylome | Shankar et al. (2015) | ||
| Not publically available | Affymetrix Human Gene 1.1 ST Array | Microarray | Primary cytotrophoblast culture | Insulin | 1st trimester placenta | Investigate the effects of obesity and insulin on trophoblast | Lassance et al. (2015) | ||
| Not publically available |
|
RNA-seq | Cytotrophoblast cell line | In vitro differentiation with forskolin | BeWo | Identify syncytialization-related genes in trophoblast | Zheng et al. (2016) | ||
|
Non-trophoblast cultures | |||||||||
| 2006 | NCBI GEO GSE5809 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Primary decidualized stromal cell culture | Conditioned medium from primary first or second trimester trophoblast | Endometrial biopsy | Determine decidual response to trophoblast secretions/paracrine signals | Hess et al. (2007) | |
| 2010 | NCBI GEO GSE21332 | Illumina HumanWG-6 v3.0 Expression Beadchip | Microarray | Primary pericyte culture | None | Term placenta | Compare pericytes by tissue source | Maier et al. (2010) | |
| 2011 | EBI ArrayExpress E-MEXP-3299 | Applied Biosystems Human Genome Survey Microarray v2.0 | Microarray | Primary placental endothelial cell culture | Foetal HDL | Term placenta | Identify apoE-HDL regulated genes in placental endothelium | Augsten et al. (2011) | |
| 2013 | NCBI GEO GSE41946 | Agilent-014850 Whole Human Genome Microarray 4 × 44K G4112F | Microarray | Primary decidual endothelial culture | None | 1st trimester placenta | Compare immunoregulatory and pro-angiogenic functions of endothelial cells relative to tissue of origin | Agostinis et al. (2019) | |
| 2016 | NCBI GEO GSE58220 | Affymetrix Human Gene 2.0 ST Array | Microarray | Primary decidual stromal cell culture | Interleukin-1-beta | Term placenta | Determine decidual response to cytokine challenge | Ibrahim et al. (2016) | |
|
| |||||||||
|
In vitro models of in vivo exposures | |||||||||
| 2012 | NCBI GEO GSE36083 | Affymetrix Human Genome U133 Plus 2.0 Array | Microarray | Primary explants | Antiphospholipid antibodies | 1st trimester placenta | Determine trophoblast response to antiphospholipid antibodies | Pantham et al. (2012) | |
| 2017 | NCBI GEO GSE104348 | Illumina HiSeq 2500-mean sequencing depth ∼65M | RNA-seq | Primary explants | Interferon-beta or interferon-lambda | 2nd trimester placenta | Assess placental response to different interferons (in the context of Zika virus infection) | Yockey et al. (2018) | |
| 2018 | NCBI GEO GSE113155 | Agilent-039494 SurePrint G3 Human GE v2 8 × 60K Microarray 039381 | Microarray | Primary explants | Infected with Trypanosoma cruzi | Term placenta | Assess placental response to Trypanosoma cruzi infection | Castillo et al. (2018) | |
| Not publically available |
|
RNA-seq | Primary decidual and chorionic villus organoid cultures | Infected with Zika virus | 1st trimester placenta | Assess decidual and chorion response to Zika virus infection | Weisblum et al. (2017) | ||
HDL, high-density lipoprotein.
Important considerations for placental transcriptome studies
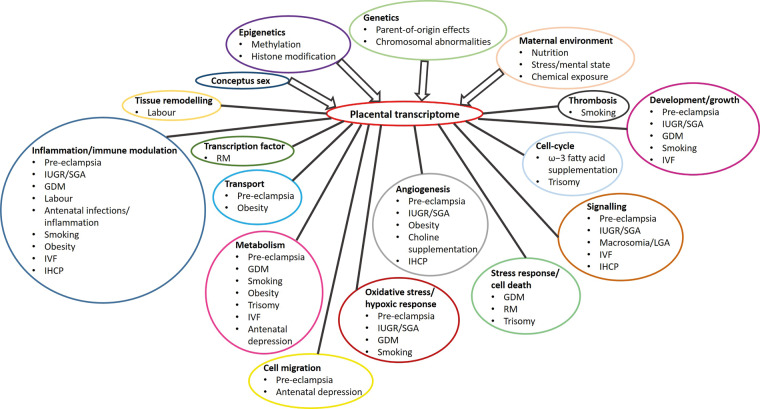
Good study design is critical to harness the potential of genome-wide transcript profiling of the placenta. Key aspects to be considered in study design are subject recruitment, sample processing at delivery, transcript profiling and data analysis methods and data validation (Fig. 1), all of which may represent potential pitfalls and limit the validity of conclusions that can be drawn.
Figure 1.
Key aspects to consider for placental transcriptome studies.
Subject recruitment
Two main points to consider in subject recruitment are selection criteria and sample size. Firstly, the selection process in case–control studies should ensure suitable controls are chosen to compare with pathological cases identified by well-defined and established clinical definitions. As will be discussed in subsequent sections, varied clinical criteria can impact study findings and reproducibility. Hence, a consensus about research definitions of common pregnancy complications should be reached to make full use of available resources. The selection process must also determine if variables, such as gestational age, sex, labour status, mode of delivery and treatment modalities, which are known to affect placental gene transcription, are part of the inclusion/exclusion criteria. Researchers should also recognize a caveat of sampling placenta from the first half of pregnancy, is that the pregnancy outcome of an electively terminated pregnancy cannot truly be guaranteed as healthy as the final outcome cannot be determined, thus interpretation of findings involving such samples must take this into account.
Secondly, inadequate sample size may affect statistical power and study reproducibility. Although the average number of placentas profiled in each study is ∼28, this is largely skewed by eight large studies of more than 100 placentas each. The median number of placentas profiled per study is merely 13 (interquartile range 8–29). It is noted that while around one in five studies used fewer than 10 placentas, a considerable number of these smaller studies were performed when the use of profiling technologies were in their infancy and they were valuable in providing an early proof of concept for application in the field. Nevertheless, meta-analysis may be a means to overcome the effects of sample size in some of these earlier studies and small studies of rare conditions. While the choice of study inclusion ultimately rests on the researchers performing the meta-analysis, we strongly recommend caution with including very small studies with fewer than five samples (Supplementary Table SI) as study batch effects are unlikely to be sufficiently corrected for in such cases. Hence, future studies should aim for much larger sample sizes that are adequately powered to answer the study question, to improve reproducibility and verify the findings of past studies going forward.
Sample processing at delivery
Another important factor is the placental sampling procedure at delivery. The region of the placenta to biopsy is dependent on the question asked (Fig. 2). For instance, studies addressing invasion of the extravillous trophoblast into the maternal decidua sample the basal plate (Winn et al., 2009), while those investigating the maternal–foetal transfer across the syncytiotrophoblast would sample the villous placenta (Bari et al., 2016). Unwanted variation may arise from inappropriate sampling of the placenta, such as non-removal of the decidua for studies of the villous placenta, inclusion of infarcted areas and insufficient cleaning to remove excess blood. Another consideration is the importance of multisite sampling of the same region as several studies established intra-placental variation of gene expression (Pidoux et al., 2004; Hughes et al., 2015). The ideal way is to run replicate samples of each placenta, although this may not always be practical given that the cost of genome-wide transcript profiling is still relatively high per sample. An alternative is to pool RNA from multiple sites of the region of interest for each placenta and to have a large enough number of different placentas, so as to ensure differential expression patterns identified are related to the condition studied, rather than normal intra- and inter-individual biological variability.
Figure 2.
Placental regions commonly sampled for transcriptome analyses.
RNA integrity is also vital for proper interpretation of transcriptome data. RNA integrity is determined by measuring the 28S to 18S rRNA ratio or assessing the RNA integrity number (RIN) by a commercially available algorithm. Some placental RNA transcripts are more susceptible to degradation than others due to differences in post-transcriptional regulation (Reiman et al., 2017). Rapidly degrading transcripts are enriched among those encoding membrane components or proteins with transporter function, while stable transcripts are primarily those involved in intracellular function (Reiman et al., 2017). A major determinant of RNA integrity is the time taken to fully immerse in RNAlater or snap-freeze placental biopsies following delivery, which shows an inverse correlation with RIN values (Fajardy et al., 2009; Jobarteh et al., 2014). Biopsies are often rinsed in ice-cold buffer to remove maternal blood contamination prior to RNA preservation, but this step may also alter the transcript profile, particularly at the villous sprouts, which are transcriptionally sensitive to mechanical disturbances (Burton et al., 2014). Storage length prior to RNA extraction may affect the RIN value as well, depending on preservation methods used (Martin et al., 2017). Flash-frozen samples appear more sensitive to RNA degradation over a long period of storage, compared with RNAlater-preserved samples (Fajardy et al., 2009; Martin et al., 2017). Therefore, RNAlater-preservation is preferential for better placental RNA quality as compared to snap-freezing in liquid nitrogen (Wolfe et al., 2014; Pisarska et al., 2016). However, if the sample is limited and there are plans to utilize the tissue for other types of analysis, a potential drawback is that preservation in RNAlater, which has a high salt content and denatures proteins, may interfere with future analysis of native proteins and other techniques. Therefore, researchers should ideally work as efficiently as possible with RNase-free equipment and consumables while handling the placenta, although it is ultimately up to the researcher how they wish to process and store their placental samples.
Transcript profiling and data analysis methods
Following sample collection, various RNA isolation and possibly enrichment methods may be required, depending on which RNA species (e.g. long non-coding RNA or mRNA) are being studied. After RNA isolation, the choice of profiling platform and data analysis is another consideration. While either microarray or RNA sequencing can be used to determine genome-wide transcriptomes, as mentioned earlier, they each have their advantages and disadvantages (thoroughly reviewed by Cox et al., 2015). Data analysis considerations include whether to identify differentially expressed genes or broad categories of dysregulated pathways in case–control studies and to ensure adequate statistical adjustment to account for multiple testing, i.e. false discovery correction. Technical notes and methods to visualize these data are further discussed in a recent review by Konwar et al. (2019).
Data validation
Another key aspect is to validate the findings from transcriptome analyses. Expression changes should ideally be confirmed at the RNA and protein levels by techniques, such as real-time qPCR, immunoblotting, enzyme-linked immunosorbent assays and immunohistochemistry. Furthermore, in vitro functional assays with primary explant cultures or isolated cells and in vivo animal models would provide deeper mechanistic insights into the role of identified genes.
Transcriptome studies of healthy placental development
Studies assessing healthy placentas (Table I) have three general aims: to investigate transcriptome changes across gestation, to characterize cell populations within the placenta or to determine the regulation and variability of gene expression across individual placentas.
Gestation-specific effects
Transcriptome datasets exist for each trimester of pregnancy, allowing identification of gestation-specific signatures (Table I: Gestation effects). Two microarray datasets contain placental expression profiles across all trimesters and serve as useful references of the temporal changes that occur during development (Mikheev et al., 2008; Soncin et al., 2018). Unsurprisingly, a common finding of first-trimester studies is an enrichment of highly expressed genes involved in cell proliferation and cell-cycle regulation (Mikheev et al., 2008; Sitras et al., 2012; Lim et al., 2017b; Soncin et al., 2018), which reflects rapid placental growth in early gestation. Interrogation of a large third-trimester microarray dataset of 157 placentas in conjunction with detailed histological analyses enabled markers of normal villous maturation to be identified, which was used to establish a method to calculate the molecular age in weeks of a given placenta, enabling a maturation measure to be assigned to placentas from pathological pregnancies (Leavey et al., 2017). However, the overall number of just under 300 placentas represented by these datasets is still relatively small, particularly for the first and second trimesters with data from only 73 and 21 placentas respectively, and expanding the number of samples profiled will enable elimination of spurious findings and further refinement of gestation-specific transcriptome signatures of placental development.
Cellular characterization and differentiation
The placenta comprises many different cell types. To improve resolution of gene expression to the cellular level, multiple studies have performed global profiling of isolated cells or primary cell cultures from the maternal–foetal interface (Table I: Cellular characterization and differentiation). However, an important caveat is that in addition to varying cell purity of the derived sample, differences may arise in the cellular response to the isolation and/or culture methods, which could become a source of bias and lead to data artefacts that are indistinguishable from naturally occurring in vivo differences. Moreover, aside from one study, datasets in this theme are derived from studies with fewer than 13 individual placentas each, with the majority having five or fewer. This is understandable, given the technical complexities and high time investment involved in isolating and culturing placental cells. However, such a limitation may lead to conclusions that may not fully reflect the natural diversity between individual placentas. Nonetheless, they can still serve as valuable baseline references for future studies in this area.
Trophoblast cells are unique to the placenta and therefore a major cell type specifically targeted for genome-wide transcript profiling. Cytotrophoblast cells are precursors of the extravillous trophoblast and syncytiotrophoblast. Extravillous trophoblast cells are responsible for maternal tissue invasion and placentation, while syncytiotrophoblast forms the maternal–foetal exchange barrier and acts as a major source of endocrine and paracrine factors for supporting pregnancy. However, common trophoblast isolation methods disrupt the multi-nucleated synctiotrophoblast layer, resulting in most studies focussing on just the cytotrophoblast and extravillous trophoblast cells. Extrapolation of their results to the understanding of syncytiotrophoblast function must, therefore, be made with extreme caution. Furthermore, one key finding is that cultured term placental trophoblast cells, as well as endothelial cells, have sex-specific expression patterns, with the male placental transcriptome enriched for pathways involved in the immune system and inflammatory response, which may partly explain the general observation of poorer outcomes for pregnancies with a male foetus (Cvitic et al., 2013). Hence, future studies should consider having a large enough sample size to stratify analyses by sex or be able to account for the possible influence of sex on the cell-specific gene expression profiles.
Recent studies have used single-cell RNA sequencing with microfluidics to characterize gene expression profiles of a variety of individual cells at the maternal–foetal interface. Although the first single-cell study only sequenced 87 cells from just two-term placentas (Pavlicev et al., 2017), it provided an important proof of concept for successfully applying this technology to interrogate the placental transcriptome. Additionally, given the limitation of microfluidics in isolating large cells, the authors utilized laser microdissection to facilitate more accurate profiling of the large multi-nucleated syncytiotrophoblast (Pavlicev et al., 2017), in sharp contrast to subsequent studies that merely dissociated placental tissues and then reported cellular characterization of the syncytiotrophoblast. Although one of these studies acknowledged the shortcomings of the cellular dissociation approach that undoubtedly alters transcript profiles, and the possible under-representation of synctiotrophoblast profiling in the data (Suryawanshi et al., 2018), this consideration was neglected and ignored by the rest (Tsang et al., 2017; Liu et al., 2018; Vento-Tormo et al., 2018). Hence, the current available data are likely biased and not fully reflective of syncytiotrophoblast gene expression across gestation, and should be viewed and used with some caution. Given the important role the syncytiotrophoblast serves as the active cellular and regulatory barrier between mother and foetus, future studies should strive to better profile the synctiotrophoblast, and ensure isolation methods are carefully justified and weaknesses accounted for to allow proper interpretation of findings.
Another methodological factor to consider is the additional mechanical stress following tissue dissociation imposed by either a pre-enrichment of cells by magnetic bead separation (Liu et al., 2018) or pre-sorting of cells by fluorescence-activated cell sorting (Vento-Tormo et al., 2018), which may potentially modify the observed gene expression patterns. A first-trimester study without prior pre-selection did, however, demonstrate a good correlation (Pearson r value of 0.86) of the multi-cell transcriptome of an average villi assessed by single-cell RNA sequencing with that assessed by bulk tissue RNA sequencing, suggesting that overall, tissue dissociation and subsequent microfluidics techniques have only minor effects on the gene expression profiles for most dissociated cells (Suryawanshi et al., 2018). Thus, these latter single-cell sequencing studies can be considered to have collectively established for the first time, at the single-cell level, cell-specific gene expression profiles of the maternal–foetal interface throughout gestation, which serve as a potential reference to deconvolute placental cellular heterogeneity in future bulk tissue studies.
Gene expression variation and regulation
Another purpose of analysing the healthy placental transcriptome is to discover how gene expression is regulated and its variability across different regions of each placenta and between individuals (Table I: Gene expression variation and regulation). One study of third-trimester placentas estimated that while more than half of term placental gene variation was due to differences between individuals, a significant third of variation was attributable to intra-individual differences, compared with only <10% of variation due to ethnic background (Hughes et al., 2015). Foetal-placental sex is a major contributor to inter-individual variation, with sexual dimorphism predominantly arising from differential expression of genes on the sex chromosomes (Gonzalez et al., 2018). Some of the observed differences between individuals and between tissue biopsies of the same placenta likely also arise from cellular heterogeneity, as evidenced by a single-cell sequencing study showing different cellular proportions between individual samples biopsied from within the same placental region and from sample replicates obtained from the same placenta (Tsang et al., 2017). Furthermore, multi-regional sampling of the placenta and associated tissues, such as the decidua and umbilical cord, highlights distinct gene expression patterns associated with each sampling region (Sood et al., 2006; Kim et al., 2012). All of these underscore the importance of sampling region selection (Fig. 2) and having a sufficiently large sample size with multi-site sampling of the same region within each placenta to overcome variations, due to sample collection and intra-individual and inter-individual placental differences, to produce robust datasets reflective of the true ‘experimental’ group differences in question.
Additionally, there are clusters of genes that demonstrate relatively consistent placental expression between individuals and throughout pregnancy regardless of gestation, while others are trimester-specific and even show conserved expression patterns between humans and mice (Buckberry et al., 2017). These consistently expressed genes are likely key regulatory genes indispensable for normal overall placental function and for directing temporal-specific changes relating to the particular requisite functions of the placenta at each trimester.
Attempts to discover what underlies placental gene transcriptional regulation have explored genotype and methylation, which are DNA-based, in association with the transcriptome. Indeed, two large RNA sequencing studies comprising a total of 239 individual placentas with genotype data showed a consistent overlap of 381 expression quantitative trait loci (eQTLs), suggesting that the placental expression of genes in these loci are under stringent genetic control (Peng et al., 2017; Delahaye et al., 2018). Methylation is also associated with variation in placental gene expression. Dual profiling of the third-trimester placental transcriptome and methylome showed that DNA methylation accounted for a greater variance of birthweight than gene expression profiles at a single timepoint, and identified MSX1 and GRB10 methylation as potential master regulators in the transcriptional control of growth-related genes in the term placenta (Turan et al., 2012). Hence, these studies highlight the complexities of placental gene expression regulation which may drive the large variation of expression observed between and within individual placentas and subsequent birth outcomes.
Placental transcriptome studies of pregnancy complications
Improved understanding of the pathophysiology of pregnancy complications is a main driver for interrogating the placental transcriptome (Table II). Studies in this research theme have compared placentas from the normal and pathological states and are discussed taking into account the considerations of study design mentioned above.
Pre-eclampsia
Pre-eclampsia, a serious hypertensive disorder of pregnancy, is the most common pathology in which the placenta has been profiled, with 40 datasets produced from a total of 1192 placentas (44% affected). These datasets represent approximately a fifth of all placental transcriptome datasets (Table II:Pre-eclampsia). Given the nature of pre-eclampsia, difficulty in prediction of its onset and challenges with sampling placenta from ongoing pregnancies, most of the placentas profiled were collected in the third trimester at delivery, during the advanced stages of the disorder. Nevertheless, there is a single study of 12 samples collected from controls and women who subsequently developed pre-eclampsia (Founds et al., 2009). This study utilized surplus tissue obtained from first-trimester chorionic villus sampling and demonstrated placental dysregulation of genes involved in immune modulation, inflammation and cell motility months before the clinical manifestation of pre-eclampsia (Founds et al., 2009), highlighting the early development of this insidious disease in the placenta. This dataset, while small, serves as a rare reference of the early placenta with known outcomes at delivery.
Expression profiling studies at the pre-eclamptic maternal–foetal interface frequently and consistently reveal dysregulated expression of genes involved in the oxidative stress and inflammatory pathways (Eide et al., 2008; Loset et al., 2011; Tsai et al., 2011; Song et al., 2013; Yong et al., 2015; Tong et al., 2018). Moreover, abnormal gene expression signatures of extravillous trophoblast cells, which was identified by the first single-cell RNA sequencing study of pre-eclamptic placenta, were also detectable in the maternal circulation in pre-eclampsia and may potentially serve as a non-invasive marker or ‘liquid biopsy’ of anomalous placental function in the future (Tsang et al., 2017). Collectively, these studies highlight how pre-eclampsia affects multiple molecular pathways and functions of cells of both maternal and foetal origin at the placental interface, thereby underscoring the complexity of disease pathophysiology and the potential to develop some of these differentially expressed genes as biomarkers of pre-eclampsia.
Attempts have been made to delineate pre-eclampsia from other complications, such as gestational diabetes mellitus (GDM), intrauterine growth restriction (IUGR) and macrosomia (Sitras et al., 2009a; Mayor-Lynn et al., 2011; Nishizawa et al., 2011; Guo et al., 2013; Sober et al., 2015; Lekva et al., 2016; Gibbs et al., 2019). Results of these studies are consistent with the notion that pre-eclampsia is heterogeneous, comprising of multiple molecular subtypes that distinctly cluster with other pregnancy pathologies (Guo et al., 2013; Gibbs et al., 2019), suggestive of a shared aetiology or pathophysiology involving the placenta (Nishizawa et al., 2011; Sober et al., 2015).
Molecular subtypes also seem to associate with gestation at onset of pre-eclampsia and placental histological findings, and efforts have been made to further characterize these subsets. Several studies sought to refine the sample population by either considering only early-onset pre-eclampsia developing before 34 weeks’ gestation (Varkonyi et al., 2011; Blair et al., 2013; Than et al., 2018), only late-onset pre-eclampsia developing from 34 weeks’ gestation (Sober et al., 2015; Lekva et al., 2016) or both (Nishizawa et al., 2007; Sitras et al., 2009b; Junus et al., 2012; Liang et al., 2016; Tong et al., 2018). A major issue in considering gestational age in placental studies is what construes an appropriate control. Of the 34 studies including preterm pre-eclampsia cases, about 21% used only preterm controls, while around 44% used only term controls, with the remainder including both preterm and term controls. Utilizing preterm controls in pre-eclampsia studies may actually confound study findings, given that preterm controls are frequently derived from those with premature rupture of membranes or preterm labour, which are also pregnancy pathologies and not an ideal reference of normal human pregnancy (Cox et al., 2015). Conversely, it could be argued that the most severe pre-eclamptic cases are usually delivered preterm and should be compared with similarly preterm but non-pre-eclamptic cases since term gestation brings in another dimension that is unrelated to the pathology of pre-eclampsia and observed differences may be due purely to the effect of gestation rather than pre-eclampsia. Hence, there remains no clear consensus within the field about the most optimal control for pre-eclamptic cases requiring preterm delivery and readers should carefully consider the interpretation of study findings whichever control is used.
Intrauterine growth restriction/small for gestational age
IUGR is the failure of a foetus to reach its full genetic growth potential in utero. Clinically, IUGR is often diagnosed with ultrasound findings of a small for gestational age (SGA) baby below the 10th centile, associated with abnormal uteroplacental blood flow (Kingdom et al., 2018). This review considers 17 datasets representing a total of 721 placentas of which 26% were either from SGA- or IUGR-affected pregnancies (Table II: IUGR/SGA). Most placental transcriptome studies utilized only the criterion of SGA (McCarthy et al., 2007; Sabri et al., 2014; Sober et al., 2015; Deyssenroth et al., 2017; Verheecke et al., 2018; Gibbs et al., 2019), while some studies excluded those who may be healthy and constitutionally small, by including an additional criterion of a small abdominal circumference below the fifth centile (Guo et al., 2013; Madeleneau et al., 2015) or ultrasound findings of abnormal uteroplacental blood flow indicative of placental insufficiency (Roh et al., 2005; Sitras et al., 2009a; Struwe et al., 2010; Nishizawa et al., 2011; Dunk et al., 2012; Majewska et al., 2019). However, while some of these IUGR cases may be explained by maternal smoking, pre-eclampsia or chemotherapy during pregnancy (Sitras et al., 2009a; Dunk et al., 2012; Verheecke et al., 2018), a considerable number of cases have no specific identifiable underlying cause and are termed idiopathic (Nishizawa et al., 2011). Hence, a common and clear research definition of IUGR is lacking within the placenta community. Moreover, as discussed earlier, gestational age is another potential confounder, with 8 out of 11 studies that included preterm IUGR cases utilizing term controls.
Nevertheless, a degree of consistency exists between these various studies, hinting at shared placental pathways involved in the pathophysiology of IUGR regardless of gestation. Dysregulated expression of genes involved in the processes of placental growth signalling, mitochondrial respiration and a hypoxic response was observed in multiple IUGR studies (Roh et al., 2005; McCarthy et al., 2007; Struwe et al., 2010; Guo et al., 2013; Sabri et al., 2014; Madeleneau et al., 2015; Deyssenroth et al., 2017; Verheecke et al., 2018). Several studies also demonstrated a substantial overlap between pre-eclampsia and normotensive growth restriction, encompassing genes involved in the molecular processes of placental angiogenesis and immune regulation (Sitras et al., 2009a; Nishizawa et al., 2011; Dunk et al., 2012; Guo et al., 2013; Sober et al., 2015; Gibbs et al., 2019; Majewska et al., 2019). Additional research to increase understanding of placental function in growth-compromised pregnancies could improve diagnosis and management of cases.
Macrosomia/large for gestational age
Conversely, some infants are macrosomic or large for gestational age (LGA). At present, four studies comprising 263 placentas (28% affected) have examined the placental transcriptome in relation to excessive intrauterine growth (Table II: Macrosomia/LGA). As with growth restriction, little consensus exists on defining excessive growth. Some studies selected cases based on an absolute birthweight cut-off of >4 kg (Song et al., 2018), while others utilize birthweight percentiles customized for sex and gestational age at a cut-off of over 90th percentile (Sabri et al., 2014; Sober et al., 2015; Deyssenroth et al., 2017); both of which do not account for the possibility of including normal constitutionally large babies. One study demonstrated that placentas of LGA babies had similar expression patterns to that of late-onset pre-eclampsia, which is often not associated with growth restriction, alluding to some common disturbances in placental function (Sober et al., 2015). Growing evidence also indicates that besides short-term morbidities of birth trauma, excessive intrauterine growth is also associated with poorer long-term cardiometabolic outcomes in offspring (Szostak-Wegierek, 2014). Thus, being large at birth should not be regarded as benign and more resources should be dedicated to understanding the possible placental mechanisms involved to prevent the potential intrauterine programming of adverse health outcomes manifesting in later life.
Gestational diabetes mellitus
Rates of GDM are rising worldwide. Follow-up studies suggest intrauterine exposure to maternal hyperglycaemia leads to poorer health outcomes in offspring (Binder et al., 2015). A total of 10 studies have profiled 282 placentas (44% affected) with respect to GDM (Table II: GDM). Again, criteria used to define GDM is varied. The World Health Organisation changed the criteria for diagnosis quite substantially between its 1999 and 2013 guidelines, while the American College of Obstetricians and Gynaecologists, UK National Institute for Health and Care Excellence and other countries continue using different diagnostic criteria (Meek, 2017). Nonetheless, most studies demonstrate that immune and inflammatory genes are among the most consistently dysregulated in placentas from GDM pregnancies (Radaelli et al., 2003; Enquobahrie et al., 2009; Zhao et al., 2011; Binder et al., 2015). In contrast, two transcriptome studies found few or no significant placental expression changes between GDM and controls (Sober et al., 2015; Lekva et al., 2016). However, these two studies had other pathological groups including pre-eclampsia, SGA or LGA, and such as the statistical power may have been insufficient to identify differentially expressed genes or clusters associated with each condition, although the overall sample size was large (Sober et al., 2015; Lekva et al., 2016).
Studies have also considered other forms of diabetes in pregnancy, such as pre-existing type I and type II diabetes (Radaelli et al., 2009; Alexander et al., 2018) or tried to account for differences in maternal obesity, which is more common in diabetic groups compared with controls (Bari et al., 2016). However, as the numbers of each diabetic type or BMI group are small, more studies are needed to confirm if the identified differences are due to diabetes or obesity. Another issue is that of selective reporting. For example, in a genome-wide transcriptome study of type I diabetes and GDM cases and controls, the authors chose to report only pre-selected genes involved in glucose and lipid metabolism based on fold changes without correcting for multiple testing (Radaelli et al., 2009), which is a biased approach and some results may turn out to be chance findings. Further studies are also needed to resolve the transcript differences associated with GDM management (diet-control, metformin or insulin treatment) or quality of glycaemic control, functionally explore the biological significance of identified placental gene expression alterations, and expand knowledge into the underlying pathophysiology of GDM and its possible implications for offspring health.
Antenatal infections and inflammation
Infections during pregnancy not only affect the mother but can also impact the foetus via the placenta. Three studies (total n = 53 placentas, 40% affected) have examined the transcriptome of placentas exposed to malaria, Trypanosoma cruzi and infections leading to preterm birth (Table II: Antenatal infections and inflammation). Expectedly, all affected placentas, regardless of infection type, show dramatic up-regulation of genes involved in the immune and inflammatory response (Muehlenbachs et al., 2007; Ackerman et al., 2016; Juiz et al., 2018). Better understanding of the functional significance of these transcriptomic alterations may lead to the identification of biomarkers or therapeutic targets for improved clinical management of infections during pregnancy, and reduce infection-induced preterm birth and adverse developmental programming.
Placental inflammation may also occur in the absence of an infection. At present, two studies (total n = 31 placentas, 52% affected) have examined cases of placental inflammation without infection (Table II: Antenatal infections and inflammations). Placentas with villitis of unknown aetiology show transcript profiles distinct from that of chorioamnionitis caused by an infection, suggesting that the pathology may arise from maternal–foetal histoincompatibility (Kim et al., 2009). Comparison of the chronically inflamed placenta with controls also suggests a key role of T cells in mediating maternal immune tolerance of foetal antigens, which may underlie chronic placental inflammation (Raman et al., 2015). Nevertheless, given the small number of studies with low sample sizes, larger studies in this area are clearly needed to confirm the findings of these past studies. It would also be interesting to explore the transcriptome profiles of autoimmune conditions, such as systemic lupus erythematosus, and those pregnancies exposed to immunosuppressive therapy (e.g. post-organ transplant), as these conditions are associated with increased risk of adverse pregnancy outcomes.
Labour
The exact mechanisms of parturition remain elusive. Presently, five datasets have examined 89 placentas and decidua in relation to labour onset (Table II: Labour), of which four involved preterm labour (27% affected). The inflammatory response is consistently identified as the most dysregulated process at the preterm labour placental interface (Chim et al., 2012; Bukowski et al., 2017; Rinaldi et al., 2017). Aberrant expression of genes involved in extracellular matrix remodelling is also implicated in preterm labour, which may in turn be affected by abnormal expression of miRNAs (Mayor-Lynn et al., 2011; Chim et al., 2012). Further refinement and validation of these labour-associated molecular factors in larger studies will enhance understanding of the normal parturition process and may lead to development of novel biomarkers to predict the onset of labour in pregnant women and interventions for either prevention of preterm labour or obstetrically indicated induction of labour.
Recurrent miscarriage
Recurrent miscarriage (RM) is the repeated loss of pregnancy in the early stages and affects around 1 in 50 couples (Rull et al., 2013). The precise cause(s) of RM is unknown for many couples, but five small studies (n = 50 placentas, 42% affected) have investigated the placental transcriptome to uncover the potential factors involved in unexplained RM (Table II: RM). It is nonetheless difficult to distinguish between transcriptomic changes that lead to the miscarriage as opposed to those that change as a result of the miscarriage process. Moreover, given that products of conception may remain in utero for days after pregnancy failure or embryo/foetal demise, placental RNA quality may be compromised by the time of collection and be another study confounder. Controls in such studies are those of elective termination of pregnancies with no known anomalies. However, the use of such controls is based on an underlying assumption that these pregnancies would have ended well without any complications, which is impossible to verify. A proposed alternative control could be first-trimester placentas collected from women who had previous successful term pregnancies, but happen to experience their first miscarriage, which presumably would likely be of a conceptus-originated aetiology rather than a parental-originated aetiology that is more likely in cases of RM.
Despite questions over suitability of controls and very small sample sizes, current studies still provide an initial insight into potential mechanisms. For instance, a microarray study identified tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) as highly expressed in RM placentas, which was detectable in the circulation of women both at the point of miscarriage and also prior to miscarriage (i.e. in women who subsequently had a miscarriage following prospective blood sampling), as compared with controls (Rull et al., 2013). Increased circulating TRAIL was verified in an independent study, which also demonstrated adverse effects of high TRAIL concentrations on trophoblast function in vitro (Agostinis et al., 2012), indicating TRAIL as a potential biomarker and determinant of pregnancy failure in early gestation. Other studies show association of RM with transcription factors, such as E2F and YY1, non-coding RNAs and DNA methylation (Sober et al., 2016; Tian et al., 2016; Huang et al., 2018; Yu et al., 2018), which are yet to be validated in larger datasets.
Chromosomal abnormalities
Profiling placentas containing chromosomal abnormalities provide a unique opportunity to study in vivo effects of genetic aberrations and inform on the pathogenesis of complications that occur as the pregnancy progresses. Just two studies have examined 38 placentas (58% affected) so far (Table II: Chromosomal abnormalities). Interrogating the trisomy 21 placental transcriptome revealed differential expression of genes associated with neurodevelopment, cancer and diabetes (Lim et al., 2017a), which may explain the disease risks of trisomy 21 individuals. Other transcriptomic investigation of placentas of either trisomy 13, 18 or 21 showed that several highly expressed genes were located on the respective trisomic chromosome, which is reflective of gene dosage (Bianco et al., 2016). However, most of the up-regulated genes were surprisingly not located on the trisomic chromosome, suggesting knock-on effects genome-wide (Bianco et al., 2016). These studies suggest that the observed phenotypes of aneuploidy, including pregnancy loss and IUGR, may arise from the collective effects of gene dosage and complex downstream genome-wide dysregulation of genes in the placenta (Bianco et al., 2016).
Intrahepatic cholestasis of pregnancy
Intrahepatic cholestasis of pregnancy (IHCP), also known as obstetric cholestasis, is associated with preterm delivery and stillbirths (Bicocca et al., 2018). The aetiology of the observed maternal pruritus and liver impairment may be associated with an enhanced response to increased oestrogen production in pregnancy resulting in abnormal hepatobiliary transport (Bicocca et al., 2018). At present, only one study of 30 placentas (67% affected) has examined the genome-wide placental transcript profile (Table II: Intrahepatic cholestasis of pregnancy). This study showed dysregulation of genes involved in vascular endothelial growth factor, G-protein-coupled receptor and immune-related signalling, with a greater dysregulation correlating with disease severity, thus, implicating abnormal placental angiogenesis and immune response in the pathophysiology of IHCP (Du et al., 2014). As with the other complications of pregnancy previously discussed, severe cases tended to deliver preterm and gestational age may be a confounder influencing the results observed since the study used term controls. Further studies are, therefore, needed to extend the knowledge of placental involvement in this complication of pregnancy.
Placental transcriptome studies of exposures during pregnancy
Many maternal sociodemographic factors, lifestyle and environmental exposures have been linked with pregnancy complications and offspring health adversity. A better understanding of how these different exposures affect the placenta will provide a greater insight into the pathophysiology and biological pathways leading to complications and abnormal programming of offspring health. As such, studies in this research theme examined placentas from pregnancies with different exposures from assisted reproduction and various clinical trial interventions to pre-existing maternal conditions (Table II).
Obesity
Alarmingly, a growing number of women are entering pregnancy in an obese state worldwide. Being obese markedly increases the risks of pregnancy complications, such as pre-eclampsia, GDM and preterm delivery (Sureshchandra et al., 2018). Transcript profiling of placentas from obese women may uncover the underlying basis of their heightened risk of pregnancy complications and the molecular mechanisms involved in programming offspring health. Five placental transcriptome studies (n = 242, 44% affected) have considered maternal obesity (Table III: Obesity). Two studies similarly found enrichment of differentially expressed genes involved in angiogenesis, lipid metabolism and the immune/inflammatory response (Saben et al., 2014a; Altmae et al., 2017). A different study identified perturbed placental nutrient transport as another consequence of exposure to the maternal obesity milieu (Sureshchandra et al., 2018), which could alter foetal growth and postnatal health trajectories. Indeed, a large study of 183 placentas examining maternal pre-pregnancy BMI as a continuous variable demonstrated that gene clusters enriched for maternal immune dysregulation were positively associated with maternal BMI and negatively associated with low birth weight, providing evidence of a molecular basis to the relationship between the two (Cox et al., 2019). Therefore, the placenta exposed to maternal obesity is characterized by immune dysregulation, which can have widespread effects on placental function and foetal growth and development.
Smoking
Tobacco smoke is an external environmental exposure that is detrimental to pregnancy. Global profiling of smoke-exposed placentas in four studies (n = 197, 30% affected, Table II: Smoking) showed harmful and dysregulating effects of tobacco smoke on different aspects of placental growth and metabolism (Huuskonen et al., 2008; Bruchova et al., 2010; Votavova et al., 2011; Votavova et al., 2012). Comparison of smoke-induced transcriptomic changes in placenta by active or passive smoking showed a substantial overlap between both groups compared with the non-smoking group in biological processes, such as lipid metabolism, oxidative stress and blood coagulation (Votavova et al., 2012), suggesting that these common molecular placental mechanisms are involved in transmitting the harm of smoking to the growing foetus be it active or passive. As such, these placental pathways could be potential intervention targets to modify the pregnancy outcomes of those exposed to any type of tobacco smoke during gestation.
Clinical trials
Various nutritional or drug interventions are being explored in clinical trials to improve pregnancy outcomes. While treatments may show promising results in vitro on placental cell cultures and in vivo in animal models, many have not shown the desired effect upon testing in most clinical trials. Transcript profiling of placentas from women treated as part of a clinical trial during pregnancy may thus provide valuable insights into the underlying molecular mechanisms affected, inter-individual variability in effects and possible explanation for trial findings. We identified three studies that analysed a total of 136 placentas from clinical trials (Table II: Clinical trials). A study conducted on placentas collected from women supplemented with a low or high dose of choline from the start of the third trimester, with the intention of improving placental vascular function and reducing the risk of developing pre-eclampsia, showed widespread effects on the placental transcriptome, particularly on processes involved in vascular regulation (Jiang et al., 2013). A promising finding was significantly reduced expression of the pre-eclampsia-associated FLT1 gene, which can induce systemic vascular dysfunction at high protein concentrations, thus providing a molecular basis for the utility of choline supplementation during pregnancy (Jiang et al., 2013). Omega-3 fatty acid supplementation altered placental expression of genes involved in cell-cycle regulation in a sexually dimorphic manner with greater changes occurring in pregnancies with female foetuses, which correlated with offspring birthweight and birthweight centiles (Sedlmeier et al., 2014). This study highlights the placental response to omega-3 supplementation and the potential mechanisms involved in modulating foetal growth and postnatal development (Sedlmeier et al., 2014). A third study examined placentas collected from obese women who were treated with metformin or a placebo (Chiswick et al., 2016), with the aim of determining if there were any changes in genes regulating foetal growth or metabolism. However, while the transcriptome dataset is publically available alongside complementary methylome data, the study findings remain unpublished. Nevertheless, this dataset serves as a valuable resource to understand the effects of metformin on the placenta and acts as a possible reference for studies involving metformin in treatment of GDM.
In vitro fertilization
With the global rise of ART, more pregnancies are now being conceived by IVF, which is associated with poorer pregnancy outcomes (Nelissen et al., 2014). Three studies have examined the IVF placental transcriptome in the first and third trimesters (n = 169 placentas, 28% exposed, Table III: In vitro fertilization). To determine which differentially expressed genes are related more specifically to IVF, the largest study of 141 first-trimester chorionic villus samples included a non-IVF ART group alongside spontaneous conceptions for comparison and identified CACNA1I, which codes for a calcium channel subunit, as one such gene (Lee et al., 2019). Further comparison between just the IVF and non-IVF ART groups showed differential expression of SLC18A2, CCL21, FXYD2, PAEP and DNER, which supports the notion that IVF also has distinct effects on the placenta compared with other types of ART (Lee et al., 2019). However, as ART are used primarily by couples who struggle to conceive naturally, discovered alterations may be due to the underlying parental factors contributing to subfertility rather than a result of ART used. Since infertility causes are so varied, such as being due to structural defects of the reproductive tract, ovulatory dysfunction, endometriosis, childhood cancer chemotherapy or unexplained maternal or paternal factors, stratifying by causes of infertility may help discriminate the unique gene signatures for infertility as compared with those that are consequential of ART in future studies.
Antenatal depression
Antenatal maternal mental health is of rising importance as cumulative evidence suggests a potent impact on pregnancy and childhood outcomes. Currently, only one study (n = 20 placentas, 50% affected, Table III: Antenatal depression) has examined the effects of maternal depression and antidepressant treatment on the placental transcriptome (Olivier et al., 2014). Most differentially expressed genes compared with controls showed limited overlap between the untreated and medically treated depression (Olivier et al., 2014). This suggests that not only does depression itself affect the placental transcriptome, but that depression and antidepressants have largely independent effects on the placenta and that those treated with antidepressants should be evaluated separately in future studies.
Transcriptome studies of in vitro placental cultures
The key advantage of culturing in vitro explants or isolated cells from the placenta is that we can determine the precise effects of altering a single factor in well-controlled experimental conditions, without having to contend with other inter-individual variability in inter-placental comparisons. Transcript profiling of cultured placental explants, primary cells and cell lines have thus also contributed to increased understanding of placental function, with many studies profiling the placental response to various agents and treatments including hypoxia, irradiation, growth factors, cytokines and infectious agents (Table III). Such in vitro models have limitations including issues with cell purity of isolated primary cells, loss of 3D cytoarchitecture and absence of cell–cell interaction in single-cell type cultures. Although placental explant and organoid cultures may overcome some of these limitations, culture conditions may not fully reflect in vivo conditions and could lead to spurious findings as a result of altered responses to the exposure of interest. Hence, transcriptome findings of in vitro models should be interpreted with some caution and further verified in ex vivo studies.
Trophoblast cultures
Most in vitro studies utilize primary trophoblast cultures from term placenta or extended trophoblast cell lines. Given the common use of immortalized cell lines in placental research, widely-used trophoblast-like cell lines were profiled to ascertain their genome-wide phenotype and how representative they were of primary trophoblast (Burleigh et al., 2007; Bilban et al., 2010; Apps et al., 2011; Takao et al., 2011). These studies found little overlap in the profiles between primary cells and cell lines, and recommended caution in use of cell lines in placental research (Burleigh et al., 2007; Bilban et al., 2010; Apps et al., 2011). Another reason for genome-wide profiling of cultured trophoblast is to characterize the differentiation process from cytotrophoblast to syncytiotrophoblast (Shankar et al., 2015; Rouault et al., 2016; Yabe et al., 2016; Zheng et al., 2016; Robinson et al., 2017; Azar et al., 2018; Gauster et al., 2018). Pairing genome-wide transcript profiling with targeted gene manipulation using small RNA or plasmid technologies allows gene regulatory effects to be determined alongside consequential effects on trophoblast cell function (Rigourd et al., 2008; Tauber et al., 2010; Xie et al., 2014; Than et al., 2018), despite the potential limitations in methodology discussed earlier.
Non-trophoblast cultures
Additionally, several studies have examined non-trophoblast cell types in culture. For example, to expand knowledge on the genetic regulation of placental vascularity, Augsten et al. (2011) performed a microarray on primary term placental endothelial cells treated with high-density lipoprotein to assess how foetal lipoprotein, which contains a considerably higher proportion of apoE than that of adults, alters placental vessel function. Another microarray study identified genes involved in the immunoregulatory and pro-angiogenic function of first-trimester decidual endothelial cells relative to another endothelial cell type from skin (Agostinis et al., 2019), providing an insight into the unique role of these maternal endothelial cells in modulating immune tolerance of the foetus at the interface. Transcriptome studies of non-trophoblastic cells remain few, and if conducted and interpreted with the caution discussed previously, further investigations utilizing cultures of these cell types may potentially enhance understanding of the complex processes occurring in pregnancy.
In vitro models of in vivo exposures
Placental-derived cultures are also used to mimic in vivo exposures. For instance, several datasets reflect the response profile of placental explants, trophoblast cell lines, primary decidual and chorion organoid cultures exposed to infectious agents – Coxiella burnetii, Trypanosoma cruzi and Zika virus (Ben Amara et al., 2010; Weisblum et al., 2017; Castillo et al., 2018), which are further informed by related studies that examine the effects of immunomodulatory cytokines on placental function (Ibrahim et al., 2016; Verma et al., 2018; Yockey et al., 2018). One study characterized the molecular signatures of the response to insulin in trophoblast cells cultured from first-trimester placentas of lean and obese women, revealing that prior exposure to obesity blunted trophoblast sensitivity to insulin (Lassance et al., 2015). Notwithstanding the technical limitations that may impact transcriptome findings, placental-derived cultures can add another dimension in expanding knowledge of placental function, which may not be fully apparent from study of tissues and immediately isolated cells.
Knowledge gaps and future directions
Addressing placental heterogeneity
Cellular heterogeneity of the human placenta is likely a major contributing factor to inconsistent transcriptome findings between studies. With the advent of single-cell transcriptomics, it may be possible to deconvolute the placental tissue transcriptome more readily and account for differences in tissue sampling in the near future. Deconvolution may also highlight differences in placental cell composition due to the underlying pathology. Single-cell human placental transcriptome profiles across all trimesters are now available and serve as a basis to develop algorithms for deconvolutions (Table I). However, sample sizes in these studies are relatively small between two and ten placentas and with the current technical limitations requiring cell dissociation, available datasets may include artefactual changes and not fully capture the transcriptome of all placental cell types, particularly the large multi-nucleated syncytiotrophoblast.
Meta-analysis of current data and ease of data access
Meta-analysis of past studies enhances statistical power, which may highlight novel molecular pathways or strengthen the evidence for previously identified genes. Indeed, multiple meta-analyses have been performed for pre-eclampsia (Kleinrouweler et al., 2013; Moslehi et al., 2013; Vaiman et al., 2013; van Uitert et al., 2015; Vaiman and Miralles, 2016; Brew et al., 2016) and preterm birth (Eidem et al., 2015; Paquette et al., 2018), as well as for investigating sexual dimorphism of the placenta (Buckberry et al., 2014) and understanding trophoblast differentiation in the context of hydatidiform moles (Desterke et al., 2018). Nevertheless, lack of data access can hamper the ability to perform powerful meta-analyses. For example, of the 10 transcript profiling studies performed for GDM, only data from three studies are publically available, representing less than a third of the profiled placentas. Efforts are ongoing to make placental data more accessible. For instance, the newly developed Placenta Atlas Tool centralized database simplifies the search for relevant placental transcriptome datasets and allows some basic analysis to be performed within the site (Ilekis et al., 2019), providing a useful starting point for researchers. Consistent and clear reporting of experimental details, such as specific microarray platforms utilized and poly A+ selection for mRNA enrichment in RNA sequencing studies, and of key clinical information to inform on disease subtypes and severity are also critical in enabling researchers to design proper integrative meta-analysis studies.
Studies on non-coding RNA
Traditionally, much focus was on protein-coding mRNA transcripts that result in functional changes. However, growing evidence implicate possible roles for non-coding RNA (e.g. long non-coding RNA, miRNA, circular RNA) in the placenta (Cox et al., 2015). Differential placental expression of non-coding RNAs has been investigated in pre-eclampsia (Gunel et al., 2017; Hu et al., 2018a; Lykoudi et al., 2018; Zhou et al., 2018), GDM (Li et al., 2015; Wang et al., 2019b), IUGR or SGA pregnancies (Wen et al., 2017; Ostling et al., 2019) and early pregnancy loss (Hosseini et al., 2018). The use of RNA sequencing technologies to further characterize these non-coding RNAs provides a tantalizing approach to identify and develop new biomarkers and therapeutic targets for pregnancy complications. As such, researchers may want to consider the different RNA extraction methods available to enable capture of all placental RNA species, both coding and non-coding, for analysis in future studies.
Further studies on other issues in pregnancy
There is a paucity of genome-wide transcriptome studies in many aspects of pregnancy (Supplementary Table SII). The pre-eclamptic placenta is disproportionately profiled as compared with other common pathologies of pregnancy including preterm labour, IUGR, GDM and stillbirth (Table II). Although an estimated 2.6 million stillbirths occur annually worldwide (Blencowe et al., 2016), of which ∼40% are unexplained (Reinebrant et al., 2018), no study has yet profiled placentas from this devastating pregnancy complication, although such studies could yield useful insights as to why a foetus dies in utero, especially in cases of unexplained stillbirth. Transcript profiling of abnormal placental development, such as placenta praevia, placenta accreta and molar pregnancy could also highlight the potential causative factors behind their pathogenesis (Desterke et al., 2018), possibly enabling the discovery of new approaches to treat or prevent the recurrence of these pathologies in future pregnancies.
Additionally, even with much ongoing research effort to minimize infectious diseases, pregnant women, being relatively immunosuppressed, remain at great risk of infections, such as malaria (Dellicour et al., 2010) and have shown increased susceptibility to recent global health emergencies, such as the swine flu pandemic and Ebola outbreak (Kourtis et al., 2014; Silasi et al., 2015). We propose that placental transcriptome studies be used to improve understanding of how maternal infections affect the placenta, so as to identify the mechanistic pathways that can be targeted to reduce the transmission of harm to the growing foetus.
Furthermore, given the increasingly recognized importance of the maternal nutritional, mental and emotional states, the rise in women exposed to harmful environmental and chemical exposures during their pregnancies and the profound impacts these can have on the pregnancy and the future health of the child (Hoirisch-Clapauch et al., 2015; Lewis et al., 2015; Unger et al., 2016; Chen et al., 2018; Henschke, 2019; Varshavsky et al., 2019), their effects on the placenta are all deserving of further investigation, so as to increase ways of promoting benefits of some lifestyles while minimizing adversity. Pre-existing medical conditions (e.g. autoimmune and endocrine diseases, thrombophilia) are strongly associated with an aberrant hormonal, metabolic and inflammatory milieu that is detrimental to placentation, and thus, such pregnancies are predisposed to significantly higher rates of complications with poorer neonatal outcomes (Ali et al., 2016; Vannuccini et al., 2016; Meakin et al., 2017; De Leo and Pearce, 2018; De Carolis et al., 2019; Mitriuc et al., 2019; Stepien and Huttner, 2019). Placental transcriptome analysis could thus reveal how such conditions heighten a woman’s susceptibility to obstetric complications, which may lead to new treatments to prevent defective placental function and improve pregnancy outcomes in affected women.
Pregnancy-specific factors can also have profound consequences on pregnancy outcomes. Given major societal changes, more women are using ART to conceive and/or entering pregnancy at an older age, both of which are associated with placental dysfunction and more obstetric problems including IUGR and stillbirth (Nelissen et al., 2014; Lean et al., 2017). Placental profiling may help reveal whether higher rates of obstetric complications observed are due to underlying subfertility or ART as suggested by animal studies (de Waal et al., 2015), and enable appropriate strategies to be developed for mitigating harms.
Frequently excluded from transcriptome studies, profiling placentas from multiple pregnancies may also demonstrate the mechanisms involved in the inherently elevated risk of obstetric complications (Witteveen et al., 2016), which could be targeted to improve outcomes for the mother and her children. Moreover, further studies of twin placental transcriptomes with disconcordant intrauterine growth, whereby the healthy twin can serve as a well-matched control (Roh et al., 2005; Wen et al., 2017), may help elucidate novel mechanisms of foetal growth that can be capitalized upon to improve IUGR outcomes.
Linkages with other ‘omics’ and long-term outcomes
Integrating placental transcriptome data with other datasets including other ‘omics’ and longitudinal data will enhance knowledge into healthy placental development and disease mechanisms. A study comparing the pre-eclamptic placental transcriptome to the blood transcriptome of cardiovascular disease identified significant overlap between the two, and provided novel insights into possible shared relationships between pregnancy complications and subsequent health in the mother or child postnatally (Sitras et al., 2015). Placental transcript profiling in birth cohorts with comprehensive longitudinal follow-up of the children may also potentially uncover new placental programming mechanisms that can influence extrauterine life in the longer term. Indeed, previously identified placental eQTLs were predictive of birthweight and subsequent childhood obesity in a cohort study, highlighting the role of placental gene expression in modulating postnatal outcomes, and such genes could serve as potential molecular targets for interventions (Peng et al., 2018). Besides profiling more placentas from additional birth cohorts, it is of great interest to examine currently available birth cohort-related placental transcriptome datasets for any possible associations with childhood outcomes (Binder et al., 2015; Cox et al., 2019). In doing so, we may be able to develop a catalogue of placental biomarkers predictive of future health, which could be used to identify offspring at high risk of subsequent poor health. Thus, the placenta may serve as a unique window into the future extrauterine life of the offspring and provide an opportunity to intervene and change the health trajectories of those exposed to an adverse intrauterine environment.
Conclusion
Placental transcript profiling presents enormous potential to enhance understanding of healthy placental development and function, highlight the possible underlying causal and consequential mechanisms of pregnancy complications (Fig. 3), and predict and improve the health outcomes of mothers and offspring from compromised pregnancies. Challenges in obtaining sufficient numbers of quality samples with clear clinical characteristics will need to be overcome to drive the field forward. Current data may also be capitalized upon by performing meta-analyses to increase statistical power, although interpretation of findings will need to carefully account for limitations, such as inconsistent clinical criteria between studies and gestational age matching of cases and controls. Furthermore, additional resources should be dedicated to analyse placentas from the understudied areas, which will enable the dynamic complexities of the placental transcriptome to be more fully appreciated.
Figure 3.
Factors that influence the placental transcriptome and the associations of altered placental molecular pathways with pregnancy complications or exposures. Dynamic transcriptional regulation of the placental interface throughout gestation is multi-factorial (arrows). Differential regulation of specific molecular pathways identified in multiple placental transcriptome studies may highlight the potential underlying causal mechanisms involved in pregnancy complications or represent the placental mechanisms affected by pregnancy exposures (connecting lines). GDM, gestational diabetes mellitus; IHCP, intrahepatic cholestasis of pregnancy; IUGR/SGA, intrauterine growth restriction/small for gestational age; LGA, large for gestational age; RM, recurrent miscarriage.
Authors’ roles
H.E.J.Y. contributed to the study design, performed the literature searches and wrote and edited the manuscript. S.Y.C. conceived the study and provided critical revision of the manuscript for intellectual content.
Funding
This work was supported by funding from the Singapore Institute for Clinical Sciences, Agency for Science, Technology and Research. S.Y.C. is supported by a Clinician Scientist Award from the Singapore National Medical Research Council (NMRC/CSA-INV/0010/2016).
Conflict of interest
H.E.J.Y. has no conflict of interest to declare. S.Y.C. is part of the EPIGEN academic consortium that has received research funding from Nestec.
Supplementary Material
Contributor Information
Hannah E J Yong, Singapore Institute for Clinical Sciences, Agency for Science, Technology and Research, Singapore, Singapore.
Shiao-Yng Chan, Singapore Institute for Clinical Sciences, Agency for Science, Technology and Research, Singapore, Singapore; Department of Obstetrics and Gynaecology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore.
References
- Ackerman WE, Buhimschi IA, Eidem HR, Rinker DC, Rokas A, Rood K, Zhao G, Summerfield TL, Landon MB, Buhimschi CS. Comprehensive RNA profiling of villous trophoblast and decidua basalis in pregnancies complicated by preterm birth following intra-amniotic infection. Placenta 2016:44;23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostinis C, Bulla R, Tisato V, De Seta F, Alberico S, Secchiero P, Zauli G. Soluble TRAIL is elevated in recurrent miscarriage and inhibits the in vitro adhesion and migration of HTR8 trophoblastic cells. Hum Reprod 2012:27;2941–2947. [DOI] [PubMed] [Google Scholar]
- Agostinis C, Masat E, Bossi F, Ricci G, Menegazzi R, Lombardelli L, Zito G, Mangogna A, Degan M, Gattei V et al. Transcriptomics and immunological analyses reveal a pro-angiogenic and anti-inflammatory phenotype for decidual endothelial cells. Int J Mol Sci 2019:20;1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J, Teague AM, Chen J, Aston CE, Leung YK, Chernausek S, Simmons RA, Pinney SE. Offspring sex impacts DNA methylation and gene expression in placentae from women with diabetes during pregnancy. PLoS One 2018:13;e0190698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z, Hansen AV, Ulrik CS. Exacerbations of asthma during pregnancy: impact on pregnancy complications and outcome. J Obstet Gynaecol 2016:36;455–461. [DOI] [PubMed] [Google Scholar]
- Altmae S, Segura MT, Esteban FJ, Bartel S, Brandi P, Irmler M, Beckers J, Demmelmair H, Lopez-Sabater C, Koletzko B et al. Maternal pre-pregnancy obesity is associated with altered placental transcriptome. PLoS One 2017:12;e0169223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apps R, Sharkey A, Gardner L, Male V, Trotter M, Miller N, North R, Founds S, Moffett A. Genome-wide expression profile of first trimester villous and extravillous human trophoblast cells. Placenta 2011:32;33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augsten M, Hackl H, Ebner B, Chemelli A, Glatter O, Marsche G, Lang U, Desoye G, Wadsack C. Fetal HDL/apoE: a novel regulator of gene expression in human placental endothelial cells. Physiol Genomics 2011:43;1255–1262. [DOI] [PubMed] [Google Scholar]
- Azar C, Valentine M, Trausch-Azar J, Druley T, Nelson DM, Schwartz AL. RNA-Seq identifies genes whose proteins are transformative in the differentiation of cytotrophoblast to syncytiotrophoblast, in human primary villous and BeWo trophoblasts. Sci Rep 2018:8;5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari MF, Ngo S, Bastie CC, Sheppard AM, Vatish M. Gestational diabetic transcriptomic profiling of microdissected human trophoblast. J Endocrinol 2016:229;47–59. [DOI] [PubMed] [Google Scholar]
- Barreto RS, Bressan FF, Oliveira LJ, Pereira FT, Perecin F, Ambrosio CE, Meirelles FV, Miglino MA. Gene expression in placentation of farm animals: an overview of gene function during development. Theriogenology 2011:76;589–597. [DOI] [PubMed] [Google Scholar]
- Ben Amara A, Ghigo E, Le Priol Y, Lepolard C, Salcedo SP, Lemichez E, Bretelle F, Capo C, Mege JL. Coxiella burnetii, the agent of Q fever, replicates within trophoblasts and induces a unique transcriptional response. PLoS One 2010:5;e15315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton SJ, Leavey K, Grynspan D, Cox BJ, Bainbridge SA. The clinical heterogeneity of preeclampsia is related to both placental gene expression and placental histopathology. Am J Obstet Gynecol 2018:219;604.e1–e25. [DOI] [PubMed] [Google Scholar]
- Bianco K, Gormley M, Farrell J, Zhou Y, Oliverio O, Tilden H, McMaster M, Fisher SJ. Placental transcriptomes in the common aneuploidies reveal critical regions on the trisomic chromosomes and genome-wide effects. Prenat Diagn 2016:36;812–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicocca MJ, Sperling JD, Chauhan SP. Intrahepatic cholestasis of pregnancy: review of six national and regional guidelines. Eur J Obstet Gynecol Reprod Biol 2018:231;180–187. [DOI] [PubMed] [Google Scholar]
- Bilban M, Tauber S, Haslinger P, Pollheimer J, Saleh L, Pehamberger H, Wagner O, Knofler M. Trophoblast invasion: assessment of cellular models using gene expression signatures. Placenta 2010:31;989–996. [DOI] [PubMed] [Google Scholar]
- Binder AM, LaRocca J, Lesseur C, Marsit CJ, Michels KB. Epigenome-wide and transcriptome-wide analyses reveal gestational diabetes is associated with alterations in the human leukocyte antigen complex. Clin Epigenetics 2015:7;79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JD, Yuen RK, Lim BK, McFadden DE, von Dadelszen P, Robinson WP. Widespread DNA hypomethylation at gene enhancer regions in placentas associated with early-onset pre-eclampsia. Mol Hum Reprod 2013:19;697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe H, Cousens S, Jassir FB, Say L, Chou D, Mathers C, Hogan D, Shiekh S, Qureshi ZU, You D et al. National, regional, and worldwide estimates of stillbirth rates in 2015, with trends from 2000: a systematic analysis. Lancet Glob Health 2016:4;e98–e108. [DOI] [PubMed] [Google Scholar]
- Brew O, Sullivan MH, Woodman A. Comparison of normal and pre-eclamptic placental gene expression: a systematic review with meta-analysis. PLoS One 2016:11;e0161504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchova H, Vasikova A, Merkerova M, Milcova A, Topinka J, Balascak I, Pastorkova A, Sram RJ, Brdicka R. Effect of maternal tobacco smoke exposure on the placental transcriptome. Placenta 2010:31;186–191. [DOI] [PubMed] [Google Scholar]
- Buckberry S, Bianco-Miotto T, Bent SJ, Clifton V, Shoubridge C, Shankar K, Roberts CT. Placental transcriptome co-expression analysis reveals conserved regulatory programs across gestation. BMC Genomics 2017:18;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckberry S, Bianco-Miotto T, Bent SJ, Dekker GA, Roberts CT. Integrative transcriptome meta-analysis reveals widespread sex-biased gene expression at the human fetal-maternal interface. Mol Hum Reprod 2014:20;810–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowski R, Sadovsky Y, Goodarzi H, Zhang H, Biggio JR, Varner M, Parry S, Xiao F, Esplin SM, Andrews W et al. Onset of human preterm and term birth is related to unique inflammatory transcriptome profiles at the maternal fetal interface. PeerJ 2017:5;e3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh DW, Kendziorski CM, Choi YJ, Grindle KM, Grendell RL, Magness RR, Golos TG. Microarray analysis of BeWo and JEG3 trophoblast cell lines: identification of differentially expressed transcripts. Placenta 2007:28;383–389. [DOI] [PubMed] [Google Scholar]
- Burton GJ, Fowden AL, Thornburg KL. Placental origins of chronic disease. Physiol Rev 2016:96;1509–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton GJ, Sebire NJ, Myatt L, Tannetta D, Wang YL, Sadovsky Y, Staff AC, Redman CW. Optimising sample collection for placental research. Placenta 2014:35;9–22. [DOI] [PubMed] [Google Scholar]
- Carter AM. Recent advances in understanding evolution of the placenta: insights from transcriptomics. F1000Res 2018:7;89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo C, Carrillo I, Libisch G, Juiz N, Schijman A, Robello C, Kemmerling U. Host-parasite interaction: changes in human placental gene expression induced by Trypanosoma cruzi. Parasit Vectors 2018:11;479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli RC, Cerdeira AS, Pernicone E, Korkes HA, Burke SD, Rajakumar A, Thadhani RI, Roberts DJ, Bhasin M, Karumanchi SA et al. Induced human decidual NK-like cells improve utero-placental perfusion in mice. PLoS One 2016:11;e0164353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centlow M, Wingren C, Borrebaeck C, Brownstein MJ, Hansson SR. Differential gene expression analysis of placentas with increased vascular resistance and pre-eclampsia using whole-genome microarrays. J Pregnancy 2011:2011;472354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Xu X, Yan Y. Estimated global overweight and obesity burden in pregnant women based on panel data model. PLoS One 2018:13;e0202183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim SS, Lee WS, Ting YH, Chan OK, Lee SW, Leung TY. Systematic identification of spontaneous preterm birth-associated RNA transcripts in maternal plasma. PLoS One 2012:7;e34328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiswick CA, Reynolds RM, Denison FC, Drake AJ, Forbes S, Newby DE, Walker BR, Quenby S, Wray S, Weeks A et al. Does Metformin Reduce Excess Birthweight in Offspring of Obese Pregnant Women? A Randomised Controlled Trial of Efficacy, Exploration of Mechanisms and Evaluation of Other Pregnancy Complications: Efficacy and Mechanism Evaluation. Southampton (UK): NIHR Journals Library; 2016. [PubMed] [Google Scholar]
- Christians JK, Leavey K, Cox BJ. Associations between imprinted gene expression in the placenta, human fetal growth and preeclampsia. Biol Lett 2017:13;20170643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa ML, Robinette ML, Bugatti M,, Longtine MS, Colvin BN, Lantelme E, Vermi W, Colonna M, Nelson DM, Cella M. Two distinct myeloid subsets at the term human fetal-maternal interface. Front Immunol 2017:8;1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B, Leavey K, Nosi U, Wong F, Kingdom J. Placental transcriptome in development and pathology: expression, function, and methods of analysis. Am J Obstet Gynecol 2015:213;S138–S151. [DOI] [PubMed] [Google Scholar]
- Cox B, Tsamou M, Vrijens K, Neven KY, Winckelmans E, de Kok TM, Plusquin M, Nawrot TS. A co-expression analysis of the placental transcriptome in association with maternal pre-pregnancy BMI and newborn birth weight. Front Genet 2019:10;354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvitic S, Longtine MS, Hackl H, Wagner K, Nelson MD, Desoye G, Hiden U. The human placental sexome differs between trophoblast epithelium and villous vessel endothelium. PLoS One 2013:8;e79233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carolis S, Moresi S, Rizzo F, Monteleone G, Tabacco S, Salvi S, Garufi C, Lanzone A. Autoimmunity in obstetrics and autoimmune diseases in pregnancy. Best Pract Res Clin Obstet Gynaecol 2019:60;66–76. [DOI] [PubMed] [Google Scholar]
- De Leo S, Pearce EN. Autoimmune thyroid disease during pregnancy. Lancet Diabetes Endocrinol 2018:6;575–586. [DOI] [PubMed] [Google Scholar]
- de Waal E, Vrooman LA, Fischer E, Ord T, Mainigi MA, Coutifaris C, Schultz RM, MS Bartolomei. The cumulative effect of assisted reproduction procedures on placental development and epigenetic perturbations in a mouse model. Hum Mol Genet 2015:24;6975–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahaye F, Do C, Kong Y, Ashkar R, Salas M, Tycko B, Wapner R, Hughes F. Genetic variants influence on the placenta regulatory landscape. PLoS Genet 2018:14;e1007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellicour S, Tatem AJ, Guerra CA, Snow RW, ter Kuile FO. Quantifying the number of pregnancies at risk of malaria in 2007: a demographic study. PLoS Med 2010:7;e1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterke C, Slim R, Candelier JJ. A bioinformatics transcriptome meta-analysis highlights the importance of trophoblast differentiation in the pathology of hydatidiform moles. Placenta 2018:65;29–36. [DOI] [PubMed] [Google Scholar]
- Deyssenroth MA, Peng S, Hao K, Lambertini L, Marsit CJ, Chen J. Whole-transcriptome analysis delineates the human placenta gene network and its associations with fetal growth. BMC Genomics 2017:18;520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, Guo F, Zhang Y, Liu XM, Xiang YQ, Zhang C, Liu ZW, Sheng JZ, Huang HF, Zhang JY et al. Integrated transcriptome sequencing analysis reveals role of miR-138-5p/TBL1X in placenta from gestational diabetes mellitus. Cell Physiol Biochem 2018:51;630–646. [DOI] [PubMed] [Google Scholar]
- Du Q, Pan Y, Zhang Y, Zhang H, Zheng Y, Lu L, Wang J, Duan T, Chen J. Placental gene-expression profiles of intrahepatic cholestasis of pregnancy reveal involvement of multiple molecular pathways in blood vessel formation and inflammation. BMC Med Genomics 2014:7;42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunk CE, Roggensack AM, Cox B, Perkins JE, Asenius F, Keating S, Weksberg R, Kingdom JC, Adamson SL. A distinct microvascular endothelial gene expression profile in severe IUGR placentas. Placenta 2012:33;285–293. [DOI] [PubMed] [Google Scholar]
- Eide IP, Isaksen CV, Salvesen KA, Langaas M, Schonberg SA, Austgulen R. Decidual expression and maternal serum levels of heme oxygenase 1 are increased in pre-eclampsia. Acta Obstet Gynecol Scand 2008:87;272–279. [DOI] [PubMed] [Google Scholar]
- Eidem HR, Ackerman WEt, McGary KL, Abbot P, Rokas A. Gestational tissue transcriptomics in term and preterm human pregnancies: a systematic review and meta-analysis. BMC Med Genomics 2015:8;27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquobahrie DA, Meller M, Rice K, Psaty BM, Siscovick DS, MA Williams. Differential placental gene expression in preeclampsia. Am J Obstet Gynecol 2008:199;566e1–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquobahrie DA, Williams MA, Qiu C, Meller M, Sorensen TK. Global placental gene expression in gestational diabetes mellitus. Am J Obstet Gynecol 2009:200;206.e1–13. [DOI] [PubMed] [Google Scholar]
- Fajardy I, Moitrot E, Vambergue A, Vandersippe-Millot M, Deruelle P, Rousseaux J. Time course analysis of RNA stability in human placenta. BMC Mol Biol 2009:10;21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Founds SA, Conley YP, Lyons-Weiler JF, Jeyabalan A, Hogge WA, Conrad KP. Altered global gene expression in first trimester placentas of women destined to develop preeclampsia. Placenta 2009:30;15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu B, Zhou Y, Ni X, Tong X, Xu X, Dong Z, Sun R, Tian Z, Wei H. Natural killer cells promote fetal development through the secretion of growth-promoting factors. Immunity 2017:47;1100–1113. [DOI] [PubMed] [Google Scholar]
- Gamliel M, Goldman-Wohl D, Isaacson B, Gur C, Stein N, Yamin R, Berger M, Grunewald M, Keshet E, Rais Y et al. Trained memory of human uterine NK cells enhances their function in subsequent pregnancies. Immunity 2018:48;951–962.e955. [DOI] [PubMed] [Google Scholar]
- Garrido-Gomez T, Dominguez F, Quinonero A, Diaz-Gimeno P, Kapidzic M, Gormley M, Ona K, Padilla-Iserte P, McMaster M, Genbacev O et al. Defective decidualization during and after severe preeclampsia reveals a possible maternal contribution to the etiology. Proc Natl Acad Sci U S A 2017:114;E8468–E8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauster M, Maninger S, Siwetz M, Deutsch A, El-Heliebi A, Kolb-Lenz D, Hiden U, Desoye G, Herse F, Prokesch A. Downregulation of p53 drives autophagy during human trophoblast differentiation. Cell Mol Life Sci 2018:75;1839–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaffari-Tabrizi-Wizsy N, Cvitic S, Tam-Amersdorfer C, Bilban M, Majali-Martinez A, Schramke K, Desoye G, Hiden U. Different preference of degradome in invasion versus angiogenesis. Cells Tissues Organs 2014:200;181–194. [DOI] [PubMed] [Google Scholar]
- Gibbs I, Leavey K, Benton SJ, Grynspan D, Bainbridge SA, BJ Cox. Placental transcriptional and histologic subtypes of normotensive fetal growth restriction are comparable to preeclampsia. Am J Obstet Gynecol 2019:220;110.e1–e21. [DOI] [PubMed] [Google Scholar]
- Gonzalez TL, Sun T, Koeppel AF, Lee B, Wang ET, Farber CR, Rich SS,, Sundheimer LW, Buttle RA, Chen YI et al. Sex differences in the late first trimester human placenta transcriptome. Biol Sex Differ 2018:9;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gormley M, Ona K, Kapidzic M, Garrido-Gomez T, Zdravkovic T, Fisher SJ. Preeclampsia: novel insights from global RNA profiling of trophoblast subpopulations. Am J Obstet Gynecol 2017:217;200.e1–e17. [DOI] [PubMed] [Google Scholar]
- Gunel T, Hosseini MK, Gumusoglu E, Kisakesen HI, Benian A, Aydinli K. Expression profiling of maternal plasma and placenta microRNAs in preeclamptic pregnancies by microarray technology. Placenta 2017:52;77–85. [DOI] [PubMed] [Google Scholar]
- Guo L, Tsai SQ, Hardison NE, James AH, Motsinger-Reif AA, Thames B, Stone EA, Deng C, Piedrahita JA. Differentially expressed microRNAs and affected biological pathways revealed by modulated modularity clustering (MMC) analysis of human preeclamptic and IUGR placentas. Placenta 2013:34;599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C, Mjosberg J, Matussek A, Geffers R, Matthiesen L, Berg G, Sharma S, Buer J, Ernerudh J. Gene expression profiling of human decidual macrophages: evidence for immunosuppressive phenotype. PLoS One 2008:3;e2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque A, Engel J, Teichmann SA, Lonnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med 2017:9;75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henschke P. Cannabis: An ancient friend or foe? What works and doesn’t work. Semin Fetal Neonatal Med 2019:24;149–154. [DOI] [PubMed] [Google Scholar]
- Herse F, Dechend R, Harsem NK, Wallukat G, Janke J, Qadri F, Hering L, Muller DN, Luft FC, Staff AC. Dysregulation of the circulating and tissue-based renin-angiotensin system in preeclampsia. Hypertension 2007:49;604–611. [DOI] [PubMed] [Google Scholar]
- Hess AP, Hamilton AE, Talbi S, Dosiou C, Nyegaard M, Nayak N, Genbecev-Krtolica O, Mavrogianis P, Ferrer K, Kruessel J et al. Decidual stromal cell response to paracrine signals from the trophoblast: amplification of immune and angiogenic modulators. Biol Reprod 2007:76;102–117. [DOI] [PubMed] [Google Scholar]
- Hoegh AM, Borup R, Nielsen FC, Sorensen S, Hviid TV. Gene expression profiling of placentas affected by pre-eclampsia. J Biomed Biotechnol 2010:2010;787545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoirisch-Clapauch S, Brenner B, Nardi AE. Adverse obstetric and neonatal outcomes in women with mental disorders. Thromb Res 2015:135 Suppl 1;S60–S63. [DOI] [PubMed] [Google Scholar]
- Hosseini MK, Gunel T, Gumusoglu E, Benian A, Aydinli K. MicroRNA expression profiling in placenta and maternal plasma in early pregnancy loss. Mol Med Rep 2018:17;4941–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Ao J, Li X, Zhang H, Wu J, Cheng W. Competing endogenous RNA expression profiling in pre-eclampsia identifies hsa_circ_0036877 as a potential novel blood biomarker for early pre-eclampsia. Clin Epigenetics 2018. a:10;48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, An Q, Sheu K, Trejo B, Fan S, Guo Y. Single cell multi-omics technology: methodology and application. Front Cell Dev Biol 2018. b:6;28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Du G, Huang X, Han L, Han X, Xu B, Zhang Y, Yu M, Qin Y, Xia Y et al. The enhancer RNA lnc-SLC4A1-1 epigenetically regulates unexplained recurrent pregnancy loss (URPL) by activating CXCL8 and NF-kB pathway. EBioMedicine 2018:38;162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DA, Kircher M, He Z, Guo S, Fairbrother GL, Moreno CS, Khaitovich P, Stoneking M. Evaluating intra- and inter-individual variation in the human placental transcriptome. Genome Biol 2015:16;54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huuskonen P, Storvik M, Reinisalo M, Honkakoski P, Rysa J, Hakkola J, Pasanen M. Microarray analysis of the global alterations in the gene expression in the placentas from cigarette-smoking mothers. Clin Pharmacol Ther 2008:83;542–550. [DOI] [PubMed] [Google Scholar]
- Ibrahim SA, WEt Ackerman, Summerfield TL, Lockwood CJ, Schatz F, DA Kniss. Inflammatory gene networks in term human decidual cells define a potential signature for cytokine-mediated parturition. Am J Obstet Gynecol 2016:214;284.e1–e47. [DOI] [PubMed] [Google Scholar]
- Ilekis JV, Keller M, Shlionskaya A, Ferguson CH, Patel B, Meitiv AL, Gorman B, Mohale A. The Placental Atlas Tool (PAT): a collaborative research and discovery platform for the placental research community. Placenta 2019:80;42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JL, Hurley DG, Gamage TK, Zhang T, Vather R, Pantham P, Murthi P, Chamley LW. Isolation and characterisation of a novel trophoblast side-population from first trimester placentae. Reproduction 2015:150;449–462. [DOI] [PubMed] [Google Scholar]
- Jarvenpaa J, Vuoristo JT, Savolainen ER, Ukkola O, Vaskivuo T, Ryynanen M. Altered expression of angiogenesis-related placental genes in pre-eclampsia associated with intrauterine growth restriction. Gynecol Endocrinol 2007:23;351–355. [DOI] [PubMed] [Google Scholar]
- Jebbink JM, Boot RG, Keijser R, Moerland PD, Aten J, Veenboer GJ, van Wely M, Buimer M, Ver Loren van Themaat E, Aerts JM et al. Increased glucocerebrosidase expression and activity in preeclamptic placenta. Placenta 2015:36;160–169. [DOI] [PubMed] [Google Scholar]
- Jiang X, Bar HY, Yan J, Jones S, Brannon PM, West AA, Perry CA, Ganti A, Pressman E, Devapatla S et al. A higher maternal choline intake among third-trimester pregnant women lowers placental and circulating concentrations of the antiangiogenic factor FMS-like tyrosine kinase-1 (sFLT1). FASEB J 2013:27;1245–1253. [DOI] [PubMed] [Google Scholar]
- Jiang X, Du MR, Li M, Wang H. Three macrophage subsets are identified in the uterus during early human pregnancy. Cell Mol Immunol 2018:15;1027–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobarteh ML, Moore SE, Kennedy C, Gambling L, McArdle HJ. The effect of delay in collection and processing on RNA integrity in human placenta: experiences from rural Africa. Placenta 2014:35;72–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juiz NA, Torrejon I, Burgos M, Torres AMF, Duffy T, Cayo NM, Tabasco A, Salvo M, Longhi SA, Schijman AG. Alterations in placental gene expression of pregnant women with chronic chagas disease. Am J Pathol 2018:188;1345–1353. [DOI] [PubMed] [Google Scholar]
- Junus K, Centlow M, Wikstrom AK, Larsson I, Hansson SR, Olovsson M. Gene expression profiling of placentae from women with early- and late-onset pre-eclampsia: down-regulation of the angiogenesis-related genes ACVRL1 and EGFL7 in early-onset disease. Mol Hum Reprod 2012:18;146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Song H, Yoon JA, Park DY,, Kim SH, Lee KJ, Farina A, Cho YK, Kim YN, Park SW et al. Preeclampsia leads to dysregulation of various signaling pathways in placenta. J Hypertens 2011:29;928–936. [DOI] [PubMed] [Google Scholar]
- Kanter DJ, O’Brien MB, Shi XH, Chu T, Mishima T, Beriwal S, Epperly MW, Wipf P, Greenberger JS, Sadovsky Y. The impact of ionizing radiation on placental trophoblasts. Placenta 2014:35;85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamichi Y, Cui CH, Toyoda M, Makino H, Horie A, Takahashi Y, Matsumoto K, Saito H, Ohta H, Saito K et al. Cells of extraembryonic mesodermal origin confer human dystrophin in the mdx model of Duchenne muscular dystrophy. J Cell Physiol 2010:223;695–702. [DOI] [PubMed] [Google Scholar]
- Kim J, Zhao K, Jiang P, Lu ZX, Wang J, Murray JC, Xing Y. Transcriptome landscape of the human placenta. BMC Genomics 2012:13;115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Romero R, Kim CJ, Tarca AL, Chhauy S, LaJeunesse C, Lee DC, Draghici S, Gotsch F, Kusanovic JP et al. Villitis of unknown etiology is associated with a distinct pattern of chemokine up-regulation in the feto-maternal and placental compartments: implications for conjoint maternal allograft rejection and maternal anti-fetal graft-versus-host disease. J Immunol 2009:182;3919–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingdom JC, Audette MC, Hobson SR, Windrim RC, Morgen E. A placenta clinic approach to the diagnosis and management of fetal growth restriction. Am J Obstet Gynecol 2018:218;S803–S817. [DOI] [PubMed] [Google Scholar]
- Kleinrouweler CE, van Uitert M, Moerland PD, Ris-Stalpers C, van der Post JA, Afink GB. Differentially expressed genes in the pre-eclamptic placenta: a systematic review and meta-analysis. PLoS One 2013:8;e68991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konwar C, Del Gobbo G, Yuan V, Robinson WP. Considerations when processing and interpreting genomics data of the placenta. Placenta 2019:84;57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopcow HD, Eriksson M, Mselle TF, Damrauer SM, Wira CR, Sentman CL, Strominger JL. Human decidual NK cells from gravid uteri and NK cells from cycling endometrium are distinct NK cell subsets. Placenta 2010:31;334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis AP, Read JS, Jamieson DJ. Pregnancy and infection. N Engl J Med 2014:370;2211–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassance L, Haghiac M, Leahy P, Basu S, Minium J, Zhou J, Reider M, Catalano PM, Hauguel-de Mouzon S. Identification of early transcriptome signatures in placenta exposed to insulin and obesity. Am J Obstet Gynecol 2015:212;64.e1–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lean SC, Heazell AEP, Dilworth MR, Mills TA, Jones RL. Placental dysfunction underlies increased risk of fetal growth restriction and stillbirth in advanced maternal age women. Sci Rep 2017:7;9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavey K, Benton SJ, Grynspan D, Bainbridge SA, Morgen EK, Cox BJ. Gene markers of normal villous maturation and their expression in placentas with maturational pathology. Placenta 2017:58;52–59. [DOI] [PubMed] [Google Scholar]
- Leavey K, Benton SJ, Grynspan D, Kingdom JC, Bainbridge SA, Cox BJ. Unsupervised placental gene expression profiling identifies clinically relevant subclasses of human preeclampsia. Hypertension 2016:68;137–147. [DOI] [PubMed] [Google Scholar]
- Leavey K, Wilson SL, Bainbridge SA, Robinson WP, Cox BJ. Epigenetic regulation of placental gene expression in transcriptional subtypes of preeclampsia. Clin Epigenetics 2018:10;28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Koeppel AF, Wang ET, Gonzalez TL, Sun T, Kroener L, Lin Y, Joshi NV, Ghadiali T, Turner SD et al. Differential gene expression during placentation in pregnancies conceived with different fertility treatments compared with spontaneous pregnancies. Fertil Steril 2019:111;535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GS,, Joe YS, Kim SJ, Shin JC. Cytokine-related genes and oxidation-related genes detected in preeclamptic placentas. Arch Gynecol Obstet 2010:282;363–369. [DOI] [PubMed] [Google Scholar]
- Lekva T, Lyle R, Roland MC, Friis C, Bianchi DW, Jaffe IZ, Norwitz ER, Bollerslev J, Henriksen T, Ueland T. Gene expression in term placentas is regulated more by spinal or epidural anesthesia than by late-onset preeclampsia or gestational diabetes mellitus. Sci Rep 2016:6;29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie K, Whitley GS, Herse F, Dechend R, Ashton SV, Laing K, Thilaganathan B, Cartwright JE. Increased apoptosis, altered oxygen signaling, antioxidant defenses in first-trimester pregnancies with high-resistance uterine artery blood flow. Am J Pathol 2015:185;2731–2741. [DOI] [PubMed] [Google Scholar]
- Lewis AJ, Austin E, Knapp R, Vaiano T, Galbally M. Perinatal maternal mental health, fetal programming and child development. Healthcare (Basel) 2015:3;1212–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Song L, Zhou L, Wu J, Sheng C, Chen H, Liu Y, Gao S, Huang W. A microRNA signature in gestational diabetes mellitus associated with risk of macrosomia. Cell Physiol Biochem 2015:37;243–252. [DOI] [PubMed] [Google Scholar]
- Liang M, Niu J, Zhang L, Deng H, Ma J, Zhou W, Duan D, Zhou Y, Xu H, Chen L. Gene expression profiling reveals different molecular patterns in G-protein coupled receptor signaling pathways between early- and late-onset preeclampsia. Placenta 2016:40;52–59. [DOI] [PubMed] [Google Scholar]
- Lim JH, Han YJ, Kim HJ, Kwak DW, Park SY, Chun SH, Ryu HM. Genome-wide gene expression analysis in the placenta from fetus with trisomy 21. BMC Genomics 2017. a:18;720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YC, Li J, Ni Y, Liang Q, Zhang J, Yeo GSH, Lyu J, Jin S, Ding C. A complex association between DNA methylation and gene expression in human placenta at first and third trimesters. PLoS One 2017. b:12;e0181155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Fan X, Wang R, Lu X, Dang YL, Wang H, Lin HY, Zhu C, Ge H, Cross JC et al. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res 2018:28;819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loset M, Mundal SB, Johnson MP, Fenstad MH, Freed KA, Lian IA, Eide IP, Bjorge L, Blangero J, Moses EK et al. A transcriptional profile of the decidua in preeclampsia. Am J Obstet Gynecol 2011:204;84.e1–e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykoudi A, Kolialexi A, Lambrou GI, Braoudaki M, Siristatidis C, Papaioanou GK, Tzetis M, Mavrou A, Papantoniou N. Dysregulated placental microRNAs in early and late onset preeclampsia. Placenta 2018:61;24–32. [DOI] [PubMed] [Google Scholar]
- Madeleneau D, Buffat C, Mondon F, Grimault H, Rigourd V, Tsatsaris V, Letourneur F, Vaiman D, Barbaux S, Gascoin G. Transcriptomic analysis of human placenta in intrauterine growth restriction. Pediatr Res 2015:77;799–807. [DOI] [PubMed] [Google Scholar]
- Maier CL, Shepherd BR, Yi T, Pober JS. Explant outgrowth, propagation and characterization of human pericytes. Microcirculation 2010:17;367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska M, Lipka A, Paukszto L, Jastrzebski JP, Gowkielewicz M, Jozwik M, Majewski MK. Preliminary RNA-seq analysis of long non-coding RNAs expressed in human term placenta. Int J Mol Sci 2018:19;1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska M, Lipka A, Paukszto L, Jastrzebski JP, Myszczynski K, Gowkielewicz M, Jozwik M, Majewski MK. Transcriptome profile of the human placenta. Funct Integr Genomics 2017:17;551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska M, Lipka A, Paukszto L, Jastrzebski JP, Szeszko K, Gowkielewicz M, Lepiarczyk E, Jozwik M, Majewski MK. Placenta transcriptome profiling in intrauterine growth restriction (IUGR). Int J Mol Sci 2019:20;1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E, Ray PD, Smeester L, Grace MR, Boggess K, Fry RC. Epigenetics and preeclampsia: defining functional epimutations in the preeclamptic placenta related to the TGF-beta pathway. PLoS One 2015:10;e0141294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NM, Cooke KM, Radford CC, Perley LE, Silasi M, Flannery CA. Time course analysis of RNA quality in placenta preserved by RNAlater or flash freezing. Am J Reprod Immunol 2017:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard SE, Min JY, Merchan J, Lim KH, Li J, Mondal S, Libermann TA, Morgan JP, Sellke FW, Stillman IE et al. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J Clin Invest 2003:111;649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor-Lynn K, Toloubeydokhti T, Cruz AC, Chegini N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod Sci 2011:18;46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy C, Cotter FE, McElwaine S, Twomey A, Mooney EE, Ryan F, Vaughan J. Altered gene expression patterns in intrauterine growth restriction: potential role of hypoxia. Am J Obstet Gynecol 2007:196;70.e1–e6. [DOI] [PubMed] [Google Scholar]
- Meakin AS, Saif Z, Jones AR, Aviles PFV, Clifton VL. Review: placental adaptations to the presence of maternal asthma during pregnancy. Placenta 2017:54;17–23. [DOI] [PubMed] [Google Scholar]
- Mecham BH, Wetmore DZ, Szallasi Z, Sadovsky Y, Kohane I, Mariani TJ. Increased measurement accuracy for sequence-verified microarray probes. Physiol Genomics 2004:18;308–315. [DOI] [PubMed] [Google Scholar]
- Meek CL. Natural selection? The evolution of diagnostic criteria for gestational diabetes. Ann Clin Biochem 2017:54;33–42. [DOI] [PubMed] [Google Scholar]
- Meng T, Chen H, Sun M, Wang H, Zhao G, Wang X. Identification of differential gene expression profiles in placentas from preeclamptic pregnancies versus normal pregnancies by DNA microarrays. OMICS 2012:16;301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsalu T, Viltrop T, Tiirats A, Rajashekar B, Reimann E, Koks S, Rull K, Milani L, Acharya G, Basnet P et al. Using RNA sequencing for identifying gene imprinting and random monoallelic expression in human placenta. Epigenetics 2014:9;1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheev AM, Nabekura T,, Kaddoumi A, Bammler TK, Govindarajan R, Hebert MF, Unadkat JD. Profiling gene expression in human placentae of different gestational ages: an OPRU Network and UW SCOR Study. Reprod Sci 2008:15;866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitriuc D, Popusoi O, Catrinici R, Friptu V. The obstetric complications in women with hereditary thrombophilia. Med Pharm Rep 2019:92;106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moslehi R, Mills JL, Signore C, Kumar A, Ambroggio X, Dzutsev A. Integrative transcriptome analysis reveals dysregulation of canonical cancer molecular pathways in placenta leading to preeclampsia. Sci Rep 2013:3;2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlenbachs A, Fried M, Lachowitzer J, Mutabingwa TK, Duffy PE. Genome-wide expression analysis of placental malaria reveals features of lymphoid neogenesis during chronic infection. J Immunol 2007:179;557–565. [DOI] [PubMed] [Google Scholar]
- Nelissen EC, Dumoulin JC, Busato F, Ponger L, Eijssen LM, Evers JL, Tost J, van Montfoort AP. Altered gene expression in human placentas after IVF/ICSI. Hum Reprod 2014:29;2821–2831. [DOI] [PubMed] [Google Scholar]
- Nishizawa H, Ota S, Suzuki M, Kato T, Sekiya T, Kurahashi H, Udagawa Y. Comparative gene expression profiling of placentas from patients with severe pre-eclampsia and unexplained fetal growth restriction. Reprod Biol Endocrinol 2011:9;107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa H, Pryor-Koishi K, Kato T, Kowa H, Kurahashi H, Udagawa Y. Microarray analysis of differentially expressed fetal genes in placental tissue derived from early and late onset severe pre-eclampsia. Placenta 2007:28;487–497. [DOI] [PubMed] [Google Scholar]
- Ohmaru-Nakanishi T, Asanoma K, Fujikawa M, Fujita Y, Yagi H, Onoyama I, Hidaka N, Sonoda K, Kato K. Fibrosis in preeclamptic placentas is associated with stromal fibroblasts activated by the transforming growth factor-beta1 signaling pathway. Am J Pathol 2018:188;683–695. [DOI] [PubMed] [Google Scholar]
- Olivier JD, Akerud H, Skalkidou A, Kaihola H, Sundstrom-Poromaa I. The effects of antenatal depression and antidepressant treatment on placental gene expression. Front Cell Neurosci 2014:8;465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostling H, Kruse R, Helenius G, Lodefalk M. Placental expression of microRNAs in infants born small for gestational age. Placenta 2019:81;46–53. [DOI] [PubMed] [Google Scholar]
- Pantham P, Rosario R, Chen Q, Print CG, Chamley LW. Transcriptomic analysis of placenta affected by antiphospholipid antibodies: following the TRAIL of trophoblast death. J Reprod Immunol 2012:94;151–154. [DOI] [PubMed] [Google Scholar]
- Paquette AG, Brockway HM, Price ND, Muglia LJ. Comparative transcriptomic analysis of human placentae at term and preterm delivery. Biol Reprod 2018:98;89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicev M, Wagner GP, Chavan AR, Owens K, Maziarz J, Dunn-Fletcher C, Kallapur SG, Muglia L, Jones H. Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Res 2017:27;349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Deyssenroth MA, Di Narzo AF, Cheng H, Zhang Z, Lambertini L, Ruusalepp A, Kovacic JC, Bjorkegren JLM, Marsit CJ et al. Genetic regulation of the placental transcriptome underlies birth weight and risk of childhood obesity. PLoS Genet 2018:14;e1007799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Deyssenroth MA, Di Narzo AF, Lambertini L, Marsit CJ, Chen J, Hao K. Expression quantitative trait loci (eQTLs) in human placentas suggest developmental origins of complex diseases. Hum Mol Genet 2017:26;3432–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux G, Gerbaud P, Laurendeau I, Guibourdenche J, Bertin G, Vidaud M, Evain-Brion D, Frendo JL. Large variability of trophoblast gene expression within and between human normal term placentae. Placenta 2004:25;469–473. [DOI] [PubMed] [Google Scholar]
- Pisarska MD, Akhlaghpour M, Lee B, Barlow GM, Xu N, Wang ET, Mackey AJ, Farber CR, Rich SS, Rotter JI et al. Optimization of techniques for multiple platform testing in small, precious samples such as human chorionic villus sampling. Prenat Diagn 2016:36;1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell RM, Lissauer D, Tamblyn J, Beggs A, Cox P, Moss P, Kilby MD. Decidual T cells exhibit a highly differentiated phenotype and demonstrate potential fetal specificity and a strong transcriptional response to IFN. J Immunol 2017:199;3406–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radaelli T, Lepercq J, Varastehpour A, Basu S, Catalano PM, Mouzon Hauguel-De S. Differential regulation of genes for fetoplacental lipid pathways in pregnancy with gestational and type 1 diabetes mellitus. Am J Obstet Gynecol 2009:201;209.e1–e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radaelli T, Varastehpour A, Catalano P, Hauguel-de Mouzon S. Gestational diabetes induces placental genes for chronic stress and inflammatory pathways. Diabetes 2003:52;2951–2958. [DOI] [PubMed] [Google Scholar]
- Raman K, Wang H, Troncone MJ, Khan WI, Pare G, Terry J. Overlap chronic placental inflammation is associated with a unique gene expression pattern. PLoS One 2015:10;e0133738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman M, Laan M, Rull K, Sober S. Effects of RNA integrity on transcript quantification by total RNA sequencing of clinically collected human placental samples. FASEB J 2017:31;3298–3308. [DOI] [PubMed] [Google Scholar]
- Reinebrant HE, Leisher SH, Coory M, Henry S, Wojcieszek AM, Gardener G, Lourie R, Ellwood D, Teoh Z, Allanson E et al. Making stillbirths visible: a systematic review of globally reported causes of stillbirth. BJOG 2018:125;212–224. [DOI] [PubMed] [Google Scholar]
- Rigourd V, Chauvet C, Chelbi ST, Rebourcet R, Mondon F, Letourneur F, Mignot TM, Barbaux S, Vaiman D. STOX1 overexpression in choriocarcinoma cells mimics transcriptional alterations observed in preeclamptic placentas. PLoS One 2008:3;e3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi SF, Makieva S, Saunders PT, Rossi AG, Norman JE. Immune cell and transcriptomic analysis of the human decidua in term and preterm parturition. Mol Hum Reprod 2017:23;708–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JF, Kapidzic M, Gormley M, Ona K, Dent T, Seifikar H, Hamilton EG, Fisher SJ. Transcriptional dynamics of cultured human villous cytotrophoblasts. Endocrinology 2017:158;1581–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh CR, Budhraja V, Kim HS, Nelson DM, Sadovsky Y. Microarray-based identification of differentially expressed genes in hypoxic term human trophoblasts and in placental villi of pregnancies with growth restricted fetuses. Placenta 2005:26;319–328. [DOI] [PubMed] [Google Scholar]
- Roost MS, van Iperen L, Ariyurek Y, Buermans HP, Arindrarto W, Devalla HD, Passier R, Mummery CL, Carlotti F, de Koning EJ et al. KeyGenes, a tool to probe tissue differentiation using a human fetal transcriptional atlas. Stem Cell Reports 2015:4;1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault C, Clement K, Guesnon M, Henegar C, Charles MA, Heude B, Evain-Brion D, Degrelle SA, Fournier T. Transcriptomic signatures of villous cytotrophoblast and syncytiotrophoblast in term human placenta. Placenta 2016:44;83–90. [DOI] [PubMed] [Google Scholar]
- Rull K, Tomberg K, Koks S, Mannik J, Mols M, Sirotkina M, Varv S, Laan M. Increased placental expression and maternal serum levels of apoptosis-inducing TRAIL in recurrent miscarriage. Placenta 2013:34;141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saben J, Lindsey F, Zhong Y, Thakali K, Badger TM, Andres A, Gomez-Acevedo H, Shankar K. Maternal obesity is associated with a lipotoxic placental environment. Placenta 2014. a:35;171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saben J, Zhong Y, McKelvey S, Dajani NK, Andres A, Badger TM, Gomez-Acevedo H, Shankar K. A comprehensive analysis of the human placenta transcriptome. Placenta 2014. b:35;125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabri A, Lai D, D’Silva A, Seeho S, Kaur J, Ng C, Hyett J. Differential placental gene expression in term pregnancies affected by fetal growth restriction and macrosomia. Fetal Diagn Ther 2014:36;173–180. [DOI] [PubMed] [Google Scholar]
- Schroeder DI, Blair JD, Lott P, Yu HO, Hong D, Crary F, Ashwood P, Walker C, Korf I, Robinson WP et al. The human placenta methylome. Proc Natl Acad Sci U S A 2013:110;6037–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlmeier EM, Brunner S, Much D, Pagel P, Ulbrich SE,, Meyer HH, Amann-Gassner U, Hauner H, Bader BL. Human placental transcriptome shows sexually dimorphic gene expression and responsiveness to maternal dietary n-3 long-chain polyunsaturated fatty acid intervention during pregnancy. BMC Genomics 2014:15;941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang H, Sun L, Braun T, Si Q, Tong J. Revealing the action mechanisms of dexamethasone on the birth weight of infant using RNA-sequencing data of trophoblast cells. Medicine (Baltimore ) 2018:97;e9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar K, Kang P, Zhong Y, Borengasser SJ, Wingfield C, Saben J, Gomez-Acevedo H, Thakali KM. Transcriptomic and epigenomic landscapes during cell fusion in BeWo trophoblast cells. Placenta 2015:36;1342–1351. [DOI] [PubMed] [Google Scholar]
- Silasi M, Cardenas I, Kwon JY, Racicot K, Aldo P, Mor G. Viral infections during pregnancy. Am J Reprod Immunol 2015:73;199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitras V, Fenton C, Acharya G. Gene expression profile in cardiovascular disease and preeclampsia: a meta-analysis of the transcriptome based on raw data from human studies deposited in Gene Expression Omnibus. Placenta 2015:36;170–178. [DOI] [PubMed] [Google Scholar]
- Sitras V, Fenton C, Paulssen R, Vartun A, Acharya G. Differences in gene expression between first and third trimester human placenta: a microarray study. PLoS One 2012:7;e33294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitras V, Paulssen R, Leirvik J, Vartun A, Acharya G. Placental gene expression profile in intrauterine growth restriction due to placental insufficiency. Reprod Sci 2009. a:16;701–711. [DOI] [PubMed] [Google Scholar]
- Sitras V, Paulssen RH, Gronaas H, Leirvik J, Hanssen TA, Vartun A, Acharya G. Differential placental gene expression in severe preeclampsia. Placenta 2009. b:30;424–433. [DOI] [PubMed] [Google Scholar]
- Sober S, Reiman M, Kikas T, Rull K, Inno R, Vaas P, Teesalu P, Marti JML, Mattila P, Laan M. Extensive shift in placental transcriptome profile in preeclampsia and placental origin of adverse pregnancy outcomes. Sci Rep 2015:5;13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sober S, Rull K, Reiman M, Ilisson P, Mattila P, Laan M. RNA sequencing of chorionic villi from recurrent pregnancy loss patients reveals impaired function of basic nuclear and cellular machinery. Sci Rep 2016:6;38439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncin F, Khater M, To C, Pizzo D, Farah O, Wakeland A, Arul Nambi Rajan K, Nelson KK, Chang CW, Moretto-Zita M et al. Comparative analysis of mouse and human placentae across gestation reveals species-specific regulators of placental development. Development 2018:145;dev156273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song GY, Na Q, Wang D, Qiao C. Microarray expression profile of lncRNAs and mRNAs in the placenta of non-diabetic macrosomia. J Dev Orig Health Dis 2018:9;191–197. [DOI] [PubMed] [Google Scholar]
- Song Y, Liu J, Huang S, Zhang L. Analysis of differentially expressed genes in placental tissues of preeclampsia patients using microarray combined with the Connectivity Map database. Placenta 2013:34;1190–1195. [DOI] [PubMed] [Google Scholar]
- Sood R, Zehnder JL, Druzin ML, Brown PO. Gene expression patterns in human placenta. Proc Natl Acad Sci U S A 2006:103;5478–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien BK, Huttner WB. Transport, metabolism, function of thyroid hormones in the developing mammalian brain. Front Endocrinol (Lausanne) 2019:10;209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe E, Berzl G, Schild R, Blessing H, Drexel L, Hauck B, Tzschoppe A, Weidinger M, Sachs M, Scheler C et al. Microarray analysis of placental tissue in intrauterine growth restriction. Clin Endocrinol (Oxf) 2010:72;241–247. [DOI] [PubMed] [Google Scholar]
- Sureshchandra S, Marshall NE, Wilson RM, Barr T, Rais M, Purnell JQ, Thornburg KL, Messaoudi I. Inflammatory determinants of pregravid obesity in placenta and peripheral blood. Front Physiol 2018:9;1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryawanshi H, Morozov P, Straus A, Sahasrabudhe N, Max KEA, Garzia A, Kustagi M, Tuschl T, Williams Z. A single-cell survey of the human first-trimester placenta and decidua. Sci Adv 2018:4;eaau4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak-Wegierek D. Intrauterine nutrition: long-term consequences for vascular health. Int J Womens Health 2014:6;647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao T, Asanoma K, Kato K, Fukushima K, Tsunematsu R, Hirakawa T, Matsumura S, Seki H, Takeda S, Wake N. Isolation and characterization of human trophoblast side-population (SP) cells in primary villous cytotrophoblasts and HTR-8/SVneo cell line. PLoS One 2011:6;e21990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber S, Jais A, Jeitler M, Haider S, Husa J, Lindroos J, Knofler M, Mayerhofer M, Pehamberger H, Wagner O et al. Transcriptome analysis of human cancer reveals a functional role of heme oxygenase-1 in tumor cell adhesion. Mol Cancer 2010:9;200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than NG, Romero R, Tarca AL, Kekesi KA, Xu Y, Xu Z, Juhasz K, Bhatti G, Leavitt RJ, Gelencser Z et al. Integrated systems biology approach identifies novel maternal and placental pathways of preeclampsia. Front Immunol 2018:9;1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian FJ, Cheng YX,, Li XC, Wang F, Qin CM, Ma XL, Yang J, Lin Y. The YY1/MMP2 axis promotes trophoblast invasion at the maternal-fetal interface. J Pathol 2016:239;36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J, Zhao W, Lv H, Li WP, Chen ZJ, Zhang C. Transcriptomic profiling in human decidua of severe preeclampsia detected by RNA sequencing. J Cell Biochem 2018:119;607–615. [DOI] [PubMed] [Google Scholar]
- Tsai S, Hardison NE, James AH, Motsinger-Reif AA, Bischoff SR, Thames BH, Piedrahita JA. Transcriptional profiling of human placentas from pregnancies complicated by preeclampsia reveals disregulation of sialic acid acetylesterase and immune signalling pathways. Placenta 2011:32;175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang JCH, Vong JSL, Ji L, Poon LCY, Jiang P, Lui KO, Ni YB, To KF, Cheng YKY, Chiu RWK et al. Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc Natl Acad Sci U S A 2017:114;E7786–E7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui NB, Chim SS, Chiu RW, Lau TK, Ng EK, Leung TN, Tong YK,, Chan KC, Lo YM. Systematic micro-array based identification of placental mRNA in maternal plasma: towards non-invasive prenatal gene expression profiling. J Med Genet 2004:41;461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turan N, Ghalwash MF, Katari S, Coutifaris C, Obradovic Z, Sapienza C. DNA methylation differences at growth related genes correlate with birth weight: a molecular signature linked to developmental origins of adult disease? BMC Med Genomics 2012:5;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco MY, Gardner L, Kay RG, Hamilton RS, Prater M, Hollinshead MS, McWhinnie A, Esposito L, Fernando R,, Skelton H et al. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 2018:564;263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger HW, Ashorn P, Cates JE, Dewey KG, Rogerson SJ. Undernutrition and malaria in pregnancy—a dangerous dyad? BMC Med 2016:14;142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uuskula L, Mannik J, Rull K, Minajeva A, Koks S, Vaas P, Teesalu P, Reimand J, Laan M. Mid-gestational gene expression profile in placenta and link to pregnancy complications. PLoS One 2012:7;e49248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaiman D, Calicchio R, Miralles F. Landscape of transcriptional deregulations in the preeclamptic placenta. PLoS One 2013:8;e65498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaiman D, Miralles F. An integrative analysis of preeclampsia based on the construction of an extended composite network featuring protein-protein physical interactions and transcriptional relationships. PLoS One 2016:11;e0165849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Uitert M, Moerland PD, Enquobahrie DA, Laivuori H, van der Post JA, Ris-Stalpers C, Afink GB. Meta-analysis of placental transcriptome data identifies a novel molecular pathway related to preeclampsia. PLoS One 2015:10;e0132468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannuccini S, Clifton VL, Fraser IS, Taylor HS, Critchley H, Giudice LC, Petraglia F. Infertility and reproductive disorders: impact of hormonal and inflammatory mechanisms on pregnancy outcome. Hum Reprod Update 2016:22;104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi T, Nagy B, Fule T, Tarca AL, Karaszi K, Schonleber J, Hupuczi P, Mihalik N, Kovalszky I, Rigo J Jr et al. Microarray profiling reveals that placental transcriptomes of early-onset HELLP syndrome and preeclampsia are similar. Placenta 2011:32 Suppl;S21–S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky J, Smith A, Wang A, Hom E, Izano M, Huang H, Padula A, Woodruff TJ. Heightened susceptibility: a review of how pregnancy and chemical exposures influence maternal health. Reprod Toxicol 2019:92;14–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park JE, Stephenson E, Polanski K, Goncalves A et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 2018:563;347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheecke M, Cortes Calabuig A, Finalet Ferreiro J, Brys V, Van Bree R, Verbist G, Everaert T, Leemans L, Gziri MM, Boere I et al. Genetic and microscopic assessment of the human chemotherapy-exposed placenta reveals possible pathways contributive to fetal growth restriction. Placenta 2018:64;61–70. [DOI] [PubMed] [Google Scholar]
- Verma S, Pal R, Gupta SK. Decrease in invasion of HTR-8/SVneo trophoblastic cells by interferon gamma involves cross-communication of STAT1 and BATF2 that regulates the expression of JUN. Cell Adh Migr 2018:12;432–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vondra S, Kunihs V, Eberhart T, Eigner K, Bauer R, Haslinger P, Haider S, Windsperger K, Klambauer G, Schutz B et al. Metabolism of cholesterol and progesterone is differentially regulated in primary trophoblastic subtypes and might be disturbed in recurrent miscarriages. J Lipid Res 2019:60;1922–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votavova H, Dostalova Merkerova M, Fejglova K, Vasikova A, Krejcik Z, Pastorkova A, Tabashidze N, Topinka J, Veleminsky M Jr, Sram RJ et al. Transcriptome alterations in maternal and fetal cells induced by tobacco smoke. Placenta 2011:32;763–770. [DOI] [PubMed] [Google Scholar]
- Votavova H, Dostalova Merkerova M, Krejcik Z, Fejglova K, Vasikova A, Pastorkova A, Tabashidze N, Topinka J, Balascak I, Sram RJ et al. Deregulation of gene expression induced by environmental tobacco smoke exposure in pregnancy. Nicotine Tob Res 2012:14;1073–1082. [DOI] [PubMed] [Google Scholar]
- Wang B, Wang P, Parobchak N, Treff N, Tao X, Wang J, Rosen T. Integrated RNA-seq and ChIP-seq analysis reveals a feed-forward loop regulating H3K9ac and key labor drivers in human placenta. Placenta 2019. a:76;40–50. [DOI] [PubMed] [Google Scholar]
- Wang F, Zhou Y, Fu B, Wu Y, Zhang R, Sun R, Tian Z, Wei H. Molecular signatures and transcriptional regulatory networks of human immature decidual NK and mature peripheral NK cells. Eur J Immunol 2014:44;2771–2784. [DOI] [PubMed] [Google Scholar]
- Wang H, She G, Zhou W, Liu K, Miao J, Yu B. Expression profile of circular RNAs in placentas of women with gestational diabetes mellitus. Endocr J 2019. b:66;431–441. [DOI] [PubMed] [Google Scholar]
- Weisblum Y, Oiknine-Djian E, Vorontsov OM, Haimov-Kochman R, Zakay-Rones Z, Meir K, Shveiky D, Elgavish S, Nevo Y, Roseman M et al. Zika virus infects early- and midgestation human maternal decidual tissues, inducing distinct innate tissue responses in the maternal-fetal interface. J Virol 2017:91;e01905–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Chen L, He J, Lin J. MicroRNA expression profiles and networks in placentas complicated with selective intrauterine growth restriction. Mol Med Rep 2017:16;6650–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn VD, Gormley M, Paquet AC, Kjaer-Sorensen K,, Kramer A, Rumer KK, Haimov-Kochman R, Yeh RF, Overgaard MT, Varki A et al. Severe preeclampsia-related changes in gene expression at the maternal-fetal interface include sialic acid-binding immunoglobulin-like lectin-6 and pappalysin-2. Endocrinology 2009:150;452–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witteveen T, Van Den Akker T, Zwart JJ, Bloemenkamp KW, Van Roosmalen J. Severe acute maternal morbidity in multiple pregnancies: a nationwide cohort study. Am J Obstet Gynecol 2016:214;641.e1–e10. [DOI] [PubMed] [Google Scholar]
- Wolfe LM, Thiagarajan RD, Boscolo F, Tache V, Coleman RL, Kim J, Kwan WK, Loring JF, Parast M, Laurent LC. Banking placental tissue: an optimized collection procedure for genome-wide analysis of nucleic acids. Placenta 2014:35;645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong FTM, Lin C, Cox BJ. Cellular systems biology identifies dynamic trophoblast populations in early human placentas. Placenta 2019:76;10–18. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Cheng Y, Li X, Li Q, Xu J, Zhang J, Liu Y, Xing Q, Wang L, He L et al. Up-regulated expression and aberrant DNA methylation of LEP and SH3PXD2A in pre-eclampsia. PLoS One 2013:8;e59753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Mouillet JF, Chu T, Parks WT, Sadovsky E, Knofler M, Sadovsky Y. C19MC microRNAs regulate the migration of human trophoblasts. Endocrinology 2014:155;4975–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe S, Alexenko AP, Amita M, Yang Y, Schust DJ, Sadovsky Y, Ezashi T, Roberts RM. Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas. Proc Natl Acad Sci U S A 2016:113;E2598–E2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yockey LJ, Jurado KA, Arora N, Millet A, Rakib T, Milano KM, Hastings AK, Fikrig E, Kong Y, Horvath TL et al. Type I interferons instigate fetal demise after Zika virus infection. Sci Immunol 2018:3;eaao1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong HE, Melton PE, Johnson MP, Freed KA, Kalionis B, Murthi P, Brennecke SP, Keogh RJ, Moses EK. Genome-wide transcriptome directed pathway analysis of maternal pre-eclampsia susceptibility genes. PLoS One 2015:10;e0128230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Du G, Xu Q, Huang Z, Huang X, Qin Y, Han L, Fan Y, Zhang Y, Han X et al. Integrated analysis of DNA methylome and transcriptome identified CREB5 as a novel risk gene contributing to recurrent pregnancy loss. EBioMedicine 2018:35;334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen RK, Chen B, Blair JD, Robinson WP, Nelson DM. Hypoxia alters the epigenetic profile in cultured human placental trophoblasts. Epigenetics 2013:8;192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisler H, Llurba E, Chantraine F, Vatish M, Staff AC, Sennstrom M, Olovsson M, Brennecke SP, Stepan H, Allegranza D et al. Predictive value of the sFlt-1:PlGF ratio in women with suspected preeclampsia. N Engl J Med 2016:374;13–22. [DOI] [PubMed] [Google Scholar]
- Zeng W, Liu Z, Liu X, Zhang S, Khanniche A, Zheng Y, Ma X, Yu T, Tian F, Liu XR et al. Distinct transcriptional and alternative splicing signatures of decidual CD4(+) T cells in early human pregnancy. Front Immunol 2017:8;682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Sun LF, Zheng XL, Liu JF, Zheng R, Wang Y, Yang R, Zhang L, Yu L, Zhang H. [In vitro fertilization-embryo transfer affects focal adhension kinase signaling pathway in early placenta]. Beijing Da Xue Xue Bao Yi Xue Ban 2019:51;151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YH, Wang DP, Zhang LL, Zhang F, Wang DM, Zhang WY. Genomic expression profiles of blood and placenta reveal significant immune-related pathways and categories in Chinese women with gestational diabetes mellitus. Diabet Med 2011:28;237–246. [DOI] [PubMed] [Google Scholar]
- Zheng R, Li Y, Sun H, Lu X, Sun BF, Wang R, Cui L, Zhu C, Lin HY, Wang H. Deep RNA sequencing analysis of syncytialization-related genes during BeWo cell fusion. Reproduction 2016:153;35–48. [DOI] [PubMed] [Google Scholar]
- Zhou R, Zhu Q, Wang Y, Ren Y, Zhang L, Zhou Y. Genomewide oligonucleotide microarray analysis on placentae of pre-eclamptic pregnancies. Gynecol Obstet Invest 2006:62;108–114. [DOI] [PubMed] [Google Scholar]
- Zhou W, Wang H, Wu X, Long W, Zheng F, Kong J, Yu B. The profile analysis of circular RNAs in human placenta of preeclampsia. Exp Biol Med (Maywood) 2018:243;1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Gormley MJ, Hunkapiller NM, Kapidzic M, Stolyarov Y, Feng V, Nishida M, Drake PM, Bianco K, Wang F et al. Reversal of gene dysregulation in cultured cytotrophoblasts reveals possible causes of preeclampsia. J Clin Invest 2013:123;2862–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.